Summary
Atherosclerosis is the disease process underlying heart attack and stroke1. Advanced lesions at risk for rupture are characterized by the pathological accumulation of diseased vascular cells and apoptotic cellular debris2. Why these cells are not cleared remains unknown3. Here we show that atherogenesis is associated with upregulation of CD47, a key ‘don’t eat me’ molecule known to render malignant cells resistant to programmed cell removal (PrCR), or ‘efferocytosis’4–7. We find that administration of CD47 blocking antibodies reverses this defect in efferocytosis, normalizes the clearance of diseased vascular tissue, and ameliorates atherosclerosis in multiple mouse models. Mechanistic studies implicate the pro-atherosclerotic factor TNF-α as a fundamental driver of impaired PrCR, explaining why this process is compromised in vascular disease. Similar to recent observations in cancer5, impaired efferocytosis appears to play a pathogenic role in cardiovascular disease, but is not a fixed defect and may represent a novel therapeutic target.
Each day the human body turns over more than 100 billion cells8. To prevent the inflammatory consequences associated with the accumulation of apoptotic debris9, these cells are rapidly and efficiently cleared through a phagocytic process known as programmed cell removal (PrCR), or ‘efferocytosis’ (Greek: To carry the dead to the grave)10. PrCR is mediated by macrophages detecting ‘eat me’ signals on the target cell surface, and can be countermanded by cell surface expression of antiphagocytic ‘don’t eat me’ signals such as CD476. While PrCR is highly conserved across almost all physiological conditions and in all tissues, it appears to be significantly impaired in atherosclerotic cardiovascular disease2, the leading cause of death worldwide11. Atherosclerosis is characterized by the accumulation of diseased macrophages and vascular smooth muscle cells (SMCs) which not only encroach on the lumen of the associated vessel, but may also undergo programmed cell death1,12. The impaired clearance of these diseased cells by lesional macrophages is thought to explain why these cells are frequently observed in the atherosclerotic necrotic core, and may potentiate vascular inflammation and risk for eventual plaque rupture3,13,14. However, the mechanism underlying this defect has not yet been identified.
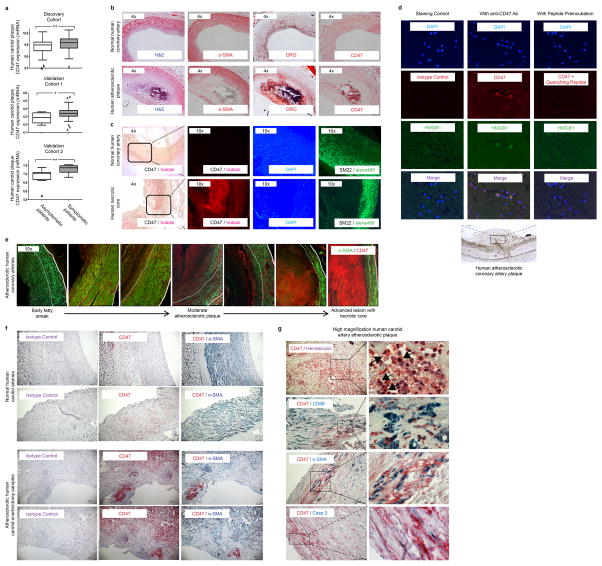
We recently found that the key ‘don’t-eat-me’ molecule, CD47, is paradoxically upregulated by a variety of cancers5,7,15. This renders malignant cells resistant to classic immune surveillance machinery such as the tumoricidal macrophage, and is now recognized as a fundamental driver of tumor growth. To determine if dysregulated CD47 may also contribute to atherogenesis, we evaluated its expression in two independent human vascular tissue biobanks16,17. We found that CD47 is consistently upregulated in human atherosclerotic plaque compared to non-atherosclerotic vascular tissue (Fig 1a), and in subjects with symptomatic cerebrovascular disease (stroke or TIA) compared to those with stable asymptomatic lesions (Extended Data Fig 1a). Because some efferocytosis molecules are known to undergo post-translational modification18, we also performed immunofluorescence and immunohistochemical staining of human coronary and carotid arteries which confirmed that CD47 is progressively upregulated during atherogenesis, and appears to localize intensely to the necrotic core (Fig 1b, Extended Data Fig 1b–g). Similar findings were observed in mouse models of atherosclerosis and other publically-available microarray datasets (Fig 1c–d, Extended Data Fig 2). Together, these data suggest that pathologic upregulation of ‘don’t-eat-me’ molecules may explain why phagocytosis is impaired within the human atherosclerotic plaque, which may in turn promote lesion expansion over time.
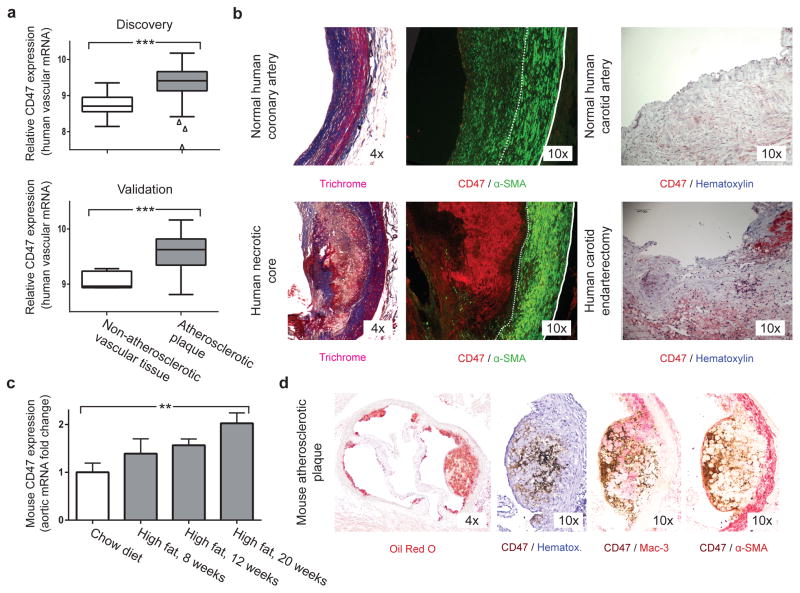
Figure 1. The ‘don’t eat me’ ligand, CD47, is upregulated in atherosclerosis.
(a). Microarray expression profiling in two carotid endarterectomy (CEA) cohorts reveals that CD47 expression is significantly increased in human atherosclerotic plaque, relative to non-diseased vascular tissue (data displayed as Tukey boxplots, n=182 subjects). (b). Immunostaining identifies intense CD47 upregulation within the necrotic core of human atherosclerotic coronary artery lesions (left) and carotid plaques (right). (c). Taqman mRNA analysis confirms that vascular CD47 expression progressively increases in a mouse model of atherosclerosis (apoE−/− mice fed hi fat diet, grey), relative to control animals (C57BL/6 mice fed chow, white, n=4 mice/timepoint). (d). Immunohistochemistry staining with a biotin labeled antibody (brown) reveals that CD47 expression co-localizes with apoptotic tissue within murine atherosclerotic plaque. *** = P < 0.001, ** = P for trend < 0.03. Error bars represent the standard error of the mean (SEM).
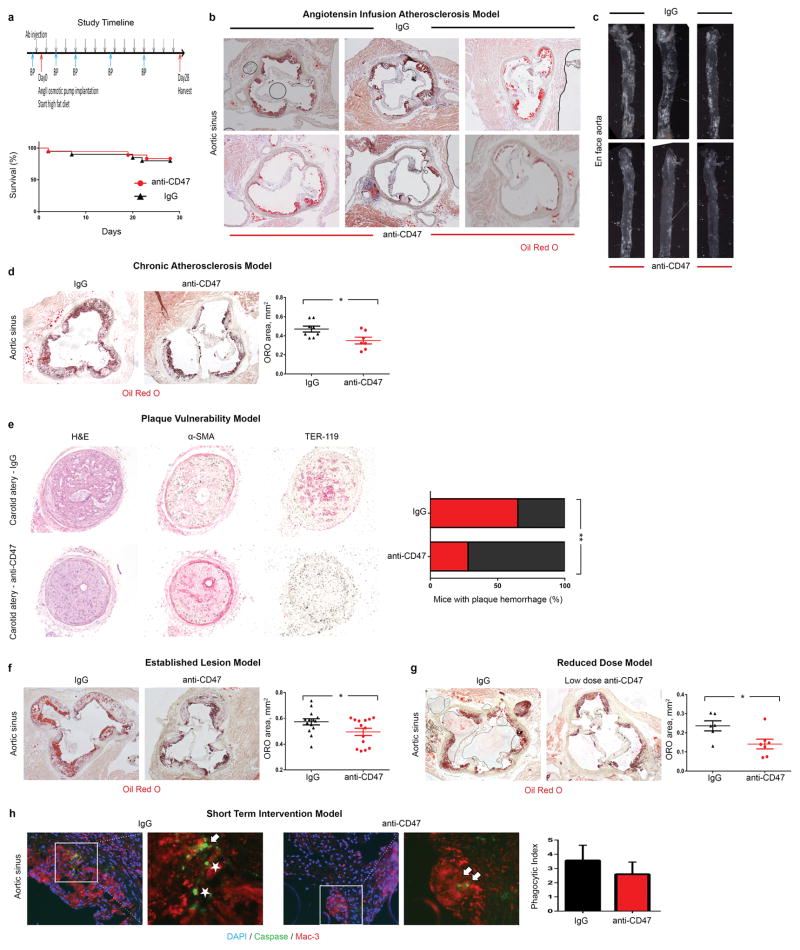
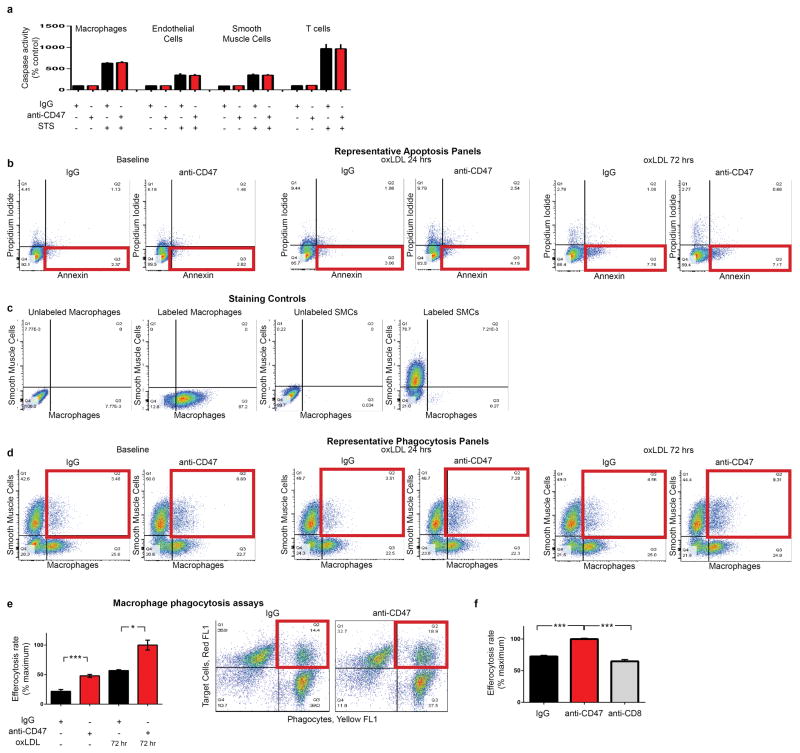
To determine if this defect could be exploited as a translational target for cardiovascular disease, we treated a cohort of atheroprone animals (apoE−/− mice implanted with Angiotensin II-infusing minipumps19) with an inhibitory antibody directed against CD47 (Extended Data Fig 3a)15. Compared to IgG control, anti-CD47 Ab treatment was associated with a dramatic reduction in atherosclerosis, both in the aortic sinus and en face in the aorta itself (Fig 2a–b, Extended Data Fig 3b–c). Similar results were observed in several additional models, including models of chronic atherosclerosis, plaque vulnerability and in mice with established lesions, as would be encountered clinically (Extended Data Fig 3d–h). Although anti-CD47 Ab had no effect on apoptosis in vitro (Fig 2c, Extended Data Fig 4a–b), we observed significantly fewer apoptotic bodies in the lesions of anti-CD47 Ab treated animals in vivo (Fig 2d). To reconcile this discrepancy, we used an established in vitro phagocytosis assay, and found that anti-CD47 Ab potently induced the clearance of diseased and apoptotic vascular smooth muscle cells and macrophages which had been exposed to oxidized phospholipids to simulate the atherosclerotic environment (Fig 2e, Extended Data Fig 4c–f). Similarly, the number of ‘free’ apoptotic bodies not associated with an intraplaque macrophage (indicative of failed efferocytosis) was reduced after anti-CD47 Ab treatment in vivo, as suggested by co-localization studies (Fig 2f, Extended Data Fig 5a) and electron microscopy (Fig 2g, Extended Data Fig 5b). Ultimately, these animals accumulated less apoptotic debris and developed lesions with smaller necrotic cores (Fig 2h, Extended Data Fig 5c). From a mechanistic perspective, anti-CD47 Ab therapy was associated with a profound suppression of intraplaque SHP1 phosphorylation, confirming interruption of the ‘don’t eat me’ axis downstream of SIRP-α, CD47’s cognate anti-phagocytic receptor (Fig 2i, Extended Data Fig 5d). These findings indicate that targeting of CD47 can reduce atherosclerosis, and appears to do so by specifically reactivating efferocytosis within the lesion, without altering programmed cell death itself.
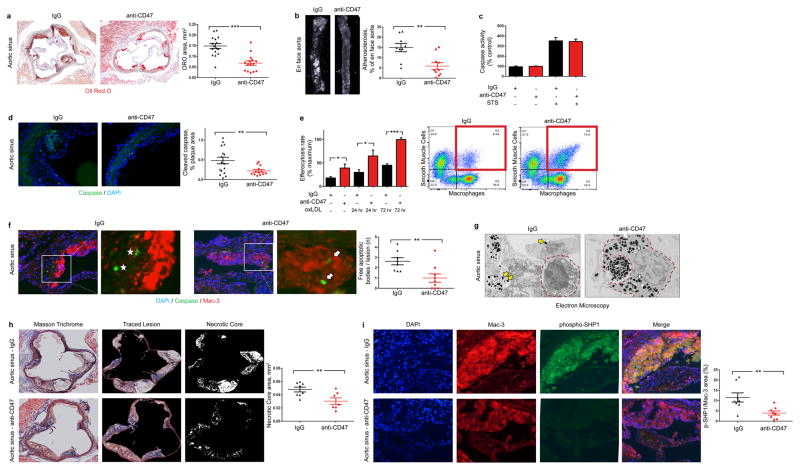
Figure 2. Inhibition of CD47 stimulates efferocytosis and prevents atherosclerosis.
(a). Compared to mice treated with a control Ab (IgG, n=16), mice treated with an inhibitory anti-CD47 Ab (n=15) develop significantly smaller atherosclerotic plaques, as measured by Oil-Red-O (ORO) content in the aortic sinus. (b). Total aortic atherosclerosis content is also reduced. Inhibition of CD47 signaling does not alter the rate of programmed cell death in vitro (c) but does reduce the accumulation of apoptotic bodies in vivo (d). (e). Anti-CD47 Ab promotes efferocytosis of vascular cells at baseline and after exposure to pro-atherosclerotic lipids. Representative FACS phagocytosis plots for lipid loaded (72hr) SMCs shown at right (all assays repeated in triplicate). (f). In vivo, anti-CD47 Ab reduces the number of ‘free’ apoptotic bodies not associated with phagocytic macrophages, potentially indicative of increased efferocytosis (stars indicate ‘free’ apoptotic bodies, arrows indicate ‘not-free’ apoptotic bodies). (g). Electron microscopy confirms that mice treated with anti-CD47 Ab display features of enhanced intraplaque efferocytosis, including an increased prevalence of macrophages which had ingested multiple apoptotic bodies (white arrows) and a reduced burden of ‘free’ apoptotic bodies (yellow arrows). (h). Mice treated with anti-CD47 Ab develop smaller necrotic cores than mice treated with IgG. (i). Anti-CD47 Ab inhibits phosphorylation of lesional SHP1, a key anti-phagocytic effector molecule known to signal downstream of CD47. STS = staurosporine. Comparisons made by two-tailed t tests. *** = P < 0.001, ** = P < 0.01, * = P < 0.05. Error bars represent the SEM.
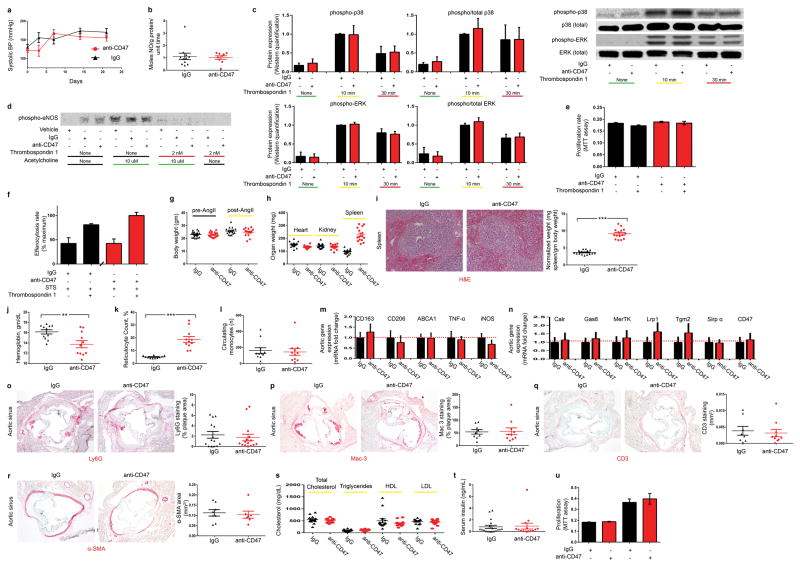
In addition to regulating phagocytosis, CD47 is also known to serve as a receptor for the vasoactive and nitric oxide (NO)-regulating cytokine, thrombospondin-1 (Tsp1)20. However, mice treated with anti-CD47 Ab and IgG control had similar systemic blood pressures (Extended Data Fig 6a) and rates of pulmonary NO elaboration (Griess reaction, Extended Data Fig 6b), arguing that anti-CD47 Ab did not alter endothelial function in vivo. Further, anti-CD47 Ab did not influence Tsp1 signaling in vitro, having no modifying effect on Tsp1-dependent MAPK signaling, eNOS phosphorylation, SMC proliferation, or phagocytosis rates (Extended Data Fig 6c–f). These data suggest that the anti-CD47 Ab employed in this study mediated its anti-atherosclerotic effects independently of Tsp1 signaling. Although full dose anti-CD47 Ab therapy was again found to promote splenic erythrophagocytosis and compensatory reticulocytosis (CD47 is a critical marker of self which is downregulated on aging RBCs)4,15, this toxicity was self-limited and anemia was not observed with chronic administration. Otherwise, this pro-efferocytic agent appeared to be well tolerated, having no discernable effect on circulating leukocytes, lipid levels, or other metabolic parameters relevant to vascular disease (Extended Data Fig 6g–u and Table 1a).
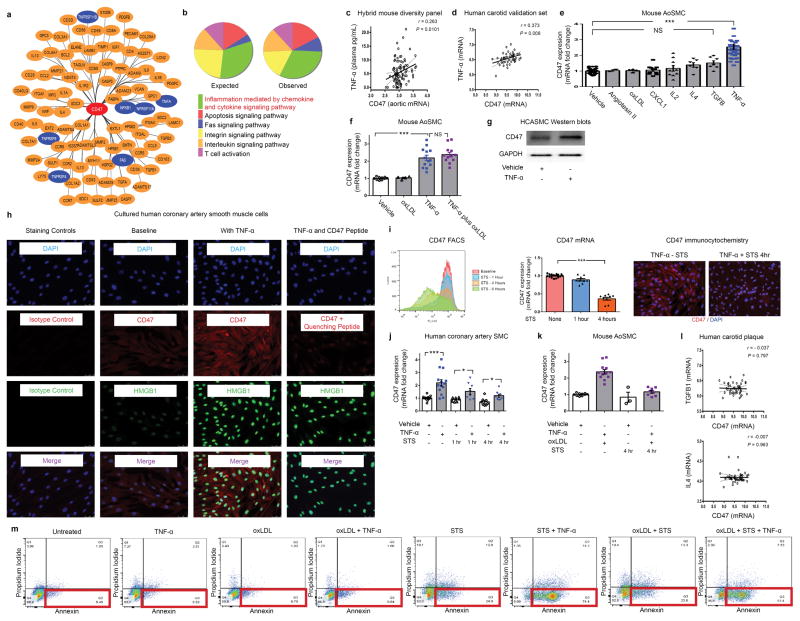
To gain fundamental insights into why CD47 is upregulated in atherosclerosis, we employed a bioinformatic approach in which we tested for genetic co-expression across panels of mouse and human vascular tissue. This yielded a list of genes which were significantly co-expressed with CD47 in vivo (Extended Data Fig 7a). Pathway analyses of these genome-wide datasets identified “inflammation mediated by chemokine and cytokine signaling pathway” as the top pathway associated with CD47 in vascular tissue (Extended Data Fig 7b and Table 1b–c), and specifically implicated “tumor necrosis factor alpha” as the factor most likely to function as its upstream regulator (Fig 3a). Subsequent correlation studies identified a strong positive association between CD47 and both the canonical TNF receptor, TNFR1 (Fig 3b), as well as TNF-α itself (Fig 3c) in human atherosclerotic vessel specimens. Similarly, CD47 expression levels were correlated with TNF-α levels in tissue from atherosclerotic mice and confirmatory human datasets (Extended Data Fig 7c–d). Together, these informatics and co-expression studies implicate TNF-α - a proinflammatory cytokine known to be upregulated in atherosclerosis - in CD47-dependent vascular disease21,22.
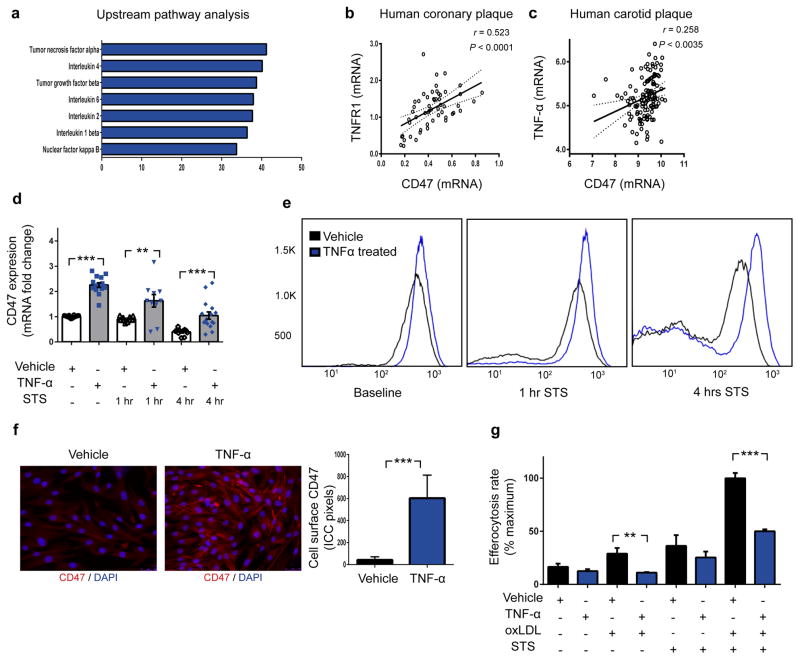
Figure 3. The pro-atherosclerotic cytokine, TNF-α, induces CD47 expression and renders vascular cells resistant to phagocytic clearance.
(a). Ingenuity Pathway Analysis identifies “Tumor necrosis factor alpha” as the regulator most likely to be upstream of genes which are co-expressed with CD47 in vascular tissue ex vivo. Co-expression studies confirm that CD47 is positively correlated with the canonical TNF-α receptor, TNFR1, in human coronary plaque (b) and TNF-α levels in human carotid plaque (c). The Pearson correlation coefficient was determined assuming a Gaussian distribution and P values were determined using a two-tailed test shown with the 95% confidence band of the best fit line. (d). In vitro, TNF-α treatment significantly increases the basal expression of CD47 in vascular SMCs, and blunts the decrease expected to occur during apoptosis. Flow cytometry (e) and fluorescent microscopy (f) confirm that TNF-α increases the cell-surface expression of CD47 on vascular cells at baseline and during programmed cell death. (g). In vitro efferocytosis assays indicate that TNF-α treatment renders vascular SMCs resistant to programmed cell clearance under a variety of pro-atherosclerotic conditions. Comparisons made by two-tailed t tests. *** = P < 0.001, ** = P < 0.01. Error bars represent the SEM.
To investigate the causality of these associations, we next tested whether CD47 is directly downstream of TNF-α. We found that treatment of vascular SMCs with recombinant TNF-α led to a consistent upregulation of cellular CD47 expression, while no effect was observed with a variety of other common pro-atherosclerotic or pro-inflammatory insults (Extended Data Fig 7e–h). Further, TNF-α blunted the progressive decrease in CD47 expression normally expected to occur during programmed cell death (Fig 3d–f, Extended Data Fig 7i–l). As a result of their higher levels of ‘don’t-eat-me’ molecules, TNF-α-treated cells were less likely to be phagocytosed by macrophages, particularly when concomitantly exposed to oxidized LDL and pro-apoptotic stimuli (Fig 3g, Extended Data Fig 7m). Because impaired efferocytosis is known to incite pro-inflammatory cytokine elaboration9, it is possible that a positive feedback loop underlies the co-localization of TNF-α, CD47, and uncleared pathological cells and apoptotic bodies within the atherosclerotic plaque22.
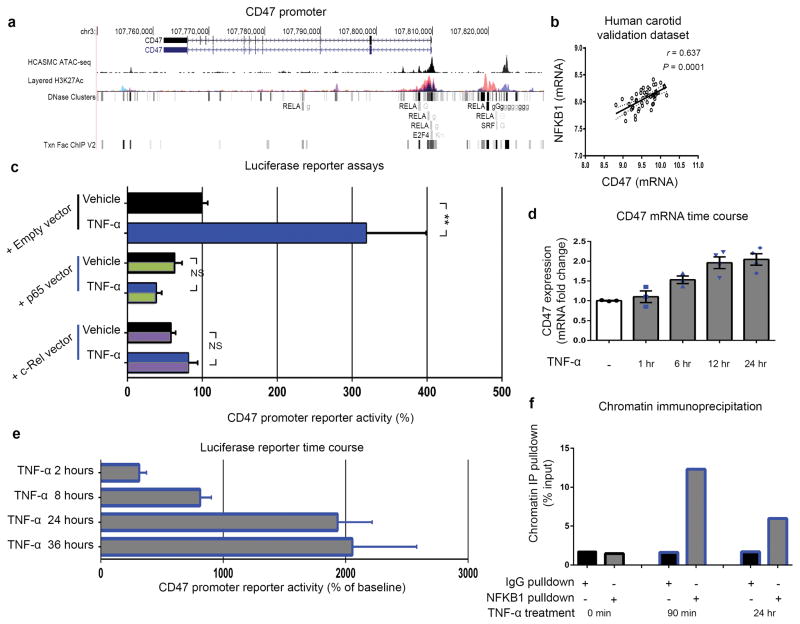
Analysis of the CD47 promoter in vascular SMCs revealed a region of open chromatin predicted to contain binding sites for several of the NFKB transcription factors known to be downstream of TNFR1 (Extended Data Fig 8a and Table 1d). Amongst these, the classical pro-inflammatory factor NFKB1 (p50) was found to be positively correlated with CD47 expression in both human coronary and carotid plaques (Fig 4a–b, Extended Data Fig 8b). Luciferase reporter assays performed with a vector containing the CD47 promoter revealed that TNF-α treatment stimulated basal CD47 expression, and that the effect was specifically enhanced when NFKB1 was simultaneously overexpressed in these cells (Fig 4c, Extended Data Fig 8c–e). Chromatin immunoprecipitation assays confirmed that NFKB1 binds to the CD47 promoter in vitro, and that this occupancy is increased several fold upon treatment with TNF-α (Fig 4d, Extended Data Fig 8f).
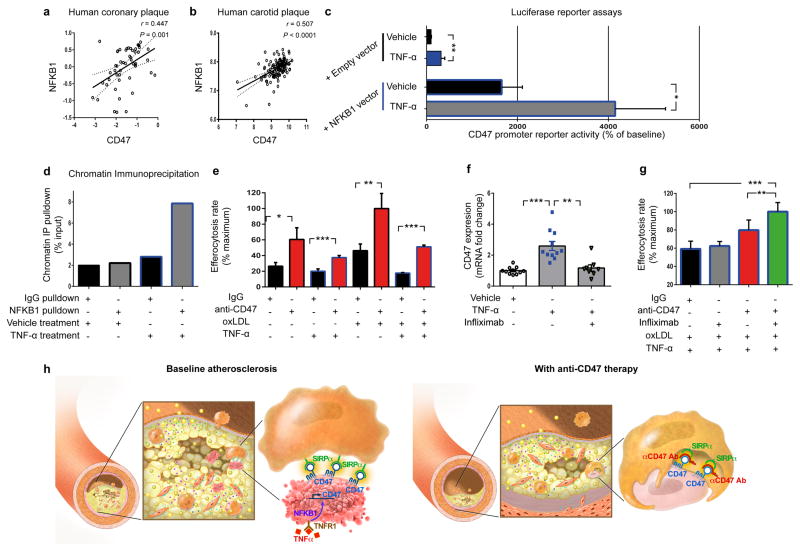
Figure 4. TNF-α promotes CD47 expression via NFKB1 and is a translational cardiovascular target.
Co-expression analyses confirm that NFKB1 is significantly correlated with CD47 expression in both human coronary (a) and carotid (b) atherosclerotic plaque. Pearson correlation coefficients were determined assuming a Gaussian distribution and P values were determined using a two-tailed test. (c). Dual luciferase reporter assays reveal that CD47 promoter activity is stimulated in cells treated with TNF-α (top) but that this effect is significantly enhanced in cells co-transfected with an NFKB1 expression vector. (d). Chromatin immunoprecipitation studies confirm significant enrichment of NFKB1 protein on the CD47 promoter in TNF-α-treated human coronary artery SMCs. (e). In vitro efferocytosis assays reveal that anti-CD47 Ab enhances the clearance of cells exposed to TNF-α, and that its pro-efferocytic capacity is enhanced under pro-atherosclerotic conditions. (f). Pretreatment with the anti-TNF-α monoclonal Ab, Infliximab, prevents the upregulation in CD47 mRNA that normally occurs in SMCs exposed to TNF-α. (g). Concomitant inhibition of CD47 and TNF-α using anti-CD47 Ab and Infliximab, respectively, produces synergistic benefit in the clearance of diseased vascular cells as assessed by ANOVA. (h). Putative mechanism explaining why efferocytosis is impaired in cardiovascular disease, and how inhibition of CD47-SIRPα signaling could represent a new therapeutic target. Comparisons made by two-tailed t tests, unless otherwise specified. *** = P < 0.001, ** = P < 0.01, * = P < 0.05. Error bars represent the SEM.
From a translational perspective, we found that anti-CD47 Ab was able to stimulate efferocytosis in TNF-α-treated cells, and that the effect was most pronounced under dyslipidemic, pro-atherosclerotic conditions (Fig 4e). A modest incremental benefit was observed when anti-CD47 Ab therapy was combined with commercially available anti-TNF-α therapies, such as infliximab or etanercept, likely because of their inhibitory influence on CD47 expression in mouse and human tissue (Fig 4f–g, Extended Data Fig 9). These data are particularly provocative given the observation that subjects prescribed TNF-α-inhibiting antibodies for inflammatory disorders such as lupus and rheumatoid arthritis appear to be protected from myocardial infarction21,23.
The finding that CD47 expression is pathologically upregulated in both cancer and cardiovascular disease suggests a commonality between these two conditions. In leukemogenesis, cancer stem cells out-compete normal hematopoietic stem cells, while countering PrCR signaling; viable MDS hematopoietic oligolineage progenitors express the ‘eat me’ signal calreticulin, but not CD47, while AML derived from MDS are CD47 positive24. Similar cellular processes in the vasculature may explain the recent observations that de-differentiated SMCs undergo clonal expansion within the atherosclerotic plaque25,26. Furthermore, the top cardiovascular GWAS locus surprisingly resides near an important tumor suppressor locus27, which in turn regulates SMC efferocytosis17. Future studies will need to examine whether expansion of CD47hi SMC clones contributes to atherosclerosis, and if their clearance can be accomplished without the induction of anemia (e.g. with a dose escalation approach which appears to be safe in non-human primates 28, and is currently being pursued in first-in-human clinical trials29).
Together, these data provide insights into why PrCR is impaired within the atherosclerotic plaque, and how this may promote lesion expansion. Our findings bolster the ‘inflammatory hypothesis’ of atherosclerosis30, and specifically link cytokine signaling with ‘don’t-eat-me’ signaling in vascular disease. Given the experimental success of pro-efferocytic therapies in the oncology field via antibodies that block the CD47 ‘don’t eat me’ signal5, it is possible that these findings will provide a novel nonsurgical treatment of cardiovascular disease (Mechanism Fig 4h).
Methods
Human cardiovascular tissue – carotid endarterectomy samples
In this study, a total of n = 182 human carotid endarterectomy samples and nonatherosclerotic control arteries were used. These include heterogeneous atherosclerotic plaque samples obtained from patients undergoing surgery for symptomatic (stroke or transient ischemic attack) or asymptomatic (no history of cerebrovascular event) high-grade carotid stenosis (>50% NASCET criteria) as part of the Biobank of Karolinska Endarterectomies (BiKE). N = 15 nonatherosclerotic control arteries (iliac artery and aorta) were obtained from organ donors without any history of cardiovascular disease. Patients were consecutively enrolled in the study, with the first n = 127 constituting the discovery cohort (40 asymptomatic, 87 symptomatic) and the next n = 50 constituting the validation cohort (10 asymptomatic, 40 symptomatic). All samples were collected with informed consent from patients, organ donors or their guardians. The BiKE study was approved by the Ethical Committee of the Northern Stockholm. DNA and RNA was extracted from these specimens and analyzed by Ilumina 610w -QuadBead SNP-chips and the Affymetrix HG-U133 plus 2.0 microarrays (discovery cohort) or the Affymetrix HG-U133a Genechip arrays (validation cohort), as previously described16,17 and deposited in Gene Expression Omnibus (accession number GSE21545). Robust multi-array average (RMA) normalization was performed and processed gene expression data was returned in log2-scale. The relative expression of each gene was determined by comparing the pixel intensity of the 11 probe pairs which correspond to each transcript to the ‘normalization control set’ specific to each array31,32. Student’s T-test with correction for multiple comparisons according to the Sidak-Bonferroni method was used for statistical analyses of microarray data. Pearson’s correlations were calculated to determine associations between expression of the gene of interest and other genes from microarrays. Publically available data from the Helsinki Carotid Endarterectomy Study (HeCES) were analyzed as a second human validation cohort33. Additional publically available vascular and nonvascular microarray data deposited in GEO were also analyzed (as indicated by GSE number in the corresponding figure legend), including studies of microdissected atherosclerotic plaques and samples taken from individuals treated with commercially available TNF-α inhibitors. A P-value <0.05 was considered to indicate significance.
Human cardiovascular tissue – coronary artery samples
In this study, a total of n = 114 human coronary artery samples were used. N = 51 atherosclerotic epicardial coronary artery segments were harvested from n = 22 orthotopic heart transplant donors, as previously described34. Additionally, n = 56 coronary artery segments were extracted from atherosclerotic intracoronary plaques in patients undergoing coronary atherectomy, as previously described35. Briefly, longitudinal atherectomy was performed with the Silverhawk atherectomy catheter in arteries with flow-limiting stenosis after diagnostic angiography. RNA was isolated from both sets of specimens and hybridized to custom dual-dye gene expression microarrays representing approximately 22,000 features34,35. Briefly, the custom probe set was identified from data mining and curation of atherosclerosis and vascular cell culture based expression analyses and combined with the Agilent Human 1A and 1B arrays. In each microarray experiment, the expression levels of CD47 were determined via array-specific hybridization probe sets (~10 per transcript). Automated feature extraction software was used to filter out background or saturated spot intensities to eliminate inherent probe and spatial biases during the fluorescent detection. This software computes the log (base2) ratios and filters features by P -value. These values were normalized according to protocol for each array, using a locally weighted linear regression curve fit (LOWESS) to correct for dye biases. This normalization procedure results in array specific log2 ratios, and provides accurate measures of relative gene expression differences36. Normalized data from candidate transcripts were then used to calculate a Pearson correlation coefficient r assuming a Gaussian distribution, and two-tailed P-values were calculated for each correlation coefficient.
In addition to those samples which were obtained for RNA analysis, an additional seven right coronary arteries were obtained from rapid autopsies from adult patients with a spectrum of coronary artery disease for histological analysis. The arteries were fixed with 4% paraformaldehyde (PFA) for several hours, and immersed in 30% sucrose at 4C° overnight. The coronaries were then serially sectioned and segments of atheroma and relatively normal coronary artery were embedded in paraffin or OCT, and sectioned at 7-μM thickness.
For immunohistochemical staining, paraffin slides were deparaffinized and rehydrated with xylene and alcohol gradients, and antigen retrieval was performed in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) using a pressure cooker. The sections were blocked with 5% goat serum, then stained with primary antibodies (anti-CD47, Abcam B6H12.2, 2 μg/ml) or mouse IgG1 kappa (eBioscience, 2 μg/ml) at 4C° overnight. The sections were washed in TBS, incubated with MACH4™ kit (Biocare Medical) according to the manufacturer’s instructions, and then detected using the Vulcan Fast Red Chromogen kit2 (Biocare Medical). Staining for SMC content was accomplished by washing sections in water and PBS, followed by incubation with anti-SM22 alpha Ab (Abcam, ab14106, 1:300). The sections were washed with PBS, incubated with Alexa Fluor 488 goat anti-Rabbit (Life technologies, 1:250), washed, and mounted with Vectashield Mounting medium with DAPI (Vector Laboratories). Pictures were taken by Nikon digital camera mounted on an inverted fluorescence microscope. Serial sections were prepared as described above, and also stained with Masson Trichrome (MT, Richard-Allan), or haematoxylin and eosin (H&E, Richard-Allan). For the staining of frozen sections, OCT was removed in water, and the sections were stained with Oil Red O (ORO, Sigma-Aldrich, O0625, 0.5%), haematoxylin and eosin (H&E, Richard-Allan), smooth muscle alpha-actin (Abcam, ab5694, 1:300), HMGB1 (Abcam, ab18256, 1:100), CD206 (Abcam, ab64693, 1:50), CD47 (Abcam B6H12.2 or Novus Biologicals, #NBP2-31106, 1:50) or mouse IgG1 kappa (eBioscience). Secondary antibodies included Alexa Fluor 594 goat anti-mouse (Life technologies, A11005, 1:300) and Alexa Fluor 488 goat anti-rabbit (Life technologies, A11034, 1:300). Antibody specificity was confirmed using isotype control and by preincubating the anti-CD47 Ab with recombinant CD47 antigen (R&D Systems) in a 1:5 ratio for 16h at 4°C before application. High resolution imaging of the carotid sections was performed as previously described37.
Murine cardiovascular tissue
Atherosclerosis models
In the atherosclerosis studies described below, a total of n = 179 male apoE deficient mice on the C57BL/6 background (apoE−/−, Jackson Laboratory, catalog #002052) were used.
In the main atherosclerosis intervention studies, 8 week old mice were implanted with subcutaneous Alzet minipumps (model 2004, Alzet Osmotic Pumps) containing Angiotensin II (AngII, Sigma-Aldrich, 1000 ng/kg/min) and initiated on a high fat Western diet (21% anhydrous milk fat, 19% casein and 0.15% cholesterol, Dyets no. 101511) for the ensuing 4 weeks, as previously described19. To determine the impact of CD47 signaling on vascular disease, mice were injected with either 200 μg of the inhibitory anti-CD47 Ab (MIAP410, BioXcell, n=18) or IgG1 control (MOPC-21, BioXcell, n=20) IP QOD, at the dose previously studied15. The Ab therapy was started one day before the pump implantation. Animals were observed daily, and in the case of premature sudden death, necropsy was performed to determine the cause of mortality. Blood pressure in conscious mice was measured at baseline (after standard acclimatization with a Visitech Systems Inc. machine), and weekly for the duration of the study. At 12 weeks of age, the mice were euthanized after an overnight fast, with serum and visceral organs (including the aortae) isolated and processed for analysis.
In the chronic atherosclerosis studies, male apoE deficient mice were weaned and initiated on a high fat diet at 4 weeks of age and maintained on this for the subsequent 12 weeks (without any Angiotensin infusion). Antibody injection was performed as described above while the high fat diet was given (n=9 per condition), and the animals were euthanized at the age of 16 weeks.
In the established disease model, apoE deficient mice were weaned onto a high fat diet at 4 weeks of age and continued on this for the ensuing 8 weeks (without any Angiotensin infusion). At 12 weeks of age (after lesions had developed), mice were initiated on 200 μg of anti-CD47 Ab (n=14) or IgG1 (n=13) IP QOD for the ensuing 6 weeks, and sacrificed at 18 weeks of age.
In the TNF-α inhibitor synergy studies 8 week old apoE deficient mice were implanted with AngII pumps and maintained on Western high fat diet, and then randomized to one of four groups, including the: (1) IgG group (mouse IgG and human Fc, n=10); (2) Etanercept group (Etanercept 0.2 mg/kg SQ weekly (Amgen) and mouse IgG IP QOD, n=8); (3) CD47 Ab group (MIAP410 50 μg IP QOD and human Fc SQ weekly, n=16); or (4) Combination group (MIAP410 and Etanercept, n=19)). In this model, the Ab was started the day before AngII pump implantation and delivered for 4 weeks, and the mice were euthanized at 12 weeks of age. Note that in this model, the anti-CD47 Ab dose was reduced by 75% to determine whether a lower dose of therapy could also impact atherogenesis.
In the short-term intervention model, 8 week old apoE deficient mice were implanted with AngII pumps and maintained on Western high fat diet without Ab therapy for the ensuing 23 days. Beginning at day 23, the mice received SQ injections of either: (1) IgG daily, n=10; (2) Etanercept (0.8 mg/kg at day 23, n=6); (3) anti-CD47 Ab (200 μg of MIAP410 daily, n=11); or (4) combination therapy, n=11 between days 23 and 27. This cohort of mice was euthanized after only 5 days of antibody treatment (at day 28) and was used to evaluate the effect of Ab therapy in established atherosclerotic plaques of identical size.
To evaluate the effect of antibody therapy on atherosclerotic plaque vulnerability, we used the recently described “Tandem Stenosis” model38. At 6 weeks of age, apoE deficient mice were initiated on high fat diet and maintained on this for ensuing 6 weeks. At 12 weeks of age, tandem stenoses were introduced in a manner shown to reproducibly alter shear stress and induce plaque rupture, as previously described38. Briefly, the mice were anesthetized by isoflurane inhalation and an incision was made to allow dissection of the right common carotid artery from the circumferential connective tissues. Serial stenosis with a 150 μm outer diameter were then introduced 1 mm and 4 mm from the carotid bifurcation. The stenosis diameter was obtained by placing a 6–0 suture around the carotid artery together with a 150 μm needle that was tied to it and later removed. Antibody therapy was started the day before the surgery and continued thereafter. Mice were euthanized 7 weeks after the surgery and intraplaque hemorrhage was quantified within the processed tissue sections.
In addition to the mice treated with anti-CD47 Ab or control Ab, a separate cohort of 12 male apoE−/− mice were fed a high fat diet for either 8, 12 or 20 weeks, but were not treated with an Ab nor implanted with osmotic minipumps. These mice were used to determine aortic gene expression changes during atherogenesis, using the RNA analysis methods described below. In these experiments, comparison was made to control C57BL/6 mice fed standard chow diet for 24 weeks.
Finally, a cohort of 3 apoE−/− mice were implanted AngII osmotic pump and fed a high fat diet for 4 weeks (but not treated with an Ab) and then injected with biotin-labeled anti-CD47 Ab 24 and 6 hours prior to sacrifice, to determine where the therapeutic antibody accumulates in vivo. Nonatherosclerotic C57BL/6 and CD47 deficient mice (CD47−/−, Jackson Laboratory, catalog #003173) were also injected with the biotin-labeled Ab injection, and served as controls. All animals were analyzed as described below.
Vascular tissue preparation, immunohistochemistry and atherosclerotic lesion quantification
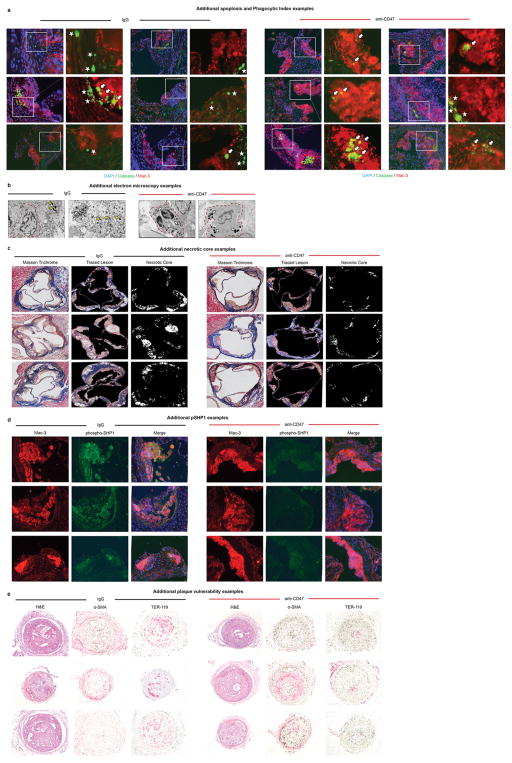
Aortic atherosclerosis lesion area was determined as described previously17. Briefly, the arterial tree was perfused with PBS and then fixed with 4% PFA. The heart and the full-length of the aorta-to-iliac bifurcation was exposed and dissected carefully from any surrounding tissues. Thoracic aortas were then opened along the ventral midline and dissected free of the animal and pinned out flat, intimal side up, onto black wax. Aortic images were captured with a digital camera mounted on a Nikon stereomicroscope and analyzed using Adobe Photoshop CS5 software. The percentage of lesion area was calculated as total lesion area divided by total surface area. The atherosclerotic lesions within the aortic valve area (aortic sinus) were analyzed as described previously17. The samples were perfused with PBS, fixed with 4% PFA, embedded in OCT, and sectioned at 7-μM thickness. Four sections at 100-μM intervals were collected from each mouse and stained with ORO, MT, H&E, smooth muscle α-actin (SMA, Abcam, ab5694, 1:300), Mac-3 (BD Sciences, BD 550292, 1:100), CD-3 (Abcam, ab5690, 1:150), and Ly-6G (BD Sciences, BD 551459, 1:300). Atherosclerosis burden was quantified from the luminal aspect of the blood vessel through the plaque to the internal elastic lamina (i.e. lipid in the neointima was quantified). Necrotic core size was quantified by calculating the area of the lesion which was acellular on Masson Trichrome staining, as previously described17,39. Plaque hemorrhage was quantified by determining the presence or absence of RBCs (TER-119, Santa Cruz Biotechnology) within the plaque, as previously described40. Subsequent immunohistochemical studies were quantified from the luminal aspect of the blood vessel through the plaque to the external elastic lamina (to assess changes which also involved the tunica media). To detect the localization of injected Biotin-labelled anti-CD47 Ab, the avidin-biotin complex technique was used. Frozen sections of aortic sinus were prepared from the mice injected with Biotin-labelled anti-CD47 Ab, as described above. Endogenous peroxide activity was blocked by incubation with 0.3% hydrogen peroxide for 30 minutes, and the sections were washed with water and PBS, followed by blocking with 5% goat serum for 30 minutes. Biotin was detected using Vecstatin ABC kit and DAB substrate kit per protocol (Vector laboratories). Corresponding aortic sinus sections from the mouse without Biotin Ab injection were used as negative controls, as were CD47−/− mice which had been injected as above. For immunofoulrescent staining of these samples, sections were blocked with 5% goat serum for 30 minutes, then incubated with Streptavidin-Alexa Fluor 546 conjugate (Life Technologies, 1:300) and Mac-3 (BD, 1:100) for 1 hour, followed by Alexa Fluor 488 donkey anti-Rat (Life Technologies, 1:300). In vivo apoptosis was assessed by staining for TUNEL positivity with the Cell Death Detection Kit (Roche), per protocol, and confirmed with Cleaved Caspase 3 (Cell Signaling #9661, 1:200) staining followed by Alexa Fluor 488 goat anti-Rabbit (Life technologies, 1:250). The cleaved caspase 3 positive area was measured and quantified using Adobe Photoshop, and the percentage of positive area was calculated as total caspase 3 positive area divided by total atherosclerotic plaque area measured by ORO staining in the serial sections. To calculate the in vivo phagocytic index, we performed double staining of Cleaved Caspase 3 (detected with Alexa Fluor 488 goat anti-Rabbit Ab) and Mac-3 (detected with Alexa Fluor 594 goat anti-Rat Ab (Life technologies, 1:250)). The number of free apoptotic cells not associated with a macrophage (indicated by a star) was manually assessed in a blinded fashion, and compared to apoptotic cells associated with a macrophage (indicated by an arrow), as previously described2. For phospho-SHP1 staining, the sections were stained with phosphor-SHP1 Ab (Abcam, ab131500, 1:50) and Mac-3 followed by Alexa Fluor. The phospho-SHP1 positive area was normalized to Mac-3 positive area. All lesion areas and indices were measured and quantified using Adobe Photoshop by a blinded observer. Samples harvested from several tissue beds were also snap frozen in liquid nitrogen for subsequent mRNA and protein expression analysis, as described below.
Vascular tissue electron microscopy
Electron microscopy was performed in the Stanford University Cell Sciences Imaging Facility, as previously described17. Briefly, samples were fixed and processed using standard histologic techniques then imaged using a JEOL JEM-1400 Transmission Electron Microscope. Ingested apoptotic bodies (indicated by white arrow), free apoptotic bodies (indicated by yellow arrow), and apoptotic bodies undergoing secondary necrosis (indicated by red arrow) were qualitatively assessed in a blinded manner, as previously described17.
Serum and plasma analysis
Serum chemistry, lipid, CBC, and differential analyses were performed by the Stanford Animal Diagnostic Laboratory, as previously described17. In brief, blood samples were collected by cardiac puncture after an overnight fast. Automated hematology was performed on the Sysmex XT-2000iV analyzer system. Blood smears were prepared for all full CBC samples and reviewed by a medical technologist. Chemistry analysis was performed on the Siemens Dimension Xpand analyzer, and included analyses of renal function, electrolyte levels, liver function tests, fasting glucose levels and fasting lipid panels. A medical technologist performed all testing, including dilutions and repeat tests as indicated, and reviewed all data. Serum insulin levels were measured by ELISA kit according to the manufacturer’s instruction (EMD Millipore).
Griess reaction
The activity of nitric oxide synthase was evaluated using a modified Griess assay. Lung samples were harvested from mice and snap frozen by liquid nitrogen prior to homogenization in PBS. The nitrate and nitrite levels were measured by Ultrasensitive Colorimetric Assay for Nitric Oxide Synthase (Oxford Biomedical Research) according to the manufacture’s instruction, and standardized by the protein amount.
Murine cardiovascular tissue – the Hybrid Mouse Diversity Panel
The Hybrid Mouse Diversity Panel (HMDP), which includes a quantitative analysis of 109 classical and recombinant inbred mouse strains41, was used to identify factors associated with vascular CD47 expression, in vivo. Briefly, whole aorta from the arch to the mid-abdomen was snap-frozen at the time of euthanasia and total RNA was isolated using the RNeasy kit (Qiagen), as described42. Genome wide expression profiles were determined by hybridization to Affymetrix HT-MG_430 PM microarrays on a subset of female mice from 104 strains (N = 2 aorta per strain). Quantification of plasma cytokines was carried out in a multiplexed immune-capture microbead system (Milliplex Mouse Cytokine / Chemokine Magnetic Bead Panel MCYTOMAG-70K, EMD Millipore) as per manufacturer’s instructions. Cytokines profiled were: G-CSF, GM-CSF, IFNr, IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-7, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IP-10, KC, MCP-1, MIP-1α, MIP-1β, M-CSF, MIP-2, MIG, RANTES, TNF-α. Plasma insulin was measured using the mouse insulin ELISA kit (80-INSMS-E01, Alpco) as per manufacturer’s instructions. Pearson’s correlations were generated to calculate transcript-transcript and transcript-trait correlations. Using these methods, the genes and plasma cytokines which were significantly associated with aortic CD47 expression levels were identified.
In Silico Bioinformatics Methods
Pathway analysis
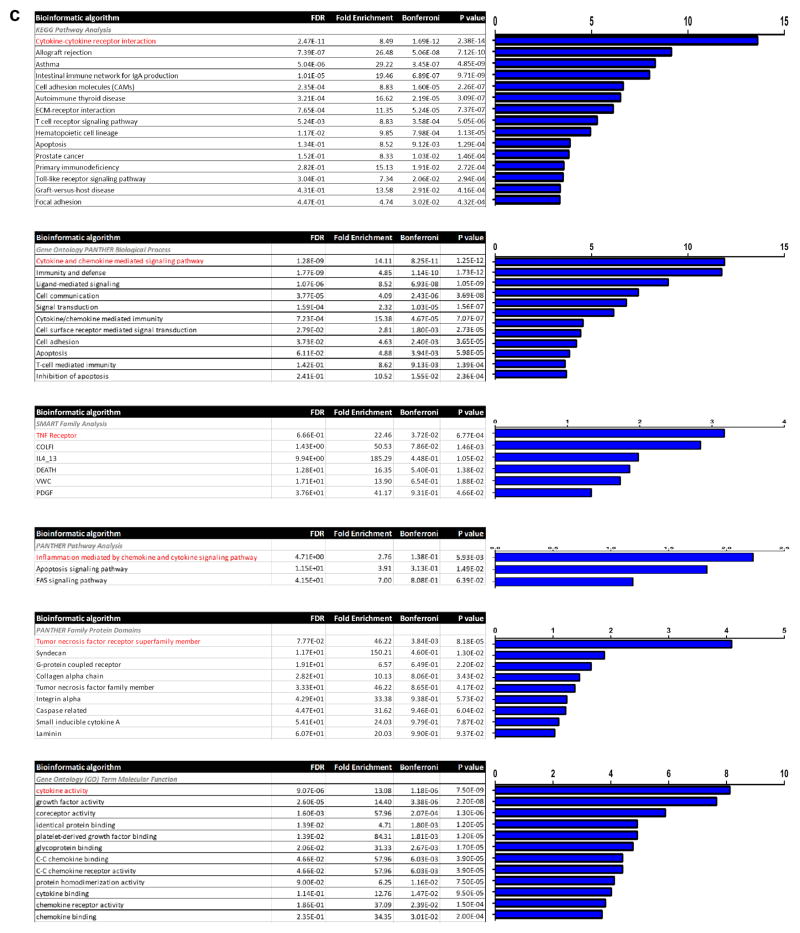
Genome-wide correlation analyses were performed to identify genes which are significantly correlated with CD47 expression in the human and murine vascular tissue collections described above. These lists were intersected to identify genes that are commonly co-expressed across multiple datasets in both species. The resulting list of genes (Extended Data Fig 7a) was subjected to a series of bioinformatics analyses including the Database for Annotation, Visualization and Integrated Discovery (DAVID), Kyoto Encyclopedia of Genomes and Genes (KEGG), Gene Ontology (GO), and Protein Analysis Through Evolutionary Relationships (PANTHER) classification. Additionally, genes were mapped to open chromatin regulatory intervals in primary human vascular cells and analyzed using the Genomic Regions Enrichment of Annotations Tool (GREAT). Pathways found to be overexpressed were ranked by P value. Statistical overrepresentation and enrichment analyses were performed using Bonferroni correction for multiple testing (P < 0.05 cutoff).
Ingenuity Pathway Analysis (IPA) was then performed on the resulting intersected CD47 co-expression gene list from human carotid atherosclerosis (BiKE study) and murine aortic atherosclerosis (HMDP study). Briefly, the 63 gene identifiers and expression values were analyzed using the Core pathway analysis after removing any duplicates and unmapped IDs. The resulting networks were then subjected to an Upstream Regulators Analysis using the Ingenuity Knowledge Base after applying a filter to include all genes, RNAs and proteins, while excluding chemicals or drugs. Overlap P-values were calculated for each upstream regulator using a Fisher’s exact test and activation z-scores were calculated by comparing observed direction of target genes with inferred literature-derived regulatory direction to identify the most significant upstream regulators for CD47.
Promoter analysis
Using the UCSC browser, the genetic sequence 1Kb upstream of the CD47 promoter was identified and analyzed, as previously described17. In addition to evaluating for open chromatin status and DNAse hypersensitivity sites, potential transcription factor binding sites (TFBS) were predicted using the following online bioinformatics tools: TRANSFAC (BIOBASE), P-Match, TFSearch, Alibaba, PROMO, and MatInspector. High confidence binding sites (85% likelihood cutoff) were accepted for additional analysis. Additionally, the predicted binding sites in the CD47 promoter region were intersected with open chromatin peaks identified from the Assay for Transpose Accessible Chromatin followed by sequencing (ATAC-seq) in primary human coronary artery smooth muscle cells (HCASMC).
In Vitro Methods
Cell culture
Primary vascular smooth muscle cells (SMCs) were harvested from the aortas of C57BL/6 mice and propagated in DMEM supplemented with 10% FBS, as previously described17,43. Human coronary artery smooth muscle cells (HCASMC, Lonza Catalog # CC-2583, passage #3–6) were propagated in SmGM-2 growth media (Lonza) containing 5% FBS. To obtain human macrophages, leukocyte reduction system (LRS) chambers were obtained from the Stanford Blood Center from anonymous donors. Monocytes were purified on an autoMACS Pro Separator (Miltenyi) using whole blood anti-CD14 microbeads (Miltenyi) and differentiated to macrophages by culture for 7–10 days in IMDM+GlutaMax (Invitrogen) supplemented with 10% AB-Human Serum (Gemini Bio-Products #100-512) and 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen). RFP+ mouse macrophages were generated and evaluated as previously described44. Briefly, bone marrow cells were isolated from C57BL/Ka Rosa26 mRFP1 transgenic mice and differentiated in IMDM+GlutaMax supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin, and 10 ng/mL murine M-CSF (Peprotech). Mouse yolk sac endothelial cell line (C166, ATCC Catalog # CRL-2581) and mouse macrophage cell line (RAW 264.7, ATCC Catalog # TIB71) were grown in DMEM-growth media containing 10% FBS, while mouse T lymphocyte cell line (EL4, ATCC Catalog # TIB-39) were grown in DMEM containing 10% horse serum. Human macrophage cell line (THP1, ATCC Catalog # TIB-202) were grown in RPMI-1640 medium containing 10% FBS and 0.05 mM 2-mercaptoethanol. Human embryonic kidney cells (HEK-293, ATCC Catalog # CRL-1573) used for luciferase reporter assays were grown in DMEM-growth media containing 10% FBS. No additional cell authentication or mycoplasma contamination testing was performed.
A variety of atherosclerosis-related or pro-apoptotic stimuli were applied to the cells in the experiments described below including: oxidized LDL (oxLDL, 50 μg/ml, Alfa Aesar), Ang II (100 nM, Sigma-Aldrich), fibroblast growth factor (FGF, 100 ng/ml, R&D), platelet-derived growth factor (PDGF, 100 ng/ml, R&D), and lipopolysaccharide (LPS, 1 μg/ml, Sigma-Aldrich). Additionally, a number of cytokines associated with CD47 expression through the HMDP Luminex array were also tested, including tumor necrosis factor-alpha (TNF-α, 50 ng/ml, R&D), interleukin 2 (IL-2, 100 ng/ml, Biolegend), chemokine receptor ligand 1 (Cxcl1, 100 ng/ml, Biolegend), Interleukin 4 (IL-4, 50 ng/ml, Biolegend), and transforming growth factor-beta (TGF-β, 50 ng/ml, R&D). Prior to experimentation, SMCs were serum-starved for 24 hours in DMEM and then stimulated with the stimuli listed above for 24 hours prior to analysis. In some experiments, the cells were stimulated with staurosporine (STS, 1 μM, Sigma-Aldrich) for 1 or 4 hours to induce apoptosis, after 24 hours of TNF-α treatment.
To inhibit TNF-α signaling, a chemical inhibitor (SPD 304, Sigma Aldrich) or a monoclonal Ab (Infliximab, Janssen) were used. Briefly, TNF-α was pre-incubated with 10 μM of SPD304 or 100 μg/ml of infliximab in serum free DMEM for 20 minutes prior to cell stimulation. To inhibit NF kappa B signaling, the cells were pre-treated with 10 μM BAY 11–7085 (Santa Cruz Biotechnologies) or DMSO, then stimulated with TNF-α.
For MAPK western blotting experiments, mouse aortic SMCs were serum starved for 48 hours, pre-treated with 10 μg/ml of CD47 Ab or IgG for 20 minutes, then stimulated with thrombospondin-1 (Tsp1, 10 μg/ml, R&D) for 10 or 30 minutes. For eNOS western blotting experiments, C166 cells were serum starved for 8 hours, pre-treated with 2 μM Tsp-1 with or without 10 μg/ml of CD47 Ab or IgG for 20 minutes, then stimulated with acetylcholine (Ach, 10 μM, Sigma-Aldrich) for 15 minutes.
mRNA isolation and quantitative reverse-transcription PCR
RNA was isolated from cell lysates using the miRNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA was isolated from murine organ samples using the Trizol method (Invitrogen). RNA was quantified with the Nanodrop machine (Agilent Technologies, Santa Clara, CA). For quantitation of gene transcription, cDNA was generated with MultiScribe reverse transcriptase (Applied Biosystems), and then amplified on the ABI PRISM 7900HT with commercially available TaqMan primers (Applied Biosystems, Foster City, CA) and normalized to 18S internal controls, as previously described17. A list of the primers and probes used in these studies is provided in Extended Data Table 1e.
Protein extraction and Western blotting
Total protein was isolated from cultured cell lines and tissue homogenates using 1X cell lysis Buffer (Cell Signaling) supplemented with 1X Halt Protease & Phosphatase Single-Use Inhibitor Cocktail (Thermo Scientific), as previously described17. The protein concentration in each sample was measured using Pierce BCA Protein Assay Kit (Thermo Scientific). Equal amounts of protein were loaded and separated on precast gels (Bio-Rad) and thereafter transferred onto PVDF membranes (Bio-Rad). Following a 1h incubation in 5% bovine albumin serum solution prepared in 1X TBST, these membranes were probed with commercially available antibodies designed to recognize endogenous P38 (Cell Signaling #9212, 1:1000), phospho-P38 (Cell Signaling #4511, 1:1000), ERK1/2 (cell signaling #9102, 1:1000), phospho-ERK1/2 (Cell Signaling #4377, 1:1000), phospho-eNOS (Cell Signaling #9571, 1:1000), CD47 (Novus Biologicals, #NBP2-31106 1:1000) and GAPD (1:1000; Cell Signaling Technologies) overnight at 4°C. Membranes were rinsed with TBST and incubated with appropriately matched HRP conjugated anti mouse (1:5000; Life Technologies) or anti rabbit (1:5000; Life Technologies) antibodies for 1 h, before protein expression was detected using SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific). Membranes were then scanned with a Licor Odyssey Fc imager for quantitative analysis. In some experiments, membranes loaded with protein were incubated with anti-CD47 Ab that had been preabsorbed with CD47 peptide (R&D systems, 1866-CD, 1:5 dilution) for 16 hours, to determine the specificity of the primary Ab.
Apoptosis assays
To evaluate apoptosis, the luminometric Caspase-Glo 3/7 Assay (Promega, G8090) was performed on cultured cells, according to the manufacturer’s protocol. Briefly, mouse aortic SMCs were seeded in 96-well plates at the density of 10,000 cells/well, grown at 37C° for 24 hours, and then serum starved for 24 hours. Apoptosis was induced with 1 μM STS treatment for 4 hours in the presence of 10 μg/ml of anti-CD47 Ab or IgG. Confirmatory assays were performed by flow cytometry, where cells where exposed to 24 or 72 hours of vehicle, 50 ng/ml of TNF-α, or 50 μg/ml of ox-LDL, then treated with 1 μM STS for 4 hours prior to being harvested in TrypLE. These cells were stained with anti-Annexin V antibody labelled with Fluorescein isothiocyanate (FITC) and Propidium Iodide (eBioscience) and analyzed by Scanford cell analyzer (Stanford Shared facility, Stanford), as previously described45. These FACS data was analyzed by FlowJo 10.1r5.
Proliferation assays
A modified MTT (3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay was performed to analyze SMC proliferation and viability. Mouse aortic SMCs were seeded in 96-well plates at the density of 6,000 cells/well, grown at 37C overnight, and then serum starved for 48 hours. The cells were stimulated with 10% serum or 10 μg/ml Tsp1 with or without 10 μg/ml of anti-CD47 Ab or IgG for 24 hours and then incubated for 4 hours in the presence of 10 μL of MTT AB solution (Millipore, Billerica, MA). The formazan product was dissolved by addition of 100 μL acidic isopropanol (0.04 N HCl) and absorbance was measured at 570 nm (reference wavelength 630 nm) on an ELISA plate reader by SpectraMax 190 Microplate Reader (Molecular Devices).
Efferocytosis assay
Standard in vitro phagocytosis assays were performed as previously described44. SMCs were labeled with 2.5 μM carboxyfluorescein succinimidyl ester (CFSE) according to the manufacturer’s protocol (Invitrogen). 100,000 SMCs were plated per well in a 96-well plate (Corning #7007) and pre-incubated with antibody (IgG1 isotype control (MOPC-21) or MIAP410 (anti-CD47)) for 30 minutes at 37 degrees. An unrelated anti-CD8 antibody was also tested as a negative control. After 30 minutes, 50,000 macrophages were added to each well and co-incubated for 2 hours in serum-free medium, then analyzed using an LSRFortessa cell analyzer with high throughput sampler (BD Biosciences). RFP+ mouse macrophages were identified by intrinsic fluorescence. Dead cells were excluded from the analysis by staining with DAPI (Sigma). Phagocytosis was evaluated as the percentage of GFP+ macrophages using FlowJo X 10.0.7r2 (Tree Star) and was normalized to the maximal response by each independent donor against each cell line. In addition to measuring basal efferocytosis rates, experiments were repeated with target cells that had been pretreated with a variety of compounds (alone or in combination) including STS, oxLDL, TNF-α, Tsp1, and Infliximab. Confirmatory assays were performed with RAW macrophages where target cells were labeled with 1 μM of CellTracker™ Deep Red dye (Life technologies) and phagocytes were labeled with 1.25 μM of CellTracker™ Orange CMRA (Life technologies) at 37°C for 30 minutes. As above, cells in these assays were treated with 50 μg/ml of ox-LDL, 50 ng/ml of TNF-α, or 100 μg/ml of Infliximab antibody for 24 hours prior to co-culture in serum-free medium with 10 μg/ml of antibody of IgG1 isotype control (MOPC-21) or anti-CD47 (MIAP410) for 2 hours at 37 degrees. Double positive cells were quantified using the Scanford cell analyzer (Stanford Shared facility) and analyzed by FlowJo 10.1r5, as previously described17. Statistical significance was determined by one- way or two-way ANOVA with Bonferroni correction using Prism 5 (Graphpad).
Flow Cytometry
To measure the cell surface expression of CD47, cells were exposed to vehicle or 50 ng/ml of TNF-α for 24 hours. In some experiments, cells were treated with 1 μM STS during the last 1 hr or 4 hours of the incubation, prior to analysis. The cells were harvested in TrypLE (Life Sciences) and stained with anti-CD47 Ab (AbD Serotec MCA2514GA, clone1/1A4, 1:400) or IgG (eBioscience), followed by Alexa Fluor 488 goat anti-mouse (Life technologies, 1:400), and then FACS sorted within one hour (BD FACSCaliber, 530 nm [FL1] and >575 nm [FL3]). Analysis was performed with FloJo 7.6.3.
Immunocytochemistry
Primary mouse aortic smooth muscle cells were seeded at approximately 60% confluence in glass bottom culture dishes (MatTek Corporation). Following treatment with TNF-α and/or STS, as described above, cells were rinsed with PBS and fixed with freshly prepared 4% PFA (Fisher Scientific). Once permeabilized with 0.1% Triton X-100 (Sigma), cells were incubated with blocking buffer (3% BSA, Cell Signaling Technology) for 1h and then incubated overnight at 4C° with a CD47 antibody (1:80; R&D Systems, Cat# AF1866-SP). After rinsing with PBS, cells were incubated in the dark with a donkey anti-goat alexa fluor 594 conjugate secondary antibody (1:1000; Life Technologies, Cat# A11058) for 1h then briefly incubated with DAPI. CD47 and DAPI localization was captured at 20x magnification using a Leica DMI3000 B microscope capable of taking fluorescent images. Studies using primary human coronary artery SMCs were also performed as above, but used 5% Goat Serum blocking buffer (ThermoFisher Scientific, PCN 5000), and the following primary and secondary antibodies: CD47 antibody (Novus Biologicals, Cat# NBP2-31106, 1:50); HMGB1 (abcam, ab18256 1:100) ); Alexa Fluor 594 goat anti-mouse (Life technologies A11005, 1:300); and Alexa Fluor 488 goat anti rabbit (Life technologies, A11034). In some studies, exogenous CD47 peptide was preincubated with the cells before the CD47 antibody was applied, as described above.
Luciferase reporter assay
CD47 LightSwitch Promoter Reporter GoClones (RenSP, S710450), Empty promoter vectors (S790005) and Cypridina TK Control constructs (pTK-Cluc, SN0322S) were obtained from SwitchGear Genomics and transfected into HEK cells using Lipofectamine 2000 (Invitrogen). For overexpression assays, expression plasmids for NFKB1 (p50), RELA (p65), NFKB2 (p52), and c-Rel were obtained from Addgene (#21965, #21966, #23289, #27256, respectively). Empty vector pCMV4 was generated by Hind III digestion of #21966, and pcDNA3.1 was obtained from life technologies. 50 ng of plasmid was co-transfected with 45ng of the RenSP reporter and 5ng of the pTK-Cluc reporter construct. Media was changed to fresh DMEM and 50 ng/ml of TNF-α was added 2, 8, 24, and 36 hours prior to harvest. The cell lysate and supernatant were harvested 48 hours after transfection and dual luciferase activity was measured with the LightSwitch Dual Assay System using a SpectraMax L luminometer (Molecular Devices), according to the manufacturer’s instructions. Relative luciferase activity (Renilla/Cypridina luciferase ratio) was quantified as the percentage change relative to the basal values obtained from control-transfected cells not exposed to TNF-α treatment.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the Millipore Magna-ChIP protocol with slight modifications. HCASMC were cultured in normal growth media until approximately 75% confluent. Cells were fixed in 1% formaldehyde for 10 minutes to cross-link chromatin, followed by quenching with glycine for 5 minutes at room temperature. 2×10^7 cells per condition were collected, and nuclear lysates were prepared according to the manufacturer’s protocol. Cross-linked chromatin nuclear extracts were sheared into approximately 500 bp fragments using a Bioruptor (Diagenode) for 3 cycles of 3 minutes (30s ON, 30s OFF). Sheared chromatin was clarified via centrifugation at 4C° for 10 minutes. 1×10^6 nuclei per condition was incubated with 2 μg Rabbit IgG or anti-NfkB p105/p50 antibody (Abcam, Ab7971) plus protein A/G magnetic beads overnight at 4C° on a rotating platform to capture the protein-DNA complexes. Complexes were washed in Low salt, High salt, LiCl, and TE buffers and then eluted with a ChIP Elution Buffer containing Proteinase K. Free DNA was subsequently purified using spin columns. Total enrichment was measured using primers designed based on the sequence of the top NFKB binding site within the CD47 promoter (F (−804 to −781): 5′ atagggaagagcagagcgagtaga 3′ and R (+627 to +609): 5′ gcgtggaccaggacaccta 3′), or a Negative Control region using the following primers (F: 5′ CCGGAAGCACTTCTCCTAGA 3′ and R: 5′ AAGAGAGAGCGGAAGTGACG 3′). Quantitative real-time PCR (ViiA 7, Life Technologies) was performed using SYBR Green (Applied Biosystems) assays and fold enrichment was calculated by measuring the delta Ct – delta Ct IgG. Melting curve analysis was also performed for each ChIP primer. Data is presented as the percentage of Input DNA and as fold enrichment of chromatin precipitated with the NFKB Ab relative to the control IgG. In some experiments, cells were treated with TNF-α for 90 minutes and 24 hours prior to isolation of nuclear lysates.
Statistical analysis
Aside from the microarray data (described above), all experimental data are presented as mean ± SEM. Data were subjected to the Kolmogorov-Smirnov test to determine distribution. Groups were compared using the Mann-Whitney U test for non-parametric data or the two-tailed Students t-test for parametric data. When comparing multiple groups, data were analyzed by analysis of variance with one way ANOVA followed by Tukey’s or Dunnett’s post-test. For multiple testing of parametric data, a value of P<0.05 was considered statistically significant. In vitro experiments were replicated at least in triplicate and all analyses were performed in a blinded fashion by two separate investigators, unless otherwise specified. In the in vivo intervention studies, comparison was made between mice treated with anti-CD47 Ab and IgG. In the in vivo synergy studies, ANOVA was performed as above with multiple comparison and linear trend post-testing across all four groups. In the in vitro studies, comparison was made between the intervention (anti-CD47 Ab) and control (IgG) arms. “Vehicle control” was only used in those experiments were an additional treatment (e.g., TNF, oxLDL, Infliximab, etc.) was studied. In the TaqMan-based CD47 expression experiments, changes were demonstrated as mRNA fold-change compared to the baseline condition (set as “1”). In the microarray-based experiments, relative expression differences were displayed across conditions (e.g. atherosclerosis vs no-atherosclerosis). In the in vitro phagocytosis assays, efferocytosis rates are displayed as percent of maximum, as previously performed44. Statistical analysis was performed with GraphPad Prism 5. Aside from the human plaque microarray studies (displayed as Tukey boxplots) and the correlation plots (displayed as the 95% confidence band of the best fit line), all error bars display the standard error of the mean.
Study approval
All animal studies were approved by the Stanford University Administrative Panel on Laboratory Animal Care (protocol 27279) and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). All human studies were performed with written informed consent and with the approval of the Ethical Committee of Northern Stockholm (BiKE).
Extended Data
Extended Data Figure 1. CD47 expression correlates with risk for clinical cardiovascular events and is progressively upregulated in the necrotic core of human blood vessels during atherogenesis.
(a). cDNA microarray expression profiling in the BiKE carotid endarterectomy biobank reveals that the relative expression of CD47 is increased in vascular homogenates taken from subjects with symptomatic disease (stroke or TIA, n=85) compared to those with stable, asymptomatic lesions (n=40). Similar findings were observed in the non-overlapping discovery and validation cohorts from BiKE (n=55), and a second validation cohort from the Helsinki Carotid Endarterectomy Study (HeCES, n=21). Data presented as Tukey boxplots. (b). Immunohistochemical staining reveals that CD47 co-localizes with lipidated plaque within human coronary lesions, as measured by Oil-Red-O (ORO) staining. (c). Immunofluorescent staining of coronary samples confirms that CD47 is upregulated within the necrotic core. (d). High magnification (40x) imaging of atherosclerotic coronary plaque confirms that CD47 expression is present on the surface of nucleated cells undergoing cell death, as indicated by HMGB1 staining. Specificity of the anti-CD47 Ab is confirmed in assays where the signal was quenched by preincubating the sections with recombinant CD47 peptide prior to primary antibody exposure. (e). Additional representative coronary artery segments spanning the spectrum of progressive coronary artery disease (non-atherosclerotic coronary, early ‘fatty streak’, inwardly remodeled plaque, and advanced ulcerated lesion with necrotic core) confirm that CD47 is progressively upregulated during the development of coronary artery disease. The tunica media is indicated by dotted lines. (f). Additional staining in human carotid artery sections confirms that CD47 expression is upregulated in atherosclerosis relative to healthy tissue, and appears most pronounced within the necrotic core. (g). High magnification (100x) imaging confirms that the CD47 expression is specific to lesional cells, including SMCs (αSMA), macrophages (CD68) and cells undergoing programmed cell death (Casp3). Comparisons made by two-tailed t tests. ** = P < 0.01, * = P < 0.05.
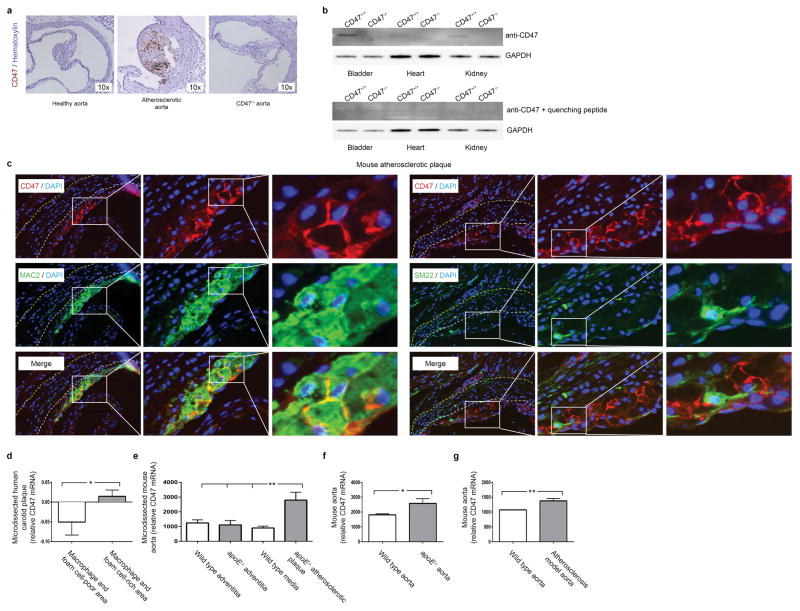
Extended Data Figure 2. CD47 expression is increased in mouse models of atherosclerosis.
(a). Mice injected with biotin labeled anti-CD47 Ab reveal that this Ab accumulates in the vasculature of atherosclerotic mice (middle), relative to non-atherosclerotic control mice (left). No staining is detected in CD47−/− mice (right), indicating specificity of the Ab. (b). Western blotting of tissue homogenates obtained from WT and CD47−/− mice (with and without quenching CD47 peptide) further confirms the specificity of the antibody. For gel source data, see Supplementary Figure 1. (c). High resolution immunofluorescent staining of murine atherosclerotic plaques indicate that CD47 is specifically expressed on the surface of lesional cells, rather than extracellular debris. (d). Publically available microarray data from laser capture microdissected (LCM) vascular tissue reveals that CD47 expression is increased within the macrophage and foam cell-rich area of human plaque, relative to macrophage and foam cell-poor areas (GSE23303). (e). Similar results were observed in LCM tissue from mouse atherosclerotic plaque tissue, relative to non-atherosclerotic medial and adventitial tissue (GSE21419). (f) and (g). Additional results from the Gene Expression Omnibus (GEO) database reveal that aortic CD47 expression is upregulated in murine models of atherosclerosis, as observed in the current study (GSE2372 and GSE19286). ** = P < 0.03, * = P < 0.05.
Extended Data Figure 3. Anti-CD47 Ab reduces atherosclerotic burden in several orthogonal in vivo models.
(a). Study timeline detailing osmotic minimpump implantation and high fat feeding to induce atherosclerosis in the apoE−/−-“Angiotensin infusion” model used herein. Kaplan Meier curves indicate no change in mortality with anti-CD47 treatment during 28 days of follow up. Additional representative examples confirm that anti-CD47 Ab: (b). Reduces atherosclerosis content in the aortic sinus; and (c). Reduces the percent of the en-face aorta covered by atherosclerotic plaque. Several additional atherosclerosis models were also used in this study to confirm the beneficial effects of anti-CD47 Ab therapy, and to model additional aspects of human cardiovascular disease. These include: (d). A “chronic atherosclerosis” model, where Ab therapy was given for 12 weeks (with no angiotensin infusion); (e). A “plaque vulnerability” model, where the impact of Ab therapy on plaque rupture and intraplaque hemorrhage was quantified; (f). An “established disease” model, where therapy was given for 7 weeks after mice had already developed advanced plaques of equivalent size; and (g). A “reduced dose” model, where the dose of anti-CD47 Ab was reduced by 75%, relative to the preceding studies. (h). Additionally, a “short term” study was performed where mice with established lesions of equivalent size and identical apoptosis rates were pulsed with only five days of anti-CD47 Ab therapy prior to harvest, to quantify the impact of therapy on efferocytosis rates, independent of lesion size (Phagocytic Index indicated by the ratio of ‘free’ (white stars) to ‘associated’ (white arrows) apoptotic bodies). Additional methodological details are provided in the Methods. Comparisons made by two-tailed t tests. ** = P < 0.03, * = P < 0.05. Error bars represent the SEM.
Extended Data Figure 4. Anti-CD47 Ab promotes the phagocytosis of diseased SMCs and macrophages, without altering apoptosis.
(a). In vitro caspase activity assays reveal that anti-CD47 Ab does not alter rates of programmed cell death in any vascular cell type (b). Flow cytometry assays confirm that anti-CD47 Ab has no effect on apoptosis at baseline, or in vascular SMCs exposed to 24 or 72 hours of oxLDL. (c). Staining controls for the in vitro phagocytosis assays. (d). Representative FACS plots for the in vitro efferocytosis conditions displayed in Figure 2E. The right upper quadrant (highlighted in red) includes double positive cells which are taken to represent a macrophage that has ingested a target cell. (e). In vitro efferocytosis assays using lipid-loaded macrophages as the target cell confirm that anti-CD47 Ab also stimulates the clearance of this vascular cell type, similar to the findings observed with SMCs. (f). Additional in vitro efferocytosis assays confirm that anti-CD47 Ab stimulates phagocytosis of vascular cells in a specific manner. Error bars represent the SEM.
Extended Data Figure 5. Additional examples confirm the pro-efferocytic properties of anti-CD47 Ab in vivo.
(a). Additional representative images detail that mice treated with anti-CD47 Ab have a lower overall burden of apoptotic debris (Caspase in green), as well as fewer examples of ‘free’ apoptotic bodies (white stars). Those apoptotic bodies that are present in these lesions are more often found in close proximity to macrophages (Mac-3 in red) and are considered ‘associated’ with a phagocyte if physically co-localized (white arrows). (b). Additional electron microscopy examples provide further qualitative evidence that phagocytes present in the lesions of mice treated with anti-CD47 Ab are more likely to have ingested several apoptotic bodies (white arrows) compared to lesions from IgG treated mice which are more likely to have a high burden of ‘free’ apoptotic bodies (yellow arrows). Additional representative examples of the necrotic core analysis, the phospho-SHP1 staining, and the plaque hemorrhage analysis are provided in (c), (d) and (e), respectively, as described in the Methods.
Extended Data Figure 6. Full dose anti-CD47 Ab induces anemia, but does not appear to alter nitric oxide (NO) elaboration, thrombospondin-1 dependent signaling, or any other processes relevant to vascular biology.
(a). No significant change in blood pressure is observed between mice treated with IgG or anti-CD47 Ab, arguing against a systemic difference in NO production due to Ab therapy. (b). Direct measurement of pulmonary NO release via the Griess reaction indicates that anti-CD47 Ab does not increase NO elaboration in vivo. (c). Western blot analysis of cultured murine vascular cells reveals that anti-CD47 Ab has no effect on the expected induction of p38 and ERK phosphorylation secondary to Tsp-1 treatment. (d). Similarly, anti-CD47 Ab has no effect on Tsp-1-dependent inhibition of eNOS phosphorylation, nor Acetylcholine-dependent induction of eNOS phosphorylation. (e). MTT assays show that anti-CD47 Ab does not affect cellular proliferation rates in the presence of Tsp-1. (f). In vitro efferocytosis assays show that the expected basal increase in phagocytosis observed after apoptotic cells are exposed to Tsp-1 (black bars) is not altered in the presence of anti-CD47 Ab (red bars). (g). Compared to mice receiving control IgG, mice receiving anti-CD47 Ab treatment have similar body weights at baseline and at sacrifice. (h). No difference is observed for the weight of any organ between groups, with the exception of splenomegaly observed in the anti-CD47 Ab treated animals. (i). Histological analysis of the explanted splenic tissue reveals an increase in the red pulp of anti-CD47 treated mice without any change in fibrosis or white pulp content, suggestive of increased erythrophagocytosis in this reticuloendothelial organ. Dot plots detail the hemoglobin count (j), reticulocyte count (k) and circulating monocyte count (l) for each animal in the acute 4 week angiotensin-infusion atherosclerosis model. Note that this anemia appears to be self-limited, and no anemia was observed in the chronic atherosclerosis model or the reduced dose model (P = 0.54 and 0.57, respectively). (m). mRNA analysis of aortic tissue reveals that anti-CD47 Ab has no significant impact on the expression of macrophage polarization factors in vivo. (n). anti-CD47 Ab also has no effect on the aortic expression of any other candidate efferocytosis gene. Additional quantitative analyses reveals that anti-CD47 Ab has no effect on in vivo: (o). Neutrophil content (as assessed by Ly6G positive area normalized to lesion size); (p) Macrophage content (as assessed by Mac-3 positive area normalized to lesion size); (q) T cell content (as assessed by CD3 positive area across the lesion and adventitia); or (r). Smooth muscle cell content (as quantified by α-SMA positive area in the aortic sinus from the external elastic lamina to the lumen). (s). anti-CD47 Ab also had no effect on lipid level or serum insulin (t). (u). MTT assays reveal that anti-CD47 Ab has no effect on the proliferation of primary aortic SMCs obtained from apoE−/− deficient mice either at baseline (left) or in the presence of 10% serum (right). Comparisons made by two-tailed t tests, unless otherwise specified. *** = P < 0.001, ** = P < 0.01, * = P < 0.05. Error bars represent the SEM. For gel source data, see Supplementary Figure 1; for detailed serological data, see Extended Data Table 1.
Extended Data Figure 7. Additional bioinformatic and experimental analyses further implicate a central role for the pro-inflammatory cytokine, TNF-α, in vascular CD47 signaling.
(a). Cytoscape network visualization of the genes which are significantly correlated with CD47 expression in both human and murine atherosclerotic plaque reveals a high number of TNF-α-related factors (indicated in blue), including ligands, receptors, and downstream signaling factors. (b). PANTHER pathway analysis of those genes which were (a) significantly associated with CD47 expression in mouse and human vascular tissue and (b) have been previously associated with atherosclerosis through the STAGE study33, identifies “inflammation mediated by chemokine and cytokine signaling pathway” as the most over-abundant pathway associated with CD47 expression in vascular tissue. (c). Using the Hybrid Mouse Diversity Panel (HMDP), which correlates aortic gene expression with Luminex cytokine array data of plasma samples from over 100 inbred strains of mice, we found that vascular CD47 expression is positively correlated with three inflammatory cytokines in vivo, including TNF-α, IL-2 and CXCL1. Correlation data shown for CD47 and TNF-α. (d). Co-expression studies confirm that TNF-α and CD47 expression are positively correlated in human carotid endarterectomy samples from the BiKE validation study. The Pearson correlation coefficient was determined assuming a Gaussian distribution and P values were determined using a two-tailed test. (e). Experiments with primarily cultured mouse aortic SMCs indicate that TNF-α reproducibly induces CD47 mRNA upregulation, while a number of other classical pro-atherosclerotic stimuli have no significant effect. Notably, CXCL1, IL4, TGFβ and IL-2 fail to induce CD47 expression in vitro, as assessed by ANOVA. (f). Additional studies suggest that the effect of TNF-α on CD47 expression persists in the presence of oxidized LDL, as occurs in the atherosclerotic plaque. (g). Western blotting confirms that TNF-α induces CD47 expression in vascular cells at the protein level. For gel source data, see Supplementary Figure 1. (h). Immunocytochemistry studies of HCASMCs confirm that CD47 expression is induced on the cell surface of TNF-α treated cells. TNF-α effect is assessed by co-staining for HMGB1, and antibody specificity is confirmed with isotype control and recombinant CD47 peptide quenching assays. (i). Multiple assays (including FACS, Taqman and immunocytochemistry studies) reveal that CD47 expression is downregulated on vascular SMCs during programmed cell death, as has previously been observed with inflammatory cells. (j). Confirmatory assays in cultured human coronary artery SMC reveal that TNF-α induces changes similar to those observed in murine cells (Fig 3D), including an induction of CD47 under physiological conditions and a blunting of its expected downregulation during apoptosis. (k). TNF-α’s capacity to impair CD47 downregulation during programmed cell death is also observed in mouse SMCs simultaneously exposed to pro-apoptotic stimuli and oxidized LDL. (l). No correlation between CD47 and other candidate cytokines was observed in the BiKE biobank, further supporting a specific relationship between CD47 and TNF-α. (m). Representative FACS-based apoptosis panels from cells exposed to the conditions used in Fig 3G confirm that TNF-α suppresses efferocytosis (Fig 3G) despite increasing programmed cell death. Comparisons made by two-tailed t tests, unless otherwise specified. *** = P < 0.001, * = P < 0.05. Error bars represent the SEM.
Extended Data Figure 8. The CD47 promoter contains predicted binding sites for the TNF-α-related transcription factor, NFKB1.
(a). UCSC genome browser screenshot showing overlay of human CD47 transcript with ENCODE transcription factor binding sites (including RELA, E2F4, and SRF), along with the active H3K27ac histone modification ChIP-seq track, and a custom track for chromatin accessibility in human coronary artery smooth muscle cells (HCASMC) using the Assay for Transposase Accessible Chromatin followed by sequencing (ATAC-seq). These chromatin, DNase hypersensitivity sites, and published ChIP-seq data suggest that members of the NFKB family of transcription factors could regulate CD47 expression in vascular tissue. (b). Additional co-expression studies in the BiKE validation study confirm that NFKB1 and CD47 expression are positively correlated in human carotid endarterectomy samples. The Pearson correlation coefficient was determined assuming a Gaussian distribution and P values were determined using a two-tailed test. (c). Additional luciferase promoter reporter assays reveal that induction of CD47 expression requires the presence of NFKB1 and cannot be induced by other NFKB co-factors such as RELA or c-REL. Time course studies confirm that CD47 expression is induced by TNF-α within 24 hours, suggesting a direct transcriptional relationship ((d) Taqman mRNA expression assays; (e) luciferase reporter assays). (f). Additional chromatin immunoprecipitation studies confirm that NFKB1 protein binds the CD47 promoter within 90 minutes of TNF-α-treatment in human coronary artery SMCs. ** = P < 0.01, * = P < 0.05. Error bars represent the SEM.
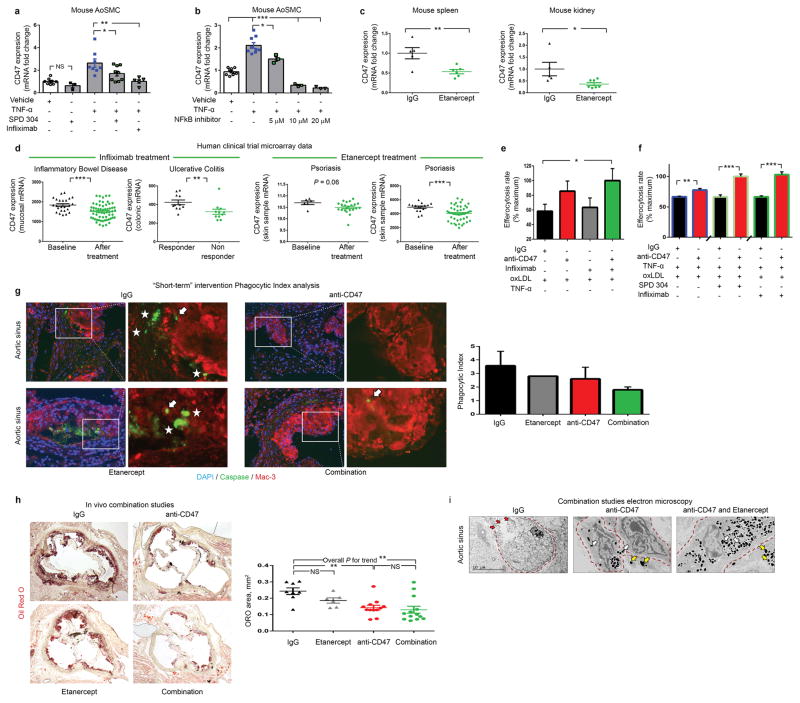
Extended Data Figure 9. Dual inhibition of CD47 and TNF-α provides a combinatorial effect.
(a). Pretreatment of mouse vascular SMCs with a chemical inhibitor (SPD 304) or a monoclonal Ab (infliximab) directed against TNF-α prevents the increase in CD47 expression normally seen after TNF-α exposure. (b). Similar effects were observed with the NFkB inhibitor, BAY 11–7085, confirming the molecular mechanism outlined in Fig 4. (c). Mice injected with four weeks of the decoy TNF-α receptor, etanercept, display a significant reduction in their in vivo expression of CD47 in splenic (left) and renal (right) tissue. (d). Publically available microarray data from human clinical trials of commercially available TNF-α inhibitors reveal that subjects treated with these agents also express lower levels of CD47 in vivo (as assessed by two-tailed t tests), confirming the mouse findings above (GSE accession #s from left to right: 16879 n=85, 12251 n=22, 47751 n=28 and 41663 n=66). Additional in vitro efferocytosis assays confirm a synergistic effect of anti-CD47 Ab with a variety of TNF-α inhibitors in both the absence (d) and presence (f) of exogenous TNF-α. (g). Mice with established plaques of identical size and with equivalent rates of apoptosis were treated with a short course (5 days) of IgG, anti-CD47 Ab, Etanercept, or combination therapy prior to harvest. As shown the phagocytic index (indicated by the ratio of ‘free’ (white stars) to ‘associated’ (white arrows) apoptotic bodies) displayed a non-significant trend toward improvement for combination therapy (P > 0.05). (h). When treated for a full 28 days in the Angiotensin-infusion model, individual comparisons showed that etanercept alone had no effect on atherosclerosis, and combination therapy was not significantly different from anti-CD47 alone, likely due to the potent effect of anti-CD47 monotherapy. ANOVA post-test analysis did identify a significant linear trend across all four groups (P for trend < 0.01). (i). Electron microscopy provides additional qualitative evidence that combination therapy may provide an incremental effect on efferocytosis, as suggested by an increased prevalence of macrophages within the plaque which had ingested a large number of apoptotic bodies (white arrows), a reduced prevalence of free apoptotic bodies (yellow arrows), and a reduced prevalence of uncleared cells undergoing secondary necrosis (red arrows). *** = P < 0.001, ** = P < 0.01, * = P < 0.05. Error bars represent the SEM.
Extended Data Table 1. In vivo serological data and additional in silico and bioinformatic data.
(a). Complete serological studies (including blood count, liver function studies, basic metabolic panel, and fasting glucose) from the 4 week apoE−/−-AngII atherosclerosis model indicate that anti-CD47 Ab induces a significant reduction in hemoglobin and compensatory reticulocytosis, consistent with prior reports4,7. The erythrophagocytosis of senescent RBCs appears to be self-limited, and no anemia was observed in the chronic atherosclerosis model or the reduced dose model (P = 0.54 and 0.57, respectively). No significant difference in any other serum marker is observed except for an increase in serum creatinine, which does not deviate outside of the reference range. Metabolic parameters and leukocyte differential data from the 12 week chronic atherosclerosis model are displayed at the bottom of the table. (b). Additional Upstream Regulator Analysis (URA) bioinformatic analyses of the Cytoscape data displayed in Extended Data Figure 7a performed within the Ingenuity Pathway Analysis (IPA) software identifies a number of TNF-α related factors (indicated in red) which are predicted to mediate transcriptional regulatory roles in the gene network shown in that panel. P-values were determined from Fisher’s Exact Test by comparing overlap of co-expressed genes with known upstream regulators from the Ingenuity Knowledge Base. (c). Several additional DAVID-based bioinformatics analyses including (KEGG, SMART, PANTHER and GO analyses) confirm the association between CD47 and inflammatory signaling related to the TNF-α pathway (indicated in red). Blue panels indicate the –logp10 value for each identified factor. (d). Transcription factor binding site prediction algorithms identify several putative NFKB family binding sites within the CD47 promoter, as displayed in Extended Data Figure 8a. (d). List of primers used in this study.
| a | ||||
|---|---|---|---|---|
| Characteristic | Reference Range | anti-CD47Ab | IgG | P value |
| Complete Blood Count | ||||
| WBC | 5.5 – 9.3 K/uL | 1.55 | 1.58 | 0.90 |
| RBC | 7.0 – 8.8 M/uL | 9.61 | 11.27 | 0.01 |
| HGB | 13.7 – 16.4 gm/dL | 13.91 | 16.15 | 0.01 |
| HCT | 39.0 – 47.0% | 46.02 | 50.97 | 0.08 |
| MCV | 52.0 – 68.7 fL | 48.02 | 45.26 | 0.00 |
| MCH | 18.4 – 19.6 pg | 14.33 | 14.37 | 0.77 |
| MCHC | 34.0 – 36.0 g/dL | 29.88 | 31.72 | 0.00 |
| Platelet count | 675 – 1338 K/uL | 793.60 | 827.36 | 0.63 |
| Reticulocyte Count | 1.0 – 2.8 % | 18.76 | 4.97 | 0.00 |
| Retic Absolute | K/uL | 1764586.40 | 553836.36 | 0.00 |
| Liver Function Studies | ||||
| AST | 192 – 388 U/L | 120.00 | 290.56 | 0.24 |
| ALT | 76 – 160 U/L | 31.57 | 39.11 | 0.57 |
| Alkaline Phosphatase | 171 – 183 IU/L | 121.43 | 135.44 | 0.54 |
| Total Protein | 5.0 – 6.2 g/dL | 5.00 | 5.30 | 0.39 |
| Albumin | 3.2 – 3.6 g/dL | 2.63 | 2.88 | 0.27 |
| Globumin | N/A | 2.37 | 2.39 | 0.96 |
| A/G Ratio | N/A | 1.11 | 1.54 | 0.38 |
| Total Bilirubin | N/A mg/dL | 0.29 | 0.19 | 0.22 |
| Serum Chemistries | ||||
| BUN | 20.3 – 24.7 mg/dL | 36.86 | 35.89 | 0.83 |
| Creatinine | 0.1 – 1.1 mg/dL | 0.30 | 0.17 | 0.04 |
| BUN/Cre | N/A | 132.93 | 205.71 | 0.22 |
| Ca | 8.9 – 9.7 mg/dL | 7.81 | 8.53 | 0.44 |
| Na | 114 – 154 mmol/L | 146.67 | 145.25 | 0.60 |
| K | 3.0 – 9.6 mmol/L | 9.47 | 9.20 | 0.80 |
| Cl | N/A mmol/L | 112.67 | 107.00 | 0.00 |
| Carbon Dioxide | N/A mmol/L | 17.60 | 18.53 | 0.38 |
| Na/K | N/A | 14.77 | 16.08 | 0.60 |
| Anion gap | N/A mEq/L | 25.87 | 28.93 | 0.20 |
| Glucose | 184 – 220 mg/dL | 116.71 | 109.44 | 0.86 |
| Metabolic Measurements | ||||
| Cholesterol | N/A mg/dL | 508.50 | 554.20 | 0.31 |
| Triglycerides | N/A mg/dL | 105.80 | 101.70 | 0.76 |
| HDL | N/A mg/dL | 391.40 | 508.80 | 0.22 |
| LDL | N/A mg/dL | 440.10 | 476.30 | 0.36 |
| Insulin | N/A ng/dL | 0.93 | 0.81 | 0.81 |
| Differential | ||||
| Neutrophils | 15 – 32% | 27.83 | 32.25 | 0.64 |
| Lymphocytes | 65 – 83% | 67.00 | 62.00 | 0.59 |
| Monocytes | 0 – 3% | 4.17 | 4.00 | 0.88 |
| Eosinophils | 0 – 3% | 3.00 | 3.50 | 0.79 |
| b | ||
|---|---|---|
| Bioinformatic algorithm | Molecule Type | P value of overlap |
| Upstream Regulator INGENUITY Pathway Analysis | ||
| TNF | cytokine | 6.50E-42 |
| IL4 | cytokine | 7.31E-41 |
| TGFB1 | growth factor | 1.97E-39 |
| IL6 | cytokine | 1.22E-38 |
| IL2 | cytokine | 2.00E-38 |
| IL1B | cytokine | 4.21E-37 |
| NFkB (complex) | complex | 1.88E-34 |
| IL17A | cytokine | 2.34E-34 |
| IL13 | cytokine | 1.28E-33 |
| CSF2 | cytokine | 3.04E-33 |
| IFNG | cytokine | 3.33E-32 |
| IL1A | cytokine | 3.09E-31 |
| STAT3 | transcription regulator | 7.23E-28 |
| RELA | transcription regulator | 2.52E-27 |
| CD3 | complex | 3.80E-27 |
| CCL2 | cytokine | 5.04E-27 |
| Immunoglobulin | complex | 8.38E-27 |
| IL12 (complex) | complex | 1.42E-24 |
| IFNB1 | cytokine | 7.90E-24 |
| STAT6 | transcription regulator | 8.32E-24 |
| IL3 | cytokine | 1.84E-23 |
| IL15 | cytokine | 9.08E-23 |
| BCL6 | transcription regulator | 2.21E-22 |
| CD40LG | cytokine | 4.94E-22 |
| KLF2 | transcription regulator | 5.16E-22 |
| TCR | complex | 2.11E-21 |
| HGF | growth factor | 2.65E-21 |
| TNFSF11 | cytokine | 3.83E-21 |
| EGR1 | transcription regulator | 1.14E-20 |
| LEP | growth factor | 3.31E-20 |
| NFKBIA | transcription regulator | 1.19E-19 |
| TGFB2 | growth factor | 1.67E-19 |
| SP1 | transcription regulator | 1.71E-19 |
| SMAD7 | transcription regulator | 2.65E-19 |
| SMAD3 | transcription regulator | 5.62E-19 |
| Ige | complex | 9.42E-19 |
| FGF2 | growth factor | 1.12E-18 |
| NFKB1 | transcription regulator | 1.34E-18 |
| TSLP | cytokine | 1.47E-18 |
| d | ||||
|---|---|---|---|---|
| Algorithm | Trans. Factor | Highest Score | Location | Consensus Sequence |
| P-Match | NFKB1 | 100.0% | 331 (−) | ggaaaTTCCC |
| p65 | 100.0% | 331 (−) | GGAAAttccc | |
| c-Rel | 100.0% | 331 (−) | GGAAAttccc | |
| TF Search | NFKB1 | 85.4% | 150 (−) | GGAAGCTCCCT |
| c-Rel | 86.8% | 150 (−) | GGAAGCTCCCT | |
| JASPAR | NFKB1 | 85.1% | 149 (−) | GGGAGCTTCCA |
| REL | 87.8% | 149 (−) | GGGAGCTTCC | |
| p65 | 86.6% | 149 (−) | GGGAGCTTCC | |
| AliBaba | NFKB1 | 783 (+) | gggagcttcc | |
| TF Bind | NFKB1 | 90.6% | 783 (−) | NGGGGAMTTTCCNN AGGGAGCTTCCACT |
| p65 | 76.0% | 785 (−) | GGGRATTTCC GGAGCTTCCA | |
| c-Rel | 90.5% | 784 (+) | SGGRNWTTCC GGGAGCTTCC | |
| e | |
|---|---|
| Primer | ID |
| Human | |
| CD47 | Hs00179953_m1 |
| Mouse | |
| TNF-α | Mm00443258_m1 |
| Cd47 | Mm00495011_m1 |
| Calreticulin (Calr) | Mm00482936_m1 |
| Gas6 | Mm00490378_m1 |
| Low density lipoprotein rece | Mm00464608_m1 |
| c-mer proto-oncogene tyros | Mm00434920_m1 |
| Milk fat globule-EGF factor | Mm00500549_m1 |
| Signal regulatory protein alp | Mm00455928_m1 |
| Transglutaminase 2 (Tgm2) | Mm00436987_m1 |
| Thrombopondin 1 (Thbs1) | Mm00449032_g1 |
| Cd163 | Mm00474091_m1 |
| Mrc1 | Mm00485148_m1 |
| Nitric oxide synthase 2 (Nos | Mm00440502_m1 |
| Abca1 | Mm00442646_m1 |
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (R01HL12522401 and R01HL12337001 to NJL and U01HL099999 to ILW) and the Ludwig Center at Stanford. The authors wish to acknowledge Josh Knowles for his critical review of the manuscript.
Footnotes
The authors declare competing financial interests: N.J.L. and I.L.W. have filed a patent describing inhibition of CD47 as a method to prevent atherosclerosis.
Supplementary Information is linked to the online version of the paper at nature.com/nature.
Author Contributions Y.K.1 designed and conducted most experiments including the mouse microsurgery, morphometric analyses, in vitro cell culture, Taqman, Western blot, ChIP and luciferase reporter experiments. V.N.1, D.D.1 and L.M.7 assisted with the electron microscopy studies and blinded histology. J.Y.1, J.P.V.2 and K.M.2 conducted the in vitro efferocytosis assays and FACS studies. L.P.8 and U.H.8 performed the carotid staining and BiKE biobank analyses. A.C.5 provided coronary samples and assisted with the histopathological studies. M.C3, A.J.L3, E.S.6, P.B.2, T.Q.4 and C.M.4 conducted the microarray, co-expression and in silico bioinformatic studies. Y.K.1, I.L.W.2 and N.J.L.1,4 conceived of the study, analyzed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
References and notes
- 1.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–25. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 2.Schrijvers DM, De Meyer GR, Kockx MM, Herman AG, Martinet W. Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:1256–61. doi: 10.1161/01.ATV.0000166517.18801.a7. [DOI] [PubMed] [Google Scholar]
- 3.Thorp E, Tabas I. Mechanisms and consequences of efferocytosis in advanced atherosclerosis. J Leukoc Biol. 2009;86:1089–95. doi: 10.1189/jlb.0209115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288:2051–4. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 5.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2012;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- 6.Gardai SJ, McPhillips KA, Frasch SC, et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–34. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinchen JM, Ravichandran KS. Phagocytic signaling: you can touch, but you can’t eat. Curr Biol. 2008;18:R521–4. doi: 10.1016/j.cub.2008.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101:890–8. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henson PM, Bratton DL, Fadok VA. Apoptotic cell removal. Curr Biol. 2001;11:R795–805. doi: 10.1016/s0960-9822(01)00474-2. [DOI] [PubMed] [Google Scholar]
- 11.www.who.int/mediacentre/factsheets/fs310/en/
- 12.Shankman LS, Gomez D, Cherepanova OA, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–37. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabas I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat Rev Immunol. 2010;10:36–46. doi: 10.1038/nri2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorp E, Cui D, Schrijvers DM, Kuriakose G, Tabas I. Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of apoe−/− mice. Arterioscler Thromb Vasc Biol. 2008;28:1421–8. doi: 10.1161/ATVBAHA.108.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willingham SB, Volkmer JP, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–7. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perisic L, Hedin E, Razuvaev A, et al. Profiling of atherosclerotic lesions by gene and tissue microarrays reveals PCSK6 as a novel protease in unstable carotid atherosclerosis. Arterioscler Thromb Vasc Biol. 2013;33:2432–43. doi: 10.1161/ATVBAHA.113.301743. [DOI] [PubMed] [Google Scholar]
- 17.Kojima Y, Downing K, Kundu R, et al. Cyclin-dependent kinase inhibitor 2B regulates efferocytosis and atherosclerosis. J Clin Invest. 2014;124:1083–97. doi: 10.1172/JCI70391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbin U, Baggio E, Stranieri C, et al. Expansion of necrotic core and shedding of Mertk receptor in human carotid plaques: a role for oxidized polyunsaturated fatty acids? Cardiovasc Res. 2013;97:125–33. doi: 10.1093/cvr/cvs301. [DOI] [PubMed] [Google Scholar]
- 19.Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J Clin Invest. 2000;105:1605–12. doi: 10.1172/JCI7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isenberg JS, Ridnour LA, Dimitry J, Frazier WA, Wink DA, Roberts DD. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J Biol Chem. 2006;281:26069–80. doi: 10.1074/jbc.M605040200. [DOI] [PubMed] [Google Scholar]
- 21.Back M, Hansson GK. Anti-inflammatory therapies for atherosclerosis. Nature reviews Cardiology. 2015;12:199–211. doi: 10.1038/nrcardio.2015.5. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins PN. Molecular biology of atherosclerosis. Physiol Rev. 2013;93:1317–542. doi: 10.1152/physrev.00004.2012. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg JD, Furer V, Farkouh ME. Cardiovascular safety of biologic therapies for the treatment of RA. Nature reviews Rheumatology. 2012;8:13–21. doi: 10.1038/nrrheum.2011.168. [DOI] [PubMed] [Google Scholar]
- 24.Pang WW, Pluvinage JV, Price EA, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110:3011–6. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil S, Fehrenbacher B, Lukowski R, et al. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014;115:662–7. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 26.Chung IM, Schwartz SM, Murry CE. Clonal architecture of normal and atherosclerotic aorta: implications for atherogenesis and vascular development. Am J Pathol. 1998;152:913–23. [PMC free article] [PubMed] [Google Scholar]
- 27.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Wang L, Zhao F, et al. Pre-Clinical Development of a Humanized Anti-CD47 Antibody with Anti-Cancer Therapeutic Potential. PLoS One. 2015;10:e0137345. doi: 10.1371/journal.pone.0137345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ClinicalTrials.gov: NCT02216409
- 30.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–74. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 31.http://www.affymetrix.com/support/technical/technotes/expression_comparison_technote.pdf.
- 32.http://www.affymetrix.com/support/technical/datasheets/hgu133arrays_datasheet.pdf.
- 33.Saksi J, Ijas P, Nuotio K, et al. Gene expression differences between stroke-associated and asymptomatic carotid plaques. J Mol Med (Berl) 2011;89:1015–26. doi: 10.1007/s00109-011-0773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King JY, Ferrara R, Tabibiazar R, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–18. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 35.Ashley EA, Ferrara R, King JY, et al. Network analysis of human in-stent restenosis. Circulation. 2006;114:2644–54. doi: 10.1161/CIRCULATIONAHA.106.637025. [DOI] [PubMed] [Google Scholar]
- 36.http://www.affymetrix.com/support/technical/datasheets/human_datasheet.pdf.
- 37.Perisic L, Aldi S, Sun Y, et al. Gene expression signatures, pathways and networks in carotid atherosclerosis. Journal of internal medicine. 2016;279:293–308. doi: 10.1111/joim.12448. [DOI] [PubMed] [Google Scholar]
- 38.Chen YC, Bui AV, Diesch J, et al. A novel mouse model of atherosclerotic plaque instability for drug testing and mechanistic/therapeutic discoveries using gene and microRNA expression profiling. Circ Res. 2013;113:252–65. doi: 10.1161/CIRCRESAHA.113.301562. [DOI] [PubMed] [Google Scholar]
- 39.Jun J, Reinke C, Bedja D, et al. Effect of intermittent hypoxia on atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis. 2010;209:381–6. doi: 10.1016/j.atherosclerosis.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander MR, Moehle CW, Johnson JL, et al. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J Clin Invest. 2012;122:70–9. doi: 10.1172/JCI43713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Ding Y, Hutchins LN, et al. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–6. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci U S A. 2003;100:13531–6. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiskopf K, Ring AM, Ho CC, et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leeper NJ, Raiesdana A, Kojima Y, et al. Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. Arterioscler Thromb Vasc Biol. 2013;33:e1–e10. doi: 10.1161/ATVBAHA.112.300399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.