Abstract
Purpose
Magnetic resonance spectroscopic imaging (MRSI) benefits from operation at 7 Tesla (7T) due to increased signal-to-noise ratio (SNR) and spectral separation. The 180° radiofrequency (RF) pulses used in the conventional MRSI sequences are particularly susceptible to the variation in the transmitted RF (B1) field and severe chemical shift localization errors at 7T. RF power deposition, as measured by specific absorption rate (SAR), also increases with field strength. Adiabatic 180° RF pulses may mitigate the effects of B1 variation. We designed and implemented a Semi-Adiabatic Spectral-spatial Spectroscopic Imaging (SASSI) pulse sequence to provide more uniform spectral data at 7T with reduced SAR.
Method
The adiabatic Shinnar Le-Roux algorithm was used to generate a high bandwidth 180° adiabatic SPSP pulse that captured a spectral range containing the main metabolites of interest. A pair of 180° SPSP pulses was used to refocus the signal excited by a 90° SPSP pulse in order to select a 3D volume of interest in the SASSI sequence
Results
The SASSI pulse sequence produced spectra with more uniform brain metabolite SNR when compared to the conventional non-adiabatic MRSI sequence.
Conclusion
SASSI achieved comparable SNR to the current adiabatic alternative, semi-LASER, but with 1/3 of the SAR.
Keywords: spectroscopic imaging, adiabatic, MRSI, chemical shift localization error, B1 sensitivity, spectral-spatial pulse, RF pulse design, high field MRI, 7 Tesla, brain, human
Introduction
Magnetic resonance spectroscopic imaging (MRSI) can be used to non-invasively investigate metabolite concentration changes in the brain 1 correlated to neurological and psychiatric diseases, brain tumors, and radiation damage 2–5. High magnetic fields, such as 7 Tesla (7T), permit us to leverage increased signal to noise ratio (SNR) and spectral separation between metabolite peaks for more sensitive metabolite detection and quantification as well as higher resolution spectral grids 6–9.
MRSI has been performed at conventional field strengths such as 1.5T and 3T 10–12. However, at higher field strengths there are challenges such as greater radiofrequency (RF) power deposition, which may approach specific absorption rate (SAR) safety limits, B1 inhomogeneity, and increased chemical shift localization (CSL) error. CSL error refers to the spatial shifts in the excited volume for metabolites resonating at different frequencies. Spectroscopic imaging is most commonly performed using a point resolved spectroscopy (PRESS) sequence 13–15 which uses a 90° RF pulse followed by at least two 180° RF pulses, which select along orthogonal dimensions and form a double spin echo over the volume of interest (VOI). In MRSI, further spatial localization is performed within this VOI through the use of phase encodes or oscillating read out gradients 16. The 180° RF pulses used in the PRESS sequence are particularly sensitive to the substantial variation in B1 at 7T resulting in signal attenuation in multiple regions of the brain. These pulses are also susceptible to CSL error, which scales with field strength, resulting in shifts in the selected volume for metabolites that are sparsely separated in frequency.
Finally, excess power deposition during the scan is a major challenge at 7T. RF power deposition, as measured by SAR, increases quadratically with field strength and 180° RF pulses are particularly SAR intensive. At 7T, RF pulse sequences containing two or more 180° pulses, such as the double spin echo PRESS sequence, may reach or exceed SAR safety limits leading to an imaging delay or termination of the acquisition.
In order to address some of these challenges, there have been several variants of the PRESS sequence proposed. Adiabatic pulses achieve B1-insensitive refocusing and reduced CSL error. A 3D adiabatic MRSI sequence, such as Localized Adiabatic SElective Refocusing (LASER) 17–19, achieves volume selection using adiabatic full passage refocusing pulses along all three spatial axes. However, adiabatic pulses are SAR-intensive and deposit non-linear phase across the spatially selective dimension which must be refocused, often with a second, identical, adiabatic full passage (AFP) pulse applied across the same axis. A fully 3D MRSI sequence, such as LASER, requires at least 6 matched adiabatic refocusing pulses, resulting in an extended minimum echo time (TE), and very high SAR. The semi-LASER sequence 20, 21 uses a non-adiabatic selective excitation, combined with two pairs of adiabatic refocusing pulses. This enables a shorter TE and results in lower SAR when compared to LASER. However, the 4 high-SAR adiabatic refocusing pulses still limit the minimum repetition time (TR) between semi-LASER MRSI pulses in vivo, extending total scan time.
Adiabatic spectral-spatial pulses (SPSP) have been used in MRSI sequences to simultaneously provide B1-insensitive selection and reduced CSL error 22–26. Since the spatial selectivity is achieved by linear-phase spatial sub-pulses, pairs of pulses are not required to refocus quadratic phase in the spatial dimensions. Quadratic phase is deposited in the spectral dimension by the first adiabatic 180°, but this quadratic phase is refocused by a second identical SPSP adiabatic 180° pulse. By obviating the need for pairs of adiabatic refocusing pulses for each spatial dimension, the use of adiabatic SPSP pulses to select the VOI may be used to reduce total SAR when compared to semi-LASER. Root-flipping has also been used as a design tool for spectral spatial pulses27 in part to reduce peak RF amplitude. However these pulses were not adiabatic and were implemented at 3T and 1.5T with long pulse lengths (~40 ms) making it difficult to capture brain metabolites with a range of T2 relaxation characteristics. In previous work with adiabatic SPSP pulses at 7T, hyperbolic secant adiabatic pulses were used as spectral envelopes to create the adiabatic SPSP pulses. This resulted in a spectral BW limited by peak RF, necessitating a spectrally interleaved approach to cover the full range of interesting brain metabolites 23, 28. Another spectral spatial pulse with adiabatic characteristics29 was developed for the 7T. However the long pulse duration of approximately 30 ms, the TE of 144, and the implemented bandwidth of 712 Hz, was insufficient to capture metabolites in the full range from MI to NAA and provide a margin for B0 shifts.
In this first application to MRSI, we used the adiabatic Shinnar Le-Roux (SLR) algorithm to create a fully adiabatic envelope with more uniform RF distribution, resulting in low peak pulse amplitudes for our two high spectral-BW 180° adiabatic SPSP pulses. These pulses can capture the main metabolites of interest at 7T, while remaining within hardware peak RF limits. The 180° SPSP pulses were paired with a 90° SPSP pulse to excite a 3D volume in a Semi-Adiabatic Spectral-spatial Spectroscopic Imaging (SASSI) sequence. The sequence was designed to generate spectroscopic grids with reduced chemical shift artifact and improved B1-immunity while having lower SAR than existing adiabatic spectroscopic imaging implementations
Methods
Pulse Design and implementation
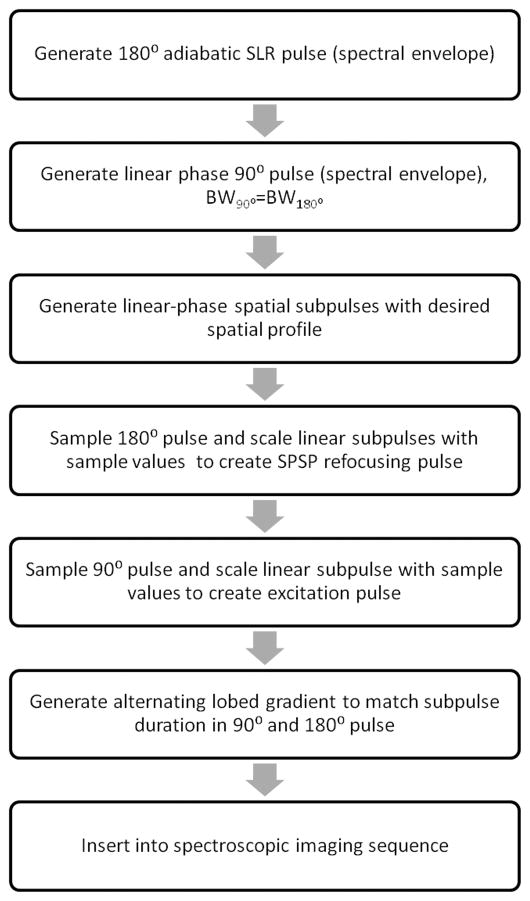
The SASSI pulses were designed using algorithms implemented in MATLAB (The Mathworks Inc., Natick, Massachusetts, USA). Figure 1 is an overview of the design path used to develop the SASSI sequences. We first designed the adiabatic 180° SPSP pulses to be used in the SASSI sequence. We used the adiabatic SLR algorithm to generate a 180° pulse capable of capturing a frequency range containing brain metabolites extending from myo-inositol (MI) to n-acetylaspartate (NAA) at 7T and to act as the spectral envelope for the SPSP pulse 30. Using a previously described adiabatic SLR algorithm 31, the frequency profile of the envelope of the refocusing pulse was the response of a least-squares linear-phase filter set to the desired bandwidth BW, which was 1.03 kHz. To introduce adiabatic behavior and distribute RF energy more uniformly, quadratic phase was applied across the frequency response for the filter; the B(z) polynomial was then calculated as the Fourier transform of the resultant frequency profile and an A(z) polynomial was calculated from B(z). The A(z) and B(z) polynomials were used as inputs for the inverse SLR transform to produce the adiabatic RF pulse envelope. The adiabatic RF envelope was then sampled at 27 evenly spaced locations and used to scale small-tip-angle, linear-phase spatial sub-pulses, creating a 180° adiabatic SPSP with a spectral BW of 1.02 kHz, a spatial BW of 9.19 kHz, duration of 9.8 ms, and peak amplitude of 14.36 μT.
Figure 1.
Flow chart showing steps in design of SASSI sequence
In the second step, a linear-phase pulse envelope was created for the 90° SPSP pulse. The spectral BW was designed be 1.05 kHz, slightly exceeding the spectral BW of the refocusing pulse. The envelope for the 90° pulse was sampled at 18 equally space locations and used to scale small-tip-angle, linear-phase spatial sub-pulses with a BW of 8.96 kHz. This created a final pulse with a total duration of 4.2 ms and peak amplitude of 16.61 μT.
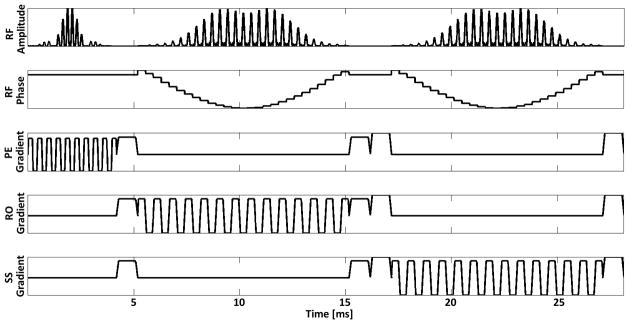
The 90° SPSP pulse and adiabatic 180° SPSP pulses were then integrated into a PRESS excitation, and coupled with oscillating slice-select gradients to create the SASSI excitation. The VERSE transformation was applied 32, 33 to compensate for the RF pulse and gradient ramp overlap. Figure 2 shows the RF, phase, and gradient waveforms for the SASSI pulse sequence which uses the SPSP 90° pulse and adiabatic SPSP 180° pulses to select along the three spatial dimensions and generate a double spin echo. The pulse sequence depicted in Figure 2 is followed by a period of signal acquisition not shown in the diagram.
Figure 2.
SASSI Pulse sequence diagram showing the amplitude, phase, and associated gradients of the SPSP excitation pulse, as well as the adiabatic SPSP refocusing pulses. In a single TR, this sequence of RF pulses would be followed by a period of signal acquisition, not shown.
Simulations
The APINS(z) and BPINS(z) polynomials were used to simulate the magnetization profiles for the SASSI excitation and refocusing pulses and used to calculate the final magnetization profile for the double spin echo generated by the pulse trio. When the initial longitudinal magnetization is assumed to be 1, Equations 1–3 are equivalent to the calculations performed by a discrete-time Bloch simulator 31.
| (1) |
| (2) |
| (3) |
Mxy is the transverse magnetization after the initial excitation and Mref is the refocusing profile for the adiabatic SPSP 180° pulses. The final magnetization (Mfin) at the second spin echo can be calculated as the effect of the two 180° pulses on the transverse magnetization produced by the 90° pulse (Equation 3).
The 2D SPSP profiles of the excitation pulse (Equation 1) and the refocusing pulses (Equation 2), as well as the final 2D SPSP profile at the second spin echo (Equation 3) were simulated as show in Figures 3A, 3B, and 3C respectively. A spectral cross-section of the profile in Figure 3C indicating coverage of major brain metabolites is shown in Figure 3D. The B1- insensitivity of the SASSI pulse was investigated in Figure 4 by simulating the spectral and spatial profiles for RF overdrive factors (ODF) ranging from 0.5 to 5, in increments of 0.25. The RF overdrive factor is equal to the RF amplitude of the applied pulse (Aapp) divided by the RF amplitude of the pulse at nominal power or adiabatic threshold (Athresh)
Figure 3.
A) Simulated 2D spectral-spatial excitation profile for the SASSI SPSP excitation pulse at ODF = 0.8 (80% of excitation threshold) and the SASSI SPSP refocusing pulse at ODF = 0.8 (80% of adiabatic threshold). B) Simulated 2D spectral-spatial excitation profile for the SASSI SPSP excitation pulse at ODF = 1.0 (100% of excitation threshold) and the SASSI SPSP refocusing pulse at ODF = 1.0 (100% of adiabatic threshold) showing both the frequency and spatial selectivity of the pulse at threshold. C) Simulated 2D spectral-spatial excitation profile for the SASSI SPSP excitation pulse at ODF = 1.2 (120% of excitation threshold) and the SASSI SPSP refocusing pulse at ODF = 1.2 (120% of adiabatic threshold). D) Simulated 2D spectral-spatial excitation profile for the SASSI SPSP excitation pulse at ODF = 1.4 (140% of excitation threshold) and the SASSI SPSP refocusing pulse at ODF = 1.4 (140% of adiabatic threshold). E) Spectral profile of the SASSI adiabatic refocusing pulses versus frequency showing relative resonant frequencies of important in vivo brain metabolites at 7T.
Figure 4.
Simulated response of the A) spectral profile and B) spatial profile of SASSI SPSP 90° excitation pulse over a range of overdrive factors. An overdrive factor (ODF) of 1 indicates nominal amplitude at which the pulse is designed to operate. The pulse exhibits behavior typical of a non-adiabatic SLR excitation pulse. C) Spectral profile and D) spatial profile of the SASSI SPSP adiabatic 180° RF pulses over a range of B1 ODFs where an ODF of 1 is at adiabatic threshold, and an ODF of 1.1 represents 10% above adiabatic threshold. The spectral profile remains fairly invariant until very high ODFs (above most RF amplifier limits) are reached, resulting in sub-pulse overdrive. Spatial profiles suffer from some ripple as B1 is increased.
| (4) |
Using Equation 4, an ODF of 1 would correspond to adiabatic threshold and an ODF of 1.1 would be 110% of adiabatic threshold (10% above adiabatic threshold).
Experiments
All experiments were performed on a 7T whole body MRI scanner (Siemens MAGNETOM 7T, Siemens, Erlangen), equipped with a SC72CD gradient coil (Gmax = 70 mT/m and max slew rate = 200 T/m/s), using a single channel transmitter and a 32-channel receive head coil (Nova Medical, Wilmington, MA). To calibrate the RF amplitude, a B1 map was acquired at the spectroscopy slice location, and RF power was selected to provide optimal flip angle at the center of the FOV.
The performance of the SASSI pulse sequence was verified using a spherical water and metabolite phantom prepared in-house following the GE “BRAINO” phantom recipe (GE Medical Systems, Milwaukee, WI USA) to mimic in vivo brain metabolite ratios. Additional gamma-amino butyric acid (GABA) was included in this phantom construction for other GABA spectroscopy experiments. However, the scope of this experiment is limited to the more prominent brain metabolites and the excess GABA was excluded from fitting in the phantom and in vivo analysis. This BRAINO phantom was imaged using three 7T spectroscopic imaging sequences: 1) a conventional double spin echo PRESS sequence; 2) a semi-LASER sequence using pairs of identical adiabatic refocusing pulses to select along two spatial dimensions (for a total of 4 adiabatic pulses); and 3) the SASSI sequence. The TR for the phantom sequences was set to be 2050 ms. This TR was chosen based on the minimum TR possible for the semi-LASER sequence and was dictated by system SAR limitations. The field of view was placed off-center in the phantom to sample the range of B1s produced in the phantom and RF power was calibrated to the center of the FOV. The water suppression provided for the semi-LASER and PRESS sequences used three CHEmical Shift Selective (CHESS) pulses34 for water suppression followed by crusher gradients. The acquisition parameters for the phantom MRSI scans were: slice thickness = 1.6 cm, FOV = 12 × 12 cm, matrix size = 32 × 32 (16×16 voxels within the VOI), voxel volume =.225 cc, TE/TR = 42/2050 ms, Navg = 1, scan time = 28:03 min with a weighted partial acquisition. For these phantom experiments, total scan time was set to be much longer (28 minutes) than in vivo applications of SASSI. Fine in-plane resolution was required to capture signal variation due to B1 and only a thin, single slice was acquired to minimize B1 variation within the through plane dimension and enable a more accurate comparison of B1 insensitivity. Chemical shift and TRs were set according to semi-LASER SAR operating limits, requiring long acquisition times. The center frequency for the pulse was set to −2.6 ppm, chosen to be between NAA and Cho resonances to distribute CSL error as evenly as possible between the metabolites of interest. Receiver gains were set to the same value for all three acquisitions in order to enable comparison of metabolite signal amplitudes between sequences.
The SASSI sequence was tested in vivo by exciting a single slice through the brain of a normal volunteer and comparing the results to those obtained using the semi-LASER sequence and a PRESS sequence. Institutional Review Board approval was obtained before commencing in vivo scans and for all human scans informed consent from the subject was obtained prior to scanning. The acquisition parameters for the in vivo MRSI scans were slice thickness = 1.6 cm, FOV = 8 × 8 cm, matrix size = 8 × 8 pixels (6×6 pixels within the VOI), pixel volume = 1.6 cc, TE/TR = 42/4080 ms, Navg = 16, scan time = 13:21 min with a weighted partial acquisition. As in the phantom experiments, the center frequency was set to −2.6 ppm.
Finally, in a set of separate scans, a higher-resolution in vivo acquisition was performed in 3 volunteers with normal MRIs. The scan was performed using only the SASSI sequence with slice thickness = 1.6 cm, FOV = 8 × 8 cm, matrix size = 10 × 10 pixels (8 × 8 pixels within the VOI), pixel volume = 1.0 cc, TE/TR = 42/1500 ms, Navg = 20, and scan time = 10:14 min. Comparative acquisitions were not obtained using semi-LASER in these last in vivo acquisitions, thus enabling the use of more reasonable TR’s, larger matrix sizes, and shorter scan times with SAR limits for the SASSI sequence.
Analysis
MRSI reconstruction, including channel-combination was performed by the scanner software (spectroscopy task card of the Siemens Syngo MR software). The MRSI data was then extracted for further spectral processing using LCModel (LCMODEL Inc, Oakville, Ontario, Canada)35. Each pixel in the field of view was corrected for frequency shift using the NAA peak at 2.02 ppm. Spectra were analyzed using a basis set to fit curves for the MI, Cho, Cr, and NAA peaks. The area under the fitted peaks was used to produce the metabolite maps shown in Figures 5 and 7 and the spectra shown in Figures 6 and 8. Line width was calculated in LCModel as full width half max (FWHM) in the spectrum for the inner 16 pixels in the phantom spectra, and the inner 4 pixels in vivo. Metabolite SNR for chosen voxels was calculated using LCModel to calculate the integral under a fitted curve and the RMS value of the residual noise after selecting the spectral range containing the metabolite as the dominant peak.
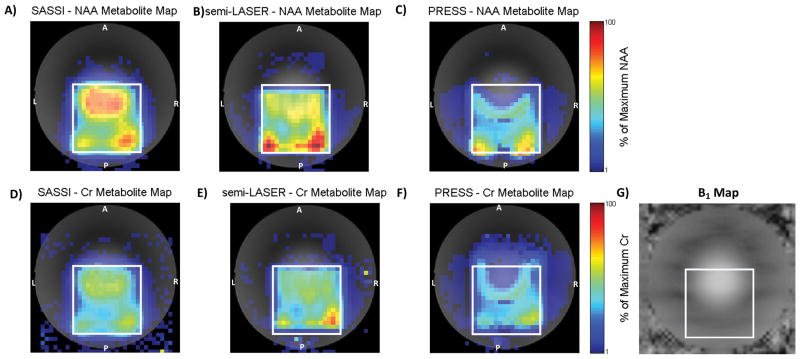
Figure 5.
NAA (A, B, and C) and Cr (D, E, and F) metabolite maps acquired on a slice of the BRAINO phantom using SASSI (A and D), semi-LASER (B and E) and PRESS (C and F). The effect of B1 inhomogeneity on PRESS can be visualized in C) and F). Over-flipping occurs in the top of the box, closest to the center of the phantom experiencing higher than optimal B1, and under-flipping occurs near the bottom of the box, near the edges of the phantom, experiencing lower than optimal B1. G) The B1 map of the phantom.
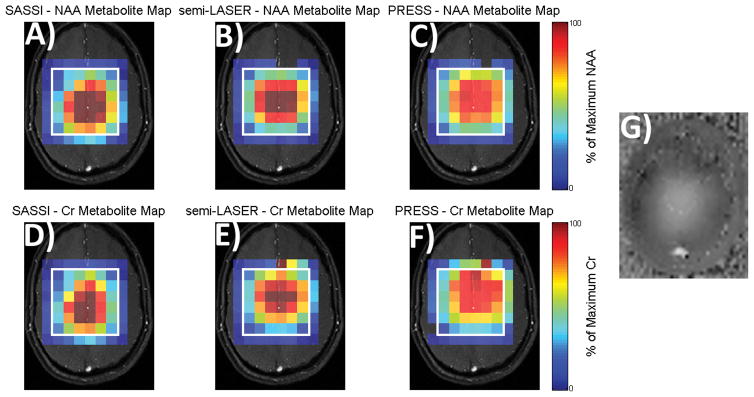
Figure 7.
NAA (A, B, and C) and Cr (D, E, and F) metabolite maps of signal acquired from a human volunteer using SASSI (A and D), semi-LASER (B and E) and PRESS (C and F). G) B1 map in vivo. Signal obtained from SASSI and semi-LASER is overall higher and less shifted than PRESS.
Figure 6.
Metabolite spectra obtained using SASSI, semi-LASER, and PRESS sequences from several locations in the selected grid. The spectra for the three sequences obtained from the A) red, B) orange, C) yellow, D) cyan, E) magenta, and F) blue voxels indicated on the H) brain metabolite phantom. G) Expanded SASSI spectra from the center of the VOI showing NAA, Cr, Cho, and MI peaks.
Figure 8.
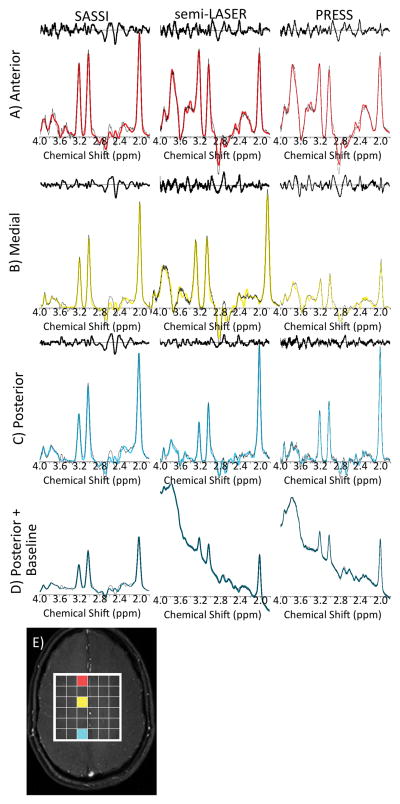
In vivo metabolite spectra fit after baseline subtraction as well as associated noise residuals for the SASSI, semi-LASER, and PRESS sequences shown for the same acquisitions and volume of the brain as the maps in Figure 7. Spectrum from the A) anterior region (red), B) medial region (yellow), C) posterior region (cyan), of the brain. D) The original spectrum from the posterior voxel, prior to baseline subtraction. E) The locations from which the spectra in A, B, C, and D are acquired.
Results
The simulated SPSP profile for the adiabatic SPSP 180° pulses used in the SASSI sequence is shown in Figure 3B. The pulses have a spectral BW of 1.02 kHz at full width half maximum and a 90%–10% spectral transition band of 150 Hz. The SASSI pulses response to increasing B1, Figure 4(C and D), shows that the magnitude of the signal increased as adiabatic threshold was approached (ODF =1) and that the spectral profile remains fairly invariant until very high overdrive factors (which are beyond many RF amplifier peak amplitude limits) start resulting in sub-pulse overdrive. The spatial profiles suffer from ripple as ODF (B1 amplitude) is increased. The response of the SPSP 90° pulse to varying B1 is shown in Figure 4(A and B). The pulse profiles vary as expected for a non-adiabatic 90° excitation pulse. However, this variation is still less severe than what would be expected for a non-adiabatic 180° pulse.
The phantom spectra obtained from all three sequences are shown in Figure 6. The SNRs of the fitted NAA and Cr peaks are listed in Table 1 for three selected voxels.
Table 1.
Relative performance of SASSI, semi-LASER, and PRESS, in phantom and in vivo experiments. For selected voxels marked in Figures 6 and 8, the SNR, calculated from fitted peak integrals and residual noise, for the NAA and Cr peaks as well as the NAA/Cr ratio is summarized for each sequence.
| Anterior | Medial | Posterior | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
NAA SNR |
Cr SNR |
NAA Cr |
NAA SNR |
Cr SNR |
NAA Cr |
NAA SNR |
Cr SNR |
NAA Cr |
||
| phantom | PRESS | 0±0% | 11±4% | 0.000 | 16±5% | 15±5% | 1.068 | 15±3% | 08±5% | 1.705 |
| semi-LASER | 05±8% | 16±8% | 0.300 | 17±5% | 13±6% | 1.269 | 17±5% | 11±8% | 1.601 | |
| SASSI | 20±5% | 16±6% | 1.225 | 18±6% | 15±5% | 1.195 | 17±4% | 14±5% | 1.173 | |
| in vivo | PRESS | 09±4% | 07±6% | 1.228 | 11±4% | 07±5% | 1.559 | 15±3% | 09±4% | 1.715 |
| semi-LASER | 09±4% | 07±4% | 1.339 | 13±4% | 08±4% | 1.537 | 18±4% | 11±3% | 1.625 | |
| SASSI | 16±2% | 11±2% | 1.482 | 18±2% | 12±3% | 1.459 | 16±2% | 12±3% | 1.366 | |
The sequence parameters were chosen to allow semi-LASER to run without interruption due to SAR safety constraints, thus the PRESS and SASSI sequences were both running below SAR constraints. For comparison, all three sequences were operated with sequence parameters, including TR, chosen such that the most SAR intensive sequence (semi-LASER) operated at the SAR limit (100%). With these parameters, the power deposited by the PRESS sequence was 17% of the SAR limit, while the power deposited by the SASSI pulse was 34% of the SAR limit in both in vivo and phantom.
Phantom Results
The line width of the spectra acquired in the phantom was similar for all 3 sequences. NAA line widths were 6 ± 1 Hz, 7 ± 1 Hz, and 8 ± 1 Hz in SASSI, semi-LASER, and PRESS, respectively; the Cr line widths were 6 ± 1 Hz, 8 ± 1 Hz, and 9 ± 1 Hz in SASSI, semi-LASER, and PRESS, respectively. The metabolite maps for NAA and Cr generated from the BRAINO phantom by the SASSI, semi-LASER, and PRESS sequences are shown in Figure 5. The sensitivity of the PRESS to B1 variation results in the signal loss seen in Figures 5C and 5F. The RF transmit voltage was set to achieve the highest average signal from the selected VOI (PRESS box in white). However, due to the significant variation in B1 over the VOI, PRESS excitation and refocusing pulses are severely overdriven in the central anterior region of the VOI, resulting in reduced metabolite SNR. Signal homogeneity is substantially improved over the VOI due to the B1-insensitivity imparted by the adiabatic refocusing pulses in SASSI (Figures 5A and 5D) and semi-LASER (Figures 5B and 5E).
A spatial shift in both the anterior-posterior (A-P) direction (vertical axis of the image) and lateral right-left (R-L) direction (horizontal axis of the image) is visible between the NAA metabolite map and the Cr metabolite map for both the semi-LASER and PRESS sequences. Therefore both sequences suffer from different degrees of CSL error due to the limited BW of the pulses used for selection in those spatial dimensions. The PRESS sequence suffered from the most severe CSL error, with spatial shifts of approximately 4 voxels (25%) in the A-P dimension and 2 voxels (12.5%) in the R-L dimension for NAA, and approximately 2 voxels (12.5%) in the A-P dimension and 1 voxel (6.25%) in the R-L dimension for Cr. Although semi-LASER uses higher-BW adiabatic pulses to select in at least one of the spatial dimensions, there is still some CSL error affecting the VOI. Spatial shifts for semi-LASER were approximately 1 voxels (6.25%) in the A-P dimension and 1 voxel (6.25%) in the R-L dimension for NAA, and approximately 1 voxel (6.25%) in the A-P dimension and 1 voxel (6.25%) in the R-L dimension for the Cr. NAA and Cr metabolites were always shifted in opposing directions. Due to the high BW (~9 kHz) spatial sub-pulses used in SASSI, the CSL error for the equivalent grid is reduced to less than 1 voxel for both NAA and Cr in both in-plane spatial dimensions.
Spectra from 6 voxels within the VOI are shown in Figure 6. These voxels are colored in red, orange, yellow, cyan, magenta, and blue in Figure 6, and represent a sampling from the anterior, medial-anterior, middle, posterior, left-lateral, and right-lateral regions of the VOI, respectively. Calculated SNR is shown in Table 1 is shown for the anterior (red) middle (yellow), and posterior (cyan) voxels. The NAA SNR was similar in the medial (16–18), and posterior voxels (15–17), for the semi-LASER, SASSI, and PRESS sequences. In the anterior (red) voxel, the NAA SNR was completely suppressed in the PRESS sequence and substantially diminished for the semi-LASER sequence (SNR = 5), when compared to the SASSI sequence (SNR = 20) due to a combination of B1 inhomogeneity and CSL error. In the posterior (cyan) voxel, the Cr SNR for the PRESS and semi-LASER sequences were reduced when compared to SASSI, also primarily due to CSL error. The SNRs for both NAA and Cr obtained using the SASSI sequence were more consistent between the edge and middle voxels than those obtained for PRESS and semi-LASER demonstrating more uniform and un-shifted coverage provided by SASSI.
In vivo Results
The line width of the spectra acquired in vivo was similar for all 3 sequences. NAA line widths were 14 ± 2 Hz, 16 ± 1 Hz, and 14 ± 1 Hz in SASSI, semi-LASER, and PRESS, respectively; the Cr line widths were 16 ± 3 Hz, 16 ± 4 Hz, and 18 ± 3 Hz in SASSI, semi-LASER, and PRESS, respectively.
Figure 7 shows in vivo metabolite maps acquired for the three sequences scaled using the same color bar. Increased SNR and less chemical shift is evident for the semi-LASER and SASSI sequences when compared to PRESS. Figure 8 show the in vivo spectra for the anterior, medial, and posterior voxels, marked in red, yellow, and cyan on the brain in Figure 8E, respectively. In Figure 8A–8C, both the spectra and the scaled residuals after the baseline and spectral fit were subtracted are shown. Spectra were scaled to the baseline noise amplitude. In Figure 8D, the spectra of a single voxel, prior to baseline subtraction, is shown. Table 1 summarizes the SNR calculated for NAA and Cr, as well as the NAA to Cr ratio for these chosen voxels.
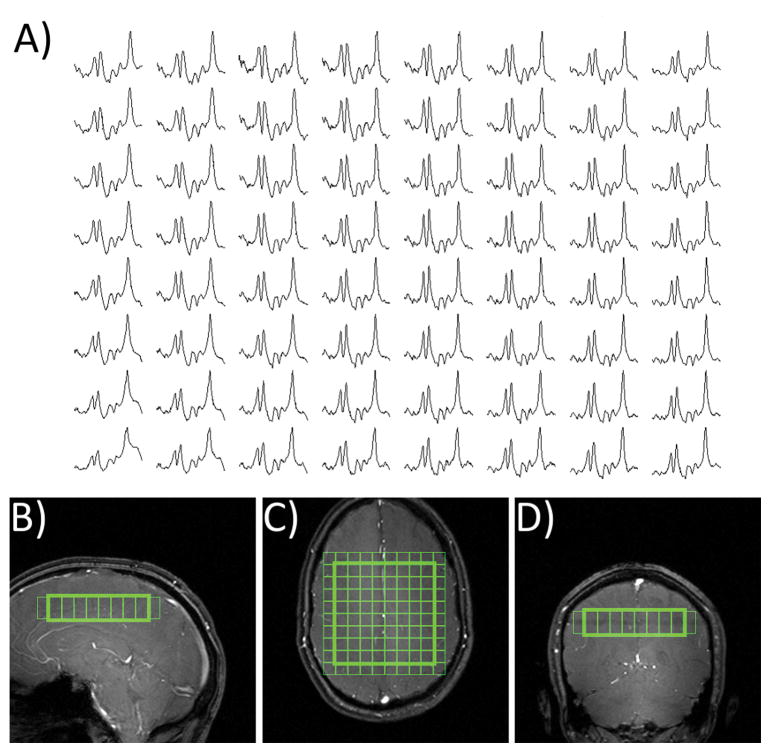
Figure 9 shows higher resolution in vivo spectral grid obtained using SASSI in the brain a new subject obtained in a separate scan. Uniform metabolite SNR is achieved in most regions of the brain. In areas of severe B0 inhomogeneity and fat content SASSI, like any other MRSI sequence, is affected by baseline artifacts, lipid contamination and line broadening. Spectral grids were not corrected for baseline artifacts to demonstrate the degree of inherent water suppression.
Figure 9.
A) In vivo spectra obtained using SASSI. B) Sagittal, C) axial, and D) coronal positioning of the spectral grid.
Discussion
Phantom and in vivo spectroscopic imaging experiments demonstrated the improved B1-insensitivity and immunity to CSL error of SASSI when compared to PRESS and substantially reduced SAR and further reduction of CSL error when compared to the currently available adiabatic alternative for MRSI, semi-LASER.
The metabolite maps obtained using the PRESS sequence shown in Figure 5 illustrates the sensitivity of standard spectroscopic imaging to varying B1. The transmit B1 amplitude was calibrated to produce as uniform a flip angle as possible over the VOI. Thus in the PRESS sequence, regions of high B1 suffer from signal dropout as the refocusing pulses exceed 180°. In regions of lower B1, the refocusing pulses fail to achieve 180° resulting in improper focusing and signal cancellation. As a result, the signal received from the PRESS sequence varies widely over the VOI. Both the semi-LASER and the SASSI sequence also show some variation in the signal intensity. This is due to a combination of the non-adiabatic excitation, and some ripple in the spatial profile of the pulses, as shown in Figures 5 and 7. The use of adiabatic 180° pulses results in much more uniform signal selection and greater SNR over the VOI. Large flip angle pulses, such as 180° pulses are much more sensitive to B1 than lower flip angle excitation pulses, making the replacement of refocusing pulses with adiabatic alternatives more critical. The in vivo B1 map can be seen in Figure 7G; signal inhomogeneity in both the semi-LASER and SASSI pulses is partially due to the non-adiabatic nature of the excitation pulse used and inhomogeneity in the coil receive sensitivity profile.
Figure 5 demonstrates the CSL error in all three sequences. Although B1 inhomogeneity accounts for much of the signal loss common to both the NAA and Cr PRESS metabolite maps, the comparison between the regions excited in the NAA and Cr maps indicate how CSL error affects the metabolites. In PRESS, there is a substantial spatial shift in the selected volume for both in-plane dimensions (~38% in AP and ~19% in R-L) between the selected volumes for NAA and Cr which are separated by 1.0 ppm. For semi-LASER, these shifts were ~12% and ~12% in the A-P and R-L dimensions, respectively. The difference in A-P and R-L shift in PRESS is primarily due to the 90° pulse used to select along one dimension and the 180° pulse used to select along the other dimension. SASSI uses the same 180° pulse to select along both in-plane dimensions. The improved performance of semi-LASER and SASSI when compared to PRESS is due to the higher bandwidth of both the adiabatic pulses, used in semi-LASER, and the spatial subpulses of the adiabatic SPSP pulses, used in SASSI.
It would be of value in many experiments to capture the range of in vivo metabolites from MI at 3.56 ppm to NAA at 2.02 ppm. As shown in the expanded spectra in Figures 6G, MI to NAA are effectively captured by the spectral passband of the SASSI pulse. The effects of SPSP pulses on strongly coupled metabolites whose coupling partners may fall outside the spectral passband was not explored, however this would be an interesting topic of future research.
In PRESS, the selected VOI for MI and NAA would be shifted by approximately 57% in the dimension selected by the 180° pulse (A-P in this experiment) in conventional PRESS. Semi-LASER would reduce this to ~19%. SASSI achieves the best CSL behavior at a less than 5% spatial shift between MI and NAA due to CSL error. To ensure that the metabolites of interest are fully excited within the PRESS box, it is possible to increase the selected VOI to larger than the desired VOI and use outer volume suppression (OVS)36 to suppress the extended regions. However, this approach can make VOIs prescribed near the edge of the brain difficult to capture, particularly when the CSL error is severe. Therefore, minimizing CSL error is critical to full brain spectral coverage.
Without OVS, semi-LASER achieves sharper spatial transition bands than SASSI as is evidenced by the sharper edges of the semi-LASER metabolite maps in Figures 5B and 5E when compared to the SASSI metabolite maps in Figures 5A and 5D. This is because the time-bandwidth product, and equivalently the selectivity, of the spatial sub-pulses used in SASSI is limited by gradient hardware limits and peripheral nerve stimulation limits. The use of OVS bands, which are available in MRSI sequences could sharpen the edges of the VOI.
Adiabatic pulses operate at higher BW and amplitude (when overdriven above adiabatic threshold) than conventional 180° pulses. Therefore semi-LASER and SASSI deposit more RF power than PRESS, with semi-LASER being the most SAR-intensive. Due to SAR safety constraints on the scanner, the TR of the semi-LASER sequence could not be reduced below 2050 ms in a phantom and 4080 ms in vivo. To compare SAR, the TR’s of SASSI and PRESS were matched to semi-LASER; although they could have been shorter. PRESS deposited the least power, 17% of the system SAR limit in both phantom and in vivo experiments. The SASSI sequence deposited 34% of the SAR limit. Since the SASSI pulses are adiabatic, they still deposit more power than the non-adiabatic pulses used in PRESS sequence. The 1/3 reduction in power deposition for SASSI, when compared to semi-LASER, is due 1) to a reduction in the number of adiabatic refocusing pulses; 2) the construction of the adiabatic pulses using the efficient adiabatic SLR transform; and 3) inherent water suppression achieved by the spectrally selective SPSP pulses, eliminating additional water suppression pulses. Not exciting, then suppressing, the water using spectrally selective pulses leaves open the possibility of quantifying metabolite concentrations by exciting and acquiring the water peak within the same TR using an interleaved sequence.
Figure 8D shows spectra from the posterior region of the brain before baseline subtraction produces the spectra shown in Figure 8C. The partial water suppression of the SASSI SPSP pulses, (spectrum before baseline subtraction shown in Figure 8D), performed better than the basic CHESS water suppression applied during the PRESS and the semi-LASER sequences. This is demonstrated by the reduced baseline artifact achieved by the inherent water suppression of the SASSI SPSP pulses when compared to PRESS and semi-LASER. The use of SPSP excitation and refocusing in SASSI ensures that only a fraction of the water is excited in the first place, as opposed to the entire water peak being excited and then suppressed in the other sequences. However, if a more complex, longer, series of CHESS pulses were used (somewhat increasing SAR), for example a variable pulse power and optimized relaxation delays (VAPOR) scheme37, more effective water suppression could be achieved for PRESS and semi-LASER. Similarly, SASSI may be combined with any of these preparatory water suppression schemes in order to achieve additional water suppression.
Other adiabatic SPSP approaches have been proposed for 7T MRSI, including an interleaved narrowband adiabatic SPSP sequence23 and a long TE SPSP sequence29. SASSI improves upon these approaches by using lower-power, higher-bandwidth adiabatic SLR spectral envelopes for the SPSP pulses in order to cover the entire spectral range from MI to NAA at 7T without interleaving. The adiabatic SLR pulse envelopes more uniformly distribute RF energy over the pulse duration resulting in SASSI’s shorter minimum TE (42 ms versus the 90 ms or 144 ms). The short TE makes it possible to capture shorter T2 coupled metabolites such as MI. SASSI can also be used to provide more B1-insensitive and CSL-insensitive voxels for single-voxel MR spectroscopy.
When applying the SASSI sequence to obtain the in vivo spectral grid shown in Figure 9, the reduced power deposition of SASSI when compared to semi-LASER, resulted in more flexible timing parameters that were dictated by metabolite T1s rather than RF duty cycles. Higher resolution grids and more averages were possible when setting acquisition parameters according to the SAR limits of SASSI rather than semi-LASER. Good coverage is obtained with high SNR, as can be seen in the spectra of Figure 9.
Conclusion
The SASSI is a low-SAR, B1-insensitive sequence for MRSI at 7T. SASSI provides greater immunity to the severe B1-inhomogeneity and CSL errors at 7T than PRESS and although the B1-insensitivity of SASSI is similar to semi-LASER, SASSI deposits a third of the RF power of the semi-LASER sequence.
Acknowledgments
Funding Sources: NIH-NINDS R00 NS070821, Icahn School of Medicine Capital Campaign, Translational and Molecular Imaging Institute and Department of Radiology, Icahn School of Medicine at Mount Sinai, Siemens Healthcare.
We would like to acknowledge helpful discussions with Nouha Salibi (Siemens Medical) and Junqian (Gordon) Xu. We would like to thank Stephen Provencher at LCMODEL Inc for the metabolite basis sets used in the spectral analysis.
References
- 1.Zhu H, Barker PB. MR spectroscopy and spectroscopic imaging of the brain. Methods Mol Biol. 711:203–226. doi: 10.1007/978-1-61737-992-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wijtenburg SA, Yang S, Fischer BA, Rowland LM. In vivo assessment of neurotransmitters and modulators with magnetic resonance spectroscopy: Application to schizophrenia. Neurosci Biobehav Rev. 2015;51:276–295. doi: 10.1016/j.neubiorev.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson M, Gill SK, MacPherson L, English M, Arvanitis TN, Peet AC. Noninvasive detection of glutamate predicts survival in pediatric medulloblastoma. Clin Cancer Res. 20:4532–4539. doi: 10.1158/1078-0432.CCR-13-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delorme S, Weber MA. Applications of MRS in the evaluation of focal malignant brain lesions. Cancer Imaging. 2006;6:95–99. doi: 10.1102/1470-7330.2006.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wijnen JP, Idema AJ, Stawicki M, Lagemaat MW, Wesseling P, Wright AJ, Scheenen TW, Heerschap A. Quantitative short echo time 1H MRSI of the peripheral edematous region of human brain tumors in the differentiation between glioblastoma, metastasis, and meningioma. J Magn Reson Imaging. 2012;36:1072–1082. doi: 10.1002/jmri.23737. [DOI] [PubMed] [Google Scholar]
- 6.Boer VO, van Lier AL, Hoogduin JM, Wijnen JP, Luijten PR, Klomp DW. 7-T (1) H MRS with adiabatic refocusing at short TE using radiofrequency focusing with a dual-channel volume transmit coil. NMR Biomed. 24:1038–1046. doi: 10.1002/nbm.1641. [DOI] [PubMed] [Google Scholar]
- 7.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62:868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61:1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 9.Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at 7 T. Magn Reson Med. 2001;46:451–456. doi: 10.1002/mrm.1213. [DOI] [PubMed] [Google Scholar]
- 10.Kreis R, Ernst T, Ross BD. Development of the human brain: in vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn Reson Med. 1993;30:424–437. doi: 10.1002/mrm.1910300405. [DOI] [PubMed] [Google Scholar]
- 11.Michaelis T, Merboldt KD, Bruhn H, Hanicke W, Frahm J. Absolute concentrations of metabolites in the adult human brain in vivo: quantification of localized proton MR spectra. Radiology. 1993;187:219–227. doi: 10.1148/radiology.187.1.8451417. [DOI] [PubMed] [Google Scholar]
- 12.Pouwels PJ, Frahm J. Regional metabolite concentrations in human brain as determined by quantitative localized proton MRS. Magn Reson Med. 1998;39:53–60. doi: 10.1002/mrm.1910390110. [DOI] [PubMed] [Google Scholar]
- 13.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 14.Moonen CT, von Kienlin M, van Zijl PC, Cohen J, Gillen J, Daly P, Wolf G. Comparison of single-shot localization methods (STEAM and PRESS) for in vivo proton NMR spectroscopy. NMR Biomed. 1989;2:201–208. doi: 10.1002/nbm.1940020506. [DOI] [PubMed] [Google Scholar]
- 15.Luyten PR, Marien AJ, den Hollander JA. Acquisition and quantitation in proton spectroscopy. NMR Biomed. 1991;4:64–69. doi: 10.1002/nbm.1940040206. [DOI] [PubMed] [Google Scholar]
- 16.Adalsteinsson E, Spielman DM. Spatially resolved two-dimensional spectroscopy. Magn Reson Med. 1999;41:8–12. doi: 10.1002/(sici)1522-2594(199901)41:1<8::aid-mrm3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Sacolick LI, Rothman DL, de Graaf RA. Adiabatic refocusing pulses for volume selection in magnetic resonance spectroscopic imaging. Magn Reson Med. 2007;57:548–553. doi: 10.1002/mrm.21162. [DOI] [PubMed] [Google Scholar]
- 18.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153:155–177. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 19.Hess AT, Andronesi OC, Tisdall MD, Sorensen AG, van der Kouwe AJ, Meintjes EM. Real-time motion and B0 correction for localized adiabatic selective refocusing (LASER) MRSI using echo planar imaging volumetric navigators. NMR Biomed. 25:347–358. doi: 10.1002/nbm.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- 21.Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. MAGMA. 2008;21:95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham CH, Vigneron DB, Marjanska M, Chen AP, Xu D, Hurd RE, Kurhanewicz J, Garwood M, Pauly JM. Sequence design for magnetic resonance spectroscopic imaging of prostate cancer at 3 T. Magn Reson Med. 2005;53:1033–1039. doi: 10.1002/mrm.20478. [DOI] [PubMed] [Google Scholar]
- 23.Balchandani P, Pauly J, Spielman D. Interleaved narrow-band PRESS sequence with adiabatic spatial-spectral refocusing pulses for 1H MRSI at 7T. Magn Reson Med. 2008;59:973–979. doi: 10.1002/mrm.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conolly SM, Pauly JM. Method and means for magnetic resonance imaging and spectroscopy using two-dimensional selective adiabatic PI pulses. 5189371. Patent US. 1993 filed August 8, 1991.
- 25.Schricker AA, Pauly JM, Kurhanewicz J, Swanson MG, Vigneron DB. Dualband spectral-spatial RF pulses for prostate MR spectroscopic imaging. Magn Reson Med. 2001;46:1079–1087. doi: 10.1002/mrm.1302. [DOI] [PubMed] [Google Scholar]
- 26.Conolly S, Pauly J, Nishimura D, Macovski A. Two-dimensional selective adiabatic pulses. Magn Reson Med. 1992;24:302–313. doi: 10.1002/mrm.1910240211. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham CH, Vigneron DB, Chen AP, Xu D, Hurd RE, Sailasuta N, Pauly JM. Design of symmetric-sweep spectral-spatial RF pulses for spectral editing. Magn Reson Med. 2004;52:147–153. doi: 10.1002/mrm.20116. [DOI] [PubMed] [Google Scholar]
- 28.Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING) Magn Reson Med. 1997;38:311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- 29.Xu D, Cunningham CH, Chen AP, Li Y, Kelley DA, Mukherjee P, Pauly JM, Nelson SJ, Vigneron DB. Phased array 3D MR spectroscopic imaging of the brain at 7 T. Magn Reson Imaging. 2008;26:1201–1206. doi: 10.1016/j.mri.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balchandani P, Pauly J, Spielman D. Designing adiabatic radio frequency pulses using the Shinnar-Le Roux algorithm. Magn Reson Med. 2010;64:843–851. doi: 10.1002/mrm.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm [NMR imaging] IEEE Trans Med Imaging. 1991;10:53–65. doi: 10.1109/42.75611. [DOI] [PubMed] [Google Scholar]
- 32.Hargreaves BA, Cunningham CH, Nishimura DG, Conolly SM. Variable-rate selective excitation for rapid MRI sequences. Magn Reson Med. 2004;52:590–597. doi: 10.1002/mrm.20168. [DOI] [PubMed] [Google Scholar]
- 33.Conolly S, Nishimura D, Macovski A, Glover G. Variable-rate selective excitation. Journal of Magnetic Resonance (1969) 1988;78:440–458. [Google Scholar]
- 34.Haase A, Frahm J, Hanicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985;30:341–344. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 35.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 36.Luo Y, de Graaf RA, DelaBarre L, Tannus A, Garwood M. BISTRO: an outer-volume suppression method that tolerates RF field inhomogeneity. Magn Reson Med. 2001;45:1095–1102. doi: 10.1002/mrm.1144. [DOI] [PubMed] [Google Scholar]
- 37.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41:649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]