Abstract
Treatment with glucocorticoids is associated with lower bone mineral density (BMD). We performed a genome-wide association study to analyze interactive effects between genotypes and cumulative dose of prednisone (PD) over 4.3 years of follow-up period on the final BMD Z-scores in 461 white children from the Childhood Asthma Management Program. No variants met the conventional criteria for genome-wide significance, and thus we looked for evidence of replication. The top 100-ranked single nucleotide polymorphisms (SNPs) were then carried forward replication in 59 children with acute lymphoblastic leukemia (ALL) exposed to large fixed doses of PD as part of their chemotherapeutic regimen. Among them, rs6461639 (interaction P = 1.88 ×10−5 in the CAMP population) showed a significant association with the final BMD Z-scores in the ALL population (P = 0.016). The association of the ALL population was only present after correction for the anti-metabolite treatment arm (high vs. low dose). We have identified a novel SNP, rs6461639, showing a significant effect on the final BMD Z-scores in two independent pediatric populations after long-term high dose PD treatment.
Keywords: Asthma, Acute lymphoblastic leukemia, Bone mineral density, Child, Glucocorticoid
INTRODUCTION
Glucocorticoids (GCs) are the core therapeutic agents in many childhood diseases. However, GCs have adverse effects on bone health as a consequence of decreased bone formation and increased bone resorption.1 It is known that long term use of GCs is associated with lower bone mineral density (BMD) compared to the population mean in children with nephrotic syndrome,2 systemic lupus erythematosus,3 acute lymphoblastic leukemia (ALL),4 and asthma.5 With ongoing skeletal development, children are more susceptible than adults to GCs effects on BMD. Various factors including gender, age, height, and pubertal stage are known to affect bone loss associated with GCs use in children.6 In terms of genetic factors, we previously reported that CRHR1 polymorphisms impacts the risk of bone density deficits in ALL children treated with GCs and antimetabolites.7 To date, there has been no study evaluating a genetic influence on GCs-induced bone loss in children using a genome-wide approach. We therefore performed a genome-wide association study (GWAS) with the hypothesis that we could identify novel genetic markers showing interactive effects with cumulative dose of GCs on the final BMD in children after a long-term GCs treatment. First, we tested our hypothesis by conducting a GWAS in white children with asthma participating in the Childhood Asthma Management Program (CAMP) trial.8 We then tested associations of the top 100-ranked SNPs in long-term survivors of childhood ALL who participated in our previous study.7
MATERIALS AND METHODS
Each study was approved by the Institutional Review Board of the corresponding institution and informed consents were obtained from all study participants.
Study populations and bone mineral density measurement
The primary group of subjects consisted of Caucasian children from the CAMP trial. The demographics of them and the study design have been described elsewhere.8 In brief, 1,041 children with mild-to-moderate asthma aged 5 to 12 years were randomized to budesonide, nedocromil, or placebo and followed for a mean of 4.3 years. For asthma exacerbations, short courses of oral prednisone (PD) were prescribed per protocol. Each burst consisted of 2 mg/kg per day (up to 60 mg) of PD for 2 days followed by 1 mg/kg per day (up to 30 mg) for 2 days and an option to continue dosing was allowed if improvement was insufficient. The CAMP population was divided into four groups according to their cumulative dose of PD over the follow-up period; PD non-takers and tertiles of PD takers [1st tertile (range: 5 ~ 330mg), 2nd tertile (range: 340 ~ 870mg) and 3rd tertile (range: 880 ~ 4,660mg)]. BMD measurements (in grams per square centimeter) of the lumbar spine (L1–L4) were performed yearly during the study period by means of dual-energy x-ray absorptiometry with the Hologic (Waltham, Mass) QDR-1500 (at 6 centers) or the Lunar (Madison, Wis) DPX (at 2 centers). Hologic DEXA machines were further divided by the use of pencilbeam or fan-beam measurements. Detailed methods to convert Lunar measures to Hologic values and to adjust deviations between pencil- and fan-beam measurements were described in our previous report.9 The BMD Z-score that represents the difference between the measured bone density in a subject and the average bone density in age- and sex-matched controls was calculated by using the CAMP internal references.5 In the present study the final BMD Z-score measured after mean follow-up duration of 4.3 years was used as a target phenotype. We confirmed that the chance of delayed puberty which might affect the final BMD Z-scores was small in the CMAP subjects.
The secondary population was drawn from participants in our previous study with ALL children.7 They were treated at St Jude Children’s Research Hospital between 1994 and 1998.10 For induction therapy, PD 40 mg per square meter of body surface area per day (mg/m2/day) was administered for one month, thereafter patients received dexamethasone 8 mg/m2/day (roughly 64 mg/m2/day in PD equivalents) for 7 days every 4 weeks for 120 weeks of continuation therapy. Based on the probability of ALL relapse, they were divided into two treatment arms; high-dose anti-metabolites + GC vs. low-dose anti-metabolites + GC. The high exposure to anti-metabolites could have obscured genetic effects on GCs-induced bone loss.7,11,12 We hypothesized that different levels of anti-metabolite administered might influence the association of GC with the final BMD, although GC dose was fixed. The final BMD measurements of the lumbar spine (L1-L2) were performed by Quantitative Computed Tomography with a Siemens Somatom-Plus spiral CT scanner (Siemens, Iselin, NY) at least 4 years after completing ALL therapy. Z-scores of the final BMD (expressed as the mean of the L1-L2 bone) were calculated for each patient. Detailed methods were described in our previous report.7
Genotyping
The CAMP population was genotyped on the HumanHap550v3 BeadChip or Infinium HD Human610-Quad BeadChip by Illumina, Inc. (San Diego, CA) and the ALL population was genotyped on the Affymetrix 5.0 chip by Affymetrix, Inc. (Santa Clara, CA). Marker quality control is detailed in the online supplement. Each single nucleotide polymorphism (SNP) had a completion rate > 95%, a minor allele frequency (MAF) of > 0.05, and a Hardy-Weinberg equilibrium P value of > 0.001. After merger with the phenotype dataset, 461 subjects with asthma and 59 subjects with ALL were included in the final analysis.
Statistical analysis
Using a linear regression gene-environment interaction model as implemented in the PLINK (http://pngu.mgh.harvard.edu/purcell/plink/),13 we performed a GWAS to test the following model: final BMD ~ genotype (additive genetic model) + cumulative dose of PD + genotype*cumulative dose of PD + covariates. Based on our previous report, age, sex, BMD (Z-score), height, body mass index, serum concentration of vitamin D, and Tanner stage measured at baseline were included as covariates.9 Since we focused on the interactive effect between genotype and cumulative dose of PD on final BMD, we reported P values from the ‘genotype*cumulative dose of PD’ term. We then selected top-ranked 100 SNPs to carry forward to test whether significant associations were found in the ALL population. Analysis was done using the following model: final BMD ~ genotype (additive genetic model) + gender + obesity + ancestry + treatment arm (high dose anti-metabolites group vs. low dose anti-metabolites group). A significant association in the secondary population was defined as a result with a nominal P value of less than 0.05. We confirmed that the assumptions of the linear regression model were met. Analysis was performed with R version 2.15.3 (www.r-project.org).
RESULTS
Table 1 summarizes the baseline characteristics of the primary (CAMP) and the secondary (ALL) populations. The final BMD Z-score of the third tertile of PD takers was significantly lower than that of PD non-takers (P = 0.015, Supplementary Figure 1).
Table 1.
Baseline characteristics of enrolled subjects
| CAMP population (n = 461)
|
ALL population (n = 59)¶ | ||||
|---|---|---|---|---|---|
| PD non-taker (n = 93) | PD taker (n = 368)
|
||||
| Tertile 1 | Tertile 2 | Tertile 3 | |||
| Male sex (%) | 62.4 | 57.7 | 66.4 | 61.0 | 52.5 |
| Age (year) | 9.7 (7.6~11.0) | 8.5 (6.8~10.6) | 8.7 (7.1~10.3) | 8.4 (7.1~10.3) | 6.9 (1.4~18.7) |
| Height (cm) | 139.7 (127.3~146.7) | 130.8 (121.0~141.2) | 131.3 (122.9~140.9) | 130.9 (122.2~142.8) | NA |
| BMI (kg/m2) | 17.7 (16.3~21.0) | 17.2 (15.7~19.3) | 17.1 (15.8~19.6) | 16.9 (15.6~18.6) | 15.8 (14.6~18.3) |
| BMD (Z-score) | −0.10 (−0.81~0.53) | −0.16 (−0.62~0.34) | 0.02 (−0.55~0.55) | −0.14 (−0.53~0.39) | NA |
| Vitamin D (ng/mL) | 35.8 (27.7~46.6) | 35.9 (28.8~47.5) | 37.8 (29.6~48.8) | 35.7 (29.3~47.2) | NA |
| Tanner stage (%) | |||||
| I/II/III/IV/V | 65.6/24.7/6.5/3.2/0 | 71.5/22.0/5.7/0/0.8 | 76.2/19.7/1.6/2.5/0 | 74.8/16.3/6.5/2.4/0 | NA |
| Cumulative PD (mg) | 0 | 180.0 (135.0~212.5) | 540.0 (420.0~660.0) | 1430.0 (1100.0~1962.0) | 14640* |
| Race/ethnicity (%) | |||||
| White | 100 | 100 | 100 | 100 | 72.9 |
| Others | 0 | 0 | 0 | 0 | 27.1 |
| Final BMD (Z-score) | −0.08 (−0.83~0.73) | −0.19 (−0.67~0.49) | −0.19 (−0.79~0.62) | −0.36 (−0.81~0.25) | −0.80 (−2.90~4.0) |
Data indicate median (IQR). NA; Not available,
High-dose anti-metabolites arm (n = 29) and low-dose anti-metabolites arm (n = 30),
mg/m2/day
No variants met the conventional criteria for genome-wide significance, and thus we looked for evidence of replication. The top 100-ranked SNPs identified in the primary GWAS are listed in Table S1 (Supplementary Table 1). Among those SNPs, we found that rs6461639 (P = 1.88 ×10−5) also showed a significant effect on the final BMD in the ALL population (P = 0.016). Power for the interaction analysis was calculated using Quanto software 1.2.4 (http://biostats.usc.edu/Quanto.html). Quanto is a program for computing either power or required sample size for association studies of genes, environmental factors, gene-environment interaction.14 An additive genetic model and alpha value (two-sided) of 0.05 were assumed. For rs6461639, an estimated power was 0.39 for main (gene) effect and 0.30 (2 df test) for interaction. The CAMP subjects carrying two variant (minor) alleles of rs6461639 showed significantly lower final BMD Z-scores compared to those with two reference (major) alleles after adjustment by covariates [P = 0.0016, odds ratio (95% confidence interval) = 0.54 (0.37 – 0.78), Figure 1a] in the third tertile of PD takers. The same trend was found in the ALL population (Figure 1b). Given that dose of PD administered in the ALL population was much higher than that of the CAMP population, we may say that the ALL population was in similar situation with the third tertile of PD takers (higher cumulative dose of PD) in the CAMP population.
Figure 1. Final bone mineral density Z-score in the CAMP and ALL population genotyped for rs6461639, stratified by genotype status.
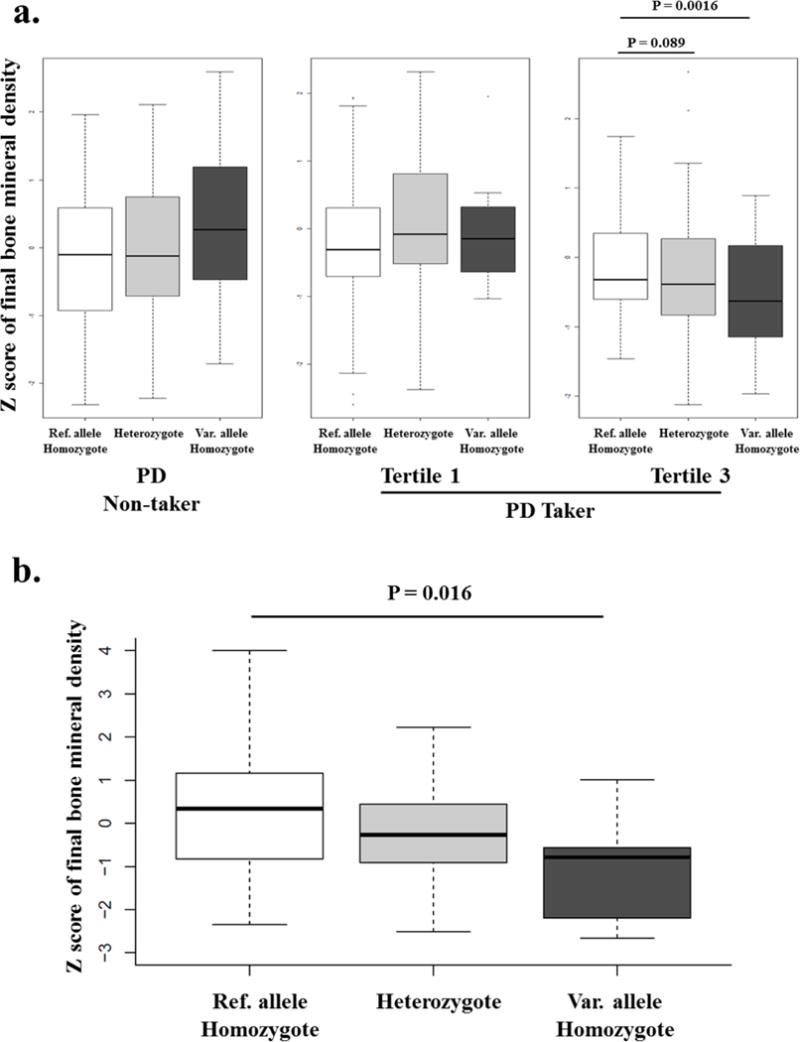
a. CAMP population
P values were obtained from regression analysis adjusted by age, sex, height, BMD Z-score, height, Tanner stage, and vitamin D level measured at baseline. Data represent median (IQR). Ref. allele; reference (major) allele, Var. allele; variant (minor) allele
b. ALL population
Ref. allele; reference (major) allele, Var. allele; variant (minor) allele
In addition to tertiles analysis, we assessed the linear association (e.g. ’final BMD Z-score = a + b * cumulative dose of PD (mg) (a; intercept, b; slope)’) for each genotype of rs6461639 in the CAMP population (Figure 2). The significant linear correlation was seen only in subjects with two variant alleles of rs6461639 (Pearson’s correlation coefficient = −0.322, P = 0.0074). To further explore potential clinical effects behind our association, we assessed the relationship between non-PD takers and a hypothetical subject with asthma who was homozygous for the mutant rs6461639 allele following 4,000 mg of corticosteroid use over time (Figure 3). Overall, this subject would demonstrate −1.25 as the final BMD Z-score of the CAMP subject vs. the non-PD counterpart.
Figure 2. Correlations between cumulative dose of prednisolone and final bone mineral density stratified by genotype of rs6461639 in the CAMP population.
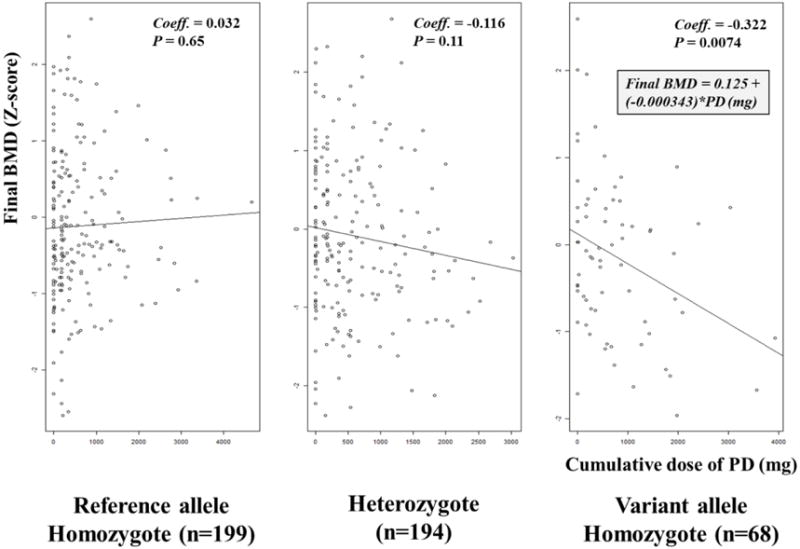
BMD; bone mineral density, Coeff; Pearson’s correlation coefficient
Figure 3. Final bone mineral density Z-score of the CAMP subject carrying two variant alleles of rs6164639 whose cumulative dose of prednisolone was 4,000mg.
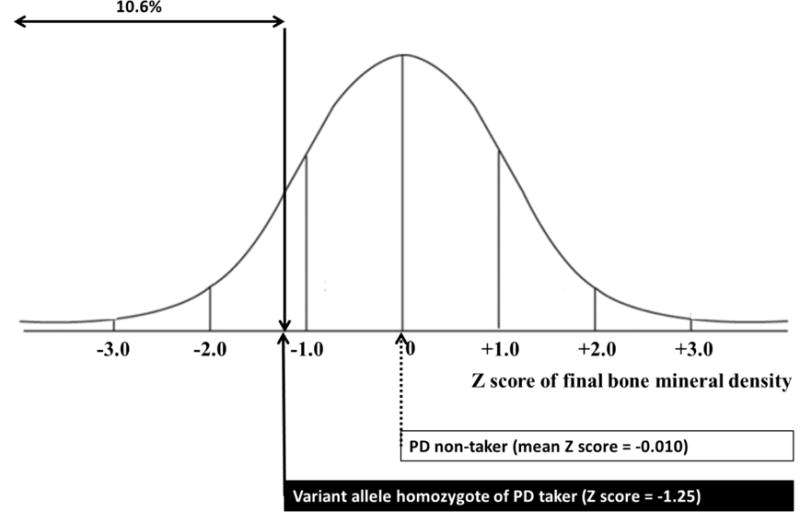
Final bone mineral density Z-score was calculated using the linear regression equation obtained from the CAMP subjects who were homozygous for variant alleles of rs6461639 (refer to Figure 2) with the assumption that the other clinical variables that may affect bone mineral density were ignored.
DISCUSSION
In the present study, we identified top-ranked 100 SNPs that showed significant interactive effects with cumulative dose of oral PD on the final BMD Z-score in childhood asthmatics. Among those 100 SNPs, we confirmed that rs6461639 located on the intron of Rap guanine nucleotide exchange factor 5 (RAPGEF5) gene was also significantly associated with the final BMD Z-score in ALL patients treated with long-term PD after adjustment of anti-metabolite effects. The significant interaction in the CAMP cohort was mainly driven by the effect observed in high dose PD takers (tertile 3 in Figure 1a). Considering that the children with ALL were treated with much higher doses of PD, the ALL cohort provides a possible evidence for replication of results shown in children taking high doses of oral GCs in the CAMP cohort. The pharmacogenomic magnitude of the clinical effect is substantial: the final BMD Z-score would be 1.25 less in CAMP subjects with two variant alleles of rs6461639, if we assumed that total dose of PD during 4.3 years of follow-up was 4,000 mg and factors other than genotype did not affect BMD. A total dose of 4,000 mg is equivalent to 5 mg of PD every other day for 4.3 years, which is not unusual in the real world setting. A Z-score of −1.25 indicates that a subject’s BMD would be within the lowest 10.6% for age. To the best of our knowledge, this is the first study showing an interactive effect on BMD between genetic factors and cumulative dose of GCs.
Bone mineralization throughout childhood and adolescence determines the peak bone mass. It is thought that heritable factors account for an estimated 60–80% of the variability in peak bone mass, with diet, physical activity and hormonal status serving as important modifiers of bone accrual.15 As discussed earlier, a chronic administration of GCs evidently hindered proper bone mineralization in children.2–6 GCs-induced bone loss is known to be associated with combined inhibition of both the osteoblast and osteoclast indicating that GCs-mediated inhibition of osteoclast function retards remodeling and thus dampens osteogenesis as shown in other states of the dynamic bone.16,17 The issue as to how GCs affect bone resorption by osteoclast has been controversial. A recent report comparing the effects of dexamethasone on wild-type osteoclasts with those derived from mice with disruption of the GC receptor in osteoclast lineage cells showed that the inhibitory effect of dexamethasone on bone resorption reflected failure of osteoclasts to organize their cytoskeleton.17 The osteoclast undergoes continuous reorganization with different phases of the resorptive cycle and thus a unique cytoskeleton is one of important characteristics of osteoclast.18 Dexamethasone in low nanomolar concentrations disarranged the osteoclast cytoskeleton, resulting in failure to spread and ineffectively resorb mineralized matrix.17 The osteoclast cytoskeleton is modulated by small GTPases like RhoA19 and Rac20 that transit to their GTP bond state under the influence of guanine nucleotide exchange factors (GEFs). Therefore, it is possible that GEFs act in the pathogenesis of GCs-induced decrease in BMD. Rs6461639 is located on the intron of RAPGEF5 gene that serves as Rap1 activators by promoting acquisition of GTP to maintain the active GTP-bound state.21 Notably, osteoclast-specific deletion of Rap1 yielded osteopetrotic mice by promoting talin/β integrin recognition and thus changing cytoskeletal organization.22 In addition, PPI finder, a web-based mining tool for human protein-protein interactions23 indicates that RAPGEF5 has a GC-regulated promoter.24 Taken together, RAPGEF5 regulated by GCs may deteriorate BMD by deranging cytoskeletal organization of osteoclast via Rap1. Recent genome-wide meta-analysis have identified several loci associated with BMD including 18p11.21 (FAM210A), 7q21.3 (SLC25A13), 11q13.2 (LRP5), 4q22.1 (MEPE), 2p16.2 (SPTBN1), 10q21.1(DKK1), 1p36.12 (ZBTB40), 1p31.3 (GPR177), and 7q.31.31 (CPED1 and WNT16).25–27 However, studies evaluating BMD decreases induced by GCs were not included, which may be the reason why RAPGEF5 was not listed in the previous results.
Rs6461639 is located outside of the coding region of RAPGEF5 gene, so we search its regulatory potential using ‘RegulomeDB’ which integrates a large collection of regulatory information from high-throughput experimental data sets and provides functional categories based on the strength of evidence.28 Rs64614639 is classified as the ‘likely to affect binding’ category (2b) based on direct evidence of binding through ChIP-seq and DNase with either a matched positional weight matrix to the ChIP-seq factor or a DNase footprint.29 We have identified 2 SNPs (rs1981597 and rs6968584) in LD with rs6461639 using the SNAP site (r2 threshold = 0.8, distant limit = 500 kb, CEU population).30 However we failed to find any functional relevancy of those two SNPs. In addition, we have identified 35 SNPs (on the same chromosome, frequency > 0.1, but not rs6461639 or in significant LD with rs6461639) showing significant (P < 0.0001, CEU population) effects on the expression of RAPGEF5 using the SCAN database.31 Although we cannot confirm a direct link between rs6461639 and RAPGEF5 gene function, rs6461639 is a functionally relevant SNP and thus may have its own role in the pathogenesis of GCs-induced decrease in BMD. Rs6461639 was significantly associated with the final BMD Z-score only in childhood asthmatics with a median cumulative dose of PD greater than 332.5 mg per year; this result was replicated in childhood ALL patients treated with cumulative dose of PD much higher than childhood asthmatics. It is not uncommon to see asthmatic children (and adults) who require equivalent doses of GC to control their disease activities in real practice. Considering that rs6461639 is a common SNP with MAF of 34% in Caucasians,32 our findings suggest that there may be clinical utility in genotyping rs6461639 in children before initiating GCs treatment, especially in cases of high dose therapy, to effectively anticipate and manage the possible decrease in BMD.
There are several potential limitations in generalizing our results. First, a P value of rs6461639 does not exceed the conventional levels for genome-wide significance (P < 5.0 × 10−8) in the first cohort and for Bonferroni correction adjusted P value (P = 0.005) in the replication cohort. Therefore, we cannot completely exclude the possibility that our results may be due to chance alone (false-positive associations). One possible explanation for these P values is the small sample size of the primary cohort was one of the contributing factors, as large sample sizes are generally needed when interactions are introduced into hypothesis testing.33 In addition, it is possible that the current GWAS design does not provide enough statistical power to detect interactions, because such a design mainly focuses on the main effect of a signal SNP on risk of common, complex diseases.34 However, it was noted that even extremely small P values provide surprisingly little information as the likelihood of subsequent replication.35 For examples, genetic variations with genome-wide significance showed poor consistency across GWAS of major depression36 and allergic rhinitis.37 Hence, we looked for evidence of replication of SNPs from our first GWAS. Secondly, the ALL population was not ideal to reveal interactive effects of SNP and the cumulative dose of GCs, because they were treated by only fixed dose of GCs, although we assumed that different levels of anti-metabolite might influence the association of GCs with the final BMD. However, it was very difficult for us to find clinical cohorts taking moderate to large doses of oral GCs with available BMD and GWAS genotype data. Therefore, it should be acknowledged that the association of the ALL population was only present after correction for the anti-metabolite treatment arm. Thirdly, the time gap between the measurement of clinical variables at baseline (covariates in our analysis) and the measurements of final BMD (target phenotype in our analysis) was considerable (over 4 years) and we could not measure the possible changes of all clinical variables that may have affected BMD during follow-up period. Nevertheless, we have adjusted our analysis for well-known potential confounders related to BMD, including age, sex, baseline BMD (Z-score), height, body mass index, serum concentration of vitamin D, and Tanner stage measured at baseline. Finally, although we found that rs64616395 has a regulatory potential, further mechanistic studies are needed to confirm the effect of rs6461639 on RAPGEF5 gene.
In conclusion, we have identified rs6461639 which demonstrates a pharmacogenomic interactive effect with cumulative dose of PD on the final BMD Z-score in children after a long-term GCs treatment. These results widen our understanding of the mechanistic basis as to how GCs induce decreases in BMD and may eventually be used as a preventive strategy in practice.
Supplementary Material
Acknowledgments
This work was supported by the NIH Pharmacogenomics Research Network: U01 HL65899 and U01 GM92666, and by NIH NHLBI R01 HL092197, NINR R01 NR013391, and NCI grants CA 142665, and CA 21765, and by the American Lebanese Syrian Associated Charities (ALSAC). The Childhood Asthma Management Program is supported by contracts NO1-HR-16044, 16045, 16046, 16047, 16048, 16049, 16050, 16051, and 16052 with the National Heart, Lung, and Blood Institute and General Clinical Research Center grants M01RR00051, M01RR0099718-24, M01RR02719-14, and RR00036 from the National Center for Research Resources.
Abbreviations
- ALL
Acute Lymphoblastic leukemia
- BMD
Bone Mineral Density
- CAMP
Childhood Asthma Management Program
- GCs
Glucocorticoids
- GWAS
Genome Wide Association Study
- PD
Prednisone
- SNP
Single Nucleotide Polymorphism
Footnotes
CONFLICT OF INTEREST
All the authors state that there are no conflicts of interest to disclose.
DISCLAIMER
The authors directed and had access to all the analyses and the full clinical and genetic database, wrote all drafts of the report, decided to publish the results and attest for the accuracy and completeness of the data.
Supplementary information is available at The Pharmacogenomics Journal’s website
References
- 1.Buehring B, Viswanathan R, Binkley N, Busse W. Glucocorticoid-induced osteoporosis: an update on effects and management. J Allergy Clin Immunol. 2013;132:1019–1030. doi: 10.1016/j.jaci.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 2.Tsampalieros A, Gupta P, Denburg MR, Shults J, Zemel BS, Mostoufi-Moab S, et al. Glucocorticoid effects on changes in bone mineral density and cortical structure in childhood nephrotic syndrome. J Bone Miner Res. 2013;28:480–488. doi: 10.1002/jbmr.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rousseau-Nepton I, Lang B, Rodd C. Long-term bone health in glucocorticoid-treated children with rheumatic diseases. Curr Rheumatol Rep. 2013;15:315. doi: 10.1007/s11926-012-0315-x. [DOI] [PubMed] [Google Scholar]
- 4.Kaste SC, Jones-Wallace D, Rose SR, Boyett JM, Lustig RH, Rivera GK, et al. Bone mineral decrements in survivors of childhood acute lymphoblastic leukemia: Frequency of occurrence and risk factors for their development. Leukemia. 2001;15:728–734. doi: 10.1038/sj.leu.2402078. [DOI] [PubMed] [Google Scholar]
- 5.Kelly HW, Van Natta ML, Covar RA, Tonascia J, Green RP, Strunk RC. Effect of long-term corticosteroid use on bone mineral density in children: a prospective longitudinal assessment in the childhood Asthma Management Program (CAMP) study. Pediatrics. 2008;122:e53–61. doi: 10.1542/peds.2007-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JJ, Zacharin MR. Proposals for prevention and management of steroid induced osteoporosis in children and adolescents. J Paediatr Child Health. 2005;41:553–557. doi: 10.1111/j.1440-1754.2005.00718.x. [DOI] [PubMed] [Google Scholar]
- 7.Jones TS, Kaste SC, Liu W, Cheng C, Yang W, Tantisira KG, et al. CRHR1 polymorphisms predict bone density in survivors of acute lymphoblastic leukemia. J Clin Oncol. 2008;26:3031–3037. doi: 10.1200/JCO.2007.14.6399. [DOI] [PubMed] [Google Scholar]
- 8.Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N Engl J Med. 2000;343:1054–63. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 9.Tse SM, Kelly HW, Litonjua AA, Van Natta ML, Weiss ST, Tantisira KG. Corticosteroid use and bone mineral accretion in children with asthma: effect modification by vitamin D. J Allergy Clin Immunol. 2012;130:53–60. doi: 10.1016/j.jaci.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104:2690–2696. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 11.Stanisavljevic S, Babcock AL. Fractures in children treated with methotrexate for leukemia. Clin Orthop Relat Res. 1977;125:139–144. [PubMed] [Google Scholar]
- 12.Sala A, Barr RD. Osteopenia and cancer in children and adolescents: The fragility of success. Cancer. 2007;109:1420–1431. doi: 10.1002/cncr.22546. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 15.Bachrach LK. Acquisition of optimal bone mass in childhood and adolescence. Trends Endocrinol Metab. 2001;12:22–28. doi: 10.1016/s1043-2760(00)00336-2. [DOI] [PubMed] [Google Scholar]
- 16.Weinstein RS. Glucocorticoid-induced osteoporosis. Rev Endocr Metab Disord. 2001;2:65–73. doi: 10.1023/a:1010007108155. [DOI] [PubMed] [Google Scholar]
- 17.Kim HJ, Zhao H, Kitaura H, Bhattacharyya S, Brewer JA, Muglia LJ, et al. Glucocorticoids suppress bone formation via the osteoclast. J Clin Invest. 2006;116:2152–2160. doi: 10.1172/JCI28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teitelbaum SL. The osteoclast and its unique cytoskeleton. Ann N Y Acad Sci. 2011;1240:14–17. doi: 10.1111/j.1749-6632.2011.06283.x. [DOI] [PubMed] [Google Scholar]
- 19.Chellaiah MA. Regulation of actin ring formation by rho GTPases in osteoclasts. J Biol Chem. 2005;280:32930–32943. doi: 10.1074/jbc.M500154200. [DOI] [PubMed] [Google Scholar]
- 20.Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, et al. Vav3 regulates osteoclast function and bone mass. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- 21.Rebhun JF, Castro AF, Quilliam LA. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J Biol Chem. 2000;275:34901–34908. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- 22.Zou W, Izawa T, Zhu T, Chappel J, Otero K, Monkley SJ, et al. Talin1 and Rap1 are critical for osteoclast function. Mol Cell Biol. 2013;33:830–844. doi: 10.1128/MCB.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He M, Wang Y, Li W. PPI finder: a mining tool for human protein-protein interactions. PLoS One. 2009;4:e4554. doi: 10.1371/journal.pone.0004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Human protein-protein interaction mining tool. Steroid-regulated promoters. http://liweilab.genetics.ac.cn/tm/search.php?st=gn&gn=steroid-regulated%20promoters&ti=9606&tn=250&sot=&pg=5.
- 25.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp JP, Medina-Gomez C, Estrada K, St Pourcain B, Heppe DH, Warrington NM, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2012;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RegulomeDB. Data supporting chr7:22186954 (rs6461639) http://regulome.stanford.edu/snp/chr7/22186954.
- 30.SNAP. SNP Annotation and Proxy Search. https://www.broadinstitute.org/mpg/snap/ldsearchpw.php.
- 31.SNAP. SNP CNV Annotation Database. http://www.scandb.org/newinterface/index_v1.html.
- 32.dbSNP. Rs6461639. http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=6461639.
- 33.Lanktree MB, Hegele RA. Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med. 2009;1:28. doi: 10.1186/gm28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller MC. Gene x environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazzeroni LC, Lu Y, Belitskaya-Lévy I. P-values in genomics: apparent precision masks high uncertainty. Mol Psychiatry. 2014;19:1336–1340. doi: 10.1038/mp.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry. 2011;16:516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson D, Andiappan AK, Halldén C, Tim CF, Säll T, Wang de Y, et al. Poor reproducibility of allergic rhinitis SNP associations. PLoS One. 2013;8:e53975. doi: 10.1371/journal.pone.0053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


