Abstract
Altered lipoprotein metabolism plays a key role during atherogenesis. For over 50 years, epidemiological data have fueled the proposal that HDL-cholesterol (HDL-c) in circulation is inversely correlated to cardiovascular risk. However, the atheroprotective role of HDL is currently the focus of much debate and remains an active field of research. The emerging picture from research in the past decade suggests that HDL function, rather than HDL-c content is important in disease. Recent developments demonstrate that miRNAs play an important role in fine-tuning the expression of key genes involved in HDL biogenesis, lipidation, and clearance, as well as in determining the amounts of HDL-c in circulation. Thus, it has been proposed that miRNAs that affect HDL metabolism might be exploited therapeutically in patients. Whether HDL-based therapies, alone or in combination with LDL-based treatments (e.g. statins), provide superior outcomes in patients has been recently questioned by human genetics studies and clinical trials. The switch in focus from “HDL-cholesterol” to “HDL function” opens a new paradigm to understand the physiology and therapeutic potential of HDL, and to find novel modulators of cardiovascular risk. In this review we summarize the current knowledge on the regulation of HDL metabolism and function by miRNAs.
1. INTRODUCTION
MicroRNAs (miRNAs) are small, ~22 nt RNAs that regulate target genes by binding to complementary sequences (generally in the 3′UTR) of target mRNA. MicroRNAs associate with the RNA-Induced Silencing Complex (RISC) and subsequently promote translational suppression and/or RNA degradation of their targets (reviewed in [1]). MicroRNAs can be intergenic, or generated from within introns and sometimes even exons of a hosting coding gene. The initial miRNA transcript (pri-miRNA) is usually long (from a few hundred to several thousand nt), and its transcription is subject to the same control mechanisms as coding mRNAs, i.e. the levels of different miRNAs can be regulated by transcription factors. The pri-miRNA transcript then undergoes sequential processing by endonucleases first to pre-miRNA, and then to the functional, mature miRNA which associates with the RISC to regulate targets [1]. In the last decade, miRNAs have been recognized as essential regulators of multiple (patho)physiological processes. A plethora of reports have recently shown that high-density lipoprotein (HDL) biogenesis, lipidation, and clearance are modulated by several miRNAs. In this review we first summarize the major proteins involved in HDL metabolism, from synthesis to clearance, then we highlight each miRNA that has been shown to control HDL metabolism, and finally we discuss our current knowledge of the mechanisms involved that lead to these changes.
Circulating HDLs are heterogeneous spherical or discoidal particles with a density > 1.063 g/mL (reviewed in [2]), and decorated at the surface with a vast array of different proteins, including several apolipoproteins and enzymes that affect HDL function. The amount of HDL in circulation has traditionally been referred to by its cholesterol content (i.e., HDL-cholesterol or HDL-c). However, HDL-c is only one measure of several important parameters of HDL particles, since cholesterol represents only 15–20% of the total mass of the lipoprotein [2]. Notably, emerging evidence suggests that HDL-c levels do not necessarily reflect particle size/number or, more importantly, biologic function. The surface of HDL particles contains phospholipids and proteins, which account for 25–30% and 45–50% of the mass of the particle, respectively [2]. The levels and composition of these HDL components now appear to be important factors that dictate HDL function. In healthy individuals, only a small amount (5%) of triacylglycerides is carried in HDL, with the bulk of the lipids that are associated with HDL being cholesterol and phospholipids [2]. The first step of HDL biogenesis requires apolipoprotein APOA1 to be lipidated at the surface of hepatocytes and enterocytes for form nascent- or preβ-HDL. A two-step model was proposed in which the sterol transporter ABCA1 promotes the initial transfer of both phospholipids and cholesterol to lipid-poor APOA1, while a second sterol transporter, ABCG1, subsequently facilitates further enrichment with cholesterol from endothelial and bone marrow-derived cells in peripheral tissues [3]. In mice, hepatic, intestinal, and adipose ABCA1 contribute approximately 70%, 15%, and 10%, respectively, of HDL-c in circulation [4–6]. Patients with Tangier disease and Abca1−/− mice have non-functional ABCA1, and show little or no circulating HDL-c [7]. Patients with familial hypoalphalipoproteinemia have reduced ABCA1 activity due to more mild mutations in ABCA1 and also exhibit abnormally low plasma HDL-c [7]. Intriguingly, although the role of ABCG1 on cholesterol efflux to HDL is well established from work in cell culture, Abcg1−/− mice do not show decreased levels of plasma HDL-c [8], and no disease-related polymorphisms have been reported in humans. ABCG1 localizes intracellularly [9], and therefore its effects on cholesterol efflux are likely secondary to a role in intracellular cholesterol mobilization.
The free cholesterol at the surface of nascent HDL is esterified by LCAT (lecithin:cholesterol acyltransferase) and translocated into the core of the particle. LCAT-deficient mice and humans have very low HDL-c (as well as very low circulating APOA1 due to rapid renal clearance of precursor HDL particles). In 1973, Glomset & Norum proposed that “the function of the LCAT reaction is to transport unesterified cholesterol synthesized in the peripheral tissues to the liver” [10]. This proposal led to the current “reverse cholesterol transport” (RCT) paradigm, which involves mobilization of extrahepatic cholesterol (e.g. from macrophages in the arterial subendothelial space) back to the liver, where cholesterol is metabolized to bile acids prior to their secretion into the bile and excretion into the feces (Figure. 1).
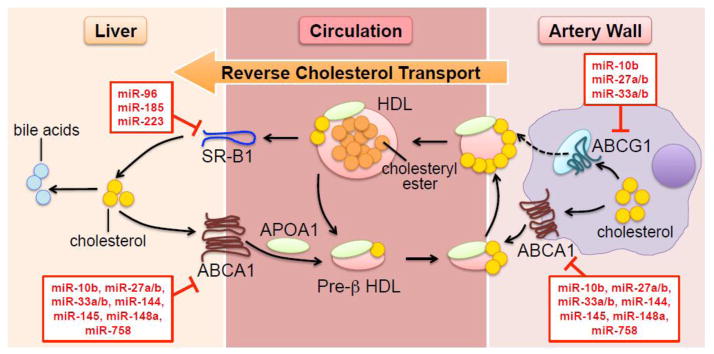
Fig. 1. Mobilization of cholesterol via HDL particles.
The lipidation of lipid-poor APOA1 in the hepatocyte (and in minor proportion in the enterocyte and adipocyte) generates pre-β HDL, which is further lipidated in the peripheral vasculature via ABCA1 and possibly ABCG1. Unesterified cholesterol in the nascent particle is esterified by LCAT. HDL exchanges lipids and apolipoproteins with APOB-containing lipoproteins (not shown in the figure). Clearance of HDL-bound cholesteryl esters is facilitated by SR-B1 in the liver. Excess hepatic cholesterol is eventually metabolized to bile acids and excreted through bile. miRNAs known to target transcripts involved in HDL biogenesis and the reverse cholesterol transport pathway are annotated in red boxes.
Another important HDL-bound protein is the enzyme CETP (cholesteryl ester:triacylglyceride transfer protein), which facilitates the exchange of HDL-derived cholesteryl esters with (V)LDL-derived triacylglycerides. The lack of CETP activity in rodents is perhaps the main reason behind the vastly different lipid lipoprotein profiles and susceptibility to atherosclerosis between humans and mice. Loss-of-function mutations in CETP in humans result in hyperalphalipoproteinemia (high HDL) [11, 12] providing further support for the idea that CETP activity and HDL-c levels are inversely correlated.
The hepatic clearance of circulating HDL is mediated by the scavenger receptor SR-B1, (gene symbol SCARB1) which was initially described as an HDL receptor in multiple tissues, especially the liver and steroidogenic tissues [13]. SR-B1 facilitates the selective uptake of HDL-derived cholesterol and cholesteryl esters. While no SR-B1-deficient patients have been identified, a number of reports have described different mutations in the human SCARB1 gene that affect HDL uptake [14]. In rodents, liver-specific Scarb1 transgenic and knock-out models revealed a critical role for hepatic SR-B1 on HDL clearance from circulation.
For the past 50 years, epidemiological data have steadily shown that circulating HDL-c is inversely correlated with cardiovascular disease and predicts cardiovascular risk. Early epidemiologic studies in the 1960’s and 1970’s revealed that low HDL-c was frequent in patients with CVD and an independent risk factor for myocardial infarction [15–18]. Consistent with these data, increasing plasma HDL-c was atheroprotective in rabbits [19], and in mice transgenic for key genes involved in HDL biogenesis [20–23]. Additionally, infusion of reconstituted HDL promoted the regression of coronary atheromata in rodents and patients [24]. The “HDL hypothesis” ascertains that HDL exerts a cardioprotective effect, and thus interventions that raise plasma HDL-c were predicted to reduce cardiovascular risk. Thus a race to develop drugs to rise circulating HDL-c began.
However, both human genome-wide association studies and clinical trials using HDL-c raising compounds (niacin, CETP inhibitors, apoA-1 transcriptional upregulators; all very effective in raising plasma HDL-c) quickly cast doubts on this simplified HDL hypothesis [25]. This led to its reformulation into the “functional HDL hypothesis”, which establishes that it is not the amount of HDL-c, but the ability of HDL particles to mobilize cholesterol through the RCT pathway, what determines their antiatherogenic properties [26, 27]. Indeed, a recent study in almost 3,000 patients that correlated the capacity of HDL to efflux cholesterol from cultured macrophages and the incidence of cardiovascular events provided clear evidence supporting the “functional HDL” hypothesis [28]. However, the relative contribution of the different biological activities of HDL towards atheroprotection (cholesterol efflux, anti-inflammatory, anti-thrombotic, anti-oxidative, pro-endothelial) remains unclear. Although HDL-c is still a useful biomarker for cardiovascular risk, current efforts in the therapeutic field are focused on improving HDL function/quality rather than HDL quantity.
2. miRNAs TARGETING HDL BIOGENESIS AND LIPIDATION
2.1. miR-33
The miR-33 family consists of two members, miR-33a and miR-33b, expressed from within an intron of SREBP-2 and SREBP-1, respectively [29–33]. SREBPs are the master regulators of sterol and fatty acid synthesis [34], and conditions that induce or repress the expression of the SREBP genes result in the concomitant change in miR-33 expression. For example, low intracellular cholesterol contents or treatment with statin drugs induce the expression of SREBP-2 and miR-33a in cells and animals [29–31]. Interestingly, the intron of SREBP-1 containing miR-33b is scrambled in mice and other rodents, thus abrogating miR-33b expression in those species. Given that the expression of the hosting SREBP-2 and SREBP-1 genes is controlled by different nutritional and hormonal stimuli, murine studies likely are missing regulatory circuits controlled by miR-33b in the primate liver, particularly in response to insulin or under lipogenic conditions (which induce SREBP-1). Both miR-33 isotypes share the same seed sequence, but differ in 2 nt in the 3′ region. Whether these 2 nt confer target specificity is currently unknown.
Initial reports showed that miR-33 represses the expression of genes involved in cholesterol efflux, including ABCA1 and ABCG1, in hepatocytes and macrophages [29–33]. Hence, enhanced expression of miR-33 in cells using miR-33 mimics or pre-miR-33-encoding adenovirus results in attenuated expression of ABCA1 and ABCG1, and led to a concomitant decrease in cholesterol efflux to exogenous ApoA1 and HDL [29–32]. Conversely, antisense oligonucleotides (ASOs) that abolished miR-33 expression increased the amounts of those transporters and cholesterol efflux to HDL both in cells and mice [29–31]. The miR-33−/− mice showed a similar phenotype, with increased hepatic ABCA1 expression and elevated plasma HDL-c [33]. Additional studies showed that miR-33 also controls hepatic biliary output, via modulation of the canalicular transporters ABCB11 (also known as BSEP) and ATP8B1 (also known as FIC1) [35]. It was hypothesized that, by controlling hepatic (and presumably also intestinal) HDL biogenesis (via ABCA1), peripheral HDL lipidation (via ABCA1 and ABCG1), and bile secretion (via ABCB11 and ATP8B1), miR-33 could modulate the flow of cholesterol through the RCT pathway. This latter proposal was validated both in chow-fed wild-type mice [35] and in western diet-fed Ldlr−/− mice [36]. Collectively, the results of these studies suggested that ASOs against miR-33 could be used therapeutically to raise HDL-c in patients and provide atheroprotection. This proposal was also fueled by studies in African green monkeys showing that the effect of anti-miR-33 ASOs on HDL-cholesterol was preserved when these monkeys were fed a variety of diets [37, 38].
In addition to its role on sterol metabolism, miR-33 also modulates lipid and glucose metabolism by directly targeting genes encoding enzymes involved in fatty acid β-oxidation (CPT1A, HADHB, CROT) [32, 39], insulin signaling (IRS2, PRKAA1, SIRT6) [39], and gluconeogenesis (PCK1, G6PC) [40]. Collectively, these studies reveal miR-33 as a regulatory hub for multiple intracellular metabolic processes, coordinating the expression of genes involved in sterol, fatty acid, and glucose homeostasis.
The functional consequences of loss of miR-33, either by genetic ablation or by therapeutic silencing with ASOs, on atherogenesis were evaluated in several studies using atherosclerosis-prone mouse models. An initial study in Ldlr−/− mice showed that a 4-week treatment with 2′-fluoro/methoxyethyl (2′F/MOE) anti-miR-33 ASOs after 14-weeks of western diet (WD) feeding increased plasma HDL-c and accelerated the regression of atheromata, compared to control ASOs [36]. In contrast, an independent study in Ldlr−/− mice using locked nucleic acid (LNA) ASOs showed that anti-miR-33 therapy failed to sustain elevated plasma HDL-c levels over the course of 12 weeks, raised the levels of plasma VLDL-tag (very low-density lipoprotein–triacylglycerides), and did not prevent the progression of atherosclerosis in WD-fed mice [41]. Two additional progression studies also reported unchanged plasma HDL-c levels, but, in contrast to the earlier study, reported a significant reduction in atheromata progression, in Ldlr−/− mice treated with 2′F/MOE anti-miR-33 ASOs [42, 43]. Paradoxically, although HDL-c was unchanged in all these progression studies, hepatic and/or macrophage Abca1 expression remained de-repressed in anti-miR-33-treated mice, compared to control ASOs.
The mechanism for the return of plasma HDL-c to control levels in these long-term experiments remains to be established. Atheroprotection following ASO treatment in the context of unchanged HDL-c [42, 43] may be the result of altered gene expression in the artery wall. Importantly, it was shown that 2′/FMOE anti-miR-33 ASOs efficiently transduce macrophages in the subendothelial space of the aortic root of Ldlr−/− mice and promote their polarization towards an anti-inflammatory M2 phenotype [36, 43] via direct targeting of PRKAA1 [43]. An additional direct target that is also de-repressed in these same macrophages was ALDH1A2 [43], which is implicated in the metabolism of retinoic acid and promotes the proliferation and differentiation of anti-inflammatory regulatory T cells [44]. These results suggest that anti-miR-33 therapy might exert its atheroprotective effects by promoting changes in the inflammatory status of the plaque, perhaps in addition to increasing ABCA1/ABCG1-dependent cholesterol efflux to HDL. Studies in ApoE−/− mice, which have little plasma HDL-c, have not addressed this postulate directly. However, ASO treatment of ApoE−/− mice resulted in atheroprotection in the absence of changes in HDL-c [45], and it was suggested that miR-33 controls mitochondrial function in the macrophage in plaques via direct targeting of PGC-1, PDK4, and SLC25A25 [45]. Finally, ApoE−/− miR-33−/− mice had decreased atherosclerotic lesions, elevated plasma HDL-c, and decreased circulating monocytes, compared to ApoE−/− mice [46]. However, ApoE−/− mice transplanted with ApoE−/− miR-33−/− bone marrow (i.e. in the absence of blood cells-derived miR-33) showed no changes in plasma HDL-c or plaque size, compared to mice transplanted with ApoE−/− [46]. Hence, these latter studies would imply that hepatic, not macrophage, miR-33 is the key therapeutic target to promote atheroprotection.
The potential of anti-miR-33 therapy was further questioned by a report showing that, compared to wild-type littermates, miR-33−/− mice gain weight at an accelerated pace and develop obesity, hepatosteatosis, and insulin resistance [47]. The same study presented SREBP1c as a direct target of miR-33, which would explain those phenotypic changes [47]. Other authors, however, have argued against SREBP1 as a direct miR-33 target [35, 48] and changes in insulin sensitivity following therapeutic silencing of miR-33 [48]. Elevated plasma TAG have been reported in high-fat diet-fed miR-33−/− mice, compared to wild-type littermates [48], and in long-term studies in wild-type mice with anti-miR-33 ASOs, compared to control ASOs [41, 49, 50]. In contrast, other similar ASO long-term studies in mice and non-human primates showed no change [38, 42, 43, 46] or even a decrease [37, 48] in plasma TAG.
The reasons behind the different outcomes among all the studies detailed above, both in terms of plasma lipoproteins levels and lesion size/regression, remain to be elucidated. It is possible that different bioavailability and/or potency of 2′F/MOE and LNA oligonucleotides, length of treatment, and interactions with dietary components or the microbiome might explain the discrepancies in HDL, VLDL, and atherogenesis among these studies. In any case, caution must be taken when translating the results from anti-miR-33 studies directly from mice to humans. As mentioned above, primates, but not rodents, express SREBP1-derived miR-33b, suggesting that miR-33 levels in mice are likely reduced compared to humans. Perhaps the recently generated miR-33b humanized transgenic mouse [51], by reintroducing the SREBP1/miR-33b–dependent regulatory circuits, will provide a better model to study the role of miR-33 on hepatic and peripheral sterol and TAG metabolism, and provide a more relevant model to human physiology and disease. In conclusion, whether anti-miR-33 therapy, alone or in combination with other drugs, will raise HDL and offer atheroprotection to dyslipidemic patients remains an open question that will need further investigation.
Finally, very recent studies have pointed to the potential of miR-33 as a biomarker for lipid-related disorders, including atherosclerosis. Hence, the levels of miR-33a and/or miR-33b were shown to be increased in: i) human atherosclerotic plaques, compared to normal artery walls [45, 52]; ii) plasma from hypercholesterolemic children, compared to healthy controls [53]; and iii) plasma from obese children, compared to lean controls [54]. The caveat of these latter reports, however, is the small number of patients involved, and the very low levels of miR-33 in the circulation. Larger population studies will be necessary to firmly establish an association between miR-33a/b in circulation, HDL-c levels, obesity, and/or cardiovascular risk.
2.2. miR-148
The newest microRNA found to regulate plasma lipid and lipoprotein metabolism is miR-148a. Two recent and independent studies found that miR-148a regulates both LDL and HDL metabolism [55, 56]. Using genome-wide association analysis, Wagschal et al. identified miR-148a as a lipid-metabolism associated miRNA that is likely the causal gene for the genetic variation known to affect plasma lipid levels in human populations [55]. Specific single nucleotide polymorphisms (SNPs) in close proximity to miR-148a showed significant association with plasma total and LDL cholesterol levels. The miR-148a locus is intergenic, and although there are protein coding genes in more distal regions, analysis of the haplotype blocks suggest that SNPs that affect miR-148a are causal for the variation in plasma lipids [55]. In a separate study, Goedeke et al. identified miR-148a as a regulator of lipid metabolism using a genome-wide screen designed to find miRNAs that regulate LDL uptake in human hepatoma cells (Huh7) [56]. Importantly, miR-148a is highly expressed in the liver in both humans and mice [57]. Given the central role of the liver in regulating plasma lipoprotein metabolism, the presence of high hepatic levels of miR-148a suggests it may play an important role in regulating plasma lipid levels.
Using UTR-luciferase reporter assays as well as RNA-RISC complex immunoprecipitation analysis, both LDLR and ABCA1 were validated as bona-fide miR-148a target genes [55, 56]. Wagschal et al. subsequently showed that overexpression of miR-148 resulted in decreased ABCA1 protein in hepatic and macrophage cells lines (HepG2 and THP1 cells, respectively), and decreased cellular cholesterol efflux to APOA1. Conversely, silencing miR-148a in cells resulted in increased ABCA1 protein and increased cholesterol efflux. Goedeke et al. demonstrated that manipulation of miR-148a expression also altered LDLR levels and LDL uptake in hepatoma cells [56]. To complement these in vitro studies, miR-148 overexpression in vivo was shown to decrease both plasma HDL-c and hepatic ABCA1 protein levels in wild-type C57BL/6 mice [55]. Additionally, silencing miR-148a expression with LNAs markedly elevated ABCA1 mRNA and protein in the livers of Western diet-fed ApoE−/− mice, and resulted in both short-term (5 days) and long-term (16 weeks) modest increases in HDL-c levels. Plasma LDL-c levels were also modestly reduced by miR-148a silencing [56]. However, it was not shown whether these changes in plasma lipid levels affected atherosclerosis, which remains an important question to be answered. The study from Goedeke et al. also used an in vivo model to show that silencing miR-148a using LNAs altered lipid metabolism. They used APOB transgenic mice with reduced LDLR levels (ApoB-Tg Ldlr+/− mice), where the plasma lipoprotein profile resembles that of hypercholesterolemic patients [56]. Silencing miR-148a in these latter mice resulted in a large increase in LDL-c, and a smaller increase in HDL-c levels.
The expression of miR-148a was also shown to be induced by high-fat diet and in obese (leptin-deficient) ob/ob mice, and following fasting and re-feeding [56]. These latter results were shown to be the consequence of miR-148a being a direct target of SREBP-1c, a transcription factor that regulates hepatic lipogenesis that is highly regulated in fasting and re-feeding conditions [58, 59]. The transcriptional regulation of miR-148a by SREBP-1c raise important questions about the physiologic setting when this miRNA is functional. Firstly, SREBP-1c is also a transcriptional target of LXRs [60], and therefore LXR activation would also increase miR-148a levels. LXRs are activated when sterol levels are high, and initiate a gene expression program the results in increased sterol efflux (via increased ABCA1 and ABCG1) as well as decreased sterol uptake by degrading the LDLR (via increased Inducible Degrader of LDL (IDOL)) [60]. In the case of LDLR, miR-148a could play a supportive role to LXRs, which mediate the post-transcriptional regulation of LDLR protein IDOL, an E3 ubiquitin ligase that directly targets the LDLR for proteasomal degradation [61]. Thus, miR-148a may complement this pathway, by also targeting the LDLR mRNA. In contrast, the regulation of ABCA1 by miR-148a would appear paradoxical, or counter productive to the role of LXRs, since ABCA1 will be induced, and then subsequently targeted by miR-148a, which would also be induced. This LXR-dependent increase in ABCA1 and miR-148a may be part of a negative feedback loop to prevent excess intracellular sterols from being lost. There may also be an additional caveat. Physiologically, but not pharmacologically, LXR activation is uncoupled from activation of SREBP-1c. LXRs are activated when cellular cholesterol levels are elevated, generating oxysterols [60]. Thus, while LXRs induce SREBP-1c expression, SREBP-1c maturation and activation is, in fact, blocked, because elevated sterols promote retention of SREBP1 in the ER via regulation of SCAP and INSIGs [34]. Thus, since miR-148a is primarily regulated by SREBP1c, the regulation of LXRs may only be important in a pharmacologic but not a physiological setting.
Whether miR-148a harbors potential to modulate cardiovascular risk in patients remains to be established. In theory, silencing miR-148a could have two obvious benefits: first, to lower circulating LDL-c by increasing LDLR-dependent uptake; and second, to increase HDL-c levels and/or raise ABCA1-mediated cholesterol efflux. However, it will also be important to determine whether the physical and functional properties of the HDL and LDL particles are also altered following miR-148a therapeutic silencing.
2.3. miR-122
The most abundant miRNA in the mouse and human liver is miR-122, comprising approximately 70% of all hepatic miRNAs [62]. Although there are extensive studies showing a role for miR-122 in regulating lipid metabolism [63], the mechanisms have been surprisingly elusive. miR-122 was one of the first miRNAs to be effectively silenced in mice and non-human primates using ASOs. The result was a dramatic reduction in plasma cholesterol and triglyceride [63]. In mice fed a high-fat diet, silencing miR-122 resulted in an even more pronounced lipid lowering effect, in both the LDL and HDL fractions [63]. To determine the potential mechanism, investigators used hepatic gene expression profiling to show that silencing miR-122 results in reduced expression of several genes involved in hepatic triglyceride metabolism genes, ultimately leading to decreased fatty acid and sterol synthesis. Such changes in gene expression do not imply these genes are directly targeted by miR-122, since the levels of direct targets would be expected to be de-repressed following miR-122 silencing. One potential target that might mediate miR-122-dependent changes in plasma lipids is AMP-activated protein kinase (AMPK) [63], a metabolic master switch that regulates several important metabolic pathways, including fatty acid oxidation and glucose uptake [64]. Additional miR-122 potential targets include PPARβ/δ and BAF60A [65]. BAF60A is part of the SWI/SNF nucleosome remodeling complex that regulates the activity of several nuclear receptors, including PPARs, which are central regulators of fatty acid metabolism [66]. The transcriptional regulation of miR-122 has also been studied, and miR-122 has been shown to be regulated by the circadian clock; however, this effect is largely observed in the pri- and pre-miR-122 transcript level, and not in the more stable mature miR-122 transcript [65].
The lipid lowering effects of miR-122 were also observed in non-human primates. Silencing miR-122 using LNAs did not result in hepatotoxic effects in either mice or non-human primates [67], consistent with the study in mice from Esau and colleagues [63]. In the primate model, the decreases in plasma lipids were more pronounced in the LDL-c fraction [67]. The role of miR-122 in regulating plasma HDL-c levels remains relatively unexplored. Targeting miR-122 therapeutically has been considered, given the data in mice discussed above. However, studies in miR-122−/− mice raised significant concerns. The latter mice were shown by two independent groups to be more prone to hepatic inflammation, fibrosis and hepatocellular carcinoma [68, 69]. These studies confirmed that miR-122 is an important regulator of plasma lipids, as miR-122−/− mice also had reduced plasma cholesterol in both LDL and HDL fractions, as well as reduced plasma triglyceride levels [68, 69]. In terms of mechanism, the two studies pointed to different sets of potential target genes, based on hepatic global gene expression, compared to wild-type mice. One group identified a set of lipid metabolism genes as direct miR-122 target genes, including members of the Agpat family and Fsp27 [68]. The other group demonstrated (likely indirect) reduced expression of lipid metabolism genes, including Mttp [69], the enzyme that is required for the lipidation and secretion of VLDL from the liver. Older miR-122−/− mice also showed a pronounced increase in hepatic triglyceride content compared to wild-type animals [68, 69], which may be secondary to a pro-inflammatory phenotype. Taken together, long-term silencing of miR-122 would seem to be associated with a significant risk of developing unwanted side effects, thus limiting their use in patients. Nevertheless, determining the molecular mechanisms for how miR-122 regulates lipid and lipoprotein metabolism remains an important, unanswered question.
2.4. miR-144
Circulating HDL-c levels have also been shown to be regulated by miR-144, which is part of a bycistronic miRNA cluster that contains miR-451. Two independent groups identified ABCA1 as a direct target of miR-144 [70, 71]. Using miRNA target prediction software, the ABCA1 3′UTR was found to contain at least two miR-144 binding sites that were tested functionally using luciferase reporter systems. Both groups also showed that increased miR-144 levels in isolated hepatocytes [70] or macrophages and THP-1 cells [71] resulted in reduced cholesterol efflux to lipid poor APOA1, as well as reducing ABCA1 protein levels. In addition, overexpression of miR-144 in the livers of mice resulted in a decrease in plasma HDL-c levels, and decreased ABCA1 protein, but not mRNA [70]. In contrast, ASO-mediated silencing of miR-144 in mice fed a high-fat diet increased both plasma HDL-c levels and hepatic ABCA1 protein levels [70, 71]. A subsequent study also confirmed that miR-144 targets ABCA1 [72]. However, compared to miR-33, the effects of miR-144 on HDL-c were more modest.
The major difference between these two miR-144 studies was the proposed mechanism governing the transcriptional regulation of the miRNA. De Aguiar Vallim and colleagues [70] identified miR-144/-451 as a direct target of the nuclear receptor FXR, a transcription factor essential for maintaining bile acid homeostasis [73]. Ramirez et al. [71] identified miR-144 as a direct target gene of LXR. These two nuclear receptors are major regulators of plasma lipid levels. FXR activation, following administration of synthetic agonists, caused pronounced hypolipidemia, with decreases in all major lipoprotein fractions, including HDL-c as well as reducing hepatic lipids [70, 73]. It has been proposed that the induction of miR-144 by FXR, and subsequent repression of ABCA1 is part of a complex pathway that involves several genes that mediate hepatic cholesterol channeling into the bile [70]. In contrast, LXR activation using synthetic agonists has been shown to be atheroprotective [74], but is associated with unwanted effects such as hepatic lipid accumulation [75]. The regulation of miR-144 by LXR is more puzzling, since LXRs also induce ABCA1. However, as we discuss above for miR-148a, miR-144 may play a role in fine tuning ABCA1 levels and preventing excess cholesterol loss from cells [60].
The therapeutic potential of silencing miR-144, by raising HDL-c levels and presumably promoting accelerated RCT remain to be determined. In a recent study, treatment of atheroscelrosis prone ApoE−/− mice with an agomir for miR-144 increased inflammation and inflammatory cytokine gene expression, reduced RCT, and increased atherosclerosis [76]. It remains to be determined whether this level of overexpression, which was presumably supra-physiologic, is relevant to patients.
2.5. miR-758
The intergenic miR-758 is abundantly expressed in the brain, heart, and aorta, with low levels in the liver. The expression of miR-758 is repressed in both livers and peritoneal macrophages in high-fat diet-fed mice, compared to chow [77]. However, the molecular mechanism governing this regulation is unknown. Interestingly, miRNomic studies using limited numbers of samples showed increased levels of miR-758 in aortic plaques from hypercholesterolemic patients, compared to those from normocholesterolemic individuals [52]. Data showed that the 3′UTR of ABCA1 contains evolutionarily conserved sequences that confer response to miR-758, and that manipulation of miR-758 levels with miR-mimics or silencing oligonucleotides modulates cholesterol efflux to APOA1, but not to HDL, in cell culture assays [77]. The consequences of therapeutic inhibition of miR-758 on RCT and atherogenesis have not yet been reported.
2.6. miR-145
The intergenic miR-143/-145 cluster is mostly expressed in the vascular smooth muscle cell (VSMC) compartment, where it regulates the expression of several key genes that control cell differentiation and contractile function, and promotes the VSMC switch from a contractile/non-proliferative to a migrating/proliferative phenotype [78]. miR-145 has been reported to control the expression of ABCA1 and cholesterol efflux hepatocytes, pancreatic islets, VSMCs, and macrophages via conserved sequences in the 3′UTR of ABCA1 [72, 79]. The same authors reported that membrane vesicles secreted by shear-stressed cultured endothelial cells were enriched with miR-143/145 and modulated gene expression in co-cultured smooth muscle cells and macrophages [79], and suggested that the transfer of miRNAs between cells within the plaque in vivo might influence gene expression and cell function (e.g. cholesterol efflux to HDL) in the subendothelial space. Genetic loss of miR-143/-145 resulted in decreased atheromata in both Ldlr−/− [79] and ApoE−/− [80] backgrounds, which would be consistent with de-repressed ABCA1 expression in plaque macrophages, although no increase in HDL-c was noted in neither study. The relative contribution of vascular ABCA1-dependent cholesterol efflux vs. vascular architectural changes to these changes in atherogenesis remains to be established. Whether miR-143/-145 might be exploited therapeutically deserves further examination, given that its levels are induced in atherosclerotic plaques from hypertensive patients [81, 82] and in plasma from patients with coronary artery disease [83].
2.7. miR-27a/b
The miR-27 family consists of two members that differ by one nucleotide outside the seed sequence, giving rise to miR-27a and miR-27b [84]. The former is an intergenic miRNA, while the latter is part of the intragenic miR-23b/-27b/-24-1 miRNA cluster expressed from within an intron of C9ORF3. The expression of miR-27 is enriched in endothelial cells, smooth muscle cells, and macrophages. Interestingly, miR-27b expression is induced in the livers of mice fed a high-fat diet [85], where it suppresses critical regulators of lipid metabolism that include PPARγ, ANGPTL3, HMGCR, SREBP-1, SREBP-2, and APOB [84, 85]. Three independent laboratories showed that ABCA1 is a conserved direct target of miR-27 in both hepatocyte and macrophage cell lines [86–88], as well as in the mouse liver [87]. Accordingly, transfection of hepatoma cells with miR-mimics or miR-inhibitors resulted in altered ABCA1-dependent cholesterol efflux to ApoA1 [86, 87]. These data suggested that a therapeutic intervention aimed at decreasing miR-27 might raise hepatic and vascular ABCA1-dependent cholesterol efflux and RCT. The interest in anti-miR-27-based therapies was enhanced by the discovery that miR-27a/b also target directly the low-density lipoprotein receptor (LDLR) and the LDLR-related genes LRP6 and LDLRAP1 [87, 89]. However, no changes in plasma total cholesterol, LDL-c or HDL-c, or in hepatic cholesterol contents were noted in mice overexpressing miR-27 via an adeno-associated virus or following therapeutic silencing with ASOs [87]. Additionally, clinical data [90] and murine models [91, 92] show that loss of ANGPTL3 (another miR-27 target) activity is correlated with decreased HDL-c, LDL-c, and VLDL-triglycerides in plasma, further suggesting that the miR-27/ANGPTL3/ABCA1/LDLR axis might be of interest to curb cardiovascular risk. Similar to miR-148 discussed above, perhaps anti-miR-27 ASOs might represent a better therapeutic approach for hypercholesterolemic patients by increasing both LDL-c clearance (via LDLR) and HDL biogenesis (via ABCA1).
2.8. miR-10b
Both epidemiological and animals studies have suggested that anthocyanins (polyphenolic compounds commonly found in certain fruits, berries, and red wine) reduce the risk of atherosclerosis and cardiovascular disease [93]. Whether anthocyanins are bioactive per se, or they need to be metabolized to exert these protective effects has not been definitively established. It was recently reported that the anthocyanin Cy-3-G is metabolized to protocatechuic acid (PCA) by the intestinal flora [94]. These same authors found that the expression of the intergenic miR-10b was induced in peritoneal macrophages following exposure to PCA, compared to vehicle. Bioinformatics analysis revealed that conserved responsive sequences for miR-10b are present in the 3′UTR of both ABCA1 and ABCG1. The authors showed that PCA promoted accelerated RCT in ApoE−/− mice by increasing both ABCA1- and ABCG1-dependent cholesterol efflux from macrophages and the overall RCT in vivo. Importantly, the effects of PCA on cholesterol efflux and RCT were mimicked by miR-10b overexpression, and abrogated by miR-10b inhibitors. Finally, Cy-3-G feeding raised HDL-c and accelerated the regression of atheromata in ApoE−/− mice in a PCA-dependent manner [94]. While the mechanism by which PCA induces the expression of miR-10b in macrophages (and perhaps other tissues) is still obscure, these studies showed for the first time that the intestinal microbiome can exert important modulatory effects on the RCT pathway and might be exploited therapeutically.
2.9. Additional miRNAs targeting ABCA1
Multiple studies in the past 5 years showed that ABCA1 is targeted by a multiplicity of miRNAs, besides the ones discussed above. Additional miRNAs that target ABCA1 include miR-128 [55] and miR-19b [95, 96] (which also modulates plasma HDL-c levels in mice), miR-26 [97], miR-106b [98], miR-93 [99], and miR-101 [100]. The reasons why ABCA1 seems to be a “hot spot” for post-transcriptional regulation are unknown, but is likely in part due to the large, broadly conserved 3′ UTR of the ABCA1 mRNA. Whether there are additive/synergistic effects among these miRNAs on ABCA1-dependent cholesterol efflux and HDL function in response to a variety of physiological stimuli, and their role on atherogenesis is unknown.
3. miRNAs TARGETING SR-B1
The expression of SR-B1 in HepG2 hepatoma cells was found to be significantly increased following silencing of Drosha and Dicer [101], suggesting that SR-B1 levels might be regulated post-transcriptionally by miRNAs. The same authors defined conserved sequences in the 3′UTR of SR-B1 that confer response to miR-185, miR-96, and miR-223 [101]. However, overexpression or silencing each miRNA had modest effects on HDL-c uptake in HepG2 and CHO cells (which express SR-B1). An additional study found conserved sites in in the 3′ UTR of SR-B1 mRNA that conferred response to miRNA-125a and miRNA-455 [102]. Both miR-125a and miR-455 are abundant in liver and steroidogenic tissues, where their expression is down-regulated by corticotrophin and cAMP [102]. Following manipulation of either miR-125a or miR-455 in murine Leydig tumor cells with miR-mimics or miR-inhibitors, SR-B1 mRNA and protein levels, SR-B1-mediated HDL-c uptake, and SR-B1-supported steroidogenesis were inhibited or stimulated, respectively [102]. Whether manipulation of any of these miRNAs (alone or in combination) results in changes in HDL-c in vivo has yet to be tested.
4. miRNAs TARGETING THE HDL PROTEOME
There has been surprisingly little attention given to miRNAs that target the various proteins that decorate the HDL particle. Of the major apolipoproteins associated with HDL, which include APOA1, APOA2, APOA4, APOC1, APOC2, APOC3, APOC4, APOD, APOF, APOH, APOJ, APOL1 and APOM [2], only APOM was shown to be a direct target of miR-573 [103]. Perhaps the fact that most mRNAs that encode apolipoproteins have relatively short 3′ UTR regions (70–200 nt) limits the opportunity for miRNA-mediated regulation. The other major protein cargo of HDL includes enzymes and other proteins such as LCAT, PLTP, CETP, PON1, PAF-AH (gene symbol PLA2G7), SAA1, SAA4 [2]. PON1 was shown to be directly regulated by miR-616, and a SNP at the binding site of miR-616 in PON1 is associated with stroke and myocardial infarction in patients [104]. The expression of PLA2G7 was induced in the livers of miR-155−/− mice [105], but whether PLA2G7 is a true physiologic target of miR-155 has not yet been determined experimentally. Importantly, the role of many of these HDL-associated proteins on RCT and atherosclerosis remains to be elucidated. As we identify additional genes that affect HDL function, we will likely recognize new roles for miRNAs in regulating these genes and pathways.
5. CIRCULATING HDL-ASSOCIATED miRNAs
The original observations that cells can secrete miRNAs and that miRNAs are present in human biological fluids, including blood, were made almost 10 years ago [106, 107]. It is possible that a significant share of miRNAs found in circulation is the result of non-selective microvesicle secretion and/or cell death. However, a pioneering report showed that HDL particles transport specific miRNAs [108]. These same authors showed that HDL-associated miRNAs could be delivered to and alter target gene expression in recipient cells and tissues [108, 109]. Interestingly, the signature of HDL-associated miRNAs was different between normal and familial hypercholesterolemia patients [108]. An independent study confirmed changes in specific miRNAs in HDL isolated from healthy subjects and patients with stable coronary artery disease or acute coronary syndrome [110]. Although in these studies HDL-mediated miRNA delivery to cells appeared dependent on the cell surface expression of SR-B1, other reports showed that miRNAs circulating in blood [111] and cerebrospinal fluid [112] may act as ligands for cell surface Toll-like receptors. The proposal that free or lipoprotein-associated miRNAs serve as second messengers to provide long-distance communication and homeostatic regulation between tissues is a new frontier in the field, with profound implications for diagnostic and therapeutic purposes. For a comprehensive review on lipoprotein-associated miRNAs, please see the article by Kasey Vickers in this Special Issue of BBA.
6. CONCLUSIONS
In the past few years, miRNAs have emerged as physiologic regulators of HDL metabolism. The traditional view that raising HDL-c levels in blood reduces cardiovascular risk has been lately challenged by both human genetic data and pharmacologic intervention studies. The new “functional HDL” paradigm states that it is the ability to mobilize cholesterol through the RCT pathway for subsequent hepatobiliary excretion, coupled to anti-inflammatory and anti-thrombotic (and perhaps other) activities, that determines HDL’s atheroprotective properties. It should be noted that the majority of the miRNA studies that affect HDL discussed in the review report on changes in HDL-c, but the role of these miRNAs on HDL functionality is unknown. Additionally, despite the plethora of reports showing miRNAs targeting one or several transcripts involved in HDL lipid metabolism, a neglected area in the field has been the description of miRNAs that impact HDL-associated proteins and non-lipid-related HDL functions. Whether miRNAs that modulate HDL biogenesis, peripheral lipidation, remodeling, or clearance can be exploited therapeutically to improve HDL function (beyond changes in HDL-c) and provide atheroprotection is a new frontier ripe for exploration.
Highlights.
In this review, we summarize the major miRNAs reported to be in involved in HDL metabolism
HDL mediate the reverse cholesterol transport pathway, and are a candidate for therapeutic intervention in cardiovascular disease.
Multiple miRNAs target key transcripts involved in HDL biosynthesis, lipidation, and clearance.
Reports in mice and non-human primates show that silencing specific miRNAs raises HDL-c in circulation.
Whether these miRNAs can be exploited to improve HDL function and offer atheroprotection is currently under investigation.
Acknowledgments
We thank Drs. Elizabeth Tarling for help with the Figure, and Peter Edwards for the critical reading of the manuscript. Á.B. is supported in part by NIH Grant HL107794 and American Heart Association (AHA) Grant GRNT20460189. T.Q.d.A.V. is supported in part by NIH Grants DK102559 and HL028481, AHA Grant SDG18440015; University of California Los Angeles (UCLA) Clinical and Translational Science Institute grant UL1TR000124; and UCLA Diabetes Research Center Grant DK063491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ángel Baldán, Email: abaldan1@slu.edu.
Thomas Q. de Aguiar Vallim, Email: tvallim@mednet.ucla.edu.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontush A, Lindahl M, Lhomme M, Calabresi L, Chapman MJ, Davidson WS. Structure of HDL: particle subclasses and molecular components. Handb Exp Pharmacol. 2015;224:3–51. doi: 10.1007/978-3-319-09665-0_1. [DOI] [PubMed] [Google Scholar]
- 3.Cavelier C, Lorenzi I, Rohrer L, von Eckardstein A. Lipid efflux by the ATP-binding cassette transporters ABCA1 and ABCG1. Biochim Biophys Acta. 2006;1761:655–666. doi: 10.1016/j.bbalip.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Timmins JM, Lee JY, Boudyguina E, Kluckman KD, Brunham LR, Mulya A, Gebre AK, Coutinho JM, Colvin PL, Smith TL, Hayden MR, Maeda N, Parks JS. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J Clin Invest. 2005;115:1333–1342. doi: 10.1172/JCI23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunham LR, Kruit JK, Iqbal J, Fievet C, Timmins JM, Pape TD, Coburn BA, Bissada N, Staels B, Groen AK, Hussain MM, Parks JS, Kuipers F, Hayden MR. Intestinal ABCA1 directly contributes to HDL biogenesis in vivo. J Clin Invest. 2006;116:1052–1062. doi: 10.1172/JCI27352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung S, Sawyer JK, Gebre AK, Maeda N, Parks JS. Adipose tissue ATP binding cassette transporter A1 contributes to high-density lipoprotein biogenesis in vivo. Circulation. 2011;124:1663–1672. doi: 10.1161/CIRCULATIONAHA.111.025445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attie AD. ABCA1: at the nexus of cholesterol, HDL and atherosclerosis. Trends Biochem Sci. 2007;32:172–179. doi: 10.1016/j.tibs.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy MA, Barrera GC, Nakamura K, Baldan A, Tarr P, Fishbein MC, Frank J, Francone OL, Edwards PA. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Tarling EJ, Edwards PA. ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter. Proc Natl Acad Sci U S A. 2011;108:19719–19724. doi: 10.1073/pnas.1113021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glomset JA, Norum KR. The metabolic role of lecithin: cholesterol acyltransferase: perspectives form pathology. Adv Lipid Res. 1973;11:1–65. [PubMed] [Google Scholar]
- 11.Koizumi J, Mabuchi H, Yoshimura A, Michishita I, Takeda M, Itoh H, Sakai Y, Sakai T, Ueda K, Takeda R. Deficiency of serum cholesteryl-ester transfer activity in patients with familial hyperalphalipoproteinaemia. Atherosclerosis. 1985;58:175–186. doi: 10.1016/0021-9150(85)90064-4. [DOI] [PubMed] [Google Scholar]
- 12.Kurasawa T, Yokoyama S, Miyake Y, Yamamura T, Yamamoto A. Rate of cholesteryl ester transfer between high and low density lipoproteins in human serum and a case with decreased transfer rate in association with hyperalphalipoproteinemia. J Biochem. 1985;98:1499–1508. doi: 10.1093/oxfordjournals.jbchem.a135418. [DOI] [PubMed] [Google Scholar]
- 13.Valacchi G, Sticozzi C, Lim Y, Pecorelli A. Scavenger receptor class B type I: a multifunctional receptor. Ann N Y Acad Sci. 2011;1229:E1–7. doi: 10.1111/j.1749-6632.2011.06205.x. [DOI] [PubMed] [Google Scholar]
- 14.Chadwick AC, Sahoo D. Functional genomics of the human high-density lipoprotein receptor scavenger receptor BI: an old dog with new tricks. Curr Opin Endocrinol Diabetes Obes. 2013;20:124–131. doi: 10.1097/MED.0b013e32835ed575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gofman JW, Young W, Tandy R. Ischemic heart disease, atherosclerosis, and longevity. Circulation. 1966;34:679–697. doi: 10.1161/01.cir.34.4.679. [DOI] [PubMed] [Google Scholar]
- 16.Miller GJ, Miller NE. Plasma-high-density-lipoprotein concentration and development of ischaemic heart-disease. Lancet. 1975;1:16–19. doi: 10.1016/s0140-6736(75)92376-4. [DOI] [PubMed] [Google Scholar]
- 17.Rhoads GG, Gulbrandsen CL, Kagan A. Serum lipoproteins and coronary heart disease in a population study of Hawaii Japanese men. N Engl J Med. 1976;294:293–298. doi: 10.1056/NEJM197602052940601. [DOI] [PubMed] [Google Scholar]
- 18.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 19.Badimon JJ, Badimon L, Fuster V. Regression of atherosclerotic lesions by high density lipoprotein plasma fraction in the cholesterol-fed rabbit. J Clin Invest. 1990;85:1234–1241. doi: 10.1172/JCI114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin EM, Ishida BY, Clift SM, Krauss RM. Expression of human apolipoprotein A-I in transgenic mice results in reduced plasma levels of murine apolipoprotein A-I and the appearance of two new high density lipoprotein size subclasses. Proc Natl Acad Sci U S A. 1991;88:434–438. doi: 10.1073/pnas.88.2.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tangirala RK, Tsukamoto K, Chun SH, Usher D, Pure E, Rader DJ. Regression of atherosclerosis induced by liver-directed gene transfer of apolipoprotein A-I in mice. Circulation. 1999;100:1816–1822. doi: 10.1161/01.cir.100.17.1816. [DOI] [PubMed] [Google Scholar]
- 22.Joyce CW, Amar MJ, Lambert G, Vaisman BL, Paigen B, Najib-Fruchart J, Hoyt RF, Jr, Neufeld ED, Remaley AT, Fredrickson DS, Brewer HB, Jr, Santamarina-Fojo S. The ATP binding cassette transporter A1 (ABCA1) modulates the development of aortic atherosclerosis in C57BL/6 and apoE-knockout mice. Proc Natl Acad Sci U S A. 2002;99:407–412. doi: 10.1073/pnas.012587699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singaraja RR, Fievet C, Castro G, James ER, Hennuyer N, Clee SM, Bissada N, Choy JC, Fruchart JC, McManus BM, Staels B, Hayden MR. Increased ABCA1 activity protects against atherosclerosis. J Clin Invest. 2002;110:35–42. doi: 10.1172/JCI15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tardif JC, Heinonen T, Noble S. High-density lipoprotein/apolipoprotein A-I infusion therapy. Curr Atheroscler Rep. 2009;11:58–63. doi: 10.1007/s11883-009-0009-7. [DOI] [PubMed] [Google Scholar]
- 25.Tariq SM, Sidhu MS, Toth PP, Boden WE. HDL hypothesis: where do we stand now? Curr Atheroscler Rep. 2014;16:398. doi: 10.1007/s11883-014-0398-0. [DOI] [PubMed] [Google Scholar]
- 26.Rosenson RS, Brewer HB, Jr, Ansell BJ, Barter P, Chapman MJ, Heinecke JW, Kontush A, Tall AR, Webb NR. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat Rev Cardiol. 2015 doi: 10.1038/nrcardio.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Triolo M, Annema W, Dullaart RP, Tietge UJ. Assessing the functional properties of high-density lipoproteins: an emerging concept in cardiovascular research. Biomark Med. 2013;7:457–472. doi: 10.2217/bmm.13.35. [DOI] [PubMed] [Google Scholar]
- 28.Rohatgi A, Khera A, Berry JD, Givens EG, Ayers CR, Wedin KE, Neeland IJ, Yuhanna IS, Rader DR, de Lemos JA, Shaul PW. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371:2383–2393. doi: 10.1056/NEJMoa1409065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science. 2010;238:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. miR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science. 2010;238:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO molecular medicine. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suarez Y, de Cabo R, Gorospe M, Fernandez-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquart TJ, Wu J, Lusis AJ, Baldan A. AntimiR-33 Therapy Does Not Alter the Progression of Atherosclerosis in Low-Density Lipoprotein Receptor-Deficient Mice. Arteriosclerosis, thrombosis and vascular biology. 2013;33:455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arterioscler Thromb Vasc Biol. 2013;33:1973–1977. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;2015 doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, Moore KJ, Perisic L, Maegdefessel L, Hedin U, Harper ME, Rayner KJ. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ Res. 2015;117:266–278. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 Deficiency Reduces the Progression of Atherosclerotic Plaque in ApoE−/− Mice. J Am Heart Assoc. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Sowa N, Yahagi N, Shimano H, Matsumura S, Inoue K, Marusawa H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun. 2013;4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karunakaran D, Richards L, Geoffrion M, Barrette D, Gotfrit RJ, Harper ME, Rayner KJ. Therapeutic Inhibition of miR-33 Promotes Fatty Acid Oxidation but Does Not Ameliorate Metabolic Dysfunction in Diet-Induced Obesity. Arterioscler Thromb Vasc Biol. 2015 doi: 10.1161/ATVBAHA.115.306404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen RM, Marquart TJ, Jesse JJ, Baldan A. Control of very low-density lipoprotein secretion by N-ethylmaleimide-sensitive factor and miR-33. Circ Res. 2014;115:10–22. doi: 10.1161/CIRCRESAHA.115.303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goedeke L, Salerno A, Ramirez CM, Guo L, Allen RM, Yin X, Langley SR, Esau C, Wanschel A, Fisher EA, Suarez Y, Baldan A, Mayr M, Fernandez-Hernando C. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med. 2014;6:1133–1141. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Nakazeki F, Ide Y, Koyama S, Sowa N, Yahagi N, Shimano H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo. Sci Rep. 2014;4:5312. doi: 10.1038/srep05312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandolini C, Santovito D, Marcantonio P, Buttitta F, Bucci M, Ucchino S, Mezzetti A, Cipollone F. Identification of microRNAs 758 and 33b as potential modulators of ABCA1 expression in human atherosclerotic plaques. Nutr Metab Cardiovasc Dis. 2015;25:202–209. doi: 10.1016/j.numecd.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Martino F, Carlomosti F, Avitabile D, Persico L, Picozza M, Barilla F, Arca M, Montali A, Martino E, Zanoni C, Parrotto S, Magenta A. Circulating miR-33a and miR-33b are up-regulated in familial hypercholesterolaemia in paediatric age. Clin Sci (Lond) 2015;129:963–972. doi: 10.1042/CS20150235. [DOI] [PubMed] [Google Scholar]
- 54.Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes. 2015 doi: 10.1111/ijpo.12050. [DOI] [PubMed] [Google Scholar]
- 55.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med. 2015;21:1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 63.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen W, Roeder RG. Mediator-dependent nuclear receptor function. Semin Cell Dev Biol. 2011;22:749–758. doi: 10.1016/j.semcdb.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 68.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Aguiar Vallim T, Tarling E, Kim T, Civelek M, Baldan A, Esau C, Edwards P. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circulation research. 2013 doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suarez Y, Fernandez-Hernando C. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang MH, Zhang LH, Wijesekara N, de Haan W, Butland S, Bhattacharjee A, Hayden MR. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler Thromb Vasc Biol. 2013;33:2724–2732. doi: 10.1161/ATVBAHA.113.302004. [DOI] [PubMed] [Google Scholar]
- 73.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joseph SB, McKilligin E, Pei L, Watson MA, Collins AR, Laffitte BA, Chen M, Noh G, Goodman J, Hagger GN, Tran J, Tippin TK, Wang X, Lusis AJ, Hsueh WA, Law RE, Collins JL, Willson TM, Tontonoz P. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc Natl Acad Sci U S A. 2002;99:7604–7609. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu YW, Hu YR, Zhao JY, Li SF, Ma X, Wu SG, Lu JB, Qiu YR, Sha YH, Wang YC, Gao JJ, Zheng L, Wang Q. An agomir of miR-144–3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS One. 2014;9:e94997. doi: 10.1371/journal.pone.0094997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sala F, Aranda JF, Rotllan N, Ramirez CM, Aryal B, Elia L, Condorelli G, Catapano AL, Fernandez-Hernando C, Norata GD. MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr−/−mice. Thromb Haemost. 2014;112:796–802. doi: 10.1160/TH13-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santovito D, Mandolini C, Marcantonio P, De Nardis V, Bucci M, Paganelli C, Magnacca F, Ucchino S, Mastroiacovo D, Desideri G, Mezzetti A, Cipollone F. Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert Opin Ther Targets. 2013;17:217–223. doi: 10.1517/14728222.2013.745512. [DOI] [PubMed] [Google Scholar]
- 82.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M, Upton P, Xin M, van Rooij E, Olson EN, Morrell NW, MacLean MR, Baker AH. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 83.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 84.Chen WJ, Yin K, Zhao GJ, Fu YC, Tang CK. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis. 2012;222:314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 85.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang M, Wu JF, Chen WJ, Tang SL, Mo ZC, Tang YY, Li Y, Wang JL, Liu XY, Peng J, Chen K, He PP, Lv YC, Ouyang XP, Yao F, Tang DP, Cayabyab FS, Zhang DW, Zheng XL, Tian GP, Tang CK. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 87.Goedeke L, Rotllan N, Ramirez CM, Aranda JF, Canfran-Duque A, Araldi E, Fernandez-Hernando A, Langhi C, de Cabo R, Baldan A, Suarez Y, Fernandez-Hernando C. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015;243:499–509. doi: 10.1016/j.atherosclerosis.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S, Yi M, Lemon SM, Kaneko S. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alvarez ML, Khosroheidari M, Eddy E, Done SC. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis. 2015;242:595–604. doi: 10.1016/j.atherosclerosis.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, Ando Y, Koishi R, Kohama T, Sakai N, Kotani K, Komuro R, Ishida T, Hirata K, Yamashita S, Furukawa H, Shimomura I. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 92.Jin W, Wang X, Millar JS, Quertermous T, Rothblat GH, Glick JM, Rader DJ. Hepatic Proprotein Convertases Modulate HDL Metabolism. Cell Metab. 2007;6:129–136. doi: 10.1016/j.cmet.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuda T. Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol Nutr Food Res. 2012;56:159–170. doi: 10.1002/mnfr.201100526. [DOI] [PubMed] [Google Scholar]
- 94.Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res. 2012;111:967–981. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 95.Lv YC, Yang J, Yao F, Xie W, Tang YY, Ouyang XP, He PP, Tan YL, Li L, Zhang M, Liu D, Cayabyab FS, Zheng XL, Tang CK. Diosgenin inhibits atherosclerosis via suppressing the MiR-19b-induced downregulation of ATP-binding cassette transporter A1. Atherosclerosis. 2015;240:80–89. doi: 10.1016/j.atherosclerosis.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 96.Lv YC, Tang YY, Peng J, Zhao GJ, Yang J, Yao F, Ouyang XP, He PP, Xie W, Tan YL, Zhang M, Liu D, Tang DP, Cayabyab FS, Zheng XL, Zhang DW, Tian GP, Tang CK. MicroRNA-19b promotes macrophage cholesterol accumulation and aortic atherosclerosis by targeting ATP-binding cassette transporter A1. Atherosclerosis. 2014;236:215–226. doi: 10.1016/j.atherosclerosis.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 97.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 98.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He Y, Lin L, Cao J, Mao X, Qu Y, Xi B. Up-regulated miR-93 contributes to coronary atherosclerosis pathogenesis through targeting ABCA1. Int J Clin Exp Med. 2015;8:674–681. [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang N, Lei J, Lei H, Ruan X, Liu Q, Chen Y, Huang W. MicroRNA-101 overexpression by IL-6 and TNF-alpha inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp Cell Res. 2015;336:33–42. doi: 10.1016/j.yexcr.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu YW, Chen ZP, Hu XM, Zhao JY, Huang JL, Ma X, Li SF, Qiu YR, Wu XJ, Sha YH, Gao JJ, Wang YC, Zheng L, Wang Q. The miR-573/apoM/Bcl2A1-dependent signal transduction pathway is essential for hepatocyte apoptosis and hepatocarcinogenesis. Apoptosis. 2015;20:1321–1337. doi: 10.1007/s10495-015-1153-x. [DOI] [PubMed] [Google Scholar]
- 104.Liu ME, Liao YC, Lin RT, Wang YS, Hsi E, Lin HF, Chen KC, Juo SH. A functional polymorphism of PON1 interferes with microRNA binding to increase the risk of ischemic stroke and carotid atherosclerosis. Atherosclerosis. 2013;228:161–167. doi: 10.1016/j.atherosclerosis.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 105.Miller AM, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA, Fernandez-Hernando C, McInnes IB, Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 107.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 108.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, Catherinet C, Prado-Lourenco L, Levin MG, Thacker S, Sethupathy P, Barter PJ, Remaley AT, Rye KA. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, Zeiher AM, Landmesser U, Dimmeler S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 111.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kalin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, Golenbock D, Schott E, Ploegh HL, Wulczyn FG, Lehnardt S. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]