Abstract
Hepatitis B virus (HBV) reverse transcription requires coordinated function of the reverse transcriptase and ribonuclease H (RNaseH) activities of the viral polymerase protein. The reverse transcriptase has been biochemically characterized, but technical difficulties have prevented both assessment of the RNaseH and development of high throughput inhibitor screens against the RNaseH. Expressing the HBV RNaseH domain with both maltose binding protein and hexahistidine tags led to stable, high-level accumulation of the RNaseH in bacteria. Nickel-affinity purification in the presence of Mg2+ and ATP removed co-purifying bacterial chaperones and yielded nearly pure monomeric recombinant enzyme. The endonucleolytic RNaseH activity required an DNA:RNA duplex ≥14 nt, could not tolerate a stem-loop in either the RNA or DNA strands, and could tolerate a nick in the DNA strand but not a gap. The RNaseH had no obvious sequence specificity or positional dependence within the RNA, and it cut the RNA at multiple positions even within the minimal 14 nt duplex. The RNaseH also possesses a processive 3’-5’ exoribonuclease activity that is slower than the endonucleolytic reaction. These results are consistent with the HBV reverse transcription mechanism that features an initial endoribonucleolytic cut, 3’-5’ degradation of RNA, and a sequence-independent terminal RNA cleavage. These data provide support for ongoing anti-RNaseH drug discovery efforts.
Keywords: Hepatitis B virus, DNA virus, RNA processing, ribonuclease, endoribonuclease
1. Introduction
Hepatitis type B is caused by hepatitis B virus (HBV), a hepatotropic DNA virus that replicates by reverse transcription (Seeger et al., 2013). HBV kills over 600,000 patients annually (Ganem and Prince, 2004; Lavanchy, 2004; Shepard et al., 2006; Sorrell et al., 2009) and chronically infects up to 350 million people worldwide. Therapy primarily employs nucleos(t)ide analog drugs that suppress the virus to near or below the limit of detection but only rarely cure the infection (Cox and Tillmann, 2011; Kwon and Lok, 2011; Marcellin et al., 2008; van Bommel et al., 2010; Woo et al., 2010; Wursthorn et al., 2010). Failure to eliminate HBV is in part due to incomplete suppression of viral genomic synthesis, leading to ongoing infection of new cells and maintenance of the viral genome in infected cells (Coffin et al., 2011; Zoulim, 2004). Therefore, virus replication resurges when the nucleos(t)ide analogs are withdrawn and therapy is essentially life-long. Greater suppression of HBV will require new drugs that will probably be used in combination with the nucleos(t)ide analogs (Block et al., 2013; Tavis et al., 2013b).
Two viral enzymatic activities are necessary for HBV reverse transcription. The DNA polymerase activity (reverse transcriptase) synthesizes new DNA from the viral pregenomic RNA. The ribonuclease H (RNaseH) activity cleaves RNA when it is in a DNA:RNA heteroduplex and destroys the viral RNA after it has been copied into DNA. These two activities are catalyzed by adjacent domains in the HBV polymerase protein. RNaseH action begins after the first strand transfer during HBV reverse transcription (Seeger et al., 2013) with an endonucleolytic cleavage near the 3’ end of the template RNA. The RNA is subsequently degraded in a 3’ to 5’ direction during elongation of the minus-polarity DNA strand (Mason et al., 1982). RNA cleavage terminates 15–18 nt prior to the 5’ end of the template RNA to form the primer for synthesis of the plus-polarity DNA strand (Loeb et al., 1991). Inhibiting the RNaseH causes viral genomic replication to stall (Chen and Marion, 1996; Chen et al., 1994; Gerelsaikhan et al., 1996), so it is a logical target for anti-HBV drugs (Tavis and Lomonosova, 2015).
RNaseH enzymes (Hostomsky et al., 1993) belong to the nucleotidyl transferase superfamily (Nowotny, 2009; Yang and Steitz, 1995) whose members share a similar protein fold and catalytic mechanism. This family includes human RNaseH 1 and 2 (Lima et al., 2001) plus the retroviral RNaseH and integrases, including the human immunodeficiency virus (HIV) enzymes (Dyda et al., 1994). The RNaseH active site contains four conserved carboxylates (the "DEDD" motif) that coordinate two divalent cations, usually Mg2+ (Nowotny et al., 2005). The RNA cleavage mechanism requires both cations to promote a hydroxyl-mediated nucleophilic scission reaction (Klumpp et al., 2003; Nowotny and Yang, 2006; Yang and Steitz, 1995). The human immunodeficiency virus (HIV) RNaseH molecular structure is known, but no structural information exists for the HBV RNaseH. The first three carboxylates in the HBV DEDD motif can be identified by sequence alignments (HBV residues D702, E731, and D750), and we have confirmed these residues by mutating them in context of the full viral genome and observing an RNaseH-deficient phenotype (Tavis et al., 2013a). The fourth carboxylate cannot be unambiguously identified from sequence alignments, but mutating D790 leads to an RNaseH-deficient phenotype (Ko et al., 2014).
The HBV RNaseH and the more extensively studied HIV enzyme are distantly related, sharing just 23% identity (Tavis et al., 2013a). Although both enzymes form the C-terminal domains of their respective polymerase proteins, the HBV RNaseH acts within an uncleaved monomer of the polymerase protein (Zhang and Tavis, 2006) whereas the HIV RNaseH acts within a proteolytically cleaved heterodimer. Despite the large genetic distance between them, the HBV and HIV RNaseHs both degrade the RNA template during viral replication to enable plus-polarity DNA synthesis (Freed and Martin, 2007; Tavis and Badtke, 2009).
We recently produced active recombinant hexahistidine-tagged HBV RNaseH that was suitable for low-throughput drug screening and used it to discover all of the currently known inhibitors of the enzyme, many of which work against viral replication in cells (Cai et al., 2014; Hu et al., 2013; Lu et al., 2015; Tavis et al., 2013a; Tavis and Lomonosova, 2015). However, the RNaseH was not pure enough for biochemical analyses or high throughput screening. Here, we report purification of soluble HBV RNaseH at high yield and provide a basic biochemical characterization of the enzyme as a step towards developing a high throughput inhibitor screen.
2. Material and methods
2.1 Expression and purification of the HBV RNaseH domain
HBV RNaseH sequences from genotype C isolate V01460 encoding residues corresponding to amino acids 688 to 844 of the HBV polymerase (reference strain ADW2, genotype A) were cloned by gene synthesis into pMAL-c5X-His with sequences encoding a hexahistidine tag appended to the 3’ end of the RNaseH gene to create pMal-HRHgtC. Sequences encoding Ala-Gly-Ala were inserted between the maltose binding protein (MBP) and HBV RNaseH sequences, and sequences encoding Gly-Ala-Gly were inserted between sequences for the RNaseH and the hexahistidine tag. Sequences encoding HBV RNaseH residues 809–844 were deleted from pMal-HRHgtC to create pMal-HRHgtCΔ5. Active site residues D702 and E731 were mutated to alanines to create pMAL-HRHgtC(D702A/E731A) which encodes an inactive RNaseH.
An overnight culture of E. coli LOBSTR-BL21(DE3) (Andersen et al., 2013) cells harboring the RNaseH expression plasmids was diluted 20-fold in 1 liter of LB broth in the presence of 50 µg/ml of ampicillin and incubated at 37°C with shaking until OD600=0.6 was reached. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.25 mM, and after 3 h of incubation at 16°C, cells were pelleted and frozen at −80°C. The pellet was suspended in 30 ml lysis buffer (buffer A: 50 mM HEPES pH 8.0, 0.1 M NaCl, 2% Tween20, 30% glycerol, 25 mM imidazole) plus 1 mM protease inhibitor cocktail (Sigma), 5 mM ATP, 1 mM MgCl2, 0.5 mM EDTA and the cells were disrupted by sonication. Debris was eliminated by centrifugation at 54,000 g for 45 min. The supernatant was loaded for 1 hour onto a 5-ml His-Trap column (GE Healthcare) equilibrated with buffer A. The column was washed with 50 ml of buffer A for 2 hours. Bound proteins were eluted with a linear gradient of lysis buffer A: buffer B (buffer B: 50 mM HEPES pH 8.0, 0.1 M NaCl, 2% Tween20, 30% glycerol, 0.5 M imidazole) in 25 column volumes. The products were evaluated by 10% SDS-PAGE and Coomassie brilliant blue staining. Samples were dialyzed into 50 mM HEPES pH 7.3, 300 mM NaCl, 20% glycerol, and 5 mM DTT, and stored in liquid nitrogen. Multimerization status and solubility of the RNaseH were evaluated by size exclusion chromatography on a Superdex 200 column (GE Healthcare) equilibrated with buffer C (50 mM HEPES pH 7.3, 0.3 M NaCl, 20% glycerol, 5 mM DTT) and eluted with buffer C.
2.2 Purification of recombinant human RNaseH1
Human RNaseH1 was cloned into pRSETb between the BamHI and XhoI sites to create pHuRH1. This appended a hexahistidine tag to the N terminus of the RNaseH. Human RNaseH1 expression was induced with the same protocol as HBV RNaseH. Purification followed the same protocol except for buffer A (buffer A’: 50 mM HEPES pH 8.0, 0.3 M NaCl, 1% Tween20, 30% glycerol, 25 mM imidazole).
2.3 Oligonucleotide-directed RNA cleavage assay
DNA oligonucleotide (ODN)-directed RNA cleavage assays were conducted as previously described (Hu et al., 2013; Tavis et al., 2013a) employing DRF+ (a 264 nucleotide RNA derived from the duck hepatitis B virus genome) or usRNA1 (a 196 nt synthetic unstructured RNA). Briefly, a uniformly 32P-labeled RNA was combined with a complementary ODN or a non-complementary control ODN; ODN and RNA sequences are in Supplementary Table 1. These substrates were incubated with the RNaseH at a final concentration of 50 mM Tris pH 8.0, 190 mM NaCl, 5 mM MgCl2, 3.5 mM DTT, 0.05% NP40, 6% glycerol, and 1% DMSO at 42 °C for 90 min. The products were resolved by 6 or 7% denaturing polyacrylamide gel electrophoresis, detected by autoradiography, and quantified using ImageJ.
3. Results
3.1 Purification of MBP-HRHgtC
We previously expressed the HBV RNaseH with a hexahistidine tag at the C-terminus in E. coli (Fig. 1, HRHPLgtD) (Tavis et al., 2013a). Purification of this enzyme by nickel-affinity chromatography led to recovery of a small quantity of the HBV RNaseH that was detectable only by western blotting (Fig. 2A). Altering the induction and purification conditions failed to increase yield or improve the integrity of the enzyme. However, adding the maltose binding protein (MBP) to the N-terminus of the hexahistidine-tagged RNaseH domain (Fig. 1) led to recovery of Coomassie-stainable levels of the full-length protein without degradation products following nickel-affinity chromatography (Fig. 2B). Typical recovery was >5 mg from two liters of growth medium
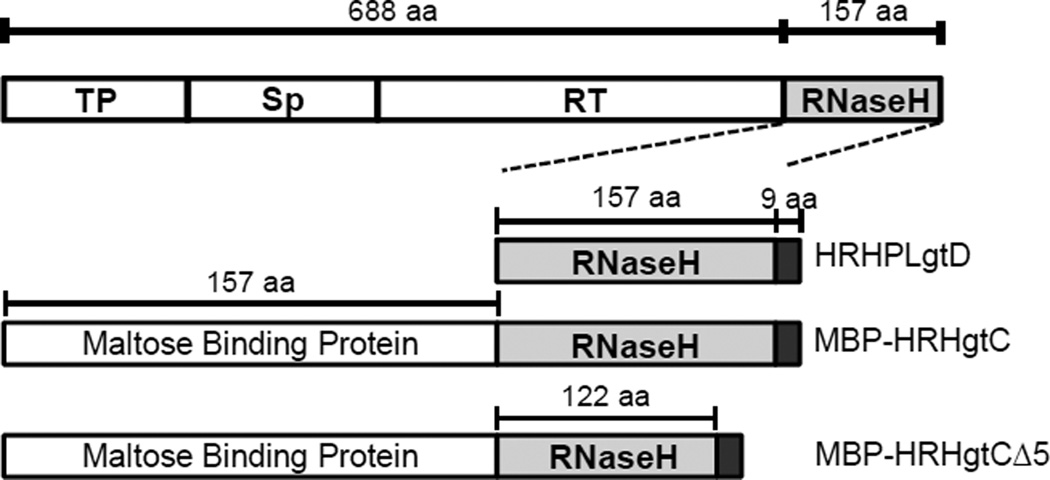
Figure 1. Structure of the HBV RNaseH constructs used.
The full-length HBV polymerase with its domains labeled is at top. The recombinant RNaseH derivatives HRHPLgtD, MBP-HRHgtC, and MBP-HRHgtCΔ5 are shown below with the hexahistidine tag indicated in black and the maltose binding protein tag in white. TP, terminal protein domain; Sp, spacer domain; RT, reverse transcriptase domain; RNaseH, RNaseH domain.
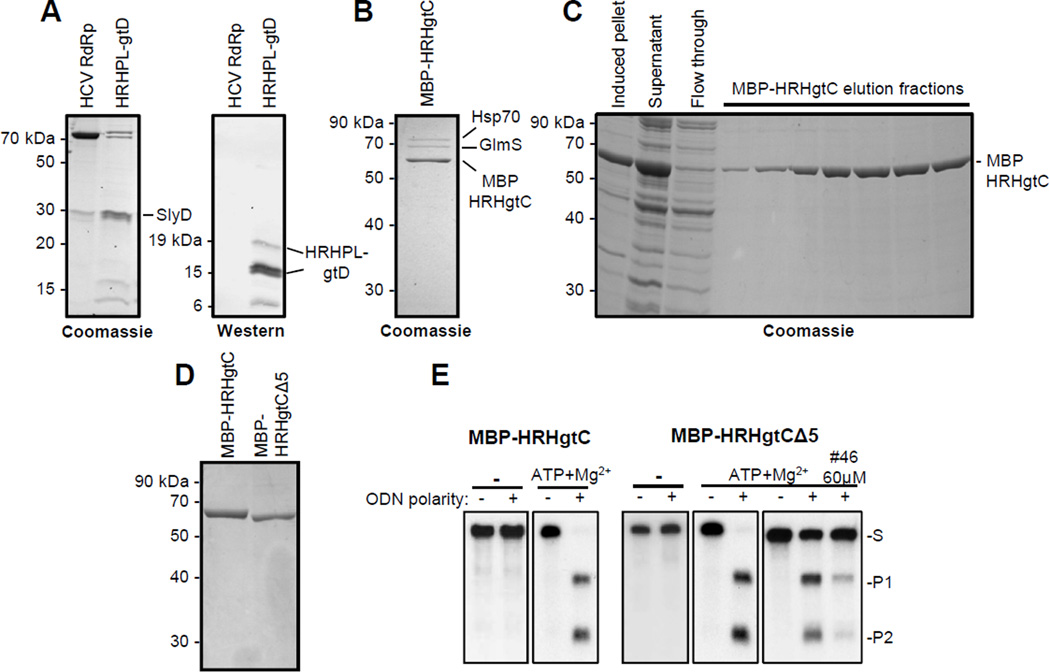
Figure 2. Purification of dual-tagged recombinant HBV RNaseH.
A. Enriched extracts of HRHPLgtD following nickel-affinity chromatography were analyzed by Coomassie-blue staining an SDS-PAGE gel and by western blot employing monoclonal antibody 9F9 which recognizes an epitope at the C-terminus of the HBV polymerase. This panel is modified from (Tavis et al., 2013a) under the Creative Commons Attribution license. B. Coomassie staining of an SDS-PAGE analysis of the final nickel-affinity chromatography elution fraction for MBP-HRHgtC purification done in the absence of ATP and Mg2+. C. Purification of HBV MBP-HRHgtC in the presence of ATP and Mg2+. D. Comparison of the final purification products for MBP-HRHgtC and MBP-HRHgtCΔ5 isolated in the presence of ATP and Mg2+ by SDS-PAGE and Coomassie staining. E. RNaseH activity in the ODN-directed RNaseH assay for MBP-HRHgtC (2.4 µg/reaction) and MPB-HRHgtCΔ5 (1.3 µg/reaction) purified in the absence (−) or presence of ATP and Mg2+. Inhibition of MBP-HRHgtCΔ5 (0.26 µg/reaction) purified in the presence of ATP and Mg2+ is shown at right; #46 is the HBV RNaseH inhibitor β-thujaplicinol. S, substrate; P1, product 1; P2, product 2.
Mass spectrometry revealed the major co-purifying protein contaminants to be DnaK and GlmS. GlmS was removed by increasing the washing time in the presence of a higher a concentration of imidazole. DnaK was eliminated by adding ATP and Mg2+ to the buffers in every step of the purification (Bolanos-Garcia and Davies, 2006) (Fig. 2C and D). We also purified a truncated version of MBP-HRHgtC (MBP-HRHgtCΔ5) that removed the C-terminal 35 amino acids that cannot be aligned with confidence against other RNaseHs (Figs. 1 and 2D).
Purification in the presence of ATP and Mg2+ greatly increased the specific activity of the RNaseH in an ODN-directed RNA cleavage assay (Fig. 2E). In this assay, a 20 mer ODN is annealed to the 264 nucleotide (nt) 32P-labeled DRF+ RNA to create an RNA:DNA heteroduplex. Cleavage of the RNA in the heteroduplex yields fragments of about 160 and 100 nts that are resolved by electrophoresis. Titrating the new enzyme preparations to obtain the same degree of cleavage observed with the old preparations indicates that 12-fold more activity per volume of bacterial culture was obtained with the new purification method.
MBP-HRHgtCΔ5 was also active in the ODN-directed RNA cleavage assay. This demonstrates that the C-terminal 35 residues are dispensable for RNaseH activity. The activity of MBP-HRHgtC (not shown) and MBP-HRHgtCΔ5 (Fig. 2E) could be suppressed with β-thujaplicinol [compound #46 (Hu et al., 2013)], indicating that the RNaseH purified by this protocol retained sensitivity to a known RNaseH inhibitor.
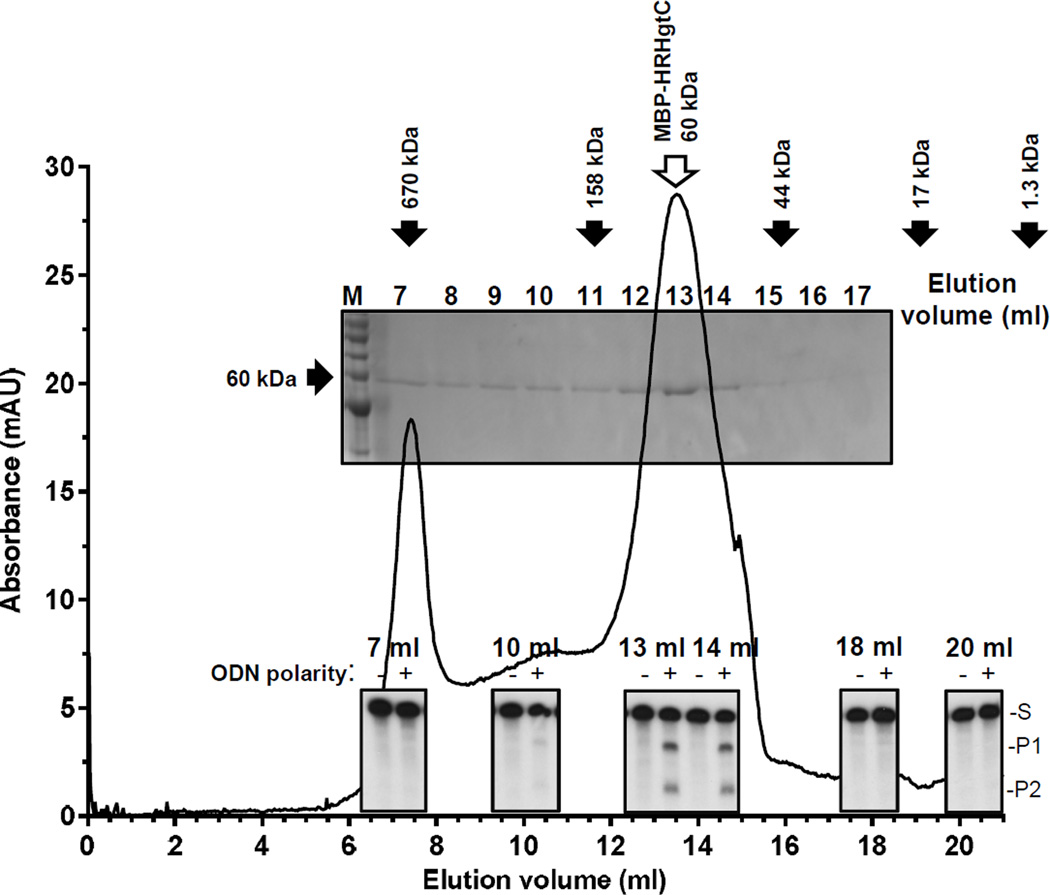
Oligomerization of MBP-HRHgtC was evaluated by size-exclusion chromatography on a Superdex 200 10/300 GL column (Fig. 3). The enzyme migrated primarily as a monomer (fractions 12–15), with minor fractions eluting as low order multimers (fractions 8–11) or higher-order multimers and aggregates (fractions 6–7). Evaluating the elution fractions for enzymatic activity revealed that the monomer was the primary active form of the HBV RNaseH, with the low-order multimers also retaining some activity.
Figure 3. MBP-HRHgtC is primarily a monomer in solution.
MBP-HRHgtC was assessed by size exclusion chromatography on a Superdex 200 10/300 GL column. A Coomassie-blue stained gel of the eluate and an ODN-directed RNA cleavage assay are overlaid at their relative positions on the elution profile from the Superdex column. The activity images were derived from a single autoradiograph. S, substrate; P1, product 1; P2, product 2. Molecular standards were: thyroglobulin (670 kDa) in the void volume, IgG (158 kDa), ovalbumin (44 kDa), myoglobulin (17 kDa), and vitamin B12 (1.5 kDa). The mass of MBP-HRHgtC is 60 kDa.
3.2 The HBV RNaseH has no strong sequence- or position-dependence for RNA cleavage
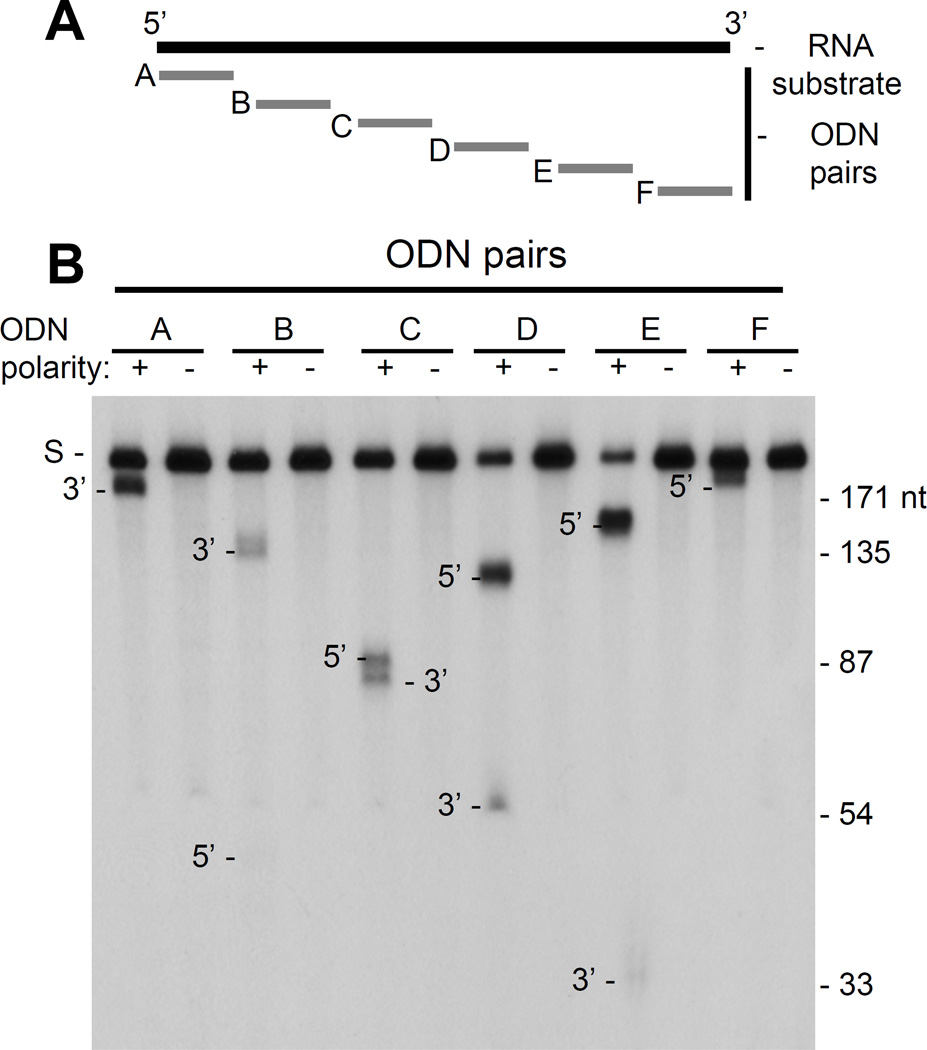
The sequence- and position-dependencies of the HBV RNaseH for RNA cleavage were evaluated using an ODN-directed RNaseH assay in which the heteroduplex substrate was formed by a set of six 20 mer DNA ODNs spaced along the length of a radiolabeled RNA (Fig. 4A). The RNA employed (usRNA1) was a 196 nt synthetic sequence with minimal secondary structure (−2.1 kCal/mol at 42°C). Each ODN was synthesized in both complementary and non-complementary polarities relative to usRNA1, with the non-complementary ODN being a specificity control that could not form an RNA:DNA heteroduplex. Specific cleavage of usRNA1 was detected with all complementary ODNs, regardless of their position on usRNA1 or their sequence (Fig. 4B). This included placing the ODN either 5 nts from the 5’ end (ODN A) or 3 nts of the 3’ end (ODN F). Therefore, there is no strong sequence or position preference of the HBV RNaseH for RNA cleavage.
Figure 4. Sequence- and positional-dependence of HBV RNaseH endonucleolytic cleavage.
An ODN-directed RNaseH assay was conducted with complementary ODNs (A+, B+, C+, D+, E+ and F+) and non-complementary ODNs (A−, B−, C−, D−, E− and F−) distributed along the length of the unstructured usRNA1. The experiment was repeated three times with very similar results and the 5’ cleavage product formed by ODN A was clearly visible on longer autoradiographic exposures. S, usRNA1 substrate; 3’, 3’ product fragment of the RNA; 5’, 5’ product fragment of the RNA.
3.3 HBV RNaseH activity requires an DNA:RNA heteroduplex of ≥ 14 bp for efficient cleavage
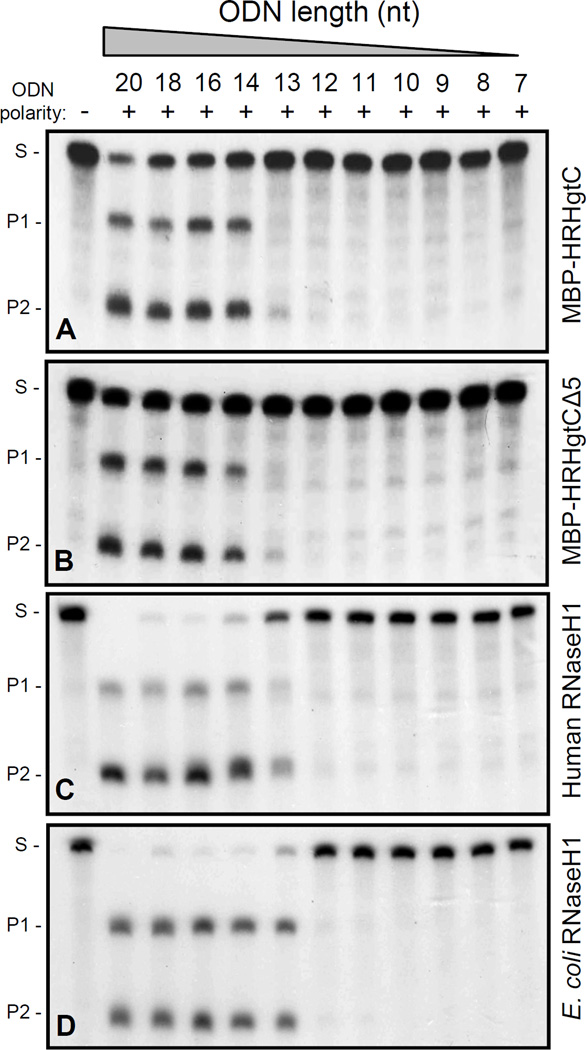
An ODN-directed cleavage assay employing ODNs of progressively shorter lengths was used to determine the minimal length of the DNA:RNA heteroduplex required for HBV RNaseH cleavage. ODNs from 20 to 7 nt whose annealing sites were centered on the same positon in the DRF(+) RNA were designed. An ODN-directed cleavage assay was conducted with these truncated ODNs using both the full-length recombinant enzyme (MBP-HRHgtC) and its C-terminal truncation derivative (MBP-HRHgtCΔ5). Recombinant human RNaseH1 and E. coli RNaseH1 (Invitrogen) enzymes were included as reference enzymes. The full-length HBV RNaseH efficiently cleaved the RNA when the ODN ranged from 20–14 nt, but little to no cleavage was observed with the 13 mer, and cleavage above background was not observed by the 12 to 7 mers (Fig. 5A). Very similar results were obtained with MBP-HRHgtCΔ5 (Fig. 5B). In contrast, human RNaseH1 and E. coli RNaseH1 cleaved the 13 nucleotide-long heteroduplex more efficiently (Figs. 5C–D). These RNaseH assays were done at the optimal temperature for the HBV enzyme of 42 °C. To exclude the possibility that the short heteroduplexes were unstable at this temperature, the experiments were repeated at 37 °C. The HBV enzymes were less active at the lower temperature, but the cleavage patterns were the same for all four enzymes (data not shown). Instability of the 13 mer that was poorly cleaved by the HBV RNaseH was also excluded because the 13 mer hybridized well enough to support RNA cleavage by the human and E. coli RNaseHs.
Figure 5. Minimal heteroduplex size recognized by the HBV RNaseH.
ODN-directed cleavage assays were performed with ODNs of different lengths (20 nt to 7 nt) for: A, HBV MBP-HRHgtC; B, HBV MBP-HRHgtCΔ5; C, Human RNaseH1; D, E. coli RNaseH1. −, non-complementary control ODN; +, complementary ODN; S, substrate; P1, product 1; P2, product 2.
3.4 Substrate specificity of the HBV RNaseH
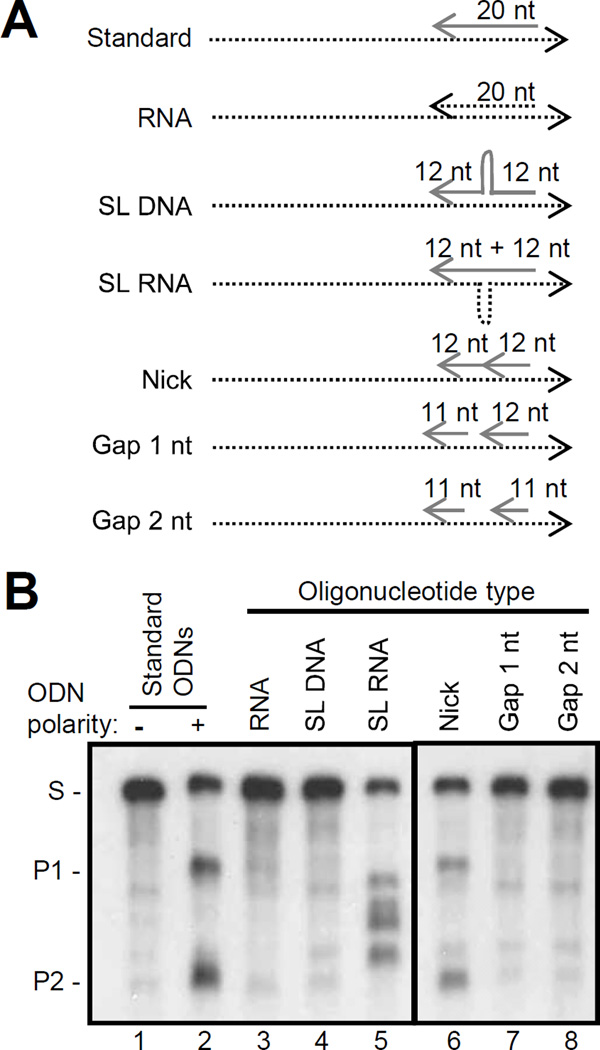
The structure of the DNA:RNA heteroduplex was varied to evaluate the types of substrates the full length HBV RNaseH could cleave (Fig. 6A). First, an RNA:RNA homoduplex was tested using a 20 mer RNA olignonucleotide that annealed to the same site as our standard ODN. No cleavage above background was detected (Fig. 6B lane 3), demonstrating that the HBV RNaseH cannot cut RNA:RNA duplexes. Introducing a 10 nucleotide stem-loop (GGGGTTCCCC) in the ODN blocked RNA cleavage (Fig. 6B lane 4), whereas introducing a stem-loop in the RNA strand (AATATATATT) led to aberrant nicking of the RNA (Fig. 6B lane 5), presumably at the base and apex of the RNA palindrome. Therefore, the RNaseH cannot tolerate a stem-loop in the DNA but it may be able to cut sharp bends in an RNA strand. The RNaseH was able to cut the RNA when a heteroduplex containing a nick was formed with two adjacent 12 mer ODNs, each of which was too short to support cleavage by itself (Fig. 6B lane 6). However, cleavage was lost when the nick between the oligos was expanded to a gap of 1 or 2 nucleotides (Fig. 6B lanes 7 and 8). Therefore, the RNaseH could cut RNA when there was a nick in the DNA but not when there was a gap.
Figure 6. Substrate specificity of the HBV RNaseH the ODN-directed RNA cleavage assay.
A. Schematic of the substrates employed. B. Oligonucleotide-directed cleavage assay with different primer structures. The figure is derived from a single autoradiograph. Dashed line, RNA; solid line, DNAs; −, non-complementary control ODN; +, complementary ODN; S, substrate; P1, product 1; P2, product 2.
3.5 RNA cleavage pattern
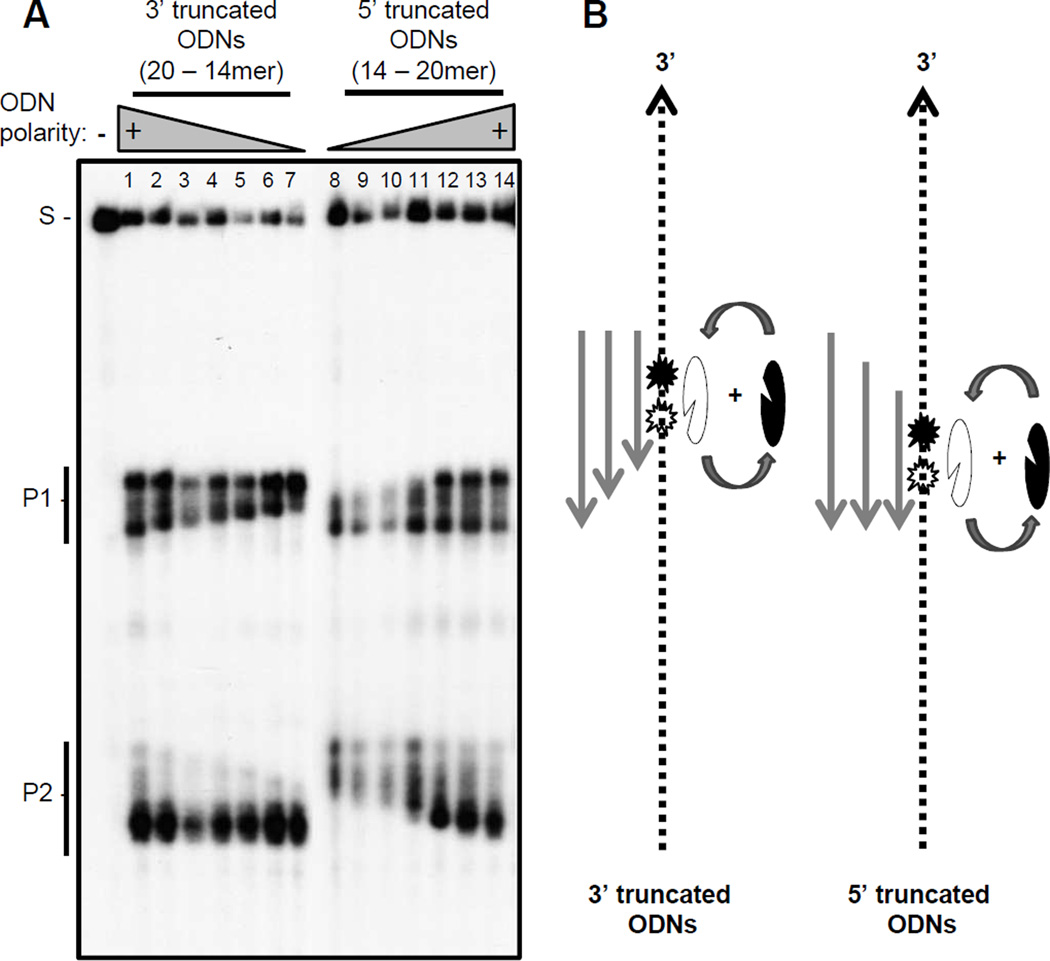
To evaluate the detailed RNA cleavage pattern, an ODN-directed RNaseH assay was conducted with 5’-or 3’-truncated ODNs ranging from 20 to 14 mers and the RNA products were resolved by high resolution denaturing polyacrylamide gel electrophoresis. The RNA was cleaved at multiple positions within the heterduplexes formed by the 20 mer ODNs (Fig. 7 lane 1 and 14). More than one band was also produced for both the P1 and P2 products even with the shortest ODNs that promote RNA cleavage (14 mers). The pattern of cleavage products contracted from a single end of each cleavage spectrum with the predicted pattern when the DNA ODNs were shorted from a 20 mer to a 14 mer. The spectrum of cleavage products produced by the shortest 3’- and 5’-truncated ODNs overlapped by one or two nucleotides (Fig. 7 lanes 7 and 8). Therefore, the RNaseH can cleave RNA at more than one position even within the shortest RNA:DNA heteroduplex it can cut.
Figure 7. Detailed RNA cleavage pattern of the HBV RNaseH.
A. An ODN-directed cleavage assay employing the DRF+ RNA substrate plus 3’- and 5’-truncated ODNs resolved by high-resolution denaturing electrophoresis was conducted. B. Interpretation of the data. Grey lines, ODNs; dashed lines, DRF+ RNA; notched ovals, MBP-HRHgtC with the notch representing the active site and the black and white shading indicating binding in the 5’-3’ and 3’-5’ orientations relative to the RNA strand of the heteroduplex; stars, cleavage sites on the RNA with the shading corresponding to the two enzyme orientations; S, substrate; P1, product 1; P2, product 2.
3.6 Identification of a 3’–5’ exoribonuclease activity of the HBV RNaseH
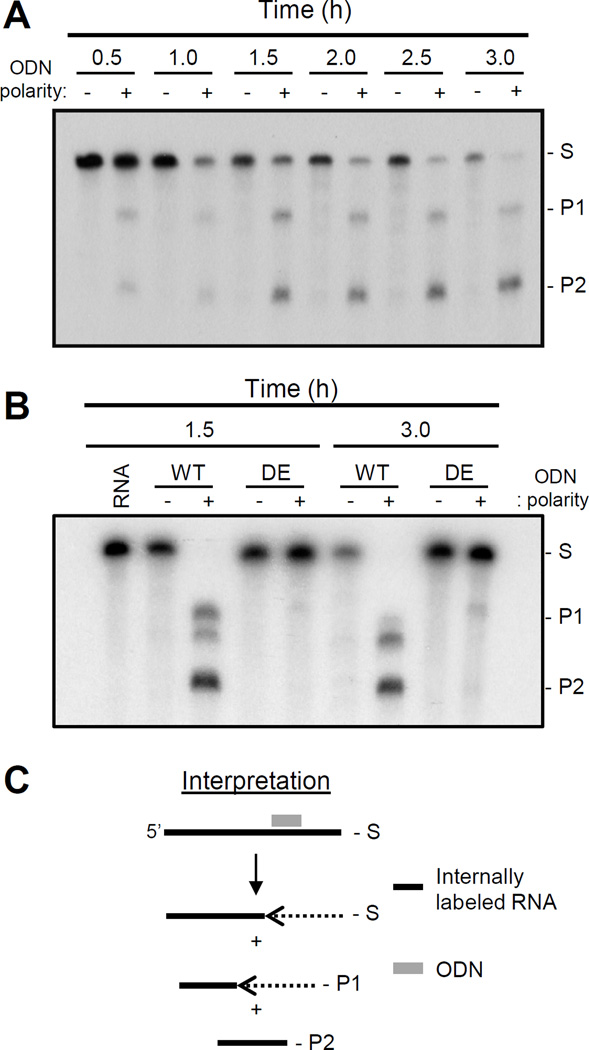
We noticed that the stoichiometry between the P1 and P2 products changed during time course studies of the HBV RNaseH activity, with the P1 product decreasing in abundance compared to the P2 product (Fig. 8A). Furthermore, the amount of substrate RNA also gradually declined without accumulation of a corresponding level of degradation intermediates even in reactions containing a non-complementary control ODN. To determine if this degradation was due to the HBV RNaseH, the stability of the RNA was evaluated in the presence of the inactive enzyme MBP-HRHgtC(D702A/E731A) that carries mutations in two of the key DEDD residues. The RNA molecule was stable in the presence of the inactive RNaseH enzyme (Fig. 8B), demonstrating that the loss of the RNA was not due to a contaminant in the enzyme preparations. This implies that the HBV RNaseH has a previously unknown 3’-5’ processive exonuclease activity in addition to its endonucleolytic activity (Fig. 8C).
Figure 8. Exonuclease activity of the HBV RNaseH.
A. Time course for an ODN-directed RNA cleavage assay with MBP-HRHgtC employing the standard ODNs that bind internally in the DRF+ substrate. B. RNA stability during an RNaseH assay employing wildtype MBP-HRHgtC and its active site mutant MBP-HRHgtC(D702A/E731A) employing the standard ODNs that bind internally in the DRF+ substrate. C. Interpretation of the data. S, substrate; P1, product 1; P2, product 2; DE, MPB-HRHgtC(D702A/E731A); −, non-complementary control ODN; +, complementary ODN; grey line, ODN; black line, DRF+ RNA; dashed line, degraded RNA.
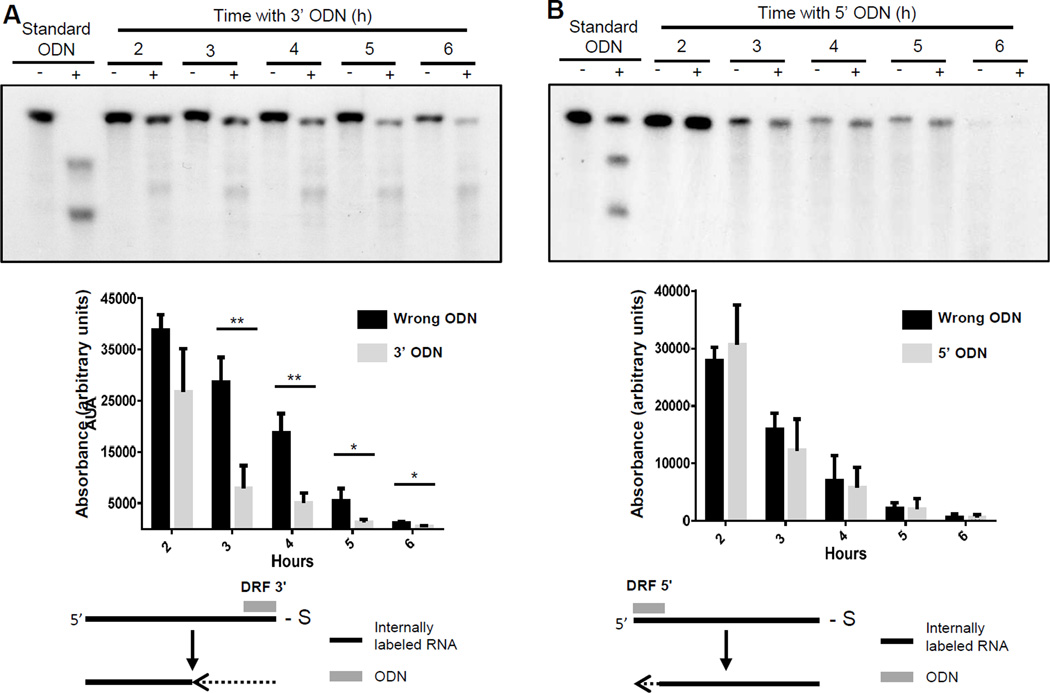
To test the possibility that local RNA features contributed to the loss of the P1 product, we conducted time courses for RNaseH assays using ODNs that annealed at the 3’ or 5’ ends of the DRF+ RNA. This led the larger of the two cleavage products to be as similar as possible to the full-length RNA substrate. As before, incubation of DRF+ RNA in the presence of non-complementary ODNs led to decay of the RNA without appreciable degradation intermediates (Fig. 9A and B). Inclusion of an ODN that bound to the 3’ end of the DNA would result in an endonucleolytic cleavage reaction in which the P1 product (5’ end of the RNA) was not shortened enough to be resolved from the substrate in the gel system employed. When this ODN was used, the P1 RNA decayed significantly faster than in the control reaction in which the non-complementary ODN was employed (Fig. 9A). In contrast, placing the ODN at the 5’ end of the RNA would yield a P2 product (the 3’ RNA fragment) almost the same size as DRF+. In this case, the RNA decayed with the same kinetics as when the non-complementary ODN was employed (Fig. 9B). These results preclude the possibility that local RNA features were responsible for the preferential loss of the P1 product. We conclude that the HBV RNaseH has a processive 3’-5’ exonuclease activity that is stimulated by the endonuclease activity.
Figure 9. Evaluation of directional preferentiality of the HBV RNA exonuclease activity.
A. ODN-directed RNaseH assay employing DRF+ RNA and a ODN that binds to the extreme 3’ end of the RNA. B. ODN-directed RNaseH assay employing DRF+ RNA and an ODN that binds to the extreme 5’ end of the RNA. Top: RNA cleavage assays. Middle: quantification of three replicate assays with error bars representing 1 standard deviation. Bottom: Interpretations. +, complementary ODNs; −, non-complementary control ODNs; standard ODN, an ODN that binds to an internal site in the RNA; 3’ ODN, an ODN that binds to the 3’ end of the RNA; 5’ ODN, ODN that binds to the 5’ end of the RNA; black line, RNA; dashed line, degraded RNA; **, p ≤0.01 by T-test; *, p ≤ 0.05 by T-test.
4. Discussion
The HBV polymerase protein is notoriously difficult to express in recombinant systems, in part due to its existence in an complex with Hsp90 chaperones in its native state (Hu and Boyer, 2006; Hu and Seeger, 1996). Our initial attempts to express active recombinant HBV RNaseH (Tavis et al., 2013a) reflected these difficulties (Fig. 2A). The few other previous reports of recombinant hepadnaviral RNaseH have also not led to enzymes suitable for biochemical characterization or inhibitor screening (Choi et al., 2002; Lee et al., 1997; Potenza et al., 2007; Wei and Peterson, 1996). Here, we found that appending MBP to the N-terminus of the RNaseH led to high yield and good stability of the dual-tagged protein in E. coli (Fig. 2B–D). The specific activity of the purified MBP fusion enzyme was initially barely detectable and was much lower than that of our original recombinant protein, HRHPL (Fig. 2). One of the main contaminants following nickel-affinity purification was DnaK, an Hsp70 chaperone. DnaK could not be removed by extensive washing or with a second step purification on an α-amylose affinity column (Fig. 2B). However, it was removed by adding ATP and Mg2+ to all buffers during purification, presumably by inducing the chaperone’s protein binding and release cycle (Bolanos-Garcia and Davies, 2006). Removing DnaK led to a large increase in the activity and specificity of the RNaseH (Fig. 2E). This increase is consistent either with the bound DnaK directly inhibiting the RNaseH and/or with improving the conformation of the RNaseH through the folding activity of DnaK induced by addition of ATP and Mg2+. Regardless of the mechanism, these improvements enabled biochemical characterization of the HBV RNaseH for the first time.
Varying the structure and length of the oligonucleotide annealed to the RNA revealed that the enzyme requires an RNA:DNA heteroduplex of ≥14 nts long without an internal stem-loop, corresponding to a heteroduplex of about 48Å (Figs. 5 and 6). This is slightly longer than the 13 mer duplexes necessary for cleavage by the human RNaseH1 or the E. coli RNaseH1 (Fig. 5C and D). The identical heteroduplex lengths required for RNA cleavage by the full-length and Δ5 versions of the HBV RNaseH imply that the C-terminal 35 residues do not play a major role in interaction of the enzyme with the substrate. The HBV RNaseH could tolerate a nick but not a gap in the DNA strand (Fig. 6). These experiments also revealed that the HBV RNaseH has little to no intrinsic sequence or positional preference for endonucleolytic RNA cleavage. Five different ODNs placed along the length of the unstructured RNA substrate usRNA1 all supported efficient cleavage of the RNA (Fig. 4), as did six different ODNs complementary to the DRF+ RNA (Figs. 2–9). This lack of sequence specificity is similar to other RNaseHs, such as the E. coli RNaseH1 and human RNaseH1 enzyme. It is also consistent with the biological role of the enzyme in degrading the 3200 nt-long HBV pregenomic RNA after it has been copied into DNA.
A symmetrical pattern of RNA cleavage products was seen when the enzyme was presented with an RNA substrate containing a stem-loop induced by annealing a 24 mer ODN homologous to two discontinuous 12 nt motifs on the DRF+ RNA (Fig. 6 lane 5). This symmetry and the sizes of the products are consistent with cleavage of the RNA at the sharp bends in the RNA. Therefore, the HBV RNase activity is not completely restricted to simple RNA:DNA heteroduplexes. There is no obvious role for this activity during HBV genomic replication (Seeger et al., 2013; Tavis and Badtke, 2009), but as the majority of the HBV polymerase produced in cells exists as a non-encapsidated, membrane-associated protein (Cao and Tavis, 2004; Yao et al., 2000), it is possible that it may cleave structured regions of an unknown cellular RNA(s).
Detailed assessment of the RNA cleavage pattern employing high-resolution electrophoresis revealed that the HBV RNaseH cleaves the minimal 14 mer heteroduplex in more than one location, and that the sizes of the cleavage bands overlap in the products from the smallest of the 3’ and 5’ truncated ODNs (Fig. 7). These data are consistent with the enzyme not needing to be in direct contact with the full 14 mer heteroduplex for cleavage. However, this explanation is unlikely because it does not explain why a 14 mer heteroduplex is needed if the heteroduplex is not fully in contact with the enzyme. The more likely explanation is that the RNaseH can bind to the heteroduplex in both possible orientations relative to the RNA strand (indicated by the black and white ovals in Fig. 7), and that cleavage occurs at a set position relative to the heteroduplex end. Counting the nucleotide positions in Fig.7 implies that the cleavage occurs about 7 nucleotides from the end of the heteroduplex. The possibility of different binding orientations during RNaseH cleavage has been described for the HIV RNaseH (Abbondanzieri et al., 2008; Arnold and Sarafianos, 2008). Although these experiments reveal that the HBV RNaseH probably resembles other RNaseHs in lacking an orientational preference on the heteroduplex, it is improbable that the enzyme acts in both orientations during HBV reverse transcription because the RNaseH is part of the HBV polymerase protein. The polymerase appears to be locked onto the RNA in a defined complex and to act in a processive manner during the coupled reverse transcription/RNaseH reactions (Gong et al., 2001; Radziwill et al., 1988).
The time course experiments in Figs 8 and 9 revealed that the amount of RNA substrate declined during incubation with the HBV RNaseH even in the presence of a non-complementary ODN, that the P1 product formed from the 5’ end of the substrate by endonucleolytic RNaseH reaction was less stable than the P2 product derived from the 3’ end of the RNA, and that this RNA degradation occurred without appreciable accumulation of breakdown intermediates. Together, these observations indicate that the HBV RNaseH has a 3’ to 5’ exonuclease activity in addition to the endonucleolytic activity measured by the ODN-directed RNA cleavage assay used in most of our experiments. The acceleration of the exonuclease activity by the endonuclease reaction is consistent with at least a large fraction of the RNaseH remaining bound to the 5’ fragment, followed by directional exonuclease activity. The relative stability of the 3’ fragment could be due to either failure of the enzyme to remain bound to the 3’ fragment following endonucleolytic cleavage and/or to the absence of a 5’-3’ exonuclease activity. The presence of the 3’ to 5’ exonuclease activity indicates that all RNA digestion following the initial endonucleolytic cleavage of the HBV pregenomic RNA (Seeger et al., 2013; Tavis and Badtke, 2009) could occur by a series of endonucleolytic cuts, by exonucleolytic degradation and/or a combination of both mechanisms. This implies that inhibitor screens designed to detect both activities may provide the best opportunity for identifying clinically useful drugs.
An exoribonuclease activity associated with an RNaseH has been reported for viral RNaseHs, including the HIV (Schatz et al., 1990), herpes simplex virus 1 (Crute and Lehman, 1989), avian myoblastosis virus (Grandgenett and Green, 1974), and bacteriophage T4 (Kholod et al., 2015) enzymes. The HIV reverse transcriptase and RNaseH domains are spatially arranged such that the terminal ribonucleotides cannot be efficiently hydrolyzed because the polymerase domain is covering them when the reverse transcriptase-RNaseH holoenzyme has migrated to the end of the template. The exonuclease activity is believed to eliminate these residues (Hansen et al., 1988; Lapkouski et al., 2013; Schatz et al., 1990; Woehrl and Moelling, 1990), but the mechanism by which the HIV RNaseH exonuclease activity gains access to the residual RNA fragment is incompletely understood.
Production of the HBV RNaseH in high yield, purity, and specific activity has permitted its basic enzymatic characterization for the first time. The enyzme’s lack of sequence or positional specificity and its possession of both endoncleolytic and 3’-5’ exonucleolytic activities are consistent with its known role in HBV reverse transcription. The discovery of a previously unknown ability to cut RNA at sharp bends raises the possibility that the large excess of non-encapsidated HBV polymerase that accumulates in the cytoplasm (Cao and Tavis, 2004; Yao et al., 2000) may regulate cellular function by cleaving a subset of cellular RNAs. Finally, producing large amounts of pure HBV RNaseH and understanding its substrate requirements will facilitate our ongoing efforts to develop a high throughput antiviral drug screening assay similar to the fluorescence-based assays that have been widely used for the HIV enzyme [example in (Wendeler et al., 2008). Such an assay would accelerate the search for clinically relevant inhibitors of the HBV RNaseH that could be added to the current treatment regimens.
Supplementary Material
Highlights.
Appending an MBP tag to the HBV RNaseH domain allows it accumulate to high levels in E. coli.
Appending a HisX6 tag to the RNaseH allows single-stage purification of monomeric soluble protein to homogeneity.
The HBV RNaseH requires a unbulged RNA:DNA heteroduplex of ≥ 14 nt for cleavage.
The enzyme has no strong positional or sequence specificity for NRA cleavage.
The HBV RNaseH has both endoribonucleolytic and 3’-5’ exoribonucleolytic activities.
Acknowledgments
This work was supported by NIH grants R01 AI104494, R03 AI109460, and U01 DK082871. We thank Duane Grandgenett for helpful discussions, Brent Znosko for assistance in designing usRNA1, and David Wood for technical assistance.
Abbreviations
- bp
Base pair
- DEDD
Active center formed by aspartic, glutamic, aspartic and aspartic
- DMSO
Dimethyl sulfoxide
- DnaK
Chaperone E. coli
- DTT
Dithiothreitol
- EDTA
Ethylenediaminetetraacetic acid
- GlmS
Glucosamine-6-phosphate synthase
- gt
Genotype
- HBV
Hepatitis B virus
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HIV
Human immunodeficiency virus
- IPTG
Isopropyl-β-D-thiogalactopyranoside
- MBP
Maltose binding protein
- nt
Nucleotide
- RNaseH
RNA nuclease H
- RT
Reverse transcriptase
- ODN
DNA oligonucleotide
- P1
product 1
- P2
product 2
- S
substrate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
JET is an inventor on US patent application 14/647,331 which covers use of recombinant HBV RNaseH for antiviral screening.
Supplementary Table 1 contains the sequences for the oligonucleotides and RNAs employed in this study.
References
- Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KR, Leksa NC, Schwartz TU. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins: Structure, Function, and Bioinformatics. 2013;81:1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E, Sarafianos SG. Molecular biology: An HIV secret uncovered. Nature. 2008;453:169–170. doi: 10.1038/453169b. [DOI] [PubMed] [Google Scholar]
- Block TM, Gish R, Guo H, Mehta A, Cuconati A, Thomas London W, Guo JT. Chronic hepatitis B: What should be the goal for new therapies? Antiviral Res. 2013;98:27–34. doi: 10.1016/j.antiviral.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Davies OR. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochimica et Biophysica Acta (BBA)-General Subjects. 2006;1760:1304–1313. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Cai CW, Lomonosova E, Moran EA, Cheng X, Patel KB, Bailly F, Cotelle P, Meyers MJ, Tavis JE. Hepatitis B virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) inhibitor of the viral ribonuclease H activity. Antiviral Res. 2014;108:48–55. doi: 10.1016/j.antiviral.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Tavis JE. Detection and characterization of cytoplasmic hepatitis B virus reverse transcriptase. J. Gen. Virol. 2004;85:3353–3360. doi: 10.1099/vir.0.80297-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Marion PL. Amino Acids Essential for RNAse H Activity of Hepadnaviruses Are Also Required for Efficient Elongation of Minus-Strand DNA. J. Virol. 1996;70:6151–6156. doi: 10.1128/jvi.70.9.6151-6156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Robinson WS, Marion PL. Selected Mutations of the Duck Hepatitis B Virus P Gene RNase H Domain Affect both RNA Packaging and Priming of Minus-Strand DNA Synthesis. J. Virol. 1994;68:5232–5238. doi: 10.1128/jvi.68.8.5232-5238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Kim EE, Park YI, Han YS. Expression of the active human and duck hepatitis B virus polymerases in heterologous system of Pichia methanolica. Antiviral Res. 2002;55:279–290. doi: 10.1016/s0166-3542(02)00023-2. [DOI] [PubMed] [Google Scholar]
- Coffin CS, Mulrooney-Cousins PM, Peters MG, van MG, Roberts JP, Michalak TI, Terrault NA. Molecular characterization of intrahepatic and extrahepatic hepatitis B virus (HBV) reservoirs in patients on suppressive antiviral therapy. J. Viral Hepat. 2011;18:415–423. doi: 10.1111/j.1365-2893.2010.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N, Tillmann H. Emerging pipeline drugs for hepatitis B infection. Expert. Opin. Emerg. Drugs. 2011;16:713–729. doi: 10.1517/14728214.2011.646260. [DOI] [PubMed] [Google Scholar]
- Crute J, Lehman I. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5'----3'exonuclease with ribonuclease H activity. Journal of Biological Chemistry. 1989;264:19266–19270. [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. HIVs and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2107–2185. [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N. Engl. J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B Virus Nucleocapsid Envelopment Does Not Occur without Genomic DNA Synthesis. J. Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Yao E, Tavis JE. Evidence that the RNAseH activity of the duck hepatitis B virus is unable to act on exogenous substrates. BMC Microbiology. 2001;1:12. doi: 10.1186/1471-2180-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett DP, Green M. Different mode of action of ribonuclease H in purified alpha and alpha beta ribonucleic acid-directed deoxyribonucleic acid polymerase from avian myeloblastosis virus. The Journal of biological chemistry. 1974;249:5148–5152. [PubMed] [Google Scholar]
- Hansen J, Schultze T, Mellert W, Moelling K. Identification and characterization of HIV-specific RNAseH by monoclonal antibody. EMBO Journal. 1988;7:239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomsky Z, Hostomska Z, Matthews DA. Ribonuclease H. In: Linn SM, Lloyd RS, Roberts RJ, editors. Nulceases. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 341–376. [Google Scholar]
- Hu J, Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis b virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cheng X, Cao F, Huang A, Tavis JE. beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res. 2013;99:221–229. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Kholod N, Sivogrivov D, Latypov O, Mayorov S, Kuznitsyn R, Kajava AV, Shlyapnikov M, Granovsky I. Single substitution in bacteriophage T4 RNase H alters the ratio between its exo-and endonuclease activities. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2015;781:49–57. doi: 10.1016/j.mrfmmm.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Klumpp K, Hang JQ, Rajendran S, Yang Y, Derosier A, Wong KI, Overton H, Parkes KE, Cammack N, Martin JA. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C, Shin YC, Park WJ, Kim S, Kim J, Ryu WS. Residues Arg703, Asp777, and Arg781 of the RNase H domain of hepatitis B virus polymerase are critical for viral DNA synthesis. J Virol. 2014;88:154–163. doi: 10.1128/JVI.01916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Lok AS. Hepatitis B therapy. Nat. Rev. Gastroenterol. Hepatol. 2011;8:275–284. doi: 10.1038/nrgastro.2011.33. [DOI] [PubMed] [Google Scholar]
- Lapkouski M, Tian L, Miller JT, Le Grice SF, Yang W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nature structural & molecular biology. 2013;20:230–236. doi: 10.1038/nsmb.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- Lee IL, Hong YB, Kim Y, Rho HM, Jung G. RNaseH Activity of Human Hepatitis B Virus Polymerase Expressed in Escherichia coli. Biochemical and Biophysical Research Communications. 1997;233:401–407. doi: 10.1006/bbrc.1997.6467. [DOI] [PubMed] [Google Scholar]
- Lima WF, Wu H, Crooke ST. Human RNases H. Methods Enzymol. 2001;341:430–440. doi: 10.1016/s0076-6879(01)41168-2. [DOI] [PubMed] [Google Scholar]
- Loeb DD, Hirsch RC, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Lomonosova E, Cheng X, Moran EA, Meyers MJ, Le Grice SF, Thomas CJ, Jiang JK, Meck C, Hirsch DR, D'Erasmo MP, Suyabatmaz DM, Murelli RP, Tavis JE. Hydroxylated Tropolones Inhibit Hepatitis B Virus Replication by Blocking the Viral Ribonuclease H Activity. Antimicrob Agents Chemother. 2015;59:1070–1079. doi: 10.1128/AAC.04617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellin P, Heathcote EJ, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, Manns M, Kotzev I, Tchernev K, Buggisch P, Weilert F, Kurdas OO, Shiffman ML, Trinh H, Washington MK, Sorbel J, Anderson J, Snow-Lampart A, Mondou E, Quinn J, Rousseau F. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N. Engl. J. Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- Mason WS, Aldrich C, Summers J, Taylor JM. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus strand DNA. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza N, Salvatore V, Raimondo D, Falanga D, Nobile V, Peterson DL, Russo A. Optimized expression from a synthetic gene of an untagged RNase H domain of human hepatitis B virus polymerase which is enzymatically active. Protein Expr. Purif. 2007;55:93–99. doi: 10.1016/j.pep.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Radziwill G, Zentgraf H, Schaller H, Bosch V. The duck hepatitis B virus DNA polymerase is tightly associated with the viral core structure and unable to switch to an exogenous template. Virology. 1988;163:123–132. doi: 10.1016/0042-6822(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Schatz O, Mous J, Le Grice S. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5'exonuclease activity. The EMBO journal. 1990;9:1171. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Philadelphia PA: Lippincott Williams & Wilkins; 2013. pp. 2185–2221. [Google Scholar]
- Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol. Rev. 2006;28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- Sorrell MF, Belongia EA, Costa J, Gareen IF, Grem JL, Inadomi JM, Kern ER, McHugh JA, Petersen GM, Rein MF, Strader DB, Trotter HT. National Institutes of Health Consensus Development Conference Statement: management of hepatitis B. Ann. Intern. Med. 2009;150:104–110. doi: 10.7326/0003-4819-150-2-200901200-00100. [DOI] [PubMed] [Google Scholar]
- Tavis JE, Badtke MP. Hepadnaviral Genomic Replication. In: Cameron CE, Gotte M, Raney KD, editors. Viral Genome Replication. New York: Springer Science+Business Media, LLC; 2009. pp. 129–143. [Google Scholar]
- Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, Sarafianos SG. The hepatitis B virus ribonuclease h is sensitive to inhibitors of the human immunodeficiency virus ribonuclease h and integrase enzymes. PLoS pathogens. 2013a;9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Gehring AJ, Hu Y. How further suppression of virus replication could improve current HBV treatment. Expert review of anti-infective therapy. 2013b;11:755–757. doi: 10.1586/14787210.2013.814846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Lomonosova E. The Hepatitis B Virus Ribonuclease H as a Drug Target. Antiviral Res. 2015;118:132–138. doi: 10.1016/j.antiviral.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bommel F, De Man RA, Wedemeyer H, Deterding K, Petersen J, Buggisch P, Erhardt A, Huppe D, Stein K, Trojan J, Sarrazin C, Bocher WO, Spengler U, Wasmuth HE, Reinders JG, Moller B, Rhode P, Feucht HH, Wiedenmann B, Berg T. Long-term efficacy of tenofovir monotherapy for hepatitis B virus-monoinfected patients after failure of nucleoside/nucleotide analogues. Hepatology. 2010;51:73–80. doi: 10.1002/hep.23246. [DOI] [PubMed] [Google Scholar]
- Wei X, Peterson DL. Expression, purification, and characterization of an active RNase H domain of the hepatitis B viral polymerase. J. Biol. Chem. 1996;271:32617–32622. doi: 10.1074/jbc.271.51.32617. [DOI] [PubMed] [Google Scholar]
- Wendeler M, Lee HF, Bermingham A, Miller JT, Chertov O, Bona MK, Baichoo NS, Ehteshami M, Beutler J, O'Keefe BR, Gotte M, Kvaratskhelia M, Le GS. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem. Biol. 2008;3:635–644. doi: 10.1021/cb8001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehrl BM, Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- Woo G, Tomlinson G, Nishikawa Y, Kowgier M, Sherman M, Wong DK, Pham B, Ungar WJ, Einarson TR, Heathcote EJ, Krahn M. Tenofovir and entecavir are the most effective antiviral agents for chronic hepatitis B: a systematic review and Bayesian meta-analyses. Gastroenterology. 2010;139:1218–1229. doi: 10.1053/j.gastro.2010.06.042. [DOI] [PubMed] [Google Scholar]
- Wursthorn K, Jung M, Riva A, Goodman ZD, Lopez P, Bao W, Manns MP, Wedemeyer H, Naoumov NV. Kinetics of hepatitis B surface antigen decline during 3 years of telbivudine treatment in hepatitis B e antigen-positive patients. Hepatology. 2010;52:1611–1620. doi: 10.1002/hep.23905. [DOI] [PubMed] [Google Scholar]
- Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- Yao E, Gong Y, Chen N, Tavis JE. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J. Virol. 2000;74:8648–8657. doi: 10.1128/jvi.74.18.8648-8657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tavis JE. The Duck Hepatitis B Virus Reverse Transcriptase Functions as a Full-length Monomer. J. Biol. Chem. 2006;281:35794–35801. doi: 10.1074/jbc.M608031200. [DOI] [PubMed] [Google Scholar]
- Zoulim F. Antiviral therapy of chronic hepatitis B: can we clear the virus and prevent drug resistance? Antivir. Chem. Chemother. 2004;15:299–305. doi: 10.1177/095632020401500602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.