Abstract
MicroRNAs (miRNAs) have recently emerged as a novel class of epigenetic regulators of gene expression. They are systemically involved in the control of lipid metabolism through a complex interactive mechanism that involves gene regulatory networks. Hence, they can contribute to defective lipid metabolism and metabolic diseases. Here, we review recent advances in the roles of lipid-sensing transcription factors in regulating miRNA gene networks, as well as miRNA expression and function in the regulation of cholesterol metabolism.
Keywords: MicroRNA, Cholesterol metabolism, Transcription factor, Metabolic disease
1. Introduction
As one of the two major cellular lipid species, cholesterol is an essential structural component of eukaryotic cell membranes. It is a precursor for the synthesis of steroid hormones and bile acids, which are natural detergents and participate as signaling molecules that contribute to a wide variety of cellular functions [1,2]. Given these diverse yet fundamental roles, it is not surprising that small perturbations in regulatory networks that modulate cholesterol homeostasis can lead to severe clinical conditions, such as atherosclerosis, heart disease, and diabetes [3]. Hence, exquisite regulatory mechanisms have evolved to balance both intracellular and membrane levels of cholesterol in response to changing physiologic demand. Cholesterol homeostasis is maintained through the feedback regulation of de novo biosynthesis, lipoprotein uptake, sterol mobilization through lipophagy, and efflux/transport, which are regulated at the transcriptional and posttranscriptional levels [4]. A key transcriptional pathway for regulating cholesterol metabolism is through the sterol regulatory element-binding proteins (SREBPs). There are three major SREBP isoforms; SREBP-1a and 1c are encoded from the SREBF1 gene on chromosome 11 from 2 separate promoters to generate mRNAs with unique 5′ termini that encode proteins that only differ at their amino-temini. A singular SREBP-2 is encoded from the SREBF2 gene that is encoded on chromosome 15 [4]. SREBPs coordinately regulate cholesterol and fatty acid biosynthesis and the published data suggest that SREBP-1c, which is a relatively weak transcriptional activator, preferentially activates genes involved in fatty acid metabolism whereas SREBP-1a activates genes of both cholesterol and fatty acid metabolism. SREBP-2 preferentially activates genes of cholesterol metabolism [5].
The different SREBP isoforms are independently regulated at the transcriptional level through different tissue-specific signaling pathways and the proteins are expressed as large 1100 amino acid precursors that are tethered to the endoplasmic reticulum (ER) membrane. When cellular sterol levels are low, the full-length endoplasmic reticulum (ER) membrane-associated SREBP precursor forms a complex with SREBP cleavage-activating protein (SCAP) and the complex trafficks to the Golgi apparatus where the amino-terminal mature SREBP transcription factor is proteolytically released from its membrane tether. The active transcription factor then translocates to the nucleus, where it induces the transcription of numerous genes involved in cholesterol biosynthesis, lipoprotein uptake, and lipophagy [6]. Evidence suggests that ER to golgi trafficking of all SREBPs is regulated by cholesterol but additional signals including polyunsaturated fatty acids also regulate the maturation of SREBP-1 isoforms [4].
A thorough review of the exquisite feedback mechanism for SREBP processing and nuclear translocation is beyond the scope of the current review and can be found elsewhere [4]. Instead, this review is focused on the emerging roles for micro-RNAs (miRNAs) in regulating cholesterol metabolism. miRNAs are a unique class of small noncoding RNAs that function as key regulators of fundamental cellular processes that modulate protein expression by hybridizing to their respective mRNA targets and increasing mRNA turnover or inhibiting translation or both [7]. miRNAs have been shown to regulate key proteins of cholesterol homeostasis, and altered expression of the implicated miRNAs is highly associated with metabolic disorders [3]. Studies have revealed that single miRNAs cannot only have multiple target sites in the 3′-un-translated region (UTR) of an mRNA, but a single mRNA molecule is also predicted to be the target of many distinct miRNAs, suggesting that miRNAs act in a concerted manner to regulate gene expression [8]. Thus, this posttranscriptional regulation action by miRNAs represents another critical layer of intricate regulatory networks, in addition to the complex regulatory layer of transcription factors and co-activators, for maintaining the intracellular cholesterol homeostasis effectively.
2. Basic biogenesis and transcription of miRNA
In mammalian miRNA biogenesis, most miRNA genes are transcribed into long primary miRNA transcripts (pri-miRNA, often several kb long) by RNA polymerase II (RNA Pol II) then capped, polyadenylated, and spliced to generate a pre-miRNA [9]. Some miRNAs are also transcribed by pol III [10]. The transient pre-miRNA intermediate is then cleaved into a hairpin-containing stem-loop precursor (pre-miRNAs, ~70–90 nucleotides [nt]) by the nuclear RNase-III DROSHA. This is followed by export to the cytoplasm, which is mediated by EXPORTIN-5, a Ran-GTP-dependent nuclear transport receptor, and further processing occurs by the cytosolic DICER, another RNase-III related enzyme, to generate the mature miRNA (~22 nt). Mature miRNAs assemble with an Argonaute family protein (AGO) to form the RNA-induced-silencing-complex (RISC), which then binds to specific sites usually within the 3'UTR of protein-coding mRNA transcripts by complementary base-pairing through a small “seed” region in the miRNA followed by enhanced mRNA turnover and/or translational repression [11].
Current mammalian genome annotation records suggest that miRNA genes are one of the most abundant gene classes, many of which are highly conserved even between distantly related species, indicating miRNAs have important roles throughout animal evolution. miRNAs are classified either as intragenic (intronic or exonic) or intergenic, based on their genomic location. Intergenic miRNAs are transcribed from their own promoters, whereas, intragenic miRNAs can be expressed from their own promoters or processed from the primary transcript of the host gene and are dependent on the transcriptional regulatory mechanisms that govern expression of the host gene. In the latter case, there is a strong correlation between expression of the host mRNA and the intragenic miRNA expression [12,13].
miRNAs gene expression is controlled through tissue-specific or developmental-stage-specific processes [14]. Since transcription of most miRNA genes is mediated by RNA Pol II, their promoters are perceived to have sequence features and transcriptional regulatory mechanisms like protein-coding genes [15]. Indeed, high throughput sequencing and bioinformatics approaches have revealed that the proximal upstream region of miRNA genes has a TATA box, initiator element, TFIIB recognition element (BRE), and a large number of transcription factor-binding motifs [12,16,17]. Moreover, recent studies have predicted miRNA promoter regions close to transcription start sites (TSSs) by the presence of CpG islands and the trimethylation of Lys4 of histone 3 (H3Kme3), suggesting the contribution of epigenetic mechanisms, such as DNA methylation and histone modification to the regulation of miRNA expression [12,18]. More detailed information on alternative promoters, splicing, and processing of miRNA genes are required for a better understanding of the diverse structures, functions, and biogenesis of miRNA genes.
3. Roles of miRNAs in cholesterol biosynthesis
3.1. Regulatory loop for cholesterol homeostasis: SREBP and miRNAs
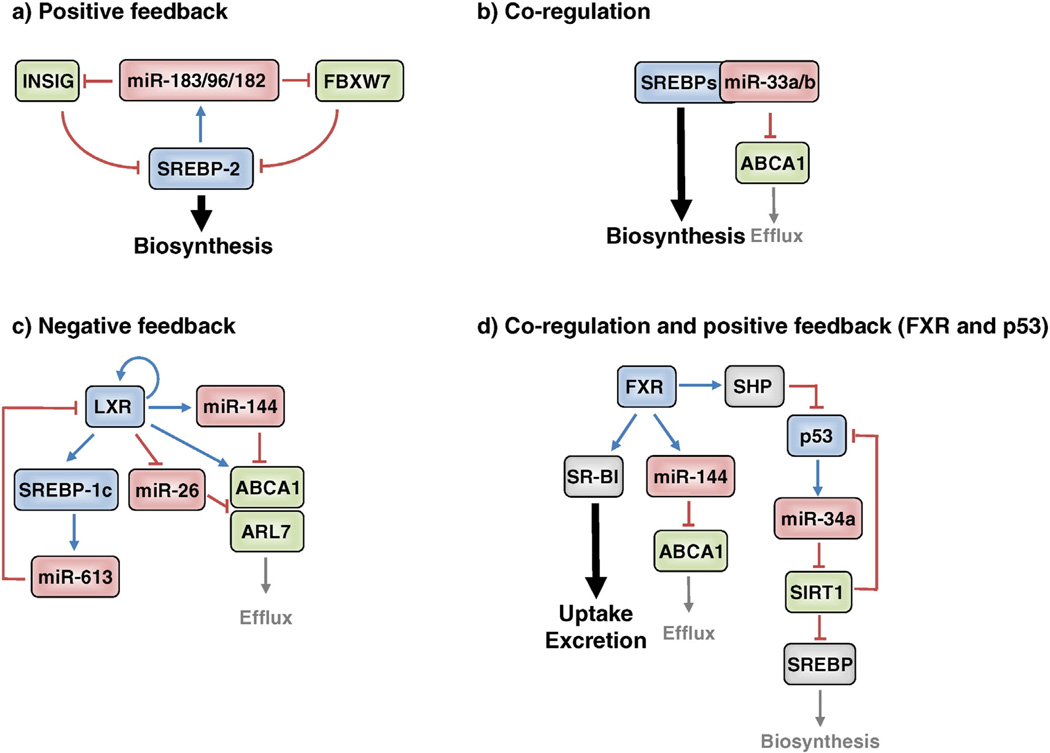
As mentioned above, SREBP-1a and SREBP-1c modulate the expression of genes involved in energy metabolism, including fatty acid and glucose metabolism, whereas SREBP-2 is more specific to the transcriptional regulation of cholesterol metabolism genes [4]. The inactive SREBP precursor in the ER interacts directly with SCAP, which is the primary membrane cholesterol sensor. When the ER membrane is above a threshold level of 5 mol/%, SCAP binds directly to cholesterol and maintains a conformation that binds a third protein, INSIG, and this interaction traps SREBP in the ER membrane. However, when the ER membrane cholesterol falls below 5%, the conformation of SCAP changes and no longer interacts with INSIG [19]. This allows SCAP to interact with the COP II trafficking complex which moves the SCAP-SREBP complex to the golgi apparatus where a concerted two-step proteolytic attack by site-1 and site-2 proteases (S1P and S2P) on SREBP releases the amino-terminal domain representing the mature transcription factor that migrates to the nucleus and activates the expression of SREBP target genes. Nuclear SREBPs are unstable and are degraded by the ubiquitin-proteasome system via targeting of the E3 ubiquitin ligase F-box and WD repeat domain containing 7 (FBXW7) [6]. Recently, our group revealed that hepatic expression of the miR-183/96/182 operon is directly activated by SREBP-2 in mice and human cells. SREBP-2 interacts with a conserved binding site (E-box) that is located near the TSS for the miR-183/96/182 transcription unit and promotes polycistronic transcription of the three miRs. miR-182 targets FBXW7 and miR-96 targets INSIG2 thereby creating a positive-feedback loop to regulate SREBP activity for cholesterol homeostasis [20,21] (Fig. 1a).
Fig. 1.
Transcription factors and miRNAs regulatory network in cholesterol homeostasis. a. SREBP-2 increased miR-183/96/182 cluster that targets INSIG2 and Fbxw7, constituting a positive-feedback loop to regulate SREBP activity. b. miR-33a/b locate within the introns of SREBP genes, and these transcripts coordinate to regulate cholesterol transport and synthesis/uptake to maintain cellular cholesterol homeostasis. c. LXR downregulates miR-26 that targets ABCA1 and ARL7, enhancing cholesterol transport, whereas miR-144 and miR613 are upregulated by LXR and target ABCA1 and LXR, suggesting the presence of negative-feedback regulation in the cholesterol efflux process. d. FXR-regulated miR-144 suppresses hepatic ABCA1 and plasma HDL-C levels, but increased hepatic SR-BI enhances biliary excretion in the RCT process. FXR also inhibits miR-34a through SHP activation that inactivates p53, which results in a positive regulation of SIRT1.
Another recent report suggests that miR-24 targets INSIG1 which would also have a positive effect on SREBP levels [22] but how regulation of miR-24 might be linked to SREBP homeostasis remains to be determined. Even so, the authors showed that antagonizing miR-24 in diet-induced obese mice significantly reduced plasma and hepatic lipid levels through down-regulation of lipogenic gene expression. Higher levels of miR-24 and lower levels of INSIG1 were also observed in non-alcoholic fatty liver disease (NAFLD) or steatohepatitis (NASH) patients [23,24], suggesting that crosstalk between miR-24 and INSIG1 may be important for controlling lipid homeostasis in metabolic diseases [22].
Expression of another microRNA, miR-185, is regulated by SREBP-1c, which binds to a specific motif within the promoter region of miR-185. In this case, gain-of-function results in decreased expression of cholesterol metabolism genes, such as hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and low density lipoprotein receptor (LDLR) by targeting the 3'UTR of SREBP-2 both in vitro and in vivo. SREBP-1c mainly regulates the production of fatty acids that are needed for cholesteryl ester (CE) synthesis, suggesting that the SREBP-1c/miR-185 axis constitutes a cholesterol-responsive feedback loop that maintains the ratio of free cholesterol and CE [25]. Further studies are needed for understanding the physiological role of miR-185 in metabolic diseases.
3.2. Other miRNAs in cholesterol biosynthesis
Knockdown studies on the liver-restricted miRNA, miR-122, suggests that it has a major effect on lipogenic and fatty acid oxidation genes, thereby causing reduction in plasma and hepatic cholesterol and triglyceride levels in mice [26,27]. Follow-up studies also showed that reduced miR-122 reduced circulating cholesterol levels in non-human primates [28]. Thus, miR-122 is considered a potential novel lipid-lowering agent suggesting it may be a therapeutic target. However, another study showed that miR-122 expression is significantly downregulated in NASH patients, and inhibition of miR-122 promotes the expression of fatty acid and cholesterol biosynthesis genes in human liver cell lines [23]. These seemingly conflicting studies are likely due to the fact that miR-122 is one of the most highly expressed miRNAs in liver, and likely has major effects on many pathways that are critical to normal hepatic function.
miR-34a levels are elevated in livers of humans with NASH and this is associated with increased SREBP-2 and HMGCR [29]. This may be through miR34a regulation of sirtuin-1 NAD-dependent deacetylase (SIRT1) as an earlier study reported that miR-34a also suppresses SIRT1 [30] which could lead to stabilization of SREBP-2 as reported previously [31]. Lee et al. [32] reported that the farnesoid X receptor (FXR) inhibits miR-34a expression through activation of the transcriptional repressor, small heterodimer partner (SHP), which results in regulation of SIRT1. Elevated p53 and miR-34a are associated with suppressed SIRT1 in NAFLD [33,34]. Ursodeoxycholic acid is a natural bile acid that reduces lipotoxicity and NAFLD possibly through activation of the bile acid receptor FXR, and Castro et al. reported that this may be through effects of FXR on the miR-34a/SIRT1/p53 axis [35] (Fig. 1d). Hence, upregulation of miR-34a may contribute to the development and progression of hepatosteatosis, suggesting anti miR-34a compounds may be beneficial for NAFLD and NASH.
Recent studies have revealed that miR-223 transcription is increased by cellular cholesterol levels. Overexpression of miR-223 in human liver cell lines suppresses cholesterol uptake and biosynthesis by inhibiting the scavenger receptor BI (SR-BI), hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), and methylsterol monooxygenase 1 (SC4MOL), as well as enhances cholesterol efflux by promoting ABCA1. Moreover, miR-223 knockout mice exhibit high plasma and hepatic cholesterol levels by inducing cholesterol biosynthesis genes, including HMGCS1 [36]. These results suggest that miR-223 plays a critical role in the regulation of systemic cholesterol homeostasis.
4. miRNA regulation of reverse cholesterol transport
4.1. Cholesterol efflux and miRNAs
In addition to biosynthesis, regulated efflux of excess cholesterol is also essential for maintaining cholesterol homeostasis. The cholesterol transporter, ATP binding cassette transporter A1 (ABCA1) promotes cholesterol efflux to extracellular acceptors such as lipid-poor apolipo-proteins A-I (APOA1) to form nascent high-density lipoprotein (HDL) particles. This process is the initial step in reverse cholesterol transport (RCT), a process by which excess cholesterol is transported from peripheral cells, such as macrophages to the liver for excretion into bile and feces [37]. Mutation of the ABCA1 gene causes Tangier disease, which is characterized by a physiological absence of HDL-cholesterol (HDL-C) [38], and inactivation of hepatic ABCA1 leads to a severe reduction of total HDL-C [39] in mice. Since plasma HDL-C levels are inversely related to cardiovascular disease (CVD), ABCA1-mediated cholesterol efflux is critical in the maintenance of cholesterol homeostasis and protection against atherosclerosis. Recent studies have demonstrated that several miRNAs target ABCA1, thereby regulating plasma levels of HDL-C. Among them, the most extensively studied miRNA, miR-33a, is located within an intron of the SREBF2 gene, and because of co-expression, miR-33a and SREBP-2 function coordinately to regulate cholesterol transport and synthesis/uptake to maintain cellular cholesterol homeostasis [40–44] (Fig. 1b). Moreover, inhibition of miR-33a leads to a significant increase in hepatic ABCA1 expression, thereby increasing circulating HDL-C in mice [40–43,45]. miR-33a is highly conserved across species, whereas miR-33b, which is encoded within an intron of the human SREBF1 gene is absent in rodents. Since hyperinsulinemia caused by insulin resistance persistently induces SREBP-1c expression, miR-33b would markedly increase under these conditions in humans. This could contribute to hyperlipidemia or CVD through dysregulation of lipid biosynthesis and cholesterol efflux. Indeed, both miR-33b and SREBP-1c are significantly increased in insulin resistant non-human primates [46], and miR-33 antagonism remarkably lowers VLDL and increases HDL through modulation of miR-33 target genes, including ABCA1 in these animals [46,47]. Although the effect of antagonists on glucose homeostasis or insulin sensitivity has not been observed, these results suggest a therapeutic potential for miR-33a/b targeting in the treatment of human metabolic disease. The roles of miR-33a and miR-33b are discussed in detail in other companion reviews in this volume.
miR-758 was also identified as a post-transcriptional regulator of ABCA1 by bioinformatics-based target prediction tools and genome-wide miRNAs analysis. This miRNA, similar to the regulation of miR-33a/b, is significantly downregulated by a cholesterol overload, increasing cholesterol efflux to APOA1 in macrophages, hepatocytes, and astrocytes [48]. In the brain, ABCA1 regulates cholesterol homeostasis and apolipoprotein metabolism, and is highly associated with the pathogenesis of Alzheimer's disease (AD) [49]. ABCA1 deficiency increases accumulation of amyloid β (Aβ), which leads to AD, and a Liver X receptor (LXR) agonist treatment reduced Aβ deposition at least partly through ABCA1 induction in mice [50]. However, human studies on the association of ABCA1 function and the risk of AD are complicated and have provided conflicting results [51]. Another recent study reported that miR-106b regulates Aβ metabolism by targeting ABCA1 in neuronal cells, but the physiological or pathological relevance of miR-106b remains to be determined [52]. Interestingly, miR-33a/b, miR-758, and miR-106b are all highly expressed in the brain, suggesting that these miRNAs may coordinately target ABCA1 regulation of neuronal cholesterol efflux with significant physiological effects on neuronal development and disease.
LXRs are master regulators of cholesterol metabolism because they activate many genes involved in cholesterol transport including ABCA1, ABCG1, ABCG5, and ABCG8 in response to endogenous oxysterol ligands [53]. A recent study showed that part of this regulation is through miR-26 which is activated by oxysterols and targets ABCA1 and the intracellular cholesterol trafficking protein ADP-ribosylation factor-like 7 (ARL7), in macrophages [54]. Conversely, miR-144 is up-regulated by LXR under similar conditions and targets ABCA1, suggesting the presence of negative-feedback regulation in the cholesterol efflux process [55]. LXR upregulation of hepatic miR-613 involves a negative auto-regulatory feedback loop by targeting the 3'UTR of LXR [56] (Fig. 1c). Interestingly, the activation of nuclear receptor FXR promotes miR-144 expression, which controls genes of bile acid synthesis, excretion, and transport. miR-144 also suppresses hepatic ABCA1 and reduces plasma HDL-C levels. Treatment with an FXR agonist also increases the expression of hepatic SR-BI, which mediates uptake of HDL-C into the liver, thereby repressing plasma HDL-C, suggesting that hepatic FXR induction of miR-144 is a complementary pathway for efficient channeling of cholesterol into bile in the RCT process [57] (Fig. 1d).
4.2. Uptake of HDL-C and miRNAs
SR-BI, is highly expressed in liver as mentioned above and in steroidogenic tissues, where it delivers cholesterol derived from HDL-C for steroid hormone synthesis [58]. Although low plasma HDL-C level is a hallmark of atherosclerosis, the benefit of raising HDL-C is still controversial. Indeed, hepatic overexpression of SR-BI in mice reduces atherosclerosis despite extremely low levels of plasma HDL-C. Gain-of-function hepatic SR-BI increases selective uptake of HDL-C and increases levels of remnant HDL particles that could be more efficient acceptors of cholesterol efflux from macrophages, thereby increasing the cholesterol excretion in bile and the overall rate of macrophage cholesterol efflux. On the other hand, SR-BI knockout mice exhibit markedly increased HDL-C levels due to impaired hepatic catabolism of HDL cholesteryl ester, but these mice are more susceptible to atherosclerosis in APOE- or LDLR-deficient mice because of low rates of macrophage RCT [59, 60]. These results suggest that hepatic SR-BI can influence overall rates of RCT, and regulation of its expression is significantly important in the maintenance of cholesterol homeostasis. Recent studies indicate that miR-185, miR-96, and miR-223 all repress SR-BI and HDL-C uptake by coordinately targeting its 3'UTR in human hepatic cell lines. Moreover, hepatic expressions of miR-96 and miR-185 were significantly downregulated in high-fat diet-fed APOE knockout mice, whereas SR-BI expression was increased, suggesting that both miRNAs may regulate plasma HDL-C levels and RCT [61]. Consistent with this finding, our recent study showed a dramatic downregulation of miR-96 in livers of high cholesterol diet-fed mice, and thus repression of SREBP-2-dependent cholesterol synthesis [20]. Therefore, miR-96 inhibition could potentially result in a reversal of atherogenic progression through suppression of cholesterol synthesis and activation of RCT. Other studies showed that miR-125a, miR-455, and SR-BI are inversely regulated by the adrenocorticotropic hormone (ACTH) in steroidogenic cells, and gain-of-function for both miR-125a, miR-455 blunts the ACTH dependent stimulation of SR-BI expression and HDL-C uptake, and thus attenuates HDL-stimulated progesterone production in steroidogenic cells [62].
5. Conclusion
miRNAs regulate cholesterol homeostasis by coordinating pathways involved in, de novo biosynthesis, lipoprotein uptake, free cholesterol efflux, and biliary excretion (Fig. 1 and Table 1). Several miRNAs are involved in this pathway coordination so dysregulation of miRNA expression could contribute significantly to the pathogenesis of cholesterol metabolism disorders. In fact, the recent evidence reviewed here points to the existence of co-regulatory networks of transcription factors and miRNAs that balance cholesterol metabolism. In this regard, intracellular sterol sensors, such as SREBPs, LXR, and FXR have been identified as factors that directly regulate transcription of miRNAs, and many of these participate in the maintenance of cholesterol homeostasis by feedback or feedforward regulatory circuits. Understanding these transcription factor-miRNA co-regulatory networks will reveal complex homeostatic mechanisms and help more fully understand how they contribute to metabolic diseases.
Table 1.
miRNAs targeting genes of cholesterol homeostasis.
| Biosynthesis | Efflux | Uptake | Trafficking |
|---|---|---|---|
| miRNA Target gene | miRNA Target gene | miRNA Target gene | miRNA Target gene |
|
|
|
|
Acknowledgments
We thank Ryan Esquejo for comments on the manuscript. Studies in T.O.'s lab are supported by the NIH (HL48044) and studies in T.I.J.’s lab are supported by Chonnam National University, 2012, and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2014R1A1A2057994).
Abbreviations
- miRNA
microRNA
- SREBP
sterol regulatory element-binding protein
- SCAP
SREBP cleavage-activating protein
- Ago
Argonaute family protein
- RISC
RNA-induced-silencing-complex
- Fbxw7
F-box and WD repeat domain containing 7
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- HMGCR
hydroxy-3-methylglutaryl-CoA reductase
- LDLR
low density lipoprotein receptor
- CE
cholesteryl ester
- FXR
farnesoid X receptor
- SHP
small heterodimer partner
- SR-BI
scavenger receptor BI
- HMGCS1
hydroxy-3-methylglutaryl-CoA synthase 1
- SC4MOL
methylsterol monooxygenase 1
- ABCA1
ATP binding cassette transporter A1
- ApoAI
apolipoproteins A-I
- HDL
high-density lipoprotein
- RCT
reverse cholesterol transport
- Aβ
amyloid β
- LXR
Liver X receptor
- ARL7
ADP-ribosylation factor-like 7
- PPAR
peroxisome proliferator-activated receptors
- LRH-1
liver receptor homolog 1
- ACTH
adrenocorticotropic hormone
Footnotes
Conflict of interest
We have no conflicts of interest to disclose.
Transparency document
The Transparency document associated with this article can be found in the online version.
References
- 1.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 2.Luu W, Sharpe LJ, Gelissen IC, Brown AJ. The role of signalling in cellular cholesterol homeostasis. IUBMB Life. 2013;65:675–684. doi: 10.1002/iub.1182. [DOI] [PubMed] [Google Scholar]
- 3.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 6.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 11.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 12.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput. Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, Osborne TF. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao J, Ling AV, Manthena PV, Gearing ME, Graham MJ, Crooke RM, Croce KJ, Esquejo RM, Clish CB, G. Morbid Obesity Study. Vicent D, Biddinger SB. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Commun. 2015;6:6498. doi: 10.1038/ncomms7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng R, Wu H, Xiao H, Chen X, Willenbring H, Steer CJ, Song G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology. 2014;60:554–564. doi: 10.1002/hep.27153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith EM, Zhang Y, Baye TM, Gawrieh S, Cole R, Blangero J, Carless MA, Curran JE, Dyer TD, Abraham LJ, Moses EK, Kissebah AH, Martin LJ, Olivier M. INSIG1 influences obesity-related hypertriglyceridemia in humans. J. Lipid Res. 2010;51:701–708. doi: 10.1194/jlr.M001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M, Liu W, Pellicane C, Sahyoun C, Joseph BK, Gallo-Ebert C, Donigan M, Pandya D, Giordano C, Bata A, Nickels JT., Jr Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J. Lipid Res. 2014;55:226–238. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 27.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:U896–U810. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 29.Min HK, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, Kellum J, Warnick R, Contos MJ, Sanyal AJ. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yahagi N, Shimano H, Matsuzaka T, Sekiya M, Najima Y, Okazaki S, Okazaki H, Tamura Y, Iizuka Y, Inoue N, Nakagawa Y, Takeuchi Y, Ohashi K, Harada K, Gotoda T, Nagai R, Kadowaki T, Ishibashi S, Osuga J, Yamada N. p53 involvement in the pathogenesis of fatty liver disease. J. Biol. Chem. 2004;279:20571–20575. doi: 10.1074/jbc.M400884200. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhary N, Pfluger PT. Metabolic benefits from Sirt1 and Sirt1 activators. Curr. Opin. Clin. Nutr. Metab. Care. 2009;12:431–437. doi: 10.1097/MCO.0b013e32832cdaae. [DOI] [PubMed] [Google Scholar]
- 35.Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 2013;58:119–125. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, Sethupathy P, Remaley AT. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14518–14523. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 39.Timmins JM, Lee JY, Boudyguina E, Kluckman KD, Brunham LR, Mulya A, Gebre AK, Coutinho JM, Colvin PL, Smith TL, Hayden MR, Maeda N, Parks JS. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin Invest. 2005;115:1333–1342. doi: 10.1172/JCI23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci. Transl. Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirsch-Reinshagen V, Wellington CL. Cholesterol metabolism, apolipoprotein E, adenosine triphosphate-binding cassette transporters, and Alzheimer's disease. Curr. Opin. Lipidol. 2007;18:325–332. doi: 10.1097/MOL.0b013e32813aeabf. [DOI] [PubMed] [Google Scholar]
- 50.Donkin JJ, Stukas S, Hirsch-Reinshagen V, Namjoshi D, Wilkinson A, May S, Chan J, Fan J, Collins J, Wellington CL. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J. Biol. Chem. 2010;285:34144–34154. doi: 10.1074/jbc.M110.108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitali C, Wellington CL, Calabresi L. HDL and cholesterol handling in the brain. Cardiovasc. Res. 2014;103:405–413. doi: 10.1093/cvr/cvu148. [DOI] [PubMed] [Google Scholar]
- 52.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases abeta levels by repressing ABCA1 expression. Exp. Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 55.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suarez Y, Fernandez-Hernando C. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ou Z, Wada T, Gramignoli R, Li S, Strom SC, Huang M, Xie W. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Mol. Endocrinol. 2011;25:584–596. doi: 10.1210/me.2010-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldan A, Esau C, Edwards PA. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 2013;112:1602–1612. doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 59.Rader DJ, Alexander ET, Weibel GL, Billheimer J, Rothblat GH. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 2009;50(Suppl):S189–S194. doi: 10.1194/jlr.R800088-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Da Silva JR, Reilly M, Billheimer JT, Rothblat GH, Rader DJ. Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J. Clin. Invest. 2005;115:2870–2874. doi: 10.1172/JCI25327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol. Cell. Biol. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]