Abstract
Six lactic acid bacterial (LAB) strains were isolated from traditionally fermented Xinjiang cheese and evaluated for functional and probiotic properties and potentials as starter cultures. The isolated six LAB strains comprised Lactobacillus rhamnosus (one strain), Lactobacillus helveticus (one strain), and Enterococcus hirae (four strains). All of the six strains were tolerant to acidic and bile salt conditions. Among which, the L. rhamnosus R4 strain showed more desirable antimicrobial, auto-aggregation, and hydrophobic activity. In addition, the strain L. rhamnosus R4 exhibited the highest level of free radical scavenging activity (53.78% of 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radicals and 45.79% of hydroxyl radicals). L. rhamnosus R4 also demonstrated cholesterol and triglyceride degradation by 50.97% and 28.92%, respectively. To further examine the health-promoting effects of these LAB strains on host lifespan, Caenorhabditis elegans was used as an in vivo model. Worms fed LAB as a food source had significant differences in lifespan compared to those fed Escherichia coli OP50 (as a negative control). Feeding of L. rhamnosus R4 extended the mean lifespan of C. elegans by up to 36.1% compared to that of the control. The results suggest that the strains isolated from Xinjiang fermented dairy products have high potential as starter cultures in the cheese industry.
Keywords: Xinjiang cheese, Lactic acid bacterial (LAB) strains, Probiotic properties, Caenorhabditis elegans, Lifespan
1. Introduction
Lactic acid bacterial (LAB) strains belong to beneficial bacteria. They are normal inhabitants of the healthy gut microbiota and present in numerous fermented dairy products such as cheese and fermented milk. Some like Lactobacillus, Streptococcus, Pediococcus, and Enterococcus are cultivable and predominant microbes in fermented dairy products, and exert a positive role in human health following oral administration. They improve the balance of the microbial community in the intestine, confer protection against potential pathogenic bacteria, and prevent and/or cure intestinal diseases (Schiffrin et al., 1997; Gionchetti et al., 2000). These effects are mediated by production of antimicrobial metabolites such as organic acids (for example lactate, acetate, and butyrate), hydrogen peroxide, and bacteriocins, and competition with harmful bacteria for nutrients or adhesion receptors (Hudault et al., 1997; Fons et al., 2000). The therapeutic effects of the beneficial microorganisms include, but are not limited to, enhanced immune function, maintenance of anti-tumor activity, reduction of the population of harmful microorganisms, improvement of the balance of microflora in the colon, prevention against some intestinal infection, increased tolerance to lactose-containing foods, and possibly prevention of cancer (Martini et al., 1987; Wang et al., 2010; Amadou et al., 2013).
The local fermented dairy products in Xinjiang Uygur Autonomous Region of China are made by nomadic people with a natural fermentation process without any starter culture. The production of Xinjiang cheese includes fermenting, drying, and forming into blocks. One of the oldest and most popular cheeses in Xinjiang is the Kurut. As a semi-hard regional cheese, the production environment of Kurut is very similar to that of a Polish cheese, Oscypek (Alegria et al., 2012), which is frequently produced not with the use of starter cultures, but only by the autochthonous microbiota present in milk and the environment. Kurut is manufactured from unpasteurized cow’s milk, and fermented spontaneously by microorganisms from the air, raw milk, and the equipment used during the production. The main process of Kurut production is to increase the acidity of milk by cold maturation at room temperature. The soured milk is then mixed with sweet milk and rennet is added. The formed coagulum is beaten and settled in order to remove the whey after coagulation. The cheese knots are then pressed into forms and smoothed. Finally the cheese is dried with facultative cold-smoking (Sip et al., 2012).
Xinjiang cheese is often a dry cheese with a rough skin. The texture can be hard, semi-hard, or soft with a sour or sweet taste. The sweet type is made of milk fat with much aroma while the sour type is less fatty and fragrant but still milky. Sour cheeses are commonly used to make local flour food, such as noodles and bread. The Xinjiang cheese has a high-acid, sharp milky flavor and is thicker and darker than ordinary cheese.
Xinjiang cheese is distinctive, high in nutrients, and always is considered a good staple food. However, the cheese is only consumed in limited areas in China, such as Xinjiang Uygur, Inner Mongolia, and Tibet Autonomous Regions. Furthermore, microbial resources, especially the LAB, in various dairy products have not been well characterized, which potentially negatively impacts cheese industries and application in human health promotion.
Caenorhabditis elegans is a small, soil nematode, which has been extensively used as a model organism to evaluate longevity extension of LAB strains (Kurz and Tan, 2004). Ikeda et al. (2007) have reported that LAB may contribute to host defenses and prolong the lifespan of C. elegans. In this study, we wished to evaluate potential traits of probiotic strains selected from Xinjiang cheese products using in vitro tests and the in vivo C. elegans model.
2. Materials and methods
2.1. Sample collection
Four traditionally fermented cheese products were collected from Xinjiang Uygur Autonomous Region of China to isolate the lactic acid bacteria in MC, M17, and MRS (de Man, Rogosa, and Sharpe, Sinopharm Chemical Reagent Company (SCRC), Shanghai, China) broth or MRS agar plates. All four cheese samples (30 d ripened) were made of raw cow’s milk and made by the nomadic people in Xinjiang with natural fermentation processes without any starter culture. Samples were transported at 4 °C to the laboratory for analysis.
2.2. Isolation and identification of lactic acid bacteria
We used the MC, M17, and MRS agar plates for microflora composition analysis. Appropriate ten-fold dilutions of the samples were prepared with sterile water. The MC, M17, and MRS agar plates supplemented with 0.01 g/ml CaCO3 were mixed with 100 μl of sample. The plates were incubated at 37 °C in an anaerobic incubator for 2‒3 d. Single colonies were picked up based on the calcium-dissolving ring (Tamang et al., 2005). Strains were identified morphologically and biochemically, e.g. the morphology, Gram staining, catalase activity, growth rate at 45 °C, and carbohydrate fermentation.
The genomic DNA of each isolate was extracted from pure overnight cultures grown in MRS broth at 37 °C by the phenol-chloroform extraction method (Schmidt et al., 1991). Each extracted DNA was dissolved in 50 μl distilled water and stored at −20 °C for further analysis.
All cultivable isolates were identified by 16S rDNA sequencing analysis. The genomic DNA was used as a template for polymerase chain reaction (PCR) amplification of a segment of its 16S rDNA gene for each isolate. Two primers were designed as described previously (Mora et al., 2003; Yu et al., 2011), namely 27f (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492r (5'-CTACGGCTACCTTGTTACGA-3'). The amplification products obtained from the PCR reaction were analyzed by 1.0% (0.01 g/ml) agarose (Hydragene, Xiamen, China) gel electrophoresis before they were sequenced. DNA sequencing was performed by the Shanghai Sangon Bio-sciences Corporation (China). The sequencing data were aligned using DNAMAN software (ver. 7.0 DNAMAN, Lynnon Biosoft Company, Foster City, CA, USA), and sequence homology was examined by comparing the obtained sequences by the BLAST program in the National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The accession numbers of these six strains were as follows: Enterococcus hirae H4, KU198310; E. hirae K4-8, KU198311; E. hirae K4-9, KU198312; E. hirae R5, KU198313; Lactobacillus rhamnosus R4, KU198314; and, Lactobacillus helveticus S4, KU198315.
2.3. Acid and bile tolerance tests
The acid resistance was performed by the viable plate count method (Maragkoudakis et al., 2006). Bacterial cells grown in MRS broth at 37 °C overnight were harvested (10 000 r/min, 10 min, 4 °C), washed twice with phosphate buffer saline (PBS) of pH 7.2 and resuspended in PBS of pH 3.0. The viable colony counts were enumerated on MRS agar after incubation at 37 °C for 0 and 3 h simulating the acidic environment in the human stomach. The experiment was repeated three times. The survival rate (SR) was calculated according to the equation below:
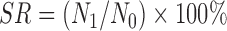 , ,
|
where N 1 (log CFU/ml) is the total viable count of selected strains after treatment and N 0 (log CFU/ml) represents the total viable count of selected strains before treatment. CFU is colony-forming unit.
The bile tolerance test was carried out in 96-well plates (Walker and Gilliland, 1993). Briefly, cells of the selected strains were grown in MRS broth at 37 °C overnight, and then subcultured in MRS broth containing 0.1%, 0.2%, and 0.3% (1%=0.01 g/ml) bile salts (SCRC, Shanghai, China). The growth rate of each strain was measured by its absorbance at 600 nm after 24 h.
The ability of isolates to tolerate the bile salt condition was also determined by the viable plate count method. Bacterial cultures were grown at 37 °C overnight and the bacterial cells were harvested (10 000 r/min, 10 min, 4 °C), washed twice with PBS of pH 7.2 before being resuspended in PBS solution containing 0.3% bile salts. The viable counts were enumerated on MRS agar after incubation at 37 °C for 0 and 4 h. The survival rate is calculated according to the above-mentioned equation.
2.4. Antimicrobial activity
The Oxford cup assay was used as reported (Wang et al., 2009). The indicator strains of antimicrobial activity included Escherichia coli O157 882364, Salmonella typhimurium S50333, Shigella flexneri CMCC (B)51592, Staphylococcus aureus ATCC 1448, and Listeria monocytogenes C53-3, which were obtained from our laboratory and cultured in Luria-Bertani (LB) medium, nutrient broth (NB) medium, tryptic soytone broth (TSB), and broth brain-heart infusion agar (BHI), respectively. All LAB strains were subcultured twice prior to being used in the experiments. Strains were incubated with the MRS medium for 24 h at 37 °C. The bacterial cultures were centrifuged at 10 000 r/min for 10 min. Supernatants were filtered through a 0.22-μm filter. The inoculum of indicator (pathogen) strains was then spread on the surface of solid nutrient agar (NA) plates. A total of 200 μl bacterial cultures of each LAB strain were added to the Oxford cups which were placed on the agar plates. The plates were incubated for 24 h at 37 °C and the zone of inhibition was determined. The inhibitory effect of MRS was tested as a negative control on each plate. The inhibition zones were measured in millimeter (mm).
2.5. Auto-aggregation activity and hydrophobicity
Auto-aggregation activity was measured as described by Collado et al. (2008). Overnight cultures were centrifuged and washed twice with PBS and resuspended in the same buffer. Equal volumes of each of the microbial suspensions were combined in sterile test tubes and mixed for 10 s on a vortex mixer, and then incubated anaerobically at 37 °C for 24 h without agitation. The auto-aggregation activity (AA) was calculated using the equation:
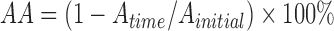 , ,
|
where A initial and A time are the culture absorbance before and after incubation, respectively.
Hydrophobicity assay was carried out according to the study of Collado et al. (2008). Overnight bacterial cultures were harvested by centrifugation at 5000 r/min for 15 min at 4 °C and washed twice in PBS. The absorbance of cell suspensions was measured at 600 nm. A total of 1 ml cell suspension was mixed with an equal proportion of hydrocarbons (xylene), and the aqueous phase was removed after 30 min at room temperature for the determination of absorbance (at 600 nm). The hydrophobicity (H) was calculated according to the following formula:
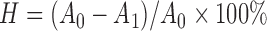 , ,
|
where A 0 and A 1 represent the absorbance before and after mixing with xylene, respectively.
2.6. Antioxidant activity
2.6.1. Preparation of the intact cells and the cell-free extracts
The strain was grown in MRS broth at 37 °C for 18–24 h. Cells were suspended in PBS. The bacterial cell pellets were adjusted to 108 CFU/ml, which was determined as the preparation of intact cells. Cell-free extracts were prepared according to the method described by Lin and Yen (1999). The cells were adjusted to 108 CFU/ml followed by ultrasonic disruption three times, and had 2-min intervals in an ice bath using an ultrasonic cell crusher (Ningbo Scientz biotechnology Company, China). After centrifugation (10 000 r/min, 15 min, 4 °C), the supernatant was obtained as the cell-free extract of the strains.
2.6.2. DPPH radical scavenging activity
The 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging activity was measured by the method of Kao and Chen (2006) with modifications. Reaction mixtures containing 1.0 ml of 108 CFU/ml bacterial cells and 1.0 ml of 50 μmol/L DPPH (Aladdin Industrial Corporation, Aladdin, Reagent Company, China) in ethanol (SCRC, Shanghai, China) were incubated at 37 °C in the dark for 30 min. The absorbance of the resulting solution at 517 nm was measured in triplicate. The DPPH scavenging activity (SADPPH) was defined as:
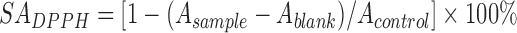 , ,
|
where A sample is the absorbance of bacterial cells, A blank is the absorbance of only cells and ethanol, and A control is the absorbance of the deionized water and DPPH solutions.
2.6.3. Hydroxyl radical scavenging activity
The hydroxyl radical scavenging activity was determined by a sulfosalicylic acid method (He et al., 2004), also known as the Fenton type reaction method. The reaction mixture including 1.0 ml of salicylic acid, FeSO4, and H2O2 (6, 2, and 6 mmol/L, respectively, all from SCRC, Shanghai, China), and 1.0 ml of bacterial cell or cell-free extract with the concentration of 108 CFU/ml was incubated at room temperature for 20 min, and the absorbance was determined at 510 nm. Hydroxyl radical scavenging activity (SAhydroxyl) is expressed as:
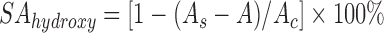 , ,
|
where A s is the absorbance in the presence of the sample, A is the absorbance without H2O2, and A c is the absorbance of the control in the absence of the strain sample.
2.7. Determination of cholesterol and triglycerides
Bacterial cholesterol lowering capability was determined by the ferric ammonium sulfate method with slight modification (Feng et al., 1973). MRS broth containing 0.1% (1 g/L) cholesterol (Solarbio, Beijing, China) was prepared as MRS-CHOL broth. Overnight cultures were inoculated (2%, v/v) into the MRS-CHOL broth and incubated anaerobically at 37 °C for 48 h. An uninoculated sample was used as a control. After incubation, 0.2 ml cultures were mixed with 4.8 ml ethanol. Then the cells were removed after centrifugation (3000 r/min, 10 min, 4 °C). A total of 2 ml of ferric ammonium sulfate color reagent (Aladdin, Shanghai, China) was slowly added to the 2 ml supernatant. The absorbance was determined at 560 nm. The cholesterol removal rate (CRR) from growth media was calculated according to the following formula:
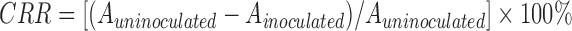 , ,
|
where A uninoculated and A inoculated are the absorbance of uninoculated and inoculated samples, respectively.
Triglyceride micelles were prepared by mixing Tween-80 and triglycerides (both from SCRC, Shanghai, China) at a ratio of 1:5 (v/v), and sterilized for 15 min at 121 °C before determination. The micelles were added to the MRS broth containing bile salt. Isolates were inoculated (2%, v/v) into 5 ml of the above MRS broth and incubated at 37 °C for 36 h. A culture without triglyceridewas used as a control. After centrifugation for 10 min at 3000 r/min, the supernatant of the cultures was used for testing the triglyceride reducing activity using the acetylacetone method (Fossati and Prencipe, 1982; Saravanan and Ponmurugan, 2012).
Three isolates were selected for further investigation to see their anti-aging effects on C. elegans.
2.8. C. elegans killing assay
The effect of selected LAB strains on longevity in C. elegans was examined using the killing assay as described by Ikeda et al. (2007), with slight modifications. Briefly, the synchronized worms were grown at 20 °C until they reached L4 stage. The worms were then transferred onto treatment plates. The treatment plates were prepared using nematode growth medium (NGM) covered with lawns of standard food resource E. coli OP50 (control group) and LAB strains (experimental group) at the concentration of 108 CFU/ml. The worms were transferred to fresh treatment plates every 2–4 d and grown at 20 °C. The number of live worms was counted every other day. Worms that failed to respond to a light touch with a platinum wire would be scored as dead and excluded from the lifespan assay. In the present study, all treatment plates were prepared on Day 0 of adulthood of the worms. This test was carried out at least three times.
2.9. Statistical analysis
Statistical analysis of the obtained data was carried out using SPSS (ver. 16.0 SPSS, Chicago, IL, USA). The comparisons of differences between the means of the treatments were tested by one-way analysis of variance (ANOVA) at a significance level of P<0.05.
3. Results
3.1. Isolation and identification of strains
More than 40 strains were preliminarily isolated from four traditionally fermented Xinjiang cheese products, and six LAB strains from them were selected for further analysis. Based on the biochemical tests, those six isolates showed Gram-positive, catalase-negative characteristics, and colony morphologies of the isolates were rod and coccus (Table S1). The species of the isolates were determined on NCBI by comparing with 16S rDNA sequences of the standard strains. The representative strains and their related type strains were selected to construct a phylogenic tree using the DNAMAN software to show their identities and diversities (data not shown). R4 and S4 were classified as L. rhamnosus and L. helveticus, respectively, while H4, K4-8, K4-9, and R5 were classified as E. hirae. The strains named with H and K refer to the samples collected from Narat Pastureland, S refers to Ili City, and R refers to Urumqi City.
3.2. Acid tolerance test
The viable counts and survival rates of the selected six LAB strains are shown in Table 1. All tested strains showed resistance to low pH. The viable counts of all strains were found to be >106 CFU/ml after incubation at pH 3.0 for 3 h and in the presence of bile salt at 0.3%. Overall, all strains showed higher resistance to low pH at the range from 74.6% to 87.1%. The differential survival rates of LAB strains suggested that the survival activity was strain-specific.
Table 1.
Survival rate of six strains at pH 3.0 and 0.3% bile salt in MRS medium
| Strain | Acid tolerance at pH 3.0 |
Bile tolerance with 0.3% bile salt |
||||
| Viable count after 0 h (log CFU/ml) | Viable count after 3 h (log CFU/ml) | Survival rate (%) | Viable count after 0 h (log CFU/ml) | Viable count after 3 h (log CFU/ml) | Survival rate (%) | |
| R4 | 8.29±0.22a | 7.22±0.19a | 87.1 | 8.99±0.13a | 6.73±0.03e | 74.9 |
| H4 | 7.92±0.27c | 6.89±0.11c | 87.0 | 8.84±0.20b | 6.77±0.21d | 76.5 |
| S4 | 7.86±0.37d | 6.72±0.35d | 85.5 | 8.20±0.14c | 6.99±0.02a | 85.2 |
| R5 | 8.11±0.32b | 6.92±0.53b | 85.3 | 8.13±0.07d | 6.96±0.03a | 85.1 |
| K4-9 | 8.22±0.63a | 6.59±0.01e | 80.2 | 8.14±0.01d | 6.86±0.04b | 84.2 |
| K4-8 | 7.21±0.30e | 5.38±0.12f | 74.6 | 7.88±0.06e | 6.79±0.08c | 86.1 |
Viable counts are presented as the mean±SD from the mean of triplicate determinations, and the values followed by different lowercases in the same column are significantly different (P<0.05)
3.3. Bile tolerance test
The growth capabilities of six previously selected strains under 0.1%, 0.2%, and 0.3% bile salt conditions were determined by the absorbance at 600 nm after 24 h incubation. As shown in Fig. 1, four strains E. hirae K4-8, K4-9, L. helveticus S4, and L. rhamnosus R4 showed good growth rates at all bile salt concentrations. The results of determination of viable counts and survival rates also showed that all tested strains were found to be resistant to bile salt of high concentration (Table 1). The strains L. helveticus S4 and E. hirae K4-8, K4-9, R5 had higher bile salt resistance.
Fig. 1.
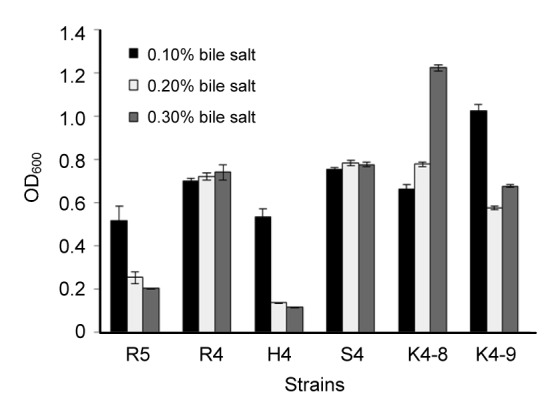
Tolerance of six strains to different bile salt conditions
Each value is expressed as mean±standard deviation (SD) of three replicates. There were significant differences (P<0.05) among different strains under three bile salt concentrations
3.4. Antimicrobial activity
All six strains presented antimicrobial activity against Escherichia coli (with an inhibition zone 6.3–10.5 mm in diameter), Staphylococcus aureus (4.0–8.2 mm), Salmonella typhimurium (2.5–8.5 mm), Listeria monocytogenes (4.0–8.5 mm), and Enterococcus faecalis (5.0–7.1 mm). However, L. rhamnosus R4 exhibited the most effective inhibition (Table 2), while E. hirae K4-8 and K4-9 strains did not inhibit the growth of Listeria monocytogenes and Enterococcus faecalis, respectively. The control (MRS media alone) showed no inhibitory effect against the pathogens tested (data not shown).
Table 2.
Antimicrobial activities of six selected strains on pathogens
| Strain | Inhibition zone (mm) |
||||
| Escherichia coli | Staphylococcus aureus | Salmonella typhimurium | Listeria monocytogenes | Enterococcus faecalis | |
| L. rhamnosus R4 | 10.5±0.5a | 8.2±0.5a | 8.5±0.0a | 8.5±0.3a | 7.1±0.0a |
| L. helveticus S4 | 10.0±0.2b | 8.0±0.3a | 8.0±0.1b | 7.2±0.0b | 6.8±0.4b |
| E. hirae H4 | 8.0±0.3c | 6.5±0.4b | 6.2±0.3d | 6.2±0.1c | 6.5±0.1c |
| E. hirae R5 | 8.2±0.2c | 6.4±0.1b | 7.1±0.0c | 7.0±0.2b | 5.5±0.3d |
| E. hirae K4-8 | 6.5±0.0d | 4.3±0.0c | 5.0±0.2e | NI | 5.0±0.2e |
| E. hirae K4-9 | 6.3±0.5d | 4.0±0.2d | 2.5±0.5f | 4.0±0.5d | NI |
NI: no inhibition. Data are presented as the mean±SD from the mean of triplicate determinations. The values followed by different lowercases in the same column are significantly different (P<0.05)
3.5. Auto-aggregation and hydrophobicity analysis
As shown in Table 3, all tested strains showed increased auto-aggregation activity ranging from 23.33% to 45.83%. L. rhamnosus R4 was the best auto-aggregating strain among those tested, resulting in 45.83% auto-aggregation over a period of 24 h incubation at 37 °C. Additionally, the cell surface hydrophobicity of all strains was tested, and the results (Table 3) showed that L. rhamnosus R4 exhibited the highest cell surface hydrophobicity in xylene (83.22%).
Table 3.
Auto-aggregation and hydrophobicity activities of six selected strains
| Strain | Auto-aggregation (%) | Hydrophobicity (%) |
| L. rhamnosus R4 | 45.83±5.60a | 83.22±0.57a |
| L. helveticus S4 | 34.55±0.88b | 75.42±1.35b |
| E. hirae H4 | 30.48±4.39c | 67.08±5.42d |
| E. hirae R5 | 33.65±2.82b | 70.98±3.53c |
| E. hirae K4-8 | 26.83±5.37d | 54.14±8.16e |
| E. hirae K4-9 | 23.33±1.46e | 48.57±6.16f |
Values are expressed as mean±SD of three replicates. The values followed by different lowercases in the same column are significantly different (P<0.05)
3.6. DPPH radical scavenging activity
The DPPH scavenging activities of the intact cells and cell-free extracts of selected six strains are shown in Fig. 2. L. rhamnosus R4, L. helveticus S4, and E. hirae H4, R5 had higher scavenging activity than others by measuring the intact cells. Among them, L. rhamnosus R4 showed the highest scavenging activity (53.78%), whereas the scavenging activity of the cell-free extract was significantly lower than that of the intact cells. However, E. hirae K4-8 showed a different behavior in that the scavenging activity of cell-free extract (32.11%) was higher than that of intact cells (18.95%).
Fig. 2.
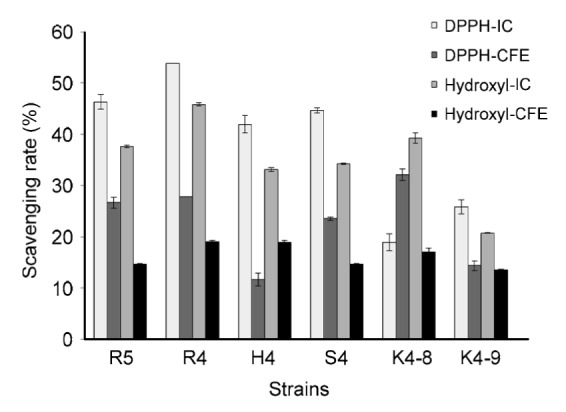
DPPH free radical and hydroxyl radical scavenging abilities of six strains
IC: intact cell; CFE: cell-free extract. Each value is expressed as mean±SD of three replicates. There were significant differences (P<0.05) among the DPPH and hydroxyl radical scavenging rates of IC and CFE of different strains
3.7. Scavenging of hydroxyl radicals
Among the six tested strains, L. rhamnosus R4 showed the highest hydroxyl radical scavenging activity (45.79%), verifying the result of DPPH scavenging activity (Fig. 2). All measured strains exhibited higher scavenging activity of the intact cells than that of the cell-free extract. As can be seen from the results, both DPPH radicals and hydroxyl radicals can be scavenged by these tested strains, which showed the significant free radical degradation activity of these strains.
3.8. Degradation of cholesterol and triglycerides
The degradation rates of cholesterol and triglycerides of the isolated strains are shown in Fig. 3. The results revealed that all examined strains were able to degrade cholesterol and triglycerides, though the cholesterol and triglyceride degradation rates of supernatants of strains were significantly higher than that of intact cells of strains. Among the tested strains, the degradation rates of the supernatants of L. rhamnosus R4 (50.97%, 28.92%) and L. helveticus S4 (52.85%, 24.56%) showed the highest cholesterol and triglyceride reducing activities compared to those of intact cells during growth. The supernatant of E. hirae H4, R5 also exhibited better degradation activity than the intact cells.
Fig. 3.
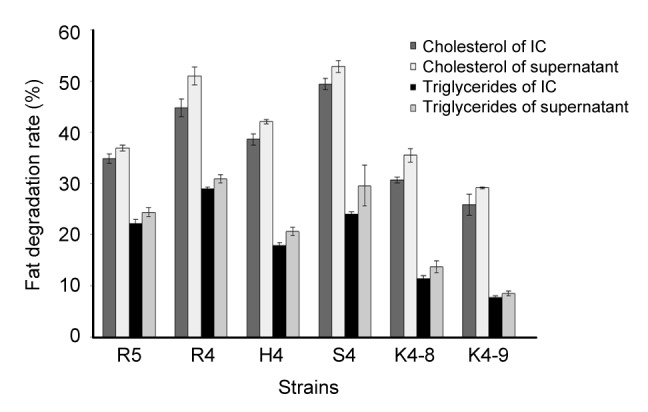
Cholesterol and triglyceride degradation activities of six selected strains
IC: intact cell. Each value is expressed as mean±SD of three replicates. There were significant differences (P<0.05) among cholesterol and triglyceride degradation rates of IC and supernatants of different strains
3.9. C. elegans killing assay
Wild-type adult C. elegans have an average lifespan of approximately 20 d at 20 °C. In our laboratory conditions, wild-type control worms lived for an average of (18.47±0.30) d (maximum of 25 d).
Here, the effect of LAB strains on lifespan of C. elegans was determined to further evaluate the health promoting effects of the isolates. Three out of the six strains were able to enhance the health-span of C. elegans. The results (Fig. 4) showed that feeding worms with L. rhamnosus R4 increased the average lifespan compared to the E. coli OP50 feeding group. The lifespan of worms supplied with L. rhamnosus R4 was (31.05±0.54) d, which was 36.1% greater than that of the control worms ((22.81±0.62) d; P<0.05).
Fig. 4.
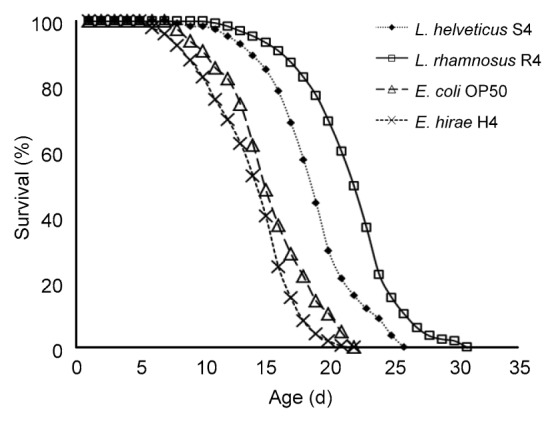
Survival curves of C. elegans fed with L. rhamnosus R4, L. helveticus S4, E. hirae H4, and E. coli OP50, respectively
Day 0 represents the first day that the L4 stage worms were transferred to NGM plates
4. Discussion
In this study, six LAB strains isolated from cheese samples were investigated for their probiotic properties. Tolerance to acidic condition is the most commonly used method to detect the viability and activity of probiotic bacteria in the small intestine and stomach. According to a previous study (Usman and Hosono, 1999), the survival rate at pH 3.0 is considered as optimal acid tolerance for selected probiotic strains. All strains from this study were able to tolerate pH 3.0 with a survival percentage of more than 70%, and therefore they can be considered as acid-tolerant LAB strains. This tolerance capacity of lactic acid bacteria warranted the production of various antimicrobial substances which might create unfavorable conditions for the growth of pathogens as well as toxigenic and spoilage organisms in humans and/or animals (van der Meulen et al., 2007).
Tolerance to bile salts is usually considered a basic property for LAB strains to survive in the small intestine. In the human gastrointestinal tract, the mean bile concentration is about 0.3% and is considered crucial and high enough to screen resistant strains (Gilliland et al., 1985). In this study, there was extreme variability of resistance to bile salts in E. hirae K4-8, R5 and L. rhamnosus R4 isolates. Lactobacillus strains, which grow and metabolize in normal physical bile concentration, could survive in gastrointestinal transit (Sanders et al., 1996). All six tested strains indicated a proportion of growth above 65% in the presence of bile salt, which demonstrated good bile salt tolerance. These results were consistent with a previous study (Turchi et al., 2013).
To further investigate the safety and functional characteristics of these strains as probiotics, several tests were also conducted to assess their antimicrobial activity, auto-aggregation capability, and cell surface hydrophobicity.
As a potential probiotic, antimicrobial activity is one important property to avoid gastrointestinal infection (Kanmani et al., 2013). In the present study, the antimicrobial activities of six selected strains were assayed. Isolate L. rhamnosus R4 inhibited the growth of all tested pathogens, and these pathogens are commonly found in fermented foods, making these products potentially hazardous (Ferreira et al., 2007). The ability to inhibit the pathogens of tested strains could be explained by the production of antimicrobial compounds. According to the study of Amenu (2013), the antimicrobial effect of LAB is due to the production of metabolites, such as lactic acid, acetic acid, diacetyl, fatty acids, aldehydes, bacteriocins, and other compounds, among which, lactic acid, acetic acid, and bacteriocins are the most powerful antimicrobial agents and are the production of the probiotics. The inhibition of pathogens’ growth suggests that the presence of LAB microorganisms in the Xinjiang cheese might influence the reduced survival of bacterial pathogens in spite of the simple and crude home-based fermenting preparation process, and these tested isolates may also have potential application in food preservation.
The aggregation capability of LAB may be of interest for both food preservation and therapeutic impact on gut microbiota. In the auto-aggregation test, all tested strains indicated various auto-aggregation abilities. L. rhamnosus R4 strain showed the highest percentage of auto-aggregation, namely 45.83%. This result suggested that the R4 strain possessed a proportionally high potential capability to adhere to epithelial cells and mucosal surfaces. Previous studies (Pelletier et al., 1997; Dunne et al., 2001) stated that the aggregation abilities of LAB might enable them to form a barrier that prevents pathogenic colonization.
Cell surface hydrophobicity analysis is also a key step for selection of functional probiotic LAB strains. For all tested strains, a significant difference in cell surface hydrophobicity was observed, ranging from 48.57% to 83.22%. Cell surface hydrophobic activity is one of the physical-chemical characteristics that promote the first contact among the host tissues and microorganisms. Hydrophobicity was found to be correlated to auto-aggregation properties as most of the strains with higher adhesion to hydrocarbons showed high auto-aggregation activity. The present study was consistent with the results of Collado et al. (2008), which showed that bacterial strains with higher adhesion to hydrocarbon (xylene) showed high auto-aggregation activity. The hydrophobic properties of the cell surface of microorganisms have been related to the attachment of bacteria to host cell. This may confer a competitive advantage and be important for bacterial maintenance in the human gastrointestinal tract (Schillinger et al., 2005).
After screening a number of potential bacteria based on their above-mentioned important selection criteria, we tested the antioxidant activity of the six isolates using free radical scavenging assays. The DPPH radicals were generally used to determine the scavenging activities of natural compounds. All tested six strains exhibited potent scavenging activities of cellular toxic substances DPPH and hydroxyl radicals to various degrees, indicating detoxificative effects by reactive oxygen species (ROS) removal. The intact cell of L. rhamnosus R4 was found to be the most effective in removal of the DPPH radicals. The scavenging activities of the intact cells of L. helveticus S4, E. hirae H4, R5 exhibited better behavior for scavenging the DPPH radicals than those of cell-free extracts, indicating that cell integrity was a factor that affected scavenging activity.
The intact cells exhibited higher hydroxyl radical scavenging activity than the cell-free extracts. It is probably due to their extracellular antioxidant components, such as polysaccharide, peptidoglycan, and teichoic acid. These basic cell-wall components play an essential role when the host suffers oxidative damage by the free radicals. Their main function is to preserve cell integrity so that the damage of extracellular antioxidant composition can be avoided.
Interestingly, E. hirae K4-8 showed a different behavior in DPPH radical scavenging activity of the cell-free extract (32.11%) compared to that of intact cells (18.95%). We inferred that some intracellular enzymes such as NADH-oxidase, NADH-peroxidase, and superoxide dismutase (SOD) were likely to be obtained after breaking up the bacterial cells into cell-free extracts that indicated the antioxidant activity of lactic acid bacteria (Kullisaar et al., 2002).
The antioxidant activities of lactic acid bacteria would be helpful in the dairy food industry. They could beneficially influence the customer by providing LAB with the potential of producing antioxidants during the period of growth in the intestinal tract or providing another dietary source of antioxidants.
In this study, in vitro assay results showed that cultures of L. rhamnosus R4, L. helveticus S4, and E. hirae H4, R5 lowered both cholesterol and triglyceride levels. It is interesting to note that the supernatant of the bacterial cells reduced more cholesterol compared to the intact cells of the bacteria during its growth, especially the strains of L. rhamnosus R4 and L. helveticus S4. This result implies that the cholesterol reduction activities of L. rhamnosus R4 and L. helveticus S4 may be due to the activity of certain components in the supernatant of the bacterial culture, in addition to those existing in the live cells.
There have been a few reports regarding cholesterol reduction (Ooi and Liong, 2010) by the extracellular components generated by LAB, which indicated that certain components or specific extracts in the supernatants of LAB were responsible for the reduction of cholesterol (Nguyen et al., 2007; Kim et al., 2008).
Our data in Fig. 3 may explain that supernatant-related cholesterol degradation was not induced by acidic co-precipitation and support a previous study reported by Liong and Shah (2005) who proposed that the co-precipitation of cholesterol was less than 5% of the total cholesterol utilized. Therefore, in this study, one possible mode for the cholesterol degradation activity of the supernatant produced from the tested strains could be mostly mediated by direct activity, including action for cholesterol degradation or inactivation, rather than by an indirect mechanism such as the deconjugation of bile salts and co-precipitation of cholesterol with the bile salts (Kim et al., 2008).
Triglycerides were also decreased by LAB strains in this study. The results revealed that triglycerides inhibited the growth of the strains. This may be due to the fact that triglycerides can be combined with peripheral bacteria for enhancing the peripheral hydrophobicity of bacteria to restrict the entry of some essential nutritive elements into the cell. From this study, we found that not only the intact cell but also the supernatant of the strains had a triglycerides’ degrading activity in a strain-specific manner, which is consistent with the research of Kim et al. (2008).
Our results also demonstrated the anti-aging role of LAB in the C. elegans model system. Feeding of L. rhamnosus R4 has effectively increased the mean lifespan of C. elegans, and the life extension rate reached a maximum of 36.1%. This is in accordance with the results described by Ikeda et al. (2007), in which feeding nematodes with bifidobacteria or lactobacilli caused increased average lifespans compared to those fed with OP50 and the life extension rates ranged from 17% to 33%. These results suggested that different strains show diverse impacts on lifespan extension. Further molecular research is required to elucidate the precise mechanism of the longevity in C. elegans. Nevertheless, our results suggested that these newly isolated LAB strains may have health promoting impacts to the host. They also offer additional support for the use of the nematode as an appropriate in vivo model system for screening functional probiotics.
The six strains in this study were also selected through the evaluation of acidification capacity, diacetyle-producing activity and autolysis according to the method described by Scatassa et al. (2015). The strains of L. rhamnosus R4 and L. helveticus S4 were those which displayed the highest acidifying activity, showing an acidity of around 0.40 g/100 ml lactic acid after 6 h and 0.58–0.72 g/100 ml after 12 h (Table S2). After 24 h, the acidity reached values around 0.73–0.86 g/100 ml. L. rhamnosus R4 showed an acidifying capacity similar to that of L. helveticus S4 strain throughout the incubation time. These LAB strains isolated from Xinjiang cheese showed an acidifying activity after 24 h of incubation similar to that detected for LAB strains isolated from Tunisian dairy products (Fguiri et al., 2015).
L. rhamnosus is a heterolactic acid bacterium, which can be used to produce flavor compounds like diacetyl and acetoin (Jyoti et al., 2003). The results on the diacetyl-producing activities of these six strains showed that during the cold storage step, the desired fermentation temperature and storage time for reaching a maximum of the diacetyl concentration varied in the final product of cheese. The maximum diacetyl concentration is between 0.68 and 1.76 μg/ml (Table S3), and the diacetyl-producing activities of Lactobacillus strains were stronger than those of Enterococcus strains. The active strong-to-weak sequence of producing diacetyl of these six strains is L. rhamnosus R4>L. helveticus S4>E. hirae H4>E. hirae R5>E. hirae K4-8>E. hirae K4-9, which indicated that these strains possess better aroma-producing capability.
The autolysis of the Lactobacillus strain has been studied in Cheddar cheese (Hannon et al., 2003), showing increased proteolysis and improved flavor. In this study, with the extension of incubation time, the autolysis of these six strains has increased. After the incubation of 4 h, the autolysis of L. rhamnosus R4 and L. helveticus S4 reached 12.70% and 11.80%, respectively (Table S4), which were higher than that of the other four strains. The L. rhamnosus R4 strain showed high autolysis during all three time periods. There was a great difference of autolysis among these six strains even after 48 h of inoculation. The L. rhamnosus R4 strain had a higher rate of autolysis. This is in agreement with the result reported by Kennya et al. (2006) who demonstrated that the presence of Lactobacillus strains resulted in a better flavor in cheese.
Salmonella sp., Listeria sp., and Shigella sp. were not detected in the cheese samples, which indicated that the local cheese samples from Xinjiang are generally safe to eat. Moreover, the nutritional value of cheese varies according to the fat content of the liquid used (milk, cream). Xinjiang cheeses always use cows’ milk as the liquid. We have determined the nutrient constituents of Xinjiang cheese (data not shown) and compared that with raw milk according to the “National Food Safety Standard of Raw Milk (GB 19301-2010)” (Ministry of Health of the People’s Republic of China, 2010; Jia et al., 2014).
In this study, Xinjiang cheeses contain 23.00% of fat and 25.02% of protein, which were 7.4 times the fat and 8.9 times the protein of raw milk, respectively. In addition, Xinjiang cheeses contain 471.2 mg of calcium per 100 g, and also contain 438.5 mg of phosphorus per 100 g, which were more than 4 times of both the calcium and phosphorus in raw milk. The total sugar content (2.17%) of Xinjiang cheeses is less than 50% compared to that of raw milk. Therefore, Xinjiang cheeses are nutritious foods with very low sugar content, high acidity, and various probiotic microorganisms.
5. Conclusions
In summary, of the six different tested strains, L. rhamnosus R4 and L. helveticus S4 were the more desirable health-promoting bacteria with better acid and bile resistance, antimicrobial activity and aggregation, hydrophobic capability, prominent antioxidant activity, and cholesterol and triglyceride reduction activity. Moreover, these strains were also able to extend the lifespan of C. elegans, and the possible mechanism might be that LAB strains could directly scavenge free radicals and protect the C. elegans from exogenous stress. This study provides some important knowledge for functional properties of probiotics in Xinjiang cheeses. Therefore, this conventionally fermented dairy product could be considered a valuable resource for probiotic strain screening and starter culture application.
List of electronic supplementary materials
Physiological and biochemical characteristics of six strains
Acidifying activity of six strains of lactic acid bacteria isolated from Xinjiang cheese
Diacetyl-producing abilities of six strains of lactic acid bacteria isolated from Xinjiang cheese
Autolysis activities of six strains of lactic acid bacteria isolated from Xinjiang cheese
Footnotes
Project supported by the National Key Technology R & D Program of China (No. 2012BAD33B08), the Zhejiang Provincial Natural Science Foundation of China (No. LY12B06006), the Research Program of Education Department of Zhejiang Province (No. Y201122061), and the National Natural Science Foundation of China (No. 20906060)
Electronic supplementary materials: The online version of this article (http://dx.doi.org/10.1631/jzus.B1500250) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Ramila AZAT, Yan LIU, Wei LI, Abdurihim KAYIR, Ding-bo LIN, Wen-wen ZHOU, and Xiao-dong ZHENG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Alegria A, Szczesny P, Mayo B, et al. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl Environ Microbiol. 2012;78(6):1890–1898. doi: 10.1128/AEM.06081-11. (Available from: http://dx.doi.org/10.1128/AEM.06081-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amadou I, Le GW, Shi YH. Evaluation of antimicrobial, antioxidant activities, and nutritional values of fermented foxtail millet extracts by Lactobacillus paracasei Fn032. Int J Food Prop. 2013;16(6):1179–1190. (Available from: http://dx.doi.org/10.1080/10942912.2011.579673) [Google Scholar]
- 3.Amenu D. Antimicrobial activity of lactic acid bacteria isolated from “Ergo”, Ethiopian traditional fermented milk. Curr Res Microbiol Biotechnol. 2013;1(6):278–284. [Google Scholar]
- 4.Collado MC, Meriluoto J, Salminen S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol. 2008;226(5):1065–1073. (Available from: http://dx.doi.org/10.1007/s00217-007-0632-x) [Google Scholar]
- 5.Dunne C, O'Mahony L, Murphy L, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73(2 Suppl.):386S–392S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 6.Feng HW, Pi CM, Wang RB, et al. Use of ferric ammonium sulfate in serum cholesterol determination. Clin Chem. 1973;19(1):121–122. [PubMed] [Google Scholar]
- 7.Ferreira V, Barbosa J, Silva J, et al. Chemical and microbiological characterisation of “Salpicao de vinhais” and “Chourica de vinhais”: traditional dry sausages produced in the North of Portugal. Food Microbiol. 2007;24(6):618–623. doi: 10.1016/j.fm.2006.12.007. (Available from: http://dx.doi.org/10.1016/j.fm.2006.12.007) [DOI] [PubMed] [Google Scholar]
- 8.Fguiri I, Ziadi M, Atigui M, et al. Isolation and characterisation of lactic acid bacteria strains from raw camel milk for potential use in the production of fermented Tunisian dairy products. Int J Dairy Technol. 2015;69(1):103–113. (Available from: http://dx.doi.org/10.1111/1471-0307.12226) [Google Scholar]
- 9.Fons M, Gomez A, Karjalainen T. Mechanisms of colonisation and colonisation resistance of the digestive tract part 2: bacteria/bacteria interactions. Microb Ecol Health Dis. 2000;12(2):240–246. (Available from: http://dx.doi.org/10.1080/089106000750060495) [Google Scholar]
- 10.Fossati P, Prencipe L. Serum triglycerides determined colorimetrically with an enzyme that produces hydrogen peroxide. Clin Chem. 1982;28(10):2077–2080. [PubMed] [Google Scholar]
- 11.Gilliland SE, Nelson CR, Maxwell C. Assimilation of cholesterol by Lactobacillus acidophilus . Appl Environ Microbiol. 1985;49(2):377–381. doi: 10.1128/aem.49.2.377-381.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gionchetti P, Rizzello F, Venturi A, et al. Probiotics in infective diarrhoea and inflammatory bowel diseases. J Gastroenterol Hepatol. 2000;15(5):489–493. doi: 10.1046/j.1440-1746.2000.02162.x. (Available from: http://dx.doi.org/10.1046/j.1440-1746.2000.02162.x) [DOI] [PubMed] [Google Scholar]
- 13.Hannon JA, Wilkinson MG, Delahunty CM, et al. Use of autolytic starter systems to accelerate the ripening of Cheddar cheese. Int Dairy J. 2003;13(4):313–323. (Available from: http://dx.doi.org/10.1016/S0958-6946(02)00178-4) [Google Scholar]
- 14.He ZS, Luo H, Cao CH, et al. Photometric determination of hydroxyl free radical in Fenton system by brilliant green. Am J Chin Med. 2004;6:236–237. [Google Scholar]
- 15.Hudault S, Liévin V, Bernet-Camard MF, et al. Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella typhimurium C5 infection. Appl Environ Microbiol. 1997;63(2):513–518. doi: 10.1128/aem.63.2.513-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda T, Yasui C, Hoshino K, et al. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar enteritidis. Appl Environ Microbiol. 2007;73(20):6404–6409. doi: 10.1128/AEM.00704-07. (Available from: http://dx.doi.org/10.1128/AEM.00704-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia X, Luan H, Huang J, et al. Marketing raw milk from dairy farmers before and after the 2008 milk scandal in China: evidence from greater Beijing. Agribusiness. 2014;30(4):410–423. (Available from: http://dx.doi.org/10.1002/agr.21375) [Google Scholar]
- 18.Jyoti BD, Suresh AK, Venkatesh KV. Diacetyl production and growth of Lactobacillus rhamnosus on multiple substrates. World J Microbiol Biotechnol. 2003;19(5):509–514. [Google Scholar]
- 19.Kanmani P, Satish Kumar R, Yuvaraj N, et al. Probiotics and its functionally valuable products–a review. Crit Rev Food Sci Nutr. 2013;53(6):641–658. doi: 10.1080/10408398.2011.553752. (Available from: http://dx.doi.org/10.1080/10408398.2011.553752) [DOI] [PubMed] [Google Scholar]
- 20.Kao TH, Chen BH. Functional components in soybean cake and their effects on antioxidant activity. J Agric Food Chem. 2006;54(20):7544–7555. doi: 10.1021/jf061586x. (Available from: http://dx.doi.org/10.1021/jf061586x) [DOI] [PubMed] [Google Scholar]
- 21.Kennya O, Fitzgerald RJ, O'Cuinnc G, et al. Autolysis of selected Lactobacillus helveticus adjunct strains during Cheddar cheese ripening. Int Dairy J. 2006;16(7):797–804. (Available from: http://dx.doi.org/10.1016/j.idairyj.2005.07.008) [Google Scholar]
- 22.Kim Y, Whang JY, Whang KY, et al. Characterization of the cholesterol-reducing activity in a cell-free supernatant of Lactobacillus acidophilus ATCC 43121. Biosci Biotechnol Biochem. 2008;72(6):1483–1490. doi: 10.1271/bbb.70802. (Available from: http://dx.doi.org/10.1271/bbb.70802) [DOI] [PubMed] [Google Scholar]
- 23.Kullisaar T, Zilmer M, Mikelsaar M, et al. Two antioxidative lactobacilli strains as promising probiotics. Int J Food Microbiol. 2002;72(3):215–224. doi: 10.1016/s0168-1605(01)00674-2. (Available from: http://dx.doi.org/10.1016/S0168-1605(01)00674-2) [DOI] [PubMed] [Google Scholar]
- 24.Kurz CL, Tan MW. Regulation of aging and innate immunity in C. elegans . Aging Cell. 2004;3(4):185–193. doi: 10.1111/j.1474-9728.2004.00108.x. (Available from: http://dx.doi.org/10.1111/j.1474-9728.2004.00108.x) [DOI] [PubMed] [Google Scholar]
- 25.Lin MY, Yen CL. Antioxidative ability of lactic acid bacteria. J Agric Food Chem. 1999;47(4):1460–1466. doi: 10.1021/jf981149l. (Available from: http://dx.doi.org/10.1021/Jf981149l) [DOI] [PubMed] [Google Scholar]
- 26.Liong MT, Shah NP. Bile salt deconjugation ability, bile salt hydrolase activity and cholesterol co-precipitation ability of lactobacilli strains. Int Dairy J. 2005;15(4):391–398. (Available from: http://dx.doi.org/10.1016/j.idairyj.2004.08.007) [Google Scholar]
- 27.Maragkoudakis PA, Zoumpopoulou G, Miaris C, et al. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J. 2006;16(3):189–199. (Available from: http://dx.doi.org/10.1016/j.idairyj.2005.02.009) [Google Scholar]
- 28.Martini MC, Bollweg GL, Levitt MD, et al. Lactose digestion by yogurt β-galactosidase: influence of pH and microbial cell integrity. Am J Clin Nutr. 1987;45(2):432–436. doi: 10.1093/ajcn/45.2.432. [DOI] [PubMed] [Google Scholar]
- 29.Ministry of Health of the People’s Republic of China. GB 19301-2010: National Food Safety Standard. Raw Milk. National Standard of the People’s Republic of China; 2010. (in Chinese) [Google Scholar]
- 30.Mora D, Scarpellini M, Franzetti L, et al. Reclassification of Lactobacillus maltaromicus (Miller et al. 1974) DSM 20342T and DSM 20344 and Carnobacterium piscicola (Collins et al. 1987) DSM 20730T and DSM 20722 as Carnobacterium maltaromaticum comb. nov. Int J Syst Evol Microbiol. 2003;53(Pt. 3):675–678. doi: 10.1099/ijs.0.02405-0. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen TD, Kang JH, Lee MS. Characterization of Lactobacillus plantarum PH04, a potential probiotic bacterium with cholesterol-lowering effects. Int J Food Microbiol. 2007;113(3):358–361. doi: 10.1016/j.ijfoodmicro.2006.08.015. (Available from: http://dx.doi.org/10.1016/j.ijfoodmicro.2006.08.015) [DOI] [PubMed] [Google Scholar]
- 32.Ooi LG, Liong MT. Cholesterol-lowering effects of probiotics and prebiotics: a review of in vivo and in vitro findings. Int J Mol Sci. 2010;11(6):2499–2522. doi: 10.3390/ijms11062499. (Available from: http://dx.doi.org/10.3390/ijms11062499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier C, Bouley C, Cayuela C, et al. Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl Environ Microbiol. 1997;63(5):1725–1731. doi: 10.1128/aem.63.5.1725-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders ME, Walker DC, Walker KM, et al. Performance of commercial cultures in fluid milk applications. J Dairy Sci. 1996;79(6):943–955. doi: 10.3168/jds.S0022-0302(96)76445-7. (Available from: http://dx.doi.org/10.3168/jds.S0022-0302(96)76445-7) [DOI] [PubMed] [Google Scholar]
- 35.Saravanan G, Ponmurugan P. Ameliorative potential of S-allylcysteine: effect on lipid profile and changes in tissue fatty acid composition in experimental diabetes. Exp Toxicol Pathol. 2012;64(6):639–644. doi: 10.1016/j.etp.2010.12.007. (Available from: http://dx.doi.org/10.1016/j.etp.2010.12.007) [DOI] [PubMed] [Google Scholar]
- 36.Scatassa ML, Gaglio R, Macaluso G, et al. Transfer, composition and technological characterization of the lactic acid bacterial populations of the wooden vats used to produce traditional stretched cheeses. Food Microbiol. 2015;52:31–41. doi: 10.1016/j.fm.2015.06.008. (Available from: http://dx.doi.org/10.1016/j.fm.2015.06.008) [DOI] [PubMed] [Google Scholar]
- 37.Schiffrin EJ, Brassart D, Servin AL, et al. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am J Clin Nutr. 1997;66(2):515S–520S. doi: 10.1093/ajcn/66.2.515S. [DOI] [PubMed] [Google Scholar]
- 38.Schillinger U, Guigas C, Heinrich Holzapfel W. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int Dairy J. 2005;15(12):1289–1297. (Available from: http://dx.doi.org/10.1016/j.idairyj.2004.12.008) [Google Scholar]
- 39.Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173(14):4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sip A, Więckowicz M, Olejnik-Schmidt A, et al. Anti-Listeria activity of lactic acid bacteria isolated from golka, a regional cheese produced in Poland. Food Control. 2012;26(1):117–124. (Available from: http://dx.doi.org/10.1016/j.foodcont.2012.01.014) [Google Scholar]
- 41.Tamang JP, Tamang B, Schillinger U, et al. Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the Eastern Himalayas. Int J Food Microbiol. 2005;105(3):347–356. doi: 10.1016/j.ijfoodmicro.2005.04.024. (Available from: http://dx.doi.org/10.1016/j.ijfoodmicro.2005.04.024) [DOI] [PubMed] [Google Scholar]
- 42.Turchi B, Mancini S, Fratini F, et al. Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J Microbiol Biotechnol. 2013;29(10):1913–1922. doi: 10.1007/s11274-013-1356-7. (Available from: http://dx.doi.org/10.1007/s11274-013-1356-7) [DOI] [PubMed] [Google Scholar]
- 43.Usman Hosono, A. Bile tolerance, taurocholate deconjugation, and binding of cholesterol by Lactobacillus gasseri strains. J Dairy Sci. 1999;82(2):243–248. doi: 10.3168/jds.S0022-0302(99)75229-X. [DOI] [PubMed] [Google Scholar]
- 44.van der Meulen R, Scheirlinck I, van Schoor A, et al. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl Environ Microbiol. 2007;73(15):4741–4750. doi: 10.1128/AEM.00315-07. (Available from: http://dx.doi.org/10.1128/Aem.00315-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker DK, Gilliland SE. Relationships among bile tolerance, bile salt deconjugation, and assimilation of cholesterol by Lactobacillus acidophilus . J Dairy Sci. 1993;76(4):956–961. doi: 10.3168/jds.s0022-0302(93)77422-6. (Available from: http://dx.doi.org/10.3168/jds.S0022-0302(93)77422-6) [DOI] [PubMed] [Google Scholar]
- 46.Wang CY, Wu SC, Ng CC, et al. Effect of Lactobacillus-fermented adlay-based milk on lipid metabolism of hamsters fed cholesterol-enriched diet. Food Res Int. 2010;43(3):819–824. (Available from: http://dx.doi.org/10.1016/j.foodres.2009.11.020) [Google Scholar]
- 47.Wang Y, Lu Z, Wu H, et al. Study on the antibiotic activity of microcapsule curcumin against foodborne pathogens. Int J Food Microbiol. 2009;136(1):71–74. doi: 10.1016/j.ijfoodmicro.2009.09.001. (Available from: http://dx.doi.org/10.1016/j.ijfoodmicro.2009.09.001) [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Wang WH, Menghe BLG, et al. Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J Dairy Sci. 2011;94(7):3229–3241. doi: 10.3168/jds.2010-3727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physiological and biochemical characteristics of six strains
Acidifying activity of six strains of lactic acid bacteria isolated from Xinjiang cheese
Diacetyl-producing abilities of six strains of lactic acid bacteria isolated from Xinjiang cheese
Autolysis activities of six strains of lactic acid bacteria isolated from Xinjiang cheese


