Abstract
Purpose
Dual human epidermal growth factor receptor 2 (HER2) targeting can increase pathologic complete response rates (pCRs) to neoadjuvant therapy and improve progression-free survival in metastatic disease. CALGB 40601 examined the impact of dual HER2 blockade consisting of trastuzumab and lapatinib added to paclitaxel, considering tumor and microenvironment molecular features.
Patients and Methods
Patients with stage II to III HER2-positive breast cancer underwent tumor biopsy followed by random assignment to paclitaxel plus trastuzumab alone (TH) or with the addition of lapatinib (THL) for 16 weeks before surgery. An investigational arm of paclitaxel plus lapatinib (TL) was closed early. The primary end point was pCR in the breast; correlative end points focused on molecular features identified by gene expression–based assays.
Results
Among 305 randomly assigned patients (THL, n = 118; TH, n = 120; TL, n = 67), the pCR rate was 56% (95% CI, 47% to 65%) with THL and 46% (95% CI, 37% to 55%) with TH (P = .13), with no effect of dual therapy in the hormone receptor–positive subset but a significant increase in pCR with dual therapy in those with hormone receptor–negative disease (P = .01). The tumors were molecularly heterogeneous by gene expression analysis using mRNA sequencing (mRNAseq). pCR rates significantly differed by intrinsic subtype (HER2 enriched, 70%; luminal A, 34%; luminal B, 36%; P < .001). In multivariable analysis treatment arm, intrinsic subtype, HER2 amplicon gene expression, p53 mutation signature, and immune cell signatures were independently associated with pCR. Post-treatment residual disease was largely luminal A (69%).
Conclusion
pCR to dual HER2-targeted therapy was not significantly higher than single HER2 targeting. Tissue analysis demonstrated a high degree of intertumoral heterogeneity with respect to both tumor genomics and tumor microenvironment that significantly affected pCR rates. These factors should be considered when interpreting and designing trials in HER2-positive disease.
INTRODUCTION
Untreated human epidermal growth factor receptor 2 (HER2) –positive disease is the most aggressive breast cancer phenotype, but its prognosis has been transformed by HER2-targeting drugs. The anti-HER2 monoclonal antibody trastuzumab has reduced mortality in stage I to III disease by 37% when combined with adjuvant chemotherapy.1 Other HER2-targeting drugs approved for metastatic disease include the small-molecule inhibitor lapatinib, the anti-HER2 heterodimerization domain antibody pertuzumab, and the antibody–drug conjugate trastuzumab emtansine. In patients with metastatic HER2-positive disease, the use of two HER2-targeted drugs (pertuzumab and trastuzumab administered with chemotherapy v trastuzumab alone2 or lapatinib and trastuzumab v lapatinib alone3) has improved survival.
Neoadjuvant (preoperative) trials deliver a potential surrogate end point (pathologic complete response [pCR]); such trials are proposed as guides in the design of adjuvant trials and, more recently, as bases for accelerated drug approval.4 In randomized neoadjuvant trials, dual HER2 targeting generally results in higher pCR rates, but the magnitude of this effect has varied.5-7 However, the extent to which an increase in pCR will improve overall outcomes remains uncertain; a recent large adjuvant trial of dual targeting with trastuzumab and lapatinib reported a nonsignificant 16% lower relapse rate in the dual targeting arm8 and no impact on overall survival.
In addition to treatment, several biologic features are implicated in response heterogeneity to HER2 targeting, including tumor intrinsic subtype,9 hormone receptor status,5,6,9,10 alterations in signaling pathways such as phosphatidylinositol 3-kinase (PI3K) and HER family members, estrogen receptor pathways,10-14 and host factors such as antitumor immune response.15,16 Recent advances in molecular biology allow practical assessments of these newly defined and evolving subtypes of cancer, and this may inform more efficient drug development as we pursue the practical deployment of precision medicine.
Cancer and Leukemia Group B (CALGB) 40601 was a three-arm randomized phase III trial in operable HER2-positive breast cancer of preoperative chemotherapy comparing paclitaxel with the addition of trastuzumab alone or lapatinib alone or dual HER2 blockade with both drugs. Dedicated research biopsies were obtained from all participants before initiation of therapy and permitted simultaneous examination of drug effect, the impact of tumor and host factors on response to therapy, and molecular profile of residual disease.
PATIENTS AND METHODS
Study Design and Patients
Patients eligible for CALGB 40601 had newly diagnosed, histologically confirmed, untreated clinical stage II to III HER2-positive disease. HER2 positivity was determined locally by immunohistochemistry or fluorescence in situ hybridization according to American Society of Clinical Oncology/College of American Pathology guidelines.17 Patients were age ≥ 18 years, had tumors ≥ 1 cm in size, and had a pretreatment left ventricular ejection fraction ≥ 50%. Patients with multicentric or bilateral disease were eligible if the target lesion met other eligibility criteria. Each participant signed an institutional review board–approved, protocol-specific informed consent, in accordance with federal and institutional guidelines.
Treatment
Patients received paclitaxel intravenously at 80 mg/m2 once per week for 16 weeks, with the addition of trastuzumab (TH), lapatinib (TL), or both (THL). One experimental arm (ie, THL) included all three drugs; the control arm included TH; the other experimental arm (TL) substituted lapatinib for trastuzumab. Trastuzumab was administered intravenously with a loading dose of 4 mg/kg in week 1 and a dose of 2 mg/kg afterward. Lapatinib was administered orally at 1,500 mg per day alone or 1,000 mg per day when administered concurrently with trastuzumab. Because of emerging data from several trials regarding excessive diarrhea, lapatinib was reduced from 1,000 to 750 mg in the THL arm in April 2010, after 34 patients were accrued.
Left ventricular ejection fraction was measured every 8 weeks during therapy. Drug toxicities were assessed and managed using protocol-directed dose interruptions and dose reductions.
Surgery was required within 42 days of last dose. Sentinel lymphadenectomy was permitted before or after neoadjuvant therapy. Other elements of postneoadjuvant therapy management, such as axillary dissection in the event of positive sentinel lymph nodes, tumor-free surgical margins, and radiation therapy for appropriate clinical circumstances, were specified by the protocol.
Protocol-defined therapy ended at surgery. Postsurgery, patients were recommended to receive adjuvant chemotherapy with doxorubicin 60 mg/m2 plus cyclophosphamide 600 mg/m2 administered every 14 to 21 days for four cycles and 36 additional weeks of trastuzumab. Endocrine therapy was recommended for patients with estrogen receptor (ER)– or progesterone receptor–positive tumors. Patients are being observed for locoregional and distant recurrence for up to 10 years from registration.
Tumor Genomic Analyses
All enrolled patients consented to undergo four pretreatment 16-gauge core biopsies using prefabricated collection and shipping kits: two immediately placed into RNA stabilization product (RNALater, Qiagen, Hilden, Germany) and two placed into 10% neutral buffered formalin. They were shipped to the CALGB Pathology Coordinating Office for distribution to approved laboratories. Samples from the surgical specimen were requested but optional.
Genomic analyses and comparison of pretreatment and post-treatment tissues are detailed in the Appendix (online only) and used predetermined RNA-based signatures.18-23 Gene expression profiles were generated by mRNAseq using an Illumina HiSeq 2000 (San Diego, CA).24 Six-level subtype classification (basal-like, luminal A, luminal B, HER2 enriched, Claudin low, and normal-like) involved a two-step normalization process using data from The Cancer Genome Atlas. Other gene expression signatures related to HER2 signaling and response to therapy that were tested included proliferation,25 fibroblasts,19 HER2 amplicon genes,19 epidermal growth factor receptor signaling,20 ER signaling,26 p53 mutation,27 KRAS amplicon,21 PI3K pathway,22 hypoxia/vascular endothelial growth factor,23 five signatures of immune-cell infiltration (immunoglobulin G [IgG], B cells, T cells, CD8, and immune cells),18,19 and correlation with the HER2-enriched centroid.25
Data Analysis and Interpretation
Patients were initially randomly assigned with equal probability to the three study arms. Randomization was stratified by hormone receptor status (ER or progesterone receptor positive v both negative) and pretreatment clinical stage (II v III). In July 2011, based on reports of inferiority and greater toxicity of lapatinib-only regimens, the TL arm was closed; accrual continued to the THL and TH arms.
The primary end point was pCR in the breast, defined as the absence of residual invasive carcinoma. A one-df χ2 test was used for separate pairwise pCR comparisons between TH and each experimental arm; exact binomial methods were used for 95% CIs. With 300 patients, the study had 87% power to detect an increase in pCR from 30% to 50% in each experimental arm (two-sided α = 0.05). After closure of the TL arm, target accrual was revised to 230 patients in the THL and TH arms, with 85% power for the THL versus TH comparison. Secondary end points included pCR in breast and ipsilateral axillary lymph nodes (defined as no invasive tumor by hematoxylin and eosin staining in any lymph node) and adverse events. Patients who did not undergo surgery were considered non-pCR. Exploratory logistic regression included an interaction term for arm by hormone receptor status or stage. All analyses used a modified intent-to-treat approach that included only patients who began protocol therapy analyzed according to the randomly assigned arm.
pCR rate by intrinsic subtype was the primary correlative objective. Pearson's and Cochran-Mantel-Haenszel χ2 tests were used to assess the association of pCR and intrinsic subtype overall and stratified within THL and TH arms; those within the TL arm are descriptive. Secondary correlative analyses included 15 established signatures reflecting cell cycle, pathway signaling, and microenvironment, and association with pCR was considered as a continuous variable in a logistic regression model. Significance of univariable tests was considered using a Bonferroni correction (P < .0033) for multiple comparisons. A multivariable model of genomic signatures was derived in stepwise fashion by adding terms to a base model of treatment arm for genomic features and clinical tumor characteristics that reached nominal significance (LRT P < .1). Relative changes in gene expression pre- to post-treatment were explored using paired t tests.
This phase III therapeutic trial was monitored at least twice per year by the data and safety monitoring board, a standing committee composed of individuals from within and outside the Alliance. Data were collected and stored by the CALGB (Alliance) Statistics and Data Center, and quality was ensured through data review by the Data Center, the study chairperson, and the surgical co-chairperson. Data collection and statistical analyses were conducted by the CALGB (Alliance) Statistics and Data Center by Alliance statisticians using SAS software (version 9.2; SAS Institute, Cary, NC) and R software (version 3.0.1; https://www.r-project.org). All analyses were based on the study database frozen on November 1, 2013.
RESULTS
Clinical
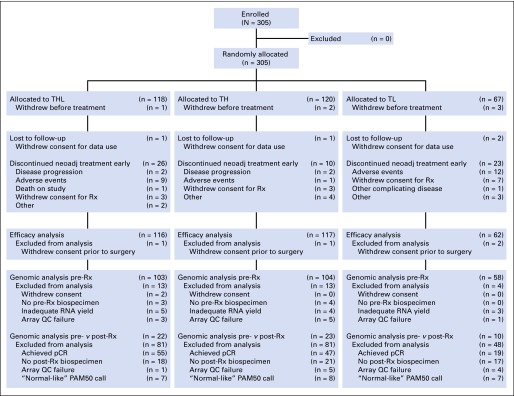
Between December 2008 and February 2012, 305 patients were enrolled, of whom 299 began protocol treatment (Fig 1). Baseline characteristics were similar across treatment arms (Table 1). Type and severity of grade ≥ 3 adverse events differed significantly by treatment, with more toxicity (particularly diarrhea and rash) and early discontinuation in the lapatinib-containing arms (Appendix Table A1, online only). There were no treatment-related deaths or episodes of symptomatic congestive heart failure.
Fig 1.
CONSORT diagram. pCR, pathologic complete response; QC, quality control; Rx, treatment; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Table 1.
Baseline Characteristics of Study Population (N = 305)
| Characteristic | No. (%) |
||
|---|---|---|---|
| THL Arm (n = 117) | TH Arm (n = 118) | TL Arm (n = 64) | |
| Age, years | |||
| Median | 48 | 50 | 50 |
| Range | 24-70 | 30-75 | 25-74 |
| Menopausal status | |||
| Pre | 72 (62) | 63 (53) | 36 (56) |
| Post | 41 (35) | 52 (44) | 27 (42) |
| Missing | 4 (3) | 3 (3) | 1 (2) |
| Racial or ethnic group | |||
| Black | 12 (10) | 7 (6) | 7 (11) |
| White | 94 (80) | 96 (81) | 48 (75) |
| Other | 11 (9) | 15 (13) | 9 (14) |
| ECOG performance status | |||
| 0 | 109 (93) | 107 (91) | 60 (94) |
| 1 | 5 (4) | 9 (8) | 3 (5) |
| Missing | 3 (3) | 2 (2) | 1 (2) |
| Hormone receptor status | |||
| ER positive, PR positive, or both | 69 (59) | 70 (59) | 37 (58) |
| ER negative and PR negative | 48 (41) | 48 (41) | 27 (42) |
| Clinical tumor size on physical examination, cm | |||
| Median | 4.0 | 4.0 | 4.0 |
| Range | 1.4-22.0 | 1.2-15.0 | 1.1-12.0 |
| Clinical stage | |||
| II | 80 (68) | 80 (68) | 47 (73) |
| III | 37 (32) | 38 (32) | 17 (27) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; ER, estrogen receptor; PR, progesterone receptor; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
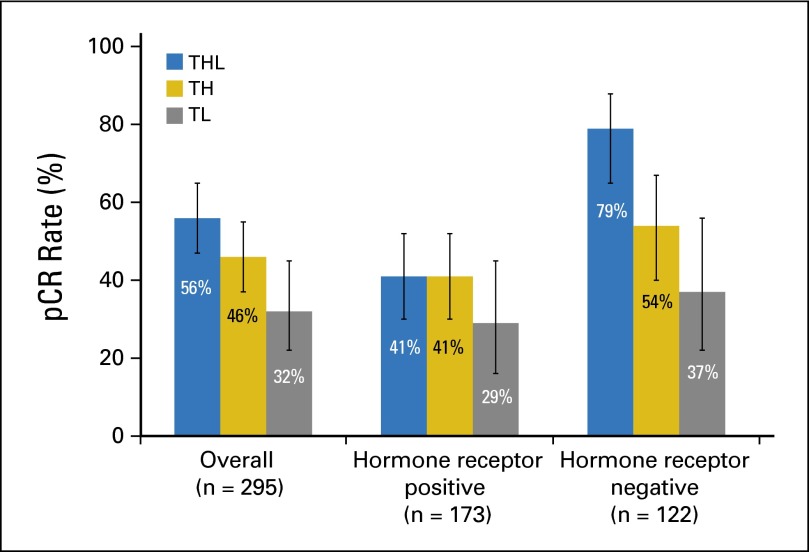
Of 295 evaluable patients, 139 experienced pCR in the breast: 56% (95% CI, 47% to 65%) in the THL arm versus 46% (95% CI, 37% to 55%) in the TH arm (P = .13); the pCR rate in the TL arm was 32% (95% CI, 22% to 45%; Fig 2). Response did not differ by treatment arm for hormone receptor–positive tumors, but it did within the receptor-negative subset; pCR in the THL arm (79%) was significantly higher than that in the TH control arm (54%; P = .01); pCR in the TL arm was the lowest of the arms (37%), with a trend toward an interaction between receptor status and treatment arm (P = .09). There was no interaction between clinical stage and treatment (Appendix Tables A2 and A3, online only).
Fig 2.
Pathologic complete response (pCR) rates in breast by treatment arm, stratified by hormone receptor status. Error bars represent 95% confidence limits. TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Gene Expression Signatures and Response
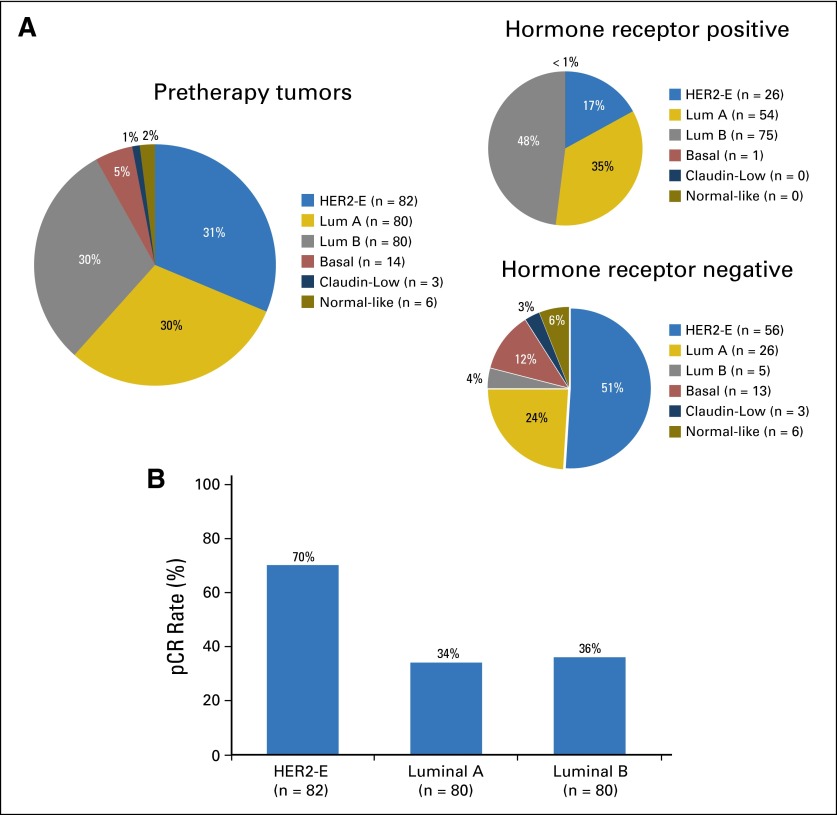
Baseline characteristics of the 265 tumors (90%) that underwent gene expression profiling did not differ from those the overall study cohort. Intrinsic subtype differed between hormone receptor–negative and –positive tumors (P < .001; Fig 3A); the largest subset of receptor-negative tumors was classified as HER2 enriched (56 [51%] of 109), whereas luminal subtypes predominated among receptor-positive tumors (129 [83%] of 156). Response varied significantly by intrinsic subtype (P < .001) and was approximately double among HER2-enriched (70%) compared with luminal A and B (34% and 36% respectively) tumors, regardless of treatment arm (Fig 3B; Appendix Tables A2 and A3) or hormone receptor status.
Fig 3.
Treatment implications of molecular heterogeneity of human epidermal growth factor receptor 2 (HER2) –positive breast cancer. (A) Intrinsic subtype overall and by hormone receptor status among study population of clinically HER2-positive tumors, demonstrating that intrinsic subtype differed between hormone receptor–negative and –positive tumors (P < .001). (B) Pathologic complete response (pCR) rates by intrinsic subtype, demonstrating significant variation by intrinsic subtype (P < .001). Uncommon subtypes not shown. HER2-E, HER2 enriched; Lum, luminal.
Genomic signatures reflecting low ER signaling, p53 mutation, high PI3K pathway signaling, high expression of HER2 amplicon genes, and correlation with the HER2-enriched centroid were significantly associated with pCR after correction for multiple testing. We also examined five published immune-cell signatures,18,19,28 finding that all of these immune signatures were highly correlated with one another (data not shown), and one (IgG signature18) remained associated with pCR after correction for multiple testing (Table 2). In multivariable analysis, treatment arm, intrinsic subtype, high HER2 amplicon expression, p53 mutation signature, and IgG immune-cell expression signature were each independently associated with pCR; clinical hormone receptor status was not (Table 2; Appendix Table A4, online only). When considered as continuous variables, ESR1 and ERBB2 gene expression as determined from the mRNAseq data was individually highly associated with pCR. In an exploratory multivariable modeling including these two genes alone as well as the predetermined signatures, genomic variables remaining significantly associated with pCR included ESR1, ERBB2, p53 signature, and IgG signature (Appendix Table A4), whereas intrinsic subtype as an overall signature, HER2 amplicon signature, and clinical assays for ER or HER2 were not.
Table 2.
Predictors of pCR
| Variable | Univariable Model |
||
|---|---|---|---|
| OR | 95% CI | P* | |
| Treatment arm | .0392 | ||
| THL v TH | 1.39 | 0.81 to 2.41 | |
| TL v TH | 0.59 | 0.3 to 1.15 | |
| Intrinsic subtype† | < .001 | ||
| Luminal A v HER2-E | 0.22 | 0.11 to 0.43 | |
| Basal v HER2-E | 0.24 | 0.07 to 0.78 | |
| Luminal B v HER2-E | 0.25 | 0.13 to 0.48 | |
| Normal v HER2-E | 0.44 | 0.08 to 2.51 | |
| Gene expression signature | |||
| p53 mutation | 2.4 | 1.69 to 3.5 | < .001 |
| IgG | 1.65 | 1.3 to 2.12 | < .001 |
| HER2 amplicon | 1.54 | 1.23 to 1.93 | < .001 |
| HER2-E correlation | 1.98 | 1.50 to 2.68 | < .001 |
| ER signaling | 0.47 | 0.33 to 0.66 | < .001 |
| B cell | 1.49 | 1.18 to 1.90 | < .001 |
| PI3K signaling | 1.72 | 1.25 to 2.41 | < .001 |
NOTE. Variables that remained significantly associated with pCR after adjusting for multiple comparisons are shown. Full list of variables and nominal and adjusted ORs from multivariable logistic regression models are included in Appendix Table A3 (online only).
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HER2-E, HER2 enriched; IgG, immunoglobulin G; OR, odds ratio; pCR, pathologic complete response; PI3K, phosphatidylinositol 3-kinase; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Derived from likelihood ratio test for each variable in logistic regression model.
Intrinsic subtype was evaluated as six-level factor (five df) in logistic regression models; zero of three Claudin-low samples achieved pCR, so CIs around OR of zero were not estimable.
DNA sequencing was successful in 181 (68%) of 265 tumors. Mutations in PIK3CA were detected in 36 (20%), including 14 (25%) of 57 HER2-enriched, four (7%) of 55 luminal A, and 16 (31%) of 51 luminal B tumors; 93% of the mutations were in exons 9 and 20. The pCR rate was 39% (14 of 36) among tumors with PIK3CA mutations and 47% (68 of 145) among those with wild-type PIK3CA (P = .5).
Residual Disease Biology
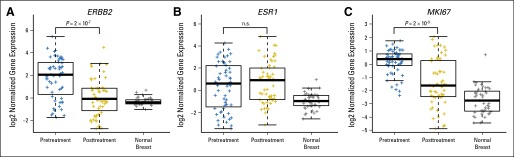
Of 144 tumors that had residual disease after treatment, 78 (54%) had successful mRNAseq on matched pre- and post-treatment tumor samples. This cohort differed from the overall cohort, with fewer (n = 36; 27%) HER2-enriched pretreatment tumors (as expected, given high pCR rates in this subtype) and more (n = 85; 63%) luminal tumors. Comparison of matched pre- and post-treatment tumors demonstrated differences in intrinsic subtype. Excluding 23 normal-like post-treatment samples, the most frequent post-treatment subtype alteration among the 55 remaining paired samples was to the luminal A subtype, occurring in 71% (12 of 17) of luminal B and 67% (six of nine) of HER2-enriched tumors (Appendix Table A5, online only) and was seen in both hormone receptor–positive (29 [78%] of 37) and –negative (nine [50%] of 18) tumors. Other studies have found intrinsic subtype maintained between samples from the same tumor29 and in matched tumor and metastatic pairs30; neither cold nor warm ischemia significantly affects subtype.31 To study whether this observation was more likely the result of treatment effect or tumor heterogeneity, we examined expression of genes implicated in both HER2-targeting response and the luminal A subtype, which revealed a significant decrease in expression of HER2 (P < .001) and the proliferation gene MKI67 (P < .001) but not in ER expression, making it less likely to represent a previously undetected subclone (Appendix Fig A1, online only), although this is speculative.
DISCUSSION
In 2013, the US Food and Drug Administration used pCR advantage in a neoadjuvant trial as the basis for accelerated approval of pertuzumab added to trastuzumab plus chemotherapy in early HER2-positive breast cancer.6 CALGB 40601 and three other trials have examined the neoadjuvant addition of lapatinib administered in a similar fashion.5,7,32 All have demonstrated numerically higher pCR rates in the dual HER2-targeting arm compared with single targeting. The results from two trials were statistically significant,5,32 whereas those of two other trials were not, including our study.7 These trials were similar in many respects, although they differed in design, including whether the regimen included a single chemotherapy drug or several drugs and the duration of the chemotherapy plus HER2-targeting regimen. The absolute pCR rates varied across these trials, including a relatively unexpectedly high pCR rate in the control arm of our trial of 46% (Appendix Table A6, online only). In CALGB 40601, the addition of lapatinib to 16 weeks of treatment with trastuzumab and paclitaxel resulted in a numeric increase in the pCR rate that did not reach statistical significance. There was no effect in hormone receptor–positive tumors, but there was a statistically significant increase in pCR in hormone receptor–negative, HER2-positive tumors. Response to therapy was associated with a number of biologic variables, including tumor molecular subtype as well as microenvironmental factors such as immune-cell gene expression. These biomarker associations have clear implications for how HER2-positive disease is viewed and need to be validated in independent data sets or a prospective trial.
This study suggests that biologic heterogeneity within HER2-positive breast cancer plays an important role in determining response to treatment. For each of the treatment approaches used in this trial, pCR was markedly higher among HER2-enriched tumors than among HER2-positive tumors of any other subtype. Among the HER2-enriched group, the TH control arm experienced a pCR of 70% in the breast, which is among the highest pCR rates ever reported in HER2-positive breast cancer. Intrinsic subtype was more important than hormone receptor status in predicting pCR; in multivariable analyses including tumor subtype, hormone receptor status was no longer significant. We also found that immune-cell gene expression predicted response to HER2 targeting that was independent of other clinical and genomic factors; this may provide additional means of identifying highly responsive tumors. This finding is consistent with reports that tumor-infiltrating lymphocytes are prognostic and predictive in HER2-positive breast cancer.15,33-36 Low B-cell receptor diversity in these tumors suggests that this is an antigen-specific response.18 Expression of the HER2 amplicon genes retained independent predictive value for pCR even after controlling for the HER2-enriched subtype, suggesting, as others have,10 that variable HER2 expression within other subtypes influences response. Within this population of clinically HER2-positive breast cancer, dominated by the luminal and HER2-enriched subtypes, we controlled for clinical ER status, which by itself was not a significant factor for predicting pCR. However, we also evaluated the quantitative expression of ESR1 mRNA and ERBB2 mRNA from our mRNAseq data treated as continuous variables, alone and in a multivariable analysis; after adjusting for the mRNA expression of these two genes, the categorical intrinsic subtype distinction was no longer a significant independent predictor of pCR. We note that all of these multivariable analyses should be considered exploratory, including the ESR1 and ERBB2 mRNA variables that have never to our knowledge been measured in this way before (ie, mRNAseq), but are encouraging and suggest that additional studies with larger sample sizes will be needed for full evaluation. In addition, if variability in pCR based on tumor and microenvironment factors translates into differences in long-term outcome, upfront assessment of these parameters may be critical.
The distribution of molecular subtypes of residual disease differed from that of untreated tumors. Under the selective pressure of combined HER2 targeting and chemotherapy, a high proportion of tumors that were not eradicated demonstrated the luminal A subtype, which is characterized by lower expression of proliferation-related genes and high expression of hormone receptor signaling–related genes. Whether this reflects stromal alterations, tumor reprogramming, intratumoral heterogeneity, decreased proliferation, and/or decreased HER2 or other specific pathway signaling cannot be adequately addressed here. We did see highly significant effects on both HER2 mRNA levels and proliferation gene expression in post-treatment tumors, but no effect on ER expression. The luminal A profile is heavily influenced by ER signaling, so this suggests a specific treatment effect rather than unmasking of a luminal A subclone, although this requires further study. Neither the treatment implications of altered biology in post-treatment tumors nor whether these changes persist over time are known, and this should not be used to make treatment decisions.
This trial demonstrates the benefits of the neoadjuvant research approach in terms of size, speed, and capacity to achieve both clinical and scientific findings. Nonetheless, there are clear limitations. Although CALGB 40601 will collect long-term outcomes of relapse-free and overall survival, it was not designed or powered for these secondary end points. Although the subtype-specific and genomic profiling analyses are intriguing, a far larger prospective trial is needed to evaluate dual versus single HER2 targeting or any specific drug combination within individual subtypes. For this reason, a pooled analysis of molecular plus clinical data from additional large randomized trials is in development. Although the intrinsic subtypes of breast cancer are increasingly familiar to practicing clinicians, additional studies are needed to clarify their role in response to HER2 targeting, as well as to examine potential interactions with antitumor immunity.
HER2-targeted drugs are among the most expensive cancer drugs. Trastuzumab-based regimens often exceed $5,000 per month, and dual therapy regimens can exceed $10,000 per month. Patients with stage I to III breast cancers receive 1 year of HER2 targeting, and treatment is generally lifelong for metastatic disease.37 Optimizing the selection of HER2-targeted regimens by identifying subpopulations of patients with HER2-positive disease who need more or less therapy could be cost effective and would spare some patients unnecessary exposure to ineffective treatments. We found substantial molecular heterogeneity of HER2-positive breast cancer that was strongly associated with variable treatment effect regardless of drug regimen, which supports a new generation of studies exploring the importance of intrinsic subtype as well as other tumor and microenvironmental variables in HER-positive disease and the implication of biologic shift in response to therapy.
Acknowledgment
We acknowledge the efforts of the Alliance for Clinical Trials in Oncology, including the Statistics and Data Center.
GLOSSARY TERMS
- genomic signatures:
the expression of a set of genes in a biologic sample (eg, blood, tissue) using microarray technology.
- HER2neu:
(human epidermal growth factor receptor 2) also called ErbB2. HER2neu belongs to the epidermal growth factor receptor (EGFR) family and is overexpressed in several solid tumors. Like EGFR, it is a tyrosine kinase receptor whose activation leads to proliferative signals within the cells. On activation, the human epidermal growth factor family of receptors are known to form homodimers and heterodimers, each with a distinct signaling activity. Because HER2 is the preferred dimerization partner when heterodimers are formed, it is important for signaling through ligands specific for any members of the family. It is typically overexpressed in several epithelial tumors.
- intrinsic subtype:
a subset of tumors that share similarities in their gene expression profile. Subtypes are identified by unsupervised analysis of gene expression.
Appendix
Methods
All of the RNA sequencing (RNAseq) and gene expression analyses were performed in the Genomics Core High Throughput Sequencing Facility and analyzed by the University of North Carolina Lineberger Comprehensive Cancer Center Bioinformatics Core at the University of North Carolina.
Intrinsic Subtyping
Gene-level abundances were estimated using the RNAseq by expectation maximization algorithm and then normalized within sample to the upper quartile of total reads, as previously described.24 For the breast tumors in Cancer and Leukemia Group B (CALGB) 40601, the intrinsic subtyping classifications were performed through a two-step normalization process with a series of 728 breast tumors from The Cancer Genome Atlas (TCGA) project (Nature 490:61-70, 2012). The mRNA sequencing data are posted at the TCGA data portal (https://tcga-data.nci.nih.gov/tcga). Briefly, an adjustment was first made to account for differences in the clinically defined human epidermal growth factor receptor 2 (HER2) –positive populations present in the two data sets by adjusting pretreatment CALGB 40601 samples to clinical HER2-positive TCGA cases. Then, the CALGB data were normalized to the TCGA-derived RNAseq correction factor; this step was needed to adjust RNAseq data to a subset of the data with the proportion of estrogen receptor (ER) –positive and ER-negative samples similar to the training data sets for PAM5025 and Claudin-low (Prat A, et al: Breast Cancer Res 12:R68, 2010) predictors.
Normalization and Intrinsic Subtyping
The goal of normalization was to correct bias that may have resulted from technical factors independent of the patient cohort differences. In the absence of controls, differences in gene-level summary measures (means or medians) may be used to estimate bias in relative expression measures. This method assumes that the same or similar population was sampled by both technologies.
For CALGB 40601, all tumors were classified as HER2 positive by clinical assay, and the gene expression data set was derived from 265 pretreatment and 55 matched post-treatment samples (Fig 1). For the TCGA data, there were 115 HER2-positive tumors defined by clinical assay, comprising 16% (115 of 728) of the TCGA population. To avoid possible confounding of treatment effect on gene expression patterns, the CALGB-to-TCGA cohort adjustment factor for each gene was calculated by taking the difference in gene summary measures (median) between the 271 pretreatment CALGB 40601 samples and the 115 clinical HER2-positive TCGA samples. For each CALGB 40601 tumor sample, the gene expression estimates were adjusted by subtracting each of the adjustment factors from its corresponding gene measurement. As shown in the principal component analysis plots of all genes, there was a good general overlap between the two data sets after this correction.
The intrinsic subtyping was then performed using the published PAM50 classifier25 and Claudin-low predictor (Prat A, et al: Breast Cancer Res 12:R68, 2010). The training sets used for both PAM50 and Claudin-low predictors were derived from microarrays. In addition, breast tumors with ER-positive status comprised 50% of the training data sets, so it was also important to normalize the CALGB 40601 data to a subset of TCGA data balanced for ER status, which we called our TCGA-derived RNAseq correction factor. TCGA data were first subsampled for a set of cases that was 50% ER positive to match the ER distribution of the PAM50 training set; this subset contained all 157 ER-negative and another randomly selected 157 ER-positive tumors. The median gene expression for the genes used in PAM50 and Claudin-low predictors was calculated, and the CALGB 40601 data were then adjusted to this median, followed by intrinsic subtyping. Principal component analysis plots illustrated a good general overlap between the CALGB 40601 and PAM50 training data sets.
Gene Expression Signatures
Gene expression signatures reflecting pathways implicated in therapeutic sensitivity were investigated in this data set. Using the combined normalized data set with the TCGA data, we applied 15 signatures to the data set in a manner consistent with their derivation. For signatures with homogeneous expression across genes, we used the median value from all genes; this included the five immune signatures, fibroblast, hypoxia or vascular endothelial growth factor, HER1, KRAS amplicon, HER2 amplicon, and phosphatidylinositol 3-kinase signature.18-23
Three of the signatures were correlated with predetermined gene centroids, including ER signaling,26 p53 mutation,27 and HER2 enriched.25 For the 11-gene proliferation signature,25 we used the mean value of the genes, as is consistent with the output from the PAM50 algorithm.
Participating Institutions
The following institutions participated in this study: Bay Area Tumor Institute Community Clinical Oncology Program (CCOP), Oakland, CA, Jon M Grief, MD; Cancer Centers of the Carolinas, Greenville, SC, Jeffrey K. Giguere, MD (supported by National Cancer Institute [NCI] Grant No. CA29165); Christiana Care Health Services CCOP, Wilmington, DE, Stephen Grubbs, MD (supported by NCI Grant No. CA45418); Dartmouth Medical School–Norris Cotton Cancer Center, Lebanon, NH, Konstantin Dragnev, MD (supported by NCI Grant No. CA04326); Dana-Farber Cancer Institute, Boston, MA, Harold J. Burstein, MD, PhD (supported by NCI Grant No. CA32291); Georgetown University Medical Center, Washington, DC, Bruce Cheson, MD (supported by NCI Grant No. CA77597); Hematology-Oncology Associates of Central New York CCOP, Syracuse, NY, Jeffrey Kirshner, MD (supported by NCI Grant No. CA45389); Massachusetts General Hospital, Boston, MA, Jeffrey W. Clark, MD (supported by NCI Grant No. CA32291); Memorial Sloan-Kettering Cancer Center, New York, NY, Clifford A. Hudis, MD (supported by NCI Grant No. CA77651); Mount Sinai Medical Center, Miami, FL, Michael A. Schwartz, MD (supported by NCI Grant No. CA45564); Mount Sinai School of Medicine, New York, NY, Lewis R. Silverman, MD (supported by NCI Grant No. CA04457); Nevada Cancer Research Foundation CCOP, Las Vegas, NV, John A. Ellerton, MD (supported by NCI Grant No. CA35421); New Hampshire Oncology-Hematology, Concord, NH, Douglas J. Weckstein, MD; Roswell Park Cancer Institute, Buffalo, NY, Ellis Levine, MD (supported by NCI Grant No. CA59518); State University of New York Upstate Medical University, Syracuse, NY, Stephen L. Graziano, MD (supported by NCI Grant No. CA21060); Ohio State University Medical Center, Columbus, OH, Clara D. Bloomfield, MD (supported by NCI Grant No. CA77658); University of California at San Francisco, San Francisco, CA, Charles J. Ryan, MD (supported by NCI Grant No. CA60138); University of Chicago, Chicago, IL, Hedy L. Kindler, MD (supported by NCI Grant No. CA41287); University of Iowa, Iowa City, IA, Daniel A. Vaena, MD (supported by NCI Grant No. CA47642); University of North Carolina at Chapel Hill, Chapel Hill, NC, Thomas C. Shea, MD (supported by NCI Grant No. CA47559); Wake Forest University School of Medicine, Winston-Salem, NC, David D. Hurd, MD (supported by NCI Grant No. CA03927); Walter Reed Army Medical Center, Washington, DC, David C. Van Echo, MD (supported by NCI Grant No. CA26806); and Washington University School of Medicine, St Louis, MO, Nancy Bartlett, MD (supported by NCI Grant No. CA77440).
Table A1.
Reasons for Patient Protocol Treatment Discontinuation and Incidence of Selected Grade ≥ 3 AEs by Arm
| Treatment Status | No. (%) |
||
|---|---|---|---|
| THL Arm | TH Arm | TL Arm | |
| Began neoadjuvant treatment | 117 (100) | 118 (100) | 64 (100) |
| Completed per protocol | 100 (85) | 108 (92) | 41 (64) |
| Ended for toxicity | 9 (8) | 1 (1) | 12 (19) |
| Ended for progression | 2 (2) | 2 (2) | 0 (0) |
| Ended for other reason | 6 (5) | 7 (6) | 11 (17) |
| Patients with delayed doses | 82 (70) | 34 (29) | 53 (83) |
| Paclitaxel | 39 (33) | 32 (27) | 33 (52) |
| Trastuzumab | 20 (17) | 25 (21) | NA |
| Lapatinib | 73 (62) | NA | 40 (63) |
| AEs* | |||
| Neutrophils† | 8 (7) | 2 (2) | 8 (12) |
| Rash‡ | 16 (14) | 2 (2) | 10 (15) |
| Diarrhea‡ | 25 (22) | 2 (2) | 14 (21) |
| Febrile neutropenia | 0 (0) | 0 (0) | 1 (2) |
| ALT† | 6 (5) | 1 (1) | 3 (5) |
| AST† | 4 (4) | 0 (0) | 3 (5) |
| Sensory neuropathy† | 11 (10) | 4 (3) | 3 (5) |
| Thrombosis | 1 (1) | 0 (0) | 0 (0) |
| Hypokalemia§ | 7 (6) | 0 (0) | 1 (2) |
Abbreviations: AE, adverse event; NA, not applicable; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Comparison between THL and TH arms.
P ≥ .10 but < .14.
P ≤ .001.
P = .021.
Table A2.
pCR Rates by Hormone Receptor Status and Stage
| No. (%) |
|||
|---|---|---|---|
| Variable |
THL Arm |
TH Arm |
TL Arm |
| Breast Only | |||
| Overall | 116 | 117 | 62 |
| pCR | 65 (56) | 54 (46) | 20 (32) |
| 95% CI, % | 47 to 65 | 37 to 55 | 22 to 45 |
| Hormone receptor positive | 69 | 69 | 35 |
| pCR | 28 (41) | 28 (41) | 10 (29) |
| 95% CI, % | 30 to 52 | 30 to 52 | 16 to 45 |
| Hormone receptor negative | 47 | 48 | 27 |
| pCR | 37 (79) | 26 (54) | 10 (37) |
| 95% CI, % | 65 to 88 | 40 to 67 | 22 to 56 |
| Stage II | 79 | 79 | 45 |
| pCR | 44 (56) | 33 (42) | 14 (31) |
| 95% CI, % | 45 to 66 | 32 to 53 | 20 to 46 |
| Stage III | 37 | 38 | 17 |
| pCR | 21 (57) | 21 (55) | 6 (35) |
| 95% CI, % | 41 to 71 | 40 to 70 | 17 to 59 |
| Breast and Axillary Lymph Nodes | |||
| Overall | 116 | 117 | 62 |
| pCR | 60 (52) | 51 (44) | 17 (27) |
| 95% CI, % | 43 to 61 | 35 to 53 | 18 to 40 |
| Hormone receptor positive | 69 | 69 | 35 |
| pCR | 28 (41) | 27 (39) | 9 (26) |
| 95% CI, % | 30 to 52 | 28 to 51 | 14 to 42 |
| Hormone receptor negative | 47 | 48 | 27 |
| pCR | 32 (68) | 24 (50) | 8 (30) |
| 95% CI, % | 54 to 80 | 36 to 64 | 16 to 49 |
Abbreviations: pCR, pathologic complete response; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Table A3.
pCR Rates by Intrinsic Subtype
| No. (%) |
||||
|---|---|---|---|---|
| Variable |
Overall (n = 265) |
THL (n = 103) |
TH (n = 104) |
TL (n = 58) |
| Breast Only | ||||
| Basal-like | 14 | 7 | 4 | 3 |
| pCR | 5 (36) | 4 (57) | 0 (0) | 1 (33) |
| 95% CI, % | 11 to 61 | 20 to 94 | NA | 0 to 87 |
| Claudin low | 3 | 1 | 1 | 1 |
| pCR | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 95% CI, % | NA | NA | NA | NA |
| HER2 enriched | 82 | 35 | 24 | 23 |
| pCR | 57 (70) | 28 (80) | 17 (71) | 12 (52) |
| 95% CI, % | 60 to 79 | 67 to 93 | 53 to 89 | 32 to 73 |
| Luminal A | 80 | 30 | 39 | 11 |
| pCR | 27 (34) | 11 (37) | 15 (38) | 1 (9) |
| 95% CI, % | 24 to 44 | 19 to 54 | 23 to 54 | 0 to 26 |
| Luminal B | 80 | 30 | 32 | 18 |
| pCR | 29 (36) | 12 (40) | 13 (41) | 4 (22) |
| 95% CI, % | 26 to 47 | 22 to 58 | 24 to 58 | 3 to 41 |
| Normal-like | 6 | 0 | 4 | 2 |
| pCR | 3 (50) | NA | 2 (50) | 1 (50) |
| 95% CI, % | 10 to 90 | NA | 1 to 99 | 1 to 100 |
Abbreviations: HER2, human epidermal growth factor receptor 2; NA, not applicable; pCR, pathologic complete response; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel plus lapatinib.
Table A4.
All Variables Considered in Univariable and Multivariable Logistic Regression Modeling of Predictors of pCR
| Variable | Univariable Model |
Multivariable |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Model One* |
Model Two† |
||||||||
| OR | 95% CI | P | OR | 95% CI | P‡ | OR | 95% CI | P‡ | |
| Treatment arm | .0392 | .0077 | .0114 | ||||||
| THL v TH | 1.39 | 0.81 to 2.41 | 1.43 | 0.76 to 2.71 | 1.5 | 0.8 to 2.85 | |||
| TL v TH | 0.59 | 0.30 to 1.15 | 0.43 | 0.19 to 0.93 | 0.48 | 0.21 to 1.03 | |||
| Hormone receptor§ | 2.17 | 1.33 to 3.59 | < .001 | NC | NS | ||||
| Clinical stage II v III | 0.67 | 0.40 to 1.13 | .6548 | NC | NS | ||||
| Intrinsic subtype‖ | < .001 | .0264 | |||||||
| Luminal A v HER2-E | 0.22 | 0.11 to 0.43 | 0.61 | 0.22 to 1.66 | NS | ||||
| Basal v HER2-E | 0.24 | 0.07 to 0.78 | 0.24 | 0.06 to 0.90 | NS | ||||
| Luminal B v HER2-E | 0.25 | 0.13 to 0.48 | 0.39 | 0.18 to 0.81 | NS | ||||
| Normal v HER2-E | 0.44 | 0.08 to 2.51 | 1.66 | 0.21 to 14.02 | NS | ||||
| Gene expression¶ | |||||||||
| HER2 | 2.2 | 1.68 to 2.93 | < .001 | NC | 1.68 | 1.25 to 2.28 | < .001 | ||
| ESR1 | 0.54 | 0.43 to 0.67 | < .001 | NC | 0.71 | 0.54 to 0.93 | .0139 | ||
| Gene expression signature | |||||||||
| p53 mutation | 2.40 | 1.69 to 3.50 | < .001 | 2.06 | 1.17 to 3.70 | .0119 | 2.33 | 1.18 to 4.71 | .014 |
| IgG | 1.65 | 1.30 to 2.12 | < .001 | 1.54 | 1.16 to 2.05 | .0024 | 1.43 | 1.08 to 1.92 | .0112 |
| HER2 amplicon | 1.54 | 1.23 to 1.93 | < .001 | 1.35 | 1.04 to 1.77 | .0252 | NS | ||
| HER2-E correlation | 1.98 | 1.50 to 2.68 | < .001 | NS | NS | ||||
| ER signaling | 0.47 | 0.33 to 0.66 | < .001 | NS | NS | ||||
| B cell | 1.49 | 1.18 to 1.90 | < .001 | NS | NS | ||||
| PI3K signaling | 1.72 | 1.25 to 2.41 | < .001 | NS | NS | ||||
| T cell | 1.39 | 1.09 to 1.79 | .0073 | NS | NS | ||||
| HER1 | 1.50 | 1.10 to 2.07 | .0103 | NS | NS | ||||
| CD8 | 1.37 | 1.07 to 1.76 | .0115 | NS | NS | ||||
| Proliferation | 1.43 | 1.07 to 1.93 | .0153 | NS | NS | ||||
| Immune cell | 1.34 | 1.05 to 1.70 | .0161 | NS | NS | ||||
| Hypoxia/VEGF | 1.26 | 0.98 to 1.64 | .0717 | NS | NS | ||||
| Fibroblast | 0.84 | 0.64 to 1.09 | .1852 | NS | NS | ||||
| KRAS amplicon | 1.11 | 0.87 to 1.43 | .4144 | NS | NS | ||||
Abbreviations: HER2, human epidermal growth factor receptor 2; HER2-E, HER2 enriched; IgG, immunoglobulin G; NC, not considered; NS, not selected; OR, odds ratio; pCR, pathologic complete response; PI3K, phosphatidylinositol 3-kinase.
Using preplanned genomic signatures.
Using same as model one, including clinical factors and with ESR1 and ERBB2 gene expression considered as continuous variables.
Derived from likelihood ratio test for each variable in logistic regression model.
ER and progesterone receptor negative versus either receptor positive.
Intrinsic subtype was evaluated as six-level factor (five df) in logistic regression models; zero of three Claudin-low samples achieved pCR, so CIs around OR of zero were not estimable.
mRNA expression levels of ESR1 and HER2 determined by mRNA sequencing and treated as continuous variables.
Table A5.
Classification of Samples Post-Treatment
| Pretreatment Subtype | No. (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| pCR | Residual Disease Post-Treatment Subtype |
|||||||
| HER2 Enriched | Luminal A | Luminal B | Basal-Like | Claudin Low | Normal-Like | NA* | ||
| HER2 enriched (n = 82) | 57 (70) | 3 (4) | 6 (7) | 0 | 0 | 0 | 3 (4) | 13 (16) |
| Luminal A (n = 80) | 27 (34) | 3 (4) | 20 (25) | 1 (1) | 0 | 0 | 7 (9) | 22 (28) |
| Luminal B (n = 80) | 29 (36) | 0 | 12 (15) | 3 (4) | 0 | 2 (3) | 9 (11) | 25 (31) |
| Basal-like (n = 14) | 5 (36) | 0 | 0 | 0 | 3 (21) | 0 | 1 (7) | 5 (36) |
| Claudin low (n = 3) | 0 | 1 (33) | 0 | 0 | 0 | 1 (33) | 1 (33) | 0 |
| Normal-like (n = 6) | 3 (50) | 0 | 0 | 0 | 0 | 0 | 2 (33) | 1 (16) |
NOTE. Pretreatment samples with intrinsic subtype information (n = 265) were compared with post-treatment intrinsic subtype for samples with residual disease. Main change with treatment seen in these paired samples was to luminal A phenotype, confirming that biology of residual disease differs from that of treatment-naive tumors.
Abbreviations: HER2, human epidermal growth factor receptor 2; NA, not available; pCR, pathologic complete response.
Lack of consent for post-treatment sample, too little RNA for RNA sequencing, or RNA sequencing failed.
Table A6.
Randomized Neoadjuvant Studies in HER2-Positive Disease Testing Dual HER2 Targeting (v single anti-HER2 drug) in Combination With Chemotherapy
| Study | No. of Patients | Chemotherapy | HER2-Targeting Drug | HER2-Targeting Duration (weeks) | pCR in Breast (%) |
|---|---|---|---|---|---|
| C40601 | 117 | Paclitaxel | Trastuzumab | 16 | 46 |
| 118 | Paclitaxel | Trastuzumab + lapatinib | 16 | 56 | |
| NeoSPHERE5,6 | 107 | Docetaxel | Trastuzumab | 12 | 29 |
| 107 | Docetaxel | Trastuzumab + pertuzumab | 12 | 46* | |
| 107 | None | Trastuzumab + pertuzumab | 12 | 17 | |
| B-418 | 176 | AC then paclitaxel | Trastuzumab | 16 | 53 |
| 165 | AC then paclitaxel | Trastuzumab + pertuzumab | 62 | ||
| NeoALTTO2 | 149 | Paclitaxel | Trastuzumab | 18 | 30 |
| 152 | Paclitaxel | Trastuzumab + lapatinib | (12 with chemotherapy) | 51* | |
| CHER-LOB32 | 36 | Paclitaxel then FEC | Trastuzumab | 26 | 25 |
| Paclitaxel then FEC | Lapatinib | 26 | 26 | ||
| Paclitaxel then FEC | Trastuzumab + lapatinib | 26 | 47* |
Abbreviations: AC, doxorubicin plus cyclophosphamide; CHER-LOB, Chemotherapy Plus Lapatinib or Trastuzumab or Both in HER2-Positive Operable Breast Cancer; FEC, fluorouracil, epirubicin, and cyclophosphamide; HER2, human epidermal growth factor receptor 2; NeoALLTO, Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimisation; NeoSPHERE, Neoadjuvant Study of Pertuzumab and Herceptin in an Early Regimen Evaluation; pCR, pathologic complete response.
Statistically significant increase in pCR according to trial prespecifications. Of note, CHER-LOB (Chemotherapy, Herceptin and Lapatinib in Operable Breast Cancer) only reported pCR in breast and axillary lymph nodes (in breast not reported).
Fig A1.
Expression of target genes HER2 (ERBB2), estrogen receptor (ESR1), and proliferation gene Ki67 (MKI67) with treatment among 55 tumors with paired pretherapy and residual disease samples, demonstrating significant decrease in human epidermal growth factor receptor 2 and proliferation gene expression, but no change in estrogen receptor expression. Normal breast gene expression is provided to illustrate baseline expression patterns.
Footnotes
Processed as a Rapid Communication manuscript.
See accompanying editorial on page 521
Written on behalf of the Alliance for Clinical Trials in Oncology.
Supported by the National Cancer Institute (NCI), National Institutes of Health, under Awards No. U10CA180821 and U10CA180882 to the Alliance for Clinical Trials in Oncology, as well as NCI Grants No. 1U10CA180838, 1U10CA180867, 1U10CA180801, and 1U10CA180791, and in part by grants from the Breast Cancer Research Foundation (Alliance, L.A.C., C.M.P.), Susan G. Komen (L.A.C.), GlaxoSmithKline, and the University of North Carolina Specialized Program of Research Excellence in Breast Cancer (NCI Grant No. P50-CA58223). Alliance/Cancer and Leukemia Group B is supported by NCI Grant No. CA31946 and Alliance Statistics and Data Center by NCI Grant No. CA33601.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Presented in part at the 49th and 50th Annual Meetings of the American Society of Clinical Oncology, Chicago, IL, May 31-June 4, 2013, and May 30-June 3, 2014, respectively.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
Clinical trial information: NCT00770809.
AUTHOR CONTRIBUTIONS
Conception and design: Lisa A. Carey, Donald A. Berry, Lyndsay N. Harris, David W. Ollila, Baljit Singh, Charles M. Perou, Eric P. Winer, Clifford A. Hudis
Financial support: Lisa A. Carey, Charles M. Perou, Clifford A. Hudis
Administrative support: Clifford A. Hudis
Provision of study materials or patients: Lisa A. Carey, David W. Ollila, Ian Krop, Clifford A. Hudis
Collection and assembly of data: Lisa A. Carey, Ian E. Krop, Norah Lynn Henry, Douglas J. Weckstein, Carey K. Anders, Katherine A. Hoadley, Charles M. Perou
Data analysis and interpretation: Lisa A. Carey, Donald A. Berry, Constance T. Cirrincione, William T. Barry, Brandelyn N. Pitcher, Katherine A. Hoadley, Michael Iglesia, Maggie Chon U. Cheang, Charles M. Perou, Eric P. Winer, Clifford A. Hudis
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Lisa A. Carey
Research Funding: GlaxoSmithKline, Genentech/Roche
Donald A. Berry
Employment: Berry Consultants
Leadership: Berry Consultants
Stock or Other Ownership: Berry Consultants
Consulting or Advisory Role: Berry Consultants
Travel, Accommodations, Expenses: Berry Consultants
Constance T. Cirrincione
No relationship to disclose
William T. Barry
No relationship to disclose
Brandelyn N. Pitcher
No relationship to disclose
Lyndsay N. Harris
No relationship to disclose
David W. Ollila
No relationship to disclose
Ian E. Krop
Employment: Vertex Pharmaceuticals (I), AMAG Pharmaceuticals (I)
Leadership: AMAG Pharmaceuticals (I)
Stock or Other Ownership: Vertex Pharmaceuticals (I), AMAG Pharmaceuticals (I)
Research Funding: Genentech
Travel, Accommodations, Expenses: Bayer
Norah Lynn Henry
Research Funding: sanofi-aventis, BioMarin, Celldex
Travel, Accommodations, Expenses: Celldex
Douglas J. Weckstein
No relationship to disclose
Carey K. Anders
Consulting or Advisory Role: sanofi-aventis, BiPar Sciences, Novartis, Merrimack, Genentech Roche, GERON, to-BBB technologies BV, Eli Lilly, Nektar
Research Funding: sanofi-aventis, BiPar Sciences, Novartis, Bristol-Myers Squibb, Geron, to-BBB technologies BV, Puma, Angiochem, Merrimack
Patents, Royalties, Other Intellectual Property: UptoDate.com, Jones and Bartlell
Baljit Singh
No relationship to disclose
Katherine A. Hoadley
No relationship to disclose
Michael Iglesia
No relationship to disclose
Maggie Chon U. Cheang
Patents, Royalties, Other Intellectual Property: PAM50 patent (no financial gain in past 2 years)
Charles M. Perou
Leadership: Bioclassifier, GenCentric Diagnostics
Stock or Other Ownership: Bioclassifier, GeneCentric Diagnostics
Consulting or Advisory Role: Bioclassifier, GeneCentric Diagnostics
Patents, Royalties, Other Intellectual Property: royalties from PAM50 breast cancer gene patent application and from lung gene signature patent
Eric P. Winer
Consulting or Advisory Role: Verastem
Research Funding: Genentech/Roche (Inst), Novartis (Inst), Merck (Inst)
Travel, Accommodations, Expenses: Genentech/Roche
Clifford A. Hudis
Consulting or Advisory Role: Roche/Genentech, Novartis
REFERENCES
- 1.Perez EA, Romond EH, Suman VJ, et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2–positive breast cancer: Planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32:3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baselga J, Cortés J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2–positive metastatic breast cancer: Final results from the EGF104900 study. J Clin Oncol. 2012;30:2585–2592. doi: 10.1200/JCO.2011.35.6725. [DOI] [PubMed] [Google Scholar]
- 4.Prowell TM, Pazdur R. Pathological complete response and accelerated drug approval in early breast cancer. N Engl J Med. 2012;366:2438–2441. doi: 10.1056/NEJMp1205737. [DOI] [PubMed] [Google Scholar]
- 5.Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 7.Robidoux A, Tang G, Rastogi P, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): An open-label, randomised phase 3 trial. Lancet Oncol. 2013;14:1183–1192. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 8.Piccart-Gebhart MJ, Holmes AP, Baselga J, et al. First results from the phase III ALTTO trial (BIG 2-06; NCCTG [Alliance] N063D) comparing one year of anti-HER2 therapy with lapatinib alone (L), trastuzumab alone (T), their sequence (T-L), or their combination (T+L) in the adjuvant treatment of HER2-positive early breast cancer (EBC) J Clin Oncol. 2014;32(suppl 15s):4s. (suppl 15s; abstr LBA4) [Google Scholar]
- 9.Montemurro F, Prat A, Rossi V, et al. Potential biomarkers of long-term benefit from single-agent trastuzumab or lapatinib in HER2-positive metastatic breast cancer. Mol Oncol. 2014;8:20–26. doi: 10.1016/j.molonc.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pogue-Geile KL, Kim C, Jeong JH, et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105:1782–1788. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dave B, Migliaccio I, Gutierrez MC, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2–overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Nagata Y, Lan K, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:17–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Zhang Q, Zhang J, et al. P13K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer. 2011;11:248. doi: 10.1186/1471-2407-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loi S, Michiels S, Salgado R, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 16.Bianchini G, Gianni L. The immune system and response to HER2-targeted treatment in breast cancer. Lancet Oncol. 2014;15:e58–e68. doi: 10.1016/S1470-2045(13)70477-7. [DOI] [PubMed] [Google Scholar]
- 17.Wolff A, Hammond M, Schwartz J, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 18.Iglesia MD, Vincent BG, Parker JS, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20:3818–3829. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan C, Prat A, Parker JS, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoadley KA, Weigman VJ, Fan C, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutti JE, Pfefferle AD, Russell SC, et al. Oncogenic PI3K mutations lead to NF-kappaB-dependent cytokine expression following growth factor deprivation. Cancer Res. 2012;72:3260–3269. doi: 10.1158/0008-5472.CAN-11-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z, Fan C, Livasy C, et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 2009;7:9. doi: 10.1186/1741-7015-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammerman PS, Lawrence MS, Voet D, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh DS, Troester MA, Usary J, et al. Estrogen-regulated genes predict survival in hormone receptor–positive breast cancers. J Clin Oncol. 2006;24:1656–1664. doi: 10.1200/JCO.2005.03.2755. [DOI] [PubMed] [Google Scholar]
- 27.Troester MA, Herschkowitz JI, Oh DS, et al. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer. 2006;6:276. doi: 10.1186/1471-2407-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexe G, Dalgin GS, Scanfeld D, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67:10669–10676. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 29.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 30.Weigelt B, Mackay A, A'Hern R, et al. Breast cancer molecular profiling with single sample predictors: A retrospective analysis. Lancet Oncol. 2010;11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 31.Hatzis C, Sun H, Yao H, et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J Natl Cancer Inst. 2011;103:1871–1883. doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarneri V, Frassoldati A, Bottini A, et al. Preoperative chemotherapy plus trastuzumab, lapatinib, or both in human epidermal growth factor receptor 2–positive operable breast cancer: Results of the randomized phase II CHER-LOB study. J Clin Oncol. 2012;30:1989–1995. doi: 10.1200/JCO.2011.39.0823. [DOI] [PubMed] [Google Scholar]
- 33.Bellati F, Napoletano C, Ruscito I, et al. Cellular adaptive immune system plays a crucial role in trastuzumab clinical efficacy. J Clin Oncol. 2010;28:e369–e370. doi: 10.1200/JCO.2010.28.6922. author reply e371. [DOI] [PubMed] [Google Scholar]
- 34.Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: Clinical response, cellular immunity, and immunoescape. J Clin Oncol. 2010;28:4390–4399. doi: 10.1200/JCO.2009.27.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gianni L, Bianchini G, Valagussa P, et al. Adaptive immune system and immune checkpoints are associated with response to pertuzumab (P) and trastuzumab (H) in the NeoSphere study. Cancer Res. 2012;72(suppl 24) 72, 2012 (suppl 24; abstr s6-7) [Google Scholar]
- 36.Perez EA, Thompson EA, Ballman KV, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group N9831 adjuvant trastuzumab trial. J Clin Oncol. 2015;33:701–708. doi: 10.1200/JCO.2014.57.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theriault RL, Carlson RW, Allred C, et al. Breast cancer, version 3.2013: Featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2013;11:753–761. doi: 10.6004/jnccn.2013.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]