Abstract
Copy number variation (CNV) is a newly discovered form of intra-species genetic polymorphism that is defined as deletions or duplications of genome segments ranging from 1 kbp to several Mbp. CNV accounts for the majority of the genetic variation observed in humans (CNV regions cover more than 10% of the human genome); therefore, it may significantly influence both the phenotype and susceptibility to various diseases. Unfortunately, the impact of CNV on a number of diseases, including hepatitis C virus (HCV) infection, remains largely unexplored. Here, we analyzed 421 human genes encoding proteins that have been shown to interact with HCV proteins or genomic RNA (proteins from the HCV-human interactome). We found that 19 of the 421 candidate genes are located in putative CNV regions. For all of these genes, copy numbers were determined for European, Asiatic and African populations using the multiplex ligation-dependent amplification (MLPA) method. As a result, we identified 4 genes, IGLL1, MLLT4, PDPK1, PPP1R13L, for which the CN-genotype ranged from 1 to 6. All of these genes are involved in host-virus interaction; thus, their polymorphism has a potential impact on the development of HCV infection and/or therapy outcome.
The hepatitis C virus (HCV), discovered in 1989, is a hepatotropic, positive-sense, single-stranded (ss) RNA virus that belongs to the family Flaviviridae1. The HCV genome encodes 10 mature viral proteins, including 3 structural proteins (C, E1 and E2) and 7 nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B)2. The potential consequences of HCV infection vary in severity from a transient and symptomless illness to a mortal, lifelong disease. Approximately 15–45% of infected persons spontaneously clear the virus without treatment during the acute phase of the infection. The remaining individuals develop chronic hepatitis C (CHC). Globally, an estimated 130–150 million people are chronically infected with HCV. Approximately 15–30% of patients with CHC develop cirrhosis over a period of 20 years. Those with cirrhosis have a poor prognosis due to a high risk of liver failure and hepatocellular carcinoma (HCC). Consequently, nearly 500 000 people die each year from hepatitis C-related liver diseases3.
Since the discovery of HCV, a number of studies have aimed to identify human- and virus-specific factors that affect the course of HCV infection and patient response to antiviral treatment. The majority of these studies have focused on the three general issues: (i) the genetic variability of HCV, (ii) the clinical status of the patient, and (iii) the genetic variation of the human population.
HCV, similar to other RNA viruses, displays an extremely high level of genetic variability. HCV is classified into 7 genotypes (numbered 1–7), which are further divided into more than 60 subtypes4,5. Genotype 1 is the most common worldwide (46.2% of infected individuals), followed by genotype 3 (30.1%) and genotypes 2, 4 and 6 (22.8% collectively). Genotypes 5 and 7 comprise the remaining 0.9%6. The major source of HCV genetic variability is the highly efficient (1012 virions per day) but low-fidelity replication5 by viral RNA-dependent RNA polymerase (NS5B) that lacks proofreading activity (this enzyme exhibits an error rate of 8.7 × 10−3−1.4 × 10−6 per site)2,7,8. The consequence of this continuous variation in the HCV genome is a presence in a single host of a collection of closely related, but genetically divergent, viral variants called a quasispecies9,10. It has been shown that susceptibility to antiviral treatment correlates with HCV variability on both the genotype and quasispecies level. Furthermore, there are a number of reports that show a genotype-specific response in patients treated with interferon and ribavirin (IFN-RBV)5,11 a standard HCV treatment before 201312. The effect of genotype, however, seems to be less prominent in the case of currently used therapies that are based on direct-acting antiviral agents (DAAs)13. In addition, analyses of viral populations isolated from individual patients have revealed a correlation between the complexity of HCV quasispecies and the outcome of CHC therapy14,15,16,17,18. Other studies have indicated that the course of viral infection and the patient’s response to the antiviral treatment may be influenced by specific mutations within the viral genome19,20. Moreover, previous data from our laboratory indicated the existence of HCV variants with higher fitness, which are capable of persisting in epidemiologically unrelated hosts14,21.
The clinical status of the patient is the second important determinant of the course of HCV infection and therapeutic outcome. The factors that have the most significant association with poor prognosis in HCV infection are HCV/HIV co-infection, alcoholism, immune system disorders as well as HCV-related and non-related oncogenesis22,23,24,25,26.
Human genetic variation is also considered one of the most significant factors that underlie the unique susceptibility of each person to HCV infection and antiviral treatment. Genome-wide association studies (GWAS) have identified that single nucleotide polymorphisms (SNPs) near the IL28B gene are strongly associated with both the spontaneous and treatment-induced clearance of HCV infection27,28,29. SNPs of this gene are also one of the factors underlying the observed differences in sustained virological response (SVR) rate to IFN-RBV treatment in various human populations27. Recently, another type of intra-species polymorphism with significant biological implications, called copy number variation (CNV), has been discovered. CNV is caused by DNA alterations that result in copy number (CN) changes of a particular DNA segment larger than 1 kb. The location of the protein-coding gene within CNV region (which is frequently observed in humans) may affect gene dosage and/or alternate gene structure. Consequently, CNV may significantly influence the phenotypes30,31,32,33 and contribute to individual variation in drug response, immune defense, disease resistance or susceptibility. The most prominent example of a CNV-based gene dosage variation with clinical implication are CN changes of CCL3L1 (chemokine (C-C motif) ligand 3-like 1) – a gene encoding chemokine that is a ligand to the co-receptor for human immunodeficiency virus (HIV). It has been found that the CN of this gene ranges from 0 to 14 in diploid human genomes34,35. In addition, it has been reported that the lower CN of CCL3L1 is an important genetic determinant for increased HIV-1 susceptibility and faster AIDS progression36. However, there are many controversies and conflicting results regarding this association37,38,39,40 (discussed in ref. 41). It was suggested that observed discordances result mostly from the lack of appropriate CNV genotyping method allowing unequivocal separation of CN-genotypes. It has also been shown that lower CN of the CCL3L1 gene, compared to the population median, is associated with susceptibility to CHC32,42. According to our best knowledge, this is the only known example of an association between a course of HCV infection and CN changes of a human gene.
In this study, we identified genes that encode proteins belonging to the HCV-human interactome that undergo CN changes and whose dosage effect could potentially impact the course of HCV infection and/or treatment outcome. After gene identification, we experimentally characterized 19 of them in individuals from different human populations and proved the polymorphic nature of four of them. Our data also confirmed previous reports regarding CCL3L1 CN distribution among different populations30,32,35.
Results
Selection of genes
To identify candidate human genes overlapping with CNVs for which the dosage effect could correlate with a course of HCV infection and/or treatment outcome, we focused on 421 genes (Supplementary Table S1) encoding proteins that have been shown to interact with HCV proteins or HCV genome43 (the most comprehensive dataset of HCV-human interactome available when we began our studies). To determine whether these genes co-localize with known CNV regions, we searched the Database of Genomic Variants (DGV)44, which collects the data regarding structural variation that have been reported to date. For our analysis, we selected only the genes for which the overlaps with CNV regions were confirmed in at least 3 independent publications (Fig. 1a). As a result, we identified 19 candidate genes (Supplementary Table S2) and selected them for further experimental studies. Additionally, we added CCL3L1, a highly polymorphic gene, to the group of the selected genes to serve as a positive control.
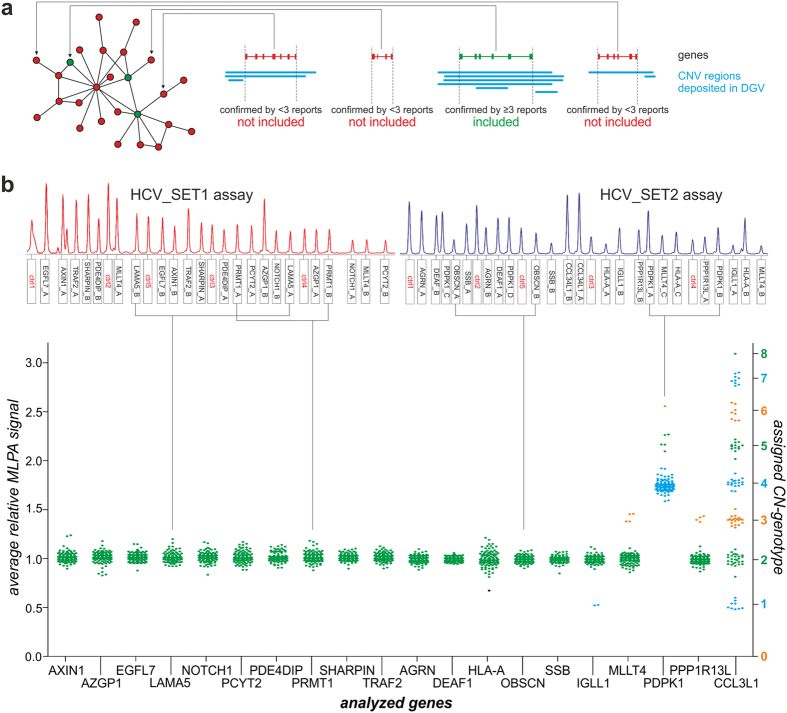
Figure 1. MLPA analysis of selected genes.
(a) Gene selection strategy. Schematic representation (on the left) shows examples of human genes involved in the HCV-human interactome network43. The gene was included in our analysis if the overlap between this gene (green) and CNV region deposited in DGV (blue) was confirmed by at least 3 independent reports. Other genes (red) were not selected for analysis in this study. (b) MLPA analysis. Presented electropherograms (upper panel) show two MLPA assays designed for analysis of selected genes: HCV_SET1 assay (red) and HCV_SET2 assay (blue). Probe IDs are indicated under the electropherograms. Control probes are indicated in red. One-dimensional signal scatter-plot (bottom panel) shows an average relative signal of gene specific MLPA probes (left y-axis) and assigned CN-genotypes (right y-axis) for selected genes (IDs indicated on x-axis) in all analyzed samples. One dot represents one sample, colored in accordance with assigned CN-genotype (right y-axis). A black dot represents a sample with no CN-genotype assigned.
CNV analysis
To analyze the 20 selected genes, we applied the multiplex ligation-dependent amplification (MLPA)45 method and the strategy of probes and assays design that we successfully used before46,47. For each gene, we designed two MLPA probes. For genes localized in more complex genomic regions, we designed additional probes: (i) one for HLA-A (major histocompatibility complex, class I, A) and MLLT4 (myeloid/lymphoid or mixed-lineage leukemia; translocated to, 4) and (ii) two for PDPK1 (3-phosphoinositide dependent protein kinase 1). All probes were split between two MLPA assays named: HCV_SET1 and HCV_SET2. The HCV_SET1 assay consisted of 22 probes that were specific for 11 of the selected genes, and the HCV_SET2 assay included 23 probes that were specific for 10 of the selected genes. Each assay also contained five control probes, specific to CN stable regions, which were used for the normalization of the run-to-run MLPA probe signal variation. The detailed features of all MLPA probes and assays layout are shown in Supplementary Table S3.
Using both assays, we analyzed the CN changes of the selected genes in 106 DNA samples from three different human populations: 31 European (CEU) samples, 48 Asiatic (CHB/JPT) samples and 27 African (YRI) samples.
The representative results of MLPA analysis (MLPA signal electropherograms) are presented in Fig. 1b (upper panel). The results obtained for all of the analyzed genes in all samples are summarized in signal scatter plot (Fig. 1b, bottom panel). The data presented in Fig. 1b indicated that the following four of the 19 total analyzed human genes showed variation in CN: (i) IGLL1 (immunoglobulin lambda like polypeptide 1), (ii) MLLT4, (iii) PDPK1, and (iv) PPP1R13L (protein phosphatase 1 regulatory subunit 13 like); the observed CN-genotypes (distinct signal clusters) ranged from 1 to 6 copies. The selected positive control gene, CCL3L1, also proved to be polymorphic, with CN range from 1 to 8, which was consistent with our previous results35. Other genes proved to be non-polymorphic and showed only one cluster corresponding to 2 CN-genotype. The list of the assigned CN-genotypes for all of the samples is shown in Supplementary Table S4.
The extent of CNV of polymorphic genes
For all polymorphic genes we observed the occurrence of several (≥2) CN-genotypes, one of which was predominant (major CN-genotype); the alternative genotypes were represented only by a small number of samples (minor CN-genotypes). For three polymorphic genes, we observed duplications (PPP1R13L and MLLT4 genes) or deletions (IGLL1 gene) of one of the gene copies, resulting in a higher (3) and lower (1) CN compared to major CN-genotype, respectively. For these genes, the European population was the most variable, with 7 (22.6%) samples presenting the minor CN-genotype. For the PDPK1 gene, we observed an increased CN for the major genotype (CN=4) with additional duplications that resulted in minor CN-genotypes of 5 and 6 copies. There was no sample demonstrating the minor CN-genotype for more than one gene.
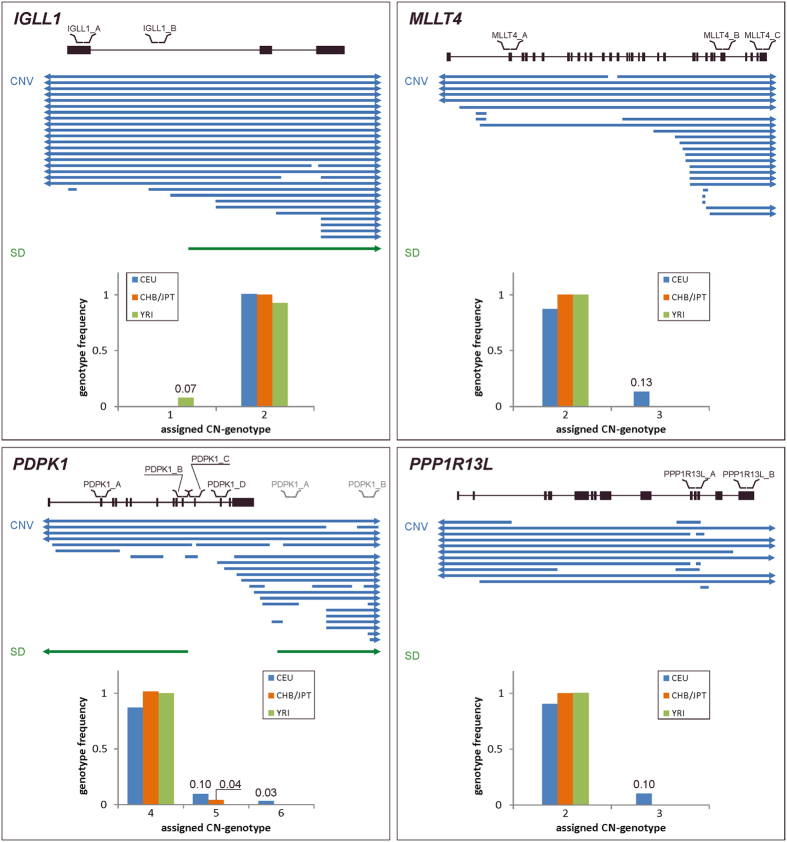
According to the DGV, all of the designed MLPA probes were located within the CNV regions overlapping selected genes. However, a detailed analysis of these regions revealed that two of the regions have more complex structures (Fig. 2). The results obtained for the three probes designed for the MLLT4 gene showed that two of the probes (MLLT4_B and MLLT4_C) targeted the polymorphic region and that the third one (MLLT4_A) targeted a non-polymorphic fragment of the gene. This result suggested that not the entire gene is located within the polymorphic region but only several exons at its 3′ end and only that part of the gene underwent CN changes in the studied samples. The structure of the region where the PDPK1 gene is localized is even more complex. Two of the probes designed for this gene (PDPK1_C and PDPK1_D) showed no polymorphism, whereas the other two probes (PDPK1_A and PDPK1_B) showed a variable number of copies. This result suggested that the polymorphic region covers only several exons at the 5′ end and that the exons at the 3′ end appear to be located outside this region. Additionally, probes PDPK1_A and PDPK1_B co-localized with segmental duplication (SD), therefore, during the MLPA experiments they hybridized to two sites, within and downstream of PDPK1 gene, thus producing a higher MLPA signal and, consequently, higher CN-genotypes.
Figure 2. Detailed characteristics of all polymorphic genes identified in this study.
Each panel shows the map of the gene with localization of all probes indicated over the map. Below the map, common structural variants available in DGV database are shown: CNV (blue) and SD (green). Arrowheads indicate that CNV/SD exceed the area shown in the figure. Bar graphs (bottom) show the frequency (y-axis) of a particular CN-genotype (x-axis) in analyzed samples from different human populations: CEU (blue), CHB/JPT (orange) and YRI (green). Note that due to the PDPK1 gene overlap with SD, two probes (PDPK1_A and PDPK1_B) map to two genomic locations, within and downstream of PDPK1. The probes mapping out of the gene sites are shown in grey to distinguish them from those mapping within the gene.
Discussion
In this study, we used bioinformatics methods to identify all human genes that both (i) encode proteins that belong to the HCV-human interactome43 and (ii) according to at least 3 independent reports co-localize with known CNV regions (listed in the DGV44). It is expected that the structural polymorphism of these genes may impact the course of HCV infection and/or treatment outcome. The above criterion was fulfilled by 19 genes, for which the changes in CN were determined using the MLPA in a cohort of 106 individuals from 3 different populations. As a result, we identified 4 polymorphic genes (IGLL1, MLLT4, PDPK1, PPP1R13L) that showed variations in CN ranging from 1 to 6 copies among the tested samples. The detailed characteristics of the structure of the polymorphic genes revealed that two of them (MLLT4 and PDPK1) were partially overlapped by CNV regions. For MLLT4, our analysis suggested that only terminal exons at the 3′ end were located within the CNV region. In the case of PDPK1, the CNV region covered ten initial gene exons at the 5′ end but not the 3′ terminal ones. IGLL1 and PPP1R13L were overlapped by CNV regions, at least in the parts where the probes were located. Three of the four identified polymorphic genes (IGLL1, PPP1R13L, MLLT4) showed CN=2 for the major CN-genotype (predominant genotype among populations). The polymorphic region of the PDPK1 gene co-localized with segmental duplication, thus giving a higher MLPA signal equal to CN=4 for the major CN-genotype. The genetic variability identified for IGLL1 was a deletion of one gene copy in samples from the African population. For the MLLT4 and PPP1R13L genes, we observed duplication - one additional copy in samples from the European population. For PDPK1, additional one or two copies resulted in 5 or 6 CN-genotype for samples from the Asiatic and European population.
The functions of the identified polymorphic genes suggest that they may have an impact on the individual susceptibility to develop HCV infection. In the case of genes that are completely overlapped by polymorphic regions, changes in CN may influence the phenotype by affecting the gene dosage, whereas a partial overlap between the gene and CNV region likely does not directly influence the full transcript dosage. The latter, however, may cause changes in the transcription regulation (altered number of regulatory elements) or induce the formation of alternative transcript variants. Moreover, each CNV may affect the phenotype, regardless of gene dosage, through a position effect or a change in the genome structure.
IGLL1 encodes immunoglobulin lambda-like polypeptide 1 protein, which is a critical B cell development receptor found on the surface of pro-B and pre-B cells. It has been shown that IGLL1 interacts with the HCV NS5A protein43. Dysfunctions of the IGLL1 gene result in a primary immunodeficiency caused by poorer proliferation and differentiation of pro-B cells and consequently, lower levels of serum antibodies and circulating B cells48,49,50. Therefore, the observed deletion of one copy of the gene in the African population suggests that the dosage effect may potentially result in poorer immunological response.
The PPP1R13L gene encodes a protein phosphatase 1 regulatory subunit 13-like protein, called iASPP protein, which inhibits apoptosis and regulates transcription by interacting with NF-kappa-B and p53/TP53 proteins51,52. It has also been shown that PPP1R13L blocks the transcription of HIV-1 by inhibiting the action of both NF-kappa-B and constitutive transcription factor SP153. Interestingly, HCV represses apoptosis54, and the activation of NF-kappa-B55 and SP156,57 in CHC is significantly modulated by viral proteins. Moreover, the direct interaction of iASPP protein and HCV NS5A (a phosphoprotein that plays a key role in HCV RNA replication58) has been shown43. The effects of viral proteins on apoptosis pathways, NF-kappa-B and SP1 functions in relation to the course of HCV infection are complex. Nevertheless, in this context, the additional copy of the PPP1R13L gene observed in samples from the European population may be significant for the progression of liver injury and the severity of CHC.
Among the four identified polymorphic genes, the initial exons at 5′ end of PDPK1 showed the highest variability in the tested populations, ranging from 4 to 6 copies. PDPK1 encodes 3-phosphoinositide-dependent protein kinase 1 (hPDK1), a serine/threonine kinase that acts as a master kinase that phosphorylates and activates a subgroup of the AGC family of protein kinases (PKA, PKC, PKG). Thus, hPDK1 plays a central role in the transduction of signals to downstream targets controlling cell proliferation and survival. For example, it negatively regulates the TGF-beta-induced signaling and activates the NF-kappa-B pathway59,60. Its interaction with the HCV NS5A protein has been shown43. Moreover, hPDK1 is an upstream kinase of the protein kinase C-related kinase 2 (PRK2) that is responsible for the phosphorylation of HCV RNA polymerase. Destabilization of hPDK1 suppresses hepatitis C virus replication61. Our results demonstrated that the CNV region overlaps the initial exons of the PDPK1 gene (and potentially the promoter region). Duplications of this fragment of the PDPK1 gene may result in the expression of alternative (shorter) transcripts or have other indirect functional impacts. The high variability of the PDPK1 gene-overlapping CNV region among populations and functional relevance of hPDK1 for HCV replication strongly encourages more detailed structural and functional studies.
The last identified polymorphic gene, MLLT4, encodes Afadin (AF6) a multi-domain protein involved in signaling and organization of cell junctions. Together with the E-cadherin-catenin system, Afadin belongs to the adhesion system that plays a role in the organization of homotypic, interneuronal and heterotypic cell-cell adherens junctions62. It has been shown that Afadin interacts with HCV nonstructural NS3 protein43. The observed additional copy of the terminal exons of the gene in samples from the European population would not directly affect the gene dosage but may have an indirect phenotypic consequence via a position effect or the altered regulation of the gene expression.
CN of the CCL3L1 gene has been previously shown to differ significantly between populations32,34,35,36. Therefore, we included CCL3L1 to the group of studied genes and used it as a positive control. Our experiments confirmed extensive CNV of CCL3L1 and specific distribution of CN-genotypes among different population. We noted a CCL3L1 CN-genotype range of 1–3 copies in the European population, 2–8 copies in the Asiatic population and 2–7 copies in the African population.
The impact of CNV on phenotype and its association with diseases remain poorly understood. To date, transcriptome analyses and single-gene expression studies are appropriate proxies to study the physiological and pathological consequences of CNV. Here, we present a selection of candidate human genes overlapped by CNV regions for further functional investigations as potential genetic markers. CNVs of these four newly identified polymorphic genes may serve in the future as potential predictors of CHC development, spontaneous virus clearance or response to treatment. These results also enhance our understanding of host genetic factors that influence pathogenesis of HCV infection.
Summarizing, in this paper we present a simple approach that permits to identify genes whose CNV may affect host-virus interactions. The proposed approach combines the data on CNV regions present in the human genome and host-virus interactome to select candidate genes. In the next step, variation in CN of each selected gene is experimentally verified by the MLPA method. The analysis described here focused on HCV, a model human RNA virus, however, there is no doubt that the same procedure can be used to determine the influence of CNV on other viral infections.
Our analysis revealed also some weaknesses of the data sets being at our disposal. Although, we applied very restrictive selection criteria only 4 out of 19 candidate genes from HCV-host interactome showed CNV in the tested set of DNA samples. Thus, more precise characterization of both CNV regions in human genome and HCV-human interactome is still necessary to better elucidate the associations between CNV and the course of HCV infection. The problem of HCV-host interactions has been recently addressed by several research groups63,64,65. As a result, the size of the interactome has been increased from 421 to 969 human proteins64. Considering the rapid progress currently observed in the studies on human genome polymorphism one can expect that the approach presented here, will be broadly applied to identify new genetic factors shaping host-virus interactions.
Methods
Gene selection
To identify the human genes whose overlap with CNV regions could possibly correlate with a course of HCV infection and/or treatment outcome, we compared the genomic localization of 421 genes encoding proteins belonging to human-HCV interactome43 and 202 430 CNV regions from the Database of Genomic Variants (updated July 23, 2013; www.dgv.tcag.ca). As genes that are potentially affected by CNV, we selected genes that overlapped with at least three CNV regions deposited in the DGV database.
In addition to genes from the human-HCV interactome, we selected one gene (CCL3L1) as a positive control that fulfilled the following criteria: (i) it has been proven to be highly polymorphic among different human populations35, and (ii) its CNV was shown to be associated with the course of HCV infection42.
Samples
In this study, 106 DNA samples from three different worldwide human populations were used: 31 European (CEU) samples from CEPH/Utah Collection, 48 Asiatic (CHB/JPT) samples from Han Chinese in Beijing (China) and Japanese in Tokyo (Japan), and 27 African (YRI) samples from Yoruba in Ibadan (Nigeria). All samples belong to the HapMap Project samples and were obtained from Coriell Institute (www.coriell.org).
MLPA probes and assays design
For the CNV analyses of 20 selected genes, two custom-made assays (sets of MLPA probes designed for the analyzed genes) named: HCV_SET1 and HCV_SET2, were designed. The HCV_SET1 assay contained 22 probes specific for 11 selected genes, and the HCV_SET2 assay contained 23 probes specific for 10 genes. Both assays also contained five control probes designed for CN stable regions of known number of copies (CN=2). The total probe length ranged from 93 to 198 and from 93 to 192 nucleotides in the HCV_SET1 and HCV_SET2 assay, respectively. All MLPA probes were designed according to a previously described strategy46,66 to avoid known SNPs, repeat elements and sequences of extremely high or low GC content. Probes were predominantly designed to be located in exons. For each gene, at least two MLPA probes were designed. For MLLT4 and HLA-A genes, one additional probe was designed, and for the PDPK1 gene, two additional probes were designed due to the complex genomic structure of the regions where these genes are located. For PDPK1 two probes were designed in a unique part of the gene (PDPK1_C, PDPK1_D), while the other two (PDPK1_A, PDPK1_B) were located in the part that overlaps with segmental duplication (SD) (Fig. 2). The latter two, due to the high homology of present SDs, were designed as universal, i.e. they recognize both sequences, within and downstream of the gene.
The detailed characteristics and sequences of all probes used in this study are presented in Supplementary data (Table S3). All probes were generated by Integrated DNA Technologies Inc. (IDT, Coralville, IA, USA; www.idtdna.com) in 100 nmole scale and purified by PAGE.
MLPA analysis
MLPA reactions were performed according to published results45 and the manufacturer’s protocol (MRC-Holland, Amsterdam, Netherlands; www.mrc-holland.com). All of the MLPA reagents, except for probe-mixes (which were prepared separately from designed and generated oligonucleotides), were purchased from MRC-Holland.
In brief, 100 ng of sample DNA was denatured and hybridized with MLPA probe-mix for 16 hours. All probes that correctly hybridized to their targets were ligated and amplified with the use of universal primer pair. PCR products were separated by capillary electrophoresis on ABI Prism 3130XL Analyzer (Applied Biosystems, Carlsbad, CA, USA). Obtained electropherograms were analyzed with the use of GeneMarker software v1.91 (SoftGenetics, State College, PA, USA; www.softgenetics.com) and signal intensity values (peak heights) were transferred to prepared Microsoft Excel sheets. The signals of all probes were first normalized. To this end, each individual signal was divided by the average signal of control probes to avoid run-to-run signal variation. Then, the normalized MLPA signal of all probes specific for a particular gene was averaged and presented in one-dimensional (1D) signal scatter plot. Because the MLPA signal is proportional to CN of tested region/gene, the plot signals from all analyzed samples formed distinct clusters representing CN-genotypes, which were assigned according to previously described method35,47.
All graphs and statistical analyses were performed using Microsoft Excel and GraphPad Prism v.5.00.
Additional Information
How to cite this article: Budzko, L. et al. Copy number variation of genes involved in the hepatitis C virus-human interactome. Sci. Rep. 6, 31340; doi: 10.1038/srep31340 (2016).
Supplementary Material
Acknowledgments
The research described in this work was funded by the European Union within the European Regional Development Fund through the MPD program. This publication was also supported by the Polish Ministry of Science and Higher Education, under the KNOW program. This work was partially financed by the Polish Ministry of Science and Higher Education grant IP2012 014972 to PJ and National Science Centre grant NCN 2011/01/B/NZ5/02773.
Footnotes
Author Contributions L.B. performed gene selection and designed MLPA probes. M.M.-S. designed and conducted the MLPA experiments and computer analysis of the data, and prepared figures. Both M.M.-S. and L.B. drafted the manuscript, prepared Supplementary materials, and participated in preparation of the manuscript’s final version. P.J. conceived the study, drafted the manuscript and participated in preparation of its final version. P.K. designed the experiments, analyzed all data, gave conceptual advice and reviewed the manuscript. M.F. conceived and supervised the study, designed the experiments, analyzed all data, and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
References
- Choo Q. L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989). [DOI] [PubMed] [Google Scholar]
- Moradpour D., Penin F. & Rice C. M. Replication of hepatitis C virus. Nat Rev Microbiol 5, 453–463 (2007). [DOI] [PubMed] [Google Scholar]
- Lozano R. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B. et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59, 318–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowiak P. et al. Phylogeny and molecular evolution of the hepatitis C virus. Infect Genet Evol 21, 67–82 (2013). [DOI] [PubMed] [Google Scholar]
- Messina J. P. et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61, 77–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powdrill M. H. et al. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc Natl Acad Sci USA 108, 20509–20513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowiak P., Figlerowicz M., Kurzynska-Kokorniak A. & Figlerowicz M. Mechanisms involved in the development of chronic hepatitis C as potential targets of antiviral therapy. Curr Pharm Biotechnol 12, 1774–1780 (2011). [DOI] [PubMed] [Google Scholar]
- Fishman S. L. & Branch A. D. The quasispecies nature and biological implications of the hepatitis C virus. Infect Genet Evol 9, 1158–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Sheldon J. & Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev 76, 159–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein N. N. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 13, 223–235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey L. C. & Lee W. M. Hepatitis C virus therapy update 2013. Curr Opin Gastroenterol 29, 243–249 (2013). [DOI] [PubMed] [Google Scholar]
- Muir A. J. & Naggie S. Hepatitis C Virus Treatment: Is It Possible To Cure All Hepatitis C Virus Patients? Clin Gastroenterol Hepatol 13, 2166–2172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlerowicz M. et al. Hepatitis C virus quasispecies in chronically infected children subjected to interferon-ribavirin therapy. Arch Virol 155, 1977–1987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittar C. et al. Genetic diversity of NS5A protein from hepatitis C virus genotype 3a and its relationship to therapy response. BMC Infect Dis 10, 36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saludes V. et al. Baseline prediction of combination therapy outcome in hepatitis C virus 1b infected patients by discriminant analysis using viral and host factors. PLoS One 5, e14132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu A. et al. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One 6, e24907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim A. C. et al. Analysis of HCV quasispecies dynamic under selective pressure of combined therapy. BMC Infect Dis 13, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon B. R. et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med 364, 1207–1217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbinnen T. et al. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J Virol 84, 11124–11133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowiak P. et al. Evolution of hepatitis C virus hypervariable region 1 in chronically infected children. Virus Res 167, 380–384 (2012). [DOI] [PubMed] [Google Scholar]
- Clausen L. N., Lundbo L. F. & Benfield T. Hepatitis C virus infection in the human immunodeficiency virus infected patient. World J Gastroenterol 20, 12132–12143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Li Y. & Ho W. Z. Drug abuse, innate immunity and hepatitis C virus. Rev Med Virol 16, 311–327 (2006). [DOI] [PubMed] [Google Scholar]
- Bhattacharya R. & Shuhart M. C. Hepatitis C and alcohol: interactions, outcomes, and implications. J Clin Gastroenterol 36, 242–252 (2003). [DOI] [PubMed] [Google Scholar]
- Fiorilli M., Mecucci C., Farci P. & Casato M. HCV-associated lymphomas. Rev Clin Exp Hematol 7, 406–423 (2003). [PubMed] [Google Scholar]
- Jeong S. W., Jang J. Y. & Chung R. T. Hepatitis C virus and hepatocarcinogenesis. Clin Mol Hepatol 18, 347–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009). [DOI] [PubMed] [Google Scholar]
- Rauch A. et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138, 1338–1345, 1345 e1-7 (2010). [DOI] [PubMed] [Google Scholar]
- Lange C. M. & Zeuzem S. IL28B single nucleotide polymorphisms in the treatment of hepatitis C. J Hepatol 55, 692–701 (2011). [DOI] [PubMed] [Google Scholar]
- McCarroll S. A. & Altshuler D. M. Copy-number variation and association studies of human disease. Nat Genet 39, S37–S42 (2007). [DOI] [PubMed] [Google Scholar]
- Stankiewicz P. & Lupski J. R. Structural variation in the human genome and its role in disease. Annu Rev Med 61, 437–455 (2010). [DOI] [PubMed] [Google Scholar]
- Conrad D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J. L. et al. Copy number variation: new insights in genome diversity. Genome Res 16, 949–961 (2006). [DOI] [PubMed] [Google Scholar]
- McCarroll S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40, 1166–1174 (2008). [DOI] [PubMed] [Google Scholar]
- Marcinkowska-Swojak M., Klonowska K., Figlerowicz M. & Kozlowski P. An MLPA-based approach for high-resolution genotyping of disease-related multi-allelic CNVs. Gene 546, 257–262 (2014). [DOI] [PubMed] [Google Scholar]
- Gonzalez E. et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307, 1434–1440 (2005). [DOI] [PubMed] [Google Scholar]
- Aklillu E. et al. CCL3L1 copy number, HIV load, and immune reconstitution in sub-Saharan Africans. BMC Infect Dis 13, 536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya T. et al. CCL3L1 and HIV/AIDS susceptibility. Nat Med 15, 1112–1115 (2009). [DOI] [PubMed] [Google Scholar]
- Field S. F. et al. Experimental aspects of copy number variant assays at CCL3L1. Nat Med 15, 1115–1117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban T. J. et al. CCL3L1 and HIV/AIDS susceptibility. Nat Med 15, 1110–1112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson L. M. & Holmdahl R. Copy number variation in autoimmunity–importance hidden in complexity? Eur J Immunol 42, 1969–1976 (2012). [DOI] [PubMed] [Google Scholar]
- Grunhage F. et al. Lower copy numbers of the chemokine CCL3L1 gene in patients with chronic hepatitis C. J Hepatol 52, 153–159 (2010). [DOI] [PubMed] [Google Scholar]
- de Chassey B. et al. Hepatitis C virus infection protein network. Mol Syst Biol 4, 230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J. R., Ziman R., Yuen R. K., Feuk L. & Scherer S. W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42, D986–D992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten J. P. et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30, e57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski P. et al. Identification of 54 large deletions/duplications in TSC1 and TSC2 using MLPA, and genotype-phenotype correlations. Hum Genet 121, 389–400 (2007). [DOI] [PubMed] [Google Scholar]
- Marcinkowska-Swojak M., Uszczynska B., Figlerowicz M. & Kozlowski P. An MLPA-based strategy for discrete CNV genotyping: CNV-miRNAs as an example. Hum Mutat 34, 763–773 (2013). [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Evans R. J., Stafford-Hollis J. M., Korsmeyer S. J. & McKearn J. P. Immunoglobulin lambda light-chain-related genes 14.1 and 16.1 are expressed in pre-B cells and may encode the human immunoglobulin omega light-chain protein. Proc Natl Acad Sci U S A 86, 5552–5556 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y. et al. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med 187, 71–77 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankovich A. J. et al. Structural insight into pre-B cell receptor function. Science 316, 291–294 (2007). [DOI] [PubMed] [Google Scholar]
- Yang J. P., Hori M., Sanda T. & Okamoto T. Identification of a novel inhibitor of nuclear factor-kappaB, RelA-associated inhibitor. J Biol Chem 274, 15662–15670 (1999). [DOI] [PubMed] [Google Scholar]
- Bergamaschi D. et al. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet 33, 162–167 (2003). [DOI] [PubMed] [Google Scholar]
- Takada N. et al. RelA-associated inhibitor blocks transcription of human immunodeficiency virus type 1 by inhibiting NF-kappaB and Sp1 actions. J Virol 76, 8019–8030 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Qu A., Han X. & Wang Y. HCV core protein represses the apoptosis and improves the autophagy of human hepatocytes. Int J Clin Exp Med 8, 15787–15793 (2015). [PMC free article] [PubMed] [Google Scholar]
- Arsura M. & Cavin L. G. Nuclear factor-kappaB and liver carcinogenesis. Cancer Lett 229, 157–169 (2005). [DOI] [PubMed] [Google Scholar]
- Presser L. D., McRae S. & Waris G. Activation of TGF-beta1 promoter by hepatitis C virus-induced AP-1 and Sp1: role of TGF-beta1 in hepatic stellate cell activation and invasion. PLoS One 8, e56367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiang Z., Qiao L., Zhou Y., Babiuk L. A. & Liu Q. Hepatitis C virus nonstructural protein-5A activates sterol regulatory element-binding protein-1c through transcription factor Sp1. Biochem Biophys Res Commun 402, 549–553 (2010). [DOI] [PubMed] [Google Scholar]
- Chong W. M. et al. Phosphoproteomics Identified an NS5A Phosphorylation Site Involved in Hepatitis C Virus Replication. J Biol Chem 291, 3918–3931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayascas J. R. Dissecting the role of the 3-phosphoinositide-dependent protein kinase-1 (PDK1) signalling pathways. Cell Cycle 7, 2978–2982 (2008). [DOI] [PubMed] [Google Scholar]
- Mora A., Komander D., van Aalten D. M. & Alessi D. R. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol 15, 161–170 (2004). [DOI] [PubMed] [Google Scholar]
- Kim M. G., Moon J. S., Kim E. J., Lee S. H. & Oh J. W. Destabilization of PDK1 by Hsp90 inactivation suppresses hepatitis C virus replication through inhibition of PRK2-mediated viral RNA polymerase phosphorylation. Biochem Biophys Res Commun 421, 112–118 (2012). [DOI] [PubMed] [Google Scholar]
- Takai Y. & Nakanishi H. Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci 116, 17–27 (2003). [DOI] [PubMed] [Google Scholar]
- Mosca E., Alfieri R. & Milanesi L. Diffusion of information throughout the host interactome reveals gene expression variations in network proximity to target proteins of hepatitis C virus. PLoS One 9, e113660 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saik O. V., Ivanisenko T. V., Demenkov P. S. & Ivanisenko V. A. Interactome of the hepatitis C virus: Literature mining with ANDSystem. Virus Res (2015). [DOI] [PubMed] [Google Scholar]
- Colpitts C. C., El-Saghire H., Pochet N., Schuster C. & Baumert T. F. High-throughput approaches to unravel hepatitis C virus-host interactions. Virus Res (2015). [DOI] [PubMed] [Google Scholar]
- Marcinkowska M., Wong K. K., Kwiatkowski D. J. & Kozlowski P. Design and generation of MLPA probe sets for combined copy number and small-mutation analysis of human genes: EGFR as an example. Scientific World Journal 10, 2003–2018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.