Abstract
Since aptamers bind their targets with high affinity and specificity, they are promising alternative ligands in protein affinity purification. As aptamers are chemically synthesized oligonucleotides, they can be easily produced in large quantities regarding GMP conditions allowing their application in protein production for therapeutic purposes. Several advantages of aptamers compared to antibodies are described in general within this paper. Here, an aptamer directed against the human Vascular Endothelial Growth Factor (VEGF) was used as affinity ligand for establishing a purification platform for VEGF in small scale. The aptamer was covalently immobilized on magnetic beads in a controlled orientation resulting in a functional active affinity matrix. Target binding was optimized by introduction of spacer molecules and variation of aptamer density. Further, salt-induced target elution was demonstrated as well as VEGF purification from a complex protein mixture proving the specificity of protein-aptamer binding.
Keywords: VEGF, Affinity separation, Protein purification, Aptamer immobilization
1. Introduction
Affinity separation is a frequently used method in downstream processing of recombinant proteins for biomedical or pharmaceutical application because it offers a high product quality and purity. Currently, natural or recombinant affinity ligands from animal or bacterial source including antibodies, Protein A or heparin are commonly used as capture molecules linked to the stationary chromatography phase [6]. However, especially the production of proteins for therapeutic purposes demands purification methods without any cell-based material to avoid potential contaminations with endotoxins, viruses or other pathogens associated with these components. In contrast to antibodies or heparin, aptamers are in vitro selected and chemically synthesized single-stranded DNA or RNA oligonucleotides and therefore fulfill this requirement.
In 1990, nucleic acid oligomers were first time selected in a process named SELEX (systematic evolution of ligands by exponential enrichment) by their ability to bind a certain molecule with high affinity [3], [26]. Due to intramolecular interactions between their nucleobases, these oligonucleotides, named aptamers, fold into distinct three-dimensional structures which enable them to recognize and to bind their corresponding target molecules. Aptamers bind their targets with affinity and specificity comparable to those of antibodies making them an attractive alternative in affinity chromatography [29], sensing applications [8], [14], [16], [25], [27] and also for therapeutic purposes [2]. Furthermore, aptamers exhibit many advantages over antibodies: They are cheap in production, have a high thermal stability, can be produced regarding GMP conditions and offer the potential for regeneration after denaturation [9], [13]. Aptamers can be chemically modified at either 3′ or 5′-terminus enabling a covalent immobilization in a controlled orientation on the solid phase. This may prevent loss of aptamer binding activity and aptamer leakage during the purification process [28]. If necessary, it is also possible to introduce molecular spacers at the aptamers' termini to avoid interactions with the solid surface that may interfere with the correct three-dimensional aptamer conformation and in consequence target binding [30]. Aptamer folding is highly influenced by surrounding buffer conditions such as pH, ionic strength or presence of specific ions. Thus, buffer exchange is an appropriate strategy not only for aptamer folding but also for unfolding and therefore target elution as well as aptamer regeneration. Moreover, during the SELEX process the selection conditions can be adapted with respect to the application to obtain aptamers with desirable properties for protein purification [9], [29].
In 1999, the application of aptamer affinity purification was firstly reported for an l-selectin-immunoglobulin fusion protein [21]. Over the past few years, similar methods utilizing DNA aptamers have been developed for many other proteins, including Taq-polymerase [19], thyroid transcription factor [17], lysozyme [7] and also histidine affinity tag [11], [31]). Although, since 1994 several aptamers binding to the human vascular endothelial growth factor (VEGF) have been selected [1], [4], [5], [10], [18], [22], there are no reports about aptamer-based VEGF affinity purification, yet. Instead, heparin is used commonly as affinity ligand for VEGF purification.
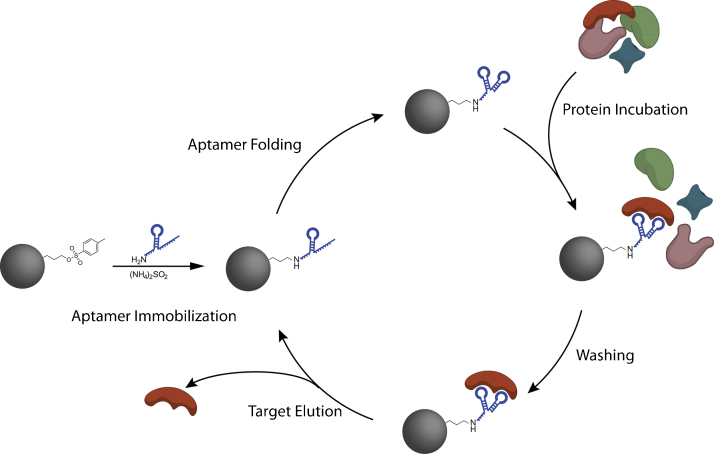
In this work the VEGF-binding DNA aptamer V7t1 [18] was covalently coupled to magnetic beads, generating a functionalized affinity matrix phase for VEGF purification (Fig. 1). The aptamer is directed against the receptor-binding domain of the protein. Since different VEGF isoforms share a common receptor binding domain, the aptamer is able to bind the different isoforms which has been demonstrated for VEGF121 [18]. While this cross-reactivity might be limiting for diagnostic application of the aptamer, it is not restrictive for the downstream processing of recombinant proteins. Oriented immobilization of the aptamer via 3′-terminal amino groups was chosen for maintenance of molecular flexibility and three-dimensional aptamer folding. Furthermore the aptamer immobilization was optimized with regard to aptamer density and the influence of spacers between aptamer and surface was investigated. The functionalized magnetic beads were utilized in protein binding experiments for characterization of VEGF binding.
Fig. 1.
Principle of protein purification process using aptamer-modified magnetic beads.
2. Material and methods
All buffer salts were purchased from Fluka (Switzerland) and Sigma (Germany) and were of per analysis quality. All solutions were prepared with deionized water (ARIUM, Sartorius Stedim Biotech, Germany) and were filtered (0.2 μm) prior to use.
2.1. Aptamers
All DNA oligonucleotides were purchased from IDT Integrated DNA Technologies BVBA, Belgium. The following DNA sequences were used:
V7t1: 5′-TGT GGG GGT GGA CGG GCC GGG TAG A-3′
V7t1_14B: 5′-TGT GGG GGT GGA CGG GCC GGG TAG ATA GTA TGT GCA ATC-3′
V7t1_14T: 5′-TGT GGG GGT GGA CGG GCC GGG TAG ATT TTT TTT TTT TTT-3′
V7t1_12EG: 5′-TGT GGG GGT GGA CGG GCC GGG TAG A/dodecaethylene glycol/-3′
Random DNA: 5′-AAA CCG CGT CTC TAC GAC CGG TGC TCG ATT TAA TTT CGC TGA CGT GA-3′
The sequences of VEGF binding aptamers V7t1 and V7t1_14B were taken from literature [18]. V7t1_14B corresponds to the minimal aptamer V7t1 which is 3′-terminal elongated with 14 bases of the originally selected aptamer Vap7. V7t1_14T accords to the V7t1 sequence elongated with 14 thymine nucleotides at 3′-terminus and V7t1_12EG accords to the V7t1 sequence expanded with a dodecaethylene glycol spacer at 3′-terminus. The spacer length of 14B, 14T and 12EG are estimated to be 5.29 nm, 7.28 nm and 4.58 nm, respectively (calculation available in the supporting information). The random DNA oligonucleotide used as negative control is a randomized oligonucleotide sequence with statistical base distribution. Oligonucleotides used for immobilization carry a 3′ terminal amino group, those for electrophoresis mobility shift assay and microscale electrophoresis experiments are labeled with cyanine 3 (Cy3) at the 3′-terminus.
2.2. Proteins
Bovine serum albumin (BSA), α-chymotrypsin and myoglobin were obtained from Sigma–Aldrich GmbH, Germany. Human VEGF165 was produced as fusion protein with an N-terminal His-tag. Gene sequence coding for the mature VEGF (amino acids 27-191 according to Acc. No.: AAA35789) was cloned into pET16b plasmid (Merck KG, Germany) and afterwards brought into E. coli BL21(DE3). Cultivation was performed in 2 L baffled shake flasks at 37 °C and 150 rpm using terrific broth medium (12 g/L tryptone, 24 g/L yeast extract, 4 mL/L glycerol, 100 mL/L potassium phosphate buffer) supplemented with 100 μg/mL carbenicillin. For main-cultivation, 500 mL medium were inoculated with E. coli cell suspension to a starting OD600 of 0.05. At OD600 ≈ 0.5 protein expression was induced with 1 mM IPTG before the culture was grown for at least further four hours at 37 °C. Protocols for isolation and resolubilization of inclusion bodies (IB) were developed on the basis of procedures described in the literature [20], [24]). Cells were harvested by centrifugation (6000 g, 4 °C, 15 min), resuspended in lysis buffer (50 mM Tris/HCl, 1 mM EDTA, 1% Triton X-100, 10 mM MgCl2, 10 μg/mL DNase II, pH 8), incubated at 20 °C for 30 min and disrupted by ultra-sonication (Labsonic M, Sartorius AG, Germany; parameters: 3 × 30 s at 70% amplitude and 0.6 s cycle). Lysates were mixed with an equal volume IB wash buffer (50 mM Tris/HCl, 10 mM 2-mercaptoethanol, 2 mM EDTA, 5% glycerol, 0.05% sodium deoxycholate, 1% Nonidet P-40, pH 8) and centrifuged at 6000 × g and 4 °C for 60 min. The insoluble protein pellet was resuspended in denaturing solubilization buffer (10 mM CHES, 1 M urea, 100 mM l-arginine, 15 mM l-cysteine, 2 mM DTT, 0.05% Tween 20, pH 9.8) and cell debris was removed by centrifugation (6000 × g, 4 °C, 120 min). VEGF was purified by Ni-IMAC chromatography using HisTrap FF chromatography column (GE Healthcare Europe GmbH, Germany) and 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 300 mM NaCl, 0.05% Tween 20, pH 8 as binding buffer. VEGF was eluted by addition of 250 mM imidazole. To separate the correct refolded VEGF, heparin affinity chromatography (HiTrap Heparin HP column; GE Healthcare Europe GmbH, Germany) was performed using 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 138 mM NaCl, 0.05% Tween 20, pH 8 as binding buffer. The VEGF containing elution fractions of the Ni-IMAC were pooled and diluted with two volumes of heparin binding buffer. VEGF was eluted by addition of 800 mM NaCl. Afterwards, buffer exchange to V7t1 binding buffer (10 mM Tris/HCl, 100 mM NaCl, 0.05 mM EDTA, 50 mM KCl, 0.05% Tween 20 pH 7.0) was performed via ultrafiltration using Vivaspin 10 kDa centrifugal concentrators (Sartorius AG, Germany). Purified proteins were stored at 4 °C.
2.3. Electrophoresis mobility shift assay
For electrophoresis mobility shift assay, 40 pmol of Cy3-labeled V7t1 aptamer were incubated with 300 pmol VEGF, myoglobin, BSA or α-chymotrypsin in V7t1 binding buffer for 30 min at 4 °C, respectively. Samples were mixed with an equal volume of loading buffer (20 mM Tris/HCl, 10% glycerol, 0.02% bromophenol blue, pH 6.8) and separated via electrophoresis utilizing a 1.5% agarose gel and 25 mM Tris/HCl, 50 mM glycine, pH 9.5 as running buffer. DNA-bands were visualized with a gel documentation system (Ge iX Imager, Intas, Germany).
2.4. Microscale thermophoresis
Microscale thermophoresis experiments were kindly enabled by NanoTemper Technologies GmbH (Germany). A final concentration of 50 nM Cy3-labeled aptamer V7t1 was incubated with a two-fold dilution series of VEGF in a final concentration range of 1250–0.61 nM for 10 min at 4 °C in V7t1 binding buffer. Afterwards, each sample was loaded into a glass capillary (K004 hydrophilic capillaries) and was subsequently analyzed by Monolith NT.115 MST system (NanoTemper Technologies GmbH, Germany) at 90% LED power and 20% MST. The KD-value was calculated from triplicate experiments by non-linear curve fitting based on the law of mass action with KD as single free parameter.
2.5. Characterization of nonspecific VEGF-binding to different surface materials
For characterization of nonspecific VEGF binding to different surface materials three types of magnetic beads were used: polystyrene beads (Dynabeads M-270Carboxyl, Life Technologies Inc., CA, USA), silane beads (BioMag Carboxyl, Polysciences Inc., PA, USA) and agarose beads (NHS Mag Sepharose, GE Healthcare Europe GmbH, Germany). In order to block reactive groups of the magnetic beads, 1.5 mg of polystyrene and silane beads or 100 μL of agarose bead suspension were coupled with Tris according to the manufacturers’ instructions, respectively. Afterwards, the beads were washed three times with V7t1 binding buffer and incubated with 100 μL purified VEGF (200 μg/mL, corresponding to 4.4 nM) in V7t1 binding buffer for one hour at 4 °C. The beads were washed three times with 100 μL V7t1 binding buffer and were finally resuspended in 50 μL V7t1 binding buffer. All fractions, including the magnetic beads were incubated with SDS sample buffer (275 mM sodium dodecyl sulfate (SDS), 500 mM DTT, 30% glycerol, 50% 2-mercaptoethanol) for 10 min at 95 °C and subsequently analyzed by SDS-PAGE analysis utilizing a 16% polyacrylamid gel under denaturing conditions. SDS-PAGE gels were either silver stained or further used for western blot analysis using WesternBreeze Chromogenic Immunodetection Kit (Life Technologies Inc., CA, USA) and anti VEGF antibody (Abgent, #AJ1813). PageRuler Unstained Protein Ladder and PageRuler Plus Prestained Protein Ladder (Thermo Scientific, MA, USA) were used as molecular weight markers.
2.6. Aptamer immobilization
Aptamers were immobilized to tosyl-activated polystyrene magnetic beads (Dynabeads M-280 Tosyl, Life Technologies Inc., CA, USA) in a EDC-free coupling reaction. Amino-modified oligonucleotides were diluted in 100 mM sodium borate buffer (SBB), pH 9.5 to a final concentration in a range between 1 and 50 μM. 1.2 mg magnetic beads were washed three times with 100 mM SBB, pH 9.5 using a magnetic stand for bead separation. Afterwards, the buffer was removed and 100 μL of oligonucleotide solution were added. After short mixing, 70 μL of ammonium sulfate solution (3 M in 100 mM SBB, pH 9.5) were added. Coupling was performed under rotation overnight at 37 °C. The supernatant was removed and 100 μL blocking buffer (50 mM Tris in 100 mM SBB pH 9.5) were added for blocking the uncoupled tosyl groups. Blocking was performed under rotation for three hours at 37 °C. The supernatant was removed, the beads were washed three times with bead wash buffer (50 mM Tris/HCl, 300 mM NaCl, 0.05% Tween 20, pH 7) and additionally three times with V7t1 binding buffer. Aptamer concentrations in the initial solutions, supernatants and washing fractions were determined with a NanoDrop™ 1000 spectrophotometer (Thermo Scientific, MA, USA) and Lambert–Beer equation utilizing the associated extinction coefficients. The amount of bound aptamer and the immobilization efficiency were calculated by subtraction of the aptamer amount in the supernatants and washing fractions from that in the initial solution.
2.7. Characterization of VEGF-binding to aptamer-modified magnetic beads
Polystyrene magnetic beads modified with V7t1 aptamer in a density of 230 pmol/mg were used for characterization of VEGF binding. Beads were washed three times for 5 min with 100 μL V7t1 binding buffer, heated for 5 min to 95 °C and afterwards cooled to room temperature to achieve proper aptamer folding. In total, 250 pmol immobilized aptamer (1.1 mg beads) were incubated with 90 μL VEGF (300 μg/mL; 6.7 μM) at 4 °C for 1 h. The beads were washed three times for 5 min with 100 μL V7t1 binding buffer and were finally resuspended in 50 μL V7t1 binding buffer. All fractions, including the magnetic beads were analyzed by SDS-PAGE as described above.
2.8. Comparison of aptamer spacers
Different aptamer variants (V7t1, V7t1_14B, V7t_14T, V7t1_12EG, Random DNA) were immobilized to polystyrene magnetic beads as described above in a density of approximately 230 pmol/mg. Beads were washed three times for 5 min with 100 μL V7t1 binding buffer, were heated for 5 min to 95 °C and afterwards cooled to room temperature. In total, 235 pmol immobilized aptamer (0.9–1.1 mg beads) were incubated with 85 μL VEGF (300 μg/mL; 6.7 μM) at 4 °C for 1 h. The beads were washed three times for 5 min at 4 °C with 100 μL V7t1 binding buffer and afterwards three times for 5 min at 4 °C with 50 μL V7t1 elution buffer (V7t1 binding buffer + 500 mM NaCl). VEGF concentration of all fractions as well as in the initial sample was analyzed by VEGF ELISA (Thermo Scientific, #EH2VEGF) according to the manufacturer’s instructions and the absolute amount of bound and eluted VEGF was calculated for each aptamer variant.
2.9. Optimization of aptamer density
Polystyrene magnetic beads conjugated with V7t1_14B aptamer in different densities between 50 and 500 pmol/mg were washed three times for 5 min with 100 μL V7t1 binding buffer. They were heated for 5 min to 95 °C and afterwards cooled to room temperature. In total, 270 pmol immobilized aptamer (0.55–4.75 mg beads) were incubated with 100 μL VEGF (250 μg/mL; 5.6 μM) at 4 °C for 1 h. Beads were washed three times for 5 min at 4 °C with 100 μL V7t1 binding buffer. VEGF concentration in all fractions as well as the initial sample was analyzed by VEGF ELISA (Thermo Scientific, #EH2VEGF) and the absolute amount of bound VEGF was calculated for each immobilization density.
2.10. VEGF purification from a complex protein mixture
For creating a complex protein mixture, fetal calf serum was diluted in V7t1 binding buffer to a total protein concentration of 1 mg/mL and supplemented with VEGF (200 μg/mL). V7t1_14B and Random DNA modified polystyrene magnetic beads were heated to 95 °C for 5 min and subsequently cooled to room temperature. The magnetic beads were incubated with 100 μL protein mixture in V7t1 binding buffer at 4 °C for 1 h. Afterwards, the beads were washed three times for 5 min at 4 °C with 100 μL V7t1 binding buffer. VEGF was eluted by incubating three times with 50 μL V7t1 elution buffer. All fractions, including the magnetic beads were analyzed by SDS-PAGE followed by silver staining and densitometric analysis using AlphaEaseFC image analysis software.
3. Results
3.1. Investigation of aptamer-target-binding
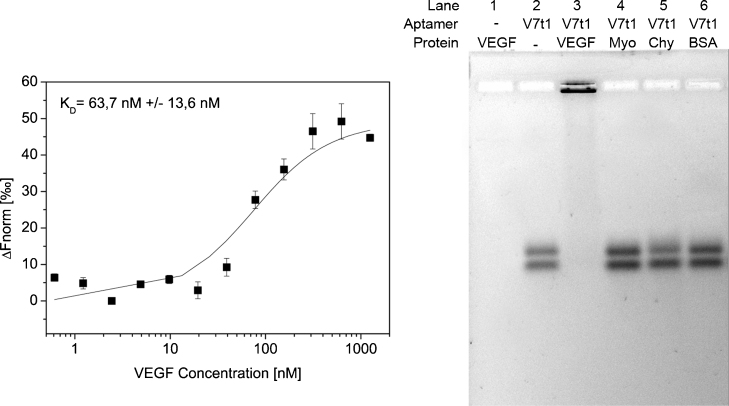
Utilization of aptamers in downstream processing requires their immobilization for example by covalent attachment using a terminal amino linker. Since aptamer modification may result in misfolding and thus reduced binding affinity, we first investigated binding of the Cy3-labeled aptamer V7t1 to the target protein VEGF. For determination of the binding affinity, V7t1 was incubated with different concentrations of VEGF and analyzed by microscale thermophoresis. Based on the resultant saturation curve (Fig. 2, left) a KD-value of 75.9 nM ± 13.0 nM was calculated. In comparison to the KD-value of 1.4 nM, that has been published for the unmodified aptamer V7t1 [18], this may indicate a diminished VEGF binding but may also be a consequence of different methods used for the determination of the KD (MST versus SPR). Nevertheless, the binding affinity in a low nanomolar range is adequate for the application of protein purification.
Fig. 2.
Characterization of aptamer target binding via microscale thermophoresis and electrophoresis mobility shift assay (EMSA). Left: saturation curve of V7t1 aptamer and VEGF. Cy3-labeled V7t1 was incubated with different concentrations of VEGF and afterwards analyzed by microscale thermophoresis as three-fold replicates. A KD of 75.9 nM ± 13.0 nM was determined for this interaction. Right: analysis of aptamer binding to the target protein VEGF and the non-target proteins myoglobin (Myo), α-chymotrypsin (Chy) and bovine serum albumin (BSA) via EMSA. The Cy3-labeled aptamer V7t1 was incubated with an excess of protein, respectively. Afterwards the samples were analyzed by agarose gel electrophoresis. Pure VEGF and V7t1 were used as negative controls, respectively. Results of the EMSA demonstrate a specific binding of V7t1 to VEGF in comparison to the other tested proteins.
For demonstration of target binding specificity and investigation of unspecific binding to other proteins based on electrostatic interactions, different aptamer–protein mixtures were analyzed by electrophoresis mobility shift assay (EMSA). The Cy3-labeled aptamer V7t1 was incubated with VEGF and with three non-target proteins offering different isoelectric points (pI), respectively: BSA (pI 4.7), myoglobin (pI 7.2) and α-chymotrypsin (pI 8.8). The protein–V7t1-mixtures were analyzed by agarose gel electrophoresis as well as pure VEGF and V7t1 as negative controls. The unbound aptamers are able to penetrate the gel, whereas the proteins and protein–aptamer complexes remain near to the loading pocket (Fig. 2, right). Two bands were found for the unbound V7t1 aptamer, indicating a variable three dimensional aptamer folding which has been described previously by the group of Plavec [15]. The EMSA confirms the formation of a VEGF-V7t1-complex (Fig. 2, right, lane 3) in contrast to V7t1 incubated with myoglobin, α-chymotrypsin or BSA (lanes 4, 5, 6). Here, the bands appear at the same height as the one of the free aptamer V7t1 (lane 2), indicating no binding between these proteins and the V7t1 even though the α-chymotrypsin has a basic isoelectric point and is consequently positively charged under assay conditions (pH 7). Nonetheless, there is no unspecific electrostatic binding to the negatively charged oligonucleotide backbone. Therefore, the results prove that protein binding is mediated by specific affinity recognition rather than electrostatic interactions.
3.2. Development of aptamer-based affinity purification
3.2.1. Choice of a suitable surface for aptamer immobilization
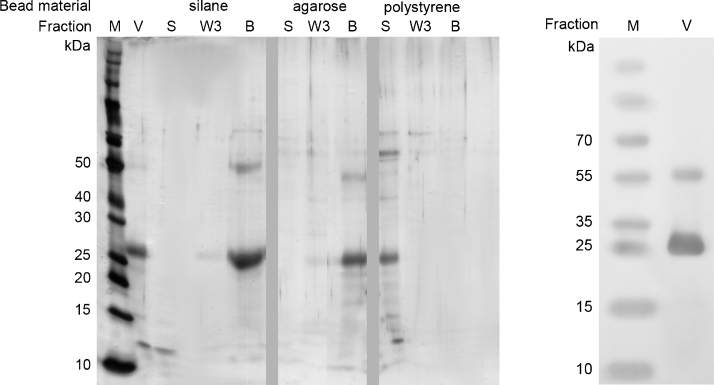
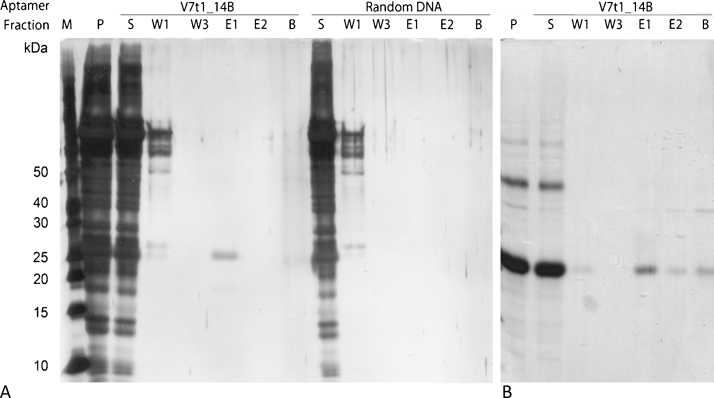
For the development of aptamer-based affinity purification, aptamer immobilization on a solid stationary phase is a prerequisite. Covalent binding of amino-modified aptamers to carboxyl-functionalized surfaces can be mediated by EDC or achieved by direct coupling to preactivated surfaces e.g. by tosyl- or N-hydroxysuccinimid (NHS) groups. For VEGF purification, functionalized magnetic beads were used as solid stationary phase material. As the VEGF tends to adsorb to solid surfaces, we primarily analyzed VEGF binding to unmodified magnetic beads. In order to minimize unspecific binding towards the stationary phase, three types of carboxyl-functionalized magnetic beads were compared, consisting of different surface materials: polystyrene (carboxyl-functionalized), silane (carboxyl-functionalized) and agarose (NHS-activated carboxyl group). Functional groups on the magnetic beads’ surfaces were blocked with Tris to avoid electrostatic interactions and the magnetic beads were incubated with purified VEGF in V7t1 binding buffer. SDS-PAGE analysis (Fig. 3, left) revealed strong bands of approximately 26 kDa (around the predicted size of monomeric VEGF) and weak bands of approximately 54 kDa (around the predicted size of dimeric VEGF) that were verified to be VEGF by western blot analysis (Fig. 3, right). Protein incubation experiments demonstrate a strong adsorption of VEGF to silane and agarose magnetic beads, as there remained no VEGF in the supernatant after protein incubation (lane S), whereas VEGF was eluted from the magnetic beads by boiling them in SDS sample buffer (lane B). Silane and agarose both possess hydrophilic surfaces offering multiple hydroxyl groups which may interact as hydrogen bond donators with carbonyl or amino groups of the VEGF. In contrast, no binding to hydrophobic polystyrene particles was observed, as hydrophobic interactions between the surface and VEGF could be prohibited by Tween 20 which is a component of the binding buffer. The results indicate the particular suitability of polystyrene matrices for VEGF purification. Therefore, they were chosen as stationary phase for further experiments.
Fig. 3.
SDS-PAGE analysis of VEGF adsorption to magnetic beads and western blot analysis of VEGF. Left: the following samples were applied to the SDS-PAGE and stained by silver staining: (M) molecular weight marker; (V) initial VEGF-sample; (S) supernatant after incubation with magnetic beads; (W3) last washing fraction; (B) VEGF eluted from the magnetic beads by boiling them in SDS sample buffer (bead fraction). VEGF (27 kDa band) adsorbed to Tris-blocked silane and agarose particles but not to polystyrene magnetic beads. Right: the initial VEGF-sample was analyzed by western blot and chromogenic immunodetection utilizing an anti-VEGF antibody. Two bands at approximately 26 kDa and 54 kDa were verified to be VEGF.
3.2.2. Binding of VEGF to aptamer-modified magnetic beads
Demonstrating the principle of aptamer-based purification, VEGF binding to aptamer-modified magnetic beads was investigated. Therefore, the aptamer V7t1 was immobilized on polystyrene particles. In total, 250 pmol immobilized aptamer V7t1 were incubated with an excess of VEGF (600 pmol) in V7t1 binding buffer. SDS-PAGE analysis reveals a VEGF band in the supernatant obtained after incubation with aptamer-modified magnetic beads (Fig. 4, lane S), no bands in the washing fractions (lanes W3, W5), but a VEGF band after boiling the magnetic beads in SDS sample buffer (bead fraction, lane B). Thus, VEGF was bound by the aptamer V7t1 proving that despite of aptamer immobilization correct aptamer folding is still possible and as a result specific target binding is achieved
Fig. 4.
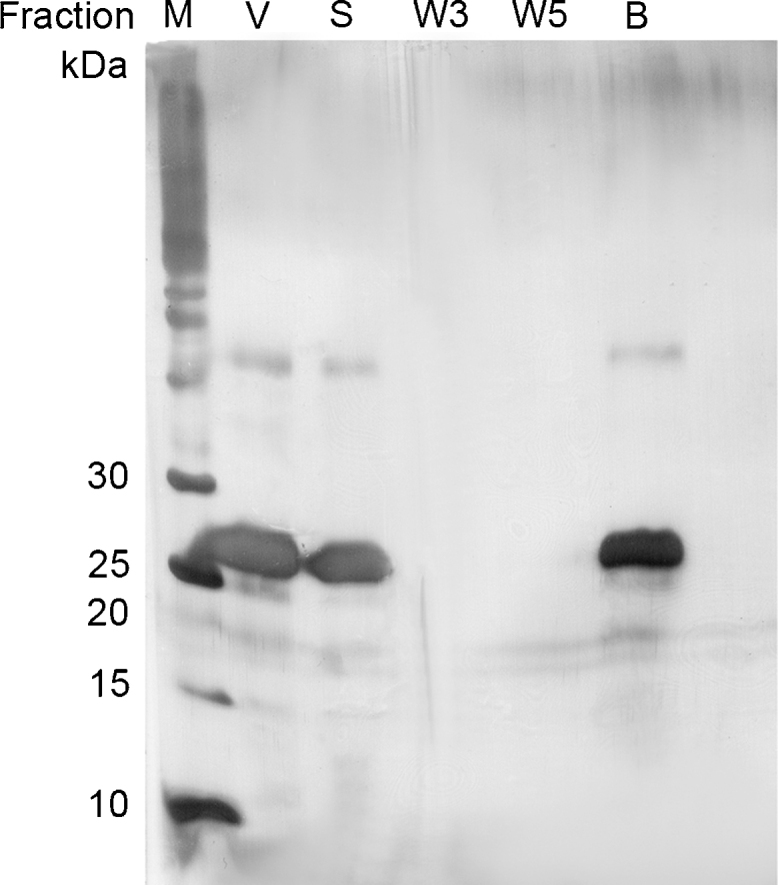
SDS-PAGE analysis of VEGF adsorption to magnetic beads. The following samples were applied to the SDS-PAGE and stained by silver staining: (M) molecular weight marker; (V) initial VEGF-sample; (S) supernatant after incubation with magnetic beads; (W3, W5) washing fractions; (B) VEGF eluted from the magnetic beads by boiling them in SDS sample buffer (bead fraction). VEGF (27 kDa band) binds to V7t1 aptamer which was immobilized on polystyrene magnetic beads.
3.2.3. Introduction of aptamer spacers
Aptamer folding into defined three-dimensional conformations depends on the molecular flexibility which may be restricted by aptamer immobilization. The forced contiguousness of the aptamer to the surface and to other aptamer molecules leads to intermolecular interactions which may disturb aptamer folding leading to inhibition of target recognition. Steric hindrance can be prohibited by introducing spacer molecules between aptamer and surface [28], [30]. The molecular spacers work like flexible arms which enlarge the distance between aptamer and surface and hence, support aptamer folding.
To study the influence of molecular spacers, V7t1 variants modified with different types of spacer molecules at the 3′-terminus were immobilized on polystyrene magnetic beads and their VEGF binding capacities were compared with those of V7t1 without any spacer and a random DNA sequence. A 36-atom dodeca-ethyleneglycol spacer (12EG spacer), an aptamer sequence that was elongated with 14 thymine nucleotides (14T spacer) and the V7t1 precursor sequence which contains 14 additional bases of the original selected aptamer Vap7 (14B spacer) were used. All aptamers were immobilized in a density of approximately 230 pmol per mg beads. Table 1 presents the amount of bound and eluted VEGF in this binding assay. All V7t1 aptamer variants were able to bind at least 96 pmol VEGF per mg magnetic beads, whereas the random DNA sequence bound less than 40 pmol VEGF. The introduction of a molecular spacer raised the VEGF binding capacity up to approximately 245 pmol per mg beads. However, not all types of spacers enhanced VEGF binding. Adding a dodeca-ethyleneglycol spacer to the V7t1 barely raises the VEGF binding capacity (1.1 times) whereas aptamer elongation with 14B spacer increases it up to 2.5 times. The 14B spacer provides a theoretic distance of 5.29 nm between aptamer and bead surface which is marginally longer than the distance spanned by the 12EG-spacer (4.58 nm). The 14T spacer is estimated to be the longest spacer molecule (7.28 nm) but enhances VEGF binding capacity only 1.8 times. Even though additional room leads to a higher molecular flexibility this effect does not seem to improve target binding significantly. Rather, neighbored nucleobases support the target binding, presumably by facilitating the target-induced aptamer folding as already described for microarray application [12]. Thus, the V7t1 aptamer with 14B spacer which contains additional bases of the originally selected aptamer Vap7 is best suited for VEGF binding. Our results suggest that these bases may additionally contribute to either target binding or stabilization of aptamer folding.
Table 1.
Effect of aptamer spacers on the VEGF binding capacity and target elution. Aptamer V7t1 with different spacer modifications was immobilized on magnetic beads in a density of app. 230 pmol/mg beads, respectively and afterwards incubated with VEGF. The following spacers were analyzed: dodeca-ethyleneglycol spacer (12EG spacer), 14 thymine nucleotides spacer (14T spacer) and an additional sequence of 14 defined nucleotides originating from the full length aptamer Vap7 (14B spacer).
| Spacer | Aptamer density (pmol/mg) | Spacer length (nm) | Bound VEGF (pmol/mg) | Eluted VEGF (pmol/mg) |
|---|---|---|---|---|
| No spacer | 230 | – | 96.06 | 82.84 |
| 12EG spacer | 247 | 4.58 | 103.23 | 81.40 |
| 14T spacer | 211 | 7.28 | 175.37 | 151.14 |
| 14B spacer | 231 | 5.29 | 244.85 | 226.78 |
| Random DNA | 207 | – | 35.17 | 30.29 |
Moreover, quantification of VEGF in the elution fractions demonstrates a successful VEGF elution by addition of 500 mM NaCl to the binding buffer. The ions may interfere with either intermolecular electrostatic interactions between aptamer and protein or with intramolecular interactions between the aptamer’s nucleobases resulting in destabilization of aptamer folding. Salt-dependent protein elution is a mild elution strategy which may avoid protein denaturation [23]. For all aptamer variants it was possible to elute at least 75% of the bound protein which is a promising basis for VEGF purification. Further, sodium chloride can be easily removed from the immobilized aptamers by thoroughly washing indicating that the reuse of aptamer-modified magnetic beads is possible.
3.2.4. Optimization of aptamer density
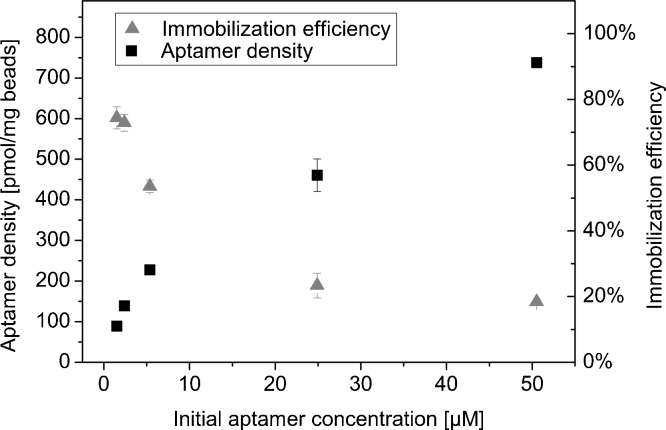
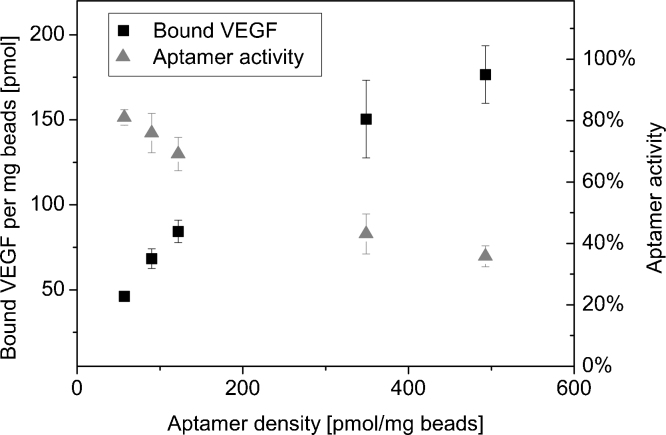
Aptamer density on the magnetic beads’ surfaces determines their maximum target binding capacity. However, a high aptamer density may also lead to steric hindrance and therefore inhibited target binding due to interactions between neighbored aptamers. For investigating the optimal aptamer density, the aptamer V7t1_14B was immobilized on the magnetic beads in different densities and VEGF binding capacity was compared. Regulation of aptamer density was achieved by variation of aptamer concentration in the immobilization reaction mixture and therefore variation of the aptamer-to-magnetic bead ratio (Fig. 5). Depending on the initial aptamer concentration, different immobilization efficiencies were achieved. Immobilization efficiency was maximal (75%) at low initial aptamer concentration of 2 μM and decreases to 18% with raising initial aptamer concentration. Concurrently, aptamer densities were in a range between 90 and 740 pmol per mg.
Fig. 5.
Immobilization of V7t1_14B on tosyl-activated magnetic beads. Different initial aptamer concentrations were applied to the immobilization process resulting in different aptamer densities on the beads’ surfaces. Furthermore, the immobilization efficiency as a function of initial aptamer concentration is presented.
For investigation of VEGF binding capacity, magnetic beads with different densities of V7t1_14B were incubated with VEGF and the amount of bound VEGF was compared. Fig. 6 presents the analysis of VEGF binding capacity as a function of aptamer density. Further, the percentage of aptamer which has bound a VEGF molecule is shown as aptamer activity. Maximum aptamer activity (81%) was achieved at an aptamer density of 55 pmol per mg beads. With increasing aptamer density the aptamer activity decreases to 36%. Thus, high aptamer densities seem to impair VEGF binding which can be a consequence of either inhibited aptamer folding or a restricted accessibility of immobilized aptamers. Nevertheless, the amount of bound VEGF per mg beads increases with raising aptamer density in a non-linear manner up to nearly 180 pmol per mg beads for an aptamer density of 493 pmol per mg beads. For optimal VEGF purification a low aptamer density of approximately 100 pmol per mg is recommended, as in this case at least 75% of the aptamer are functionally active. Further, due to the high immobilization efficiency of approximately 73% at these concentrations, using a low aptamer density allows a more cost-efficient bead preparation leading to an economic purification process. Therefore, using polystyrene magnetic beads and the aptamer V7t1_14B immobilized in a density of approximately 100 pmol per mg beads is recommended for VEGF purification.
Fig. 6.
Analysis of VEGF binding capacity as a function of aptamer density. V7t1_14B modified magnetic beads with different aptamer densities were incubated with VEGF. The amount of bound VEGF and the percentage of aptamer which has bound a VEGF-molecule (aptamer activity) were calculated.
3.3. VEGF purification from a complex protein mixture
Based on the optimized parameters for VEGF binding, we investigated VEGF purification from a complex protein mixture imitating a protein rich cell lysate fraction consisting of FCS (1 mg/mL) and VEGF (200 μg/mL). Aptamer-modified magnetic beads were incubated with the protein mixture, beads were washed with V7t1 binding buffer and finally VEGF was eluted by addition of 500 mM NaCl. SDS-PAGE analysis demonstrates a successful purification of VEGF without any visible contaminations (Fig. 7A). The purity was estimated to be 91% by densitometry.
Fig. 7.
SDS-PAGE analysis of VEGF purification from a complex protein mixture.
(A) A mixture of VEGF and FCS in V7t1 binding buffer was incubated with V7t1_14B- and random DNA-modified magnetic beads. VEGF bound to V7t1_14B but not to the random DNA. The following samples were applied to the SDS-PAGE and stained by silver staining: (M) molecular weight marker; (P) initial protein sample; (S) supernatant after incubation with magnetic beads; (W1, W3) washing fractions; (E1, E2) elution fractions; (B) VEGF eluted from the magnetic beads by boiling them in SDS sample buffer (bead fraction).
(B) VEGF was purified from solubilized VEGF inclusion bodies, diluted in V7t1 binding buffer using V7t1_14B- modified magnetic beads.
Most of the bound VEGF was eluted under the elected elution conditions (500 mM NaCl), Only a small amount VEGF remained on the magnetic beads which could be eluted by boiling them in SDS sample buffer (lane B). In contrast, no unspecific binding towards the random DNA-modified beads was observed.
To prove the applicability of the aptamer-based VEGF purification to the E. coli protein extract, solubilized inclusion bodies were diluted with equal volume of V7t1 binding buffer and afterwards VEGF purification was performed as described above (Fig. 7B). The purity estimated by densitometry was 91% and therefore is comparable to the purity achieved by combination of Ni-IMAC and Heparin-Affinity chromatography which was about 93%.
4. Conclusions
A new aptamer-based affinity purification platform for the Vascular Endothelial Growth Factor was developed and optimized in a small scale by utilizing magnetic beads. The aptamer V7t1 was covalently immobilized on polystyrene magnetic beads in a controlled orientation resulting in a functionally active stationary affinity matrix which concomitantly exhibits low unspecific binding. The aptamers’ binding activity was optimized by diminishing the aptamer density and by introducing a spacer molecule which consists of 14 additional nucleotides. The platform permits target elution by enhanced ionic strength due to NaCl addition. Those mild elution conditions prevent protein denaturation and facilitate a high protein recovery of at least 75%. In summary, this work demonstrates that the aptamer V7t1 is a promising alternative to heparin for VEGF affinity purification.
Acknowledgement
We thank Moran Jerabek-Willemsen (NanoTemper Technologies GmbH) for the advice and for the opportunity for the MST measurements. This work is supported by funding from the Deutsche Forschungsgemeinschaft (DFG) for the Cluster of Excellence REBIRTH (From Regenerative Biology to Reconstructive Therapy).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2015.08.006.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Burmeister P.E., Lewis S.D., Silva R.F., Preiss J.R., Horwitz L.R., Pendergrast P.S., McCauley T.G., Kurz J.C., Epstein D.M., Wilson C. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005;12(1):25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Dausse E., Da Rocha Gomes Sonia, Toulmé J.-J. Aptamers: a new class of oligonucleotides in the drug discovery pipeline? Curr. Opin. Pharmacol. 2009;9(5):602–607. doi: 10.1016/j.coph.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.L., Gold, N., Janjic, US patent 669625 (1998).
- 5.Green L.S., Jellinek D., Bell C., Beebe L.A., Feistner B.D., Gill S.C., Jucker F.M., Janjić N. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem. Biol. 1995;2(10):683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 6.Hage D.S., Anguizola J.A., Bi C., Li R., Matsuda R., Papastavros E., Pfaunmiller E., Vargas J., Zheng X. Pharmaceutical and biomedical applications of affinity chromatography: recent trends and developments. J. Pharm. Biomed. Anal. 2012;69:93–105. doi: 10.1016/j.jpba.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han B., Zhao C., Yin J., Wang H. High performance aptamer affinity chromatography for single-step selective extraction and screening of basic protein lysozyme. J. Chromatogr. B. 2012;903:112–117. doi: 10.1016/j.jchromb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Hianik T., Wang J. Electrochemical aptasensors—recent achievements and perspectives. Electroanalysis. 2009;21(11):1223–1235. [Google Scholar]
- 9.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45(9):1628–1650. [PubMed] [Google Scholar]
- 10.Jellinek D., Green L.S., Bell C., Janjić N. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry. 1994;33(34):10450–10456. doi: 10.1021/bi00200a028. [DOI] [PubMed] [Google Scholar]
- 11.Kökpinar O., Walter J.-G., Shoham Y., Stahl F., Scheper T. Aptamer-based downstream processing of his-tagged proteins utilizing magnetic beads. Biotechnol. Bioeng. 2011 doi: 10.1002/bit.23191. [DOI] [PubMed] [Google Scholar]
- 12.Lao Y.-H., Peck K., Chen L.-C. Enhancement of aptamer microarray sensitivity through spacer optimization and avidity effect. Anal. Chem. 2009;81(5):1747–1754. doi: 10.1021/ac801285a. [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109(5):1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lönne M., Zhu G., Stahl F., Walter J.-G. Aptamer-modified nanoparticles as biosensors. Adv. Biochem. Eng. Biotechnol. 2014;140:121–154. doi: 10.1007/10_2013_231. [DOI] [PubMed] [Google Scholar]
- 15.Marusic M., Veedu R.N., Wengel J., Plavec J. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013;41(20):9524–9536. doi: 10.1093/nar/gkt697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer M., Scheper T., Walter J.-G. Aptamers: versatile probes for flow cytometry. Appl. Microbiol. Biotechnol. 2013;97(16):7097–7109. doi: 10.1007/s00253-013-5070-z. [DOI] [PubMed] [Google Scholar]
- 17.Murphy M.B., Fuller S.T., Richardson P.M., Doyle S.A. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res. 2003;31(18) doi: 10.1093/nar/gng110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nonaka Y., Sode K., Ikebukuro K. Screening and improvement of an anti-VEGF DNA aptamer. Molecules. 2010;15(1):215–225. doi: 10.3390/molecules15010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oktem H.A., Bayramoglu G., Ozalp V.C., Arica M.Y. Single-step purification of recombinant Thermus aquaticus DNA polymerase using DNA-aptamer immobilized novel affinity magnetic beads. Biotechnol. Prog. 2007;23(1):146–154. doi: 10.1021/bp0602505. [DOI] [PubMed] [Google Scholar]
- 20.Pizarro S.A., Gunson J., Field M.J., Dinges R., Khoo S., Dalal M., Lee M., Kaleas K.A., Moiseff K., Garnick S., Reilly D.E., Laird M.W., Schmelzer C.H. High-yield expression of human vascular endothelial growth factor VEGF(165) in E. coli and purification for therapeutic applications. Protein Expr. Purif. 2010;72(2):184–193. doi: 10.1016/j.pep.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Romig T.S., Bell C., Drolet D.W. Aptamer affinity chromatography combinatorial chemistry applied to protein purification. J. Chromatogr. B. 1999;731(2):275–284. [PubMed] [Google Scholar]
- 22.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjić N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273(32):20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 23.Sibler A.-P., Baltzinger M., Choulier L., Desplancq D., Altschuh D. SPR identification of mild elution conditions for affinity purification of E6 oncoprotein, using a multivariate experimental design. J. Mol. Recognit. 2008;21(1):46–54. doi: 10.1002/jmr.865. [DOI] [PubMed] [Google Scholar]
- 24.Siemeister G., Schnurr B., Mohrs K., Schächtele C., Marmé D., Martiny-Baron G. Expression of biologically active isoforms of the tumor angiogenesis factor VEGF in E. coli. Biochem. Biophys. Res. Commun. 1996;222(2):249–255. doi: 10.1006/bbrc.1996.0730. [DOI] [PubMed] [Google Scholar]
- 25.Strehlitz B., Nikolaus N., Stoltenburg R. Protein detection with aptamer biosensors. Sensors. 2008;8(7):4296–4307. doi: 10.3390/s8074296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 27.Walter J.-G., Heilkenbrinker A., Austerjost J., Timur S., Stahl F., Scheper T. Aptasensors for small molecule detection. Z. Naturforsch. B. 2012;67b:976–986. [Google Scholar]
- 28.Walter J.-G., Koekpinar O., Friehs K., Stahl F., Scheper T. Systematic investigation of optimal aptamer immobilization for protein-microarray applications. Anal. Chem. 2008;80(19):7372–7378. doi: 10.1021/ac801081v. [DOI] [PubMed] [Google Scholar]
- 29.Walter J.-G., Stahl F., Scheper T. Aptamers as affinity ligands for downstream processing. Eng. Life Sci. 2012;12(5):496–506. [Google Scholar]
- 30.Zhu G., Lübbecke M., Walter J.-G., Stahl F., Scheper T. Characterization of optimal aptamer-microarray binding chemistry and spacer design. Chem. Eng. Technol. 2011;34(12):2022–2028. [Google Scholar]
- 31.Zhu G., Walter J.-G. Aptamer-modified magnetic beads in affinity separation of proteins. Methods Mol. Biol. 2015;1286:67–82. doi: 10.1007/978-1-4939-2447-9_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.