Abstract
The genetic modification of primary bacterial disease isolates is challenging due to the lack of highly efficient genetic tools. Herein we describe the development of a modified PCR-based, λ Red-mediated recombineering system for efficient deletion of genes in Gram-negative bacteria. A series of conjugally transferrable plasmids were constructed by cloning an oriT sequence and different antibiotic resistance genes into recombinogenic plasmid pKD46. Using this system we deleted ten different genes from the genomes of Edwardsiella ictaluri and Aeromonas hydrophila. A temperature sensitive and conjugally transferable flp recombinase plasmid was developed to generate markerless gene deletion mutants. We also developed an efficient cloning system to capture larger bacterial genetic elements and clone them into a conjugally transferrable plasmid for facile transferring to Gram-negative bacteria. This system should be applicable in diverse Gram-negative bacteria to modify and complement genomic elements in bacteria that cannot be manipulated using available genetic tools.
Keywords: Recombineering, Genetic modification, Bacterial pathogens
1. Introduction
Genetic manipulation of bacterial strains provides critical information on the contributions of specific loci to virulence or other cellular functions, and many systems have been developed to achieve genetic knockouts and modifications [4], [5], [18]. The modification of bacterial genomes using counter-selectable double-crossover methods are labor intensive and sometimes very difficult to achieve due to the low frequency of recombination events [21], [26], [31]. In contrast, the λ Red recombineering system [39], [41] has many advantages as a fast, efficient and reliable means of generating targeted genetic modifications in prokaryotes [11], [61] and eukaryotes [7]. The λ Red system expresses Exo, Beta and Gam proteins that work coordinately to recombine single and double stranded DNA [11], [38], [61], and has been exploited for genome modifications in Escherichia coli, Salmonella enterica and other Gram-negative bacteria [9], [11], [40], [61]. Exo has a 5′–3′ double stranded DNA (dsDNA)-dependent exonuclease activity for generating 3′ single stranded DNA (ssDNA) overhangs [6], [32], [34] which then serve as a substrate for ssDNA-binding protein Beta to anneal complementary DNA strands for recombination [8], [28], [38]. Gam, an inhibitor of host exonuclease activity due to RecBCD [44], helps to improve the efficiency of λ Red-mediated recombination with linear double-strand DNA. Unlike recA-dependent homologous recombination which requires longer regions of sequence homology with the targeted genetic region [25], the λ Red apparatus can efficiently recombine DNA with homologous regions as short as 30–50 bp which can directly be incorporated into oligonucleotide primers in a PCR [11], [61]. The recombineering technique is widely used to generate precise deletions [11], substitutions [33], insertions [36] or tagging [57] of targeted genes. One of the biggest advantages of the recombineering method is that modifying DNA can precisely eliminate the antibiotic selection markers for subsequent modification of the targeted DNA [11], [42].
While this recombineering system works well in a model bacterium such as E. coli [37], [39], bacteria often express restriction endonucleases that make them recalcitrant to foreign DNA even among naturally competent strains [1], [3]. In fact, it was the study of experimental infections of E. coli strains with bacteriophage λ that led to the discovery of restriction-modification (RM) systems [2]. Overcoming host RM systems can be accomplished via the passage of plasmids through a methylation-minus E. coli strain [51], but in highly methylated bacterial strains it may be necessary to use an in vitro or in vivo methylation strategy to achieve more efficient electroporation [12], [13], [29]. However, modulating the plasmid DNA methylation status is inefficient and labor-intensive compared to using conjugal transfer to introduce foreign DNA into a bacterial strain using a broad host range plasmid like IncP when electroporation is problematic [14], [15], [17].
Our need to generate targeted genetic deletions in Gram-negative bacterial pathogens of farmed catfish led to the development of recombinogenic plasmids that could be introduced into Gram-negative bacteria via conjugation. Our studies focused on two bacterial pathogens, including motile Aeromonas septicemia (MAS) and enteric septicemia of catfish (ESC) caused by Aeromonas hydrophila and Edwardsiella ictaluri, respectively, which are responsible for significant economic losses to the channel catfish industry in the Southeastern United States [56]. Fish diseases caused by strains of E. ictaluri are also frequently reported in catfish farming in Asia [46]. While E. ictaluri was formerly the most important bacterial pathogen in farmed US catfish, in 2009 US catfish farmers experienced epidemic disease outbreaks of motile Aeromonas septicemia (MAS) caused by a highly virulent Aeromonas hydrophila strain [20]. This newly emergent and virulent A. hydrophila strain, which has been implicated to have an Asian origin [23], is responsible for the death of millions of pounds of food-sized channel catfish in the US [23]. Though both E. ictaluri and A. hydrophila pose serious threats to the US catfish industry [24], [45], [56] as well as global fish farming [46], [62], highly efficient genome modification techniques have not been developed yet to study the virulence mechanisms and permit generation of avirulent vaccines for these two pathogens.
Though recombineering techniques are widely being used for genome modification of domesticated laboratory isolates such as E. coli strains, the implementation of these techniques for primary pathogenic isolates is quite challenging. In this study, we modified the available λ Red recombination tools [11], [54] to generate markerless mutants of E. ictaluri and A. hydrophila. Several conjugally transferable and temperature-sensitive plasmids were constructed to facilitate the genome modification by recombineering and removal of antibiotic resistance marker followed by recombineering. In addition, we also developed a novel in vivo error-free cloning system that can be used to clone large fragments of genomic DNA without PCR amplification of the inserts and used to complement larger genomic regions.
2. Materials and methods
2.1. Bacterial strains and plasmids
The list of bacterial strains and plasmids used in this study is presented in Table 1. E. ictaluri and A. hydrophila strains were routinely grown on Trypticase Soy Broth (TSB) or Agar (TSA) medium at 28 °C and 30 °C, respectively. E. coli SM10λpir [50] was routinely used for the conjugal transfer of mobilizable plasmids to strains of E. ictaluri and A. hydrophila as previously described. E. coli BW25141 and BT340 [11] were received from the Yale University Genetic Stock Center. When antibiotic selection was required, bacterial growth media were supplemented with kanamycin (50 μg/ml), chloramphenicol (15 and 25 μg/ml for strains of E. ictaluri and A. hydrophila, respectively), tetracycline (10 μg/ml) and/or colistin (10 μg/ml).
Table 1.
List of bacterial strains and plasmids used in this study.
| Bacterial strains or plasmid | Features | References |
|---|---|---|
| E. coli | ||
| SM10λpir | thi-1thr leutonAlacYsupE recA::RP4-2-TcT::Mu Kmrλpir | [50] |
| BW25113/pKD46 | F-, Δ(araD-araB) 567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB) 568, hsdR514, pKD46 | [11] |
| BT340 | F-, Δ(argF-lac) 169, ϕ80dlacZ58(M15), glnV44(AS), λ−, rfbC1, gyrA96(NalR), recA1, endA1, spoT1, thiE1, hsdR17, pCP20 | [11] |
| BW25141/pKD4 | F-, Δ(araD-araB) 567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR) 580, λ−, galU95, ΔuidA3::pir+, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB) 568, hsdR514, pKD4 | [11] |
| “E. cloni” 10G | F−mcrAΔ(mrr-hsdRMS-mcrBC) endA1 recA ϕ80dlacZΔM15ΔlacX74 araD139 Δ (ara,leu) 7697 galU galK rpsL (StrR) nupG λ−tonA | Lucigen Corp. WI |
| E. ictaluri | ||
| Alg-08-183 | Pathogenic isolates from diseased catfish | [22] |
| Alg-08-183 (pMJH46) | E. ictaluri strain Alg-08-183 with plasmid pMJH46 | This study |
| R4383 | Highly hemolytic E. ictaluri strain from diseased catfish | [59] |
| R4383 (pMJH46) | E. ictaluri strain R4383 with plasmid pMJH46 | This study |
| Alg-08-183ompLC::kanR | Replacement of hemolysin ompLC gene with kanR gene | This study |
| Alg-08-183ompLC::kanR (pCP20) | E. ictaluri Alg-08-183ompLC::kanR with pCP20 | This study |
| Alg-08-183 drtA::kanR | Replacement of hemolysin dtrA gene with kanR gene | This study |
| Alg-08-183 drtA::kanR (pCP20) | E. ictaluri Alg-08-183 drtA::kanR with pCP20 | This study |
| Alg-08-183ΔompLC | In-frame deletion of ompLC gene | This study |
| Alg-08-183ΔdrtA | In-frame deletion of dtrA gene | This study |
| R4383eihA::kanR | Replacement of hemolysin eihA gene with kanR gene | This study |
| R4383eihA::kanR (pCP20) | E. ictaluri R4383eihA::kanR with pCP20 | This study |
| R4383ΔeihA | In-frame deletion of hemolysin gene eihA | This study |
| A. hydrophila | ||
| Ml09-119(pMJH46) | A. hydrophila ML09-119 with pMJH46 | This study |
| Ml09-119(pMJH65) | A. hydrophila ML09-119 with pMJH65 | This study |
| ML09-119ymcC:cat (pCMT-flp) | A. hydrophila ML09-119ymcC:cat with pCMT-flp | This study |
| ML09-119ymcC:cat | Replacement of ymcA gene with cat gene | This study |
| ML09-119ΔymcC | Unmarked deletion of ymcC gene | This study |
| ML09-119waaL::cat | Replacement of waaL gene with cat gene | This study |
| ML09-119iolA::cat | Unmarked deletion of iolA gene | This study |
| ML09-119hlyA::cat | Replacement of hlyA gene with cat gene | This study |
| ML09-119ΔhlyA | Unmarked deletion of hly gene | This study |
| ML09-119aerA::cat | Replacement of aerA gene with cat gene | This study |
| ML09-119 vgr3::cat | Replacement of vgr3 gene with cat gene | This study |
| ML09-119Δvgr3 | Unmarked deletion of vgr3gene | This study |
| ML09-1193,822,477 | Deletion of genetic region 3822,477..3,822,683 of ML09-119 | This study |
| ML09-119 (pBBC2) | A. hydrophila ML09-119 with pBBC2 | This study |
| Plasmids | ||
| pACYC184 | Cloning vector with p15A origin of replication | [63] |
| pKD46 | Temperature-sensitive recombinogenic plasmid | [11] |
| pKD4 | Template for recombineering substrate | [11] |
| pMJH46 | Conjugally transferrable recombinogenic plasmid | This Study |
| pMJH65 | Conjugally transferrable recombinogenic plasmid | This Study |
| pCMT-flp | Temperature-sensitive Flp recombinase plasmid | This Study |
| pMJH97 | cat-oriT-oriR backbone plasmid for PCR-free cloning | This Study |
| pCP20 | Temperature-sensitive Flp recombinase plasmid | [7] |
| pGNS-BAC | Conjugally transferable BAC vector | [27] |
2.2. Recombinant DNA techniques and conjugal transfer of recombinogenic plasmids
The list of primers used in this study is presented in Table 2. All oligonucleotides were purchased from Eurofins MWG Operon (Huntsville, AL). For cloning purposes, we routinely used electrocompetent E. coli (“E. cloni 10G”, Lucigen Corp., Middleton, WI). PCR amplifications were carried out using EconoTaq DNA polymerase (Lucigen Corp.), Pfu DNA polymerase (Life Technologies, Grand Island, NY) and TaKaRa Ex Taq (Clontech, Mountain View, CA) as appropriate. Genomic DNAs and plasmids were extracted using the E.Z.N.A. DNA Isolation Kit (Omega Biotek, Atlanta, GA) and FastPlasmid Mini Kit (5 Prime, Gaithersburg, MD), respectively. Restriction enzymes and T4 DNA Ligase (Quick ligase) used for restriction digestion of DNAs and cloning, respectively, were purchased from New England Biolabs (Ipswich, MA). Restriction digested DNAs with sticky ends were blunt-ended using a DNA Terminator kit (Lucigen Corp.). Digested DNAs and ligation mix were purified using DNA Clean and Concentrator-5 (Zymo Research, Irvine, CA). DNA concentrations were quantified using a Qubit 2.0 Fluorometer (Life Technologies). The mobilizable recombinogenic plasmids pMJH46 and pMJH65, and flp recombinase plasmid pCMT-flp were introduced into E. coli SM10λpir by electroporation according to a previously published method [47]. Plasmids were conjugally transferred into E. ictaluri and A. hydrophila by filter mating experiments according to the methods described previously [35]. E. ictaluri and A. hydrophila transconjugants were selected on LB plates supplemented with chloramphenicol and colistin, or tetracycline and colistin, respectively. The introduction of plasmids into E. ictaluri or A. hydrophila was confirmed by their growth in the presence of appropriate antibiotics and by conducting PCR with a plasmid-specific primer set.
Table 2.
List of primers used in this study.
| Primer Name | Sequence in 5′ to 3′ direction |
|---|---|
| pKD4-ompLCf | AACTGGTAGATCATACCAACGCCAACGATGTTGTCGGTGCTGATACCGGCGTGTAGGCTGGAGCTGCTTC |
| pKD4-ompLCr | GTTCAAAAAATTCCCGATGGAATCAAATTAGGCAGTGGCAGGTGTCAAAACATATGAATATCCTCCTTAGT |
| ML44-RedF | ATGCTTACAACAAAAAATATGCCAGCCAATGCTGGGCTGGCAGCGTTTTCTGGTGTAGGCTGGAGCTGCTTC |
| ML44-RedR | TTAGCAAGGGGGAAGATGCTCTGGTGGTGATGGTCTGTTTTTCTGATGATAGCATATGAATATCCTCCTTAGT |
| ML-44R | TATGCAAGCTTATATAAGTGTAGTGCAGTATG-3 |
| 44expandedF | TATGCTCTAG AACTTAACTGTTGGTCATAG-3' |
| 44expandedR | TATGCTCTAG AATATTCAACGGCATTAC-3' |
| Hemo-redF | TTCCTTTTAACTCTGCTTTGGCGCCCATGGGCGCTGATATGAGGCAATCTCTGTGTAGGCTGGAGCTGCTTC |
| Hemo-redR | ACGGCGGCCCGCAGGCCGCCGTTGAGGATGGATAACGTCGCCACTATCCGGTCATATGAATATCCTCCTTAGT |
| Takara-hemoF | TATGCAAGCTTCTCCTCATAGTGTGTCCGCAGT |
| Takara-hemoR | TATGCAAGCTTGCATTGACATAGGCGTTCATCT |
| H-RedtrackF | GATGTCTATCTGTTCAGCTC |
| H-RedtrackR | GTACGCAATACCAATAGTG |
| RE33-165F | TATGCAAGCTTGTAGTTCTTGCTGGTCTC |
| RE33-165R | TATGCAAGCTTGTAACGCAACATTCTAAC |
| k1 | CAGTCATAGCCGAATAGCCT |
| k2 | CGGTGCCCTGAATGAACTGC |
| kt | CGGCCACAGTCGATGAATCC |
| CatF | TATCGTGACTGACTGCTGCGTGTAGACTTCCGTTGAACT |
| CatR | ATGCAGATATCGCCTAATGAGTGAGCTAA |
| MobicatF | AGAGTGCTGACAGATGAG |
| MobicatR | ACGCAGCAGTCAGTCACGATAATGATGTGGTCTGTCCT |
| tetAR | CGACAGGAGCACGATCAT |
| tetAF | TGTAGCACCTGAAGTCAGC |
| Flp-pRhamF | CGC GAA CAG ATT GGA GGTCCACAATTTGGTATATTATGTA |
| Flp-pRhamR | GTG GCG GCC GCT CTA TTATATGCGTCTATTTATGTAGGA |
| UP-F-flp-oriR | ATGGCTTCCATGTCGGCAGAAT |
| DN-R-oriT | TTGGTGTATCCAACGGCGTCAGCCGGGCAGGATAGGTGAAGTAGGCCCACCCGCGAGCGGGTGTTCCTTCTTCACTGTCCCTTATTCGTTCCACTGAGCGTCAGACC-3′ |
| Li-CCatF | T*G*G*G*GCAGTTGATGAAACATCGCGCAGCCTGCCGGCCCCACATGGCCTCGACAGCCGCTAGGTACC CGCTCCATGAGCTTATCGCGAAT |
| Li-AAAAR | A*T*G*C*ACTTTTTCATGCACAACCCCGGTGGGGCCGGGCTCTATCTGCCGTTCAACGCCTGGGGC CCTCCTGTTCAGCTACTGACG |
| CCatR-oriT | TTGGTGTATCCAACGGCGTCAGCCGGGCAGGATAGGTGAAGTAGGCCCACCCGCGAGCGGGTGTTCCTTCTTCACTGTCCCTTATTCGGCCGTCGACCAATTCTCATGTT |
| CatFseq | CTGGTTGCTACGCCTGAATAAGTG |
| p15AF | TCACATATTCTGCTGACGCACC |
| Li234R-HindII | AGT CTA AGC TTG CTC AAG CCA ACA ACC GCG AA |
| CCatR | GGC CGT CGA CCA ATT CTC ATG TT |
| ymcA-CM-1F | GCGACAAAAATAAGGCTGCCA |
| pMJH46SeqF | CGTCTACTCCGTTACAA |
| Mob-seqR | GGCTTCACCTTCAACC |
| pMJH46SeqR | AGTATGATCTCAATGGTTCG |
| Cat-SeqF | CAGAATGCTTAATGAATTAC |
| CatR-int | CATGCGATATCTAATGAATCGGCCAAC |
| FlpF1 | CGCATTCACAGTTCTCCGCAAG |
| FlpR1 | GTGCCTACTAACGCTTGTCT |
| FlpF2 | CTTCGATCATTGGACCGCTG |
| FlpR2 | CGAATCATCGGAAGAAGCAG |
| 97seq1F | ACAAGACGTTGAGGCCACTATC |
| 97seq2F | TTGGTCTGCGCGTAATCTCTTG |
| 97seq3F | GGAACTGAGTGTCAGGCGTGGA |
| 97seq4R | GGAGGCCAGATGTTGAGTCGCA |
| 97Seq8R | GCAGCAGCCACTGGTAATTGA |
| 97Seq7R | CAAGAGATTACGCGCAGACCA |
| 97Seq5F | CAAGATGTGGCGTGTTACGGT |
| 97Seq6F | GGACAGTGAAGAAGGAACACC |
| CCatF | CGCTCCATGAGCTTATCGCGAAT |
| AAAAF | CCTCCTGTTCAGCTACTGACG |
| BBBBR | TATCGATGATAAGCTGTCAA |
| Vgr3F | TCACCCGGCTTGCAGTGCCCCGCCTGATGGGGCTACACGACATCTCAGAGGCGCTCAGCGGTGTAGGCTGGAGCTGCTTC |
| Vgr3R | GATGGCACGAAAAAGGTCTGCGCAGGCCCTTGCTCCTTGAGCAGCGCCTCCATCGCCTTCATATGAATATCCTCCTTAGT |
| vgR3outR | GCATGCCGATGAACTCTTCAAGTG |
| vgR3outF | ATCCTGCGAAGTCTGACTTCACC |
| hlyA-RedF | T*A*A*T*ATGGTTATGCCGTGTTCGTTCATTGTTTAAATAGCTTGGCGTGATTCGACAAGGAGATAACAGTGTAGGCTGGAGCTGCTTC |
| hlyA-RedR | C*C*C*T*GCTCTGTCAGTGACTGGCCGGTGGCCCGAAGATGCGGGTGTAGGAGGTCAGGGTCCGTACGCCATATGAATATCCTCCTTAGT |
| hlyoutF | GCATGCCGAATCATCCACCTTAGA |
| hlyoutR | CAGACCTTCTACAAGCTGGCGGAG |
| aero-RedF | T*G*C*C*GATATATAAGCGCTGGTGAATGTATGTCAATGTTCAATATATTGGGGTTGCTGTGTAGGCTGGAGCTGCTTC |
| aero-RedR | C*A*G*T*GCAAACAAAAACCGGGCCAGAGGCCCGGTTCCATCACTACAACGCACTGCCGATGGGAATTAGCCATGGTCC |
| Li234R | GCTCAAGCCAACAACCGCGAA |
| Li234*R | G*C*T*C*AAGCCAACAACCGCGAA |
| Ligase*F | G*A*C*C*AGCGCATTGAGAGAGAGG |
| Liup*F | A*C*T*T*AAGCTCGCCGAACTC |
| Lidn*R | T*G*A*T*TATGATGTAATGACTGG |
| Ligase-catF | T*G*G*G*GCAGTTGATGAAACATCGCGCAGCCTGCCGGCCCCACATGGCCTCGACAGCCGCTAGTAGACTTCCGTTGAACT |
| Ligase-catR | C*C*C*T*TTTATTATCTACCCAAGATATATGGTAATCTGCAGAAATTATGCTAGGAATGCATGGCCTAATGAGTGAGCTAA |
| Li-CatF | TGGGGCAGTTGATGAAACATCGCGCAGCCTGCCGGCCCCACATGGCCTCGACAGCCGCTAGTAGACTTCCGTTGAACT |
| Li-CatR | CCCTTTTATTATCTACCCAAGATATATGGTAATCTGCAGAAATTATGCTAGGAATGCATGGCCTAATGAGTGAGCTAA |
| O_2RecF | T*C*T*G*AGCGTAATCCATAGTCAAACCAGAAATTTTAAATTTAAGGATGTTGAATTTTGTAGACTTCCGTTGAACT |
| O_2RecR | T*A*G*A*GGAGGTATTACCCTCCTCTACTCCAAATTTTTATAAAAAATTCCGAATGTGAGCCTAATGAGTGAGCTAA |
2.3. Construction of broad host range recombinogenic plasmids
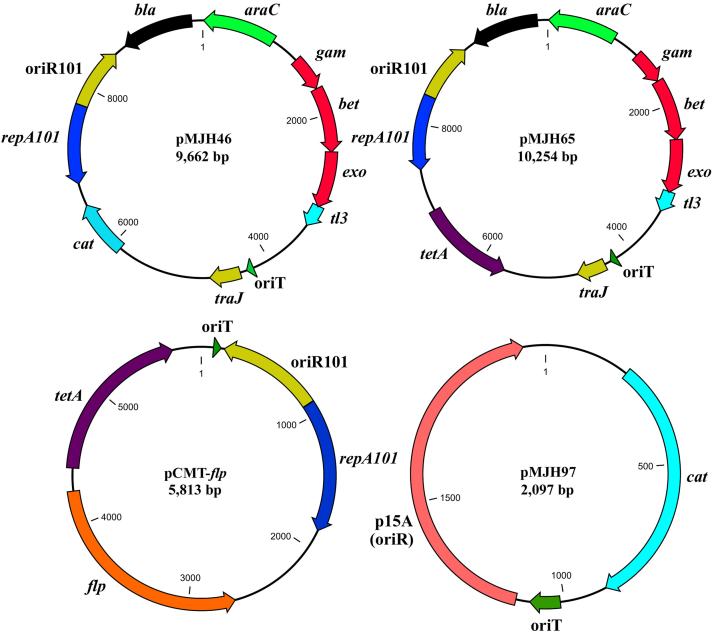
A list of plasmids used in this study is presented in Table 1. The mobilizable plasmid pMJH46 was constructed by introducing the oriT sequence and chloramphenicol acetyltransferase (cat) gene into the recombinogenic plasmid pKD46 [19] which contains an arabinose-inducible λ-Red cassette (exo, bet and gam genes) required for recombineering (Fig. 1). The oriT sequence and cat gene were PCR amplified from pGNS-BAC [27] using primers MobicatF and MobicatR, and CatF and CatR, respectively. Amplicons for the oriT sequence and cat gene were fused by splicing by overlap extension (SOE) PCR [52] using primers MobicatF (forward) and CatR (reverse). The oriT-cat cassette and pKD46 plasmid were digested with EcoRV and NcoI, respectively. NcoI digested pKD46 plasmid was blunt-ended and ligated to oriT-cat cassette using a DNA Terminator kit (Lucigen Corp., Middleton, WI) and T4 DNA ligase (Promega, WI), respectively. The ligation mixture was then transformed into electrocompetent E. coli (E. cloni 10G, Lucigen Corp.) for cloning. Transformants were selected on 2 × YT medium supplemented with ampicillin and chloramphenicol after incubation overnight at 30 °C. The introduction of the oriT-cat cassette into pKD46, resulting in pMJH46, was confirmed by PCR and sequencing as described below. To construct the recombinogenic plasmid pMJH65, plasmid pMJH46 was digested with BstZ17I and SfiI, and blunt-ended using the DNA Terminator kit. A tetracycline resistance gene (tetA) cassette was PCR amplified from pACYC184 using primers tetAF and tetAR and ligated to blunt-ended pMJH46 using T4 DNA ligase. The ligation mixture was then transformed into electrocompetent E. coli (E. cloni 10G, Lucigen Corp.) for cloning. Transformants were selected on 2×YT medium supplemented with tetracycline after overnight incubation at 30 °C. The construction of plasmid pMJH65 was confirmed by PCR and sequencing as described below.
Fig. 1.
Schematic maps of conjugally transferable recombinogenic and flp recombinase plasmids constructed in this study. The oriT sequence cloned into these plasmids facilitates the conjugal transfer of these plasmids using appropriate donor E. coli strain. Red recombinogenic plasmids pMJH46, pMJH65 and flp recombinase plasmid pCMT-flp are easily cured after heat induction at 37 °C due to temperature sensitive repA101 gene. Plasmid maps were generated by CLC Genomics Workbench (version 4.9).
2.4. Construction of conjugally transferable Flp plasmid pCMT-flp
The flp gene, which is required for FRT mediated site-specific recombination [7], was PCR amplified from pCP20 using primers Flp-pRhamF and Flp-pRhamR and was cloned into pRham N-His SUMO vector (Lucigen Corp.) under the control of the rhaPBAD promoter. The resulting plasmid pRham-flp was then digested with XbaI and blunt-ended in order to insert a tetracycline resistance gene (tetA) which was PCR amplified from pMJH65 using primers tetAF and tetAR. After cloning this tetA cassette into the pRham-flp plasmid, resulting in plasmid pRham-flp-tetA, the flp-tetA cassette was digested with AlwNI and BsaAI, and blunt-ended for cloning into repA101-oriR101 cassette which was PCR amplified from pMJH65 using primers UP-F-flp-oriR and DN-R-oriT. After cloning flp-tetA into repA101-oriR101 cassette, the construction of the resulting plasmid pCMT-flp was confirmed by sequencing as described below. To determine the efficacy of pCMT-flp plasmid in excision of an antibiotic resistance cassette flanked by FRT sequences, pCMT-flp was transferred into strains of A. hydrophila mutants by conjugation as described above.
2.5. Preparation of linear double stranded DNA (dsDNA) substrate for recombineering
The linear dsDNA fragments used for deletion of the ompLC gene from E. ictaluri with recombineering were generated by PCR amplification of the kanamycin resistance gene (kanR) cassette with its flanking FRT sequences using plasmid pKD4 as a template [11]. All other linear dsDNA used for deletion of E. ictaluri genes eihA and dtrA were PCR amplified from a kanR cassette located within the genome of E. ictaluri Alg-08-183ompLC::kanR mutant generated in this study by recombineering. Likewise, the linear dsDNA substrate used for recombineering in A. hydrophila was generated by PCR amplification of the cat gene with its flanking FRT sequences integrated within the genome of A. hydrophila ML09-119 (see below). Recombineering primers contained 50–60 bp of homology to the targeted genes at their 5′ ends and 20–22 bp of homology to the cat cassette at their 3′ ends. Primers were modified with four consecutive 5′ phosphorothioates bonds when appropriate to reduce the chance of degradation by exonucleases during recombination. To introduce ∼250 and ∼500 bp homologous arms on either ends of the recombineering substrates for the determination of the effect of length homology in recombination frequency, primers were designed to anneal ∼250 and ∼500 bp upstream and downstream, respectively, of the cat gene of A. hydrophila ML09-119 waaL::cat mutant generated by recombineering in this study. PCR amplification of the respective antibiotic resistance gene cassette using these gene-targeted primers was performed using high fidelity Takara Ex Taq Polymerase (Clontech) and EconoTaq PLUS GREEN (Lucigen Corp.). At least 10 positive PCR amplicons of 50 μl volume were pooled together and purified by phenol–chloroform extraction followed by ethanol precipitation [47] or using the Wizard DNA Clean-Up System (Promega, Madison, WI). Purified PCR products were resuspended in nuclease-free water and used for transformation into electrocompetent E. ictaluri and A. hydrophila strains harboring recombinogenic plasmids pMJH46 and pMJH65, respectively.
2.6. Deletion of E. ictaluri and A. hydrophila genes by recombineering
Electrocompetent E. ictaluri and A. hydrophila harboring recombinogenic plasmids pMJH46 and pMJH65, respectively, were prepared as described follows. E. ictaluri strains were grown in TSB at 28 °C for overnight in the presence of chloramphenicol, whereas A. hydrophila was grown at 30 °C for overnight in TSB supplemented with tetracycline. Cultures were then diluted 1:70 in 40 ml of Super Optimal broth (SOB) medium supplemented with appropriate antibiotics and 10 mM L-arabinose, and grown with vigorous shaking until the OD600 reached to 0.45 or 0.6 for E. ictaluri and A. hydrophila, respectively. Cells were harvested by centrifugation at 5000 × g for 8 min at 4 °C, washed three times with ice-cold 10% glycerol and finally cells were concentrated 400-fold by resuspending with 100 μl of ice-cold GYT (10% glycerol, 0.125% yeast extract and 0.25% tryptone) medium or 10% glycerol. Freshly prepared electrocompetent cells were immediately used for electroporation. For deletion of targeted genes from E. ictaluri using recombineering, a dsDNA substrate of 10 μg were mixed with 50–55 μl of electrocompetent cells in a pre-chilled electroporation cuvette (0.1-cm gap), and pulsed at 1.8 kV with 25 μF and 200 W using an Eppendorf Electroporator 2510 (Hamburg, Germany). For A. hydrophila, the same electroporation procedures were followed with the exception that cells were pulsed at 1.2 kV. Immediately after electroporation, 950 μl of SOC supplemented with 10 mM l-arabinose was added and incubated at an appropriate temperature with vigorous shaking for at least 4 hrs for E. ictaluri and overnight for A. hydrophila. Cells were then spread onto 2 × YT agar plates supplemented with kanamycin and chloramphenicol for E. ictaluri and A. hydrophila, respectively, and incubated at an appropriate temperature to obtain mutants with the targeted deletions. Mutants grown on antibiotic selective plates were purified by streaking on TSA plates for isolated colonies. The correct deletions of the targeted genes were confirmed by PCR and/or sequencing as previously described [11]. To determine the effect of (1) phosphorothioate-modified primers, (2) the size of the gene-specific region of homology and (3) the concentration of the dsDNA substrates on recombination frequencies, each experiment was repeated independently at least three times.
2.7. Flp-mediated excision of antibiotic resistance gene cassettes to generate unmarked mutants
Before removal of the antibiotic resistance gene cassettes using Flp/FRT mediated recombination, recombinogenic plasmids were cured from the mutants of E. ictaluri and A. hydrophila. Plasmid pMJH46 was cured from E. ictaluri mutants by growing cells on TSB medium at 28 °C until the OD600 reached to 1.0 and then cells were subjected to heat induction at 43 °C for 1 h with shaking at 250 rpm. Heat-induced cultures were serially diluted in sterile water and spread for isolated colonies onto BHI Blood Agar plates that were then incubated at 28 °C for 36 h. To cure plasmid pMJH65 from A. hydrophila mutants, cultures were grown in TSB broth at 37 °C overnight and streaked onto TSA plates for isolated colonies. The loss of plasmid pMJH46 and pMJH65 from E. ictaluri and A. hydrophila mutants were confirmed by determining the lack of ability of individual mutant colonies to grow on TSA plates supplemented with chloramphenicol and tetracycline, respectively. Plasmid pCP20 that contains the Flp recombinase [7] required for FRT sequence-specific recombination was electroporated into E. ictaluri mutants according to the methods described above. E. ictaluri mutants harboring pCP20 were selected on 2 × YT agar plates supplemented with chloramphenicol. These E. ictaluri mutants were grown in TSB at 28 °C until OD600 of 1.0 and temperature was shifted by incubating at 37 °C for 1 h with shaking at 250 rpm to induce the removal of kanamycin resistance gene cassette by FLP recombinase. To obtain isolated colonies diluted cultures were plated onto BHI Blood Agar plates and incubated at 28 °C for up to 36 h. Flp recombinase plasmid pCMT-flp constructed in this study was conjugally transferred to A. hydrophila mutants as described above and induced for the removal of chloramphenicol resistance gene cassette by incubating at 37 °C. Induced cultures were streaked onto TSA plates and colonies grown on non-selective plates that subsequently failed to grow on antibiotic selective plates were tested by PCR and sequencing to confirm the Flp-mediated excision of antibiotic resistance gene cassette which was introduced by recombineering.
2.8. Cloning large genomic inserts without PCR amplification of the targeted genetic locus
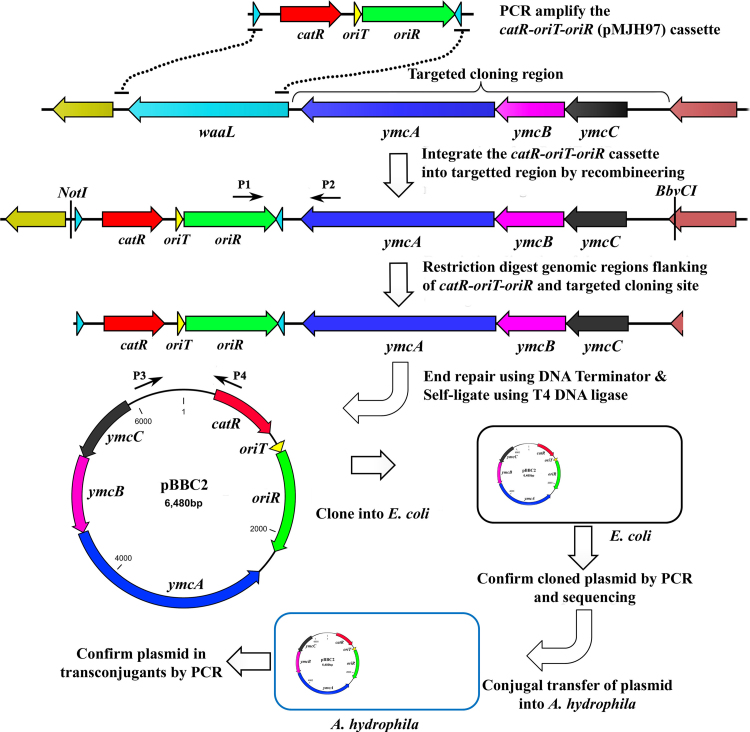
To construct a small, conjugally transferrable, and low copy-number plasmid backbone, the cat gene and p15A origin of replication (oriR) were PCR amplified using primers Li-CCatF and CCatR-oriT and CatFseq and Li-AAAAR, respectively, from the genome of A. hydrophila ML09-119hlyA::cat (generated in this study) and plasmid p1R17 (unpublished) with p15A of pACYC184 origin, respectively. The reverse primer CCatR-oriT used for amplification of the cat gene contains 87 bp of oriT sequence (Table 2) to facilitate the conjugal transfer of large insert clones to Gram-negative bacteria. The amplicons of cat-oriT cassette and p15A (oriR) were fused together to construct a 2097 bp plasmid backbone cat-oriT-oriR (pMJH97) using SOE PCR with outermost primers Li-CCatF and Li-AAAAR. To clone the ymcABC genetic cluster (unpublished data, manuscript in preparation) of A. hydrophila ML09-119, the pMJH97 plasmid backbone was PCR amplified using primers Li-CCatF and Li-AAAR that are homologous to the nucleotide regions 3,497,544-3497603 and 3,499,203-3499265, respectively, of the A. hydrophila ML09-119 genome [53]. These regions correspond to a specific region which is upstream of the ymcABC genetic cluster in the A. hydrophila ML09-119 genome. Purified PCR products were electroporated into A. hydrophila ML09-119 harboring plasmid pMJH65 for genomic integration into the targeted regions by recombineering. Colonies selected on 2 × YT plates containing chloramphenicol were subjected to PCR to confirm the correct integration of the pMJH97 backbone plasmid into the genome using primers p15AF and Li234R-HindII, and amplicons of the expected size were selected for sequencing. Once the correct integration of pMJH97 into the genome of A. hydrophila ML09-119 was confirmed by PCR and sequencing, genomic DNA was extracted from ML09-119::cat-oriT-oriR and restriction digested with BbvCI and NotI. Blunt-ended and purified genomic DNA fragments were self-ligated using T4 DNA ligase and electroporated into E. coli (E. cloni 10G, Lucigen Corp.) for cloning. Clones were selected on 2 × YT plates with chloramphenicol and the cloned plasmid pBBC2 was verified by PCR and sequencing using primers CCatR and ymcA-CM-1F for the presence of the complete ymcABC genetic cluster as an insert. Once the complete ymcABC cloning was confirmed, the pBBC2 was introduced into E. coli SM10λpir by electroporation. The plasmid was conjugally transferred into A. hydrophila ML09-119 as described above. Ten transconjugants which were grown on 2 × YT plates supplemented with chloramphenicol and colistin were double purified and subjected to PCR to confirm pBBC2 mobilization into A. hydrophila ML09-119 using primers CCatR and ymcA-CM-1F.
2.9. Sequencing of conjugally transferable recombinogenic and flp plasmids
The constructions of plasmids pMJH46 and pMJH65 were confirmed by PCR and sequencing using primers pMJH46SeqF, Mob-seqR, pMJH46SeqR and Cat-SeqF for plasmid pMJH46 and primers CatF and CatR-int for plasmid pMJH65 (Table 1). Plasmid pCMT-flp was sequenced using Illumina MiSeq according to methods described previously [43]. The gaps between the contigs obtained after assembling of Illumina MiSeq sequence reads of pCMT-flp were filled by PCR and sequencing using primers FlpFa, FlpR1, FlpF2 and FlpR2 (Table 2). The sequencing of the linear cassette pMJH97 (catR-oriT-oriR) integrated into the A. hydrophila genome was confirmed by PCR and sequencing using primers 97Seq1F, 97Seq2F, 97Seq3F, 97Seq4R, 97Seq8R, 97Seq7R, 97Seq5F, 97Seq6F, p15AF, CCatF, AAAAF and BBBBR (listed in Table 2).
2.10. Nucleotide sequence accession and Addgene deposition ID numbers
The sequences of plasmids pMJH46, pMJH65, pMJH97 and pCMT-flp were deposited to the NCBI GenBank sequence database under accession numbers JQ070344, KF195927, KT072897, and KT072898, respectively. Plasmids pMJH46, pMJH65 and pCMT-flp were deposited with Addgene (https://www.addgene.org/) with the plasmid numbers 67,272, 67,273 and 67,274, respectively.
3. Results
3.1. Construction of conjugally transferable recombinogenic plasmids
The expression of exo, bet and gam within bacterial cells substantially improves their recombination frequencies that can be exploited to modify bacterial genomes by recombineering [11]. Though published reports indicate that some E. ictaluri strains are capable of accepting foreign DNA of up to 45 kb by electroporation [23], our repeated attempts failed to introduce the recombinogenic plasmid pKD46 [11] into primary disease isolates of E. ictaluri or A. hydrophila. To introduce the recombinogenic λ-Red cassette into E. ictaluri, a mobilizable plasmid was constructed by introducing the ‘mob cassette’ (oriT region, traJ and traK) along with a chloramphenicol resistance (cat) gene into pKD46, resulting in plasmid pMJH46 (Fig. 1, accession no. JQ070344). The cat gene introduction broadens the applicability of this plasmid since some E. ictaluri strains are intrinsically resistant to ampicillin [58]; therefore, the original plasmid pKD46 expressing the bla gene is incompatible for these E. ictaluri isolates. In this study, we successfully transferred recombinogenic plasmid pMJH46 into different E. ictaluri strains by conjugation with E. coli SM10λpir. In subsequent studies, the pMJH46 plasmid was modified by replacing the cat gene with tetA to construct recombinogenic plasmid pMJH65 (Fig. 1, accession no. KF195927) which allows the use of the cat gene as a recombineering substrate. The plasmid pMJH65 was successfully introduced into highly virulent catfish isolate A. hydrophila ML09-119 [53] in order to generate genomic modifications through recombineering.
3.2. Deletion of E. ictaluri and A. hydrophila genes by recombineering
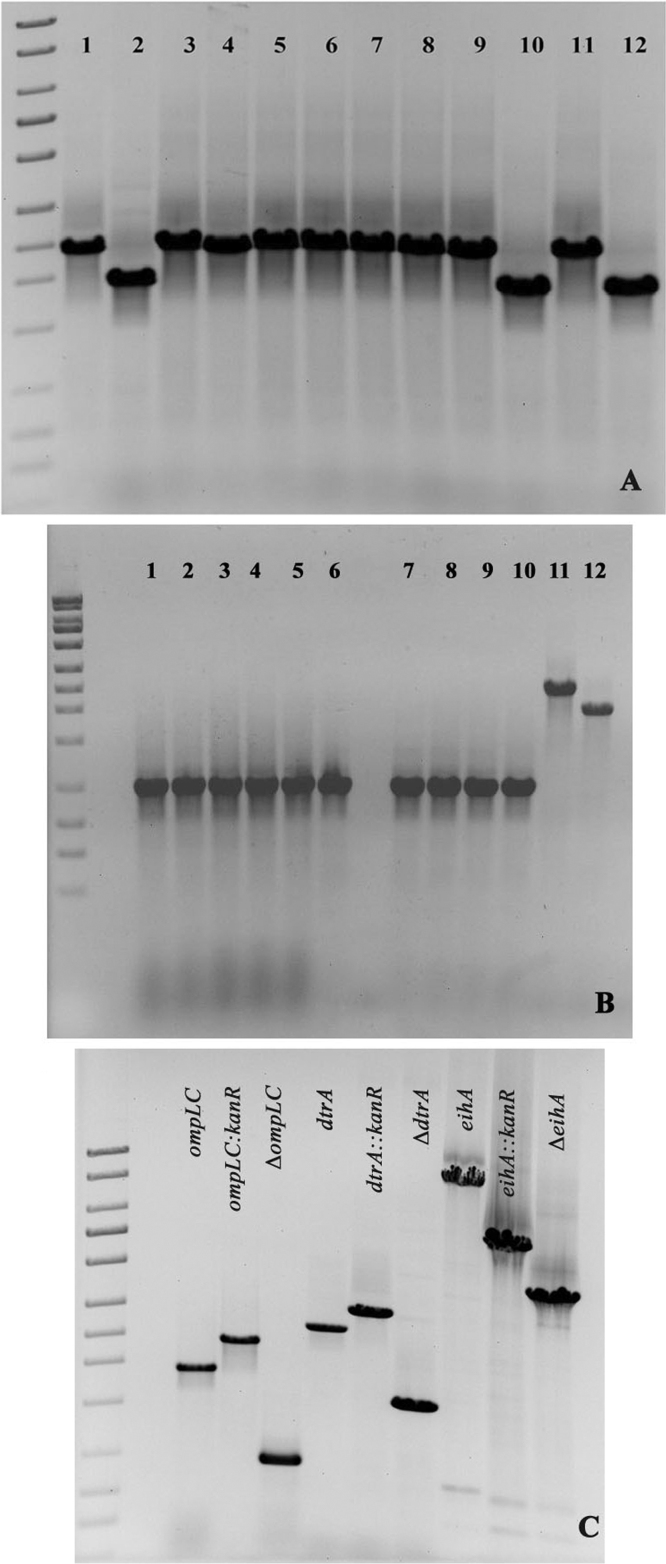
To determine the feasibility of using this recombineering system in E. ictaluri, we deleted the ompLC gene that is required for phage ΦeiAU-183 attachment to E. ictaluri strain Alg-08-183 [22]. The PCR screening of colonies grown on antibiotic selection plates showed that 1% colonies were true mutants (data not shown). Unfortunately, a large number of colonies grown on 2 × YT plates supplemented with kanamycin were determined to be false positive even though the suicide plasmid pKD4 [10] used as template was treated with DpnI before electroporation into E. ictaluri. To avoid the occurrence of background colonies, we subsequently used the genomic DNA of the E. ictaluri Alg-08-183 ompLC::kanR mutant as a PCR template for amplification of the kanamycin resistance gene cassette. Using this chromosomal template to prepare amplicons, we obtained at least ten colonies per experiment, of which ∼80% of them were true mutants (Fig. 2). In addition to ompLC of E. ictaluri Alg-08-183, we deleted two additional genes that included dtrA of E. ictaluri Alg-08-183, and eihA of E. ictaluri R4383 [59] (Fig. 2). In this study, using a recombineering approach, we also deleted seven different genes from the primary disease isolate A. hydrophila ML09-119 (Table 1). PCR and sequencing confirmed that all genes that were targeted for deletion from E. ictaluri and A. hydrophila strains were successfully deleted by recombineering. As a control experiment, A. hydrophila ML09-119 (pMJH65) and this same strain without the presence of the recombineering plasmid were both subjected to electroporation with equal amounts (900 ng) of a waaL::cat PCR construct (Table 1), and only in the presence of pMJH65 were any transformants obtained at a frequency of 0.45 ± 0.27 transformants per ng of amplicon DNA.
Fig. 2.
Targeted deletion of E. ictaluri genes ompLC, dtrA and eihA by recombineering. (Panel A) Colonies gown on 2 × YT plates supplemented with kanamycin were selected for PCR screening of ompLC gene deleted mutants. Lanes 1, 3–9 and 11 represent the PCR products of ompLC gene mutants disrupted with the kanR gene (ompLC::kanR) and lanes 2, 10 and 12 represents the PCR product of wild type ompLC gene of E. ictaluri strain Alg-08-183. (Panel B) Removal of the kanamycin resistance marker using the Flp recombinase of plasmid pCP20. PCR screening of E. ictaluri mutants plated after temperature induction showed that all tested mutants had lost the antibiotic resistance marker. (Panel C) PCR confirmation of deletion of the ompLC and drtA genes from E. ictaluri strain Alg-08-183 and eihA from E. ictaluri strain R4383.
3.3. Effects of primer modification, length of homology and dsDNA substrate concentration on recombination frequency
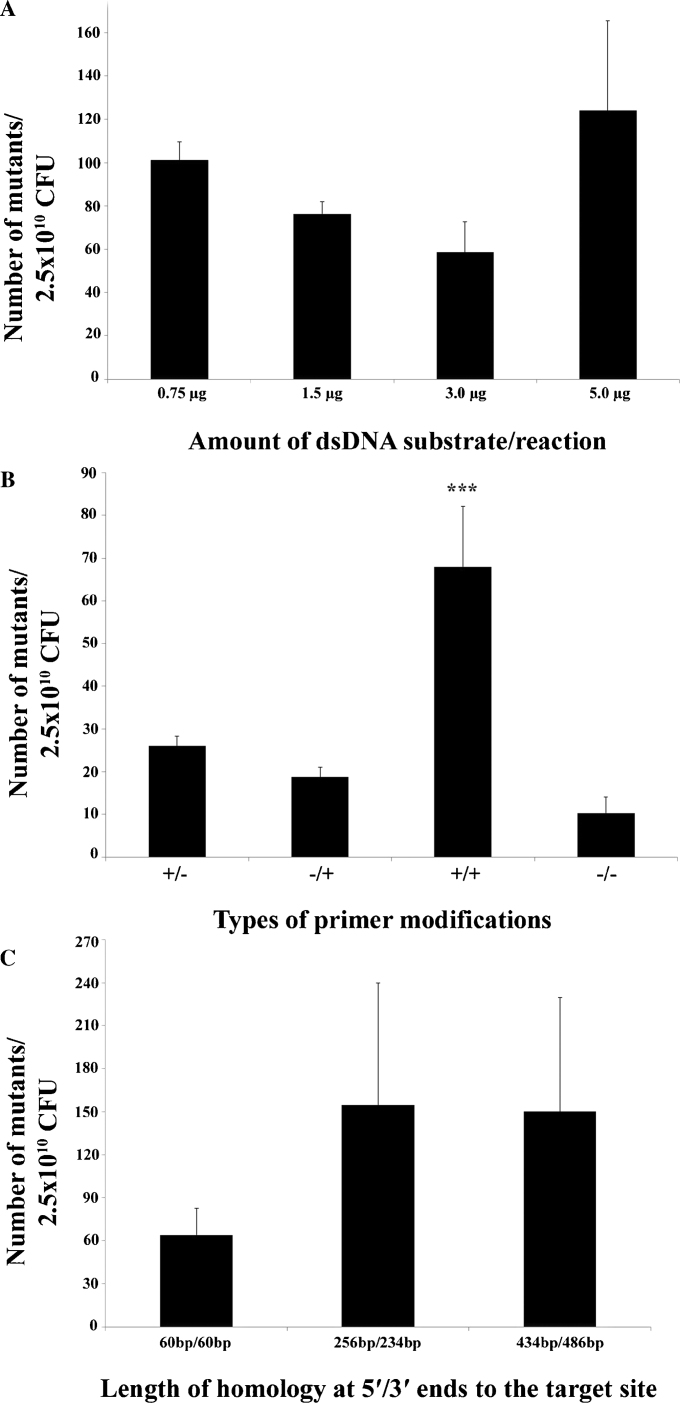
To determine the effect of strand protection through primer modifications on recombination frequencies in A. hydrophila ML09-119, four different primers combinations were used for the preparation of dsDNA substrates to delete the waaL gene of A. hydrophila ML09-119 [53]. In the type “+/+”primer combination, both the forward and reverse primers (Ligase-catF and Ligase-catR, in Table 2) were modified with four consecutive 5′-phosphorothioate bonds, whereas in the type “−/−” primer combination both the forward and reverse primers (Li-catF and Li-catR) were unmodified. In the type “+/−” primer combination, only the forward primer (Ligase-catF) was modified, whereas in the type “−/+” primer combination only the reverse primer (Ligase-catR) was modified with four consecutive 5′-phosphorothioate bonds. In the latter two cases, the alternative primers were unmodified. We found that dsDNA substrate prepared with both the leading and lagging strand-specific phosphorothioate modified primers (type “+/+”in Fig. 3B) provided significantly more mutants, whereas three other combinations did not affect recombination frequency (Fig. 3B). Once we determined that modified primers provided significantly more mutants, all of our subsequent recombineering experiments in A. hydrophila were carried out using both modified primers.
Fig. 3.
Determination of recombination frequency in A. hydrophila. (Panel A) The effect of dsDNA substrate concentration on recombination frequency in A. hydrophila was determined using four different dsDNA substrate concentration ranging from 0.75 μg to 5.0 μg per recombineering experiment. (Panel B) Four different primer combinations were generated using modified and unmodified primers. Modified primers included four consecutive phosphorothioate bonds at the 5′ end of the primers. Type “−/−” used unmodified primers as a negative control, type “+/−” included modification of the forward primer but not the reverse primers, type “−/+” included modification to the reverse but not forward primer, and type “+/+” included phosphorothioate bonds in both primers. The latter condition in which both primers were modified provided significantly more mutants than any other types of dsDNA substrates used for recombineering (***p-value = 0.0026). (Panel C) The effect of varying the length of the homologous regions of the dsDNA substrate to the targeted chromosomal site on the recombination frequency was determined using approximately 60 bp, 250 bp and 500 bp of homologous sequence at both the 5′ and 3′ ends. The average number of mutants obtained was derived from three independent recombineering experiments.
To determine the effect of the length of the gene-specific regions of homology of the dsDNA substrate on recombination efficiency, three different dsDNA substrates that included approximately 60 bp, 250 bp and 500 bp of homologous sequence at both the 5′ and 3′ ends were used for targeted deletion of the waaL gene of A. hydrophila ML09-119 [53]. The number of mutants obtained from this experiment demonstrated that the recombination frequencies were not significantly different in A. hydrophila ML09-119 due to the varying length of homologous arms flanking to the targeted gene (Fig. 3C).
To determine the effect of dsDNA substrate concentration on recombination frequencies in A. hydrophila, we used four different concentrations of dsDNA substrate that included 0.75, 1.5, 3.0 and 5.0 μg of dsDNA substrate for each recombineering experiment. Our findings demonstrated that increasing the dsDNA substrate concentrations did not change the recombination frequency significantly in A. hydrophila ML09-119 (Fig. 3A). The number of mutants we routinely obtained in this experiment was within the range of approximately 30–200 per recombineering reaction.
3.4. Removal of antibiotic resistance cassette by Flp recombinase
The temperature induction of E. ictaluri Alg-08-183ompLC::kanR, dtrA::kanR and E. ictaluri R4383 eihA::kanR mutant at 43 °C for 1 h followed by plating on BHI blood agar plates resulted in the curing of the recombinogenic plasmid pMJH46 (data not shown). We found that only highly rich medium such as BHI supplemented with 5% Sheep Blood, unlike TSA, supported the growth of the high temperature-induced E. ictaluri strains. The introduction of plasmid pCP20 containing the Flp recombinase by electroporation [7] followed by their growth at 37 °C resulted in removal of the antibiotic marker from the E. ictaluri ompLC mutant (Fig. 2B). PCR amplification of the targeted genes with their flanking primers indicated a 100% frequency for removal of the antibiotic selection marker. The antibiotic resistance markers from the E. ictaluri dtrA and eihA mutants were also removed using the Flp recombinase (Fig. 2C). We found that, in addition to the removal of the antibiotic resistance marker, heat induction efficiently cured the plasmid pCP20 from all mutant colonies tested. Cured mutants lacking the antibiotic resistance cassette could be subsequently targeted for deletion of additional genes. Since genes from A. hydrophila were replaced using the cat gene cassette, plasmid pCP20 containing the cat gene was not compatible for conducting Flp/FRT mediated recombination in A. hydrophila mutants. Therefore, we constructed a new flp recombinase plasmid pCMT-flp (Fig. 1D) with a tetracycline selectable marker. This plasmid was conjugally transferred into A. hydrophila mutants for markerless mutant construction. The screening of A. hydrophila mutants harboring the pCMT-flp plasmids for lack of growth in the presence of chloramphenicol resulted in more than 10% of the mutants with documented loss of the antibiotic resistance cassette (data not shown).
3.5. Cloning without PCR amplification of large inserts
Since the cloning of large inserts using traditional cloning techniques is challenging and PCR amplification of the targeted inserts can introduce unwanted mutations, we developed a novel technique to clone large genomic inserts of A. hydrophila that does not require PCR amplification of the targeted insert (Fig. 4). As a proof of concept of this technique, we targeted for cloning the 3.6 kb ymcABC operon of A. hydrophila strain ML09-119 for cloning. For this purpose, we constructed a small conjugally transferrable low copy-number plasmid backbone (pMJH97) which was integrated contiguous to the ymcABC operon of A. hydrophila ML09-119 by recombineering (data not shown). We confirmed the correct integration of the plasmid backbone (pMJH97) upstream of the ymcABC operon by PCR and sequencing. Restriction digestion of the genomic DNA isolated from the integrant and self-ligation followed by electroporation resulted in hundreds of chloramphenicol-resistant E. coli clones on selective plates. Two of the clones selected for PCR and sequencing confirmation demonstrated that the intact ymcABC operon was cloned into the plasmid pMJH97 (data not shown). This plasmid was conjugally transferred into A. hydrophila ML09-119 to determine its conjugal transferability; screening of ten transconjugants using PCR demonstrated that all of the transconjugants harbored plasmid with an intact ymcABC operon insert (data not shown).
Fig. 4.
Strategy for PCR-free cloning of large bacterial genetic regions. The major steps of cloning large genetic inserts are indicated. The catR-oriT-oriR (pMJH97) cassette was PCR amplified using primer pairs with 50–60 bp homologous sequence at their 5′-ends specific to the targetred site. Depending on the choice of restriction enzymes, the resulting dsDNA substrate can be integrated upstream or downstream of the targeted site of the genome using the recombineering system. Once the catR-oriT-oriR (pMJH97) cassette integration into the genome was confirmed by PCR and sequencing using primers P1 and P2, the genomic DNA of integrants was restriction digested with an appropriate restriction enzyme to clone into E. coli after self-ligation using T4 DNA ligase. The cloning of the correct insert into the plasmid pMJH97 was verified by PCR and sequencing using vector and insert specific primers P3 and P4, respectively. The plasmids with cloned inserts were then readily transfered to other Gram-negative bacterial strain by oriT sequence-mediated conjugal transfer using an appropriate donor strain.
4. Discussion
The genetic manipulation of primary pathogenic isolates, compare to domesticated laboratory isolates, can be challenging due to many factors including antibiotic resistance [16], [30], poor recombination efficiency and wide-spread occurrence of restriction-modification systems [37], [54]. Our attempts to genetically modify the fish pathogens E. ictaluri and A. hydrophila were inhibited due to our inability to introduce the λ Red recombineering system into these bacterial isolates. Similar difficulties were observed by several other researchers who reported reduced transformation efficiency of pKD46 in E. coli by electroporation [48], demonstrating the need for an alternative route to introduce the recombineering system, i.e., via conjugation. In this study we describe the development of a fast, efficient, and reliable technique for genetic modification of E. ictaluri and A. hydrophila (and presumably other Gram-negative bacteria) using a recombineering system that is readily transferrable by conjugation. The introduction of a mob cassette to pKD46 [11] permitted the resulting plasmid pMJH46 to transfer into different E. ictaluri strains by conjugation. Additional modified recombinogenic plasmids were constructed to make it compatible for gene deletion in a highly virulent strain of A. hydrophila. Furthermore, we demonstrated the applicability of this method by creating multiple mutants in E. ictaluri and A. hydrophila.
Our first experiments using recombineering in E. ictaluri unfortunately were plagued by a large number of background colonies on the antibiotic selection plates that were not successful recombinants. These results were obtained even though we used suicide plasmid pKD4 as a template for PCR amplification of the antibiotic cassette and treated the DNA with DpnI treatment, as had been shown to reduce the number of background colonies [49]. An alternative solution to reducing the high background of antibiotic resistant colonies was to use genomic DNA isolated from a successful genomic integrant (E. ictaluri Alg-08-183ompLC::kanR) constructed in this study as a template for PCR of the recombineering construct. Therefore, all of our subsequent recombineering experiments for gene deletion in E. ictaluri and A. hydrophila used genomic DNA as template for PCR amplification of the respective antibiotic resistance gene cassettes.
We were able to use the Flp recombinase encoded on the temperature-sensitive plasmid pCP20 [7] to successfully remove a FRT-flanked antibiotic resistance cassette used for genome modification in E. ictaluri. Before introducing pCP20 into E. ictaluri mutants, pMJH46 was cured by heat induction since both plasmids contain the cat gene. Unlike E. coli [11], E. ictaluri mutants required a highly rich medium (BHI supplemented with 5% sheep blood) to recover after heat-induction at 43 °C, which may be due to the mesophilic growth temperature (28 °C) of E. ictaluri. Because of antibiotic resistance marker incompatibility, a new conjugally transferable flp recombinase plasmid, pCMT-flp, was constructed that can efficiently remove FRT-flanked antibiotic resistance gene cassettes from mutants of A. hydrophila.
In addition to developing techniques for genetic modification in E. ictaluri and A. hydrophila, we devised a technique for cloning large fragments of bacterial genomes without PCR amplification of the targeted region. Similar in concept to the VEX-capture system that uses a lox/Cre site-specific recombination system [60], or the use of an in vivo recombineering method [55], these cloning systems are advantageous in allowing the cloning of larger fragments of genomic DNA without the need for PCR amplification, given the difficulties in producing larger amplicons and the potential for incorporating PCR-mediated errors. This method was validated by the cloning of the A. hydrophila genetic operon ymcABC, as an example of this method that can overcome the shortcomings of PCR-based methods for the cloning and conjugal transfer of genetic elements. The maximum possible size of the cloned region will depend on multiple factors, such as the presence of suitable restriction sites and the efficiency of conjugal transfer, but would be expected to be theoretically suitable for genomic regions such as genomic islands, prophages, and other genetic clusters.
We have described a highly efficient and rapid procedure for the generation of markerless mutants in E. ictaluri and A. hydrophila by recombineering. The newly constructed conjugally transferable recombinogenic plasmids pMJH46 and pMJH65 and recombinase plasmid pCMT-flp can presumably be used for other Gram-negative bacteria for generating markerless mutants, especially for bacterial isolates that are recalcitrant to electroporation. Finally, the development of a PCR-free system for cloning and transfer will facilitate cloning and complementation of much larger genetic elements.
Acknowledgements
This work was supported by a grant from the United States Department of Agriculture’s Agriculture and Food Research Initiative (#2013-67015-21313). We thank the members of the Liles laboratory, in particular Ms. Nancy Capps and Mr. Cody Rasmussen-Ivey, for their support of this research.
References
- 1.Ando T., Xu Q., Torres M., Kusugami K., Israel D.A., Blaser M.J. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 2000;37:1052–1065. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 2.Arber W., Dussoix D. Host specificity of DNA produced by E. coli I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- 3.Arber W., Linn S. DNA modification and restriction. Annu. Rev. Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 4.Aubert D.F., Hamad M.A., Valvano M.A. A markerless deletion method for genetic manipulation of Burkholderia cenocepacia and other multidrug-resistant gram-negative bacteria. Methods Mol. Biol. 2014;1197:311–327. doi: 10.1007/978-1-4939-1261-2_18. [DOI] [PubMed] [Google Scholar]
- 5.Cartman S.T., Minton N.P. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 2010;76:1103–1109. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassuto E., Lash T., Sriprakash K.S., Radding C.M. Role of exonuclease and β protein of phage λ in Genetic Recombination, V. recombination of λ DNA in vitro. Proc. Natl. Acad. Sci. 1971;68:1639–1643. doi: 10.1073/pnas.68.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov P.P., Wackernagel W. Gene disruption in E. coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 8.Copeland N.G., Jenkins N.A., Court D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 9.Court D.L., Sawitzke J.A., Thomason L.C. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 10.Czarniak F., Hensel M. Red-mediated recombineering of Salmonella enterica genomes. Methods Mol. Biol. 2015;1225:63–79. doi: 10.1007/978-1-4939-1625-2_4. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in E. coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawoud T.M., Jiang T., Mandal R.K., Ricke S.C., Kwon Y.M. Improving the efficiency of transposon mutagenesis in Salmonella enteritidis by overcoming host-restriction barriers. Mol. Biotechnol. 2014;56:1004–1010. doi: 10.1007/s12033-014-9779-4. [DOI] [PubMed] [Google Scholar]
- 13.Donahue J.P., Israel D.A., Peek R.M., Blaser M.J., Miller G.G. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 2000;37:1066–1074. doi: 10.1046/j.1365-2958.2000.02036.x. [DOI] [PubMed] [Google Scholar]
- 14.Eden P.A., Blakemore R.P. Electroporation and conjugal plasmid transfer to members of the genus Aquaspirillum. Arch. Microbiol. 1991;155:449–452. doi: 10.1007/BF00244960. [DOI] [PubMed] [Google Scholar]
- 15.Elhai J., Wolk C.P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 16.Esteve C., Alcaide E., Blasco M.D. Aeromonas hydrophila subsp. dhakensis Isolated from Feces, Water and Fish in Mediterranean Spain. Microbes Environ. 2012;27:367–373. doi: 10.1264/jsme2.ME12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flett F., Mersinias V., Smith C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from E. coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 18.Hadjifrangiskou M., Gu A.P., Pinkner J.S., Kostakioti M., Zhang E.W., Greene S.E., Hultgren S.J. Transposon mutagenesis identifies uropathogenic E. coli biofilm factors. J. Bacteriol. 2012;194:6195–6205. doi: 10.1128/JB.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He P., Hao K., Blom J., Ruckert C., Vater J., Mao Z., Wu Y., Hou M., He P., He Y., Borriss R. Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601-Y2 and expression of mersacidin and other secondary metabolites. J. Biotechnol. 2012;164:281–291. doi: 10.1016/j.jbiotec.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Hemstreet B. An update on Aeromonas hydrophila from a fish health specialist for summer 2010. Catfish J. 2010;24 [Google Scholar]
- 21.Hirayama Y., Sakanaka M., Fukuma H., Murayama H., Kano Y., Fukiya S., Yokota A. Development of a double-crossover markerless gene deletion system in bifidobacterium longum: functional analysis of the α-Galactosidase gene for raffinose assimilation. Appl. Environ. Microbiol. 2012;78:4984–4994. doi: 10.1128/AEM.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hossain M.J., Rahman Kh S., Terhune J.S., Liles M.R. An outer membrane porin protein modulates phage susceptibility in Edwardsiella ictaluri. Microbiology. 2012;158:474–487. doi: 10.1099/mic.0.054866-0. [DOI] [PubMed] [Google Scholar]
- 23.Hossain M.J., Sun D., McGarey D.J., Wrenn S., Alexander L.M., Martino M.E., Xing Y., Terhune J.S., Liles M.R. An asian origin of virulent aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. mBio. 2014;5 doi: 10.1128/mBio.00848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hossain M.J., Waldbieser G.C., Sun D., Capps N.K., Hemstreet W.B., Carlisle K., Griffin M.J., Khoo L., Goodwin A.E., Sonstegard T.S., Schroeder S., Hayden K., Newton J.C., Terhune J.S., Liles M.R. Implication of lateral genetic transfer in the emergence of aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS One. 2013;8:e80943. doi: 10.1371/journal.pone.0080943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasin M., Schimmel P. Deletion of an essential gene in E. coli by site-specific recombination with linear DNA fragments. J. Bacteriol. 1984;159:783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jost B.H., Homchampa P., Strugnell R.A., Adler B. The sacB gene cannot be used as a counter-selectable marker in Pasteurella multocida. Mol. Biotechnol. 1997;8:189–191. doi: 10.1007/BF02752263. [DOI] [PubMed] [Google Scholar]
- 27.Kakirde K.S., Wild J., Godiska R., Mead D.A., Wiggins A.G., Goodman R.M., Szybalski W., Liles M.R. Gram negative shuttle BAC vector for heterologous expression of metagenomic libraries. Gene. 2011;475:57–62. doi: 10.1016/j.gene.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karakousis G., Ye N., Li Z., Chiu S.K., Reddy G., Radding C.M. The beta protein of phage binds preferentially to an intermediate in DNA renaturation. J. Mol. Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 29.Kurosawa N., Grogan D.W. Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol. Lett. 2005;253:141–149. doi: 10.1016/j.femsle.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 30.Lee D.J., Bingle L.E., Heurlier K., Pallen M.J., Penn C.W., Busby S.J., Hobman J.L. Gene doctoring: a method for recombineering in laboratory and pathogenic E. coli strains. BMC Microbiol. 2009;9:252. doi: 10.1186/1471-2180-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X.-t., Thomason L.C., Sawitzke J.A., Costantino N., Court D.L. Positive and negative selection using the tetA-sacB cassette: recombineering and P1 transduction in E. coli. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little J.W. An Exonuclease Induced by Bacteriophage. J. Biol. Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 33.Matsuda T., Freeman T.A., Hilbert D.W., Duff M., Fuortes M., Stapleton P.P., Daly J.M. Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery. 2005;137:639–646. doi: 10.1016/j.surg.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Matsuura S.-i, Komatsu J., Hirano K., Yasuda H., Takashima K., Katsura S., Mizuno A. Real-time observation of a single DNA digestion by λ exonuclease under a fluorescence microscope field. Nucleic Acids Res. 2001;29:e79. doi: 10.1093/nar/29.16.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurer K.J., Lawrence M.L., Fernandez D.H., Thune R.L. Evaluation and Optimization of a DNA Transfer System for Edwardsiella ictaluri. J. Aquat. Anim. Health. 2001;13:163–167. [Google Scholar]
- 36.Merabishvili M., Pirnay J.-P., Verbeken G., Chanishvili N., Tediashvili M., Lashkhi N., Glonti T., Krylov V., Mast J., Van Parys L., Lavigne R., Volckaert G., Mattheus W., Verween G., De Corte P., Rose T., Jennes S., Zizi M., De Vos D., Vaneechoutte M. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monk I.R., Shah I.M., Xu M., Tan M.W., Foster T.J. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012;3 doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muniyappa K., Radding C.M. The homologous recombination system of phage lambda. Pairing activities of beta protein. J. Biol. Chem . 1986;261:7472–7478. [PubMed] [Google Scholar]
- 39.Murphy K.C. Use of bacteriophage lambda recombination functions to promote gene replacement in E. coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy K.C. Use of bacteriophage λ recombination functions to promote gene replacement in E. coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy K.C., Campellone K.G., Poteete A.R. PCR-mediated gene replacement in E. coli. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 42.Muyrers J.P., Zhang Y., Benes V., Testa G., Ansorge W., Stewart A.F. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 2000;1:239–243. doi: 10.1093/embo-reports/kvd049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasrin S., Hossain M.J., Liles M.R. Draft Genome Sequence of Bacillus amyloliquefaciens AP18 with Antibacterial Activity against Methicillin-Resistant Staphylococcus aureus. Genome Announc. 2015;3 doi: 10.1128/genomeA.00162-15. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poteete A.R., Fenton A.C., Murphy K.C. Modulation of E. coli RecBCD activity by the bacteriophage lambda Gam and P22 Abc functions. J. Bacteriol. 1988;170:2012–2021. doi: 10.1128/jb.170.5.2012-2021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pridgeon J.W., Klesius P.H. Molecular identification and virulence of three Aeromonas hydrophila isolates cultured from infected channel catfish during a disease outbreak in west Alabama (USA) in 2009. Dis. Aquat. Organ. 2011;94:249–253. doi: 10.3354/dao02332. [DOI] [PubMed] [Google Scholar]
- 46.Rogge M.L., Dubytska L., Jung T.S., Wiles J., Elkamel A.A., Rennhoff A., Oanh D.T., Thune R.L. Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis. Aquat. Organ. 2013;106:17–29. doi: 10.3354/dao02620. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1998. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 48.Serra-Moreno R., Acosta S., Hernalsteens J., Jofre J., Muniesa M. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 2006;7:31. doi: 10.1186/1471-2199-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon R., Priefer U., Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1983;1:784–791. [Google Scholar]
- 51.Sitaraman R., Leppla S.H. Methylation-dependent DNA restriction in Bacillus anthracis. Gene. 2012;494:44–50. doi: 10.1016/j.gene.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szewczyk E., Nayak T., Oakley C.E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S.A., Oakley B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2007;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 53.Tekedar H.C., Waldbieser G.C., Karsi A., Liles M.R., Griffin M.J., Vamenta S., Sonstegard T., Hossain M., Schroeder S.G., Khoo L., Lawrence M.L. Complete genome sequence of a channel catfish epidemic isolate, aeromonas hydrophila strain ML09-119. Genome Announc. 2013;1 doi: 10.1128/genomeA.00755-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 55.Thomason L., Court D.L., Bubunenko M., Costantino N., Wilson H., Datta S., Oppenheim A. John Wiley & Sons, Inc; 2001. Recombineering: Genetic Engineering in Bacteria Using Homologous Recombination. Current Protocols in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 56.USDA, (2010) Catfish 2010 part I: reference of catfish health and production practices in the United States, 2009. USDA-APHIS-VS, CEAH, Ft. Collins, CO.
- 57.Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welch T.J., Evenhuis J., White D.G., McDermott P.F., Harbottle H., Miller R.A., Griffin M., Wise D. IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen edwardsiella ictaluri. Antimicrob. Agents Chemother. 2009;53:845–846. doi: 10.1128/AAC.01312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams M.L., Lawrence M.L. Identification and characterization of a two-component hemolysin from Edwardsiella ictaluri. Vet. Microbiol. 2005;108:281–289. doi: 10.1016/j.vetmic.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 60.Wilson J.W., Figurski D.H., Nickerson C.A. VEX-capture: a new technique that allows in vivo excision, cloning, and broad-host-range transfer of large bacterial genomic DNA segments. J. Microbiol. Methods. 2004;57:297–308. doi: 10.1016/j.mimet.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Yu D., Ellis H.M., Lee E.-C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in E. coli. Proc. Natl. Acad. Sci. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X.-J., Yang W.-M., Li T.-T., Li A.-H. The genetic diversity and virulence characteristics of Aeromonas hydrophila isolated from fishponds with disease outbreaks in Hubei province. Acta Hydrobiol. Sinica. 2013;37:458–466. [Google Scholar]
- 63.Rose R.E. The nucleotide sequence of pACYC184. Nucl. Acids Res. 1988;16(1):355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]