Next-generation sequencing (NGS) has made it possible to identify about 20,000 variants in the protein-coding exome of each individual, of which only a few are likely to underlie a genetic disease. Variant-level methods such as PolyPhen-2, SIFT and CADD are useful for obtaining a prediction as to whether a given variant is benign/damaging1–3 or tolerant/intolerant1–3 (we hereafter use the terms benign/deleterious). These methods are commonly interpreted in a binary manner for filtering out benign variants from NGS data, with a single significance cutoff value across all protein-coding genes. PolyPhen-2 and SIFT integrate the fixed cutoff in the software. CADD proposed (but did not recommend for categorical usage) the fixed value of 15 (or another value between 10 and 20). Gene-level methods, such as RVIS, de novo excess and GDI are also useful4–6. Combining fixed gene-level and variant-level cutoffs is also applied in the RVIS hot zone approach4. However, owing to the diversity of medical and population genetic features between human genes and across populations, a uniform cutoff is unlikely to be accurate genome-wide.
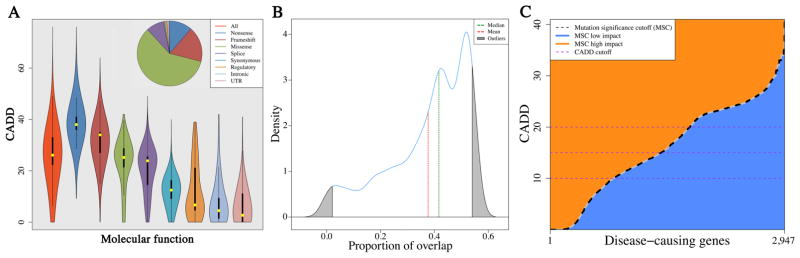
We found that CADD with fixed cutoffs outperformed PolyPhen-2 and SIFT (Fig. S1A). 40.84% of HGMD7 curated disease-associated mutations are not missense (Fig. 1A), contributing to low TP prediction with PolyPhen-2 and SIFT. We demonstrated that the 95% confidence interval (CI) of CADD scores for the disease-associated mutations of a given HGMD gene overlapped on average with only 37.63% (41.89% median) of the 95% CIs for CADD scores for the disease-associated mutations of all other HGMD genes (Fig. 1B). We then showed significantly higher CADD scores of private as compared with non-private disease-associated mutations (P<10−300, Fig. S1B), resulting in lower overall impact prediction scores when the allele frequency of a mutation was considered (Fig. S2).
Figure 1. Disease-associated mutation features.
(A) Violin plots of the CADD scores across the various HGMD molecular categories of 174,183 disease-associated deleterious mutations. (B) A density plot of the proportion of 95% CIs for HGMD disease-associated deleterious mutations overlap between a given disease-causing human gene and all other 2,946 disease-causing human genes. (C) The 95% CIs MSC scores of HGMD 2,947 disease-associated genes. Blue color (CADD score below the MSC) suggests a low impact for the gene, orange color (CADD score above or equal to the MSC) suggests high impact, and the horizontal purple dashed lines indicate current predictions based on fixed CADD scores.
We developed the mutation significance cutoff (MSC), a quantitative approach and server (http://lab.rockefeller.edu/casanova/MSC), providing gene-level and gene-specific low/high phenotypic impact cutoff values to improve the use of existing variant-level methods. We defined the MSC of a gene as the lower limit of the CI (90%, 95% or 99%) for the CADD, PolyPhen-2, or SIFT score of all its high quality mutations described as pathogenic in HGMD or ClinVar8. Remarkably, the 95% CI MSC values varied considerably, between 0.001 and 41 (Fig. 1C), with similar patterns observed for 90% and 99% CIs (Tables S1–S3). We estimated the MSC values of the remaining protein-coding genes by an extrapolation from their rare non-synonymous 1,000 Genomes Project9 alleles and validated by bootstrapping simulations (see Fig. S3, Tables S1–S9 for MSC based on CADD, PolyPhen-2 and SIFT with 90%, 95% and 99% CIs, respectively, and Fig. S4A for 95% CI MSC scores).
We found significant correlations between MSC, gene damage index (GDI, P<1.0×10−5, Fig. S4B)6 and purifying selection pressure (P<1.0×10−5, Fig. S4C). Low MSC genes were associated with immune system pathways, whereas genes with high MSC values were enriched in ribosome biology genes (Figs. S4D, S4E and Table S10). We showed by ROC curves significant improvement in distinguishing benign from deleterious alleles by CADD scores using CADD-based 99% and 95% CI HGMD-based MSCs, compared with CADD scores using fixed cutoffs of 10, 15 and 20, PolyPhen-2, SIFT, and RVIS hot zone predictions (Fig. S5, Table S11, Fig. S6). Most results obtained with MSCs generated from HGMD outperformed those with ClinVar-MSCs (Table 12). CADD-based MSC using HGMD generated with a 99% CIs achieved a 98% true positive detection rate, making MSC the first approach that enables filtering out benign variants from NGS data with little risk. See Supplementary Information for in-depth Abstract, Methods, Results and Discussion.
Supplementary Material
References
- 1.Adzhubei IA, et al. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P, et al. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 3.Kircher M, et al. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovski S, et al. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samocha KE, et al. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itan Y, et al. Proc Natl Acad Sci U S A. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenson PD, et al. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landrum MJ, et al. Nucleic Acids Res. 2014;42:D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auton A, et al. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.