Abstract
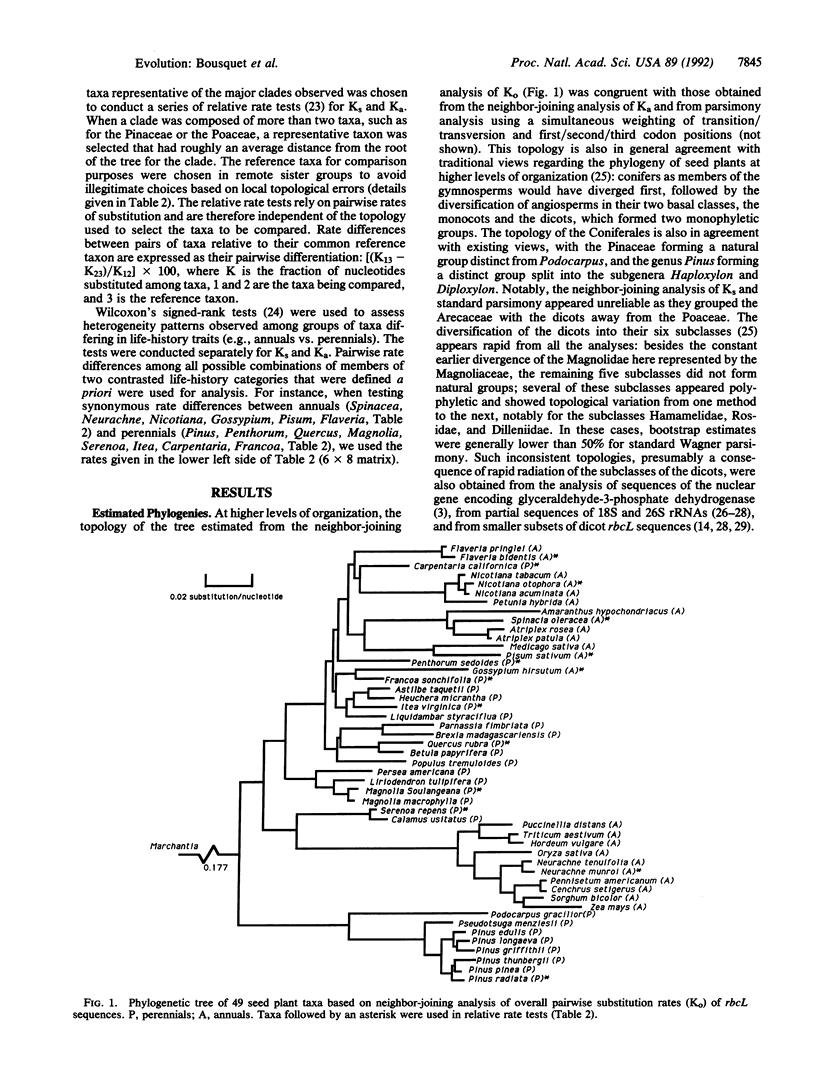
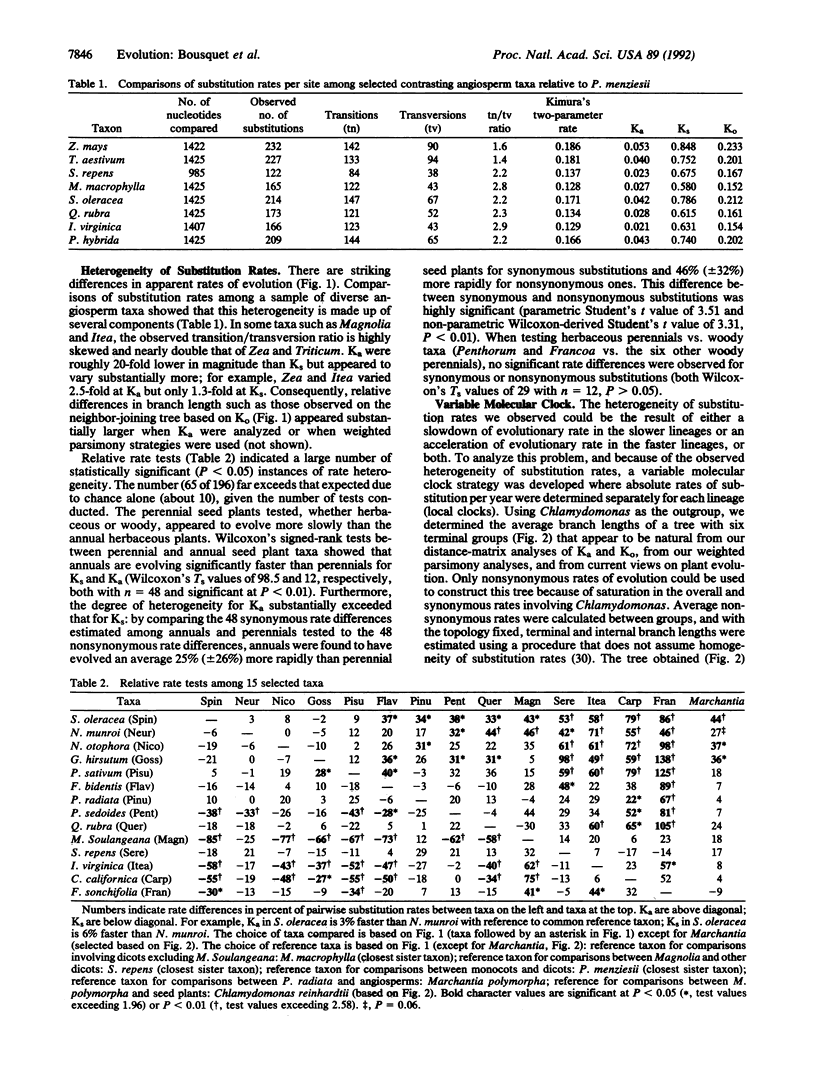
Extensive variation in synonymous and nonsynonymous rates of substitution was observed among 50 sequences of the gene coding for the large subunit of ribulose-1,5-bisphosphate carboxylase (rbcL) representing bryophyte, conifer, dicot, and monocot taxa. Relative rate tests revealed rate differences of up to 138% for nonsynonymous substitutions and up to 85% for synonymous ones. Within angiosperms, the annual forms evolved more rapidly, on average, than perennial forms. This rate heterogeneity was more extensive at nonsynonymous sites than synonymous ones, and it resulted primarily from a recent acceleration of substitution rate in many groups of angiosperms.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Higuchi R. G., Ochman H. Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res. 1989 Jul 25;17(14):5865–5865. doi: 10.1093/nar/17.14.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P., Paige K. N., Whitham T. G., Lark K. G. Genetic analysis of an interspecific hybrid swarm of Populus: occurrence of unidirectional introgression. Genetics. 1989 Nov;123(3):557–565. doi: 10.1093/genetics/123.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kohne D. E. Evolution of higher-organism DNA. Q Rev Biophys. 1970 Aug;3(3):327–375. doi: 10.1017/s0033583500004765. [DOI] [PubMed] [Google Scholar]
- Les D. H., Garvin D. K., Wimpee C. F. Molecular evolutionary history of ancient aquatic angiosperms. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10119–10123. doi: 10.1073/pnas.88.22.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. A., Wilson A. C. Rates of evolution in seed plants: Net increase in diversity of chromosome numbers and species numbers through time. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2086–2090. doi: 10.1073/pnas.73.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H. Simple method for constructing phylogenetic trees from distance matrices. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1085–1089. doi: 10.1073/pnas.78.2.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987 Mar 5;326(6108):93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wu C. I., Luo C. C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985 Mar;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Martin P. G., Dowd J. M. A comparison of 18s ribosomal RNA and rubisco large subunit sequences for studying angiosperm phylogeny. J Mol Evol. 1991 Sep;33(3):274–282. doi: 10.1007/BF02100679. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Birky C. W., Jr Effects of periodic selection on gene diversity in organelle genomes and other systems without recombination. Genetics. 1991 Feb;127(2):449–451. doi: 10.1093/genetics/127.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S. R., Bogorad L. Molecular evolution and nucleotide sequences of the maize plastid genes for the alpha subunit of CF1 (atpA) and the proteolipid subunit of CF0 (atpH). Genetics. 1987 May;116(1):127–139. doi: 10.1093/genetics/116.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runnegar B. Nucleic acid and protein clocks. Philos Trans R Soc Lond B Biol Sci. 1991 Sep 30;333(1268):391–397. doi: 10.1098/rstb.1991.0089. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Clegg M. T., Durbin M. rbcL sequence divergence and phylogenetic relationships in Saxifragaceae sensu lato. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4640–4644. doi: 10.1073/pnas.87.12.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdis J., Nei M. Relative efficiencies of the maximum parsimony and distance-matrix methods in obtaining the correct phylogenetic tree. Mol Biol Evol. 1988 May;5(3):298–311. doi: 10.1093/oxfordjournals.molbev.a040497. [DOI] [PubMed] [Google Scholar]
- Wagner D. B., Furnier G. R., Saghai-Maroof M. A., Williams S. M., Dancik B. P., Allard R. W. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc Natl Acad Sci U S A. 1987 Apr;84(7):2097–2100. doi: 10.1073/pnas.84.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore A. T., Schaal B. A. Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Gaut B., Clegg M. T. Chloroplast DNA evolves slowly in the palm family (Arecaceae). Mol Biol Evol. 1990 Jul;7(4):303–314. doi: 10.1093/oxfordjournals.molbev.a040605. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Gouy M., Yang Y. W., Sharp P. M., Li W. H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Li W. H., Sharp P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]


