Abstract
Two series with the general formula of 4,6-diaryl-2-oxo-1,2 dihydropyridine-3-carbonitriles and their isosteric 4,6-diaryl-2-imino-1,2-dihydropyridine-3-carbonitrile were synthesized through one pot reaction of the appropriate acetophenone, aldehyde, ammonium acetate with ethyl cyanoacetate or malononitrile, respectively. The synthesized compounds were evaluated for their tumor cell growth inhibitory activity against the human HT-29 colon tumor cell line, as well as their PDE3 inhibitory activity. Compound 4-(2-Ethoxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3 carbonitrile (21) showed tumor cell growth inhibitory activity with an IC50 value of 1.25 μM. Meanwhile, 4-(4-Ethoxyphenyl)-2-imino-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (26) showed inhibitory effect upon PDE3 using cAMP or cGMP as substrate. No correlation exists between PDE3 inhibition and the tumor cell growth inhibitory activity. Docking compound 21 to other possible molecular targets showed the potential to bind PIM1 Kinase.
Keywords: Multi-component reaction, Growth inhibition, Phosphodiesterase inhibition
1. Introduction
3-Cyano-2-Pyridones and 3-cyano-2-iminopyridines are known to have diverse biological and pharmacological activity, particularly antimicrobial [1], antidepressant [2], cardiotonic [3], and anticancer activity [4]. There is much interest in the anticancer activity of these compounds owing to different types of biological targets they might interfere with for this effect to occur e.g. PDE3, PIM1 Kinase, and Survivin protein.
PDE3 is a member of the phosphodiesterase super family of isozymes and is characterized by having high affinity for both cAMP and cGMP as substrates. Specific inhibition of PDE3s in different tissues leads to various biological actions such as stimulation of myocardial contractility, inhibition of platelet aggregation, and the relaxation of vascular smooth muscle, most probably due to the elevation of cyclic adenosine monophosphate levels (cAMP). Moreover, inhibition of PDE3 results in blocking both the anti-lipolytic and the induced lipogenesis actions of insulin, most probably through inhibiting PDE3B [4]. PDE3 is recently shown to be over-expressed in some malignant cells such as squamous cell carcinoma in head and neck, malignant melanoma, osteosarcoma (HOSM-1), and also in malignant salivary gland cells (HSG) [5,6]. The inhibition of PDE3 was able to inhibit the growth and proliferation of the squamous cell carcinoma cell line HeLa, and in HSG cells and further studies revealed that the pyridone derivative, cilostamide– a selective PDE3 inhibitor– has synergism action to the anti-apoptotic action of PDE4 inhibitors in leukemia cells [5–7]. This effect may be explained by the elevation of cAMP levels caused by PDE3 inhibition that leads to anti-angiogenic effect, and interfering with the activation of EGRF to inhibit DNA synthesis and cell proliferation [8,9]. However, most of the studies showed that cross inhibition of different PDEs rather than a specific isozyme is associated anti-angiogenesis or tumor cell growth inhibitory activities.
PIM1 kinase is a member of a family of proteins containing homologues PIM-2 and PIM-3, and is transcriptionally regulated by cytokines, mitogens, and numerous growth factors. PIM1 Kinase is responsible for the phosphorylation, and activation of a large number of endogenous substrates that are responsible for cell division and proliferation [10–12].
Survivin belongs to a family of proteins known as Inhibitors of Apoptosis (IAP) and several studies have shown that survivin is highly expressed in many common human cancers. Moreover, survivin has a very significant role in mitotic cell division where it displays cell cycle-dependent expression, with a pronounced accumulation at mitosis. Survinin over expression in tumor cells is often associated with poor prognosis and shorter survival rates of the patient. In addition, survivin is one of the chromosomal passenger proteins; it complexes and localizes to Aurora B kinase in the mitotic machinery [13–16].
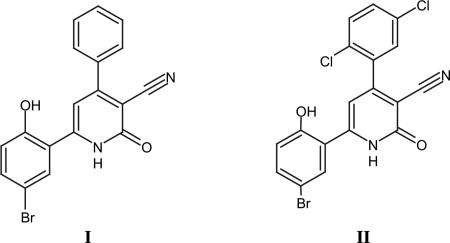
The lead compound 6-(5-bromo-2-hydroxyphenyl)-2-oxo-4-phenyl-1,2-dihydropyridine-3-carbonitrile I has been reported both as PIM1 kinase inhibitor and survivin inhibitor with IC50 of 50 nm and KD of 5.7 μM, respectively. The latter compound was successfully co-crystallized with PIM-1 kinase (PDB ID code 2OBJ). Compound I bound convincingly within the ATP-binding site of PIM-1 suggesting an ATP-competitive inhibitory mechanism. Perhaps the most significant finding from this complex structure was the prominent interaction of the carbonyl group on the pyridone ring with both the Lys67 side chain (interacts with phosphates from ATP) of PIM-1 together with a hydrogen bond to a water molecule that appeared to play an integral part of a larger H-bonding network in this region. The apparent H-bonding network further consisted of a second conserved water molecule that interacted with both the PIM-1 kinase catalytic residue Phe187. Interestingly, the only potential interaction of Compound I with the hinge region involves a weak H-bond between the main carbonyl of Glu 121 and an aromatic hydrogen, potential H-bonding between the same carbonyl of Glu 121 and the aromatic H at the C-4-position may also contribute to stabilization of I with the hinge The role of the Br atom on the phenyl at position 6 of the pyridone ring is less well defined from a SAR perspective although its importance is exemplified by the fact that replacement of the Br by a methyl group leads to a 7-fold loss of inhibition [11].
Nuclear overhauser effect experiments have shown also that this 2-pyridone derivative is also able to inhibit survivin protein through multiple interactions. Moreover, the affinity of I to survivin was enhanced by improving the liophilicity through the introduction of 2 halogen atoms to the phenyl at position 4 of the pyridone ring e.g., compound II with KD of 0.8 μM. This indicates the potentiality of 4,6-diaryl-3-cyano-2-pyridone skeleton as a template for further optimization and with multimodal anticancer molecular targets.
Herein, we report upon novel derivatives with the 3-cyano-2-pyridone skeleton and their isosteric 2-imino derivatives, their tumor cell growth inhibitory against the human HT-29 colon adenocarcinoma tumor cell line and the human PDE3A isozyme. These studies provide insight to defining the scope and limitations of this class of compounds as potential anticancer agents.
2. Chemistry
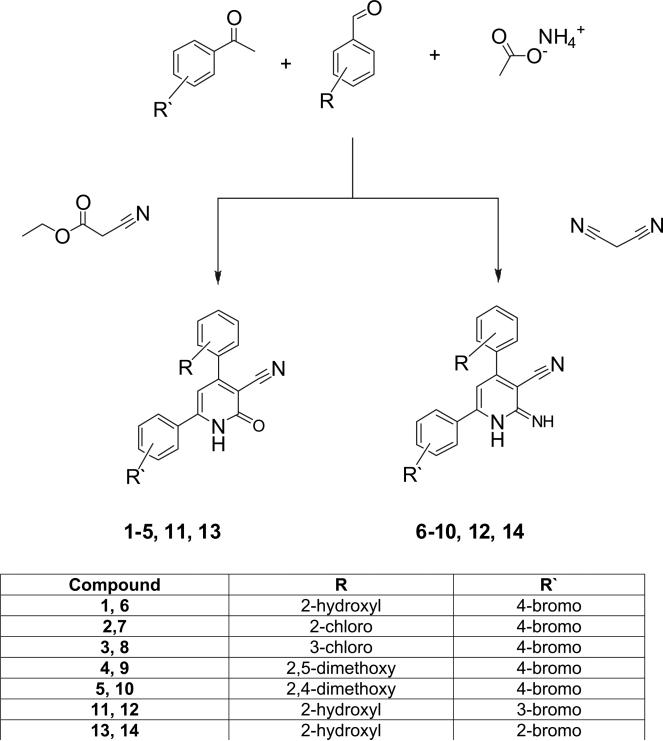
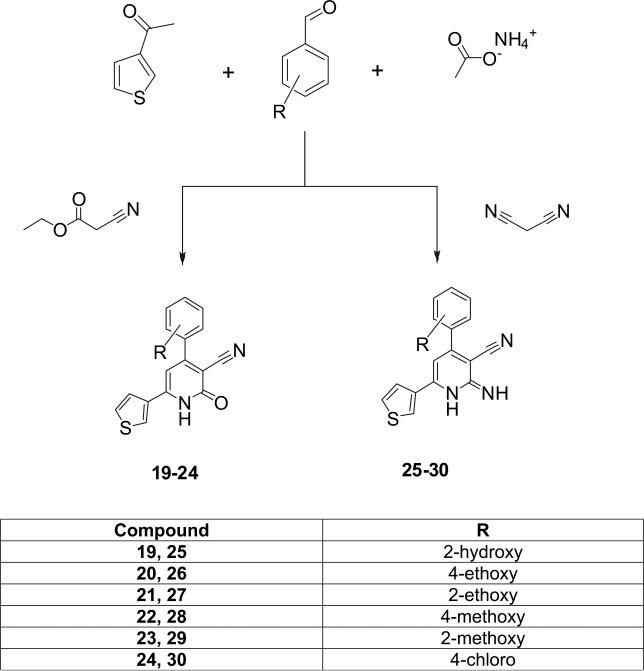
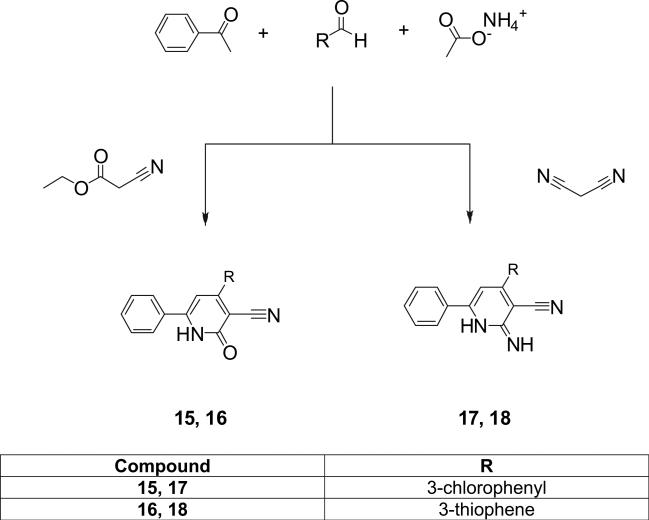
The general synthesis of the 3-cyano-2-pyridone and the 3-cyano-2-iminopyridine derivatives is illustrated in Schemes 1–3. We utilized the in-solution one-pot synthesis. Whereby, the respective aromatic ketone, the respective aromatic aldehyde, ammonium acetate and ethyl cyanoacetate or malononitrile were refluxed in ethanol [3,16]. The desired compounds were purified by re-crystallization from a mixture of DMF-Ethanol in different ratios or by column chromatography.
Scheme 1.
Scheme 3.
All the synthesized compounds are diarylpyridine derivatives. In the 1H-NMR spectra all compounds showed aromatic protons as multiplet peaks at δ 6.90–8.50 ppm. The aromatic proton at position 5 of the pyridine ring appeared as singlet peak at δ about 6.90 ppm.
Mass Spectroscopy of the synthesized compounds showed molecular ion peak [M+] corresponding to either the exact mass or the molecular weight of the target compounds. The molecular ion peaks were also the base peaks indicating the stable nature of these compounds. Bromine containing derivatives showed molecular ion peak at [M+] and [M+ + 2] in a ratio of almost1:1 due to the isotopic nature of the Bromine atom. Meanwhile, the chlorine containing derivatives showed molecular ion peak [M+] and [M+ + 2] in a ratio of 3:1 indicating the isotopic nature of the chlorine atom.
Infrared spectra of all compounds showed bands at ≈3100–3480 cm −1 (N–H) stretching and a band at ≈2200 cm −1 (CN) stretching. On the other hand, the 3-cyano 2-pyridone derivatives showed an extra band at 1640–1753 cm −1 for the pyridone carbonyl amide stretching. This indicates that the ionization status of the carbonyl is variable and the potentiality for amide-iminol equilibrium.
3. Biological results and discussion
All compounds were tested for their in vitro tumor cell growth inhibitory activity using the human HT-29 colon tumor cell line, which is known to express different PDEs, including PDE3. Most of compounds were evaluated in 2 steps, first, the percentage inhibition at a screening dose of 50 μM was performed in triplicate, then for compounds displaying a percentage of inhibition >50% were evaluated by testing a range of 10 concentrations with at least two replicates per concentration to calculate an IC50 value. The results are summarized in Table 1.
Table 1.
Inhibitory effect of the synthesized compounds upon HT29 cells and PDE3.
| Cpd # | % Growth Inhibition at 50 μM | Growth inhibition IC50 μM | % PDE3 Inhibition at 50 μM |
PDE3 inhibition IC50 μM |
||
|---|---|---|---|---|---|---|
| cAMP | cGMP | cAMP | cGMP | |||
| 1 | 94 | 21 | 10 | 28 | ND* | |
| 2 | 5 | ND | 18 | 36 | ND | |
| 3 | 7 | ND | 8 | 21 | ND | |
| 4 | 33 | ND | 8 | 17 | ND | |
| 5 | 0 | ND | 2 | –8 | ND | |
| 6 | 23 | ND | 22 | 27 | ND | |
| 7 | –16 | ND | 36 | 47 | ND | |
| 8 | 7 | ND | 10 | 13 | ND | |
| 9 | 95 | 4 | 20 | 20 | ND | |
| 10 | 2 | ND | 25 | 70 | ND | |
| 11 | –5 | ND | 20 | 45 | ND | |
| 12 | 30 | ND | 18 | 36 | ND | |
| 13 | 22 | ND | 34 | 50 | ND | |
| 14 | 28 | ND | 20 | 40 | ND | |
| 15 | 80 | 2.1 | 10 | 24 | ND | |
| 16 | –10 | ND | 7 | 9 | ND | |
| 17 | 20 | ND | 15 | 20 | ND | |
| 18 | 25 | ND | 4 | 0 | ND | |
| 19 | 79 | 46 | 21 | 37 | ND | |
| 20 | –1 | ND | 7 | 0 | ND | |
| 21 | 80 | 1.25 | 5 | 20 | ND | |
| 22 | –20 | ND | 8 | 0 | ND | |
| 23 | 20 | ND | 10 | 8 | ND | |
| 24 | 5 | ND | 5 | 14 | ND | |
| 25 | 44 | ND | 21 | 26 | ND | |
| 26 | –10 | ND | 55 | 80 | 13.3 | 13.5 |
| 27 | 92 | 6 | 37 | –10 | ND | |
| 28 | 75 | 2.7 | 10 | 30 | ND | |
| 29 | 78 | 35 | 47 | –95 | ND | |
| 30 | 65 | ND | 28 | 45 | ND | |
ND = Not determined.
Only compound 26, showed significant PDE3 inhibition (>50%) at the screening dose using either cAMP or cGMP as substrates with almost equal IC50s but no tumor cell growth inhibitory activity. This reveals no direct correlation between inhibiting PDE3 and tumor cell growth inhibitory activity.
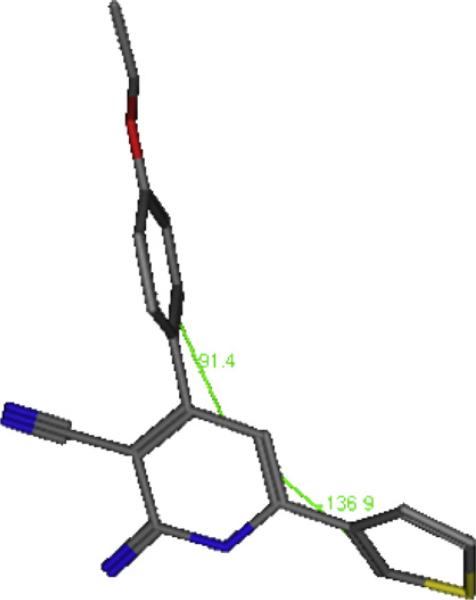
The energy minimized form of 26 (Fig. 1) showed a dihedral angle of 91° between the phenyl and pyridone, and a dihedral angle of 138° between the thiophen and pyridone, this noncoplanarity seems essential for proper interaction with the receptor and indicates the topology of the area of PDE3 with which 26 interacts.
Fig. 1.
The energy minimized form of compound 26, showing non-coplanarity of the pyridone and phenyl ring at position 4 with dihedral angle 91° and the thiophene ring to pyridone ring, dihedral angle 138°.
A series of compounds, however, displayed tumor cell growth inhibitory activity including Compounds 1, 9, 15, 19, 21, 27, 28, and 29. Analysis of SAR reveals the following conclusions: For the 2-pyridone function versus the isosteric imino, there is no preferentiality in this regard, compounds 21 and 27 are the exact isostere of each other; both were active with IC50 = 1.25 and 6 μM, respectively. In addition, four of the active compounds 1, 15, 19 and 21 were 2-pyridone derivatives, while compounds 9, 27, 28 and 29 were 2-iminopyridine derivatives. Thus, the tumor cell growth inhibitory activity of this group of derivatives appears to be modulated by the aryl substituents at position 4 and 6.
For compounds with potent tumor cell growth inhibitory activity that contained a carbonyl function, such as Compounds 1, 15, 19 and 21 we noticed the activity is relatively higher when the infrared ν cm −1 of the carbonyl is lower, specifically 1753, 1654, 1738 and 1641 cm −1 versus. 21, 2.1, 46 and 1.25 μM, respectively. This indicates that more single bond character to the –C=O (enolization) may be beneficial for tumor cell growth inhibitory activity. Compound 21 is of 2- ethoxy (electron donating) substituent on the phenyl at position 4; thus in a mesomeric fashion may increase the electron density of carbonyl oxygen (H bond acceptor).
For compounds with potent tumor cell growth inhibitory activity that contained an imino function, such as Compounds 29, 27, 9 and 28, the tumor cell growth inhibitory activity is directly proportional to the ν cm −1 of the =NH, specifically 3102, 3242, 3360 and 3477 cm −1 Versus 35, 6, 4 and 2.7 μM, respectively. This confirms the critical role of the NH acidic protons upon activity, although other substituents on the pyridone ring should be considered.
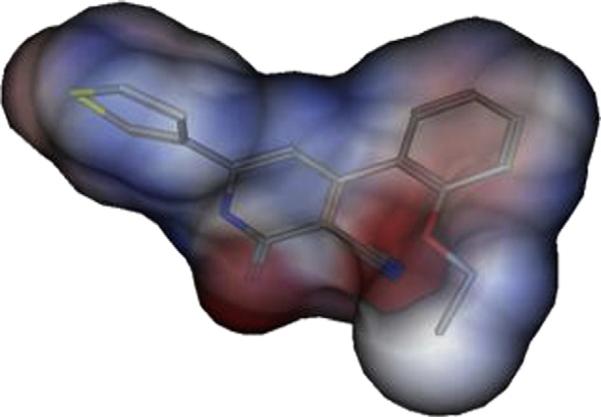
Six out of the 8 active compounds, viz 1, 9, 19, 27, 28 and 29 were substituted with an alkoxy substituent particularly at the ortho position of the phenyl at Position 4 of the pyridone, hydrogen bond formation from the receptor may be involved. It should be noted that alkoxy substituent on the phenyl and the carbonyl of the pyridone will create negative electrostatic potential that seems essential for activity as shown in Fig. 2. Moreover, this substituent at the ortho position will lead to non coplanarity of the pyridone and phenyl, a feature that appears to be essential for tumor cell growth inhibitory activity. Comparing the activity of compound 21 relative to 20 verifies the importance of the substituent positioning.
Fig. 2.
Electrostatic potential map of compound 21 on Connolly surface (red = negative); (blue = positive); (white = neutral), hydrogens removed for clarification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Position 6 of the pyridone was substituted with 4-bromophenyl, 3-bromophenyl, 2-bromophenyl, phenyl and thiophene, significant activity is obtained with those with 4-bromophenyl, phenyl and thiophene substitution.
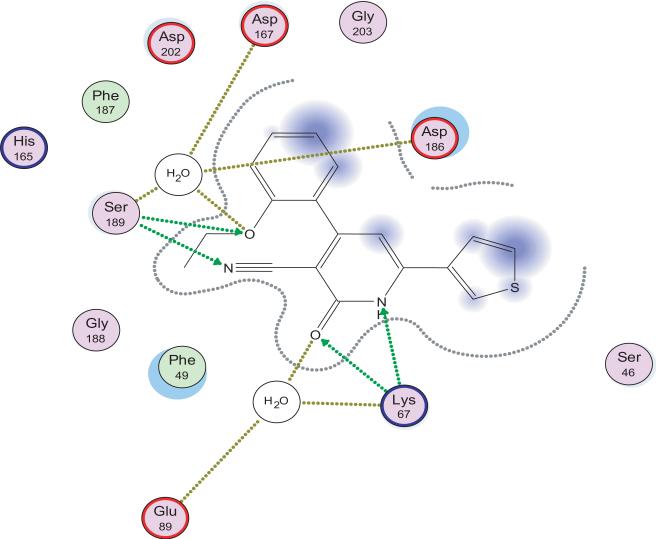
AS the anticancer activity is not correlated with PDE3 inhibition, thus other targets such as survivin protein or PIM1 kinase may be involved. As a predictive tool, docking the most active candidate 21 to PIM1 (PDB ID code 2OBJ). Fig. 3 shows hydrogen bonding interaction of the pyridine N and carbonyl with Lys67 which may also interact with the CN at position 3. Also, Ser 189 interacts via hydrogen bonding with the CN group. It is worthy to mention that compound I interacts similarly with the same protein. Interestingly, the ethoxy group of compound 21 is involved in extra hydrogen bonding through water molecule with Asp 167, Asp 186, and Ser 189. This may explain the high potency of compound 21.
Fig. 3.
2D interaction of compound 21 with PIM1 kinase.
On the other hand, docking of 21 to the binding pocket of survivin (PDB ID code 1F3H) showed no specific interaction with the residues of the binding pocket (data not shown). From these data we conclude that PIM1 kinase is the most appropriate molecular target for the tumor cell growth inhibitory activity of this series of derivatives. It should be taken into consideration that the survivin inhibitor II is of highly lipophilic substituent at position 4 and an oxygenated aryl at position 6; meanwhile our most active anticancer compound 21 is of an oxygenated aryl at position 4 and a lipophilic aryl (thiophene) at position 6. This retro order of substituents hydrophobicity nature may explain the bias towards PIM1 kinase in our case.
4. Experimental
4.1. Chemistry
All reactions were performed with commercially available reagents and they were used without further purification. Solvents were dried by standard methods and stored over molecular sieves. All reactions were monitored by thin-layer chromatography (TLC) carried on fluorescent precoated plates and detection of the components was made by short UV light. Melting points were determined in open capillaries using MEL-TEMP II and Buchi B-540 Melting Point apparatus and are uncorrected. FTIR spectra were recorded on Nicolet Avatar 380 spectrometer. 1H-NMR spectra were recorded on Varian Mercury VX-300 MHz and spectrometer. Mass spectra were obtained with Hewlett Packard GC-MS, model 5890, series II at an ionization potential of 70 ev.
Elemental analyses were performed by Institute of Organic Chemistry, Jena University and the Microanalytical Unit, Faculty of Science, Cairo University; found values were within ±0.4% of the theoretical ones, unless otherwise indicated. Yields are not optimized.
4.1.1. General procedure for the preparation of 4,6-diaryl-2-oxo-1,2 dihydropyridine-3-carbonitriles
The respective aromatic ketone (1 mmol), together with the respective aromatic aldehyde (1 mmol), ethyl cyanoacetate (1 mmol), and ammonium acetate (8 mmol) were dissolved in ethyl alcohol (30 ml) and put under reflux for 10 to 12 h. The precipitate obtained was filtered, washed with ethyl alcohol and dried. For the purification purpose, the precipitate was subjected either to re-crystallization from a mixture of DMF-Ethanol (1:10), or to column chromatography on silica gel, eluting with chloroform.
4.1.1.1. 6-(4-Bromophenyl)-4-(2-hydroxyphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (1)
Yield 92%; mp 294–296 °C; IR (cm −1): 3394, 3278, 2246, 1753; 1H-NMR (DMSO-d6): 6.90 (s, 1H, aromatic), 7.36–8.27 (m, 9H, aromatic and NH); MS (EI): m/z 369 (M+ + 2), 367 (M+); Anal (C18H11BrN2O2) C, H, N.
4.1.1.2. 6-(4-Bromophenyl)-4-(2-chlorophenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (2)
Yield 87%; mp 293–295 °C; IR (cm −1): 3300, 2223, 1640; 1H-NMR (DMSO-d6): 6.90 (s, 1H, aromatic), 7.49–7.86 (m, 9H, aromatic and NH); MS (EI): m/z 388 (M+ + 2), 386 (M+); Anal. (C18H10BrClN2O) C, H, N.
4.1.1.3. 6-(4-Bromophenyl)-4-(3-chlorophenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (3)
Yield 85%; mp 294–296 °C; IR (cm −1): 3300, 2221, 1657; 1H-NMR (DMSO-d6): 6.90 (s, 1H, aromatic), 7.57–7.90 (m, 9H, aromatic and NH); MS (EI): m/z 388 (M+ + 2), 386 (M+); Anal. (C18H10BrClN2O) C, H, N.
4.1.1.4. 6-(4-Bromophenyl)-4-(2,5-dimethoxyphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (4)
Yield 65%; mp 296–297 °C; IR (cm −1): 3476, 2218, 1683; 1H-NMR (DMSO-d6): 3.70 (s, 3H, -OCH3), 3.86 (s, 3H, -OCH3), 6.99 (s, 1H, aromatic) 7.01–7.15 (m, 4H, aromatic and NH), 7.71–7.74 (d, 2H, aromatic), 7.81–7.84 (d, 2H, aromatic); MS (EI): m/z 412 (M+ + 2), 410 (M+); Anal. (C20H15BrN2O3) C, H, N.
4.1.1.5. 6-(4-Bromophenyl)-4-(2,4-dimethoxyphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (5)
Yield 62%; mp 274–276 °C; IR (cm −1): 3436, 2215, 1710; 1H-NMR (DMSO-d6): 3.80 (s, 3H, –OCH3), 3.83 (s, 3H, –OCH3), 6.65-7.77 (m, 9H, aromatic and NH); Anal. (C20H15BrN2O3) C, H, N.
4.1.1.6. 6-(3-Bromophenyl)-4-(2-hydroxyphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (11)
Yield 88%; mp 294–296 °C; IR (cm −1): 3394, 3278, 2246,1753; 1H-NMR (DMSO-d6): 7.37-8.55 (m, 9H, aromatic), 12.35 (s, 1H, NH); MS (EI): m/z 369 (M+ + 2), 367 (M+); Anal. (C18H11BrN2O2) C, H, N.
4.1.1.7. 6-(2-Bromophenyl)-4-(2-hydroxyphenyl)-2-oxo-1,2-dihydropyridine-3-carbonitrile (13)
Yield 87%; mp 295–297 °C; IR (cm −1): 3394, 3278, 2246, 1753; 1H-NMR (DMSO-d6): 7.17-8.33 (m, 10H, aromatic and OH), 12.5 (s, 1H, NH); MS (EI): m/z 369 (M+ + 2), 367 (M+); Anal. (C18H11BrN2O2) C, H, N.
4.1.1.8. 4-(3-Chlorophenyl)-2-oxo-6-phenyl-1,2-dihydropyridine-3-carbonitrile (15)
Yield 83%; mp 244–246 °C; IR (cm −1): 3325, 2219, 1654; 1H-NMR (DMSO-d6): 6.87-7.89 (m, 10H, aromatic), 12.87 (s, 1H, NH); MS (EI): m/z 307 (M++1), 329 (M++ Na+); Anal. (C18H11ClN2O) C, H, N.
4.1.1.9. 2-Oxo-6-phenyl-4-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (16)
Yield 82%; mp 300–302 °C; IR (cm −1): 3438, 2213, 1637; 1H-NMR (DMSO-d6): 6.92 (s, 1H, aromatic), 7.52–7.89 (m, 7H aromatic), 8.35 (s, 1H, aromatic), 12.69 (s, 1H, NH); Anal (C16H10N2OS) C, H, N.
4.1.1.10. 4-(2-Hydroxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (19)
Yield 89%; mp 295–296 °C; IR (cm −1): 3441, 3330, 2224, 1738; 1H-NMR (DMSO-d6): 7.34-8.50 (m, 8H, aromatic and OH), 8.65 (s, 1H, aromatic), 12.10 (s, 1H, NH); MS (EI): m/z 294 (M+); Anal. (C16H10N2O2S): C, H, N.
4.1.1.11. 4-(4-Ethoxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (20)
Yield 85%; mp 295–297 °C; IR (cm −1): 3449, 2212, 1637; 1H-NMR (DMSO-d6): 1.32-1.38 (t, 3H, -CH3), 4.07–4.15 (q, 2H, -OCH2), 6.90-8.51 (m, 8H, aromatic), 12.56 (s, 1H, NH); Anal. (C18H14N2O2S) C, H, N.
4.1.1.12. 4-(2-Ethoxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3 carbonitrile (21)
Yield 78%; mp 280–281 °C; IR (cm −1): 3418, 2216, 1641; 1H-NMR (DMSO-d6): 1.24–1.30 (t, 3H, eCH3), 4.05–4.14 (q, 2H, –OCH2), 6.84–8.49 (m, 8H, aromatic), 12.60 (s, 1H, NH); MS (EI): m/z 322 (M+), 345 (M+ + Na+). Anal. (C18H14N2O2S) C, H, N.
4.1.1.13. 4-(4-Methoxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (22)
Yield 88%; mp 338–340 °C; IR (cm −1): 3453, 2221,1650; 1H-NMR (DMSO-d6): 3.83 (s, 3H, -OCH3), 6.90–8.51 (m, 8H, aromatic), 12.60 (s, 1H, NH); MS (EI): m/z 308 (M+), 331 (M+ + Na+, 100%). Anal. (C17H12N2O2S) C, H, N.
4.1.1.14. 4-(2-Methoxyphenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (23)
Yield 82%; mp 311–313 °C; IR (cm −1): 3356, 2220, 1648; 1H-NMR (DMSO-d6): 3.80 (s, 3H, –OCH3), 6.83–8.48 (m, 8H, aromatic), 12.62 (s, 1H, NH); MS (EI): m/z 308 (M+), 331 (M+ + Na+). Anal. (C17H12N2O2S) C, H, N.
4.1.1.15. 4-(4-Chlorophenyl)-2-oxo-6-thiophen-3-yl-1,2-dihydropyridine-3-carbonitrile (24)
Yield 78%; mp 310–312 °C; IR (cm −1): 3446, 2220, 1644; 1H-NMR (DMSO-d6): 6.94 (s, 1H, aromatic), 7.62–7.83 (m, 6H, aromatic), 8.52 (s, 1H, aromatic), 12.72 (s, 1H, NH); Anal. (C16H9Cl N2OS) C, H, N.
4.1.2. General procedure for the preparation of 4,6-diaryl-2-imino-1,2-dihydropyridine-3-carbonitrile
The respective aromatic keton (1 mmol), together with the respective aromatic aldehyde (1 mmol), malononitrile (1 mmol), and ammonium acetate (8 mmol) were dissolved in ethyl alcohol (30 ml) and put under reflux for 10–12 h. The precipitate obtained was filtered, washed with ethyl alcohol and dried. For the purification purpose, the precipitate was subjected either to re-crystallization from a mixture of DMF-Ethanol (1:10), or to column chromatography on silica gel, eluting with chloroform and/or methylene chloride.
4.1.2.1. 6-(4-Bromophenyl)-4-(2-hydroxyphenyl)-2-imino-1,2-dihydropyridine-3-carbonitrile (6)
>Yield 73%; mp 294–296 °C; IR (cm −1): 3430, 3138, 2195; 1H-NMR (DMSO-d6): 6.87–7.72 (m, 11H, aromatic and NHs), 9.86 (s, 1H, OH); MS (EI): m/z 368 (M+ + 2), 366 (M+). Anal. (C18H12BrN3O) C, H, N.
4.1.2.2. 6-(4-Bromophenyl)-4-(2-chlorophenyl)-2-imino-1,2-dihydropyridine-3-carbonitrile (7) [17]
Yield 75%; mp 255–257 °C; IR (cm −1): 3446, 2220.
4.1.2.3. 6-(4-Bromophenyl)-4-(3-chlorophenyl)-2-imino-1,2-dihydropyridine-3-carbonitrile (8) [18]
Yield 80%; mp 265–277 °C; IR (cm −1): (–NH), (–CN); 1H-NMR (DMSO-d6): 6.84–8.33 (m, 11H, aromatic and NHs); MS (EI): m/z 387 (M+ + 2), 385 (M+).
4.1.2.4. 6-(4-Bromophenyl)-4-(2,5-dimethoxyphenyl)-2-imino1,2-dihydropyridine3-carbonitrile (9)
Yield 68%; mp 284–286 °C; IR (cm −1): 3360, 2217; 1H-NMR (DMSO-d6): 3.60 (s, 3H, –OCH3), 3.86 (s, 3H, –OCH3), 7.09–7.75 (m, 10H, aromatic and NHs); MS (EI): m/z 411 (M+ + 2), 409 (M+); Anal. (C20H16BrN3O2) C, H, N.
4.1.2.5. 6-(4-Bromophenyl)-4-(2,4-dimethoxyphenyl)-2-imino-1,2-dihydropyridine3-carbonitrile (10)
Yield 76%; mp 225–227 °C; IR (cm −1): 3448, 2215; 1H-NMR (DMSO-d6): 3.82 (s, 3H, –OCH3), 3.83 (s, 3H, –OC H3), 6.60–7.81 (m, 10H, aromatic and NHs); MS (EI): m/z 432 (M+ + Na+), m/z 411 (M+ + 2), m/z 409 (M+); Anal.(C20H16BrN3O2) C, H, N.
4.1.2.6. 6-(3-Bromophenyl)-4-(2-hydroxyphenyl)-2-imino-1,2-dihydropyridine3-carbonitrile (12)
Yield 85%; mp 291–293 °C; IR (cm −1): 3415, 3316, 2195; 1H-NMR (DMSO-d6): 6.85–8.96 (m, 11H, aromatic and NHs); MS (EI): m/z 368 (M+ + 2), 366 (M+); Anal. (C18H12BrN3O) C, H, N.
4.1.2.7. 6-(2-Bromophenyl)-4-(2-hydroxyphenyl)-2-imino-1,2-dihydropyridine3-carbonitrile (14)
Yield 84%; mp 284–286 °C; IR (cm −1): 3412, 3309, 2205; 1H-NMR (DMSO-d6): 6.88-8.98 (m, 11H, aromatic and NHs); MS (EI): m/z 368 (M+ + 2), 366 (M+); Anal. (C18H12BrN3O) C, H, N.
4.1.2.8. 4-(3-Chlorophenyl)-2-imino-6-phenyl-1,2-dihydropyridine3-carbonitrile (17) [18]
Yield 70%; mp 214–216 °C; IR (cm −1): 3354, 2217.
4.1.2.9. 2-Imino-6-phenyl-4-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (18)
Yield 62%; mp 150–152 °C; IR (cm −1): 3476, 2204; 1H-NMR (DMSO-d6): 6.96–8.17 (m, 11H, aromatic and NHs); Anal. (C16H11N3S) C, H, N.
4.1.2.10. 2-Imino-4-phenyl-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (25)
Yield 84%; mp 290–292 °C; IR (cm −1): 3408, 3108, 2195; 1H-NMR (DMSO-d6):7.39–8.51 (m, 10H, aromatic and NHs); MS (EI): m/z 293 (M+); Anal. (C16H11N3OS) C, H, N.
4.1.2.11. 4-(4-Ethoxyphenyl)-2-imino-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (26)
Yield 70%; mp 220–222 °C, IR (cm −1): 3200, 2198; 1H-NMR (DMSO-d6): 1.33–1.37 (t, 3H, –CH3), 4.06–4.13 (q, 2H, –OCH2), 6.99–8.13 (m, 10H, aromatic and NHs); Anal. (C18H15N3OS) C, H, N.
4.1.2.12. 4-(2-Ethoxyphenyl)-2-imino-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (27)
Yield 65%; mp 242–244 °C; IR (cm −1): 3243, 2199; 1H-NMR (DMSO-d6): 1.38–1.43 (t, 3H, -CH3), 4.19–4.26 (q, 2H, -OCH2), 7.09–8.31 (m, 10H, aromatic and NHs); Anal. (C18H15N3OS) C, H, N.
4.1.2.13. 2-Imino-4-(4-methoxyphenyl)-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (28)
Yield 65%; mp 270–272 °C; IR (cm −1): 3477, 2203; 1H-NMR (DMSO-d6): 3.82 (s, 3H, –OCH3), 6.83–8.48 (m, 10H, aromatic and NHs); Anal. (C17H13N3OS) C, H, N.
4.1.2.14. 2-Imino-4-(2-methoxyphenyl)-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (29)
Yield 60%; mp 288–290 °C; IR (cm −1): 3102,2205; 1H-NMR(DMSO-d6): 3.94(s,3H, –OCH3),7.11–8.25(m,10H, aromatic and NHs); MS (EI): m/z 307 (M+); Anal. (C17H13N3OS) C, H, N.
4.1.2.15. 4-(4-Chlorophenyl)-2-imino-6-(thiophen-3-yl)-1,2-dihydropyridine-3-carbonitrile (30)
Yield 65%; mp 270–272 °C; IR (cm −1): 3355, 2219; 1H-NMR (DMSO-d6): 6.79–8.30 (m, 10H, aromatic and NHs); MS (EI): m/z 313 (M+ + 2), m/z 311 (M+); Anal. (C16H10ClN3S) C, H, N.
4.2. Biology
4.2.1. Cell cultures
HT-29 tumor cells were obtained from ATCC and were grown under standard cell culture conditions at 37 °C in a humidified atmosphere with 5% CO2. Cells were grown in RPMI 1640 containing 5% fetal bovine serum. Cell count and viability was determined by Trypan blue staining followed by hemocytometry. Only cultures displaying >95% cell viability were used for experiments.
4.2.2. Growth assays
Tissue culture treated microtiter 96-well plates were seeded at a density of 5000 cells/well. The plates were incubated for 18–24 h prior to any treatment. Cell viability was measured 72 h after treatment by the Cell Titer Glo Assay (Promega), which is a luminescent assay that is an indicator of live cells as a function of metabolic activity and ATP content. The assay was performed according to manufacturer's specifications [19].
4.2.3. Phosphodiesterase assay
PDE activity was measured using an adaptation of the IMAP® fluorescence polarization phosphodiesterase assay (Molecular Devices). PDE hydrolysis of the fluorescent-labeled substrate allows it to bind the IMAP® reagent, which increases fluorescence polarization (FP). The assay used fluorescein (Fl)-cAMP and tetramethylrhodamine (TAMRA)-cGMP as substrates. The different excitation and emission spectra of the substrates (485–530 nm for Fl and 530–590 nm for TAMRA) allowed for simultaneous measurement of cAMP and cGMP hydrolysis in the same well. The assays were performed in 96-well microtiter plates using a reaction buffer containing 10 mM Tris-HCl (pH 7.2) 10 mM MgCl2, 0.05% NaN3 and 0.1% phosphate-free BSA as the carrier. Each well contained 20 μl of recombinant enzyme (BPS Biosciences, San Diego, CA) and 10 μl inhibitor. The reaction was initiated by the addition of 10 μl of a substrate solution containing 50 nM Fl–cAMP and/or TAMRA–cGMP. After incubating at room temperature for 60 minutes, the reaction was terminated by adding 120 μl of binding solution. FP was measured with a BioTek Synergy 4.
4.2.4. Experimental design and data analysis
Compound effects on tumor cell growth and PDE activity were measured and potency was expressed by an IC50 value (50% inhibitory concentration). For growth assays, the IC50 value was determined by testing a range of 8 concentrations with at least four replicates per concentration. For enzyme assays, the IC50 value was determined by testing a range of 10 concentrations with at least two replicates per concentration. Dose response curves were analyzed using Prism™ 4 software (GraphPad) to calculate IC50 values using a four parameter logistic equation.
4.3. Molecular modeling
4.3.1. Molecular modeling and conformational search
The compounds were drawn on Chemoffice 2002 and saved as mol file, the latter were subjected to energy minimization using MMFF94x and systemic conformational search using MOE software [20]. Dihedral angles of the most favorable conformer were then measured.
4.3.2. Electrostatic potential map
The relevant energy minimized compounds were opened on MOE software, their partial charges were calculated, and their EPS were obtained on Connolly surface.
4.3.3. Docking
For docking on the PIM1 Kinase, the crystal structure of Human PIM1 Kinase complexed with the inhibitor was downloaded from the Protein data bank (PDB ID code 2obj) and opened with MOE software. The Molecular Operating Environment of docking was used to calculate the docking energies between ligand and the enzyme catalytic site as given in the software manual. MOE-Dock is used to search for favorable binding configurations between the ligand and the macromolecular target. The lowest energy conformation was selected as the best.
For docking on Survivin, the crystal structure of Human Survivin was downloaded from the Protein data bank (PDB ID code 1f3 h) and opened with MOE software. The protein was then edited through removing zinc and sulphate molecules, then the reported amino acid pocket namely; L 102, F 93, L 96, L 98, L 104, V 89, K 15, and F 86 was selected and the residue around the pocket was extended, and the partial charges were calculated. The Molecular Operating Environment of docking was used to calculate the docking energies between ligand and the protein catalytic site as given in the software manual. MOE-Dock is used to search for favorable binding configurations between the ligand and the macromolecular target. The lowest energy conformation was selected as the best.
Scheme 2.
Acknowledgment
The first author is grateful to the Alexander von Humboldt foundation Germany, for providing him with a research stay in Germany. The second author is grateful for the Faculty of graduate studies, German University in Cairo for partial financing of the project.
References
- 1.Abdel-Aziz A, El-Subbagh HI, Kunieda T. Bioorg. Med. Chem. 2005;13:4929–4935. doi: 10.1016/j.bmc.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Latif N. Sci. Pharm. 2005;73:195–216. [Google Scholar]
- 3.Bekhit AA, Baraka AM. Boll. Chim. Farma. 2005;145:133–138. [Google Scholar]
- 4.Thompson P, Manganiello VC, Degerman E. Curr. Top. Med. Chem. 2007;7:421–436. doi: 10.2174/156802607779941224. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu K, Murata T, Okumura K, Manganiello VC, Tagawa TT. Anticancer Drugs. 2002;13:875–880. doi: 10.1097/00001813-200209000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Murata T, Sugatani T, Shimizu K, Manganiello VC, Tagawa T. Anticancer Drugs. 2001;12:79–83. doi: 10.1097/00001813-200101000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Moon E, Lee R, Near R, Weintraub L, Wolda S, Lerner A. Clin. Cancer Res. 2002;8:589–595. [PubMed] [Google Scholar]
- 8.Stuart N, Donald M. Mol. Pharmacol. 2005;67:263–272. [Google Scholar]
- 9.Shafer SH, Phelps SH, Williams CL. Biochem. Pharmacol. 1998;56:1229–1236. doi: 10.1016/s0006-2952(98)00260-3. [DOI] [PubMed] [Google Scholar]
- 10.Cheney IW, Yan S, Appleby T, Walker H, Vo T, Yao N, Hamatake R, Hong Z, Wu JZ. Bioorg. Med. Chem. Lett. 2007;17:1679–1683. doi: 10.1016/j.bmcl.2006.12.086. [DOI] [PubMed] [Google Scholar]
- 11.Wendt MD, Sun C, Kunzer A, Sauer D, Sarris K, Hoff E, Yu L, Nettesheim DG, Chen J, Jin S, Comess KM, Fan Y, Anderson SN, Isaac B, Olejniczak ET, Hajduk PJ, Rosenberg SH, Elmore SW. Bioorg. Med. Chem. Lett. 2007;17:3122–3129. doi: 10.1016/j.bmcl.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 12.Tong Y, Stewart KD, Thomas S, Przytulinska M, Johnson EF, Klinghofer V, Leverson J, McCall O, Soni NB, Luo Y, Lin NH, Sowin TJ, Giranda VL, Penning TD. Bioorg. Med. Chem. Lett. 2008;18:5206–5208. doi: 10.1016/j.bmcl.2008.08.079. [DOI] [PubMed] [Google Scholar]
- 13.Zhonghong L, Lianjie L, Changqing Z, Ying H, Yu J, Yan L. J. Exp .Clin. Cancer Res. 2008;27:20. doi: 10.1186/1756-9966-27-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rankin WV, Henry CJ, Turnquist SE, Turk JR, Beissenherz ME, Tyler JW, Rucker EB, Knapp DW, Rodriguez CO, Green JA. Vet. Comp. Oncol. 2008;6:141–150. doi: 10.1111/j.1476-5829.2007.00150.x. [DOI] [PubMed] [Google Scholar]
- 15.Sejeong S, Byung-Je S, Yong-Soon C, Hyun-Ju K, Nam-Chul H, Jong-Ik H, Chul-Woong C, Yong-Keun J, Byung-Ha O. Biochemistry. 2001;40:1117–1123. [Google Scholar]
- 16.Abadi AA, Al-Deeb O, Al-Afify A, El-Kashef H. Farmaco. 1999;54:195–201. doi: 10.1016/s0014-827x(99)00004-x. [DOI] [PubMed] [Google Scholar]
- 17.Khatoon S, Yadav AK. Phosph. Sulf. Silicon. 2004;179:345–352. [Google Scholar]
- 18.Popat H, Kachadia V, Nimavat K, Joshi HS. J. Ind. Chem. Soc. 2004;81:157–159. [Google Scholar]
- 19.Whitehead CM, Earle KA, Fetter J, Xu S, Hartman T, Chan DC, Zhao TL, Piazza GA, Klein-Szanto AJ, Pamukcu R, Alila H, Bunn PA, Jr., Thompson WJ. Mol. Cancer Ther. 2003;2:479–488. [PubMed] [Google Scholar]
- 20.MOE . Chemical Computing Group Inc.; Montreal: http://www.chemcomp/com. [Google Scholar]