Abstract
Leptospira kirschneri is one of the pathogenic species of the Leptospira genus. Human and animal infection from L. kirschneri gained further attention over the last few decades. Here we present the isolation and characterisation of Brazilian L. kirschneri serogroup Pomona serovar Mozdok strain M36/05 and the comparative genomic analysis with Brazilian human strain 61H. The M36/05 strain caused pulmonary hemorrhagic lesions in the hamster model, showing high virulence. The studied genomes presented high symmetrical identity and the in silico multilocus sequence typing analysis resulted in a new allelic profile (ST101) that so far has only been associated with the Brazilian L. kirschneri serogroup Pomona serovar Mozdok strains. Considering the environmental conditions and high genomic similarity observed between strains, we suggest the existence of a Brazilian L. kirschneri serogroup Pomona serovar Mozdok lineage that could represent a high public health risk; further studies are necessary to confirm the lineage significance and distribution.
Keywords: L. kirschneri, serovar Mozdok, genomics
Leptospirosis is an emerging worldwide zoonosis which is caused by spirochetes of the Leptospira genus (Levett 2001). To date, the genus comprises 11 pathogenic species (Bourhy et al. 2014) of which Leptospira interrogans is the most associated with human infection. Nevertheless, L. kirschneri infection has gained further attention over the last few decades.
Masuzawa et al. (2006) reported one of the first cases of L. kirschneri human infection by direct contact with southern flying squirrels imported from the United States; two Japanese workers were infected by L. kirschneri serovar Grippotyphosa from the imported American exotic pets. The L. kirschneri serogroup Pomona serovar Mozdok has also been related to human infection in Cuba (Obregón et al. 2007). In Europe, the serovar Mozdok is described as endemic in wild rodents (Majetic et al. 2014) and has also been associated with canine leptospirosis (Renaud et al. 2013).
In Brazil, L. kirschneri serogroup Pomona serovar Mozdok has recently been described as the causative agent of human and canine infection in different time points (Cunha et al. 2016). Here we present the isolation and characterisation of L. kirschneri serogroup Pomona serovar Mozdok strain M36/05 and the comparative genomic analysis with previously described Brazilian L. kirschneri serogroup Pomona serovar Mozdok human strain 61H.
The M36/05 strain was isolated from the kidney of a captured urban black rat (Rattus rattus) in Suzano, metropolitan region of São Paulo State, Brazil, in 2005. For isolation, 5 g of kidney sample was collected and homogenised in 50 mL of Sorensen saline, and 100 µL aliquots of 10-1 to 10-3 dilutions were inoculated into duplicate tubes containing EMJH (DIFCO, USA) enriched with 15% rabbit serum, 5-fluorouracil and nalidixic acid. Once isolated, the strain was stored in Fletcher’s medium (DIFCO/USA), enriched with 15% rabbit serum and maintained in EMJH broth (DIFCO/USA) at 30ºC, as part of the Leptospira collection of the Laboratory of Bacterial Zoonosis - University of São Paulo.
Serogrouping was performed at the Laboratory of Veterinary Bacteriology - Fluminense Federal University. The isolate was subjected to microscopic agglutination test (MAT) using a panel of polyclonal rabbit antisera of 32 reference serovars representing the 24 known serogroups (provided by Royal Tropical Institute - KIT, Amsterdam). The M36/05 strain presented high agglutination rates with serogroup Pomona antisera which was then identified as the strain presumptive serogroup (Dikken & Kmety 1978).
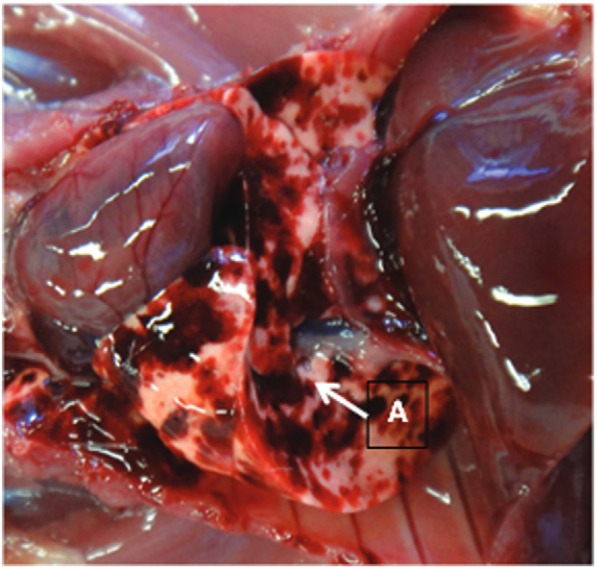
To evaluate the strain virulence, five Golden Syrian hamsters (Mesocricetus auratus) were infected with M36/05 strain (108 leptospires) through intraperitoneal route. The animal experiment was conducted with the approval of the Ethics Committee from the School of Veterinary Medicine and Animal Science - University of São Paulo (2244/2011). Clinical symptoms were checked daily. Within five days post-infection, the M36/05 strain caused lethal leptospirosis and the infected hamsters were euthanised. The inoculated animals developed acute infection characterised mainly by haemorrhagic pulmonary lesions (Fig. 1).
Fig. 1. : macroscopic lesions observed in respiratory tract of M36/05 infected hamsters. (A) The main feature was haemorrhagic pulmonary lesions.
Genomic DNA was extracted and purified with illustra™ bacteria genomicPrep Mini Spin Kit (GE Healthcare do Brasil Ltda, São Paulo, Brazil) and used for paired-end library preparation with Nextera™ DNA Sample Prep Kit (Illumina®) and sequencing through Illumina® Miseq platform. The de novo assembly was performed with Geneious 8.1.8 (Biomatters Ltd, Auckland, New Zealand) and CLC Main Workbench 7.5.1 (CLC Bio, Denmark) and resulted in 44 scaffolds with a N50 of 241,247 bp.
The M36/05 draft genome (LLJK00000000) comprises ~4.46 Mb with overall GC content of 35.9%. Automatic genome annotation was performed with NCBI Prokaryotic Genome Annotation Pipeline. The basic annotation features identified in M36/05 strain are summarised in Table. With regard to the virulence genes, M36/05 presents genes encoding the main Leptospira virulence factors as lipoproteins and immunoglobulin-like proteins (ligA, ligB, ligC lolC/D, lipA, lipL32 and loa22) and also flagellar proteins (flaA and flab subunits, fliG/F). In addition, antimicrobial resistance genes were also identified (tetA, ermA), including efflux pumps (norM, mdtA and qacA).
TABLE. Assembly statistics, basic annotation features and sequence types for respective Leptospira multilocus sequence typing (MLST) schemes observed for Brazilian L. kirschneri serogroup Pomona serovar Mozdok strains.
| Strain | Assembly statistics | Basic annotation features | Leptospira MLST schemes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| Scaffolds | N50 | Length | CG% | CDS | rRNAs | tRNAs | Ahmed et al. (2006) | Boonsilp et al. (2013) | Varni et al. (2014) | |||
| M36/05 | 44 | 241,247 | 4.46 Mb | 35.9 | 3,606 | 6 | 37 | ST98 | ST117 | ST101 | ||
| 61H | 174 | 45,311 | 4.48 Mb | 35.9 | 3,629 | 5 | 38 | ST98 | ST117 | ST101 | ||
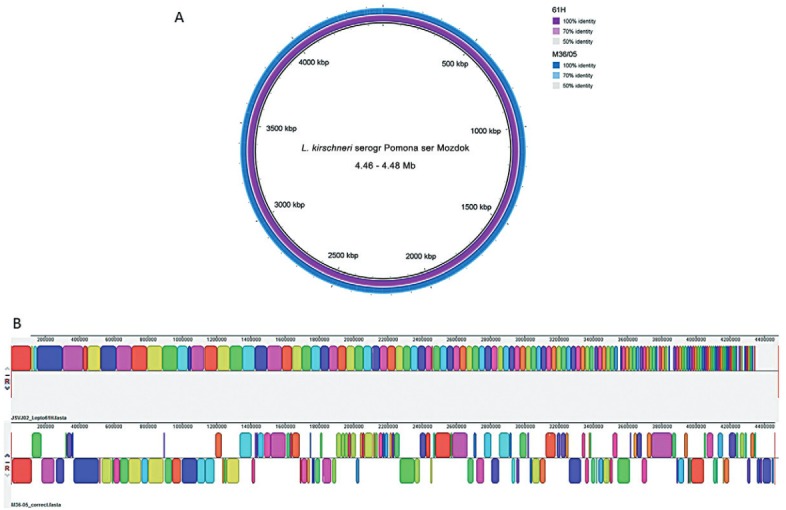
The M36/05 genome was compared to the Brazilian L. kirschneri serogroup Pomona serovar Mozdok human strain 61H (JSVJ00000000) through Mauve multiple genome aligner (Darling et al. 2004) and BLAST Ring Image Generator (BRIG) (Alikhan et al. 2011) and presented high symmetrical identity (98.86%) (Fig. 2). Due to the unavailability of a complete L. kirschneri reference genome, the chromosomes of the studied genomes were not individualised. The genetic content of strains is highly similar (Fig. 2A) and the few structural differ- ences observed (Fig. 2B) could be due to differences in the applied assembly and ordering methodologies between the draft genomes. The absence of a reference genome for L. kirschneri still poses a challenge for assertive assembly and comparative analysis.
Fig. 2. : whole-genome sequencing analysis of Brazilian Leptospira kirschneri serogroup Pomona serovar Mozdok strains. (A) BRIG plot displaying genomic similarity; (B) mauve alignment blocks.
The in silico multilocus sequence typing (MLST) analysis was performed for the three Leptospira MLST protocols available (Table). Both strains presented similar results with ST98 and ST117 for Ahmed et al. (2006) and Boonsilp et al. (2013) protocols, respectively; these sequence types had already been associated with L. kirschneri serogroup Pomona serovar Mozdok. For the Varni et al. (2014) protocol, however, both strains presented a new allelic profile (7, 5, 22, 8, 7, 7, 5) which originated a new ST101 that so far has only been associated with the Brazilian L. kirschneri serogroup Pomona serovar Mozdok strains.
Considering the geographical and chronological distance between strains, since the 61H strain was isolated from human blood sample from Pelotas, in the metropolitan region of Rio Grande do Sul State, Brazil (~1,380 Km from São Paulo), in 2013 (Cunha et al. 2016), it is possible to infer that L. kirschneri serogroup Pomona serovar Mozdok has already been circulating in the southeast and southern regions of Brazil during the last two decades. It has apparently adapted to rodents as a reservoir and presents high virulent potential to humans. L. kirschneri serogroup Pomona serovar Mozdok has also been isolated from an asymptomatic dog in Pelotas (Cunha et al. 2016), suggesting that the serovar has already adapted to different reservoir hosts in urban areas.
In view of the environmental conditions and the high genomic similarity observed between strains, this may suggest that they could be a Brazilian L. kirschneri serogroup Pomona serovar Mozdok lineage that could represent a high public health risk; further studies are necessary to confirm the lineage significance and distribution. Therefore, it should be included in the Leptospira battery of tests of national reference laboratories to enable proper identification and further epidemiological studies.
Footnotes
Financial support: CNPq, CAPES, FAPESP (2011/18290-0, 2013/17136-2).
REFERENCES
- Ahmed N, Manjulata DS, Valverde M, Vijayachari P, Machangu RS, Ellis WA, et al. Multilocus sequence typing method for identification and genetic classification of pathogenic Leptospira species. 28Ann Clin Microbiol Antimicrob. 2006;5 doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. 402BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Bailey MS, Holden MT, et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. e1954PLoS Negl Trop Dis. 2013;7(1) doi: 10.1371/journal.pntd.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P, Collet L, Brisse S, Picardeau M. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int J Syst Evol Microbiol. 2014;64(Pt 12):4061–4067. doi: 10.1099/ijs.0.066597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha CEP, Felix SR, Seixas ACP, Neto, Campello-Felix A, Kremer FS, Monte LG, et al. Infection with Leptospira kirschneri serovar Mozdok: first report from the Southern Hemisphere. Am J Trop Med Hyg. 2016;94(3):519–521. doi: 10.4269/ajtmh.15-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6): doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikken H, Kmety E. Serological typing methods of leptospires. In: Bergan T, Norris JR, editors, editors. Methods in microbiology. New York: Academic Press; 1978. pp. 268–307. [Google Scholar]
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majetic ZS, Galloway R, Sabljic ER, Milas Z, Perko VM, Habus J, et al. Epizootiological survey of small mammals as Leptospira spp. reservoirs in Eastern Croatia. Acta Trop. 2014;131:111–116. doi: 10.1016/j.actatropica.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzawa T, Okamoto Y, Une Y, Takeuchi T, Tsukagoshi K, Koizumi N, et al. Leptospirosis in squirrels imported from United States to Japan. Emerg Infect Dis. 2006;12(7):1153–1155. doi: 10.3201/eid1207.060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obregón AM, Fernández C, Rodríguez I, Rodríguez J. The application of monoclonal antibody methodology as a tool for serotyping Leptospira isolates in Cuba. Rev Cubana Med Trop. 2007;59(1):68–70. [PubMed] [Google Scholar]
- Renaud C, Andrews S, Djelouadji Z, Lecheval S, Corrao-Revol N, Buff S, et al. Prevalence of the Leptospira serovars Bratislava, Grippotyphosa, Mozdok and Pomona in French dogs. Vet J. 2013;196(1):126–127. doi: 10.1016/j.tvjl.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Varni V, Ruybal P, Lauthier JJ, Tomasini N, Brihuega B, Koval A, et al. Reassessment of MLST schemes for Leptospira spp. typing worldwide. Infect Genet Evol. 2014;22:216–222. doi: 10.1016/j.meegid.2013.08.002. [DOI] [PubMed] [Google Scholar]


