Strong changes in the light-dark regime negatively affect the circadian clock, causing circadian stress, which leads to jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants.
Abstract
The circadian clock helps plants measure daylength and adapt to changes in the day-night rhythm. We found that changes in the light-dark regime triggered stress responses, eventually leading to cell death, in leaves of Arabidopsis thaliana plants with reduced cytokinin levels or defective cytokinin signaling. Prolonged light treatment followed by a dark period induced stress and cell death marker genes while reducing photosynthetic efficiency. This response, called circadian stress, is also characterized by altered expression of clock and clock output genes. In particular, this treatment strongly reduced the expression of CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). Intriguingly, similar changes in gene expression and cell death were observed in clock mutants lacking proper CCA1 and LHY function. Circadian stress caused strong changes in reactive oxygen species- and jasmonic acid (JA)-related gene expression. The activation of the JA pathway, involving the accumulation of JA metabolites, was crucial for the induction of cell death, since the cell death phenotype was strongly reduced in the jasmonate resistant1 mutant background. We propose that adaptation to circadian stress regimes requires a normal cytokinin status which, acting primarily through the AHK3 receptor, supports circadian clock function to guard against the detrimental effects of circadian stress.
INTRODUCTION
The circadian clock, an intrinsic timekeeping mechanism, synchronizes with the periodic environment through daily entrainment, especially by light and temperature, to adjust the internal rhythm accordingly (McClung, 2011). Once synchronized with the environment, the circadian clock can anticipate daily environmental fluctuations, thus coordinating diverse physiological and developmental processes in a time-of-day-specific manner, which enhances plant fitness and survival (Green et al., 2002; Michael et al., 2003; Dodd et al., 2005; Yerushalmi et al., 2011). The circadian clock also plays an important role under adverse environmental conditions, modulating biotic and abiotic stress responses (Roden and Ingle, 2009; Sanchez et al., 2011; Seo and Más, 2015).
The circadian clock of Arabidopsis thaliana is composed of multiple components acting in interlocking transcription-translation feedback loops, which form a critical part of the oscillatory mechanism (Carré and Veflingstad, 2013; Hsu and Harmer, 2014). Two single MYB-domain transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), are expressed in the morning (Wang and Tobin, 1998; Schaffer et al., 1998) and repress the expression of the evening component TIMING OF CAB EXPRESSION1 (TOC1), also called PSEUDO RESPONSE REGULATOR1 (PRR1) (Strayer et al., 2000; Alabadí et al., 2001). In turn, TOC1 represses the transcription of CCA1 and LHY (Gendron et al., 2012; Huang et al., 2012). Furthermore, other members of the PRR family, PRR9, PRR7, and PRR5, which are expressed in consecutive waves throughout the day, also repress CCA1 and LHY expression (Nakamichi et al., 2010), while CCA1 and LHY promote PRR9 and PRR7 expression in the morning (Farré et al., 2005). The evening complex (EC), composed of EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX), represses the expression of PRR9 (Helfer et al., 2011; Nusinow et al., 2011). The EC components are crucial for high-amplitude diurnal and circadian rhythms of both CCA1 and LHY (Doyle et al., 2002; Hazen et al., 2005; Kolmos et al., 2009; Dixon et al., 2011), which is thought to be achieved indirectly through PRR9 repression and, hence, CCA1 and LHY derepression (Pokhilko et al., 2012).
The reciprocal regulation of clock genes ensures proper time-of-day-specific expression of each clock component within this circuit, as well as correct time-of-day-specific expression of clock-regulated genes, the clock output genes. Transcriptional regulation is an important means for circadian output control, since genes in many fundamental biological pathways are clock-regulated (Harmer et al., 2000; Covington et al., 2008). Interestingly, a large proportion of clock-controlled genes (68%) are linked to stress regulation, including genes with roles under cold, salt, and osmotic stress (Kreps et al., 2002). A general mechanism to control certain clock outputs is to modulate the responsiveness to intrinsic or extrinsic stimuli in a time-of-day-specific manner. This mechanism, called gating, is important for normal plant physiology and development as well as responses to environmental cues (Hotta et al., 2007; Seo and Más, 2015). One additional way in which the circadian clock influences a large number of biological processes is by modulating hormonal pathways (Robertson et al., 2009), which is well characterized for auxin (Rawat et al., 2009) and abscisic acid (ABA) signaling (Legnaioli et al., 2009).
Cytokinin is another plant hormone that regulates numerous physiological and developmental processes, including cell division, shoot and root growth, and leaf senescence (Werner and Schmülling, 2009; Kieber and Schaller, 2014). Recently, cytokinin was also shown to play various roles in the response to adverse environmental conditions (Ha et al., 2012; O’Brien and Benková, 2013). The cytokinin signal is transduced by a multistep phosphorelay mechanism, which shares commonalities with the bacterial two-component system (Hwang et al., 2002). In Arabidopsis, the cytokinin signal is perceived by three different receptors, ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1 (CRE1)/AHK4 (Inoue et al., 2001; Suzuki et al., 2001), which have distinct properties (Heyl et al., 2012). The signal, in the form of a phosphoryl group, is further transmitted via HISTIDINE PHOSPHOTRANSFER PROTEINs to B-type ARABIDOPSIS RESPONSE REGULATORs (ARRs), which, once activated by phosphorylation, act as transcription factors to regulate cytokinin response genes (Heyl and Schmülling, 2003; Kieber and Schaller, 2014). The cytokinin signal also depends on the level of cytokinins present, which is to a large extent dependent on their biosynthesis and degradation. The rate-limiting step of cytokinin biosynthesis is catalyzed by isopentenyltransferases (IPTs) (Kakimoto, 2003) and their degradation by cytokinin oxidases/dehydrogenases (CKXs) (Werner et al., 2006).
The plant hormone jasmonic acid (JA) also regulates plant growth and development as well as responses to biotic and abiotic stresses. JA plays a key role in plant defenses against herbivores and necrotrophic pathogens as well as in the regulation of wound responses (Wasternack and Hause, 2013). In the presence of the biologically active form of JA, jasmonoyl-isoleucine (JA-Ile), the F-box protein CORONATINE-INSENSITIVE1 (COI1) (Xie et al., 1998) and JASMONATE ZIM DOMAIN (JAZ) transcriptional repressor proteins (Chini et al., 2007; Thines et al., 2007) associate and form a coreceptor complex (Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). This results in the degradation of JAZ repressors by the ubiquitin-proteasome system (Chini et al., 2007; Thines et al., 2007), which releases transcription factors such as MYC2 and hence enables the transcription of JA response genes, including MYC2 itself, JAZ genes, and basically all JA biosynthesis genes (e.g., LIPOXYGENASE [LOX] genes and OXOPHYTODIENOATE-REDUCTASE3 [OPR3]) (Thines et al., 2007; Chung et al., 2008; Wasternack and Hause, 2013).
Programmed cell death (PCD) is an essential feature of plant growth and development. For example, its strict regulation is important during embryogenesis, tracheary element differentiation, and leaf senescence. However, PCD also occurs in response to environmental cues such as mechanical damage, abiotic stress, and pathogen attack and becomes visible as lesions (Jones, 2001; Lam, 2004; Coll et al., 2011). Known regulators of cell death in plants include BAX INHIBITOR1 (BI1), metacaspases such as MCP2D, reactive oxygen species (ROS), and plant hormones (Overmyer et al., 2003; Love et al., 2008; Ishikawa et al., 2011; Tsiatsiani et al., 2011; Kim et al., 2012; De Pinto et al., 2012).
Several studies point to an interplay between circadian timekeeping and cytokinin. For example, the circadian clock regulates 38 and 45%, respectively, of genes usually upregulated and downregulated by cytokinin (Covington et al., 2008). Clock mutants affected in CCA1 and/or LHY function show altered cytokinin responses in root elongation, hypocotyl growth, and tissue culture (Zheng et al., 2006). Cytokinin causes phase delays of 1 to 3 h in different circadian rhythms (Hanano et al., 2006; Zheng et al., 2006); however, circadian periodicity is not very strongly affected by cytokinin (Hanano et al., 2006; Salomé et al., 2006; Zheng et al., 2006). In general, these reports suggest that the influence of cytokinin on clock function is rather moderate. Here, we describe a type of abiotic stress, which we coined circadian stress. Specific changes in the light-dark regime negatively affected the circadian clock and caused strong JA-dependent cell death in cytokinin-deficient plants as well as in several clock mutants. These results indicate that cytokinin is required for proper clock function under these conditions, thereby preventing disproportionate stress responses and cell death. We hypothesize that cytokinin supports clock function, which is especially relevant under unfavorable conditions that potentially perturb clock function and thus negatively affect plant performance.
RESULTS
Plants with a Reduced Cytokinin Status Are Sensitive to Changes in the Light-Dark Regime
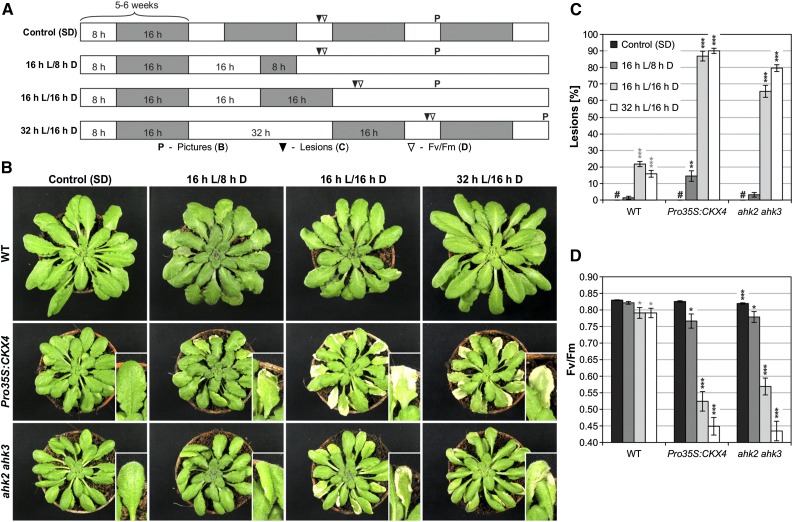
Plants with a reduced cytokinin status, i.e., Pro35S:CKX4 transgenic plants and ahk2 ahk3 receptor mutants, showed a stress phenotype upon transfer from short-day (SD; control) to long-day (LD) conditions (Figure 1A; 16 h L/8 h D). This phenotype was characterized by the formation of lesions, indicating the induction of PCD, especially in Pro35S:CKX4 plants (Figures 1B and 1C), and by a decrease in photosystem II maximum quantum efficiency (Fv/Fm), which was used as a general stress marker, in both Pro35S:CKX4 and ahk2 ahk3 plants (Figure 1D). Wild-type plants remained unaffected by the altered light regime. Additional light-dark regimes with a longer dark period (Figure 1A; 16 h L/16 h D) or longer light and dark periods (SD/32 h L/16 h D) revealed that a prolongation of the dark period with or without further extension of the light period substantially enhanced the severity of the phenotype. The percentage of mature leaves (defined as fully expanded leaves) with lesions increased up to 90% in Pro35S:CKX4 and up to 80% in ahk2 ahk3 plants (Figures 1B and 1C) and accordingly the Fv/Fm values strongly declined (Figure 1D). In contrast, the wild type was only marginally affected under these conditions. These results showed that both an extended light treatment and the length of the following long dark period influenced the severity of the stress response. For further analyses, the 32 h L/16 h D regime was used as standard stress regime, referred to as continuous light (CL) regime or CL treatment, respectively.
Figure 1.
Changes in the Light-Dark Regime Lead to Cell Death in Cytokinin-Deficient Plants.
(A) Schematic overview of light-dark regimes. Plants were grown under SD conditions for 5 to 6 weeks prior to the exposure to an altered light-dark regime. The regime shown in the lowest row was chosen as the standard stress regime (CL regime). White, light period (L); gray, dark period (D). “P” and triangles indicate time points of sampling for (B) to (D).
(B) Phenotypes of representative wild-type and cytokinin-deficient plants after respective treatments. Pictures were taken at the time points indicated in (A). Insets show representative examples of lesions.
(C) Percentage of mature leaves with lesions (n = 10; #, not detected) at the time points indicated in (A).
(D) PSII maximum quantum efficiency (Fv/Fm) in representative leaves (n = 12) at the time points indicated in (A).
Data are mean values ± se. Asterisks indicate significant differences from the respective wild type (black) and the corresponding control (gray, for wild type only). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
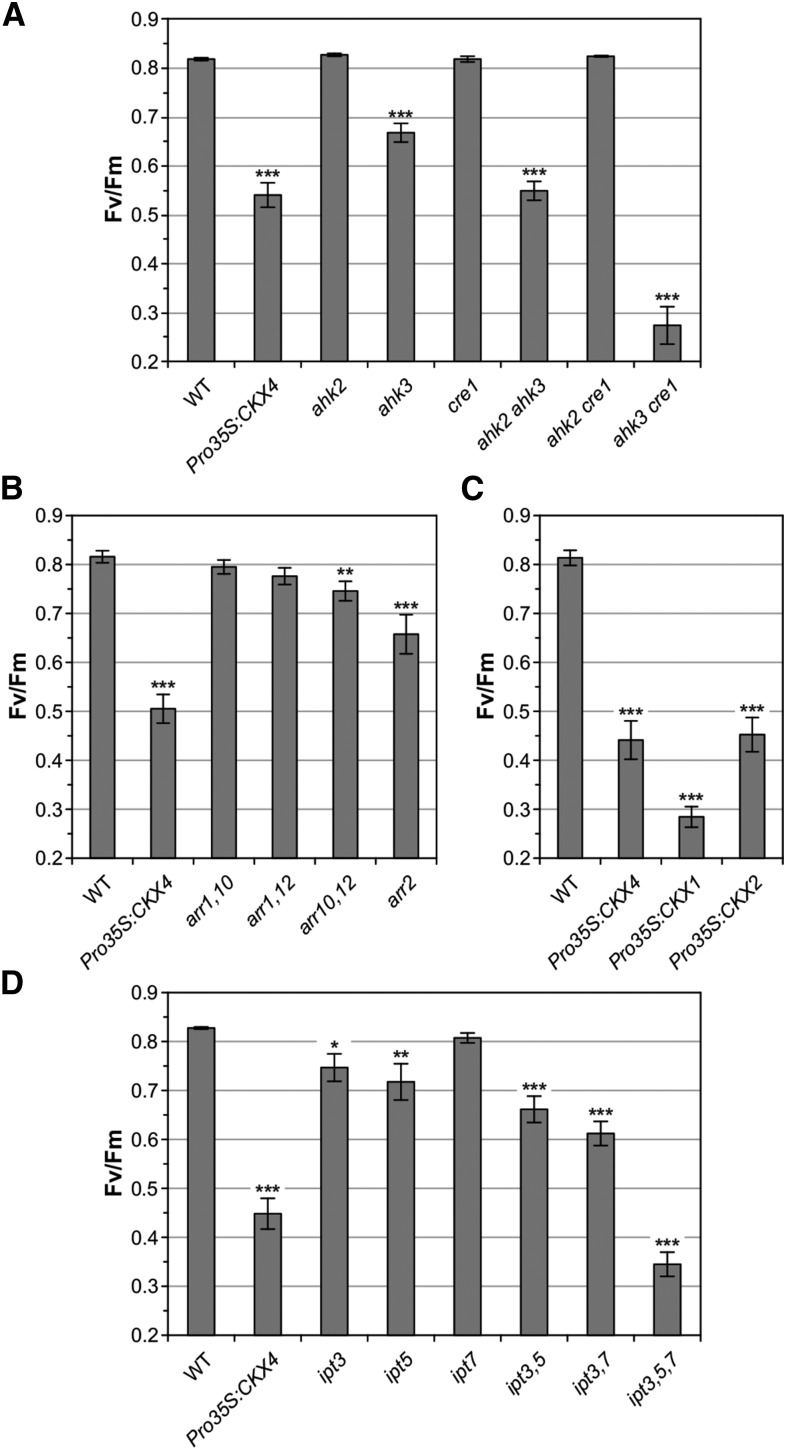
To further evaluate the hypothesis that cytokinin plays a role in the proper response to changing light-dark regimes, more cytokinin signaling mutants and plants with a lowered cytokinin status, respectively, were tested. Throughout this study, control plants that remained under SD conditions were always included in the experiments and were completely unaffected, showing no lesions and no decrease in Fv/Fm values (Fv/Fm > 0.8). Analysis of cytokinin receptor single mutants revealed that the AHK3 receptor is the key mediator in this adaptive response, since only AHK3 loss of function resulted in a stress phenotype after CL treatment (Figure 2A; Supplemental Figures 1A and 1B). However, additional mutation of AHK2 and especially of CRE1/AHK4 enhanced the stress response of ahk3 plants, indicating that both AHK2 and CRE1/AHK4 play accessory roles. Next, the response of B-type arr mutants to the CL regime was tested. The results showed that ARR2 and to a lesser extent the combined action of ARR10/ARR12 are important under these conditions, as reflected by the decline of Fv/Fm (Figure 2B) and the development of lesions (Supplemental Figures 1C and 1D). However, the overall response to the CL regime was moderate in these plants compared with Pro35S:CKX4 plants. Testing different mutants with a reduced cytokinin content confirmed the relevance of the endogenous cytokinin level for this stress response. In addition to Pro35:CKX4 plants, CKX1- and CKX2-overexpressing plants were also strongly affected by the CL regime (Figure 2C; Supplemental Figures 1E and 1F). Analysis of ipt single, double, and triple mutants carrying mutations in IPT3, IPT5, and/or IPT7 revealed that the degree of stress following CL treatment increased with the number of IPT gene mutations, resulting in the strongest stress phenotype in ipt3,5,7 mutants (Figure 2D; Supplemental Figures 1G and 1H). Taken together, these results point to a role for cytokinin in the adaptive response to changes in the light-dark regime and to the redundant roles of several cytokinin biosynthesis and signaling genes in preventing stress and cell death under these conditions.
Figure 2.
Cytokinin Signaling Mutants and Plants with a Reduced Cytokinin Content Show a Stress Phenotype upon CL Treatment.
Stress-induced decrease of Fv/Fm measured 1 d after CL treatment in cytokinin single and double receptor mutants (A), B-type arr mutants (B), plants overexpressing different CKX genes (C), and ipt mutants defective in cytokinin biosynthesis (D). Pro35S:CKX4 transgenic plants served as a positive control in all experiments. Control plants of all genotypes remained continuously in the SD rhythm and were not affected. Data are mean values ± se (n = 10 to 20). Asterisks indicate significant differences from the wild type (black). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
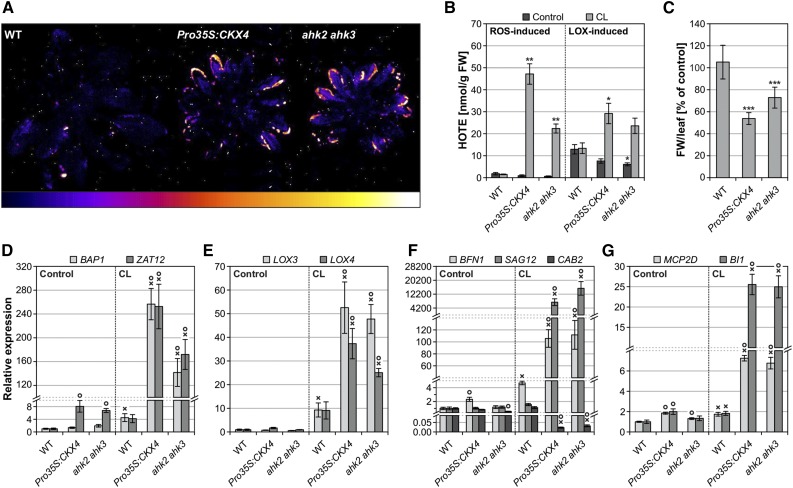
Lipid Peroxidation, Water-Soaked Lesions, and Strong Changes in the Expression of Stress- and Cell Death-Associated Genes Are Part of the Stress Response in Cytokinin-Deficient Plants after CL Treatment
Cell death progression in cytokinin-deficient plants following CL treatment was accompanied by increased lipid peroxidation (LPO), which was visualized as autoluminescence and indicates increased oxidative stress (Figure 3A). Quantification of hydroxyoctadecatrienoic acid (HOTE) isomers showed that LPO in cytokinin-deficient plants was induced by both ROS and LOXs, ROS-induced LPO being more pronounced (Figure 3B). The severe stress in cytokinin-deficient plants was also hallmarked by leaf limpness, as reflected by a strong loss of fresh weight to 55 or 70% of control levels in Pro35S:CKX4 and ahk2 ahk3, respectively (Figure 3C). In line with the phenotypically visible cell death progression, many stress- and cell death-related genes were strongly induced in cytokinin-deficient plants 1 d after CL treatment. In accordance with the increased LPO, the expression of oxidative stress marker genes BAP1 and ZAT12 as well as lipoxygenase genes LOX3 and LOX4 increased between 25- and 250-fold in these plants. For comparison, the transcript levels of these genes increased only between 5- and 10-fold in wild-type plants (Figures 3D and 3E). Furthermore, senescence marker genes exhibited strongly increased (BFN1 and SAG12) or reduced (CAB2) expression, respectively, in Pro35S:CKX4 and ahk2 ahk3 (Figure 3F). Also, cell death marker genes MCP2D and BI1 were upregulated exclusively in CL-treated cytokinin-deficient plants (Figure 3G). Taken together, a number of cellular and molecular features characterize the stress response to an altered light regime in cytokinin-deficient plants.
Figure 3.
Cell Death Progression in Cytokinin-Deficient Plants Is Accompanied by Increased Oxidative Stress, Leaf Limpness, and Induction of Stress and Cell Death Marker Genes.
(A) Autoluminescence imaging of lipid peroxidation 1 d after CL treatment revealing increased oxidative stress in cytokinin-deficient plants. Color scale indicates signal intensity from 0 (dark blue) to saturation (white).
(B) ROS- and LOX-induced lipid peroxidation 1 d after CL treatment (n = 4).
(C) Fresh weight (FW) of leaves analyzed 1 d after CL treatment. Decreased fresh weight reflects limpness of leaves (n = 10).
(D) to (G) Transcript levels of oxidative stress marker genes (D), lipoxygenase genes (E), senescence-associated genes (F), and cell death marker genes (G) 24 h after CL treatment of six biological replicates, where each replicate comprises a pool of leaves from two to three plants. For SAG12, no transcripts could be detected in the controls and in CL-treated wild type (F). Therefore, a threshold cycle (Ct) of 40 was assumed in order to calculate relative expression values for CL-treated Pro35S:CKX4 and ahk2 ahk3. Expression levels were normalized to the respective wild-type control, which was set to 1.
Data are mean values ± se. Asterisks in (B) and (C) indicate significant differences from the respective wild type. *P < 0.05, **P < 0.01, and ***P < 0.001 (t test). Symbols in (D) to (G) indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O).
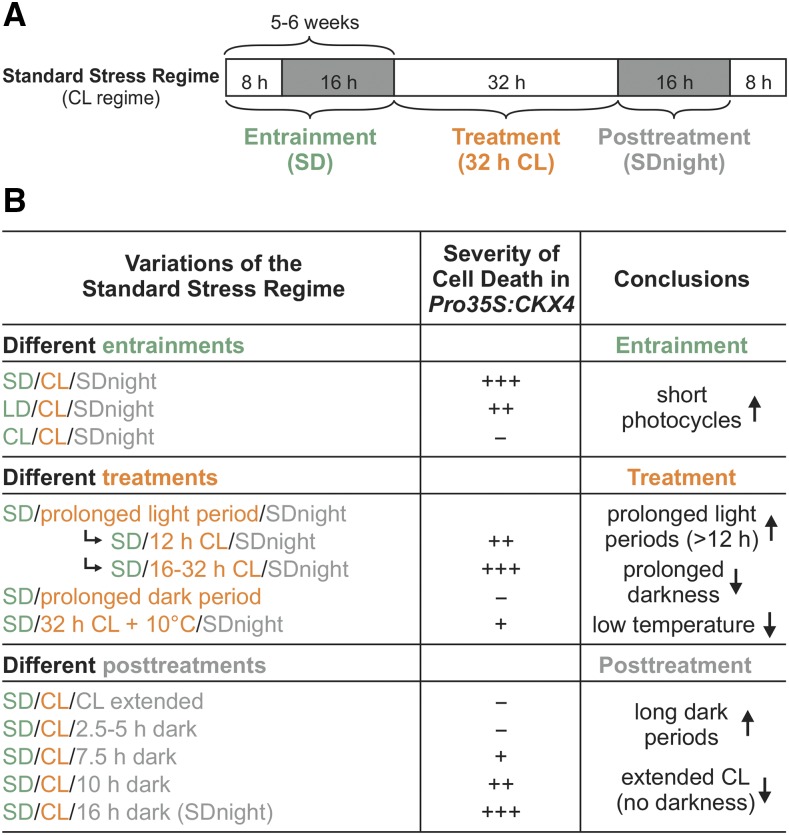
The Cell Death Phenotype in Cytokinin-Deficient Plants Is Modulated by the Interplay of Three Different Factors: Entrainment, Treatment, and Posttreatment Regime
To investigate in more detail which parameters of the light-dark regime are causally involved in provoking the stress and cell death response in plants with a lowered cytokinin status, we analyzed how alterations of entrainment, treatment, or posttreatment, respectively, influence the severity of the cell death phenotype. For comparison, Figure 4A shows the standard stress regime (CL regime) used in this study, which was composed of SD entrainment, 32 h CL treatment, followed by 16 h darkness (SD night), respectively (Figure 4A). This treatment is named SD/CL/SDnight in Figure 4B.
Figure 4.
Entrainment, Treatment, and Posttreatment Regimes Determine the Severity of the Circadian Stress Phenotype in Cytokinin-Deficient Plants.
(A) Scheme of the standard stress regime including the 32-h CL treatment. White, light period; gray, dark period.
(B) Severity of cell death in Pro35S:CKX4 plants induced by variations of the standard stress regime with changes in entrainment (green), treatment (orange), or posttreatment (gray), as indicated in (A). −, No lesions. +, ≤30%; ++, 31 to 50%; and +++, >50% of mature leaves with lesions. Arrow up/arrow down, factors and conditions promoting or noninducing/reducing the cell death phenotype, respectively.
See Supplemental Figures 2 to 4 for corresponding data.
Figure 4B summarizes the results of the experiments for Pro35S:CKX4 plants; for the corresponding data for wild type and ahk2 ahk3 plants, see Supplemental Figures 2 to 4. Solely changing the entrainment regime revealed that short photocycles (SD) prior to CL treatment caused the most severe cell death phenotype, whereas LD or CL entrainment resulted in an intermediate or no response, respectively, in cytokinin-deficient plants (Figure 4B; Supplemental Figure 2).
Light treatments of different lengths (12 to 32 h) following SD entrainment confirmed the finding reported above (Figure 1) that prolonged light treatment is important for the severity of the stress response. A comparable strong cell death phenotype was caused by light periods of 16 to 32 h, irrespective of the exact duration, while a rather weak cell death phenotype was caused by 12 h of light, revealing a 4-h prolongation of light treatment as necessary but not fully effective (Figure 4B; Supplemental Figures 3A to 3C). In contrast, prolonged dark treatment following SD entrainment without a prolonged light treatment was not capable of inducing cell death, confirming that the prolonged light treatment is a prerequisite for the phenotype (Figure 4B; Supplemental Figures 3D and 3E). Interestingly, the CL-dependent phenotype in cytokinin-deficient plants was strongly reversed when a temperature cycle was applied by lowering the temperature to 10°C during the subjective dark period under CL treatment (Figure 4B Supplemental Figures 3F to 3H). This shows that low temperature was protective during CL treatment, in contrast to the stress-enhancing effect of low temperature during light stress (Murata et al., 2007).
Lastly, different posttreatments showed that the dark period following CL treatment was equally important for the development of a stress phenotype. An extended light treatment alone did not lead to a stress phenotype, but a dark period following an extended light period was an absolute requirement for inducing cell death (Figure 4B; Supplemental Figures 4A and 4B). Strikingly, the severity of cell death gradually increased with increasing night length (Figure 4B; Supplemental Figures 4C to 4E).
In conclusion, a combination of short photocycles (entrainment), prolonged light periods (treatment), and long dark periods (posttreatment) triggered the strongest stress response in cytokinin-deficient plants. This shows that the stressor was not the prolonged light treatment alone but the overall change in the light-dark regime. The circadian clock is important for daylength measurement and, hence, for the adaptation to seasonal changes in the day-night rhythm (Imaizumi, 2010; Song et al., 2013). Therefore, we hypothesized that a perturbation of the circadian clock might be causative for the stress phenotype in response to the altered light-dark regime; therefore, we tentatively termed this phenomenon circadian stress.
Stress Responses and Subsequent Cell Death in Cytokinin-Deficient Plants upon Circadian Stress Are Initiated during the Dark Period Following CL Treatment
As summarized in Figure 4B, the dark period following CL treatment was crucial for the induction of cell death. Accordingly, increased ion leakage in cytokinin-deficient plants was already detectable during the dark phase (at 13 h) and was even more pronounced at its end (at 16 h; Figure 5A). To further trace the beginning of the stress response, the kinetics of stress and cell death marker gene expression was measured by qRT-PCR during the dark period following CL treatment (experimental scheme shown in Figure 5B). The oxidative stress marker genes BAP1 and ZAT12 were already strongly upregulated in cytokinin-deficient plants at 5 h after CL treatment (Figures 5C and 5D). In the wild type, only a weak induction of these genes was detected, which is in accordance with its weak cell death phenotype following the standard CL regime (Figure 1C). In cytokinin-deficient plants, upregulation of the cell death marker gene BI1 started a bit later at 7.5 h, while the wild-type BI1 transcript levels remained almost unchanged (Figure 5E). The induction of oxidative stress and cell death marker genes occurred only in mature (affected) leaves, while they were expressed at control levels in young (unaffected) leaves, suggesting that that the changes in gene expression are causally linked to the cell death response (Supplemental Figure 5). Taken together, circadian stress-induced changes in cytokinin-deficient plants were initiated as early as 5 h after onset of darkness following CL treatment. The first visible symptoms, in the form of leaf limpness, were detected ∼5 h later, as indicated in Figure 5B.
Figure 5.
Cell Death in Cytokinin-Deficient Plants Is Initiated during the Dark Period Following CL Treatment, as Indicated by Increased Ion Leakage and Strong Induction of Stress- and Cell Death-Associated Genes.
(A) Ion leakage indicating loss of membrane integrity due to cell death progression at different time points after CL treatment. Data are mean values ± se (n = 4). Asterisks indicate significant differences from respective wild types. **P < 0.01 and ***P < 0.001 (t test).
(B) Experimental scheme for (C) to (E). Prior to the experiment, plants were SD-entrained for 6 weeks. Leaf samples were collected in 2.5-h intervals starting at the end of a normal SD light period (controls) and the standard CL treatment, respectively. White, light period; gray, dark period; first symptoms, leaves start to go limp in cytokinin-deficient plants.
(C) to (E) Transcript levels of oxidative stress marker genes BAP1 (C), ZAT12 (D), and cell death marker gene BI1 (E) at the time points indicated in (B). Expression levels were normalized to the 0 h wild-type control, which was set to 1. Data are mean values of four biological replicates ± se. Symbols indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O).
Changes in Clock and Clock Output Gene Expression Indicate a Perturbation of the Circadian Clock in Cytokinin-Deficient Plants under Circadian Stress
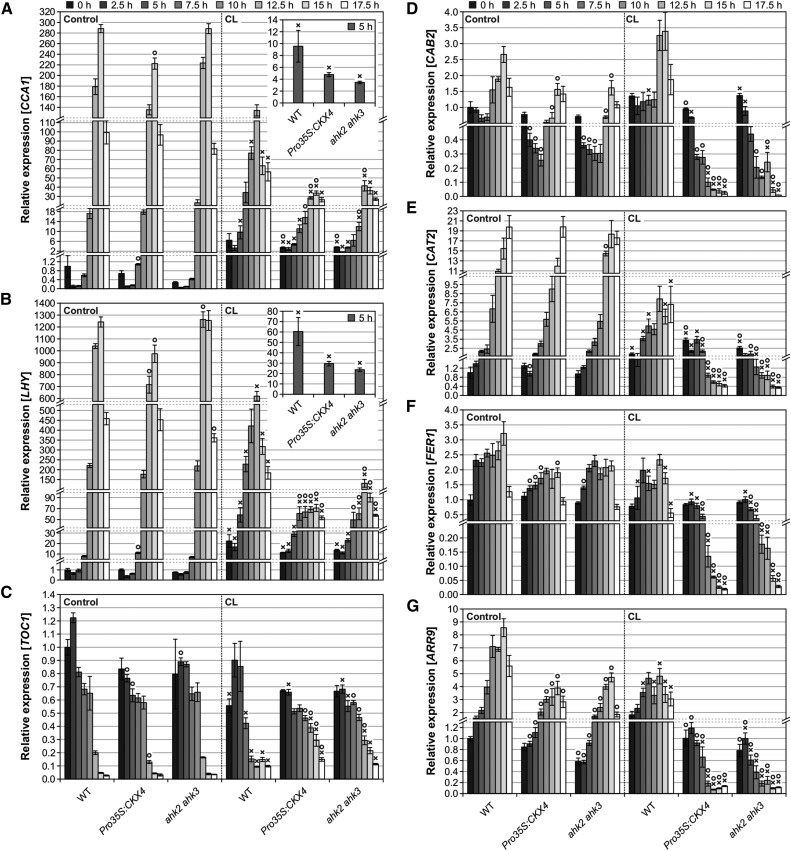
To find out whether the circadian stress regime indeed affects the performance of the circadian clock, the setup shown in Figure 5B was also used to analyze transcript levels of clock and clock output genes, respectively. Strikingly, CCA1 and LHY clock gene expression was strongly affected by the CL regime (Figures 6A and 6B). All genotypes exhibited robust oscillations of CCA1 and LHY gene expression under control conditions, showing a ≥2000-fold difference between the lowest and highest expression level for both genes. Under CL conditions, this maximum fold change was reduced in the wild type (to ∼40× for both CCA1 and LHY) and even more strongly reduced in Pro35S:CKX4 (11× for CCA1; 6× for LHY) and ahk2 ahk3 (25× for CCA1; 12× for LHY) plants. One reason for the reduced amplitudes was the higher expression of both genes compared with the controls directly after CL treatment (0 h) and during the early night (2.5 h to 7.5 h). However, this change was similar in all genotypes. Another reason was the reduced peak expression. Compared with the respective control, maximal expression after CL treatment was only reduced by about 2-fold in the wild type, while it was much more strongly reduced in Pro35S:CKX4 (7× for CCA1; 14× for LHY) and ahk2 ahk3 (7× for CCA1; 10× for LHY) plants. Hence, the CL regime negatively affected the expression of the clock genes CCA1 and LHY. This effect was particularly strong in cytokinin-deficient plants, which did not even exhibit distinct morning peak expression of CCA1 and LHY. Interestingly, the divergence between wild-type and cytokinin-deficient plants already started 5 h after CL treatment (Figures 6A and 6B, insets), coinciding with the induction of BAP1 and ZAT12 (Figures 5C and 5D).
Figure 6.
Circadian Clock Gene and Clock Output Gene Expression during the Dark Period Following CL Treatment.
Transcript abundances of circadian clock genes CCA1 (A), LHY (B), and TOC1 (C) as well as clock output genes CAB2 (D), CAT2 (E), FER1 (F), and ARR9 (G) at different time points during the dark period following CL treatment. The experimental design is described in Figure 5B. Expression levels were normalized to the 0 h wild-type control, which was set to 1. Data are mean values of four biological replicates ± se. Symbols indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O). Insets in (A) and (B) show CCA1 and LHY expression levels, respectively, at 5 h after CL on a different scale, indicating that the divergence between wild-type and cytokinin-deficient plants had already started at this time point.
The expression of the clock gene TOC1 is usually repressed by CCA1 and LHY in the morning (Alabadí et al., 2001). Consistent with the lower expression of CCA1 and LHY, TOC1 transcript levels increased in cytokinin-deficient plants starting at 10 h after CL treatment (Figure 6C). As summarized in Figure 4B, short nights following CL treatment did not induce cell death, while the severity of cell death gradually increased with increasing night length. Accordingly, CCA1, LHY, and TOC1 exhibited control-like or even slightly higher expression levels after short nights, whereas reduced (CCA1/LHY) or elevated (TOC1) expression levels were detected following intermediate and long nights, respectively (Supplemental Figure 6). Therefore, the degree of altered expression of CCA1, LHY, and TOC1 correlated well with the severity of the cell death phenotype in cytokinin-deficient plants.
Next, the expression pattern of the clock-regulated genes CAB2, CATALASE2 (CAT2), FERRETIN1 (FER1), and ARR9 (Millar and Kay, 1991; Zhong and McClung, 1996; Ishida et al., 2008a; Hong et al., 2013) was analyzed to determine potential changes in clock output. The transcript levels of these genes were especially strongly reduced in cytokinin-deficient plants in the second half of the 16-h night following CL treatment starting after 7.5 to 10 h of darkness (Figures 6D to 6G), which is consistent with their compromised clock gene expression, indicating perturbed clock function.
In conclusion, the CL regime strongly affected clock and clock output gene expression in cytokinin-deficient plants, corroborating the circadian stress hypothesis. CCA1 and LHY expression was most strongly altered by the circadian stress regime and reduced CCA1 and LHY induction correlated with a strong cell death phenotype, suggesting an important role for CCA1 and LHY.
Clock Mutants Are Also Sensitive to Circadian Stress Regimes
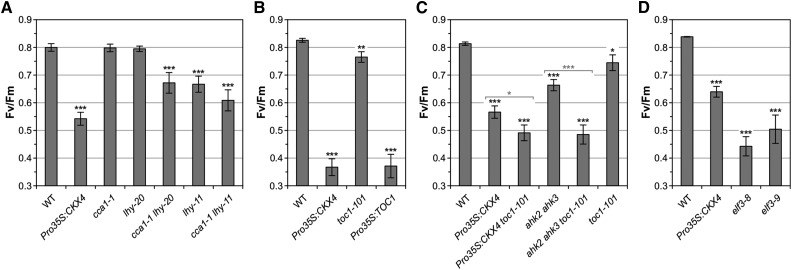
To analyze the role of specific clock components under circadian stress, we first tested the CL response of plants with reduced CCA1 and/or LHY function. Single cca1-1 and lhy-20 mutants mostly exhibited a wild-type-like stress response following the CL regime, with lhy-20 showing slightly more lesions than the wild type. In contrast, the cca1-1 lhy-20 double mutant displayed a pronounced stress and cell death phenotype (Figure 7A; Supplemental Figure 7A). This indicates functional redundancy between CCA1 and LHY in this response and supports the idea that both oscillator components are required to avoid the detrimental effects of circadian stress. A strong circadian stress phenotype was also observed in cca1-1 lhy-11, an additional double mutant tested (Figure 7A; Supplemental Figure 7A). Interestingly, the lhy-11 allele alone caused a stronger stress response than the lhy-20 allele, which could be due to the different nature of these mutations (Mizoguchi et al., 2002; Michael et al., 2003).
Figure 7.
A Defective Circadian Clock Renders Plants Sensitive to Circadian Stress.
Circadian stress-induced decrease of Fv/Fm in plants with CCA1/LHY loss of function (A), TOC1 loss of function and overexpression (B), TOC1 loss of function in the Pro35S:CKX4 and ahk2 ahk3 background, respectively (C), and elf3 mutants (D) measured 1 d after standard CL treatment. Plants were SD-entrained for 5 to 6 weeks. Data are mean values ± se (n = 11 to 20). Asterisks indicate significant differences from the wild type (black) and between cytokinin-deficient plants in the wild-type or toc1-101 background (gray, in [C] only). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test). See Supplemental Figures 7 and 8 for supporting information.
To study the contribution of TOC1, we examined the circadian response in toc1-101 and Pro35S:TOC1 plants. toc1-101 loss-of-function plants showed a weak response to the CL regime, while TOC1 overexpression resulted in a strong stress and cell death phenotype (Figure 7B; Supplemental Figure 7B). This outcome raised the question of whether the elevated expression of TOC1 in cytokinin-deficient plants (Figure 6C) contributed to their circadian stress response in addition to the reduced CCA1 and LHY expression. Therefore, we introgressed the toc1-101 allele into Pro35S:CKX4 and ahk2 ahk3, respectively, to study the role of TOC1 under circadian stress in the cytokinin-deficient background. The resulting hybrid plants showed a more pronounced circadian stress response than the parental Pro35S:CKX4 and ahk2 ahk3 lines (Figure 7C; Supplemental Figure 7C). This suggests that elevated TOC1 levels were not responsible for cell death induction in cytokinin-deficient plants.
A functional EC, comprising ELF3, ELF4, and LUX, is required to provide sufficient nighttime CCA1 and LHY expression (Doyle et al., 2002; Hazen et al., 2005; Kolmos et al., 2009; Dixon et al., 2011; Nusinow et al., 2011). Strikingly, mutation of any of the corresponding evening clock genes caused a stress and cell death phenotype following CL treatment (Figure 7D; Supplemental Figures 7D to 7F). Analysis of ELF3, ELF4, and LUX expression kinetics showed that cytokinin-deficient plants exhibited a stronger decrease in ELF3 transcript levels than wild-type plants in response to the CL regime at 5, 7.5, and 10 h (Supplemental Figures 8A to 8C). Hence, the reduced ELF3 expression temporally correlated with the lack of CCA1 and LHY induction in cytokinin-deficient plants, indicating that limited ELF3 expression during the night following CL treatment might be one factor limiting CCA1 and LHY expression in these plants.
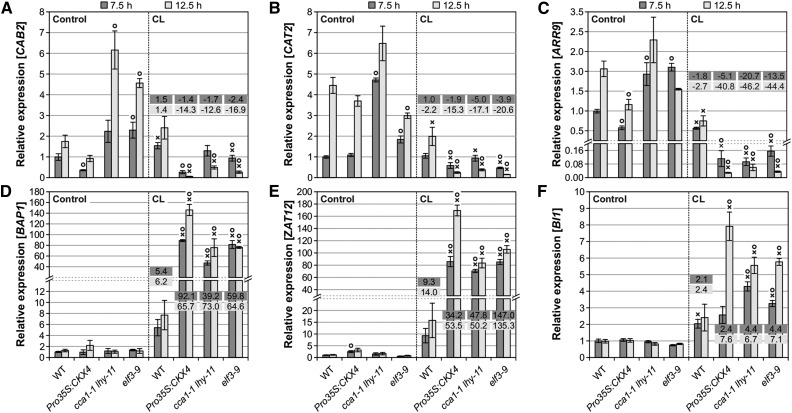
Analysis of the expression kinetics of clock output genes as well as stress and cell death marker genes after CL treatment revealed very similar changes in clock mutants (elf3-9 and cca1-1 lhy-11) compared with Pro35S:CKX4 plants (Figures 8A to 8F). This indicates that the cell death phenotype in these clock mutants is likely the consequence of similar changes at the molecular level. Interestingly, the expression of the cytokinin responsive A-type ARR genes ARR4, ARR7, ARR9, and ARR16 was not only reduced in Pro35S:CKX4 plants following CL treatment but also in the clock mutants tested (Supplemental Figure 9). This suggests that the decrease in expression results, at least in part, from the perturbed circadian clock under circadian stress and is not (or not only) due to a reduced cytokinin status, revealing a link between the clock and the regulation of these A-type ARR genes.
Figure 8.
Clock Mutants Display a Highly Similar Molecular Phenotype to That of Cytokinin-Deficient Plants in Response to Circadian Stress.
Transcript levels of clock output genes CAB2 (A), CAT2 (B), ARR9 (C), oxidative stress marker genes BAP1 (D) and ZAT12 (E), and cell death marker gene BI1 (F) during the dark period at 7.5 and 12.5 h under control conditions (left) and after CL treatment (right), respectively. Fold changes of relative expression levels (CL conditions compared with respective control conditions) are displayed in the graphs. The experimental design corresponds to the one shown in Figure 5B. Expression levels were normalized to the 7.5 h wild-type control, which was set to 1. Data are mean values of four biological replicates ± se. Symbols indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O).
Next, we analyzed the role of PRR9, PRR7, and PRR5, transcriptional repressors of CCA1 and LHY (Nakamichi et al., 2010), as well as PRR3, a positive regulator of TOC1 stability (Para et al., 2007), under circadian stress. Even higher order prr mutants such as prr9 prr7 and prr9 prr7 prr5 that exhibit distinct clock defects but very high CCA1 and LHY expression (Nakamichi et al., 2005) were not affected after CL treatment, while prr3 plants showed a cell death phenotype intermediate between Pro35S:CKX4 and the wild type (Supplemental Figure 8D). These results underline the importance of sufficient CCA1 and LHY expression, and they also indicate that PRR3 plays a protective role under circadian stress. The latter was supported by the observation that PRR3 expression was strongly reduced in cytokinin-deficient plants, also starting at 5 h after CL treatment (Supplemental Figures 8E to 8H).
Since clock mutants lacking CCA1 and LHY function also exhibited a distinct cell death phenotype after CL treatment, we conclude that the reduced CCA1 and LHY expression in cytokinin-deficient plants was likely a major cause of the circadian stress phenotype. This suggests that cytokinin supports circadian clock function, either directly or indirectly, by promoting CCA1 and LHY expression.
Circadian Stress-Induced Cell Death Is Unlikely Initiated by Increased Oxidative Stress
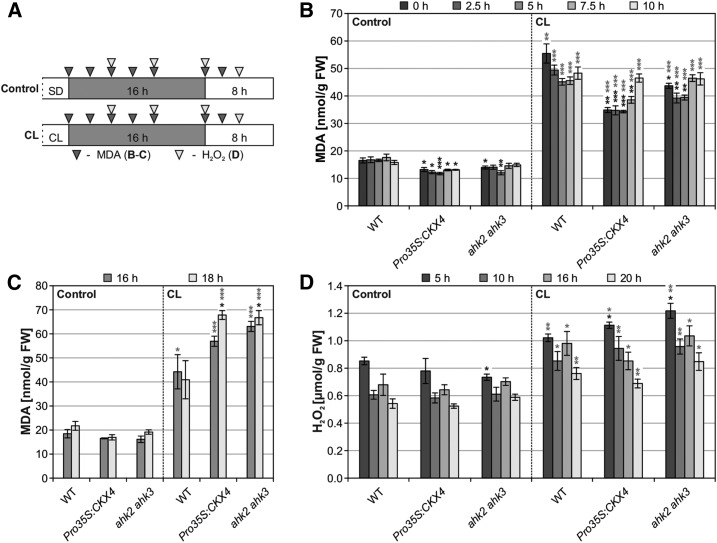
Increased oxidative stress was characteristic of late stages of cell death progression in cytokinin-deficient plants in response to CL treatment (Figures 3A, 3B, and 3D). Since oxidative stress marker genes were already induced 5 h after CL treatment (Figure 5), we measured malondialdehyde (MDA) and H2O2 levels as indicators of oxidative stress, respectively, including earlier time points (Figure 9A). These experiments revealed no increase in oxidative stress in cytokinin-deficient plants compared with the wild type directly and soon after CL treatment (Figure 9B). In contrast, MDA levels in cytokinin-deficient plants slightly exceeded wild-type levels at 16 h and even more at 18 h after CL treatment, indicating that oxidative stress occurred after cell death initiation (Figure 9C). Similarly, at 5 and 10 h after CL treatment, H2O2 levels in cytokinin-deficient plants did not strongly differ from the wild type and did not gradually increase toward 16 and 20 h after CL treatment (Figure 9D). Taken together, oxidative stress appears to be a later event that occurs after circadian stress and could not be correlated with the early induction of oxidative stress marker genes in cytokinin-deficient plants following CL treatment. Hence, the data indicate that oxidative stress accompanied cell death progression rather than being causal for cell death initiation. Moreover, the absence of strong H2O2 accumulation in cytokinin-deficient plants suggests that ROS other than H2O2 are responsible for the oxidative stress observed later during cell death progression.
Figure 9.
Oxidative Stress Is Unlikely the Reason for Cell Death Initiation in Cytokinin-Deficient Plants after CL Treatment.
(A) Experimental scheme for (B) to (D). Prior to the experiment, plants were SD-entrained for 5 to 6 weeks. Leaf samples were collected at the indicated time points (triangles). White, light period; gray, dark period.
(B) and (C) Determination of oxidative stress by quantification of MDA levels at the time points indicated in (A).
(D) H2O2 levels determined by the Amplex Red method at the time points indicated in (A).
Data are mean values ± se (n = 4). Asterisks indicate significant differences from respective wild types (black) and the corresponding controls (gray). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
Activation of the Jasmonic Acid Pathway Is One of the Consequences of Circadian Stress
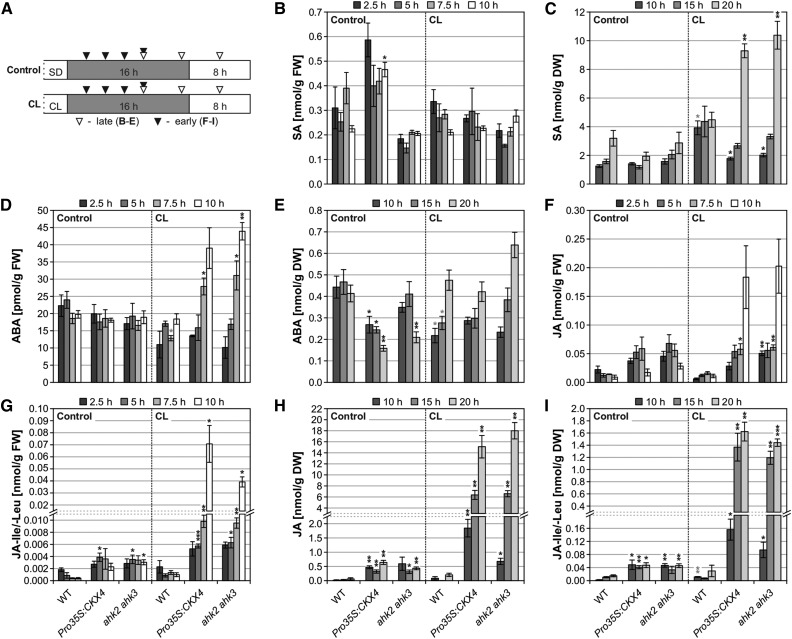
Several hormones including salicylic acid (SA), JA, and ABA are known to play important roles in various responses to stress. We therefore determined the concentrations of these hormones at different time points following CL treatment (Figure 10A). SA levels were not substantially altered and increased only late (at 20 h) following the circadian stress regime (Figures 10B and 10C). Also, ABA levels did not strongly change (Figures 10D and 10E). However, a very strong increase in JA metabolite levels occurred in cytokinin-deficient plants after CL treatment (Figures 10F to 10I). An increase in the contents of JA and its amino acid conjugates JA-Ile/-Leu was first observed 10 h after CL treatment (Figures 10F and 10G), coinciding with the appearance of the first visible symptoms in cytokinin-deficient plants at 10 h after CL treatment (Figure 5B). After 20 h, their concentrations had increased ∼30-fold (Figures 10H and 10I), and other JA metabolites including the precursor oxo-phytodienoic acid (OPDA) showed a similar pattern (Supplemental Figure 10).
Figure 10.
Cell Death Progression in Cytokinin-Deficient Plants Following the CL Regime Is Accompanied by Accumulation of JA Metabolites.
(A) Experimental scheme for (B) to (I). Plants were SD-entrained for 6 weeks. Time of sampling is indicated by triangles. White, light period; gray, dark period.
(B) to (I) Phytohormone levels measured at early (n = 3) and late (n = 4) time points shown in (A). DW, dry weight; FW, fresh weight.
Data are mean values ± se. Asterisks indicate significant differences from respective wild types (black) and the corresponding control conditions (gray; for wild type only). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
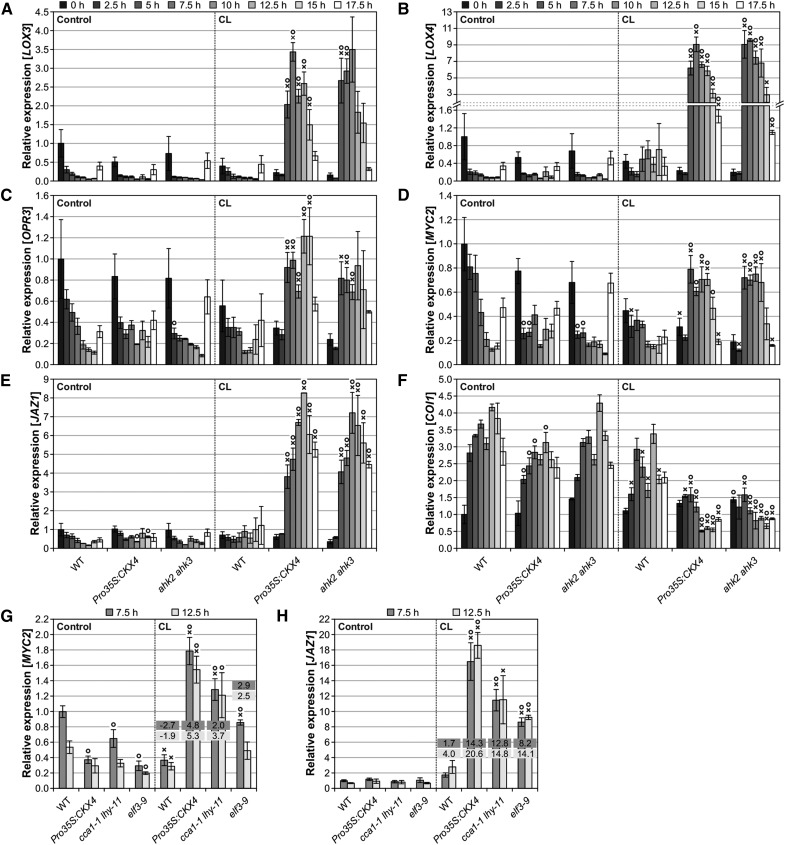
Next, we studied possible changes in expression of JA-related genes. LOX3 and LOX4, which act redundantly in JA biosynthesis to ensure male fertility (Caldelari et al., 2011), were found to be induced in cytokinin-deficient plants at a late time point after CL treatment (Figure 3E), and we therefore analyzed them in more detail together with the JA biosynthesis gene OPR3 (Figures 11A to 11C), the MYC branch marker genes MYC2 and JAZ1 (Figures 11D and 11E), and the receptor gene COI1 (Figure 11F). In wild-type plants, a diurnal expression pattern was observed for these genes, exhibiting minimal or maximal expression (in the case of COI1), respectively, during the night (Figures 11A to 11F). In contrast, in cytokinin-deficient plants, the biosynthesis genes and the JA response genes of the MYC branch were strongly induced starting at 5 h after CL treatment, whereas reduced transcript levels were found for COI1. The early induction of JA-related genes indicates that the increase in JA metabolite levels in cytokinin-deficient plants is a consequence rather than the cause of the changes in gene expression. Furthermore, the results show that circadian stress caused an activation of the JA pathway only in cytokinin-deficient plants, suggesting a repressive function of cytokinin in the wild type. Strikingly, the induction of MYC2 and JAZ1 was also detected in the clock mutants cca1-1 lhy-11 and elf3-9, indicating that a strong circadian stress response is commonly linked to an activated JA pathway (Figures 11G and 11H). These results strongly suggest that the JA pathway is functionally relevant in the response to circadian stress.
Figure 11.
JA Biosynthesis and Response Genes Are Strongly Induced in Cytokinin-Deficient Plants and Clock Mutants during the Dark Period Following CL Treatment.
(A) to (F) Transcript abundances of LOX3 (A), LOX4 (B), OPR3 (C), MYC2 (D), JAZ1 (E), and COI1 (F) in cytokinin-deficient plants compared with the wild type at different time points during the dark period under SD conditions (left) or following CL treatment (right). The experimental design is described in Figure 5B. Expression levels were normalized to the 0 h wild-type control, which was set to 1.
(G) and (H) Transcript levels of JA response genes MYC2 (G) and JAZ1 (H) in clock mutants during the dark period at 7.5 and 12.5 h under control conditions (left) and after CL treatment (right). Fold changes of relative expression levels (CL conditions compared with respective control conditions) are displayed in the graphs. The experimental design corresponds to the one described in Figure 5B. Expression levels were normalized to the 7.5 h wild-type control, which was set to 1.
Data are mean values of four biological replicates ± se. Symbols indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O).
Cell Death Development in Cytokinin-Deficient Plants upon Circadian Stress Is Promoted by JA-Dependent Signaling
To analyze the actual connection between JA and the development of circadian stress-induced cell death in more detail, we first analyzed BI1 and JAZ1 expression following different night lengths (Figures 12A and 12B). The severity of cell death in cytokinin-deficient plants gradually increased with increasing night length (Figure 4B; Supplemental Figures 4C to 4E). In line with this, BI1 and JAZ1 transcript levels were highest after 10 h of continuous darkness, while shorter dark periods did not result in an increase (2.5- and 5-h night) or caused only a smaller increase in expression (7.5-h night) (Figures 12A and 12B). Interestingly, JAZ1 transcript levels were already elevated after 5 h of darkness following CL treatment (Figure 11E), but earlier onset of light prevented further induction, leading to control-like expression levels 5 h after the restart of light treatment (see CL + 5 h D, Figure 12B). Similar to the expression of oxidative stress marker genes (Supplemental Figures 5C and 5D), JAZ1 was solely induced in mature (affected) leaves in cytokinin-deficient plants correlating with the induction of cell death (Supplemental Figure 11A).
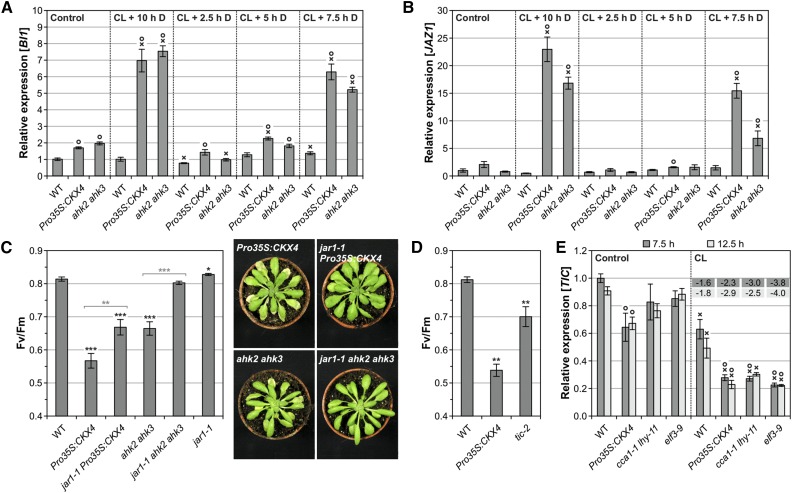
Figure 12.
Activation of the JA Pathway upon Circadian Stress Promotes Cell Death Development in Cytokinin-Deficient Plants.
(A) and (B) Transcript levels of cell death marker gene BI1 (A) and JA response gene JAZ1 (B) at 10 h after standard CL treatment following different night lengths of four biological replicates. After shorter nights (2.5, 5, and 7.5 h) plants were exposed to light until sampling. Control plants remained under SD conditions. The experimental scheme is shown in Supplemental Figure 6A. Expression levels were normalized to the wild-type control, which was set to 1.
(C) and (D) Circadian stress-induced decrease of Fv/Fm in cytokinin-deficient plants in the wild-type and jar1-1 background (C) and Pro35S:CKX4 plants in comparison with JA-hypersensitive tic-2 plants (D) 1 d after standard CL treatment (n = 15 to 16). For the setup in (C), examples of cell death phenotypes 2 d after CL treatment are shown.
(E) Transcript levels of the TIC gene in Pro35S:CKX4 and clock mutants during the dark period at 7.5 and 12.5 h under control conditions (left) and after standard CL treatment (right) of four biological replicates. Fold changes of relative expression levels (CL conditions compared with respective control conditions) are displayed in the graph. The experimental design corresponds to the one described in Figure 5B. Data are mean values ± se (n = 4). Expression levels were normalized to the 7.5 h wild-type control, which was set to 1.
Data are mean values ± se. Asterisks in (C) and (D) indicate significant differences from the wild type (black) and between cytokinin-deficient plants in the wild-type or jar1-1 background (gray, in [C] only). *P < 0.05, **P < 0.01, and ***P < 0.001 (t test). Symbols in (A), (B), and (E) indicate significant differences (P < 0.05; t test) from the corresponding control (X) and the respective wild type (O).
Genetic crosses between cytokinin-deficient plants and the JA biosynthesis mutant jar1-1 were performed to analyze the impact of the JA pathway on cell death development in response to circadian stress. Strikingly, the cell death phenotype of cytokinin-deficient plants was strongly reversed in the jar1-1 mutant background (Figure 12C; Supplemental Figure 11B). JAR1 catalyzes the last step in the formation of the biologically active JA conjugate JA-Ile; consequently, jar1-1 has a strongly reduced JA-Ile content (Suza and Staswick, 2008). Therefore, this result demonstrates that JA-Ile biosynthesis is required to promote cell death in response to circadian stress.
The clock-associated component TIME FOR COFFEE (TIC) is important for the gating of JA responses, and tic-2 is hypersensitive to JA (Shin et al., 2012). We analyzed the CL response of tic-2 mutants and found that they showed a stress and cell death response intermediate between Pro35S:CKX4 and wild-type plants (Figure 12D; Supplemental Figure 11C). This result indicates that the JA-Ile-dependent cell death phenotype in cytokinin-deficient plants might at least in part be caused by impaired gating of JA responses. Furthermore, we found that TIC expression was reduced in cytokinin-deficient plants and CL-sensitive clock mutants (Figure 12E). TIC repression was only observed in mature (affected) leaves of cytokinin-deficient plants (Supplemental Figure 11D). These data support a link between TIC and circadian stress and more specifically suggest that a deficiency in TIC expression under circadian stress might have contributed to the cell death phenotype in cytokinin-deficient plants, possibly by causing JA hypersensitivity.
DISCUSSION
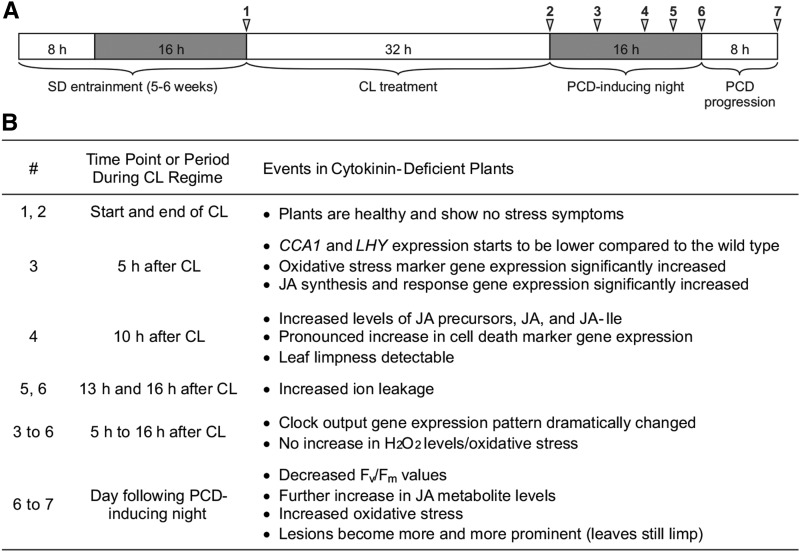
The results of this study reveal a type of abiotic stress in plants, which we coined circadian stress. Proper adaptation to changes in the light-dark regime requires a functional circadian clock and is less efficient when the robustness of the circadian system is reduced. In our study, cytokinin-deficient plants were, like some clock mutants, highly sensitive to strong changes in the light-dark regime, indicating that clock function is compromised in these plants. The model shown in Figure 13 summarizes the detrimental consequences circadian stress regimes can have, which eventually lead to the induction of PCD, and the protective role cytokinin plays under these conditions, as discussed in the following sections. In order to facilitate understanding of this phenomenon, we prepared a timeline that summarizes the most relevant events that occur in cytokinin-deficient plants under circadian stress (Figure 14).
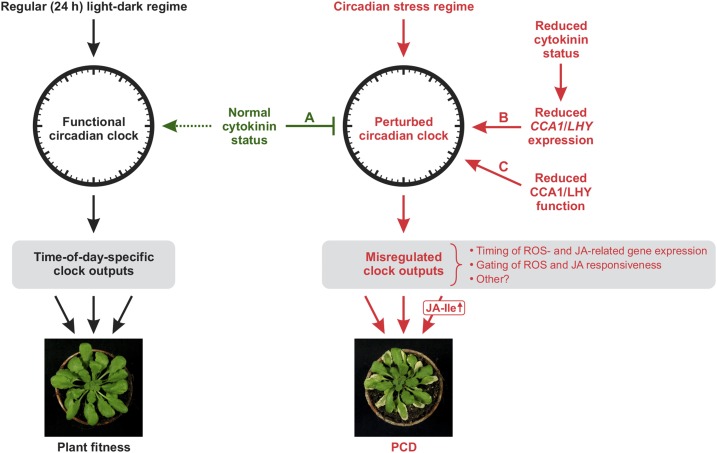
Figure 13.
Model for the Role of Cytokinin under Circadian Stress in Preventing Disproportionate Stress Responses and Cell Death.
Regular (24 h) light-dark cycles adjust circadian time on a daily basis, leading to optimal synchronization with the environment. This allows the clock to coordinate diverse physiological and developmental processes, including stress responses, in a time-of-day-specific manner, thereby enhancing plant fitness (left side). In contrast, circadian stress regimes lead to a perturbation of the circadian clock, which consequently generates altered, in part detrimental, clock outputs (right side). While a normal cytokinin status can strongly alleviate this response (A), it is particularly pronounced in plants with a reduced cytokinin status exhibiting insufficient nighttime expression of CCA1/LHY (B) and in plants with reduced CCA1/LHY function (C). In these plants, circadian stress causes disproportionate stress responses, specifically oxidative stress and JA responses, eventually leading to cell death. We showed that the activation of the JA pathway, through JA-Ile, promotes cell death development. We conclude that cytokinin, primarily by supporting nighttime CCA1 and LHY expression, sustains proper clock function under circadian stress. We propose that this function reflects an activity of the hormone in promoting clock function that is also exerted under natural favorable conditions (dashed arrow) and serves as a backup mechanism under unfavorable conditions.
Figure 14.
Timeline of Events in Cytokinin-Deficient Plants under Circadian Stress.
(A) Schematic overview of the standard stress regime. White, light period; gray, dark period.
(B) Summary of the most relevant events, occurring in cytokinin-deficient plants under circadian stress. Time points (#) are indicated in (A).
Circadian Stress Is Caused by Specific Changes in the Light-Dark Regime
The severity of PCD in cytokinin-deficient plants was determined by a specific combination of entrainment, treatment (light/temperature), and posttreatment regimes. This let us conclude that the stress phenotype is due to a perturbation of the circadian clock. Entrainment was relevant, as SD-entrained plants were more strongly affected than LD-entrained plants, showing that the stress phenotype was inversely correlated with the length of the photoperiod prior to CL treatment. In SD-entrained plants, a minimal light treatment of 12 h was necessary for inducing cell death, and the severity of cell death was positively correlated with the length of darkness following 32 h CL treatment. It is possible that prolonged light treatment let the plants adjust to longer photoperiods (long light/short dark period), which could not be reversed in cytokinin-deficient plants, making them incapable of coping with long dark periods. As a consequence, they exhibited reduced CCA1/LHY expression and the induction of cell death. In contrast, no PCD was observed after short nights. In this case, CCA1/LHY expression was similar to wild-type levels or even higher (Supplemental Figure 6). These observations could be explained by the earlier onset of light following short nights. Since light is a very strong input signal for the circadian clock (Millar, 2004; Salomé and McClung, 2005b), it might compensate for the absent cytokinin input signal in cytokinin-deficient plants, effectively resetting the circadian oscillator. Taken together, short photoperiods followed by prolonged light treatments and succeeding long dark periods affected the plants most strongly, constituting a circadian stress regime (Figure 13).
In addition to light, temperature is also an important input signal for the circadian oscillator (Salomé and McClung, 2005b; McWatters and Devlin, 2011). Interestingly, low temperature during CL treatment resulted in protection against cell death. This could be due to substitution of the missing light-dark cycle with a temperature cycle. Indeed, Bieniawska et al. (2008) showed that low temperatures under constant light conditions not only stop circadian oscillations, but they also lead to constantly high CCA1 and LHY expression, which is presumably protective under circadian stress conditions.
A Reduced Cytokinin Status Renders Plants Sensitive to Circadian Stress
The response of plants with a reduced cytokinin status showed that the adaptive response to changing light-dark regimes requires the redundant action of cytokinin biosynthesis and signaling genes. The severity of PCD increased with an increasing number of IPT gene mutations, revealing a dose–response relationship. Pro35S:CKX1 plants exhibited a very severe circadian stress phenotype, whereas Pro35S:CKX2 and Pro35S:CKX4 plants both were less affected. This outcome is in accordance with the stronger cytokinin deficiency syndrome observed in Pro35S:CKX1 plants (Werner et al., 2003) and the stronger reduction in the levels of active cytokinins in these plants (Nishiyama et al., 2011).
Among the three cytokinin receptors, AHK3 turned out to be the main player because only ahk3 single mutants exhibited a distinct cell death phenotype. The circadian stress phenotype was strongly aggravated in ahk2 ahk3 and cre1 ahk3 mutants, revealing the accessory functions of AHK2 and CRE1/AHK4. The strong phenotype of ahk2 ahk3 plants is consistent with the predominant expression and function of AHK2 and AHK3 in shoot tissues, including leaves (Higuchi et al., 2004; Nishimura et al., 2004; Kim et al., 2006; Riefler et al., 2006; Stolz et al., 2011; Cortleven et al., 2014). In contrast, the enhancing influence of cre1 on AHK3 loss of function was unexpected and uncovered a role for the CRE1/AHK4 receptor in leaves. CRE1/AHK4 is mainly expressed in the vasculature of roots and mainly functions in belowground tissues (Mähönen et al., 2000; Ueguchi et al., 2001b; Birnbaum et al., 2003; Higuchi et al., 2004). In aerial tissues, this gene is expressed in the shoot apical meristem as well as the vasculature (Ueguchi et al., 2001a; Nishimura et al., 2004; Mähönen et al., 2006; Gordon et al., 2009; Chickarmane et al., 2012), but a function in leaves has not been described (Heyl et al., 2012). Downstream of the receptors, the role of cytokinin in the circadian stress response is mediated by different B-type ARRs. Among ARR1, ARR10, and ARR12, which are the most important B-type ARRs in the shoot during vegetative growth (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008b), only the combined loss of ARR10 and ARR12 resulted in a detectable (although weak) stress response, indicating an involvement of both proteins in coping with circadian stress. However, the strongest circadian stress phenotype was observed in arr2 plants, indicating a principal role for ARR2, which is particularly known for its role in leaf senescence (Kim et al., 2006) and pathogen response (Choi et al., 2010).
Coping with Circadian Stress Requires CCA1 and LHY
Transcript analyses revealed strongly altered clock gene expression in cytokinin-deficient plants during the dark period following CL treatment. In particular, CCA1 and LHY expression was affected, with an almost complete lack of peak expression, indicating an impaired anticipation of dawn. Sufficient nighttime expression of CCA1 and LHY could be crucial for appropriate responses to circadian stress. Consistently, the earlier onset of light after short nights to prevent the detrimental impact of long dark periods following CL treatment caused resetting of CCA1 and LHY expression.
In accordance with the idea that CCA1 and LHY are crucial for the circadian stress response (Figure 13), the standard stress regime also induced cell death and similar changes in gene expression in clock mutants lacking proper CCA1 and LHY function, particularly in cca1 lhy mutants. Also, elf3-9 (also elf3-8), lux-1, and Pro35S:TOC1 plants were sensitive to circadian stress. Although these plants are impaired in different parts of the core oscillator, they all have reduced CCA1 and LHY expression in common (Doyle et al., 2002; Makino et al., 2002; Hazen et al., 2005; Kolmos et al., 2009; Dixon et al., 2011; Gendron et al., 2012; Huang et al., 2012). In addition, toc1-101 plants exhibited a weak cell death phenotype following CL treatment, which is in accordance with the decreased CCA1 and LHY expression in toc1 plants (Alabadí et al., 2001; Más et al., 2003; Kikis et al., 2005). Together, our results indicate that reduced CCA1 and LHY expression renders plants sensitive to circadian stress regimes. Their expression seems to be very relevant during the dark period following CL treatment when stress and PCD are induced. Consistent with this idea, we found that plants with high levels of CCA1 and LHY, such as prr9 prr7 prr5 triple mutant plants, were not affected by circadian stress.
A crucial question is how sufficient CCA1 and LHY expression levels might be achieved through the action of cytokinin. Cytokinin causes phase delays in different circadian rhythms, including CCA1 and LHY, which is mediated by ARR4 and PHYTOCHROME B (PHYB) (Hanano et al., 2006; Salomé et al., 2006; Zheng et al., 2006). Neither arr4 or arr3,4 mutants nor phyB mutants exhibited a pronounced stress phenotype following CL treatment (Supplemental Figure 12), suggesting that the phase adjustment by cytokinin is of minor importance for a proper circadian stress response. However, cytokinin can induce CCA1 and LHY expression (Zheng et al., 2006), and a time-of-day-dependent induction of CCA1 has been observed (Hanano et al., 2006). Therefore, it is conceivable that cytokinin acts through transcriptional regulation of these core clock genes, which would explain its functional relevance.
Circadian Stress Disrupts Time-of-Day Specificity of Clock Outputs Causing Disproportionate Oxidative Stress and JA Responses
Proper matching of internal circadian timing with the environment enhances plant fitness and survival (Green et al., 2002; Michael et al., 2003; Dodd et al., 2005; Yerushalmi et al., 2011). This fitness advantage is in part conferred by the ability to anticipate environmental challenges and, hence, to optimally respond to biotic and abiotic stresses in a time-of-day-specific manner (Seo and Más, 2015; Grundy et al., 2015; Greenham and McClung, 2015). Our circadian stress regime caused insufficient nighttime CCA1 and LHY expression in cytokinin-deficient plants. This can be interpreted as a desynchronization between endogenous rhythms and the environment, resulting in a disrupted time-of-day specificity of clock outputs. In our model (Figure 13), we propose that changes in clock output regulation probably include both incorrect timing of stress gene expression and an impaired gating of stress responsiveness, which contributed to the pronounced stress phenotype in these plants.
In cytokinin-deficient plants, circadian stress had a strong impact on the expression of the ROS-inducible genes BAP1 and ZAT12 as well as JA-related genes (e.g., JAZ1). Their early induction preceded the development of cell death and coincided with a perturbation of the circadian clock (i.e., diminished CCA1 and LHY expression). Clock mutants with reduced CCA1 and LHY function exhibited similar changes in stress-related gene expression. Rescuing short nights following CL treatment resulted in wild-type-like JAZ1 transcript levels, indicating that the resetting of the oscillator by the earlier onset of light helped to prevent stress responses. In this regard, it is interesting that both oxidative stress responses and the JA pathway are influenced by the circadian clock. The circadian clock regulates ROS homeostasis and oxidative stress responses (Lai et al., 2012). Many ROS-related genes display robust diurnal and circadian oscillations (Covington et al., 2008; Hazen et al., 2009; Lai et al., 2012), and the same is true for JA-related genes (Mizuno and Yamashino, 2008; Covington et al., 2008). Moreover, JA-mediated defense against herbivory is optimized by circadian regulation of JA biosynthesis (Goodspeed et al., 2012), and JA responsiveness is gated through the clock-associated component TIC (Shin et al., 2012).
Both BAP1 and ZAT12 expression are clock-regulated, and ZAT12 responsiveness to cold and paraquat is gated by the circadian clock (Fowler et al., 2005; Hazen et al., 2009; Lai et al., 2012). ZAT12 is a CCA1 target (Lai et al., 2012; Nagel et al., 2015), indicating that CCA1 can directly influence ZAT12 expression, which is in accordance with the importance of CCA1 under our conditions. The strong induction of BAP1 and ZAT12 in response to circadian stress could thus be directly due to the perturbed circadian clock in cytokinin-deficient plants, leading to incorrect timing of their expression and/or an increased inducibility due to impaired gating. Impaired gating might have caused hypersensitivity to oxidative stress in cytokinin-deficient plants, explaining the stronger stress response.
The responsiveness to oxidative stress is regulated by the circadian clock, with different clock components contributing differently. Therefore, clock mutants strongly differ in their sensitivity to oxidative stress. For example, gigantea mutants display a high tolerance to paraquat-induced oxidative stress (Kurepa et al., 1998), while hypersensitivity to paraquat treatment was observed in cca1 lhy, elf3, and lux mutants, as reflected by a pronounced cell death phenotype (Lai et al., 2012). Interestingly, the latter mutants were also highly vulnerable toward circadian stress. Intriguingly, the prr9 prr7 prr5 triple mutant is extremely tolerant to various stresses (Nakamichi et al., 2009), which is also consistent with the high stress tolerance of prr9 prr7 prr5 plants found in this study. Taken together, our mutant analyses support the view that circadian stress is at least in part caused by an impaired gating of oxidative stress responses, which results in an “open gate” (which would usually be “closed” at that time of day), enabling the strong induction of BAP1 and ZAT12.
Similar to the oxidative stress response, the JA response could also be due to incorrect timing of gene expression caused by a malfunctioning circadian clock. Among the tested JA-related genes, all except LOX4 were found to be clock-regulated (Covington et al., 2008; Hazen et al., 2009). We could confirm the diurnal expression pattern of MYC2 and COI1 (Shin et al., 2012) and found the same for all other tested JA-related genes. As described for MYC2 by Shin et al. (2012), they all exhibited a very low nighttime expression under control conditions, while the opposite was true for COI1. Intriguingly, circadian stress led to an inverted expression pattern. Hence, it seems that the maximal expression of these genes has been shifted to the wrong time of day. In addition, an impaired gating of JA signaling could explain why the JA pathway was initially activated without an elevation in JA levels (Figure 9). Consistent with this idea, tic-2 plants, which exhibit enhanced JA responsiveness (Shin et al., 2012), also showed a circadian stress phenotype, supporting the view that the cell death phenotype caused by circadian stress could at least in part result from an impaired gating of JA responses.
Contribution of ROS and JA Responses to PCD Following Circadian Stress
ROS play a key role in the induction, signaling, and execution of plant PCD (Van Breusegem and Dat, 2006; Gechev et al., 2006, 2010; Gadjev et al., 2008; De Pinto et al., 2012). We found that oxidative stress in cytokinin-deficient plants increased later during cell death progression, while cell death initiation in these plants did not seem to be associated with an oxidative burst. However, ROS-responsive genes such as BAP1 (op den Camp et al., 2003) and ZAT12 (Rizhsky et al., 2004; Davletova et al., 2005) were strongly upregulated in cytokinin-deficient plants early upon circadian stress. This could be interpreted as a ROS footprint, indicating active ROS signaling (Gadjev et al., 2006), which is possibly sensed as a death signal, facilitating death execution.
Although death execution pathways undoubtedly exist, little is known about the molecular mechanisms. One example is the 1O2-induced cell death in fluorescent in blue light (flu) mutants, which is dependent on the proteins EXECUTER1 (EX1) and EX2 that are hence part of this 1O2-dependent death execution pathway (Wagner et al., 2004; Lee et al., 2007). Furthermore, H2O2-induced cell death in cat2 mutants is dependent on long photoperiods, suggesting the existence of an execution pathway that is only present (or more active) in LD conditions (Queval et al., 2007). CAT2 gene expression was strongly reduced in cytokinin-deficient plants in response to circadian stress. In addition, as discussed earlier, cytokinin-deficient plants may have irreversibly adjusted to longer photoperiods during CL treatment. In the future, it would therefore be interesting to investigate whether death execution following circadian stress is enabled by a similar LD-dependent pathway. In this regard, it is striking that the circadian clock seems to be a crucial regulator of death execution pathways, as indicated by a study from Korneli et al. (2014) showing that the severity of HR cell death is dependent on the (circadian) time of infection. However, it remains to be elucidated exactly how death execution is regulated by the circadian clock, in particular under circadian stress. The modulation of ROS responses is probably only one among several possible ways to control cell death in a clock-dependent fashion, since so many pathways are under clock control. According to our results, another factor involved in controlling circadian stress-induced PCD is JA-Ile (Figure 13).
While the involvement of ROS signaling in circadian stress was mainly deduced from the transcriptional responses of indicator genes, the functional relevance of the JA pathway is well supported by genetic data. The circadian stress regime specifically activated the JA pathway, while pathways of other stress hormones such as ABA and SA remained largely unaffected. Consistent with its function in the response to circadian stress, the JA pathway was also activated in circadian stress-sensitive clock mutants and was only switched on in mature, affected leaves and not in young, unaffected leaves of the same plants. The expression levels of JAZ1 (and BI1) correlated well with the severity of cell death in cytokinin-deficient plants (Figure 12; Supplemental Figure 4). This is consistent with results showing that quantitative differences in cell death-related gene expression better correlated with the observed cell death phenotypes than qualitative changes (Brosché et al., 2014). This finding also indicates that the extent to which the JA pathway is activated is a decisive factor in determining the severity of circadian stress-induced PCD. Most importantly, cytokinin-deficient plants also carrying the jar1-1 allele causing JA-Ile deficiency (Suza and Staswick, 2008) showed a strongly attenuated cell death phenotype, which demonstrates the requirement of JA-Ile for PCD development under circadian stress. The lack of a full rescue could be due to the fact that JA-Ile is not completely eliminated in the jar1-1 mutant (Suza and Staswick, 2008) and/or that activation of the JA pathway is only one of several perturbations promoting PCD in response to circadian stress. In addition, it cannot be ruled out that oxylipins other than JA-Ile, e.g., OPDA, which is also biologically active (Taki et al., 2005; Mueller et al., 2008; Park et al., 2013), contribute to the promotion of cell death under circadian stress. OPDA levels were also strongly increased in cytokinin-deficient plants under circadian stress, and OPDA biosynthesis is not compromised in jar1-1 plants. In order to study this, it would be interesting to analyze more oxylipin (JA) biosynthesis mutants such as aos (dde2) and opr3 (dde1) in the cytokinin-deficient background, which lack both OPDA and JA-Ile or only JA-Ile, respectively. Several studies support a promoting role for JA in PCD development. For example, JA promotes 1O2-dependent cell death in the flu (Danon et al., 2005) and chlorina1 mutants (Ramel et al., 2013). Also, lesion development and spreading in the lesion mimic mutant cpr5 is JA dependent (Clarke et al., 2000). Together, our data demonstrate that JA is a positive regulator of circadian stress-induced PCD, uncovering an additional link between JA and cell death.
Although the standard stress conditions used in our study are rather extreme, we regard them as a tool to push the usually very robust and redundant circadian system to its limits. This helped us to uncover functional links between cytokinin and the circadian clock as well as between the circadian clock and JA-dependent cell death. Furthermore, it should be noted that smaller changes in the light-dark regime, such as the shift from SD to LD (Figure 1), resulted in an obvious phenotype in cytokinin-deficient plants. Therefore, it is conceivable that plants encounter circadian stress in nature, especially at high latitudes where the daylength varies more drastically throughout the year. Milder circadian stress conditions may not necessarily induce cell death. They likely cause more subtle changes, leading to reduced plant fitness similar to the consequences of circadian dissonance described by Dodd et al. (2005), compromising general plant performance. Last but not least, mild circadian stress in natural environments could negatively affect plant stress tolerance, since the circadian clock is important for the proper anticipation of stresses as well as the adequate response to stresses (Seo and Más, 2015; Grundy et al., 2015).
METHODS
Plant Material and Growth Conditions
The Columbia-0 (Col-0) ecotype of Arabidopsis thaliana was used as the wild type. The following mutant and transgenic Arabidopsis plants were used in this study: arr2 (GK-269G01); arr1-3 arr10-5, arr1-3 arr12-1, and arr10-5 arr12-1 (Mason et al., 2005; Argyros et al., 2008); arr3,4 (To et al., 2004); Pro35S:CKX1, Pro35S:CKX2, and Pro35S:CKX4 (Werner et al., 2003); ahk2-5, ahk3-7, cre1-2, and corresponding double mutants (Riefler et al., 2006); ipt3-2, ipt5-2, ipt7-1, and corresponding double and triple mutants (Miyawaki et al., 2006); cca1-1 (in Col-0) (Yakir et al., 2009); lhy-11 and cca1-1 lhy-11 (in Col-0) (Ito et al., 2007); lhy-20 (Michael et al., 2003); toc1-101 (Kikis et al., 2005); Pro35S:TOC1 (APRR1-ox) (Makino et al., 2002); elf3-8 and elf3-9 (Hicks et al., 2001); elf4-101 (Khanna et al., 2003); lux-1 (Hazen et al., 2005; corresponding wild type C24); prr9-1, prr7-3, prr5-3, prr3-1, prr9-1 prr7-3, and prr7-3 prr5-1 (Michael et al., 2003; Salomé and McClung, 2005a); phyB-9 (Reed et al., 1993); tic-2 (Ding et al., 2007); and jar1-1 (Staswick et al., 1992, 2002). Seeds were obtained from The European Arabidopsis Stock Centre (NASC; http://arabidopsis.info/) or kindly provided by Tatsuo Kakimoto, Takafumi Yamashino, Takeshi Mizuno, Rachel Green, C. Robertson McClung, Jos Schippers, and Patrice Salomé.
The following mutant and transgenic plants were generated by genetic crossing: cca1-1 lhy-20, Pro35S:CKX4 toc1-101, ahk2 ahk3 toc1-101, prr9-1 prr5-3, prr9-1 prr7-3 prr5-3, jar1-1 ahk2 ahk3, and jar1-1 Pro35S:CKX4. The genotypes were confirmed by PCR analysis.
If not stated otherwise, Arabidopsis plants were grown on soil in a growth chamber under SD conditions (8 h light/16 h dark), light intensities of 120 to 170 µmol m−2 s−1, using a combination of Philips SON-T Agros, 400 W, and Philips Master HPI-T Plus, 400 W/645 lamps, generating warm white and neutral white light, respectively, at 22°C and 60% relative humidity.
Circadian Stress Treatments
If not stated otherwise, 5- to 6-week-old SD-grown plants were used for the experimental treatments. The standard circadian stress regime was 32 h light treatment integrated into a SD regime (Figure 4A), referred to as CL regime. Control plants remained continuously in the SD rhythm and were completely unaffected.
For phenotypical analyses, affected and unaffected leaves from treated plants of the same developmental stage were chosen. For most experiments, only the distal halves of these leaves were harvested corresponding to the most affected parts in cytokinin-deficient plants. Whole leaves were used for analysis of Fv/Fm, HOTE, fresh weight, and ion leakage measurements. Harvest during the dark period was performed under green light.
Analysis of Cell Death Progression
Mature leaves, defined as fully expanded leaves, with lesions were counted 20 to 24 h after CL treatment and are given as percentage of all mature leaves. The loss of fresh weight was determined by detaching and weighing at least four leaves per sample and is expressed as percentage of control.
For ion leakage measurements, whole leaves were detached at the indicated time points and floated abaxial side up on a defined volume of distilled water in a Petri dish for 4 h at RT. After incubation, the conductivity of the bathing solution was measured using a conductivity meter (EC-Controller; Stelzner) and given as percentage of the respective control.
Chlorophyll Fluorometry
Directly before chlorophyll fluorescence measurements, leaves were detached, floated on water, and dark-adapted for 20 to 30 min. The PSII maximum quantum efficiency was determined from the ratio of variable (Fv) to maximum (Fm) fluorescence [Fv/Fm = (Fm − F0)/Fm] using the FluorCam chlorophyll fluorometer (Photon Systems Instruments). An actinic light pulse (0.2 µmol m−2 s−1) was used to measure the initial (minimum) PSII fluorescence in the dark-adapted state (F0), and Fm was determined by a saturating light pulse (1500 µmol m−2 s−1). Although in many cases we only show the Fv/Fm data for CL-treated plants, we always included control (SD) plants in our experiments. These plants were completely unaffected and showed no decrease in Fv/Fm values (Fv/Fm > 0.8).
LPO and Oxidative Stress Analysis
The photon emission associated with LPO called autoluminescence (Birtic et al., 2011) was imaged at RT using a highly sensitive CCD camera (VersArray LN/CCD 1340-1300B; Roper Scientific) with a liquid nitrogen-cooled sensor as described (Havaux et al., 2009). For these measurements, CL-treated plants were dark-adapted for 2 h before imaging to allow chlorophyll luminescence to fade away. The luminescence signal measured after this dark-adaptation period was shown to originate predominantly from the slow, spontaneous decomposition of lipid peroxides (Birtic et al., 2011).
HOTE isomers were analyzed as a measure of LPO/oxidative stress (op den Camp et al., 2003; Montillet et al., 2004). Leaf material (700 to 800 mg) was harvested, flash-frozen, and ground with a mortar and pestle. Five hundred milligrams of the ground leaf material was used for the extraction of lipids. After extraction, ROS- and LOX-induced LPO were determined as described previously (Montillet et al., 2004; Havaux et al., 2009).
MDA, an indicator of oxidative stress, was quantified according to Heath and Packer (1968).
To determine H2O2 levels, leaf material (∼100 mg) was harvested and flash-frozen at the indicated time points. The frozen samples were ground using a Retsch mill in precooled adapters. H2O2 measurements were performed using the Amplex red hydrogen peroxide/peroxidase assay kit (Invitrogen) according to the manufacturer’s instructions.
RNA Isolation and Quantitative RT-PCR
Leaf material (≤100 mg) was harvested and flash-frozen at the indicated time points. The frozen samples were ground using a Retsch mill in precooled adapters. Total RNA was extracted with the TRIzol method as described in the GIBCO TRIzol manual (Invitrogen) followed by RNA purification using the RNeasy mini kit including on-column DNase digestion (Qiagen) according to the manufacturer’s protocol (experiment in Figure 3) or using kits only (either the RNeasy plant mini kit [Qiagen] or the NucleoSpin RNA plant kit [Macherey-Nagel]) including on-column DNase digestion according to the manufacturer’s protocols.
For qRT-PCR analysis, cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen) using 1 µg of total RNA. Primer pairs were designed using Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). All primers used in this study are listed in Supplemental Table 1. qRT-PCR was performed with the 7500 Fast Real-Time PCR system (Applied Biosystems) using SYBR Green I technology and universal FAST cycling conditions (15 min at 95°C, 40 cycles of 5 s at 95°C, 15 s at 55°C, and 10 s at 72°C) followed by the generation of a dissociation curve to check for specificity of the amplification. Reactions contained SYBR Green Master Mix (Applied Biosystems), 300 nM forward and reverse primers, and 1 μL of the diluted cDNA in a 20-μL reaction. Gene expression data were normalized against reference genes according to Vandesompele et al. (2002). For most experiments, PROTEIN PHOSPHATASE2A SUBUNIT A2 (PP2AA2) and MCP2D served as reference genes; PP2AA2, SAND, and UBIQUITIN-CONJUGATING ENZYME10 (UBC10) were used for the data shown in Figure 3.
Phytohormone Analysis
For phytohormone measurements, leaf material (400 to 500 mg) was harvested and flash-frozen at the indicated time points. One hundred milligrams of fresh plant tissue (early time points) or 10 mg lyophilized plant tissue (late time points) was extracted and analyzed as described previously (Matyash et al., 2008; Ternes et al., 2011).
Accession Numbers
All gene accession numbers are provided in Supplemental Table 2.
Supplemental Data
Supplemental Figure 1. Defective Cytokinin Signaling and a Reduced Cytokinin Content Lead to Cell Death in Response to CL Treatment.
Supplemental Figure 2. The Entrainment Strongly Influences the Severity of Cell Death Following Changes in the Light-Dark Regime.
Supplemental Figure 3. The Cell Death Phenotype Depends on an Extended Light Period and Can Be Alleviated by Temperature Cycles.
Supplemental Figure 4. A Dark Period Following Extended Light Treatment Is Required to Induce the Cell Death Phenotype.
Supplemental Figure 5. Induction of Stress- and Cell Death-Associated Genes Only Occurs in Mature Leaves, Initiating Cell Death.
Supplemental Figure 6. Earlier Onset of Light, Following Short Nights after CL Treatment, Resets Circadian Clock Gene Expression, Coinciding with the Lack of Cell Death in Cytokinin-Deficient Plants.
Supplemental Figure 7. Clock Mutants Are Also Sensitive to Circadian Stress.
Supplemental Figure 8. Kinetics of Evening Complex and Pseudo-Response Regulator Gene Expression after CL Treatment and CL Responses of Different prr Mutants.
Supplemental Figure 9. A-Type ARR Gene Expression Is Decreased in Pro35S:CKX4 Plants as well as in Clock Mutants in Response to Circadian Stress.
Supplemental Figure 10. Jasmonic Acid Precursor Levels in Response to CL Treatment.
Supplemental Figure 11. Circadian Stress-Induced Cell Death in Cytokinin-Deficient Plants Is Linked to an Activated Jasmonic Acid Pathway.
Supplemental Figure 12. PHYB, ARR3, and ARR4 Have No Major Role under Circadian Stress.
Supplemental Table 1. Primers Used in the Study.
Supplemental Table 2. List of Accession Numbers.
Supplementary Material
Acknowledgments
This work was supported by funding of Deutsche Forschungsgemeinschaft in the frame of CRC 973 (project C1) (T.S.) and by Grants Schm 814/27-1 to T.S. and INST 186/822-1 to I.F. We thank Tatsuo Kakimoto, Takafumi Yamashino, Takeshi Mizuno, Rachel Green, C. Robertson McClung, Jos Schippers, and Patrice Salomé for kindly providing seeds of mutant and transgenic plants for this study. We also thank Brigitte Ksas for technical help with the lipid peroxidation measurements.
AUTHOR CONTRIBUTIONS
S.N. and T.S. designed experiments. S.N., A.C., T.I., and M.H. performed experiments. S.N., A.C., T.I., I.F., M.H., M.R., and T.S. analyzed data. S.N. and T.S. wrote the article.
Glossary
- EC
evening complex
- ABA
abscisic acid
- JA
jasmonic acid
- JA-Ile
jasmonoyl-isoleucine
- PCD
programmed cell death
- ROS
reactive oxygen species
- SD
short-day
- LD
long-day
- CL
continuous light
- LPO
lipid peroxidation
- HOTE
hydroxyoctadecatrienoic acid
- MDA
malondialdehyde
- SA
salicylic acid
- OPDA
oxo-phytodienoic acid
- FW
fresh weight
Footnotes
Articles can be viewed without a subscription.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Birtic S., Ksas B., Genty B., Mueller M.J., Triantaphylidès C., Havaux M. (2011). Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. 67: 1103–1115. [DOI] [PubMed] [Google Scholar]
- Brosché M., et al. (2014). Transcriptomics and functional genomics of ROS-induced cell death regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 10: e1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari D., Wang G., Farmer E.E., Dong X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75: 25–33. [DOI] [PubMed] [Google Scholar]
- Carré I., Veflingstad S.R. (2013). Emerging design principles in the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24: 393–398. [DOI] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Choi J., Huh S.U., Kojima M., Sakakibara H., Paek K.H., Hwang I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19: 284–295. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.D., Volko S.M., Ledford H., Ausubel F.M., Dong X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Nitschke S., Klaumünzer M., Abdelgawad H., Asard H., Grimm B., Riefler M., Schmülling T. (2014). A novel protective function for cytokinin in the light stress response is mediated by the Arabidopsis histidine kinase2 and Arabidopsis histidine kinase3 receptors. Plant Physiol. 164: 1470–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A., Miersch O., Felix G., Camp R.G., Apel K. (2005). Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 41: 68–80. [DOI] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pinto M.C., Locato V., De Gara L. (2012). Redox regulation in plant programmed cell death. Plant Cell Environ. 35: 234–244. [DOI] [PubMed] [Google Scholar]
- Ding Z., Millar A.J., Davis A.M., Davis S.J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77. [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Stone J.M., Gechev T.S. (2008). Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. In International Review of Cell and Molecular Biology, Vol. 270, K.W. Jeon, ed (Cambridge, MA: Academic Press; ), pp. 87–144. [DOI] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T., Petrov V., Minkov I. (2010). Reactive oxygen species and programmed cell death. In Reactive Oxygen Species and Antioxidants in Higher Plants, S.D. Gupta, ed (New York: CRC Press), pp. 65–78. [Google Scholar]
- Gechev T.S., Van Breusegem F., Stone J.M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28: 1091–1101. [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D., Chehab E.W., Min-Venditti A., Braam J., Covington M.F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 109: 4674–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.M., Tingay S., Wang Z.Y., Tobin E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129: 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K., McClung C.R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16: 598–610. [DOI] [PubMed] [Google Scholar]
- Grundy J., Stoker C., Carré I.A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S., Vankova R., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. (2012). Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17: 172–179. [DOI] [PubMed] [Google Scholar]
- Hanano S., Domagalska M.A., Nagy F., Davis S.J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Havaux M., Ksas B., Szewczyk A., Rumeau D., Franck F., Caffarri S., Triantaphylidès C. (2009). Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol. 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Naef F., Quisel T., Gendron J.M., Chen H., Ecker J.R., Borevitz J.O., Kay S.A. (2009). Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R.L., Packer L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125: 189–198. [DOI] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Riefler M., Romanov G.A., Schmülling T. (2012). Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. 91: 246–256. [DOI] [PubMed] [Google Scholar]
- Heyl A., Schmülling T. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6: 480–488. [DOI] [PubMed] [Google Scholar]
- Hicks K.A., Albertson T.M., Wagner D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101: 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Kim S.A., Guerinot M.L., McClung C.R. (2013). Reciprocal interaction of the circadian clock with the iron homeostasis network in Arabidopsis. Plant Physiol. 161: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta C.T., Gardner M.J., Hubbard K.E., Baek S.J., Dalchau N., Suhita D., Dodd A.N., Webb A.A. (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 30: 333–349. [DOI] [PubMed] [Google Scholar]
- Hsu P.Y., Harmer S.L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Más P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Hwang I., Chen H.C., Sheen J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Mizuno T. (2008a). Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72: 3025–3029. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. (2008b). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49: 47–57. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Watanabe N., Nagano M., Kawai-Yamada M., Lam E. (2011). Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 18: 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Nakamichi N., Nakamura Y., Niwa Y., Kato T., Murakami M., Kita M., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. (2007). Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 48: 971–983. [DOI] [PubMed] [Google Scholar]
- Jones A.M. (2001). Programmed cell death in development and defense. Plant Physiol. 125: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (2003). Biosynthesis of cytokinins. J. Plant Res. 116: 233–239. [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Kikis E.A., Quail P.H. (2003). EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 133: 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Schaller G.E. (2014). Cytokinins. Arabidopsis Book 12: e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313. [DOI] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Zhang S., Lee K.P., Lakshmanan Ashok M., Blajecka K., Herrfurth C., Feussner I., Apel K. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Ryu H., Hong S.H., Woo H.R., Lim P.O., Lee I.C., Sheen J., Nam H.G., Hwang I. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Nowak M., Werner M., Fischer K., Schwarz G., Mathews S., Schoof H., Nagy F., Bujnicki J.M., Davis S.J. (2009). Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3: 350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneli C., Danisman S., Staiger D. (2014). Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock. Plant Cell Physiol. 55: 1613–1622. [DOI] [PubMed] [Google Scholar]
- Kreps J.A., Wu Y., Chang H.S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Van Montagu M., Inzé D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14: 759–764. [DOI] [PubMed] [Google Scholar]
- Lai A.G., Doherty C.J., Mueller-Roeber B., Kay S.A., Schippers J.H., Dijkwel P.P. (2012). CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 109: 17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. (2004). Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5: 305–315. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T., Cuevas J., Más P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28: 3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A.J., Milner J.J., Sadanandom A. (2008). Timing is everything: regulatory overlap in plant cell death. Trends Plant Sci. 13: 589–595. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14: 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.P., Higuchi M., Törmäkangas K., Miyawaki K., Pischke M.S., Sussman M.R., Helariutta Y., Kakimoto T. (2006). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 16: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Makino S., Matsushika A., Kojima M., Yamashino T., Mizuno T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43: 58–69. [DOI] [PubMed] [Google Scholar]
- Más P., Alabadí D., Yanovsky M.J., Oyama T., Kay S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. (2008). Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R. (2011). The genetics of plant clocks. Adv. Genet. 74: 105–139. [DOI] [PubMed] [Google Scholar]
- McWatters H.G., Devlin P.F. (2011). Timing in plants--a rhythmic arrangement. FEBS Lett. 585: 1474–1484. [DOI] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053. [DOI] [PubMed] [Google Scholar]
- Millar A.J. (2004). Input signals to the plant circadian clock. J. Exp. Bot. 55: 277–283. [DOI] [PubMed] [Google Scholar]
- Millar A.J., Kay S.A. (1991). Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103: 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Yamashino T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49: 481–487. [DOI] [PubMed] [Google Scholar]
- Montillet J.L., Cacas J.L., Garnier L., Montané M.H., Douki T., Bessoule J.J., Polkowska-Kowalczyk L., Maciejewska U., Agnel J.P., Vial A., Triantaphylidès C. (2004). The upstream oxylipin profile of Arabidopsis thaliana: a tool to scan for oxidative stresses. Plant J. 40: 439–451. [DOI] [PubMed] [Google Scholar]
- Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Mueller M.J., Berger S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767: 414–421. [DOI] [PubMed] [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46: 686–698. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462. [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J.A., Benková E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp R.G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon, A., Wagner, D., Hideg, E., Göbel, C., Feussner, I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K., Brosché M., Kangasjärvi J. (2003). Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8: 335–342. [DOI] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A. (2007). PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., et al. (2013). Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 110: 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657. [DOI] [PubMed] [Google Scholar]
- Ramel F., Ksas B., Akkari E., Mialoundama A.S., Monnet F., Krieger-Liszkay A., Ravanat J.L., Mueller M.J., Bouvier F., Havaux M. (2013). Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25: 1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Schwartz J., Jones M.A., Sairanen I., Cheng Y., Andersson C.R., Zhao Y., Ljung K., Harmer S.L. (2009). REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 106: 16883–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M., Novák O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L., Davletova S., Liang H., Mittler R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279: 11736–11743. [DOI] [PubMed] [Google Scholar]
- Robertson F.C., Skeffington A.W., Gardner M.J., Webb A.A. (2009). Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 69: 419–427. [DOI] [PubMed] [Google Scholar]
- Roden L.C., Ingle R.A. (2009). Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21: 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005a). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005b). What makes the Arabidopsis clock tick on time? A review on entrainment. Plant Cell Environ. 28: 21–38. [Google Scholar]
- Salomé P.A., To J.P., Kieber J.J., McClung C.R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Shin J., Davis S.J. (2011). Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. [DOI] [PubMed] [Google Scholar]
- Seo P.J., Más P. (2015). STRESSing the role of the plant circadian clock. Trends Plant Sci. 20: 230–237. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J.E., Davis S.J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Riefler M., Lomin S.N., Achazi K., Romanov G.A., Schmülling T. (2011). The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 67: 157–168. [DOI] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- Suza W.P., Staswick P.E. (2008). The role of JAR1 in Jasmonoyl-L:-isoleucine production during Arabidopsis wound response. Planta 227: 1221–1232. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. (2001). The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol. 42: 107–113. [DOI] [PubMed] [Google Scholar]
- Taki N., et al. (2005). 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139: 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., Riezman H., Feussner I. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192: 841–854. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- To J.P., Haberer G., Ferreira F.J., Deruère J., Mason M.G., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P.V. (2011). Metacaspases. Cell Death Differ. 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C., Koizumi H., Suzuki T., Mizuno T. (2001a). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42: 231–235. [DOI] [PubMed] [Google Scholar]
- Ueguchi C., Sato S., Kato T., Tabata S. (2001b). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42: 751–755. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F., Dat J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Köllmer I., Bartrina I., Holst K., Schmülling T. (2006). New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381. [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Kron I., Hassidim M., Melamed-Book N., Green R.M. (2009). Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 150: 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S., Yakir E., Green R.M. (2011). Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 20: 1155–1165. [DOI] [PubMed] [Google Scholar]
- Zheng B., Deng Y., Mu J., Ji Z., Xiang T., Niu Q.-W., Chua N.-H., Zuo J. (2006). Cytokinin affects circadian-clock oscillation in a phytochrome B- and Arabidopsis response regulator 4-dependent manner. Physiol. Plant. 127: 277–292. [Google Scholar]
- Zhong H.H., McClung C.R. (1996). The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol. Gen. Genet. 251: 196–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.