A peptidoglycan wall surrounding each plastid in the moss Physcomitrella patens was visualized using metabolic labeling with a d-Ala-d-Ala dipeptide probe and click chemistry.
Abstract
It is believed that the plastids in green plants lost peptidoglycan (i.e., a bacterial cell wall-containing d-amino acids) during their evolution from an endosymbiotic cyanobacterium. Although wall-like structures could not be detected in the plastids of green plants, the moss Physcomitrella patens has the genes required to generate peptidoglycan (Mur genes), and knocking out these genes causes defects in chloroplast division. Here, we generated P. patens knockout lines (∆Pp-ddl) for a homolog of the bacterial peptidoglycan-synthetic gene encoding d-Ala:d-Ala ligase. ∆Pp-ddl had a macrochloroplast phenotype, similar to other Mur knockout lines. The addition of d-Ala-d-Ala (DA-DA) to the medium suppressed the appearance of giant chloroplasts in ∆Pp-ddl, but the addition of l-Ala-l-Ala (LA-LA), DA-LA, LA-DA, or d-Ala did not. Recently, a metabolic method for labeling bacterial peptidoglycan was established using ethynyl-DA-DA (EDA-DA) and click chemistry to attach an azide-modified fluorophore to the ethynyl group. The ∆Pp-ddl line complemented with EDA-DA showed that moss chloroplasts are completely surrounded by peptidoglycan. Our findings strongly suggest that the moss plastids have a peptidoglycan wall containing d-amino acids. By contrast, no plastid phenotypes were observed in the T-DNA tagged ddl mutant lines of Arabidopsis thaliana.
INTRODUCTION
The acquisition of two energy-supplying organelles, mitochondria and chloroplasts, were important events in the evolution of eukaryotic cells. It is now widely accepted that a symbiotic association between a cyanobacterium and a mitochondriate host led to the origin of the plastids of primary photosynthetic eukaryotes, such as glaucophytes, red algae, and green plants (Keeling, 2010). Almost all free-living bacteria, including cyanobacteria, use peptidoglycan as part of their cell wall (Leganés et al., 2005). Peptidoglycan is a continuous covalent macromolecule composed of a sugar-amino acid polymer containing d-amino acids (Typas et al., 2011). Bacterial peptidoglycan confers mechanical resistance to osmotic pressure, maintains cell shape, and functions in cell division. Therefore, the evolution of plastid morphology and division is related to modification and/or loss of peptidoglycan. In addition, the presence of peptidoglycan between plastid envelopes may impede protein transport into plastids (Steiner and Löffelhardt, 2002). However, evolution of the endosymbiont cell wall has received little attention because it is assumed that plastids have no peptidoglycan layer, with the exception of the glaucophytes (Keeling, 2010), a small group of algae comprising ∼10 species.
In bacteria, the peptidoglycan sacculus is generated in several steps (Figure 1A). Because there is no peptidoglycan layer in animal cells, the peptidoglycan biosynthesis pathway is a major target for antibiotics. If the plastids of green plants have no peptidoglycan, antibiotics that inhibit peptidoglycan biosynthesis should have no effect on their cells. However, we and others have found that antibiotics that interfere with peptidoglycan biosynthesis can inhibit plastid division, generating giant chloroplasts, in the moss Physcomitrella patens (Kasten and Reski, 1997; Katayama et al., 2003). Due to progress in the P. patens genome-sequencing project, we were able to identify 10 homologs of Mur genes: MurA to G, MraY, penicillin binding protein (PBP), and d-Ala:d-Ala ligase (DDL), which are involved in biosynthesizing peptidoglycan (Machida et al., 2006; Homi et al., 2009). Disruption of the P. patens MurA, MurE, MraY, or Pbp resulted in the appearance of a smaller number of macrochloroplasts in each protonema cell, in contrast to wild-type plant cells, which have ∼50 chloroplasts (Machida et al., 2006; Homi et al., 2009). MurE mediates the formation of UDP-N-acetylmuramic acid (MurNAc)-tripeptide in bacteria (Figure 1A). Cross-species complementation assays with the P. patens MurE-knockout line showed that cyanobacterial MurE fused to the P. patens MurE plastid-targeting signal can restore the normal chloroplast phenotype, suggesting that P. patens MurE and bacterial MurE have similar functions (Garcia et al., 2008). These results suggest that the peptidoglycan biosynthetic pathway is linked to chloroplast division in moss.
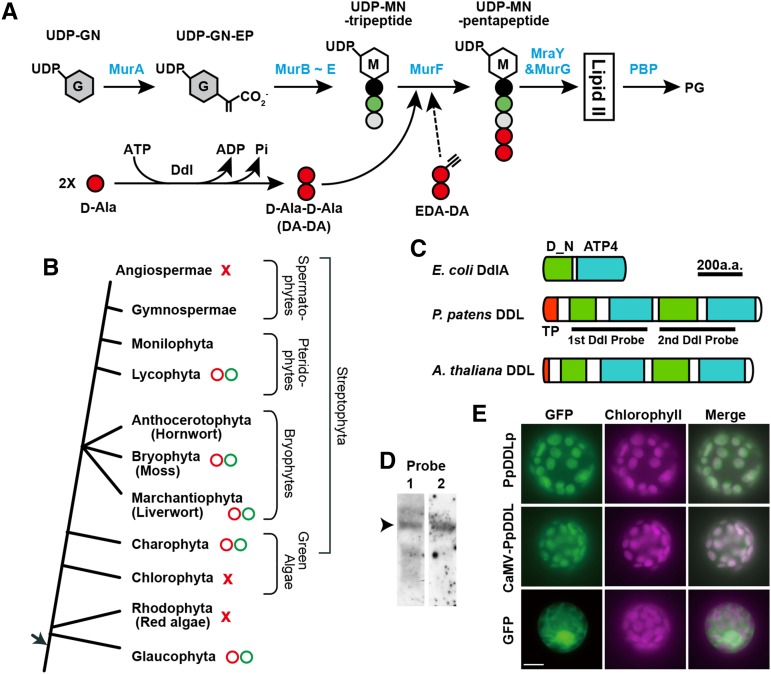
Figure 1.
Peptidoglycan Biosynthesis and d-Ala:d-Ala Ligases.
(A) The general bacterial peptidoglycan biosynthesis pathway is shown with enzyme names in blue. After conversion from UDP-N-acetylglucosamine (UDP-GN) to UDP-GN-enolpyruvate (UDP-GN-EP), UDP-MurNAc (UDP-MN)-tripeptide is generated using the enzymes MurB to E. MurF catalyzes the formation of UDP-MN pentapeptide with d-Ala-d-Ala. d-Ala:d-Ala ligase (Ddl) catalyzes ATP-dependent peptide bond formation between two d-Ala molecules. After Lipid II (undecaprenyl-pyrophosphoryl-MN-[pentapeptide]-GN) passes through the membrane, the disaccharide pentapeptide monomer unit is cross-linked to preexisting peptidoglycan by penicillin binding proteins (PBPs). EDA-DA can be inserted into peptidoglycan instead of DA-DA.
(B) Evolution of plants containing peptidoglycan. While the total genome sequences of red algae, Chlorophyta, and angiosperms indicate that they have only a few Mur genes (red crosses), Mur genes required for peptidoglycan biosynthesis have been retained in Glaucophyta, Charophyta, mosses, liverworts, and lycophytes (red circles). Moreover, treatment using antibiotics that inhibit peptidoglycan biosynthesis resulted in giant chloroplasts in the desmid of Charophyta, liverworts, mosses, and Lycophyta in addition to Glaucophyta (green circles). In the green plant lineage, basal streptophytes may use a peptidoglycan system (i.e., wall structure) for plastid division and morphology.
(C) Schematic representation of the domain structure of d-Ala:d-Ala ligases from E. coli, P. patens, and Arabidopsis (At3g08840). DdlA of E. coli contains d-Ala:d-Ala ligase N terminus (pfam 01820; D_N in green) and ATP-grasp_4 super family (cl17255; ATP4 in blue) domains. Transit peptides (red) are shown as TP. P. patens and Arabidopsis DDL are predicted to be fusion proteins consisting of two sets of Ddl-characteristic domains. The probe regions for (D) are also shown.
(D) RNA gel blot analysis of the P. patens transcripts with the 1st or 2nd DDL region indicated in (C) as a probe. The size of the bands is 3.3 kb (arrowhead).
(E) Subcellular localization of the GFP fusion proteins. P. patens DDL was predicted using TargetP software to encode a plastid-targeting sequence of 65 amino acids (score of 0.76). To analyze the subcellular localization of Pp-DDL, we constructed plasmids directing the expression of the putative transit peptide fused to GFP from its native (PpDDLp) or CaMV35S promoter (CaMV-PpDDL). After PEG-mediated transformation with the generated plasmids, fluorescence images of GFP and chlorophyll autofluorescence and merged images of the transformants were obtained. Bar = 10 µm.
d-Amino acids, commonly d-Ala and d-Glu, are basic components of bacterial peptidoglycan, although l-amino acids are the predominant form occurring in biological molecules. It was thought that the use of d-amino acids was restricted to bacteria. However, d-amino acids have been found in animals and do have physiological functions in eukaryotes (Kirschner and Green, 2009). For example, d-Ser, which is found at high levels in brain tissue, binds to the co-agonist binding site of the N-methyl-d-aspartate subtype of glutamate receptors and modulates neurotransmission (Henneberger et al., 2012). d-Amino acids, including d-Ala-d-Ala (DA-DA), were first detected in plants in the 1960s (Noma et al., 1973; Frahn and Illman, 1975; Robinson, 1976). Unlike the case of animals, however, little is known about the physiological functions of d-amino acids in plants (Michard et al., 2011). The relationship between d-amino acids and plastid division in moss is also unclear, although peptidoglycan may be necessary for this process. The formation of DA-DA is catalyzed by Ddl from two molecules of d-Ala, and this is then joined to the C terminus of the UDP-MurNAc-tripeptide in bacterial peptidoglycan (Figure 1A). Previously, we identified DDL cDNAs from an enriched, full-length cDNA library of P. patens (Machida et al., 2006). There may be a relationship between d-amino acids and plastid division because d-cycloserine, an inhibitor of both Ddl and alanine racemase, rapidly decreases the number of plastids in P. patens cells (Katayama et al., 2003).
It is informative to consider plastid peptidoglycan from an evolutionary viewpoint (Figure 1B). In the green plant lineage, chlorophytes are thought to have branched off earliest. The genome sequences of the chlorophytes Chlamydomonas reinhardtii and Volvox carteri contain no Mur genes except MurE, suggesting that chlorophytes lack plastid peptidoglycan (Merchant et al., 2007; Prochnik et al., 2010). By contrast, as in P. patens, we found that antibiotic treatment resulted in giant chloroplasts in the charophycean green alga Closterium peracerosum-strigosum-littorale complex, the liverwort Marchantia polymorpha, and the lycophyte Selaginella nipponica (Figure 1B) (Takano and Takechi, 2010; Matsumoto et al., 2012). Genomes of the charophyte Klebsormidium flaccidum, M. polymorpha, and the lycophyte Selaginella moellendorffii have many Mur genes (Hori et al., 2014; Takano and Takechi, 2010; Banks et al., 2011), suggesting that these plants, like P. patens, use peptidoglycan in plastid division. By contrast, no plastid peptidoglycan system remains in angiosperms; indeed, the angiosperm genome sequences show that many Mur genes have been lost. The Arabidopsis thaliana genome (Arabidopsis Genome Initiative, 2000) contains a gene homologous to the bacterial Ddl gene (At-DDL) and homologs of the MurE, MraY, and MurG genes. We investigated the function of the MurE gene in Arabidopsis using four T-DNA tagged lines and found that At-MurE is involved in chloroplast development, not in plastid division (Garcia et al., 2008). These results suggest that the bacterial peptidoglycan biosynthesis pathway has a conserved function in plastid division in the basal streptophytes, including charophycean algae, bryophytes, and lycophytes, but not in angiosperms (Figure 1B).
Although indirect evidence is available for the existence of plastid peptidoglycan in the basal streptophyte plants, a peptidoglycan-like structure has not been observed using electron microscopy in all streptophytes, and this includes P. patens (Takano and Takechi, 2010). In our latest review (Takano and Takechi, 2010), we mentioned that this situation is similar to the “Chlamydial anomaly.” Although the intracellular pathogen Chlamydia has all the genes required for peptidoglycan biosynthesis and exhibits susceptibility to antibiotics that target this process, attempts to detect peptidoglycan in Chlamydia have been unsuccessful (McCoy and Maurelli, 2006). However, recently, a metabolic cell wall-labeling method using click chemistry showed the existence of peptidoglycan in Chlamydia (Pilhofer et al., 2013; Liechti et al., 2014). This in vivo peptidoglycan-labeling method used a DA-DA dipeptide analog probe modified with an alkyne functional group, ethynyl-DA-DA (EDA-DA) (Liechti et al., 2014). After incubation with EDA-DA, bacterial cells were fixed, permeabilized, and subjected to click chemistry to attach an azide-modified fluorophore Alexa 488 to the EDA-DA probe. Using this method, the presence of chlamydial peptidoglycan was demonstrated, solving the “Chlamydial anomaly.” In this study, we applied the same method to visualize moss plastid peptidoglycan.
RESULTS
Characterization of d-Ala:d-Ala Ligase in the moss Physcomitrella patens
The appearance of macrochloroplasts in response to d-cycloserine treatment suggests that d-amino acids, especially DA-DA, may be involved in plastid division in moss (Katayama et al., 2003). Previously, we identified two DDL cDNAs with minor differences from the full-length cDNA library of P. patens (Machida et al., 2006). Only one gene was found in the updated P. patens genome sequence (Rensing et al., 2008; cDNA accession number AB194083). Another cDNA (AB194084) may have been generated via a PCR error. Therefore, we analyzed one DDL gene in the P. patens genome. A domain search revealed that the P. patens DDL gene encoded a protein resembling two fused bacterial Ddl proteins (Figure 1C). DDL genes with two Ddl regions each encoding the Ddl_N and ATP-grasp_4 domains are also found in the genomes of many sequenced plants, including the spikemoss S. moellendorffii (XP_002983726) (Banks et al., 2011) and Arabidopsis (At3g08840) (Thimmapuram et al., 2005). RNA gel blot analysis using the first or second DDL region as a probe indicated that major transcripts of 3.3 kb, which included the two DDL regions, were expressed (Figure 1D).
P. patens DDL was predicted to encode a plastid-targeting sequence of 65 amino acids (score of 0.76) using the program TargetP (Emanuelsson et al., 2000). To analyze the subcellular localization of Pp-DDL, we constructed plasmids that expressed the putative transit peptide fused to GFP from its native promoter or a CaMV35S promoter. Polyethylene glycol (PEG)-mediated transformation using these plasmids resulted in GFP fluorescence in the chloroplasts of P. patens, corroborating the plastid localization prediction (Figure 1E).
Knockout of DDL Results in Macrochloroplasts in P. patens
To characterize the function of the DDL gene in plastid division in moss, we generated DDL-knockout lines by targeted gene disruption via homologous recombination in P. patens (Figure 2A). The 5′ and 3′ genomic regions of Pp-DDL were amplified via genomic PCR and cloned. The Zeocin resistance gene, driven by the CaMV35S promoter and terminated using the CaMV35S polyadenylation signal, was inserted between the 5′ and 3′ genomic regions of Pp-DDL. PEG-mediated transformation of P. patens was performed using the constructed plasmids. We selected three transformants with a giant chloroplast phenotype (described below) using primary genomic PCR. DNA gel blot experiments showed that transgenic line #3 had a single insertion of the antibiotic resistance gene in the Pp-DDL gene region (Figure 2B). Based on RT-PCR analysis, Pp-DDL transcripts were not detected in transformant #3 (Figure 2C). Disruption of the P. patens Mur gene results in the appearance of a few macrochloroplasts (Takano and Takechi, 2010). Microscopy observation showed that protonemal cells in the Pp-DDL-knockout transformant (∆Pp-ddl) also contained huge chloroplasts (Figure 2D), with an average chloroplast number of 2.03 ± 1.4 (n = 100), whereas cells in wild-type plants had an average of 45 chloroplasts (43.4 ± 6.49). The chloroplasts in gametophores were also very large in the ∆Pp-ddl line (Figure 2D). Under electron microscopy, no obvious differences in the shape of the envelopes were observed between the giant chloroplasts in ∆Pp-ddl and the small ones in wild-type plants (Figure 2F). Wavy thylakoid membranes were observed in ∆Pp-ddl. This may result from the irregular shape of macrochloroplasts. Even in the wild-type chloroplasts, we did not observe peptidoglycan-like structures between the two envelopes. These results suggest that P. patens uses the DDL gene and d-amino acids in the plastid peptidoglycan biosynthesis pathway for plastid division, similar to the bacterial peptidoglycan system, and that new methods are necessary to observe plastid peptidoglycan.
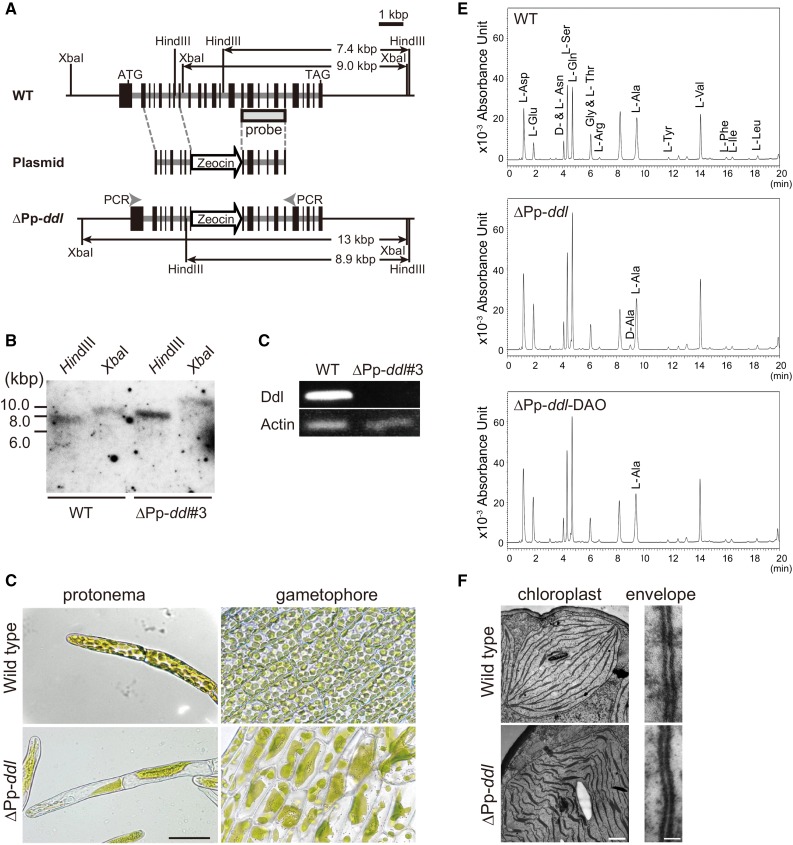
Figure 2.
Knockout of DDL Results in Macrochloroplasts in P. patens.
(A) Schematic representation of the DDL genomic region in wild-type (top) and knockout (∆Pp-ddl; bottom) P. patens. The plasmid constructed for gene disruption is shown in the middle (Plasmid), though the p35S-Zeo vector region is omitted. Start (ATG) and stop (TAG) codons are indicated. Exons are indicated by black boxes. The arrow labeled “Zeocin” indicates the Zeocin cassette. The probe region and the predicted sizes of restriction fragments detected in the DNA gel blot analyses are given. The locations of the RT-PCR primers for (C) are also indicated in the DDL knockout genome.
(B) The results of DNA gel blot analysis with the DDL probe are shown. Genomic DNA from the wild-type and ∆Pp-ddl#3-knockout plants was digested with HindIII or XbaI.
(C) Pp-DDL expression in wild-type and the knockout plants was examined by RT-PCR. Pp-Actin was used as an internal control.
(D) Phenotype of ∆Pp-ddl#3. Micrographs of protonema and leaf cells from wild-type and Pp-DDL-knockout plants are shown at the same magnification. Bar = 50 µm.
(E) UPLC analysis of P. patens cells. Protonema cells of wild-type and ∆Pp-ddl plants were used. DAO from porcine kidney was used for the degradation of d-amino acids from ∆Pp-ddl plants. For details of the chromatography procedure, see Methods.
(F) Electron micrographs of the chloroplasts (bar = 500 nm) and chloroplast envelopes (bar = 20 nm) of wild-type and ∆Pp-ddl plants. The right side of the envelopes is the cytoplasm region.
If Pp-DDL synthesizes DA-DA, the concentration of d-Ala may be increased in the ∆Pp-ddl line. We determined the free d-Ala concentration in cells using an ultraperformance liquid chromatography (UPLC) system because it is difficult to isolate intact huge chloroplasts from knockout protonemata (Figure 2E). d-Ala was not detected in wild-type cells (< 10 µM), but the accumulation of d-Ala was observed in ∆Pp-ddl line #3 (0.30 ± 0.02 mM). A peak in d-Ala detected in the knockout line disappeared after d-amino acid oxidase (DAO) treatment, confirming that this peak consisted of the d-form of the amino acid. The free l-Ala concentration in ∆Pp-ddl line #3 (4.74 ± 0.03 mM) was similar to that in the wild-type plants (4.30 ± 0.26 mM) and unchanged after DAO treatment (4.83 ± 0.10 mM). These results suggest that the generation of DA-DA was inhibited by targeted disruption of the Pp-DDL gene.
d-Ala-d-Ala Dipeptide Could Rescue the Macrochloroplast Phenotype of ∆Pp-ddl Plants
To confirm the function of Pp-DDL, complementation tests were performed using medium supplemented with DA-DA, l-Ala-l-Ala (LA-LA), or d-Ala (Figure 3A). In addition to ∆Pp-ddl knockout line #3, we examined knockout lines for Pp-MurE and Pp-Pbp (Machida et al., 2006). While MurE catalyzes the formation of the UDP-MurNAc-tripeptide in bacterial peptidoglycan biosynthesis, PBP is involved in the biosynthesis and insertion of the disaccharide pentapeptide monomer unit into the sacculus (Figure 1A). Gene targeting of Pp-MurE (∆Pp-MurE) or Pp-Pbp (∆Pp-Pbp) resulted in the macrochloroplast phenotype (Figure 3B) (Machida et al., 2006). The addition of 1 mg/mL DA-DA dipeptide to ∆Pp-ddl plants partially rescued the phenotype, with a chloroplast number of 22.9 ± 9.8 (Figure 3A), whereas DA-DA treatment did not alter the phenotype of wild-type, ∆Pp-MurE, or ∆Pp-Pbp plants (Figures 3A and 3B). In contrast to DA-DA, the addition of 1 mg/mL LA-LA did not rescue the normal chloroplast phenotype in knockout plants. It is known that 1 mM d-Ala is toxic to seed plants (Erikson et al., 2004) but nontoxic to bacteria (Wargel et al., 1970). Because the addition of 1 mg/mL (11.2 mM) d-Ala caused the death or inhibited the growth of the moss lines, we used 0.1 mg/mL d-Ala. While the addition of 0.1 mg/mL d-Ala did not rescue the phenotype of the ∆Pp-ddl plants, 0.1 mg/mL DA-DA did partially recover the phenotype, with a chloroplast number of 9.2 ± 4.6 (Figure 3A). The addition of 1 mg/mL d-Ala-l-Ala (DA-LA) or l-Ala-d-Ala (LA-DA) did not rescue the phenotype of the ∆Pp-ddl knockout plants (Figure 3C). These results confirm that Pp-DDL functions as a d-Ala:d-Ala ligase.
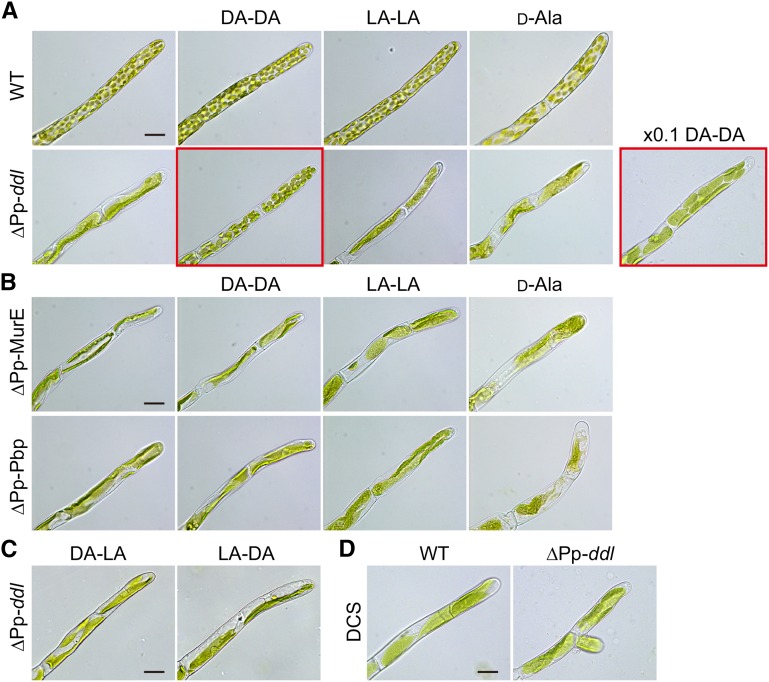
Figure 3.
d-Ala-d-Ala Dipeptide Could Rescue the Macrochloroplast Phenotype of ∆Pp-ddl Plants.
(A) Wild-type and Pp-ddl knockout (∆Pp-ddl) plants were grown on BCDAT medium supplemented with 1 mg/mL d-Ala-d-Ala (DA-DA), 1 mg/mL l-Ala-l-Ala (LA-LA), or 100 µg/mL d-Ala. ∆Pp-ddl plants were also grown with 100 µg/mL DA-DA (×0.1 DA-DA). Only DA-DA could rescue the macrochloroplast phenotype of ∆Pp-ddl plants, although chloroplast number was not recovered to that of wild-type plants (photos with red frame). The magnification is identical in all photographs. Bar = 20 µm.
(B) Pp-MurE (∆Pp-MurE) and Pp-Pbp (∆Pp-Pbp) knockout plants were grown with 1 mg/mL DA-DA, 1 mg/mL LA-LA, or 100 µg/mL d-Ala. The magnification is identical in all photographs. Bar = 20 µm.
(C) ∆Pp-ddl plants were grown with 1 mg/mL d-Ala-l-Ala (DA-LA), or l-Ala-d-Ala (LA-DA). Bar = 20 µm.
(D) Wild-type and ∆Pp-ddl plants were grown on BCDAT medium with 100 µM d-cycloserine (DCS). Bar = 20 µm.
Treatment with d-cycloserine resulted in the macrochloroplast phenotype in wild-type P. patens plants (Figure 3D) (Katayama et al., 2003). We examined the effect of this antibiotic on chloroplasts of ∆Pp-ddl (Figure 3D). The average chloroplast number of ∆Pp-ddl treated with d-cycloserine for 1 week (2.20 ± 1.7) was the same as that of this mutant in medium without antibiotic, suggesting that inhibition and disruption of Pp-DDL affected the identical peptidoglycan biosynthetic pathway.
A Metabolic Cell Wall-Labeling Method Using Click Chemistry Reveals Peptidoglycan Surrounding Moss Plastid
Our data strongly suggest that P. patens uses the peptidoglycan biosynthetic pathway with d-Ala in chloroplast division. However, electron microscopy showed no wall-like structure between the inner and outer envelopes of P. patens chloroplasts (Figure 2F) (Takano and Takechi, 2010). There are two possibilities for peptidoglycan in moss. One is that peptidoglycan surrounds the plastids, as it does in bacteria. In this case, moss plastid peptidoglycan would have to be invisible under normal electron microscopy, since it has not been observed to date. The other possibility is that peptidoglycan is present only at the plastid division plane and supports the division machinery. In this case, moss chloroplasts would not be surrounded by peptidoglycan. We applied the metabolic labeling method to EDA-DA to visualize moss plastid peptidoglycan using click reaction chemistry and an azide-modified fluorophore. To investigate the uptake and incorporation of EDA-DA, ∆Pp-ddl-knockout line #3 was inoculated in medium containing 1 mg/mL EDA-DA. EDA-DA was able to complement the giant chloroplast phenotype, similar to DA-DA, suggesting that EDA-DA was integrated in a similar manner to DA-DA (Figure 4A). Protonema cells of ∆Pp-DDL-knockout line #3 complemented by the addition of EDA-DA were fixed, permeabilized, and used for the click reaction. The azide-modified Alexa Fluor 488 used can be selectively captured to EDA-DA via a click reaction. Fluorescence was observed around each chloroplast in the protonemata, and there were no other fluorescent organelles (Figure 4B; Supplemental Movies 1 to 3). The optical slices through one dividing chloroplast clearly showed that a plastid peptidoglycan layer formed at the division plane (Figure 4C). No fluorescence was detected in the protonemata of the ∆Pp-ddl line complemented with DA-DA. The addition of 1 mg/mL EDA-DA to ∆Pp-MurE plants did not rescue the macrochloroplast phenotype and no fluorescence was detected in the protonemata used for the click reaction. These results suggest that a peptidoglycan wall surrounding the plastids is involved in plastid division in moss, similar to cyanobacteria, and indicates that moss plastid peptidoglycan is invisible under normal electron microscopy.
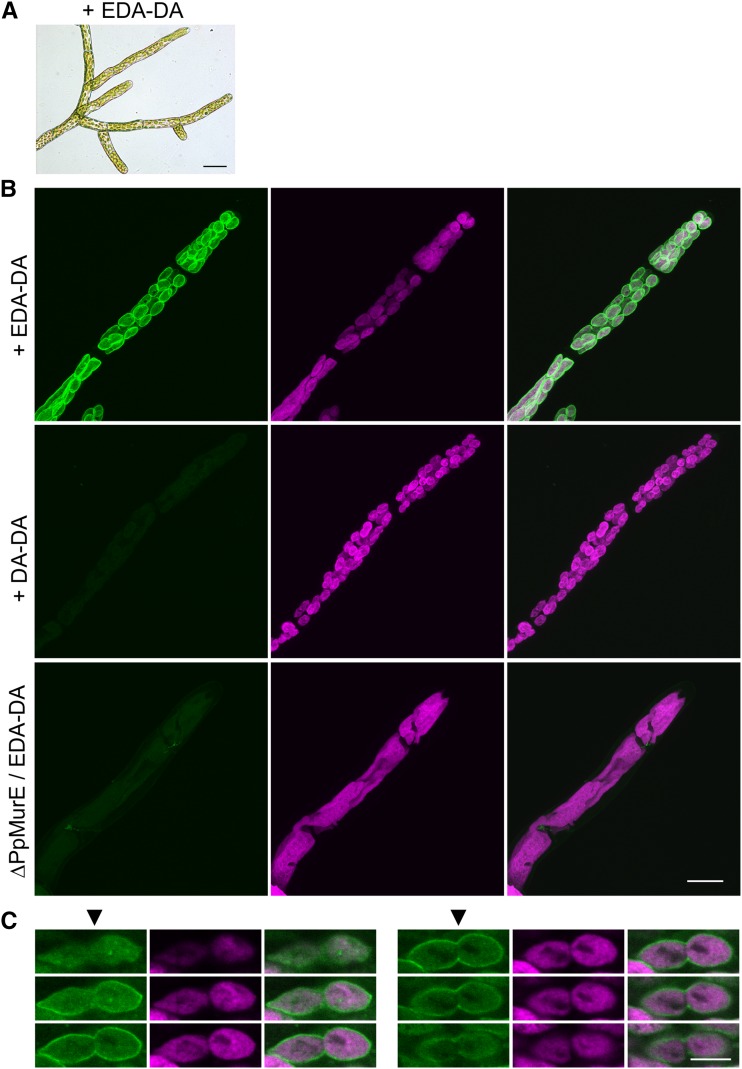
Figure 4.
Peptidoglycan Surrounds Moss Plastids.
(A) Photographs of ∆Pp-ddl-knockout plants grown in liquid BCDAT medium supplemented with 1 mg/mL EDA-DA for 9 d. Bar = 50 µm.
(B) ∆Pp-ddl plants were grown in liquid BCDAT medium supplemented with 1 mg/mL EDA-DA or DA-DA. ∆Pp-MurE plants were also cultured in medium containing 1 mg/mL EDA-DA. Protonemata were fixed and permeabilized. Azide-modified Alexa Fluor 488 was bound to EDA-DA using click chemistry and the protonemata were observed using confocal microscopy. Reconstructed three-dimensional images showing Alexa 488 fluorescence (left) were merged with images showing chlorophyll autofluorescence (middle) in the right photographs. No fluorescence was detected in the protonemata of the ∆Pp-ddl complemented with DA-DA or those of the ∆Pp-MurE cultured with EDA-DA. Bar = 20 µm.
(C) Six optical slices (0.5 µm apart) through one dividing chloroplast visualized by confocal microscopy. Alexa 488 fluorescence (left), chlorophyll autofluorescence (middle), and merged images (right) are shown. The division plane is indicated by arrowheads on the left panels. Bar = 5 µm.
Arabidopsis DDL Is Not Involved in Plastid Division
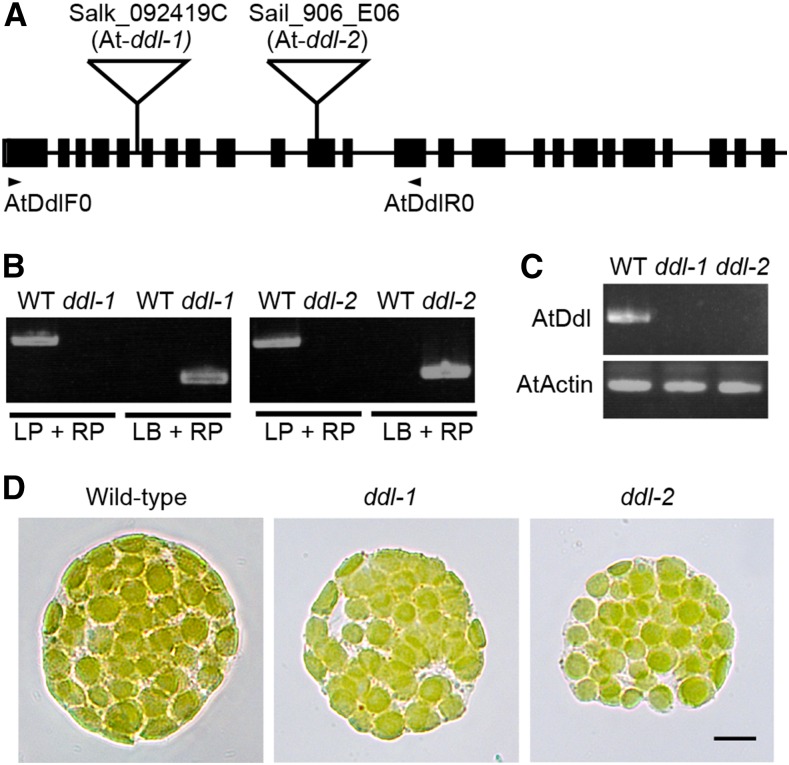
The Arabidopsis genome (Arabidopsis Genome Initiative, 2000) contains a gene homologous to bacterial Ddl (At-DDL; Figure 1C) in addition to homologs of the MurE, MraY, and MurG genes. To analyze the relationship between At-DDL and chloroplast division/morphology, we used two T-DNA tagged lines (Figures 5A and 5B), which were unable to express At-DDL (Figure 5C). Although according to mass spectrometry analyses, At-DDL localizes to the stroma of chloroplasts (Zybailov et al., 2008), we did not observe defects in chloroplast morphology in these T-DNA-tagged lines (Figure 5D), suggesting that At-DDL is not involved in plastid division in Arabidopsis. Our results confirm that the role of the bacterial peptidoglycan biosynthesis pathway in plastid division is not conserved in angiosperms.
Figure 5.
Arabidopsis DDL Is Not Involved in Plastid Division.
(A) Schematic representation of the At-DDL locus in the Arabidopsis genome. The T-DNA insertion positions in the SALK_092419C (5th intron, At-ddl-1) and Sail_906_E06 (11th exon, At-ddl-2) lines are marked with triangles. Black boxes and lines represent exons and introns, respectively. The locations of the primers (AtDdlF0 and AtDdlR0) used for RT-PCR are indicated.
(B) Genomic PCR data with a gene-specific primer (LP and RP) and a primer corresponding to the sequence of the T-DNA left border (LB).
(C) RT-PCR data derived using At-DDL primers are shown. The Arabidopsis ACTIN gene was used as an internal control.
(D) Protoplasts of mesophyll cells from wild-type, At-ddl-1, and At-ddl-2 plants. Bar = 10 µm.
DISCUSSION
Function of the DDL Protein and d-Ala in Moss
Pp-DDL consists of two fused Ddl proteins (Figure 1C). Similar fused DDL proteins have been found in many of the sequenced plant genomes, including S. moellendorffii (Banks et al., 2011) and Arabidopsis (At3g08840) (Thimmapuram et al., 2005). The physiological significance of the duplication of Ddl is unclear. A database search suggested that several bacteria, including Cytophaga hutchinsonii, Saprospira grandis, and Candidatus protochlamydia, have fused Ddl proteins. Duplication of the Ddl region may facilitate dimerization, as the crystal structures of Ddl from several bacterial species have indicated that it functions as a dimer (Bruning et al., 2011).
Our results suggest that DDL in P. patens is a d-Ala:d-Ala ligase, similar to bacterial Ddl proteins, and that DA-DA is essential for plastid division in moss. DDL proteins and DA-DA in streptophyte plants, which show defects in plastid division when treated with d-cycloserine, likely have the same function. The average chloroplast number in ∆Pp-ddl grown on the medium supplemented with 1 mg/mL DA-DA was about half of that in the wild-type moss. The import of DA-DA from outside the cell to the stroma may be limiting for recovery of chloroplast number because Pp-DDL located in the stroma (Figure 1E) has to use DA-DA for peptidoglycan biosynthesis. The growth of pollen tubes in pistils is affected by d-Ser via the activity of glutamate receptor-like channels (Michard et al., 2011). Our work reveals another function for d-amino acids in plants. Although d-Ala is involved in plastid division, it is toxic to P. patens, as it is to other land plants. However, because d-Ala is a component of peptidoglycan, it is nontoxic to bacteria (Wargel et al., 1970). The reasons for the toxicity of d-Ala in land plants are not well understood. The presence of DDL in plastids did not decrease the toxicity of d-Ala, suggesting that inhibition occurs in the cytoplasm.
Function of the DDL Protein and d-Ala in Angiosperms
The functions of DDL and d-Ala in seed plants remain unknown, although this dipeptide was detected in Nicotiana tabacum and Phalaris tuberosa as early as the 1970s (Noma et al., 1973; Frahn and Illman, 1975). The Arabidopsis genome also contains genes homologous to bacterial Ddl (At-DDL; Figures 1C and 5). Although chloroplast proteome analysis has shown that At-DDL localizes to the stroma (Zybailov et al., 2008), we observed no defects in chloroplast morphology in either of two T-DNA-tagged lines (Figure 5), confirming that DDL is not involved in plastid division in Arabidopsis. The d-Ala:d-Ala ligase activity of At-DDL also remains unconfirmed. In contrast to its function in moss, we previously found that another peptidoglycan biosynthetic protein, MurE, plays a role in chloroplast development but not plastid division in Arabidopsis (Garcia et al., 2008). Moreover, At-MurE has been found in an affinity-purified, plastid-encoded RNA polymerase (PEP) complex from tobacco chloroplasts and in transcriptionally active chromosomes of plastids isolated from mustard and Arabidopsis (Suzuki et al., 2004; Pfalz et al., 2006). In Arabidopsis murE mutants, the level of expression of the plastid genome, transcribed by PEP, is low (Garcia et al., 2008). These results indicate that At-MurE has changed its function and has become a component of the PEP complex, involved in gene expression. Similarly, it is possible that the function of DDL may have changed in Arabidopsis.
Peptidoglycan-Surrounded Plastids
Our data strongly suggest that P. patens plastids are surrounded by a peptidoglycan layer. Moreover, the process of peptidoglycan biosynthesis is important in the division of plastids in moss, as it is in bacteria. The peptidoglycan is clearly not confined to the plastid division plane. The precise localization of peptidoglycan in the plastid remains unclear, although it is probably found between the inner and outer envelopes of the chloroplast, similar to the peptidoglycan present in the cyanelles of glaucophytes. Our observations, by electron microscopy, did not show a wall-like structure between the P. patens chloroplast envelopes (Figure 2F). In the Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa, the thickness of the peptidoglycan layer is 6.35 ± 0.53 and 2.41 ± 0.54 nm, respectively (Vollmer and Seligman, 2010). Because the intermembrane space between the moss plastid envelopes is greater than 5 nm, it is possible that there is a peptidoglycan layer there. It is possible that moss plastid peptidoglycan is invisible under normal electron microscopy conditions because the wall is very thin or because there are few peptidoglycan binding proteins present. Although many previous attempts to detect peptidoglycan were unsuccessful (McCoy and Maurelli, 2006), both the metabolic cell wall labeling method using click chemistry and the electron cryotomography method have now shown the presence of peptidoglycan in Chlamydia (Pilhofer et al., 2013; Liechti et al., 2014). Similar to Chlamydia, the anammox bacteria were proposed to lack peptidoglycan because no peptidoglycan layer was observed in resin-embedded sections of either high-pressure frozen and freeze-substituted or chemically fixed anammox cells (Lindsay et al., 2001). However, cryo-transmission electron microscopy and metabolic labeling with EDA-DA revealed a peptidoglycan layer in anammox bacteria (van Teeseling et al., 2015). Therefore, cryo-transmission electron microscopy may also be capable of detecting plastid peptidoglycan in moss. In the future, the existence of plastid peptidoglycan should be confirmed using other methods, such as isolation. In this case, the metabolic labeling method may be useful for detecting plastid peptidoglycan because it involves attaching a fluoroprobe during peptidoglycan isolation.
Plant genome information, together with the observation that inhibition of peptidoglycan biosynthesis by antibiotics affects chloroplast division in glaucophytes, charophytes, liverworts, mosses, and lycophytes, suggests that the loss of plastid peptidoglycan occurred during the evolution of angiosperms in the land plant lineage (Figure 1B). The method described in this work may be capable of identifying plastid peptidoglycan in other organisms. For example, the peptidoglycan-containing plastids, called cyanelles and found in glaucophytes, might be used to confirm the applicability of metabolic labeling methods. In the near future, we will present the results of applying the metabolic labeling method to other species that have plastids thought to contain peptidoglycan.
METHODS
Moss Culture
Protonemata and gametophores of the moss Physcomitrella patens subsp patens (Gransden Wood strain) (Ashton and Cove, 1977) were grown on BCDAT medium solidified with 0.8% agar in a regulated chamber equipped with Biolux lamps (NEC Lighting) at 25°C under continuous white light (100 µmol photons m−2 s−1) (Nishiyama et al., 2000). The P. patens MurE (∆Pp-MurE) and Pbp (∆Pp-Pbp) lines were generated in a previous study (Machida et al., 2006). For the complementation test, wild-type, ∆Pp-ddl, ∆Pp-MurE, and ∆Pp-Pbp plants were grown on BCDAT solid medium supplemented with 1 mg/mL DA-DA (Sigma-Aldrich), 1 mg/mL LA-LA (Sigma-Aldrich), or 100 µg/mL d-Ala (Nacalai Tesque) for 1 week. The ∆Pp-ddl plants were also grown on BCDAT solid medium supplemented with 100 µg/mL DA-DA, 1 mg/mL DA-LA, or 1 mg/mL LA-DA for 1 week. For the antibiotic treatment, wild-type and ∆Pp-ddl plants were grown on BCDAT solid medium with 100 µM d-cycloserine for 1 week. To measure chloroplast number, we counted the number of chloroplasts in subapical cells after 1 week of culture because protonemata undergo apical growth.
The (S)-2-((R)-2-aminopropanamido)propanoic acid⋅trifluoroacetic acid salt (DA-LA⋅TFA salt) was prepared as follows. To a solution of N-(tert-butoxycarbonyl)-d-alanine (500.0 mg, 2.6 mM), l-Ala-OtBu hydrochloride (481.0 mg 1.0 equiv.), i-Pr2EtN (1.38 mL, 7.8 mM, 3.0 equiv.) in dry DMF (13.2 mL), and 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one (DEPBT, 791.5 mg, 2.6 mM, 1.0 equiv.) were added at 0°C. The reaction mixture was stirred for 18 h at 23°C under argon. The reaction mixture was slowly quenched with water at 0°C, and the resulting mixture was stirred for an additional 30 min at 23°C. The crude mixture was dissolved using EtOAc, followed by addition of aqueous 1 M HCl. The organic layer was separated, and the aqueous layer was extracted twice with EtOAc. The combined organic layer was washed with saturated aqueous NaHCO3, washed with saturated aqueous NaCl, and then dried over MgSO4 and filtered. The filtrate was concentrated under reduced pressure. Flash chromatography (SiO2, 20% EtOAc/n-hexane) provided (S)-tert-butyl 2-((R)-2-((tert-butoxycarbonyl)amino)propanamido)propanoate. Trifluoroacetic acid (TFA; 5 mL) was added to a solution of (S)-tert-butyl 2-((R)-2-((tert-butoxycarbonyl)amino)propanamido)propanoate in CH2Cl2 (5 mL), and the reaction mixture was stirred for 1 h at 23°C. The resulting mixture was concentrated under reduced pressure. An excess of CH2Cl2 was added to the residue, and the crude mixture was concentrated under reduced pressure three times to completely remove TFA. After the residue was dried under a vacuum, the DA-LA⋅TFA salts were obtained as a colorless amorphous powder (611.7 mg 84%). The spectral data were as follows: 1H NMR (500 MHz, CD3CN/D2O) δ 4.41 (q, J = 7.5 Hz, 1H), 4.09 (q, J = 7.0 Hz, 1H), 1.53 (d, J = 7.0 Hz, 3H), 1.46 (d, J = 7.5 Hz, 3H); 13C NMR (125 MHz, CD3CN/D2O) δ 176.8, 171.3, 50.0, 49.7, 17.2; ESI-MS m/z 161 [M+H]+.
The synthesis of (R)-2-((S)-2-aminopropanamido)propanoic acid⋅trifluoroacetic acid salt (LA-DA⋅TFA salt) was as follows. To a solution of N-(tert-butoxycarbonyl)-l-alanine (500.0 mg, 2.6 mM), d-Ala-OtBu hydrochloride (480.1 mg 1.0 equiv.), i-Pr2EtN (1.38 mL, 7.8 mM, 3.0 equiv.) in dry DMF (13.2 mL), and 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one (DEPBT, 791.2 mg, 2.6 mM, 1.0 equiv.) were added at 0°C. The reaction mixture was stirred for 18 h at 23°C under argon. The reaction mixture was slowly quenched with water at 0°C, and the resulting mixture was stirred for an additional 30 min at 23°C. The crude mixture was dissolved using EtOAc, followed by the addition of aqueous 1 M HCl. After the organic layer was separated, the aqueous layer was extracted twice with EtOAc. The combined organic layer was washed with saturated aqueous NaHCO3, washed with saturated aqueous NaCl, and then dried over MgSO4 and filtered. The filtrate was concentrated under reduced pressure. Flash chromatography (SiO2, 20% EtOAc/n-hexane) provided (R)-tert-butyl 2-((S)-2-((tert-butoxycarbonyl)amino)propanamido)propanoate. TFA (5 mL) was added to a solution of (R)-tert-butyl 2-((S)-2-((tert-butoxycarbonyl)amino)propanamido)propanoate in CH2Cl2 (5 mL), and the reaction mixture was stirred for 1 h at 23°C. The resulting mixture was concentrated under reduced pressure. An excess of CH2Cl2 was added to the residue, and the crude mixture was concentrated under reduced pressure three times to completely remove TFA. After the residue was dried under a vacuum, the LA-DA⋅TFA salts were obtained as a colorless amorphous powder (656.8 mg 91%). The spectral data were as follows: 1H NMR (500 MHz, CD3CN/D2O) δ 4.41 (q, J = 7.0 Hz, 1H), 4.09 (q, J = 7.0 Hz, 1H), 1.54 (d, J = 7.5 Hz, 3H), 1.46 (d, J = 7.5 Hz, 3H); 13C NMR (125 MHz, CD3CN/D2O) δ 176.9, 171.3, 50.0, 47.9, 17.5, 17.1; ESI-MS m/z 161 [M+H]+.
All reactions were monitored by thin-layer chromatography using Merck 60 F254 precoated silica gel plates (0.25 mm thickness). 1H and 13C NMR spectra were recorded on a JEOL ECX 500 FT-NMR spectrometer (500 MHz for 1H NMR, 125 MHz for 13C NMR). Data for 1H NMR are reported as chemical shift (δ ppm), multiplicity (d = doublet, q = quartet), coupling constant (Hz), integration, and assignment. Data for 13C NMR are reported as chemical shift. 1H NMR spectra recorded in CD3CN/D2O were referenced to the CH3CN peak at 2.06 ppm. 13C NMR spectra recorded in CD3CN/D2O were referenced to the CD3CN peak at 1.47 ppm. ESI-MS was recorded on a Shimadzu LCMS-2020. Flash chromatography was performed using silica gel 60N of Kanto Chemical.
Characterization of the P. patens DDL Genes
Previously, we identified two DDL cDNAs with minor differences (0.1%) (Machida et al., 2006) from the enriched, full-length cDNA library of P. patens (Nishiyama et al., 2000). To determine the number of DDL genes in the P. patens genome, the genome sequence of P. patens (http://www.phytozome.jgi.doe.gov; P. patens ver. 3.0 genome) (Rensing et al., 2008) was searched using TBLASTN and the DdlA amino acid sequence from Escherichia coli (YP_488674). The results suggested that only one gene exists in the genome (cDNA accession number AB194083). Another cDNA with an insertional mutation (AB194084) may have been generated by a PCR error.
RNA was isolated from wild-type protonemata, as described previously (Machida et al., 2006). Probes for RNA gel blot hybridization were generated using a PCR DIG Probe kit (Roche Diagnostics) with the appropriate primer sets. The primers used in this study are listed in Supplemental Table 1.
Subcellular Localization of the Pp-DDL-GFP Fusion Protein
Computer predictions of subcellular protein localization were made using TargetP software (Emanuelsson et al., 2000). To construct the PpDDLp-TP-GFP plasmid, which fused GFP to the N terminus of Pp-DDL driven from its native promoter, we used a DNA fragment that included the putative promoter sequence of 3635 bp from −3635 to 1 (A of the start codon [ATG] was defined as 1) and the coding sequence corresponding to the N-terminal 71 amino acid residues amplified by genomic PCR with the primers PpDdl2/F7-SacI and PpDdl2/R7-NcoI. Genomic DNA was isolated from the protonemata of P. patens using the cetyltrimethylammonium bromide method (Machida et al., 2006). The amplified DNA was digested with NcoI and SacI and then inserted into the SacI/NcoI-digested sGFP(S65T) plasmid (Chiu et al., 1996). P. patens was transformed as described previously (Machida et al., 2006). Bright-field and epifluorescence cell images were recorded with a CCD camera (Zeiss AxioCam) under a microscope (Zeiss Axioskop 2 plus). To construct the 35S-PpDDL-TP-GFP plasmid, in which GFP was fused to 71 amino acids from the N terminus of Pp-DDL, we amplified a DNA fragment by genomic PCR using the primers PpDdl2/F0-SalI and PpDdl2/R7-NcoI. The amplified DNA was digested with SalI and NcoI and then inserted into 35S-sGFP(S65T) digested with SalI and NcoI.
Generation of the Knockout Lines
To generate Pp-DDL-knockout plants, we constructed plasmids for gene targeting (Figure 2A). The p35S-Zeo plasmid carrying the Zeocin resistance gene expression cassette (Sakakibara et al., 2008) was used to target P. patens DDL. The Zeocin cassette consists of the CaMV35S promoter, bleomycin (Zeocin) resistance gene, and CaMV35S polyadenylation sequence. For gene disruption by homologous recombination, the 5′ region of Pp-DDL was amplified from wild-type P. patens genomic DNA via PCR using the primers PpDdl2/F3 and PpDdl2/R3, subjected to blunting using a Takara BKL kit (Takara Bio), and cloned into the blunted HindIII site located upstream of the Zeocin cassette in the p35S-Zeo plasmid. Next, the 3′ region was PCR-amplified using the primers PpDdl2/F4 and PpDdl2/R4, subjected to blunting, and inserted into the blunted XbaI site located downstream of the Zeocin cassette. The constructed plasmid was linearized by digestion with KpnI and SacI and used to transform P. patens. Primary screening for Zeocin-resistant transformants was performed using genomic PCR with a Pp-DDL gene-specific primer and a primer in the Zeocin cassette. We selected three transformants with a giant chloroplast phenotype using primary genomic PCR. DNA gel bots were used to detect additional insertions of the transformed DNA into the P. patens genome (Figure 2B). Probes for DNA gel blot hybridization were generated using a PCR DIG Probe kit (Roche Diagnostics) employing the primers PpDdl2/F4 and PpDdl2/R4. When the constructed plasmid was inserted into the Pp-DDL gene region by homologous recombination, one HindIII and one XbaI site was removed from the genome. Therefore, the sizes of the hybridized bands for transformed genomic DNA were changed from 7.4 (wild type) to 8.9 kb for HindIII digestion and from 9.0 (wild type) to 13 kb for XbaI digestion (Figure 2A). If the transformants had an additional insertion, we observed other hybridized bands. Only line #3 had no additional insertions (Figure 2B). For RT-PCR, we isolated RNA from the disrupted line and wild-type plants, treated it with DNase I, and used it to generate cDNA with oligo(dT) primers. RT-PCR was performed using the primers PpDdl2/F0 and PpDdl2/R0. Pp-Actin was used as a control.
Measurement of d-Ala
We harvested 150 mg of P. patens protonemata 5 d after transfer to new medium. Samples were frozen, ground to a powder in a liquid nitrogen-cooled mortar and pestle, and suspended in 10 mM potassium phosphate buffer (pH 7.0). After centrifugation, the supernatant was filtered through a Centricon apparatus (molecular mass cutoff, 3 kD; Merck Millipore). The samples were derivatized with o-phthaldialdehyde (OPA) and N-acetyl-l-cysteine (NAC), as described by Aswad (1984). An aliquot (1 µL) of the mixture was then subjected to UPLC. The analysis was performed using an Acquity UPLC TUV system consisting of a Waters Binary Solvent Manager, Sample Manager, FLR Detector, and Capcell Pak C18 IF2 (2.1 × 100-mm column, 2-µm particle size; Shiseido) with an eluent flow rate of 0.25 mL/min. The column temperature was 40°C, and the excitation and emission wavelengths for the fluorescent detection of amino acids were 350 and 450 nm, respectively. Data processing was performed using Empower 3 (Waters). The UPLC gradient system for OPA-NAC derivatives (A = 50 mM sodium acetate buffer, pH 6.9, and B = methanol) was 8 to 14% B for 1.5 min, 14 to 22% B for 0.1 min, 22 to 24% B for 4.4 min, 24 to 28% B for 2.0 min, 28 to 38% B for 3.0 min, 38 to 45% B for 0.5 min, 45 to 47% B for 3.5 min, 47 to 60% B for 3.0 min, and 60 to 8% B for 0.2 min. We prepared seven-point calibration curves for d- and l-Ala by plotting the peak areas of authentic d- and l-Ala against the corresponding concentration (µM) of the standard solutions. DAO from porcine kidney (Sigma-Aldrich) was used for the degradation of d-amino acids to ascertain whether the detected peak was d-Ala. The reaction mixture (0.5 mL) consisted of 59 mM pyrophosphate-HCl buffer (pH 8.5), 2.9 units of DAO, and the filtered sample. The reaction mixture was incubated at 25°C overnight. Next, the mixture was filtered through an Amicon Ultra 0.5-mL centrifugal filter 3 K device (Millipore). The flow-through was derivatized with OPA-NAC and the derivative was used for UPLC.
Electron Microscopy
For electron microscopy, samples were fixed in 2% glutaraldehyde buffered with 50 mM sodium cacodylate (pH 7.4) and exposed to a 2% osmium tetroxide aqueous solution containing 0.25% potassium hexacyanoferrate (II). Then, these were dehydrated using a series of ethanol washes and embedded in Quetol-651 resin. Thin sections were cut and stained with uranyl acetate and lead citrate and observed using a JEM-1200EX transmission electron microscope (JEOL).
Detection of Peptidoglycan
EDA was purchased from Nagase Co. The “clickable” Alexa Fluor 488 azide and Click-iT cell reaction buffer kit were purchased from Invitrogen (Life Technologies). EDA-DA was synthesized according to the methods of Liechti et al. (2014). Protonemata from the Pp-DDL-knockout transformant inoculated in BCDAT liquid medium including dipeptides for 9 d were used to detect peptidoglycan. ∆Pp-MurE plants grown in the medium supplemented with 1 mg/mL EDA-DA for 9 d were also used. The cells were washed three times with liquid medium, fixed, and permeabilized with 2.5% formalin, 0.1 M PIPES-NaOH (pH 6.8), 2.5 mM EGTA, 1 mM MgCl2, 0.01% Nonidet P-40, 1% DMSO, and 0.5 mM PMSF for 1 h and then washed once with 1% BSA in 1× PBS (8 g/L NaCl, 0.2 g/L KCl, 1.78 g/L Na2HPO4-2H2O, and 0.27 g/L KH2PO4, pH 7.4) and used for the click reaction with a Click-iT cell reaction buffer kit according to the manufacturer’s instructions. Confocal laser scanning microscopy images were collected using a Fluoview FV1200 confocal laser scanning microscope (Olympus) with a 100× objective lens.
Characterization of Arabidopsis T-DNA-Tagged Lines for DDL
The Arabidopsis plants used were wild-type (Columbia) and two T-DNA-tagged lines: SALK_092419 (At-ddl-1) and SAIL_906_E06 (At-ddl-2) of the DDL gene of Arabidopsis (McElver et al., 2001; Alonso et al., 2003). Sterilized seeds from the T-DNA-tagged plants were sown on 1× Murashige and Skoog medium containing vitamin B5 with 0.8% agar and 2% sucrose and grown in a cultivation room with a white light intensity of 100 µmol photons m−2 s−1 (Biolux lamp; NEC) at 23°C under a 9.5-h-light/14.5-h-dark cycle. The plants were later transplanted to soil and grown under the same conditions. Homozygotes of each line were obtained by self-pollination and confirmed by genomic PCR using a gene-specific primer and a primer corresponding to the sequence of the T-DNA left border (Figure 5B). For RT-PCR, we isolated RNA from mutant and wild-type plants, treated it with DNase I, and used the RNA to generate cDNA from oligo(dT) primers. RT-PCR was performed using the appropriate primer sets (AtDdlF0 and AtDdlR0). ACTIN served as a control. Arabidopsis protoplasts were isolated as described previously (Qi et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: cDNA for Pp-DDL AB184083 and AB194084.
Supplemental Data
Supplemental Table 1. Primers used in this study.
Supplemental Movie 1. Alexa 488 fluorescence in protonemal cells, as shown in Figure 4B.
Supplemental Movie 2. Chlorophyll autofluorescence in protonemal cells, as shown in Figure 4B.
Supplemental Movie 3. Fluorescent labeling of plastid peptidoglycan with chlorophyll autofluorescence.
Supplementary Material
Acknowledgments
We thank M. Hasebe (National Institute for Basic Biology, Japan) and Y. Niwa (University of Shizuoka, Japan) for providing the plasmids containing the Zeocin resistance gene and the sGFP(S65T) gene, respectively, and the ABRC for seeds from the SALK and SAIL lines. This work was supported by the Japan Society for the Promotion of Science KAKENHI (Grants 25440158, 26117720, and 15K07130 to H.T. and Ka.T.). H.I. and H.T. are members of the Research Core “Integrated Science for Molecular Chirality in Biology and Chemistry” of the Graduate School of Science and Technology of Kumamoto University.
AUTHOR CONTRIBUTIONS
T.H., Ko.T., Ka.T., and H.T. performed the experiments with P. patens. Y.S., Y.K., and T.O. carried out the measurements of d-amino acids. T.H., Ka.T., Sh.T., H.I., and H.T. observed the plastid peptidoglycan using click chemistry. M.S. performed the experiments with Arabidopsis. Su.T., Ka.T., and H.T. designed the research and analyzed the data. H.T. wrote the manuscript. All authors contributed to the preparation of the manuscript.
Glossary
- PEG
polyethylene glycol
- UPLC
ultraperformance liquid chromatography
- DAO
d-amino acid oxidase
- PEP
plastid-encoded RNA polymerase
- TFA
trifluoroacetic acid
- OPA
o-phthaldialdehyde
- NAC
N-acetyl-l-cysteine
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Ashton N.W., Cove D.J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Gen. Genet. 154: 87–95. [Google Scholar]
- Aswad D.W. (1984). Determination of d- and l-aspartate in amino acid mixtures by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. Anal. Biochem. 137: 405–409. [DOI] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning J.B., Murillo A.C., Chacon O., Barletta R.G., Sacchettini J.C. (2011). Structure of the Mycobacterium tuberculosis d-alanine:d-alanine ligase, a target of the antituberculosis drug d-cycloserine. Antimicrob. Agents Chemother. 55: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Erikson O., Hertzberg M., Näsholm T. (2004). A conditional marker gene allowing both positive and negative selection in plants. Nat. Biotechnol. 22: 455–458. [DOI] [PubMed] [Google Scholar]
- Frahn J.L., Illman R.J. (1975). The occurrence of d-alanine and d-alanyl-d-alanine in Phalaris tuberosa. Phytochemistry 14: 1464–1465. [Google Scholar]
- Garcia M., Myouga F., Takechi K., Sato H., Nabeshima K., Nagata N., Takio S., Shinozaki K., Takano H. (2008). An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 53: 924–934. [DOI] [PubMed] [Google Scholar]
- Henneberger C., Bard L., Rusakov D.A. (2012). d-Serine: a key to synaptic plasticity? Int. J. Biochem. Cell Biol. 44: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homi S., Takechi K., Tanidokoro K., Sato H., Takio S., Takano H. (2009). The peptidoglycan biosynthesis genes MurA and MraY are related to chloroplast division in the moss Physcomitrella patens. Plant Cell Physiol. 50: 2047–2056. [DOI] [PubMed] [Google Scholar]
- Hori K., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten B., Reski R. (1997). β-Lactam antibiotics inhibit chloroplast division in a moss (Physcomitrella patens) but not in tomato (Lycopersicon esculentum). Plant Physiol. 150: 137–140. [Google Scholar]
- Katayama N., Takano H., Sugiyama M., Takio S., Sakai A., Tanaka K., Kuroiwa H., Ono K. (2003). Effects of antibiotics that inhibit the bacterial peptidoglycan synthesis pathway on moss chloroplast division. Plant Cell Physiol. 44: 776–781. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner D.L., Green T.K. (2009). Separation and sensitive detection of d-amino acids in biological matrices. J. Sep. Sci. 32: 2305–2318. [DOI] [PubMed] [Google Scholar]
- Leganés F., Blanco-Rivero A., Fernández-Piñas F., Redondo M., Fernández-Valiente E., Fan Q., Lechno-Yossef S., Wolk C.P. (2005). Wide variation in the cyanobacterial complement of presumptive penicillin-binding proteins. Arch. Microbiol. 184: 234–248. [DOI] [PubMed] [Google Scholar]
- Liechti G.W., Kuru E., Hall E., Kalinda A., Brun Y.V., VanNieuwenhze M., Maurelli A.T. (2014). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506: 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M.R., Webb R.I., Strous M., Jetten M.S., Butler M.K., Forde R.J., Fuerst J.A. (2001). Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175: 413–429. [DOI] [PubMed] [Google Scholar]
- Machida M., Takechi K., Sato H., Chung S.J., Kuroiwa H., Takio S., Seki M., Shinozaki K., Fujita T., Hasebe M., Takano H. (2006). Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc. Natl. Acad. Sci. USA 103: 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H., Takechi K., Sato H., Takio S., Takano H. (2012). Treatment with antibiotics that interfere with peptidoglycan biosynthesis inhibits chloroplast division in the desmid Closterium. PLoS One 7: e40734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Maurelli A.T. (2006). Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14: 70–77. [DOI] [PubMed] [Google Scholar]
- McElver J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E., Lima P.T., Borges F., Silva A.C., Portes M.T., Carvalho J.E., Gilliham M., Liu L.-H., Obermeyer G., Feijó J.A. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 332: 434–437. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Hiwatashi Y., Sakakibara K., Kato M., Hasebe M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7: 9–17. [DOI] [PubMed] [Google Scholar]
- Noma M., Noguchi M., Tamaki E. (1973). Isolation and characterization of d-alanyl- d-alanine from tobacco leaves. Agric. Biol. Chem. 37: 2439. [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.-J., Oelmüller R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M., Aistleitner K., Biboy J., Gray J., Kuru E., Hall E., Brun Y.V., VanNieuwenhze M.S., Vollmer W., Horn M., Jensen G.J. (2013). Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4: 2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S.E., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Kishigami A., Nakagawa Y., Iida H., Sokabe M. (2004). A mechanosensitive anion channel in Arabidopsis thaliana mesophyll cells. Plant Cell Physiol. 45: 1704–1708. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Robinson T. (1976). d-amino acids in higher plants. Life Sci. 19: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Sakakibara K., Nishiyama T., Deguchi H., Hasebe M. (2008). Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol. Dev. 10: 555–566. [DOI] [PubMed] [Google Scholar]
- Steiner J.M., Löffelhardt W. (2002). Protein import into cyanelles. Trends Plant Sci. 7: 72–77. [DOI] [PubMed] [Google Scholar]
- Suzuki J.Y., Ytterberg A.J., Beardslee T.A., Allison L.A., Wijk K.J., Maliga P. (2004). Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 40: 164–172. [DOI] [PubMed] [Google Scholar]
- Takano H., Takechi K. (2010). Plastid peptidoglycan. Biochim. Biophys. Acta 1800: 144–151. [DOI] [PubMed] [Google Scholar]
- Thimmapuram J., Duan H., Liu L., Schuler M.A. (2005). Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome. RNA 11: 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Banzhaf M., Gross C.A., Vollmer W. (2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling M.C.F., Mesman R.J., Kuru E., Espaillat A., Cava F., Brun Y.V., VanNieuwenhze M.S., Kartal B., van Niftrik L. (2015). Anammox Planctomycetes have a peptidoglycan cell wall. Nat. Commun. 6: 6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W., Seligman S.J. (2010). Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18: 59–66. [DOI] [PubMed] [Google Scholar]
- Wargel R.J., Shadur C.A., Neuhaus F.C. (1970). Mechanism of d-cycloserine action: transport systems for d-alanine, d-cycloserine, l-alanine, and glycine. J. Bacteriol. 103: 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.