Arabidopsis HOOKLESS1 contributes to plant immunity and responses to the stress hormone ABA and is a critical component of the transcription complex modulating defense gene expression.
Abstract
Arabidopsis thaliana HOOKLESS1 (HLS1) encodes a putative histone acetyltransferase with known functions in seedling growth. Here, we show that HLS1 regulates plant responses to pathogens and abscisic acid (ABA) through histone acetylation at chromatin of target loci. The hls1 mutants show impaired responses to bacterial and fungal infection, accelerated senescence, and impaired responses to ABA. HLS1 modulates the expression of WRKY33 and ABA INSENSITIVE5 (ABI5), known regulators of pathogen and ABA responses, respectively, through direct association with these loci. Histone 3 acetylation (H3Ac), a positive mark of transcription, at WRKY33 and ABI5 requires HLS1 function. ABA treatment and pathogen infection enhance HLS1 recruitment and H3Ac at WRKY33. HLS1 associates with Mediator, a eukaryotic transcription coregulatory complex, through direct interaction with mediator subunit 18 (MED18), with which it shares multiple functions. HLS1 recruits MED18 to the WRKY33 promoter, boosting WKRY33 expression, suggesting the synergetic action of HLS1 and MED18. By contrast, MED18 recruitment to ABI5 and transcriptional activation are independent of HLS1. ABA-mediated priming of resistance to fungal infection was abrogated in hls1 and wrky33 mutants but correlated with ABA-induced HLS1 accumulation. In sum, HLS1 provides a regulatory node in pathogen and hormone response pathways through interaction with the Mediator complex and important transcription factors.
INTRODUCTION
Plants fend off infection by deploying multiple immune responses that correspond to the various pathogen virulence strategies. Pathogen-associated molecular patterns (PAMPs), such as components of fungal cell walls, chitin, or flagellin protein from bacterial flagella by pattern recognition receptors, stimulate PAMP-triggered immunity. A more specialized resistance, commonly known as effector-triggered immunity, is activated upon recognition of effectors by intracellular resistance (R) proteins (Jones and Dangl, 2006). Downstream of pathogen recognition, a cascade of regulatory hierarchy activates an array of molecules that counteract pathogens. Transcriptional regulations of genes encoding diverse molecules are widely recognized to be important in plant immune responses. Posttranslational modifications of histone tails are prominent mechanisms that modulate gene expression in plant responses to pathogens as well as regulate other plant functions. Posttranslational modifications of histone tails alter interactions between DNA and histones, resulting in chromatin structure that is permissive or repressive to transcription and other DNA metabolic processes (Chen and Tian, 2007). The open or permissive state of chromatin allows the recruitment of transcriptional complexes and enzymes, including DNA binding proteins, cofactors, and RNA polymerase II (RNAPII), to enhance transcription.
The acetylation of lysine residues on histones is dynamically regulated by histone acetyltransferases (HATs) and histone deacetylase that ultimately alter gene expression patterns. HATs are divided into four groups: GENERAL CONTROL OF NON-DEREPRESSIBLE 5-RELATED N-ACETYLTRANSFERASE; CREB binding protein (CBP)/p300 super families; members of MOZ-YBF2-SAS2-TIP60 (MYST); and TBP-associated factor 1, all of which have conserved catalytic domains (Pandey et al., 2002). GCN5 in Arabidopsis thaliana is implicated in plant developmental functions, such as floral meristem formation or root meristem differentiation (Bertrand et al., 2003; Kornet and Scheres, 2009), as well as responses to cold, light, or iron homeostasis (Vlachonasios et al., 2003; Benhamed et al., 2006; Xing et al., 2015). Histone acetyltransferases also play vital roles in plant resistance to the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000) (Defraia et al., 2013; Wang et al., 2013; Singh et al., 2014).
Mediator is a conserved multisubunit protein complex that functions as a cofactor between transcription factors and RNAPII (Reeves and Hahn, 2003). Through interaction with histone acetylation complexes, mediator also stimulates transcription (Liu et al., 2008). Mediator activates or represses transcription through interaction with transcription factors (An and Mou, 2013). Among 34 Mediator subunits identified in Arabidopsis, six are implicated in plant immunity functions through jasmonic acid/ethylene (JA/ET)- or salicylic acid (SA)-dependent defense pathways. For example, by interacting with MYC2, MEDIATOR25 (MED25) induces JA signaling genes to enhance resistance to fungal pathogens (Chen et al., 2012). MED16 regulates both JA/ET and SA pathways and contributes to resistance to Botrytis cinerea (Zhang et al., 2012). Recent data show that MED21 interacts with the chromatin-modifying enzyme, HISTONE MONOUBIQUITINATION1 (HUB1), affecting plant resistance to fungal infection (Dhawan et al., 2009). MED18 functions as a coactivator or repressor modulating abscisic acid (ABA) responses, flowering time, and plant defense through interactions with different transcription factors (Lai et al., 2014). Interestingly, MED18 also modulates specific gene expression and histone methylation of genes that function in pathogen and hormone responses (Lai et al., 2014). MED16 interacts with MED25 or nonmediator proteins to form a complex that regulates EIN3/EIL1-mediated pathway in iron homeostasis (Yang et al., 2014). Together, mediator subunits form a complex with RNAPII and different transcription factors or cofactors to regulate gene expression in hormone or defense signaling.
Arabidopsis HOOKLESS1 (HLS1) was previously implicated in the regulation of seedling growth responses to ethylene. However, despite its high sequence similarity to histone acetyltransferases, its molecular, biochemical, and physiological functions are still poorly understood. Here, we describe the functions of HLS1 in plant defense and the underlying molecular mechanisms. The hls1 mutant displayed enhanced disease symptoms in response to fungal and bacterial infections, accelerated senescence, and impaired responses to the plant hormone ABA, suggesting the critical role of HLS1 in regulating these processes. The hls1 mutant expressed enhanced bacterial disease symptoms regardless of the Pst strain. By contrast, the hls1 mutant supported increased bacterial growth when inoculated with Pst DC3000 (avrRpm1) with no increase in bacterial growth after inoculation with virulent and nonpathogenic strains. Ectopic expression of HLS1 enhanced resistance to B. cinerea, which was associated with B. cinerea- or ABA-induced accumulation of HLS1 protein. In the absence of HLS1, ABA promotes senescence and enhanced susceptibility to B. cinerea. In addition, HLS1 and MED18, which are both required for seedling apical hook formation, responses to ABA, and resistance to B. cinerea, physically interact. HLS1 recruits MED18 to the WRKY33 locus, which then enhances transcriptional activation of WRKY33. Interestingly, HLS1 modulates histone H3 acetylation at hormone and pathogen response regulatory loci ABI5 and WRKY33. ABA and B. cinerea enhance HLS1 transcriptional and posttranslational regulation and HLS1-mediated histone H3 acetylation at target loci. Our results shed light on molecular mechanisms underlying the multiple biological functions of HLS1 in plant hormone and defense responses.
RESULTS
Arabidopsis HLS1 Mediates Responses to Fungal and Bacterial Pathogens
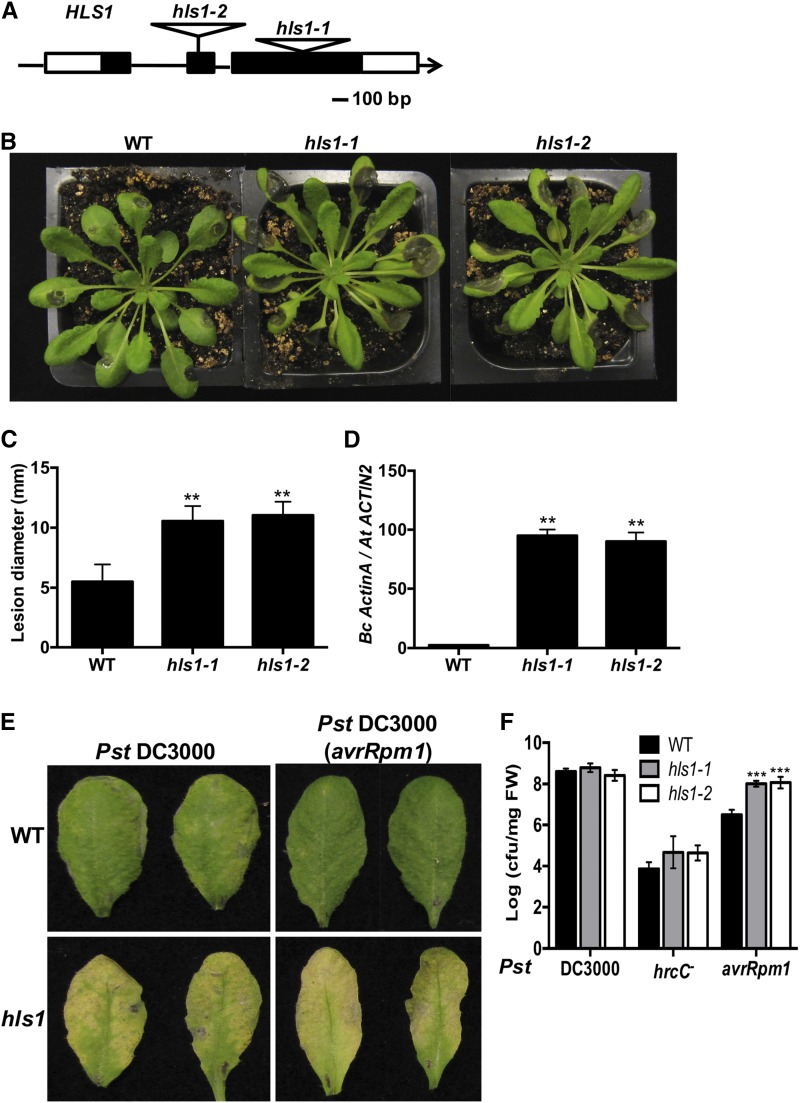
We recently described the Arabidopsis med18 mutant, which displays enhanced susceptibility to the necrotrophic fungal plant pathogen B. cinerea and hookless phenotype in seedlings (Lai et al., 2014). The hookless phenotype in the med18 mutant involves altered seedling apical hook formation in response to ethylene and is associated with impaired defense responses in adult plants. Due to the potential link between defense and the hookless phenotype, the Arabidopsis hls1 mutant was tested for altered defense responses. Two Arabidopsis T-DNA insertion alleles of HLS1 (hls1-1, SALK-136528C; hls1-2, SLAK-009473) that lacked any HLS1 transcript (Supplemental Figure 1) displayed increased susceptibility to B. cinerea with larger disease lesions, enhanced necrosis, and chlorosis compared with the wild-type Col-0 plants (Figures 1A to 1C). More fungal biomass accumulated in the hls1 mutant as measured by quantitative PCR amplification of B. cinerea ActinA DNA, confirming the role of HLS1 in suppressing fungal growth in infected plants (Figure 1D).
Figure 1.
Arabidopsis HLS1 Mediates Responses to Fungal and Bacterial Pathogens.
(A) Schematic diagram showing T-DNA insertion alleles of the HLS1 gene. HLS1 gene and T-DNA insertions are shown in the hls1 mutant alleles. Black and white squares are exons and untranslated regions, respectively.
(B) B. cinerea disease symptoms showing susceptibility of hls1 mutant and
(C) Disease lesion size in the wild-type and hls1 mutants. The data represent mean values ± sd from (n = 36).
(D) Enhanced B. cinerea growth in hls1 mutants as measured by qPCR. Fungal growth was determined by qPCR amplification of the B. cinerea ActinA gene relative to Arabidopsis ACTIN2 gene. The data represent mean values ± se (n = 3).
(E) Enhanced disease symptoms in hls1 mutant after inoculation with Pst strain. Inoculated leaves were detached for photographing at 3 days after inoculation (dai). Plants were infiltrated with a bacterial suspension (OD600 = 0.0005, ∼2.5 × 105 CFU/mL).
(F) Bacterial growth in the wild-type and hls1 mutants showing altered responses to bacterial infection. Bacterial growth is expressed in colony-forming units per mg fresh weight (cfu/mg FW). The data represent mean values ± sd (n = 24).
In (C), (D), and (F), the mean values with statistically significant differences are indicated by asterisks (ANOVA test: **P < 0.01 and ***P < 0.001). The experiment was repeated three times with similar results.
To test the role of HLS1 in bacterial resistance, hls1 mutant plants were infiltrated with different Pst DC3000 strains. Regardless of the strain, hls1 mutants displayed enhanced disease symptoms, composed primarily of chlorotic lesions (Figure 1E). Response of hls1 mutants to the nonpathogenic strain of Pst DC3000 hrcC- was comparable to wild-type plants. The Pst DC3000 hrcC- strain is defective in Type III secretion but retains PAMP molecules through which it is able to activate PAMP-triggered immunity. The hls1 mutant supported a level of bacterial growth comparable to that in wild-type plants after inoculation with the virulent bacterial strain Pst DC3000 (Figure 1F). By contrast, inoculation with the avirulent bacterial strain Pst DC3000 (avrRpm1) resulted in significantly more bacterial growth in the hls1 mutants than the wild-type plants. Thus, HLS1 suppresses disease symptom expression regardless of the bacterial strain but also is required to limit growth of Pst DC3000 (avrRpm1), suggesting that HLS1 modulates some specific effector-triggered immunity responses.
HLS1 Regulates Expression of Defense-Related Genes in Response to B. cinerea
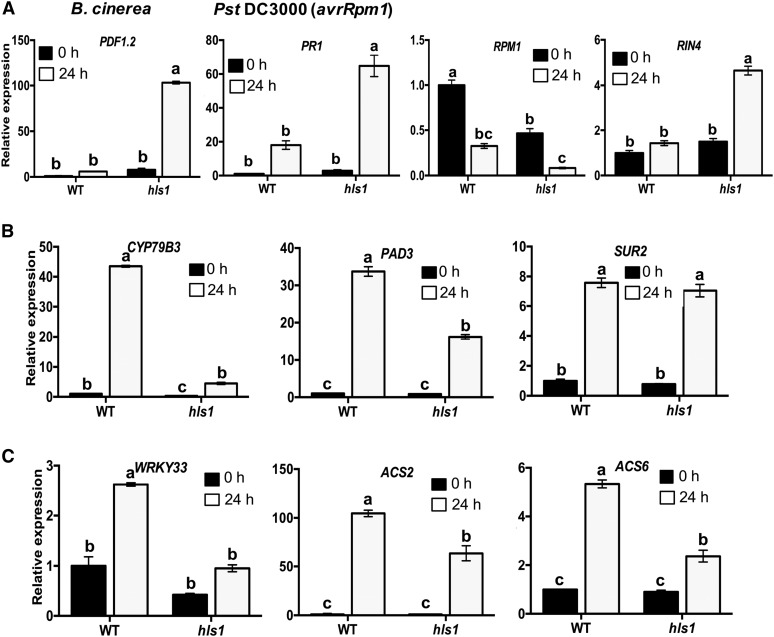
To gain insight into the molecular function of HLS1 in plant defense responses, the expressions of immune response marker genes were studied. PLANT DEFENSIN1.2 (PDF1.2) is a well-known defense-related gene induced by fungal infection in JA- and ET-dependent pathways (Penninckx et al., 1998). The expression of PDF1.2 significantly increased in the hls1 mutant after B. cinerea infection (Figure 2A), while that of PATHOGENESIS-RELATED PROTEIN1 (PR1) increased after Pst DC3000 (avrRpm1) infiltration. The expression of PR1 correlates with activation of the SA pathway that mediates responses to Pst (Delaney et al., 1994), implying HLS1-mediated Pst resistance is unlikely to be due to loss of the SA-mediated defense pathway but rather due to the increase in SA levels in response to increased bacterial growth in the hls1 mutant (Figure 2A). The R genes P. syringae pv maculicola 1 (RPM1) and RPM1-INTERACTING PROTEIN4 (RIN4) are positive and negative regulators of resistance to Pst DC3000 (avrRpm1), respectively. However, the expression of RPM1 was reduced and RIN4 expression increased after Pst DC3000 (avrRpm1) inoculation in the hls1 mutant. Together, a distinct regulation of RPM1 and RIN4 expression by HLS1 correlated with resistance to Pst DC3000 (avrRpm1).
Figure 2.
HLS1 Regulates Expression of Genes in Different Defense Pathways.
(A) The expression of defense genes: PDF1.2 after inoculation with B. cinerea and PR1, and RPM1 and RIN4 after inoculation with Pst DC3000 (avrRpm1).
(B) Phytoalexin and glucosinolate biosynthesis genes CYP79B3, PAD3, and SUR2 in B. cinerea-inoculated plants.
(C) Transcription factor WRKY33 and ethylene biosynthesis genes ACS2 and ACS6 in response to B. cinerea.
In (A) to (C), relative gene expression is normalized to Arabidopsis ACTIN2 (ACT2) gene. The data represent mean values ± se (n = 3) from at least two independent experiments. The gene expression in the wild type at 0 h after inoculation is set to 1. Statistically significant differences are marked by different letters (least squares means post hoc test: P < 0.05).
Based on genetic evidence, HLS1 is predicted to function at the boundary between ethylene and auxin pathways (Li et al., 2004). Therefore, we tested the expression of genes in auxin perception, signaling, and transport, which also affect plant immunity (Kazan and Manners, 2009). The expression of CYP79B3, encoding an enzyme that catalyzes the conversion from tryptophan to indole-3-acetaldoxime, a precursor of camalexin and indole-glucosinolates (IGs), was reduced in the hls1 mutant (Figure 2B). IG also contributes to fungal resistance and SUR2 is required for conversion of indole-3-acetaldoxime to IG. The expression of SUR2 displayed no difference between the wild type and the hls1 mutant in response to B. cinerea (Figure 2B). PHYTOALEXIN DEFICIENT3 (PAD3) is an enzyme required for camalexin biosynthesis and widely known to contribute to fungal resistance (Zhou et al., 1999). PAD3 expression was significantly reduced in the hls1 mutant consistent with the susceptibility of the hls1 mutant to fungal infection (Figure 2B) (Petersen et al., 2008). The transcription factor WRKY33 regulates expression of camalexin biosynthesis genes, including PAD3 and CYP79B3 (Zheng et al., 2006; Mao et al., 2011). Pathogen-induced WRKY33 expression was attenuated in the hls1 mutant (Figure 2C). The 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS2 and ACS6) genes, which function in the WRKY33-regulated ethylene biosynthesis pathway, also showed reduced expression in the hls1 mutant after B. cinerea inoculation (Figure 2C). The results further confirm the effects of HLS1 on WRKY33-regulated pathway. These gene expression data indicate that HLS1 regulates WRKY33 to modulate the expression of downstream genes.
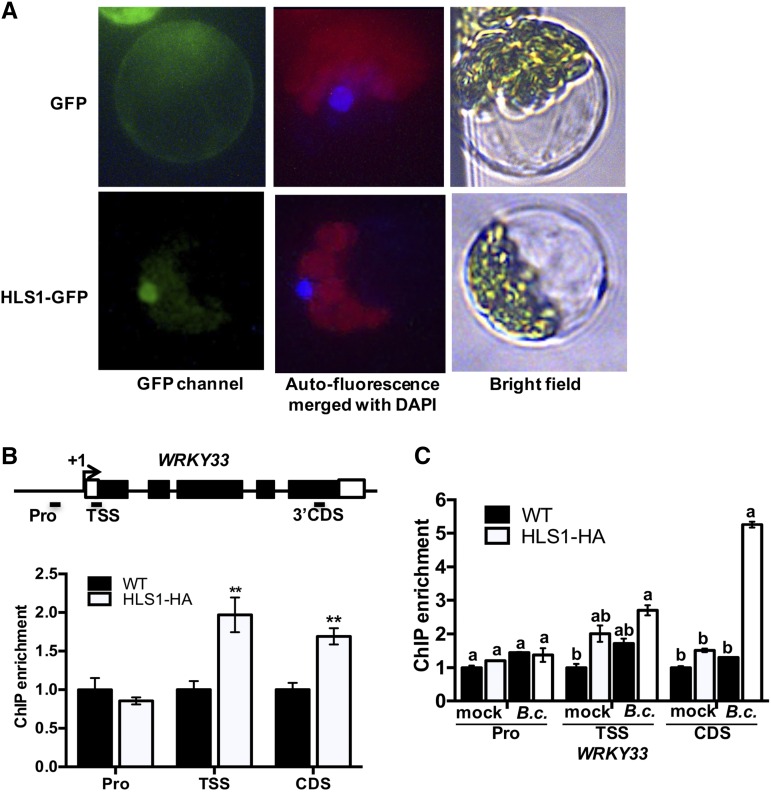
HLS1 Is a Nuclear Protein That Associates with WRKY33
HLS1 is a putative GCN5 acetyltransferase based on amino acid sequence alignment to other known acetyltransferases but its molecular function has not been determined (Lehman et al., 1996). Epitope-tagged HLS1 predominantly localized to the nucleus in Arabidopsis protoplasts consistent with its potential function as a histone acetyltransferase (Figure 3A). To determine whether HLS1-dependent expression of target genes is through direct association with target genes, the recruitment of HLS1 to specific genes was tested by chromatin immunoprecipitation-qPCR (ChIP-qPCR) experiments with transgenic plants that express hemagglutinin (HA)-tagged HLS1 under the control of the CaMV 35S promoter (HLS1-HA). WRKY33 is a potential target of HLS1 due to the loss of basal and induced WRKY33 expression in the hls1 mutant. HLS1 associated with the transcription start site (TSS) and 3′-coding sequence (CDS) regions of WRKY33 as revealed by the precipitation of HLS1 and WRKY33 protein-DNA complex (Figure 3B). Interestingly, the association between HLS1 and WRKY33 was enriched after B. cinerea inoculation consistent with their functions in regulating responses to pathogens (Figure 3C). Thus, HLS1 directly regulates WRKY33 expression through association with its regulatory regions. However, HLS1-HA failed to associate with the TSS or CDS region of the RPM1 gene despite the reduced expression of RPM1 in the hls1 mutant (Supplemental Figure 2). Hence, the effects of HLS1 on RPM1 expression and its role in Pst DC3000 (avrRpm1) resistance appear to be indirect.
Figure 3.
Subcellular Localization of HLS1 and Its Association with Target Genes.
(A) HLS1 is predominantly localized to the nucleus. A plasmid expressing HLS1-GFP fusion under the control of the CaMV 35S promoter was transfected into Arabidopsis protoplasts. The fluorescence signals were observed by epifluorescence microscopy. The red, green, and blue signals are autofluorescence from chloroplasts, GFP, and DAPI, respectively. The merged image is an overlap of DAPI staining and autofluorescence channels.
(B) HLS1 associates with the WRKY33 gene. The upper panel shows a schematic diagram of the genomic structure of the WRKY33 gene. The association of HLS1 in the wild type with WRKY33 is set to 1 as a background control. Statistically significant differences are indicated by asterisks (Student’s t test: **P < 0.01). Black squares and +1 represent exons and the transcription start site for WRKY33 gene, respectively. The short black bars represent qPCR amplicons in different regions of the WRKY33 gene.
(C) HLS1 association with the WRKY33 is enhanced in B. cinerea-inoculated plants. The association of the wild type with the WRKY33 gene under mock treatment is set to 1 as a background control. Statistically significant differences are marked by different letters (least squares means post hoc test: P < 0.05).
In the ChIP-qPCR assay ([B] and [C]), the chromatin and protein complexes were prepared from Arabidopsis wild-type and HLS1-HA-overexpressing plants. The anti-HA antibody was used for immunoprecipitation, and IgG antibody was used as a negative control for immunoprecipitation. The ACT2 gene represented an internal control for qPCR. The data represent mean values ± se (n = 3). The experiment was repeated three times with similar results. HLS1-HA, HLS1-tagged HA epitope driven by CaMV 35S promoter; Pro, promoter region; Mock, treated with 1% Sabouraud Maltose Broth; B.c., B. cinerea inoculation.
HLS1 Mediates Histone Acetylation on WRKY33 Chromatin
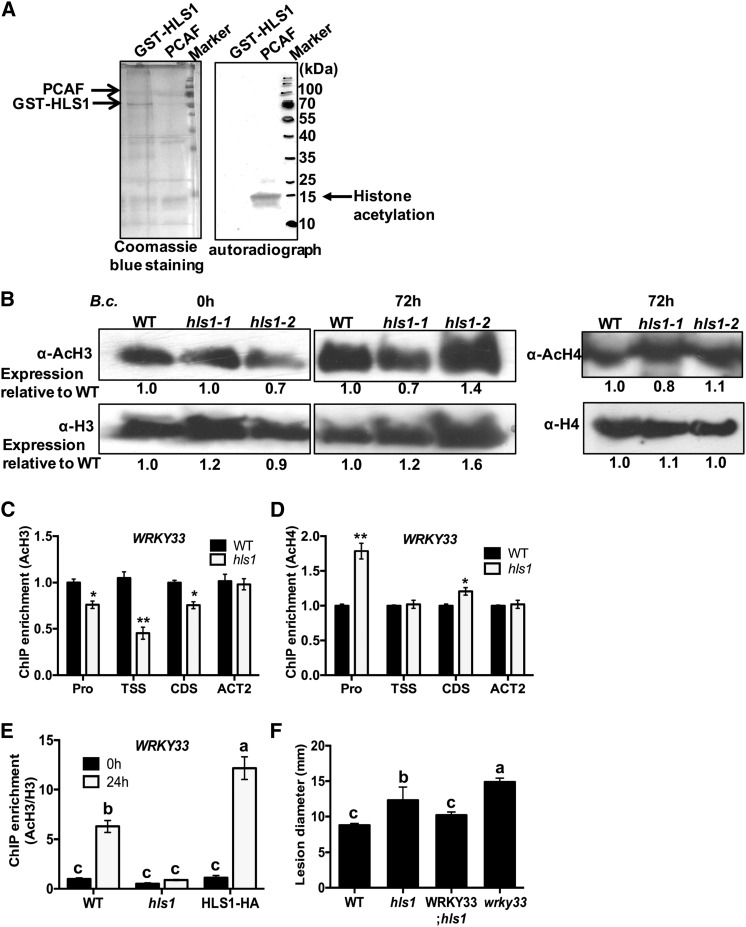
Previously, Arabidopsis HLS1 was studied in connection with plant growth, but its biochemical function was not determined. In light of the predicted HAT functions of HLS1, we tested the acetyltransferase activity of recombinant HLS1 protein tagged with glutathione S-transferase (GST-HLS1) purified from Escherichia coli. Autoradiographs labeled with tritium of the acetyl group revealed no detectable histone acetyltransferase activity from recombinant HLS1 protein when core histones were supplied as substrates (Figure 4A). The control reaction with the P300/CBP-associated factor (PCAF), a protein with known HAT activity, clearly acetylated histones. Global histone H3 and H4 acetylation was analyzed in hls1 mutants to clarify the impact of HLS1 on this process in vivo. There were no significant differences in the levels of global histone acetylation in the hls1 mutant before or after inoculation with B. cinerea (Figure 4B).
Figure 4.
HLS1 Modulates Histone H3 Acetylation on WRKY33 Chromatin.
(A) Histone acetyltransferase activity assay with recombinant GST-HLS1. The recombinant protein expressing full-length HLS1 tagged with GST (GST-HLS1) was produced in E. coli and affinity purified. The recombinant acetyltransferase PCAF protein was used as a positive control in the HAT assay. The chicken core histone used as substrate was obtained from Millipore. The recombinant GST-HLS1 protein run on 15% SDS-PAGE gel and stained with Coomassie blue is shown as a loading control (left panel). The H3-labeled acetyl-CoA was detected on acetylated histones by autoradiography. The autoradiograph shows that PCAF, but not HLS1, transferred acetyl groups labeled with H3 from acetyl-CoA to core histones.
(B) Global H3 and H4 acetylation is not altered in hls1 mutant plants. Histone proteins were extracted from plants inoculated with B. cinerea. The global histone H3, acetyl-histone H3, histone H4, and acetyl-histone H4 accumulations were detected using specific antibodies as indicated. The signals on immunoblots were quantified by ImageJ software and numbers are shown corresponding to each lane on the blot.
(C) and (D) ChIP-qPCR showing histone H3 (C) and H4 (D) acetylation at WRKY33 chromatin. ChIP-qPCR was performed using antibodies that recognize acetylated histone H3 or H4. The different regions of WRKY33 were amplified by qPCR. The enrichment of WRKY33 in wild-type plants was set to 1 as a background control in ChIP-qPCR assay. The data represent mean values ± se (n = 3). Statistically significant differences are indicated by asterisks (Student’s t test: *P < 0.05 and **P < 0.01).
(E) Histone H3 acetylation at WRKY33 chromatin is enhanced in B. cinerea-inoculated plants. The chromatin complexes were collected from wild-type, hls1 mutant, and HLS1-HA plants at 0 or 24 h after B. cinerea inoculation. The H3 acetylation status is normalized with histone H3 from each sample. The enrichment of WRKY33 in the wild type at 0 h is set to 1 as a background control in the assay. The data represent mean values ± se (n = 3). Statistically significant differences are marked by different letters (least squares means post hoc test: P < 0.05).
(F) Overexpression of WRKY33 restored the disease susceptibility of the hls1 mutant to wild-type levels. The data represent mean disease lesion size ± se (n = 12). The statistical significance of the differences in the mean values is indicated by different letters (least squares means post hoc test: P < 0.05). The experiment was repeated at least two times with similar results.
Pro, promoter region; WRKY33; hls1, overexpressing WRKY33 in the hls1 mutant background.
The histone acetylation at chromatin of the WRKY33 locus was tested in the hls1 mutant with antibodies that recognize acetylated histones (AcH3 and AcH4). Interestingly, the histone H3 acetylation at WRKY33 chromatin was reduced significantly in the hls1 mutant relative to the wild type (Figure 4C). However, histone H4 acetylation at the WRKY33 promoter region was increased significantly relative to other regions that show no difference between the wild type and the hls1 mutant. The results suggest that WRKY33 chromatin is mainly acetylated by HLS1 on histone H3 (Figure 4D). To eliminate the possibility that the reduction of histone H3 acetylation in the hls1 mutant results from the loss of histone H3 occupancy, we normalized the histone H3 acetylation to total histone H3 on the WRKY33 TSS region. The significantly increased histone H3 acetylation in wild-type plants correlated with WRKY33 induction triggered by B. cinerea (Figures 2C and 4E). Interestingly, the increased histone H3 acetylation level in wild-type plants was suppressed in the hls1 mutant. By contrast, histone H3 acetylation in the HLS1-overexpressing plants was enriched significantly more than in wild-type plants (Figure 4E; Supplemental Figure 3). These results suggest that the regulatory impact of HLS1 on WRKY33 gene expression is mediated through histone H3 acetylation. To determine the biological function of this regulation, transgenic plants overexpressing WRKY33 in hls1 mutant background were tested for their responses to B. cinerea. WRKY33 rescued the B. cinerea susceptibility of hls1 mutant to wild-type levels (Figure 4F), suggesting that WRKY33 is a direct target of HLS1 during plant responses to pathogens.
HLS1 Mediates Responses to ABA through Direct Association and Histone Acetylation of ABI5
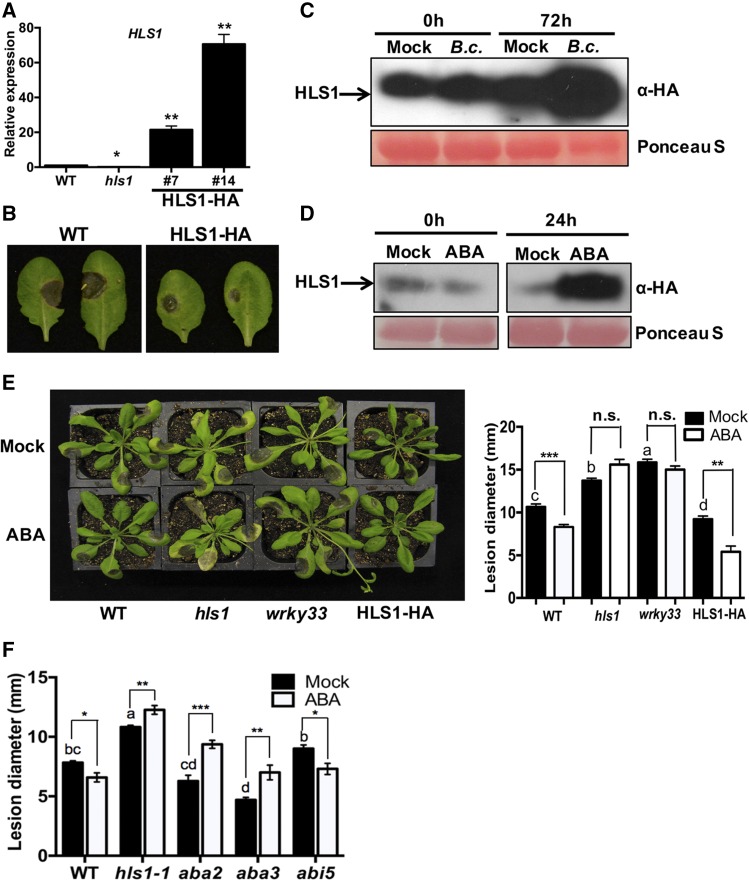
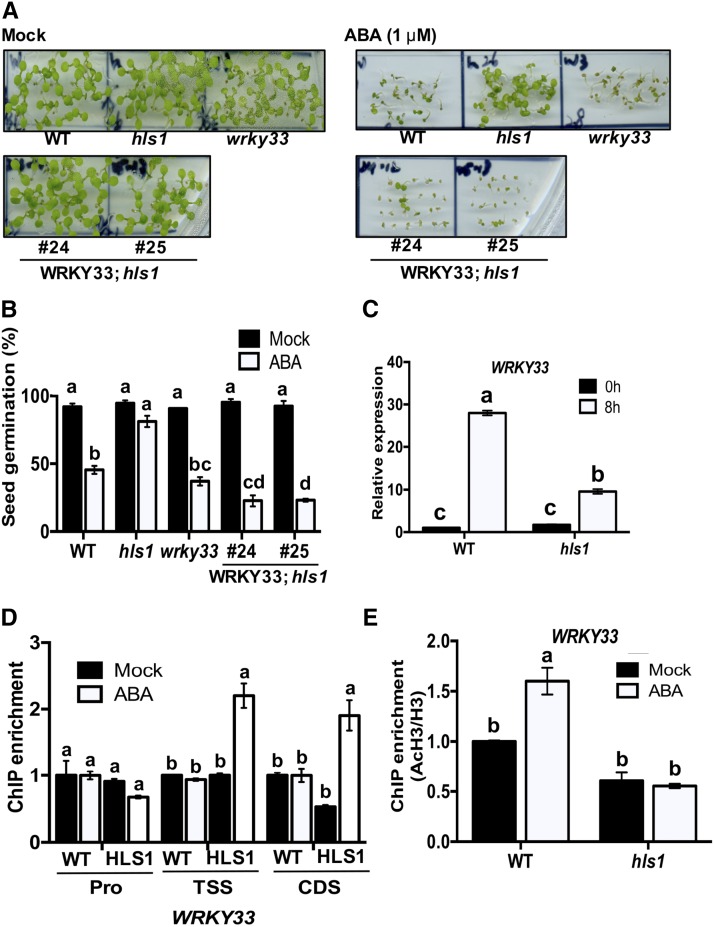
The med18 and hls1 mutants share similar phenotypes such as disease susceptibility and hookless seedlings in response to ethylene. To determine whether the functional overlap between MED18 and HLS1 extends to other biological functions, the role of HLS1 in response to ABA was tested in seedling germination and growth assays. The role of MED18 in plant response to ABA has been studied previously (Lai et al., 2014). In the presence of 0.5 or 1 µM ABA, the germination of wild-type seedlings was reduced, but med18 and hls1 mutants were less affected, exhibiting insensitivity to ABA similar to the abi5 mutant used as a positive control (Figures 5A and 5B). By contrast, transgenic seedlings expressing HLS1-HA were hypersensitive to ABA relative to the wild-type plants. In addition, HLS1 expression was induced by ABA treatment (Figure 5C), confirming that HLS1 is positively regulated by ABA and contributes to ABA response.
Figure 5.
HLS1 Associates with ABI5 through Histone Acetylation Regulating Responses to ABA.
(A) HLS1 is required for ABA responses as measured by seed germination assays on ABA-containing media. The data represent mean values ± sd (n = 30), and the statistically significant difference is indicated by an asterisk (ANOVA test: *P < 0.05).
(B) Seed germination and seedling growth on media supplemented with ABA.
(C) Expression of HLS1 is induced by ABA. The data represent mean values ± se (n = 3) from three independent biological replicates, and the statistically significant differences are indicated by asterisks (ANOVA test: **P < 0.01 and ***P < 0.001).
(D) HLS1 is required for ABA-induced expression of ABA response and regulatory genes. The expression of ABA response genes in the wild type after mock inoculation is set to 1. The data represents mean values ± se (n = 3) from three independent experiments, and statistically significant differences are indicated by different letters (least squares means post hoc test: P < 0.05).
(E) Schematic diagram showing the genomic structure of ABI5. HLS1 associates with TSS and CDS regions on the ABI5 gene. The association of the wild type with ABI5 gene is set to 1 as a background control.
(F) Histone H3 acetylation at ABI5 chromatin is reduced in the hls1 mutant. The histone H3 acetylation levels in the wild type are set to 1 as a background control.
In (E) and (F), the data represent mean values ± se (n = 3) from three independent biological replicates. The statistically significant difference is indicated by asterisks (Student’s t test: *P < 0.05 and **P < 0.01). HLS1-HA, HLS1-tagged HA epitope driven by CaMV 35S promoter; ABS, ABI4 binding region.
To assess the impact of HLS1 on the ABA response pathway at the molecular level, the expression of genes involved in the ABA response pathway was examined. Expression of WRKY40, RD29a, HY5, ABI3, and ABI5 genes that are involved in the ABA signaling pathway were attenuated in the hls1 mutant in response to ABA (Figure 5D). Interestingly, HLS1 directly associated with the ABI5 gene, particularly with the TSS and CDS regions, but the region for a transcription factor ABI4 recruitment (ABI4 binding site [ABS]) remained at background level (Figure 5E). Thus, ABI5 expression is directly regulated by HLS1 similar to our previous observation showing MED18 directly regulates ABI5 expression (Supplemental Figure 4A) (Lai et al., 2014). However, the recruitment of HLS1 to the ABI5 promoter was not enhanced by B. cinerea (Supplemental Figure 4B), which suggests that HLS1 association with ABI5 is not directly related to plant defense against B. cinerea. ChIP-qPCR experiments with AcH3 antibody revealed that the hls1 mutant has significantly reduced AcH3 levels at the ABI5 TSS and CDS regions, whereas the H3 acetylation level at Arabidopsis ACTIN2, used as a control, was comparable between hls1 and wild-type plants (Figure 5F), suggesting that HLS1 is required for histone H3 acetylation. Therefore, we concluded that HLS1-mediated ABI5 expression correlates with the HLS1-dependent histone H3 acetylation.
Functional and Molecular Convergence of HLS1 and MED18 on Target Genes ABI5 and WRKY33
Due to the overlapping biological functions of HLS1 and MED18, we tested their physical interactions by coimmunoprecipitation (co-IP) assays. MED18-MYC precipitated with HLS1-HA in Nicotiana benthamiana after transient coexpression, suggesting their presence in the same complex (Figure 6A). MED18-MYC immunoprecipitated with HLS1-HA but not with the empty vector expressing MYC alone. The interaction was confirmed in transgenic Arabidopsis plants coexpressing MED18-MYC and HLS1-HA (Figure 6B). Our current and previous data demonstrated that HLS1 and MED18 are required for responses to ABA and that both proteins associate with the ABI5 gene (Figures 6A and 6B; Lai et al., 2014). To confirm their functional interdependence, the HLS1-HA and MED18-MYC plasmids were transfected together with a reporter construct, ABI5 promoter fused with GUS (pABI5:GUS) into protoplasts. HLS1-HA or MED18-MYC induced ABI5-GUS expression in protoplast transactivation assay (Figure 6C). Furthermore, ABI5-GUS accumulation significantly increased when MED18-HA and HLS1-MYC were coexpressed, suggesting their synergistic action on ABI5 expression. Similarly, the synergistic action of HLS1 and MED18 on WRKY33 expression was determined in parallel protoplast transactivation experiments (Figure 6C). Transient HLS1 expression enhanced WRKY33-GUS accumulation, confirming the regulatory role of HLS1 on WRKY33 expression (Figure 2C). However, MED18 failed to activate WRKY33 gene expression when expressed alone. Interestingly, MED18 enhanced expression of WRKY33 when coexpressed with HLS1 in protoplasts, suggesting it enhances the function of HLS1 on transcriptional regulation of WRKY33 (Figure 6C). RT-qPCR data indicated that the B. cinerea-induced WRKY33 expression is reduced in the hls1 mutant and in MED18-MYC; hls1 transgenic plants comparable to the hls1 mutant, implying that MED18-mediated expression of WRKY33 induction requires HLS1 (Figure 6D). In addition, ChIP-qPCR assay further confirmed that MED18-MYC was associated with the WRKY33 TSS and 3′CDS regions after B. cinerea inoculation, but this association was lost in the absence of HLS1 (Figure 6E). However, disease assays on MED18-MYC; hls1 transgenic plants revealed that MED18-MYC rescued the B. cinerea susceptibility of the hls1 mutant to wild-type levels, suggesting that MED18-mediated plant immunity may be partially independent of HLS1 (Figure 6F). The expression of LOCUS OF INSENSITIVITY TO VICTORIN (LIV1, TRX-h5), a defense-associated thioredoxin and direct target for the victorin toxin (Lorang et al., 2012), was elevated in the med18 mutant, which contributes to its susceptibility (Lai et al., 2014). TRX-h5 expression is reduced in MED18 overexpression plants with a similar pattern of expression in MED18-MYC; hls1 and MED18-MYC; HLS1 plants, demonstrating that MED18-modulating plant defense may be HLS1 independent (Supplemental Figure 5). In sum, our data suggest that HLS1 and MED18 coregulate WRKY33 and ABI5 genes. Interestingly, MED18-mediated plant resistance appears to be partially dependent on HLS1 function.
Figure 6.
Molecular Interaction between MED18 and HLS1 and Their Synergistic Action on Target Gene Expression.
(A) and (B) Interaction between MED18 and HLS1 in co-IP assay in N. benthamiana (A) and transgenic Arabidopsis (B) plants. In (A), HLS1-HA was transiently coexpressed with MED18-MYC by agroinfiltration in N. benthamiana leaves. The empty vector expressing MYC was used as a negative control. In (B), transgenic Arabidopsis plants stably expressing HLS1-HA and MED18-MYC were used in the co-IP assays. Anti-HA beads were used to precipitate HLS1-HA protein. Anti-HA (α-HA) and anti-MYC (α-MYC) antibodies were used to detect protein accumulation in input or immunoprecipitated (IP) samples.
(C) Synergistic action of HLS1 and MED18 in the regulation of ABI5 or WRKY33 expression. The schematic diagram shows plasmid constructs used in transcriptional activation assay. The CaMV 35S promoter driving the luciferase reporter gene (35S:LUC) and the WRKY33 or ABI5 promoter fused with GUS reporter gene (pWRKY33/pABI5:GUS) are used as an internal control and a reporter, respectively. 35S promoter driving expression of HLS1 tagged with HA (HLS1-HA) is used as an effector. The bar graphs show the mean relative GUS activity from expression of the various plasmids depicted in the schematic. The mean values from protoplasts transfected with empty vector, pWRKY33/pABI5:GUS, and 35S:LUC were set to 1 as an internal control. The GUS signal is normalized with the LUC signal. The data represent mean values ± se (n = 3) from two independent biological replicates, and statistically significant differences are indicated by different letters (least squares means post hoc test: P < 0.05).
(D) MED18-mediated WRKY33 expression is dependent on HLS1. Relative gene expression is normalized to ACT2. The relative expression in wild-type plants at 0 h is set to 1.
(E) MED18 recruitment to transcription start site and 3′-coding regions of WRKY33 is enhanced by inoculation with B. cinerea in an HLS1-dependent manner. The enrichment of the WRKY33 gene in the wild type at 0 h is set to 1 as a background control in the ChIP-qPCR assay.
(F) Ectopic expression of MED18 rescues disease phenotype of hls1 mutant. The disease lesion size was determined after drop inoculation with B. cinerea. The data represent mean values ± se (n = 20). Statistically significant differences are indicated by asterisks compared with wild-type plants (Student’s t test: *P < 0.05 and ***P < 0.001).
In (D) and (E), the data represent mean values ± se (n = 3), and the statistically significant differences are marked by different letters (least squares means post hoc test: P < 0.05). pABI5:GUS and pWRKY33:GUS, reporter GUS fused with ABI5 or WRKY33 promoter region, respectively; HLS1, HLS1 tagged with HA; MED18, MED18 tagged with MYC. MED18; WT, overexpressing MED18 in wild-type background. MED18; hls1, overexpressing MED18 in the hls1 mutant background.
ABA-Induced HLS1 Accumulation Enhances Resistance to B. cinerea
To test whether overexpression of HLS1 is sufficient for resistance to B. cinerea, we generated transgenic Arabidopsis plants. Two independent HLS1-HA transgenic lines with increased HLS1 expression were selected (Figure 7A). The transgenic plants showed enhanced resistance to B. cinerea as well as an exaggerated hook in seedlings germinated in the dark consistent with previous reports (Lehman et al., 1996) (Figure 7B; Supplemental Figure 6A). The transgenic plants also displayed delayed flowering (Supplemental Figure 6B). Interestingly, significantly more HLS1 protein accumulated at 3 d after inoculation (dai) with B. cinerea relative to mock-inoculated plants (Figure 7C). The results suggest that either HLS1 is induced by a posttranslational mechanism involving the removal of a repressor or the rate of HLS1 turnover is decreased in response to infection.
Figure 7.
Resistance to B. cinerea Is Enhanced through ABA-Induced HLS1 Protein Accumulation.
(A) HLS1 expression in wild-type, hls1, and 35S:HLS1-HA plants.
(B) Increased disease resistance evaluated by disease symptoms in B. cinerea drop-inoculated plants.
(C) and (D) B. cinerea- (C) and ABA- (D) induced accumulation of HLS1 protein. HLS1-HA plants were inoculated with B. cinerea (top panel) or infiltrated with 100 μM ABA (bottom panel). Mock-treated plants were infiltrated with 0.5% methanol. Total protein was extracted from HLS1-HA plants at 0 or 72 h after B. cinerea inoculation or 24 h after treatment with ABA. HLS1 protein level was detected on immunoblot with anti-HA antibody. Equal loading is shown by Ponceau S staining of total protein.
(E) Disease symptoms (left panel) and disease lesion size (right panel) in ABA-treated plants. Plants were pretreated by infiltration with ABA 1 d prior to B. cinerea inoculation. Disease symptoms and lesion size were recorded at 3 d after B. cinerea inoculation. The data represent mean values ± se (n = 20). Statistically significant differences are marked by asterisks (Student’s t test: **P < 0.01 and ***P < 0.001; n.s., not significant) and by different letters (least squares means post hoc test: P < 0.05).
(F) Enhanced B. cinerea disease lesions in hls1, aba2, and aba3 mutants in response to ABA pretreatment. The disease lesions were measured at 2 dai. The aba2 and aba3 mutant plants displayed enhanced resistance to B. cinerea, while abi5 was comparable to wild-type plants. The data represent mean values ± se (n = 24). Statistically significant differences are marked by asterisks (Student’s t test: *P < 0.05, **P < 0.01, and ***P < 0.001) and by different letters (least squares means post hoc test: P < 0.05).
In addition, the transgenic HLS1-HA plants treated with ABA accumulated significantly more HLS1 protein than the mock-treated plants (Figure 7D), suggesting that ABA modulates HLS1 at the protein level. To establish a functional link between ABA-induced accumulation of HLS1 and B. cinerea resistance, we infiltrated ABA into Arabidopsis leaves 1 d prior to B. cinerea inoculation. In wild-type plants, ABA pretreatment enhanced resistance to B. cinerea, resulting in significantly reduced disease lesion size (Figures 7E and 7F). The expansion of disease lesions in the hls1 mutant was comparable in treated and nontreated plants, whereas the HLS1-HA plants exhibited further increase in resistance to B. cinerea after ABA treatment. The B. cinerea responses of the hls1 mutant after ABA treatment confirm the hls1 insensitivity to ABA on seedling germination. The wrky33 mutant also displayed a loss of ABA-induced resistance to B. cinerea similar to hls1. The ABA biosynthetic mutants aba2 and aba3 and the ABA insensitive mutant abi5, which is impaired in ABA response, were selected to determine the role of ABA in induced resistance to B. cinerea. The ABA biosynthesis mutants displayed enhanced resistance to B. cinerea consistent with previous studies (Adie et al., 2007), but abi5 was comparable to the wild type (Figure 7F). ABA increased susceptibility to B. cinerea in aba2 and aba3 relative to mock-treated plants. It has been suggested that ABA pretreatment suppresses callose deposition triggered by flagellin 22 through downregulation of gene expression in ET signaling and indole glucosinolate biosynthesis pathways (Clay et al., 2009). Consistent with this observation, ABA suppressed plant defense gene expression and increased susceptibility to Pst (Supplemental Figure 7). In our case, ABA-induced resistance to B. cinerea did not correlate with these defense pathways. Overall, plant resistance to fungal infection is enhanced through ABA-induced HLS1 protein accumulation, suggesting a priming effect of ABA though the stabilization of HLS1 protein.
HLS1-Regulated Plant Immunity Is Associated with Senescence
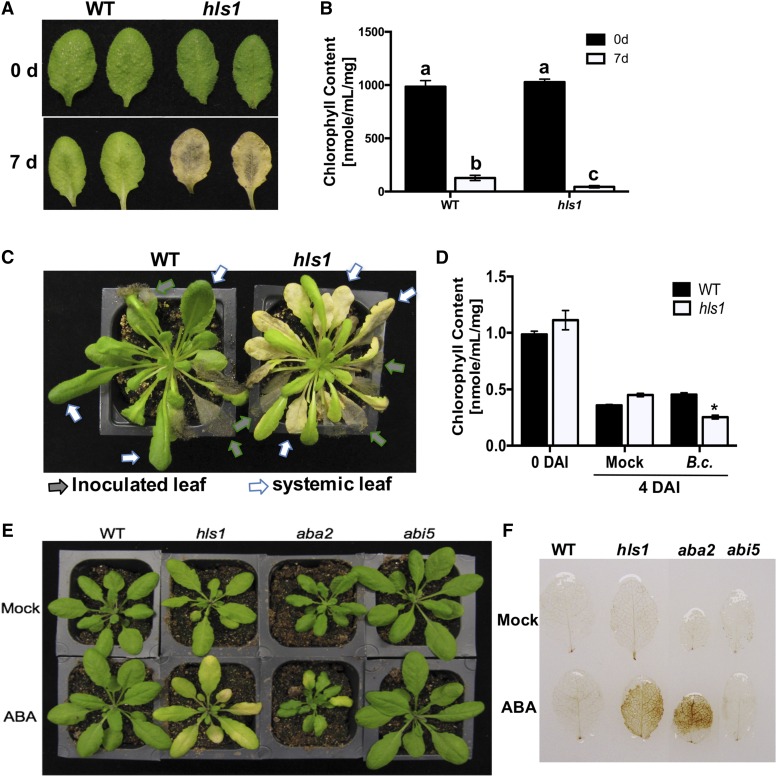
The hls1 mutant plants displayed enhanced disease symptoms independent of increased pathogen growth and flowered earlier than the wild type regardless of the duration of light. To address a potential link between senescence, early flowering, and plant immunity, we tested dark-induced leaf senescence in hls1 mutant and wild-type plants. The hls1 mutants displayed early leaf senescence marked by extensive chlorosis when detached leaves were placed in the dark for 7 d (Figure 8A). The loss of chlorophyll increased significantly in the hls1 mutant consistent with the dark-induced senescence phenotype (Figure 8B). Interestingly, senescence-like responses occurred in systemic (noninoculated) leaves of hls1 plants when lower leaves were drop-inoculated with B. cinerea (Figure 8C). Symptoms of leaf senescence were increased in hls1 mutants inoculated with B. cinerea as measured by total chlorophyll content (Figure 8D). Whether the early flowering of hls1 mutants is due to the increased senescence and disease susceptibility was also analyzed. Compared with hls1 mutants grown in long day (12 h:12 h, light:dark cycle), plants grown in short day (8 h:16 h, light:dark cycle) exhibited increased leaf number indicative of delayed flowering. However, plant susceptibility to B. cinerea was unchanged under short day, implying that the hls1 susceptibility is likely not caused by its early flowering (Supplemental Figure 8). ABA pretreatment increased leaf senescence in hls1 and aba2 mutants, which may explain the ABA-induced susceptibility to B. cinerea (Figures 7F and 8E). Staining with 3,3′-diaminobenzidine revealed that hls1 and aba2 mutants accumulated more H2O2, while wild-type plants accumulated lower levels after ABA treatment, suggesting H2O2 accumulation is associated with enhanced leaf senescence (Figure 8F). Overall, HLS1-mediated plant resistance may partially be explained by its function in maintaining normal levels of H2O2 and senescence.
Figure 8.
Accelerated Dark- or Pathogen-Induced Leaf Senescence in the hls1 Mutant.
(A) and (B) Enhanced dark-induced leaf senescence (A) and reduced chlorophyll content (B) in hls1 mutant. The photo was taken at 7 d after plants were incubated in the dark. Leaves from similar positions were detached from plants, and senescence was compared through analysis of chlorophyll content. The data represent mean values ± sd from (n = 5). The experiment was repeated two times with similar results.
(C) Enhanced systemic leaf senescence and extensive chlorosis are induced by B. cinerea in the hls1 mutant. Leaf senescence is observed on noninoculated systemic leaves of the hls1 mutant after inoculation of lower leaves with B. cinerea. The photo was taken 10 d after inoculation.
(D) Chlorophyll contents in plants showing senescence-like symptoms at 4 d after inoculation with B. cinerea. The data represent mean values ± sd from (n = 5). The experiment was repeated two times with similar results. Statistically significant differences are marked with an asterisk (ANOVA test: *P < 0.05).
(E) ABA-induced leaf senescence in aba2 and hls1 mutants. Leaf senescence in the abi5 mutant was comparable to wild-type plants. The photo was taken 24 h after ABA infiltration.
(F) Increased accumulation of H2O2 in aba2 and hls1 mutants in response to ABA. Plants were stained 24 h after ABA treatment. The leaves from similar positions were infiltrated with ABA and H2O2 was detected by 3,3′-diaminobenzidine staining.
ABA-Induced WRKY33 Expression Is Dependent on HLS1 Association and Chromatin Acetylation
We investigated genetic and molecular interactions between HLS1 and WRKY33 in ABA responses. Overexpressing WRKY33 in the hls1 mutant background restored the ABA-insensitive phenotype in the hls1 mutant to wild-type levels, suggesting that WRKY33 is a downstream target of HLS1 during ABA responses (Figures 9A and 9B). In response to ABA, the expression of WRKY33 was significantly reduced in the hls1 mutant, suggesting that ABA-induced expression of WRKY33 is also dependent on HLS1 (Figure 9C). Consistent with this, the association of HLS1 protein with WRKY33 TSS and CDS regions is enriched by ABA treatment, indicating that HLS1-dependent WRKY33 induction is also modulated by ABA (Figure 9D). Interestingly, ABA significantly enhanced the H3 acetylation at the WRKY33 locus in wild-type plants, whereas the hls1 mutant remained at the nontreated level even after ABA treatment (Figure 9E). This result suggested that HLS1-mediated WRKY33 expression responds to ABA through histone acetylation. Together, ABA-induced plant resistance to B. cinerea may go through HLS1-mediated WRKY33 activation. In sum, HLS1 association with WRKY33 is enhanced after B. cinerea inoculation, and both HLS1 and WRKY33 contribute to ABA-mediated priming of plant defense responses.
Figure 9.
ABA-Mediated WRKY33 Expression Is Modulated by HLS1.
(A) Ectopic expression of WRKY33 restores the ABA insensitivity of the hls1 mutant to the wild-type level. Seeds germinated on 1/2 MS medium supplemented with 0.005% methanol (mock) or 1 μM of ABA. The photos were taken at 5 d.
(B) Seed germination and seedling growth on media supplemented with ABA. The data represent mean values ± sd (n = 30) and the statistical significance of the difference in mock or ABA treatment is indicated by different letters (least squares means post hoc test: P < 0.05).
(C) ABA-induced expression of WRKY33 is attenuated in hls1 mutant. The expression of WRKY33 in wild-type plants at 0 h is set to 1.
(D) ABA mediates the recruitment of HLS1 to the WRKY33 transcription start and coding regions. The association of HLS1 with WRKY33 under mock treatment is set to 1 as a background control.
(E) ABA-enhanced H3 acetylation at the WRKY33 locus is HLS1 dependent.
In (C) to (E), the data represents mean values ± se (n = 3), and statistically significant differences are indicated by different letters (least squares means post hoc test: P < 0.05). HLS1, HLS1 tagged with HA; WRKY33; hls1, overexpressing WRKY33 in the hls1 mutant background; Pro, promoter region.
DISCUSSION
We describe the functions of Arabidopsis HLS1 and its role in plant immunity and responses to ABA primarily through its function in histone acetylation and interaction with the transcription coregulatory protein complex Mediator. We show that (1) HLS1 and the MED18 subunit of the Mediator complex share significant biological function and interact physically. (2) HLS1 is required for histone acetylation at the ABI5 and WRKY33 loci, critical regulators of ABA and pathogen responses, respectively. (3) Ectopic expression of HLS1 is sufficient for increased resistance to B. cinerea. Enhanced HLS1 protein accumulation in response to ABA and B. cinerea results in increased fungal resistance. ABA primes resistance to B. cinerea, likely through pathogen- and ABA-induced accumulation of HLS1 protein and upregulation of WRKY33 expression. (4) HLS1 and MED18 coregulate ABI5 and WRKY33 target genes through direct association in response to ABA and B. cinerea. This observation is further supported by synergistic actions of MED18 and HLS1 on ABI5 and WRKY33 gene expression. HLS1 is required for MED18 association with WRKY33 but not with ABI5 upstream regulatory regions, suggesting a complex relationship with these partners. (5) The hls1 mutant plants show enhanced senescence with particularly striking symptoms of senescence in systemic leaves in response to B. cinerea. (6) HLS1 is required for H3 acetylation at WRKY33 and ABI5 chromatin. The histone H3 acetylation at WRKY33 chromatin increased in response to pathogen infection and ABA treatment consistent with positive regulation of the WRKY33 and ABI5 by HLS1. In sum, HLS1 is a major regulator in priming plant immune responses through transcriptional and posttranslational mechanisms.
HLS1 Is a Critical Transcriptional and Posttranscriptional Regulator of ABA-Mediated Priming of Plant Defense
The function of HLS1 in ABA responses is supported by genetic and molecular data. HLS1 is induced by ABA, and expression of ABA-responsive genes, including ABI5, a direct target of HLS1 and a known ABA response regulator. Germination and seedling growth of the hls1 mutant are insensitivity to ABA, whereas the overexpression line is hypersensitive to ABA. Interestingly, ABA primes resistance to fungal infection in wild-type plants, whereas in hls1, ABA failed to prime resistance, consistent with the loss of ABA sensitivity. In addition, the HLS1-interacting protein MED18 and the common target gene WRKY33 were implicated in ABA responses. The overexpression of WRKY33 confers ABA hypersensitivity in Arabidopsis, similar to the overexpression of HLS1, whereas med18 and hls1 mutants are insensitive to ABA (Jiang and Deyholos, 2009). All three partners, HLS1, MED18, and WRKY33, are involved in pathogen and ABA response pathways. In particular, the critical role of HLS1 as a transcriptional activator of defense is underlined by its relationship with WRKY33, a major immune response regulator that operates through multiple mechanisms, including the regulation of phytoalexin biosynthesis, autophagy, and interaction with the MAPK pathway (Lai et al., 2011; Mao et al., 2011). WRKY33 also modulates ABA responses and biosynthesis (Liu et al., 2015) and its expression is mediated through ABA-induced HLS1 recruitment and histone acetylation. WRKY33, similar to ABI5, is regulated by direct association with HLS1 and potentially functions through ABA-dependent and ABA-independent pathways, consistent with its multifunctionality (Liu et al., 2015; Mao et al., 2011).
Multiple lines of genetic evidence suggest a link between ABA and pathogen responses. However, the role of ABA in plant defense responses is complex and varies depending on the nature of pathogens, the types of tissues, and the infection stages (Ton et al., 2009). Previous observations suggest that ABA promotes plant susceptibility to disease. For example, genes in the ABA biosynthesis pathway promote plant susceptibility to pathogens. The ABA-deficient tomato mutant sitins is resistant to B. cinerea and displayed increased basal and induced JA/ET-dependent defense gene expression and enhanced cuticle permeability (Asselbergh et al., 2007; Curvers et al., 2010). ABA-deficient mutants, such as aba1 and aba2, modulate JA/ET-responsive genes to enhance plant resistance to necrotrophs (Anderson et al., 2004). WRKY33 suppresses downstream target genes NCED3 and NCED5, thus abrogating ABA biosynthesis and increasing disease resistance (Liu et al., 2015).
In contrast to the above data, pathogens hijack ABA, either by manipulating its biosynthesis or antagonizing the SA-mediated resistance pathway to attenuate plant immunity (Xu et al., 2013; de Torres‐Zabala et al., 2007; Jiang et al., 2010). ABA signaling mutants abi1-1 and abi2-1 increase susceptibility to Ralstonia solanacearum (Hernández-Blanco et al., 2007). Arabidopsis mutants, such as med25 and med18, with enhanced disease susceptibility phenotypes also displayed altered responses to ABA (Chen et al., 2012; Lai et al., 2014), but how the ABA function relates to the pathogen response functions of the genes is unclear. The non-protein amino acid β-amino-butyric acid (BABA) primes resistance to necrotrophic pathogens (Ton et al., 2005) based on primed callose accumulation, controlled by an ABA-dependent defense pathway. BABA-induced resistance was blocked in the ABA-deficient mutant aba1-5 and the ABA-insensitive mutant abi4-1 (Ton and Mauch-Mani, 2004). Application of ABA mimicked the effects of BABA on callose accumulation and resistance. Thus, ABA is required for BABA-induced resistance to pathogens by enhancing callose deposition. The phenotypes of ABA pretreatment and plant susceptibility in ABA biosynthetic mutants are similar to those in hls1 mutants. This suggests that the plant resistance to pathogens is a consequence of ABA pretreatment. ABA suppressed expression of defense genes in wild-type plants, consistent with previous reports (Supplemental Figure 7; Clay et al., 2009). However, ABA-induced plant resistance to B. cinerea showed no correlation with ABA induced expression of defense genes. Instead, plant susceptibility in hls1 was associated with ABA-induced senescence and accumulation of H2O2.
ABA is a well-known regulator in abiotic and biotic stress responses. Many studies implicate exogenous ABA treatment in increased plant tolerance to abiotic stresses such as chilling and osmotic stress (Jiang and Zhang, 2002; Guo et al., 2012; Ozfidan et al., 2012), but ABA is often implicated as a suppressor of plant resistance (Curvers et al., 2010). The mechanism underlying this disparity between plant responses to biotic and abiotic stress responses is unclear. ABA increases reactive oxygen species (ROS) production, which then activates an antioxidative defense response in maize (Zea mays) seedlings (Jiang and Zhang, 2001). hls1 and ABA biosynthesis mutants accumulate increased ROS and display enhanced leaf senescence, likely without the concomitant increase in the appropriate antioxidant systems. This contention is consistent with the role of WRKY33 on ROS detoxification and scavenging, suggesting that WRKY33 may modulate ROS turnover in response to ABA (Jiang and Deyholos, 2009; Golldack et al., 2014). HLS1 may share the function of MED18 in the control of ROS homeostasis.
Application of ABA at the time of pathogen inoculation enhanced susceptibility (Liu et al., 2015); thus, the timing of ABA treatment may be important to determine defense functions in plants. Many genes in the ABA and defense pathways displayed altered expression in the hls1 mutant. PR1 expression is highly activated in the hls1 mutant, consistent with previous reports that ABA signaling antagonizes SA-dependent responses (Yasuda et al., 2008; Pieterse et al., 2012; Liu et al., 2015). The induction of PR1 in the hls1 mutant does not correlate with resistance but implies that some pathways leading to PR1 expression are affected. Alternatively, due to the susceptibility of the hls1 mutant and increased fungal growth, some genes displayed increased gene expression. ABA antagonizes the ETHYLENE RESPONSE FACTOR (ERF) branch of the JA pathway and regulates defense marker gene PDF1.2 (Anderson et al., 2004). However, the hls1 mutant displayed increased expression of PDF1.2, which is linked to fungal resistance (Penninckx et al., 1996), but the mutant remained susceptible to B. cinerea. HLS1-mediated gene expression is not a function of the antagonism between JA- and SA-regulated pathways, since markers of both pathways are also upregulated in the mutant. Together, the HLS1-mediated plant immunity works through the ABA signaling pathway but is independent of the ET/JA- and SA-regulated pathways as well as independent of their antagonistic interactions.
The presented data and discussions in the preceding sections imply loss of HLS1-regulated senescence, which may account for the enhanced susceptibility of the mutant. Senescence-like responses are triggered by dark or ABA treatment in the hls1 mutant. In particular, the senescence phenotype observed in secondary (noninoculated) leaves in mutant plants implies that HLS1 is important for restricting senescence-like symptoms that include extensive chlorosis and death of tissue away from the infection site. The increased susceptibility to B. cinerea may stem from impaired cell death control, including senescence.
HLS1-Mediated Histone Acetylation of Target Genes ABI5 and WRKY33
HLS1 is required for H3 acetylation at ABI5 and WRK33 chromatin based on changes in acetylation status in hls1 and HLS1-HA plants. However, acetyltransferase activity, measured through a standard HAT assay using recombinant protein, revealed no HAT activity, possibly due to either the GST tag in the GST-HLS1 fusion affecting the structure of the protein and its acetyltransferase activity, or the requirement for HLS1 to recruit other cofactors for activity. Histone acetylation alters the structure of defense and non-defense genes that underlie plant responses to the environment. Histone H4 deacetylase, HDT701, reduces global histone H4 acetylation and modulates defense-related genes in rice resistance to Magnaporthe oryzae and Xanthomonas oryzae pv oryzae (Ding et al., 2012). The elongator complex subunit 2 (ELP2) and ELP3 regulate resistance to P. syringae pv maculicola (Psm) ES4326 through their HAT activity on defense-related genes (Defraia et al., 2013; Wang et al., 2013). As shown in this study, HLS1 modulates H3 acetylation on specific loci required for plant immunity and ABA responses.
Plant defense genes are poised to counteract attempted pathogen infection through priming, which has been linked to posttranslational modification of histone tails. The promoter regions of defense-related transcription factor genes WRKY6, WRKY26, or WRKY53 were either acetylated or methylated in primed plants treated with the SA analog, benzothiadiazole (Jaskiewicz et al., 2011). Arabidopsis HAT1 mediates activation of PTI-related genes WRKY53, FRK1, or NHL10 primed by environmental stresses (Singh et al., 2014). BABA-triggered chromatin modification activates defense-related gene transcription (Po-Wen et al., 2013). HLS1 mediates WRKY33 expression through histone acetylation in response to ABA and pathogens, supporting the dynamic chromatin modification in response to stimuli. The results are consistent with previous studies demonstrating that histone modification enzymes stand by on specific target loci and modulate gene expression following attempted infection (Jaskiewicz et al., 2011).
Histone deacetylases reverse acetylation status on histones to remove acetyl group from substrates, resulting in repression of gene expression. Arabidopsis HDA6 encodes histone deacetylase and the hda6 mutation results in hypersensitivity to ABA, delayed senescence, and flowering (Wu et al., 2008). HDA6 recruits a JA-Zim domain (JAZ) protein to repress EIN3/EIL1-dependent transcription (Zhu et al., 2011b). Arabidopsis histone deacetylase HDA19 is another histone-modifying enzyme and the hda19 mutant results in early senescence, hypersensitivity to ABA, and susceptibility to Alternaria brassicicola (Wu et al., 2000; Tian et al., 2005; Zhou et al., 2005). HDA6 and HDA19 share contrasting biological functions with HLS1, consistent with their distinct roles in histone modifications. Whether HLS1, HDA6, and HDA19 target the same set of genes for reversible modification of histone acetylation is unclear.
Functional Interaction between Histone Acetylation and Mediator
Current and previous data show that HLS1 and MED18 are positive regulators of ABA signaling and resistance to B. cinerea (Lai et al., 2014). HLS1 and MED18 interact and are associated with WRKY33 and ABI5 regulatory regions. This association increases in response to ABA and B. cinerea as determined through ChIP-qPCR and transcription activation assays. Many studies in yeast and mammalian cells have shown that mediator complexes modulate histone modification. The Spt-Ada Gcn5-acetyltransferase (SAGA) complexes require a mediator complex to be recruited to the GCN4-regulated promoters of ARG1, ARG4, or SNZ1 (Yoon et al., 2003; Qiu et al., 2005). In mammalian cells, MED25 affects methylation or acetylation at H3K27 at CYP2C9 promoter region by dissociation from Polycomb repressive complex 2 and activates CYP2C9 expression (Englert et al., 2015). The Arabidopsis E3 ligase HUB1 is required for resistance to fungal pathogens and regulates ABA responses and biosynthesis (Peeters et al., 2002; Liu et al., 2007; Dhawan et al., 2009). HUB1 interacts with MED21 and activates gene transcription through H2B ubiquitination, implying it functions as a component of transcriptional activation complexes. In another report, mediator localization is determined by the interaction between mediator and histone tails. The interaction is relieved by the acetylation of H4K16 (Zhu et al., 2011a). MED18 is also associated with WRKY33 TSS and CDS regions, similar to the genomic localization of HLS1. Interestingly, MED18 is unable to associate with WRKY33 in the absence of HLS1, suggesting that HLS1 is required for MED18 recruitment to specific loci.
Proposed Model of HLS1 Function
Collectively, we demonstrate that HLS1 associates with MED18 at the ABI5 and WRKY33 loci and modulates their expression through acetylation of chromatin at these loci. Although many genes are regulated by HLS1 in response to ABA or pathogens, some of these are affected only indirectly. HLS1 associates with the WRKY33 gene and activates its expression through histone acetylation after B. cinerea inoculation or ABA treatment. HLS1 recruits MED18 to the WRKY33 locus where MED18 enhances the role of HLS1 in transcriptional activation of WRKY33 (Figure 9). MED18 also enhances the HLS1-regulated expression of ABI5, but its recruitment to the ABI5 locus is independent of HLS1. Interestingly, HLS1 is required and sufficient for the histone acetylation at WRKY33 chromatin, consistent with the high sequence similarity of HLS1 to the GCN5 histone acetyl transferase. Other components that potentially associate with the two proteins are not known, but transcription factors, coactivators, or other chromatin remodeling components may be involved to form a preinitiation complex. Other non-histone proteins may be recruited with HLS1 and MED18 for initiation of gene expression. Identifying additional proteins that interact with HLS1 will help us understand the acetylation mechanism that modulates responses to biotic and abiotic stresses. In response to ABA or B. cinerea, the complex enhances histone acetylation to remodel chromatin structure, favoring increased gene expression. The consequences of these will be enhanced transcriptional activation of genes that requires the recruitment of HLS1, which then recruits MED18 to target sites. Biologically, HLS1 participates in different response pathways (light, sugar, and pathogen) and is regulated by hormone crosstalk (JA, gibberellin, ET, and ABA). Therefore, it will be important to investigate the global targets of HLS1 through ChIP-seq analysis to identify additional targets bound by HLS1 to decipher its regulatory impact and to determine histone acetylation mechanisms dynamically responding to environmental challenges.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants used in this study are in the Col-0 ecotype background. Plants were grown in a growth chamber at 24°C, 70% relative humidity, 110 to 130 μE m−2 s−1 light intensity by fluorescence tubes (model F32T8/TL741) with a 12-h-light/12-h-dark cycle unless stated otherwise. T-DNA insertion Arabidopsis mutants hls1-1 (SALK_136528C) and hls1-2 (SALK_009473) in the Col-0 background were obtained from the ABRC and confirmed by PCR to verify their T-DNA insertion. HLS1 expression in hls1 mutant plants was confirmed in 3-d-old seedlings compared with wild-type or transgenic plants by qPCR. Transgenic plants overexpressing HLS1 tagged with HA under the CaMV 35S promoter were generated by Agrobacterium tumefaciens-mediated transformation. Plants were screened on half-strength Murashige and Skoog (1/2 MS) medium supplemented with hygromycin or the herbicide basta. Protein or mRNA levels of HLS1 were verified by immunoblotting analysis with anti-HA-specific antibody or qPCR assays, respectively. Transgenic plants coexpressing HLS1 and MED18 were generated by Agrobacterium-mediated transformation and HLS1 and MED18 expression was detected by immunoblotting. Transgenic plants overexpressing MED18-MYC or WRKY33-MYC in the hls1 mutant background were generated by Agrobacterium-mediated transformation and MED18 or WRKY33 transgenic plants were screened on 1/2 MS medium supplemented with basta and protein expression was detected by immunoblotting.
Seed Germination, Dark-Induced Senescence, ABA Treatment, and Disease Assay
For seed germination assays, Arabidopsis mutant or transgenic seedlings were germinated on 1/2 MS medium supplemented with different concentrations of ABA and grown in a room at 22°C, 110 to 130 μE m−2 s−1 light intensity with a 16-h-light/8-h-dark cycle. For dark-induced senescence, comparable leaves from 4-week-old plants were detached and placed in water-saturated plates and incubated in the dark. The total chlorophyll content was measured by absorbance at 647 and 665 nm on a NanoDrop 2000c spectrophotometer (Thermo Scientific).
Fungal and bacterial disease assays were conducted as previously described (Laluk et al., 2011). In brief, for the Botrytis cinerea disease assay, 4-week-old plants were spray or drop inoculated with a conidial suspension (2.5 × 105 spores/mL) of B. cinerea strain B05.10 in 1% Sabouraud Maltose Broth and maintained under a transparent cover at high humidity. For the Pseudomonas syringae disease assay, plants were infiltrated with the bacterial strains and bacteria were extracted from inoculated leaves. The colony growth was determined and expressed in colony forming units on King’s B medium supplemented with the antibiotics rifampicin and kanamycin. For ABA treatment, 4-week-old plants were infiltrated with 100 μM ABA. The accumulation of H2O2 in leaves was detected by 3,3′-diaminobenzidine staining (Daudi and O'Brien, 2012).
RNA Extraction and RT-qPCR Assay
Total RNA was extracted from leaves or seedlings with Trizol reagent according to the manufacturer’s instructions (Sigma-Aldrich). The procedures for RNase-free DNase I treatment (Promega) and cDNA synthesis (New England Biolabs) from total RNA were conducted following the manufacturer’s instructions. qPCR was performed with SYBR green supermix reagents (Bio-Rad) using gene-specific primers (Supplemental Table 1) and the Arabidopsis ACTIN2 gene as an internal reference for normalization.
co-IP Assay
The co-IP was conducted following the previously described procedure (Lai et al., 2014; Zhu et al., 2014). Briefly, the plasmids containing full-length HLS1-HA and MED18-MYC driven by the CaMV 35S promoter were generated and transformed into Agrobacterium. The Agrobacterium strains were then infiltrated into Nicotiana benthamiana. After 36 h, total protein was extracted from infiltrated leaves with extraction buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM EDTA, 50 mM EGTA, 25 mM NaF, 1 mM NaVO3, 50 mM β-glycerophosphate, 20% [v/v] glycerol, 1 mM PMSF, 0.1% [v/v] Triton X-100, 1 mM DTT, and 1× protease inhibitor cocktail [Sigma-Aldrich]). After removing debris by centrifugation at 12,000g for 10 min, 1 mL of supernatant mixed with anti-HA antibody-conjugated agarose beads (Sigma-Aldrich) and rotated overnight at 4°C. Then, beads with immunoprecipitates were washed four times with extraction buffer. Immunoprecipitates were detected by immunoblotting with anti-HA-specific (Covance) or anti-MYC-specific (Abcam) antibodies. A similar co-IP procedure was employed in Arabidopsis plants expressing HLS1-HA and MED18-MYC.
ChIP-qPCR Assay
ChIP assay was conducted as described previously with minor modifications (Saleh et al., 2008). Briefly, chromatin complexes with proteins were cross-linked and isolated from 4-week-old Arabidopsis plants. After sonication, protein complexes were precipitated with anti-HA (Abcam), anti-H3, anti-acetyl-H3, or anti-acetyl-H4 (Millipore) antibody at 4°C overnight and then captured with salmon sperm DNA/Protein A agarose (Millipore). Beads were washed and reverse cross-linked, and proteins were digested prior to DNA purification. The immunoprecipitated DNA was amplified with specific primers listed in Supplemental Table 1. ChIP enrichment was normalized with input from a non-precipitated sample and promoter region of ACTIN2 as an internal control. Wild-type plants treated with the same procedure were used as a background control and IgG was used for the immunoprecipitation control. Primers at transcription start site and C-terminal sequences of Arabidopsis ACTIN7 gene were used as a background control in ChIP-qPCR assay for histone H3 acetylation.
In Vitro HAT Assay
The HAT assay was conducted as described (Qian et al., 2012). The full-length HLS1 fused with GST was generated and purified in Escherichia coli. The GST-HLS1 recombinant proteins (5 μg), purified by glutathione Sepharose 4B beads (GE Healthcare Life Science), were mixed with 10 μg of chicken core histones (Millipore) and 1 μCi of H3-acetyl-CoA (Perkin-Elmer Life Science) in HAT buffer containing 50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 10 mM butyric acid, and 1 mM PMSF, and incubated at 30°C for 2 h. After incubation, samples were run on 15% SDS-PAGE gel and the gel was fixed with 40% methanol-10% acetic acid. The gel was treated with an autoradiographic enhancer (Perkin-Elmer Life Science) and vacuum dried. The signals were detected after 2 weeks of exposure at −80°C. The same amount of PCAF (Abcam) with a known acetyltransferase activity was used as a positive control in parallel with GST-HLS1 in the reaction.
Transcriptional Activation Assays
The ABI5 or WRKY33 promoter region was fused with the GUS reporter gene to generate a transcriptional fusion. Two effector plasmids, HLS1-HA and MED18-MYC, were generated and each cotransfected with the reporter construct into ∼2 × 104 protoplasts isolated from 4-week-old Arabidopsis plants as described (Yoo et al., 2007). Protoplasts were lysed in lysis buffer containing 50 mM phosphate buffer, pH 7.0, 1 mM DTT, 2 mM trans-1,2-diaminocyclohexane-N,N,N’,N’-tetraacetic acid monohydrate, 10% glycerol, and 1% Triton X-100. The lysate was mixed with 4-methylumbelliferyl-β-d-glucuronide (MUG) substrate buffer (10 mM Tris-Cl, pH 8.0, 1 mM MUG, and 2 mM MgCl2) and incubated at 37°C. The reaction was stopped by adding 0.2 M Na2CO3, and the LUC substrates (Promega) were mixed with lysates. GUS and LUC activities were detected with a VICTOR 3V Multilabel plate reader (Perkin-Elmer). LUC reading was used as an internal control in each sample normalized to GUS reading.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HLS1 (AT4G37580), WRKY33 (AT2G38470), ABI5 (AT2G36270), MED18 (AT2G22370), ACT2 (AT3G18780), ACT7 (AT5G09810), PR1 (AT2G14610), ERF1 (AT3G23240), PDF1.2 (AT5G44420), RPM1 (AT3G07040), RIN4 (AT3G25070), CYP79B3 (AT2G22330), CYP81F2 (AT5G57220), PAD3 (AT3G26830), SUR2 (AT4G31500), ACS2 (AT1G01480), ACS6 (AT4G11280), ABI3 (AT3G24650), RD29a (AT5G52310), KAT2 (AT2G33150), WRKY40 (AT1G80840), and HY5 (AT5G11260).
Supplemental Data
Supplemental Figure 1. The hls1-1 and hls1-2 mutant alleles showing lack of HLS1 transcript.
Supplemental Figure 2. HLS1 protein is not associated with resistance gene RPM1.
Supplemental Figure 3. The histone H3 acetylation at ACTIN7 chromatin shows no difference between wild-type and hls1 plants.
Supplemental Figure 4. The recruitment of HLS1 protein to the ABI5 locus is not enhanced by B. cinerea inoculation.
Supplemental Figure 5. MED18-mediated plant resistance through TRX-h5 regulation is HLS1 independent.
Supplemental Figure 6. Overexpressing HLS1-HA in Arabidopsis shows developmental phenotypes opposite to the hls1 mutant.
Supplemental Figure 7. Expression of defense related genes is induced in hls1 mutant in response to ABA.
Supplemental Figure 8. Leaf number, but not disease phenotype, is affected by HLS1 under short-day conditions.
Supplemental Table 1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
This research was funded by grants from the National Science Foundation (IOS-1456594) and the Next-Generation BioGreen 21 Program (SSAC Project No. PJ01137902), Rural Development Administration. We thank Zhixiang Chen (Purdue University) for the WRKY33-MYC construct used in our studies.
AUTHOR CONTRIBUTIONS
C.-J.L. conducted most of the experiments. C.-J.L. and T.M. designed most of the experiments and directed the project. Z.L. and S.L. performed the initial mutant screen that identified the hls1 and other mutants. T.M., C.J.L., and D.-J.Y. wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- HAT
histone acetyltransferase
- JA
jasmonic acid
- ET
ethylene
- SA
salicylic acid
- ABA
abscisic acid
- IG
indole-glucosinolate
- ChIP-qPCR
chromatin immunoprecipitation-qPCR
- CDS
coding sequence
- TSS
transcription start site
- co-IP
coimmunoprecipitation
- dai
days after inoculation
- BABA
β-amino-butyric acid
- ROS
reactive oxygen species
- 1/2 MS
half-strength Murashige and Skoog
References
- Adie B.A., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Mou Z. (2013). The function of the Mediator complex in plant immunity. Plant Signal. Behav. 8: e23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B., Curvers K., Franca S.C., Audenaert K., Vuylsteke M., Van Breusegem F., Höfte M. (2007). Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144: 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Bergounioux C., Domenichini S., Delarue M., Zhou D.X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278: 28246–28251. [DOI] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Tian L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curvers K., Seifi H., Mouille G., de Rycke R., Asselbergh B., Van Hecke A., Vanderschaeghe D., Höfte H., Callewaert N., Van Breusegem F., Höfte M. (2010). Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol. 154: 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A., O'Brien J.A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2: e263. [PMC free article] [PubMed] [Google Scholar]
- Defraia C.T., Wang Y., Yao J., Mou Z. (2013). Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 13: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E., Ryals J. (1994). A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M., Truman W., Bennett M.H., Lafforgue G., Mansfield J.W., Rodriguez Egea P., Bögre L., Grant M. (2007). Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26: 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel.R., Ning Y., Meyers B.C., Wang G.L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24: 3783–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert N.A., Luo G., Goldstein J.A., Surapureddi S. (2015). Epigenetic modification of histone 3 lysine 27: mediator subunit MED25 is required for the dissociation of polycomb repressive complex 2 from the promoter of cytochrome P450 2C9. J. Biol. Chem. 290: 2264–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 5: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.L., Chen R.G., Gong Z.H., Yin Y.X., Ahmed S.S., He Y.M. (2012). Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 11: 4063–4080. [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhänsel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.J., Shimono M., Sugano S., Kojima M., Yazawa K., Yoshida R., Inoue H., Hayashi N., Sakakibara H., Takatsuji H. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 23: 791–798. [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2001). Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 42: 1265–1273. [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2002). Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215: 1022–1030. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M.K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69: 91–105. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14: 373–382. [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968. [DOI] [PubMed] [Google Scholar]
- Lai Z., Schluttenhofer C.M., Bhide K., Shreve J., Thimmapuram J., Lee S.Y., Yun D.J., Mengiste T. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5: 3064. [DOI] [PubMed] [Google Scholar]
- Laluk K., Luo H., Chai M., Dhawan R., Lai Z., Mengiste T. (2011). Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 23: 2831–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J.M., Ecker J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Liu S., Kracher B., Ziegler J., Birkenbihl R.P., Somssich I.E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 4: e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Vorontchikhina M., Wang Y.L., Faiola F., Martinez E. (2008). STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol. Cell. Biol. 28: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J., Kidarsa T., Bradford C.S., Gilbert B., Curtis M., Tzeng S.C., Maier C.S., Wolpert T.J. (2012). Tricking the guard: exploiting plant defense for disease susceptibility. Science 338: 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozfidan C., Turkan I., Sekmen A.H., Seckin B. (2012). Abscisic acid-regulated responses of aba2-1 under osmotic stress: the abscisic acid-inducible antioxidant defence system and reactive oxygen species production. Plant Biol. (Stuttg.) 14: 337–346. [DOI] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters A.J., Blankestijn-De Vries H., Hanhart C.J., Léon-Kloosterziel K.M., Zeevaart J.A., Koornneef M. (2002). Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant. 115: 604–612. [DOI] [PubMed] [Google Scholar]
- Penninckx I.A., Thomma B.P., Buchala A., Métraux J.P., Broekaert W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Eggermont K., Terras F.R., Thomma B.P., De Samblanx G.W., Buchala A., Métraux J.P., Manners J.M., Broekaert W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K., Fiil B.K., Mundy J., Petersen M. (2008). Downstream targets of WRKY33. Plant Signal. Behav. 3: 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Po-Wen C., Singh P., Zimmerli L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Zhang F., Hwang G.J., Swanson M.J., Boonchird C., Hinnebusch A.G. (2005). Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves W.M., Hahn S. (2003). Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Singh P., Yekondi S., Chen P.W., Tsai C.H., Yu C.W., Wu K., Zimmerli L. (2014). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26: 2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M.P., Wang J.J., Wei N.E., Jiang H., Doerge R.W., Chen Z.J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38: 119–130. [DOI] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14: 310–317. [DOI] [PubMed] [Google Scholar]
- Ton J., Jakab G., Toquin V., Flors V., Iavicoli A., Maeder M.N., Métraux J.P., Mauch-Mani B. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17: 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios K.E., Thomashow M.F., Triezenberg S.J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., An C., Zhang X., Yao J., Zhang Y., Sun Y., Yu F., Amador D.M., Mou Z. (2013). The Arabidopsis elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell 25: 762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Malik K., Tian L., Brown D., Miki B. (2000). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44: 167–176. [DOI] [PubMed] [Google Scholar]
- Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59: 225–234. [DOI] [PubMed] [Google Scholar]
- Xing J., Wang T., Liu Z., Xu J., Yao Y., Hu Z., Peng H., Xin M., Yu F., Zhou D.X., Ni Z. (2015). GCN5-mediated histone acetylation of FRD3 contributes to iron homeostasis in Arabidopsis thaliana. Plant Physiol. 168: 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Audenaert K., Hofte M., De Vleesschauwer D. (2013) Abscisic acid promotes susceptibility to the rice leaf blight pathogen pv by suppressing salicylic acid-mediated defenses. PLoS One 8: e67413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ou B., Zhang J., Si W., Gu H., Qin G., Qu L.J. (2014). The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77: 838–851. [DOI] [PubMed] [Google Scholar]
- Yasuda M., Ishikawa A., Jikumaru Y., Seki M., Umezawa T., Asami T., Maruyama-Nakashita A., Kudo T., Shinozaki K., Yoshida S., Nakashita H. (2008). Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoon S., Qiu H., Swanson M.J., Hinnebusch A.G. (2003). Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 23: 8829–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605. [DOI] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhang Y., Bjornsdottir G., Liu Z., Quan A., Costanzo M., Dávila López M., Westholm J.O., Ronne H., Boone C., Gustafsson C.M., Myers L.C. (2011a). Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 39: 8342–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Schluttenhoffer C.M., Wang P., Fu F., Thimmapuram J., Zhu J.K., Lee S.Y., Yun D.J., Mengiste T. (2014). CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26: 4149–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011b). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.