Amplified short interfering RNAs are made in the absence of RNA cleavage activity of ARGONAUTE1, implying that recruitment of RNA-dependent RNA Polymerase 6 does not require RNA cleavage fragments.
Abstract
ARGONAUTE1 (AGO1) mediates posttranscriptional silencing by microRNAs (miRNAs) and short interfering RNAS (siRNAs). AGO1-catalyzed RNA cleavage (slicing) represses miRNA targets, but current models also highlight the roles of slicing in formation of siRNAs and siRNA-AGO1 complexes. miRNA-guided slicing is required for biogenesis of phased, trans-acting siRNAs (tasiRNAs), whose cleaved precursor fragments are converted to double-stranded RNA by RNA-dependent RNA polymerase 6 (RDR6). In addition, unwinding of duplex siRNA bound to AGO1 requires passenger strand cleavage in vitro. In this study, we analyze how mutation of four metal ion-coordinating residues of Arabidopsis thaliana AGO1 affects slicer activity in vitro and siRNA function in vivo. We show that while all four residues are required for slicer activity, they do not contribute equally to catalysis. Moreover, passenger strand cleavage is required for assembly of active AGO1-siRNA complexes in vivo, and many AGO1-bound siRNAs are trimmed in the absence of slicer activity. Remarkably, seedlings defective in AGO1 slicer activity produce abundant siRNAs from tasiRNA loci in vivo. These siRNAs depend on RDR6 and SUPPRESSOR OF GENE SILENCING3, but unlike wild-type tasiRNAs, they are unphased. These results demonstrate that slicing is solely required for phase definition of tasiRNAs, and they strongly support recruitment of RDR6 by AGO1 rather than by cleavage fragments.
INTRODUCTION
MicroRNAs (miRNAs) and short interfering RNAs (siRNAs) are 20- to 24-nucleotide small RNAs that regulate genes at the transcriptional or posttranscriptional levels. Both types of small RNA derive from longer precursors with double-stranded features, but while miRNAs are typically excised as single species of mismatch-containing duplexes from pre-miRNAs, a single long, double-stranded RNA (dsRNA) precursor gives rise to many distinct and perfectly base-paired siRNA duplexes (Bologna and Voinnet, 2014; see Zhai et al. [2015] for an exception). Consequently, miRNA and siRNA biogenesis involve different protein factors (Chapman and Carrington, 2007). Small RNAs exert their functions in RNA-induced silencing complexes (RISCs) whose central components are proteins of the ARGONAUTE (AGO) family. They use base pairing to target specific, complementary mRNA for repression via recruitment of AGO and associated factors. AGO proteins initially bind small RNA duplexes, and a mature RISC composed of AGO and the guide RNA is subsequently formed by duplex unwinding and degradation of the so-called passenger strand (Carthew and Sontheimer, 2009; Kawamata and Tomari, 2010).
In plants, AGO1 exerts posttranscriptional gene regulation by many siRNAs and nearly all miRNAs (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005). AGO1 can catalyze cleavage (“slicing”) of RNA base-paired to an AGO1-bound guide RNA (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005). The mechanism of slicing is reasonably well understood, in particular through biochemical and structural analyses of bacterial, animal, and yeast AGO proteins. Early studies identified three metal-coordinating residues required for slicer activity (Song et al., 2004; Rivas et al., 2005), and analysis of the structure of the yeast (Kluyveromyces polysporus) Ago suggested that a fourth metal-coordinating residue can be recruited to the active site to constitute a catalytic tetrad similar to that observed in RNaseH (Nakanishi et al., 2012). The requirement for base pairing around the cleavage site is explained by the necessity of correct steric positioning of the metal ion catalysts relative to the scissile phosphodiester bond (Wang et al., 2008; Schirle et al., 2014; Sheng et al., 2014). Slicer activity is indeed exceedingly sensitive to steric changes because even in the presence of all four metal-coordinating residues, mammalian Ago1, Ago3, and Ago4 require additional corrective amino acid changes to achieve slicer activity (Hauptmann et al., 2013; Nakanishi et al., 2013; Schürmann et al., 2013).
Slicer activity has been proposed to play several biological roles in plants. The most intuitive is degradation of mRNA targets of miRNAs and siRNAs (Llave et al., 2002; Kasschau et al., 2003). In addition, slicer activity has been implicated in more specific functions of siRNAs. First, in all organisms tested, it is required for loading of siRNAs because strand separation upon association of the siRNA duplex with AGO requires slicing of the passenger strand (Matranga et al., 2005; Miyoshi et al., 2005; Rand et al., 2005; Leuschner et al., 2006; Iki et al., 2010). Second, in plants, it is implicated in the biogenesis of trans-acting siRNAs (tasiRNAs), which play fundamental roles in plant development and stress adaptation (Allen et al., 2005; Fahlgren et al., 2006; Luo et al., 2012). TasiRNA biogenesis involves miRNA-guided slicing of a precursor transcript, classically a long noncoding RNA (Allen et al., 2005; Yoshikawa et al., 2005), but more recent studies have also revealed tasiRNA-like species derived from mRNAs (Axtell et al., 2006; Howell et al., 2007; Zhai et al., 2011; Shivaprasad et al., 2012; Boccara et al., 2014). One of the cleavage fragments is then converted into dsRNA by the RNA-dependent RNA polymerase RDR6 and its requisite accessory factor SUPPRESSOR OF GENE SILENCING3 (SGS3) (Peragine et al., 2004; Vazquez et al., 2004). Ultimately, DICER-LIKE4 (DCL4) processes the dsRNA into mature tasiRNA species in a 21-nucleotide phase with the miRNA cleavage site (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). In Arabidopsis thaliana, four tasiRNA families are well defined: tasiRNAs generated from TAS1abc, TAS2, and TAS4 precursor transcripts involve AGO1-mediated cleavage guided by miR173 (TAS1abc, TAS2) and miR828 (TAS4) and derive from the 3′-cleavage fragment. In contrast, tasiRNA generation from TAS3 precursors involves AGO7-mediated cleavage, and the tasiRNAs derive from the 5′-cleavage fragment (Allen et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Rajagopalan et al., 2006; Montgomery et al., 2008). TAS3 precursors contain two miR390 target sites that define the region producing tasiRNAs (Axtell et al., 2006). This observation gave rise to the two-hit hypothesis, which states that siRNAs are formed particularly efficiently in regions between two small RNA target sites (Axtell et al., 2006). TasiRNA production from TAS1abc and TAS2 precursors may also involve double hits because the RNA intervals spawning tasiRNAs are defined on one side by the miR173 cleavage site and on the other side by cleavage guided by a “master siRNA” generated from TAS1c (Rajeswaran et al., 2012). Consistent with the two-hit trigger hypothesis, a number of transcripts targeted by both a miRNA and one or more TAS1/TAS2 tasiRNAs also spawn siRNAs (Howell et al., 2007), but the involvement of slicing in the generation of these siRNAs has not been examined. For tasiRNAs, the role of slicing guided by the trigger miRNA could in principle be limited to the definition of the phase of tasiRNAs, but current models hold that slicing is absolutely required for siRNA production from TAS precursor transcripts (Yoshikawa et al., 2005; Carbonell et al., 2012).
Previous studies have analyzed some aspects of the slicer activity of plant AGO1. Using in vitro cleavage assays, two of the three classically defined metal-coordinating residues (Song et al., 2004) have been shown to be required for catalysis (Baumberger and Baulcombe, 2005; Qi et al., 2005; Iki et al., 2010), and analyses of AGO1 mutants in each of the three residues expressed in vivo suggested that slicer activity is required for the release of miRNA targets from AGO1 (Carbonell et al., 2012). This observation probably explains the enhancement of developmental phenotypes of a hypomorphic ago1 allele by expression of AGO1 slicer-deficient mutants (Carbonell et al., 2012). Critically, however, molecular analyses of mutants expressing slicer deficient AGO1 in a null background have not been reported.
In this study, we conduct a thorough biochemical investigation of the four amino acid residues believed to constitute the catalytic center in Arabidopsis AGO1. We show that each of the tetrad residues is required for efficient slicing, but that mutations in them have different effects on catalysis that are consistent with their differential involvement in coordination of the two catalytic MgII ions proposed earlier (Wang et al., 2008; Nakanishi et al., 2012). We analyze the implication of slicer activity in vivo in RISC assembly with miRNAs and siRNAs, and in the biogenesis of siRNAs, in particular tasiRNAs. Our results show that slicing is critical for the formation of siRNA-RISC, but not for miRNA-RISC, in vivo. In the absence of slicing, complexes of AGO1 with both guide and passenger siRNAs remain detectable, but many AGO1-bound duplex siRNAs are trimmed. Our comparative analysis of tasiRNA production in wild-type, ago1 null, and ago1 slicer-deficient mutants expressed in a null background shows that slicing by AGO1 is required only for definition of the phase, but not for the production of RDR6/SGS3-dependent siRNAs derived from TAS precursors. This observation calls into question a role of RNA cleavage fragments as mandatory triggers of RDR6 activity, and it strongly supports the idea that AGO1 can directly recruit RDR6 to amplify small silencing RNAs.
RESULTS
Different Catalytic Roles of Mg2+-Coordinating Active-Site Residues
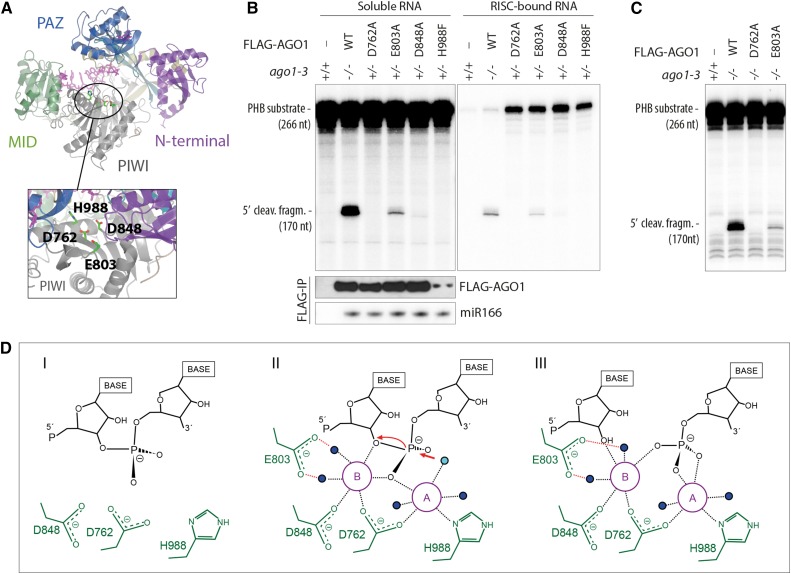
To analyze the contribution of each of the tetrad residues to AGO1-catalyzed target RNA slicing, we produced FLAG-tagged AGO1WT and point mutants in predicted Mg2+-coordinating residues (D762A, E803A, D848A, and H988F) (Figure 1A; Poulsen et al., 2013) and isolated stable transgenic lines in a genetic background heterozygous for the Arabidopsis ago1-3 null allele (G42STOP; Bohmert et al., 1998). This approach enabled us to FLAG affinity-purify AGO1 protein for biochemical analysis from inflorescence tissue of ago1-3 heterozygotes and to analyze phenotypes in ago1-3 homozygotes expressing the different wild-type or point mutant constructs. FLAG-AGO1WT fully complemented ago1-3, while the expression of each of the four point mutants in ago1-3 caused strong developmental defects similar to, albeit slightly distinct from, the null (Arribas-Hernández et al., 2016). The residues Asp-762 and Asp-848 have been shown directly to be required for slicing (Baumberger and Baulcombe, 2005; Iki et al., 2010), and indirect tests indicate a requirement for His-988 (Carbonell et al., 2012). The contribution of Glu-803 to catalysis has not been tested in any AGO protein, although the equivalent E1013A mutant in K. polysporus Ago is RNAi deficient (Nakanishi et al., 2012). In this study, FLAG-AGO1WT, FLAG-AGO1D762A, FLAG-AGO1E803A, FLAG-AGO1D848A, or FLAG-AGO1H988F immuno-affinity purified from inflorescences was used in RNA cleavage assays with cap-labeled target RNAs containing either miR159 or miR166 binding sites. These analyses showed that while no cleavage activity was detectable for FLAG-AGO1D762A and FLAG-AGO1H988F, residual activity was clearly detectable for FLAG-AGO1E803A with substrate RNAs corresponding to fragments of the miR166 target PHABULOSA (PHB) (Kidner and Martienssen, 2004) and the miR159 target MYB65 (Achard et al., 2004) (Figure 1B, left panels; Supplemental Figure 1A). The residual activity was not due to copurification of endogenous AGO1 specifically with the FLAG-AGO1E803A mutant because similar residual activity was observed when the protein was immunopurified from ago1-3 homozygous seedlings expressing FLAG-AGO1E803A (Figure 1C). Thus, slicer activity is strongly decreased, but not abolished, in E803A mutants. We also observed a very low residual level of activity in the FLAG-AGO1D848A mutant (Figure 1B; Supplemental Figures 1A and 1B).
Figure 1.
Different Catalytic Roles of the Four Mg2+-Coordinating Active-Site Residues.
(A) Structural model of AGO1 based on the crystal structure of human Ago2 (PDB ID 4EI3). The locations of the four residues involved in coordination of Mg2+-ions are indicated.
(B) Slicer activity of immuno-affinity-purified FLAG-AGO1 from Arabidopsis inflorescences of stable transgenic lines expressing wild-type and slicer-deficient FLAG-AGO1. A 266-nucleotide 32P cap-labeled PHB single-stranded RNA bearing the endogenous miR166 target site was used as substrate. FLAG-AGO1 was immobilized on M2-conjugated beads during the reaction. The left panel shows RNA extracted from the supernatant of the reaction. The right panel shows RNA extracted from the agarose beads that contain immobilized FLAG-AGO1. Cleavage activity is revealed by the accumulation of the 5′ cleavage fragment. Nontransgenic Col-0 was used to control for unspecific binding of endogenous AGO1 to M2-conjugated beads (left lanes). A reproduction of this panel with higher contrast can be found in Supplemental Figure 1 to facilitate visualization of the 5′-cleavage fragment produced by FLAG-AGO1D848A. FLAG IPs were split into three parts for analysis of cleavage activity, FLAG-AGO1 levels and miR166 levels. Lower panels show the amount of FLAG-AGO1 and miR166 purified in the IP and used in the slicer assay by protein and RNA gel blot analyses, respectively. The weak FLAG signal for FLAG-AGO1H988F detected on this membrane was in part due to sample loss during gel loading and was not observed in other experiments (see Supplemental Figure 1 for an example). This is consistent with the nearly equal amount of miR166 immunoprecipitated from FLAG-AGO1H988F compared with the other lines. Additional controls show that slicer activity is detectable in less than 1% of the amount of FLAG-AGO1WT used here (Supplemental Figure 1), corroborating the conclusion that FLAG-AGO1H988F is defective in slicer activity.
(C) Slicer activity of immuno-affinity-purified FLAG-AGO1 from Arabidopsis seedlings of stable transgenic lines expressing wild-type and slicer-deficient FLAG-AGO1 in an ago1-3 homozygous background. The assay was performed as in (B), and RNA was extracted from the whole reaction.
(D) Proposed slicing mechanism of Arabidopsis AGO1. Steps in RNA cleavage (I, II, and III) follow the previously proposed structure-based model for slicing by Thermus thermophilus Ago (Sheng et al., 2014). (I) Precleavage state: Three active site residues in close proximity to the scissile phosphate bond of the RNA target. Absence of the glutamic finger (E803) and probably also of metal ions. (II) Cleavage state: An activated water molecule (cyan) coordinated by Mg2+ ion A (magenta) is positioned to perform a nucleophile attack on the scissile phosphodiester. Both ions A and B are involved in coordinating the pentavalent hydrolysis intermediate. Note the presence of the glutamic finger and its role in coordinating metal ion B through two water molecules. (III) Postcleavage state before product release: All active-site residues and metal ions are present. Water molecules not directly involved in catalysis are colored blue.
FLAG-AGO1 protein levels were roughly equal in the different transgenic lines, and miR159 and miR166 levels were equal in all FLAG immunopurified fractions (Figure 1B, bottom left panels; Supplemental Figure 1C). This suggests that the contribution of Glu-803 to catalysis may be less important than that of the canonical triad residues. This result is consistent with the two metal ion reaction mechanism proposed based on the structure of bacterial, fungal, and human AGOs (Wang et al., 2008; Nakanishi et al., 2012, 2013; Sheng et al., 2014). In this model, Asp-762 and His-988 interact directly with the metal ion that coordinates the water molecule responsible for hydrolysis, while Asp-848 and Glu-803 coordinate the second ion responsible for stabilization of the 3′-O− leaving group. Glu-803 only does so indirectly through water molecules (Figure 1D), which is consistent with this residue having the least strong effect on catalysis. Nonetheless, catalysis is severely impaired in E803A mutants, and there were no clear phenotypic differences between null mutants expressing FLAG-AGO1E803A and mutants in canonical triad residues (Arribas-Hernández et al., 2016).
Slicer-Deficient AGO1 Is Defective in Substrate Release
A previous study used AGO1 RNA immunoprecipitations from total plant lysates to show that slicer-deficient AGO1 mutants bind stably to miRNA targets in vivo (Carbonell et al., 2012). Thus, one explanation for the lack of detectable cleavage activity in slicer-deficient mutants could be that miRNA-loaded AGO1 is base-paired to endogenous plant RNA targets and hence cannot bind the radiolabeled substrate provided in vitro. To test this, we performed the slicer assay with FLAG-AGO1 immobilized on the FLAG immuno-affinity resin and analyzed separately the RNA released into the supernatant (Figure 1B, left panel) and retained on the AGO1 protein (Figure 1B, right panel). The experiment showed that substantially higher amounts of uncleaved substrate RNA were bound to the point mutants than to FLAG-AGO1WT. This analysis indicates that the four mutants are capable of binding to substrate RNA and, hence, that they are genuinely deficient in target RNA cleavage. In addition, the higher level of associated, intact target RNA with the point mutants compared with FLAG-AGO1WT provides direct evidence that they are defective in target RNA release upon base pairing, as suggested by Carbonell et al. (2012).
Slicer-Deficient AGO1 Loads miRNAs Normally in Vivo
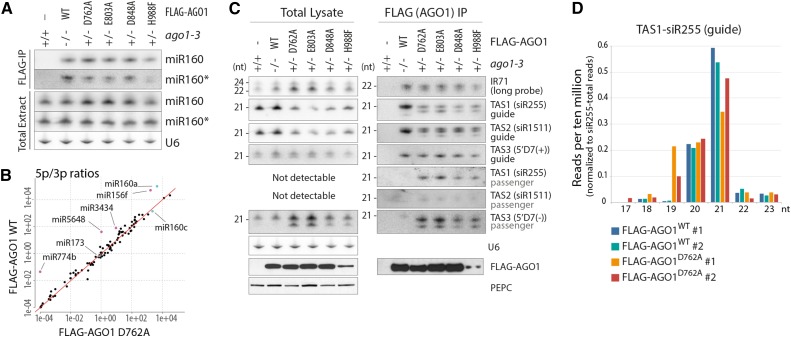
We next analyzed the relevance of slicer activity for small RNA association with AGO1 in vivo. Biochemical analysis in plant and animal lysates have indicated that loading of miRNAs occurs independently of slicer activity (Matranga et al., 2005; Iki et al., 2010), probably because they are imperfectly base paired to their corresponding passenger strands (also known as miRNA*s). Our in vivo analysis corroborated this result, since we observed comparable association with FLAG-AGO1WT and slicer-deficient AGO1 alleles for miR166 (Figure 1B), miR159 (Supplemental Figure 1C), miR160, and miR160* (Figure 2A). Profiling of small RNAs in FLAG affinity-purified AGO1WT and AGO1D762A also confirmed that miR/miR* read count ratios were similar (Figure 2B).
Figure 2.
miRNA and siRNA Loading in Slicer-Deficient AGO1 in Vivo.
(A) RNA gel blot analyses of total or FLAG-AGO1-bound miR160 and miR160* from inflorescences of stable transgenic lines expressing FLAG-AGO1 wild-type or slicer-deficient mutants. U6 is used as loading control for total extracts. The same membranes were rehybridized with miR160, miR160*, and U6 probes and were also used for siRNA analysis in (C) and, in the case of FLAG-IP, for miR166 analysis in Figure 1B.
(B) Plot of ratios between reads mapping the 5p and the 3p strands of miRNAs bound to wild-type and slicer-deficient (D762A) FLAG-AGO1 purified as in (A). Red line illustrates the diagonal. miRNAs of particular relevance for this study (miR173 and miR160) are highlighted in cyan; outliers with higher 5p/3p ratios in FLAG-AGO1WT than in FLAG-AGO1D762A are highlighted in magenta.
(C) RNA gel blot analyses of total or FLAG-AGO1-bound siRNAs from inflorescences of stable transgenic lines expressing FLAG-AGO1 wild-type or slicer-deficient mutants. The same membranes were used to rehybridize different siRNA probes. Lower panels show expression and purification of FLAG-AGO1 by protein gel blot. PEPC is used as loading control for total protein. The results shown are from the same immunoprecipitation as the one used for the cleavage activity assay reported in Figure 1B.
(D) Reads per 10 million of siR255 (guide) sorted by size and normalized to total number of reads mapping to siR255, from RNA bound to affinity-purified FLAG-AGO1WT or FLAG-AGO1D762A. Two replicates of each genotype are shown. Only siRNAs that start with the same 5′ nucleotide as canonical 21-nucleotide siR255 (UUCUAAGUCCAACAUAGCGUA) have been considered, so that differences in size reflect 3′-truncations or 3′-extensions.
Slicer-Deficient AGO1 Is Defective in siRNA Duplex Unwinding in Vivo
In contrast to miRNAs, siRNAs derive from perfectly base paired duplexes. In vitro analysis of evacuolated tobacco (Nicotiana tabacum) BY-2 lysates indicated that AGO1 slicer activity is mandatory for strand separation of such duplexes (Iki et al., 2010), which is consistent with what was previously shown in Drosophila melanogaster (Matranga et al., 2005). It remains unclear, however, whether such a requirement also exists in vivo. Indeed, a recent study of human Ago2 suggested the existence of a slicer-independent pathway for duplex siRNA unwinding in human cells (Park and Shin, 2015). It is also unclear what the fate of a stalled AGO1-duplex siRNA loading intermediate would be in vivo. To address these issues, we analyzed endogenous hairpin-derived siRNA and tasiRNA populations bound to FLAG immuno-affinity-purified mutant and wild-type AGO1 by RNA gel blot analysis. In all cases, we observed siRNAs bound to AGO1, and mutant AGO1 fractions were strongly enriched for tasiRNA passenger strands (Figure 2C). These data are consistent with defective duplex siRNA separation in slicer-deficient mutants in vivo, and they argue that a loading checkpoint to rid cells of stalled intermediates containing duplex siRNA bound to AGO1 does not exist. Nonetheless, we did observe that slicer-deficient AGO1 complexes contained siRNA species that migrated faster than the ones bound to wild-type AGO1 (Figure 2C). This was particularly clear for the tasiRNA guides siR255 (TAS1) and siR1511 (TAS2) as well as the passenger strand TAS3 5′D7(−). For IR71 siRNAs, which are derived from an endogenous inverted repeat transcript, a faster migrating species could also be seen, although it was less well separated from the wild-type species compared with the tasiRNAs (Figure 2C). We note that the shorter siRNAs bound to slicer-deficient AGO1 were observed in inflorescences, but not in seedlings (Supplemental Figure 2).
The Shorter siR255 Species Arises by 3′-Trimming
To identify the nature of the faster migrating siRNA species, we sequenced small RNA populations bound to FLAG-AGO1WT and FLAG-AGO1D762A. The sizes of siRNAs starting with the same 5′-nucleotide were compared between the two samples. For siR255, these analyses showed that a 19-nucleotide species was present in addition to the canonical 21-nucleotide species (Figure 2D). This is consistent with trimming of the 3′-overhang of the AGO1-bound guide strand in an siRNA duplex. AGO1-dependent 3′-modifications, including trimming and uridylation, have previously been observed (Zhai et al., 2013; Ren et al., 2014), and it is possible that the trimming observed here is a manifestation of the same phenomenon that produces the previously described AGO1-dependent 3′-trimming of small RNA. We could not clearly assign the faster migrating species of siRNAs other than siR255 to reads detected by deep sequencing.
TasiRNAs Are Produced in Slicer-Deficient AGO1 Mutants
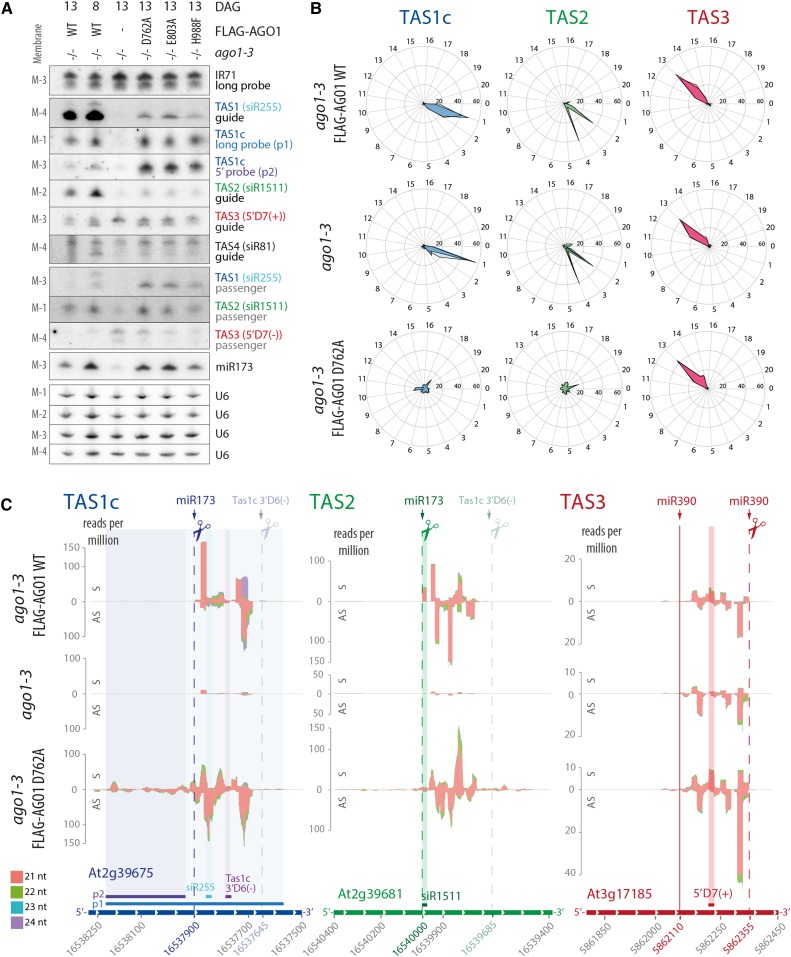
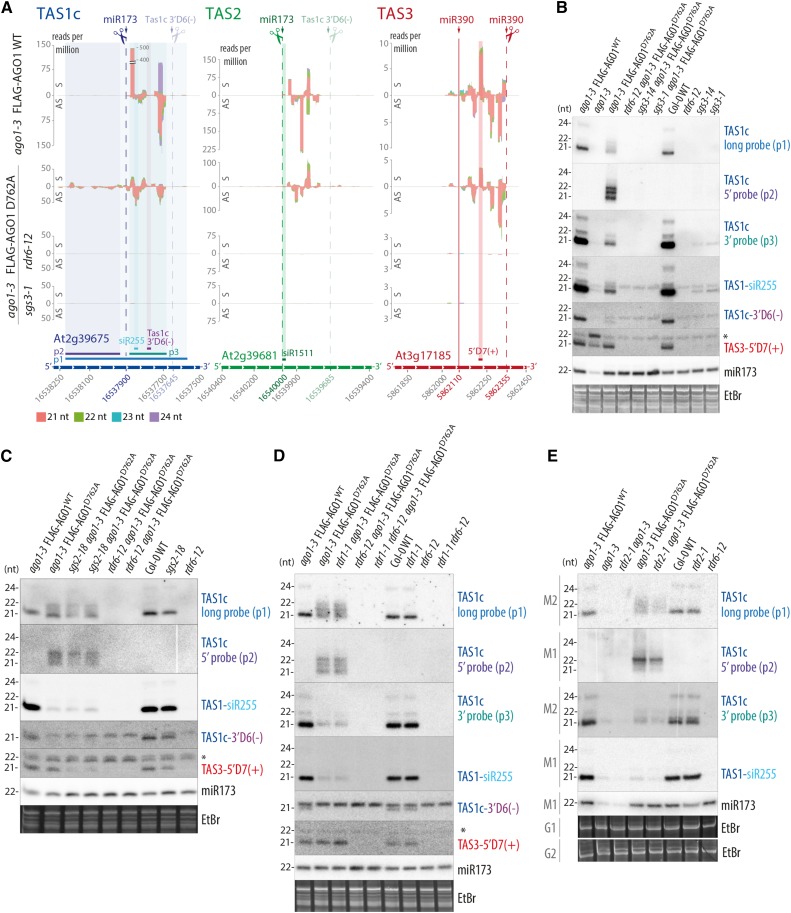
We next analyzed the requirement of slicing for production of siRNAs, in particular tasiRNAs. Previous work using transient coexpression of TAS1 precursor and wild-type or catalytically inactive AGO1 in Nicotiana benthamiana indicated that AGO1 slicer activity was required for production of the tasiRNA species siR255 (Carbonell et al., 2012). In addition, stable expression of slicer-deficient AGO1 in the hypomorphic ago1-25 mutant background led to decreased accumulation of siR255 (Carbonell et al., 2012). We compared IR71 siRNA and tasiRNA populations in ago1-3 null mutant seedlings with those in ago1-3 null expressing FLAG-AGO1WT and each of three slicer deficient AGO1 mutants (D762A, E803A, and H988F). As expected, IR71 siRNA levels were unaffected by loss of AGO1 slicer activity (Figure 3A). Surprisingly, however, 21-nucleotide signals were also clearly detectable with siR255 (TAS1), siR1511 (TAS2), and siR81 (TAS4) probes in slicer-deficient mutants, contrary to ago1-3 null mutants in which only very faint 21-nucleotide siR255 or siR81 signals were detected (Figure 3A). Remarkably, hybridization to a probe covering a 600-nucleotide region of the TAS1c transcript (p1; Figures 3C and 4A) gave comparable signals for wild-type and slicer-deficient mutants (Figure 3A). Furthermore, a probe 5′ to the miR173 target site of TAS1c (p2; Figures 3C and 4A) produced a clear signal in slicer-deficient mutants, in contrast to wild-type and ago1-3 null seedlings (Figure 3A). On the other hand, the AGO7-miR390-dependent 21-nucleotide TAS3 siRNA 5′D7(+) showed accumulation similar to the wild type in all three slicer-deficient AGO1 mutants, while the 21-nucleotide TAS3 5′D7(+) band (lower band) was undetectable in ago1-3 null mutants (Figure 3A). These RNA gel blot results indicate that secondary siRNA production from TAS1, TAS2, and TAS4 precursor transcripts depends on AGO1 protein, but not on its slicer activity.
Figure 3.
AGO1 Slicer Activity Is Required for Phasing but Not for Production of TasiRNAs.
Analysis of siRNAs in total RNA extracted from 8- or 13-d-old seedlings of Arabidopsis ago1-3 and stable transgenic lines expressing wild-type or slicer-deficient FLAG-AGO1 in the ago1-3 null background.
(A) RNA gel blot analysis. Four membranes (M-1, -2, -3, and -4) with identical loading were rehybridized with the indicated probes. Positions of probes matching Tas1c, Tas2, and Tas3 are indicated in (C). U6 was used as loading control. DAG, days after germination.
(B) Percentages of TAS1c, TAS2, and TAS3a tasiRNA reads for each of the 21 possible phases are shown for wild-type, ago1-3 null, or AGO1 slicer-deficient (D762A) 13-d-old seedlings. Data from the two biological replicates of every genotype are depicted with different shades of color and overlaid on the same diagram.
(C) Small RNA reads mapping to TAS1c, TAS2, and TAS3 transcripts. Abscissae, TAIR9 coordinates; ordinates, reads per million. S indicates sense siRNAs, and AS indicates antisense siRNAs, relative to the TAS precursor transcript. Cleavage sites are indicated by dashed lines, and the noncleavable miR390-site in TAS3 is indicated by a solid line. Positions of probes detecting specific siRNAs or populations of siRNAs in defined intervals are indicated.
Data for additional TAS transcripts can be found in Supplemental Figure 4, and counts of tasiRNAs are detailed in Supplemental Data Set 1. Membranes 1 to 4 that were used to hybridize siRNA probes in Figure 3A are the same as the ones used for miRNA analysis in Figure 2A of Arribas-Hernández et al. (2016). Accordingly, the U6 loading controls are the same in the corresponding panels.
Figure 4.
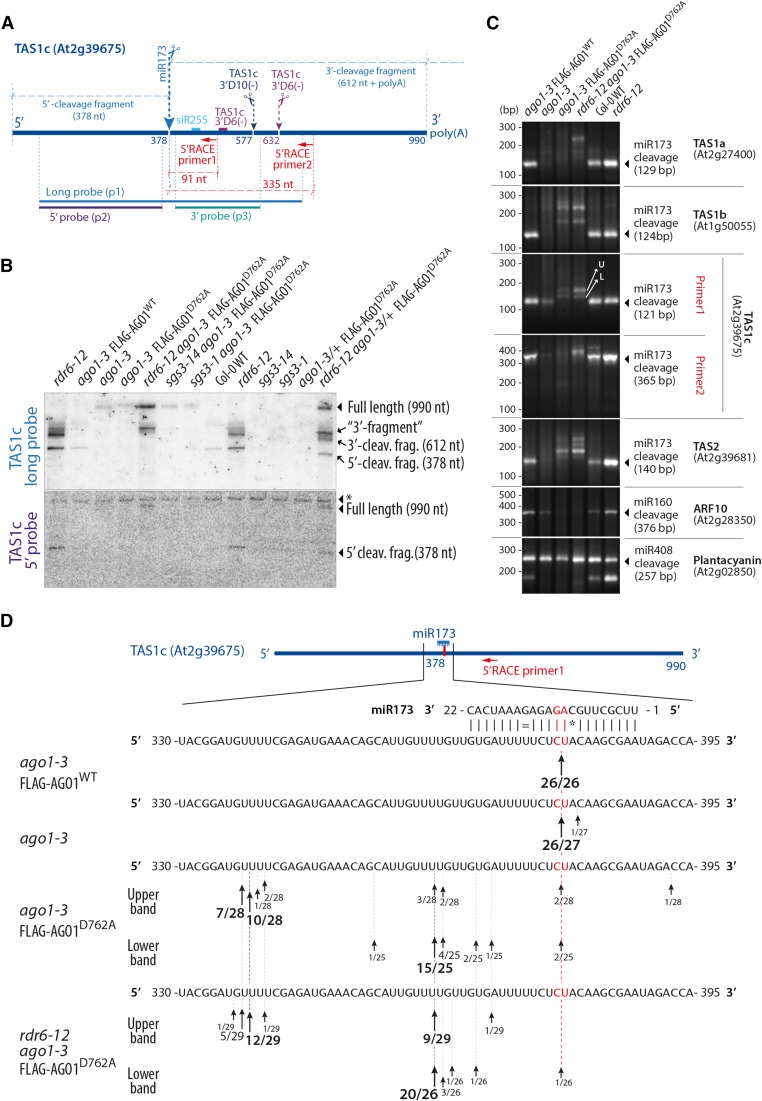
miR173 Does Not Guide TAS1/2 Precursor Cleavage in the Absence of AGO1 Slicer Activity.
(A) Schematic representation of the TAS1c precursor transcript. Positions of cleavage sites of miR173, TAS1c 3′D10(−), and TAS1c 3′D6(−) are indicated, and sizes of cleavage fragments [excluding poly(A) tails] are given. The positions of probes used in (B) and of 5′-RACE primers used in (C) are also indicated.
(B) RNA gel blot of total RNA separated by electrophoresis in a denaturing 5% polyacrylamide gel to analyze high molecular weight RNA. The asterisk in the lower panel indicates a band produced by unspecific hybridization that serves as a loading control. Results of an independent experiment in which the 5′-cleavage is more clearly detectable in the wild type are shown in Supplemental Figure 5.
(C) PCR fragments amplified by a modified 5′-RACE procedure that allows isolation of 5′-ends of 5′-phosphorylated, polyadenylated RNA. All gene-specific primers were placed 3′ to predicted cleavage sites. Arrows in the TAS1c panel indicate the excised bands used for sequence analysis in (D). U, upper band; L, lower band. Notice that the predicted sizes of PCR products are 30 bp longer than the distance between the gene specific primer and the free 5′-end, corresponding to the sequence in the RNA adaptor oligonucleotide from the 5′ nested primer.
(D) Sequence analysis of 5′-ends of cloned DNA fragments shown in (C) (TAS1c, Primer 1). Arrows indicate positions of identified 5′-ends. The number of clones containing a fragment with the given 5′-end and the total number of clones sequenced are indicated.
AGO1 Slicer Activity Is Required for Phasing of TasiRNAs
To analyze tasiRNA populations in more detail, we sequenced total small RNA libraries from ago1-3 null mutants and from ago1-3 lines expressing wild-type or slicer-deficient AGO1. The analysis confirmed that TAS1 and TAS2 precursors produce abundant siRNAs in AGO1 slicer-deficient mutants, while very low numbers of siRNA reads were detected in ago1-3 null (Supplemental Data Set 1). Interestingly, tasiRNAs produced in all three slicer-deficient mutants exhibited two properties not seen in the wild type. First, they nearly completely lost the phased register of accumulation (Figure 3B; Supplemental Figure 3). Second, the region spawning siRNAs were extended in the 5′-direction, particularly in TAS1c (Figure 3C; Supplemental Figure 3). The 3′-borders of the main siRNA-spawning intervals were largely retained, although in TAS1a/b and TAS2, low-abundance reads extended beyond the position of the 3′-border in the wild type (Figure 3C; Supplemental Figures 3 and 4). In all cases, the 3′-border of the main siRNA-spawning intervals was found 40 to 45 nucleotides upstream of the cleavage site of the TAS1c 3′D6(−) siRNA. We could not complete similar analyses for TAS4 because of the insufficient number of sequence reads recovered on that locus (Supplemental Figure 4 and Supplemental Data Set 1). TAS3 tasiRNAs retained their phased register in all ago1 slicer-deficient mutants (Figures 3B and 3C; Supplemental Figure 3), which is consistent with the dependence of AGO7-miR390 for biogenesis of these tasiRNAs (Adenot et al., 2006; Montgomery et al., 2008). We note that in contrast to slicer-deficient mutants, residual TAS1/TAS2 tasiRNAs in ago1-3 were phased (Figure 3B), suggesting that other AGO proteins, perhaps AGO10, might bind miR173 and cleave the TAS1/TAS2 transcripts in the absence of AGO1 protein. We also note that the nearly complete loss of TAS1/TAS2, but not TAS3, tasiRNAs in ago1-3 supports a role of AGO1 protein in tasiRNA biogenesis and not merely in stabilization. TasiRNAs are sorted into different AGOs depending on the identity of the 5′-terminal nucleotide, and numerous TAS1/TAS2/TAS3 tasiRNAs species associate with both AGO1 and AGO2 (Mi et al., 2008; Takeda et al., 2008). If TAS1/TAS2 tasiRNAs were produced in significant amounts in the absence of AGO1, loading into AGO2 would stabilize several species, allowing detection by RNA-seq as is the case for many TAS3 tasiRNAs. Indeed, the absence of TAS3 5′D7(+) signal in ago1-3 by RNA gel blot analysis probably reflects rapid degradation of this species because it associates nearly exclusively with AGO1 (Mi et al., 2008), strongly suggesting that the remaining TAS3 siRNAs in ago1-3 are those species that load into AGOs other than AGO1 in the wild type. Conversely, TAS1c 3′D6(−) was undetectable in ago1-3 (Figure 3C; see Supplemental Figure 5 for gel blot analysis), despite the fact that it associates equally efficiently with AGO1, AGO2, and AGO4 (Rajeswaran et al., 2012), indicating that it is not produced in ago1-3. We conclude from these analyses that the production of TAS1/TAS2 tasiRNAs requires AGO1 protein, but not its slicer activity, and that AGO1 slicer activity is specifically required for definition of the phase of tasiRNAs.
miR173 Does Not Guide Cleavage of TAS1/2 Precursors in Seedlings Specifically Defective in Slicer Activity of AGO1
The production of tasiRNAs in the absence of slicer activity of AGO1 does not necessarily exclude slicing of TAS1/2 precursor transcripts by other AGO proteins guided by miR173. To test whether miR173-guided cleavage occurs in the absence of AGO1 activity, we first used the finding that TAS1/2 cleavage fragments diagnostic of miR173-guided slicing are detectable in the wild type (Yoshikawa et al., 2005). RNA gel blots showed the presence of TAS1c 5′- and 3′-cleavage fragments in the wild type, but not in ago1-3 null or in the slicer-deficient D762A mutant (Figures 4A and 4B; Supplemental Figure 5). Since cleavage fragments are stabilized in rdr6 mutants (Yoshikawa et al., 2005), we also included rdr6-12/ago1-3/FLAG-AGO1D762A in our analyses. These samples confirmed the absence of a 5′-cleavage fragment upon loss of slicer activity, but they did reveal a TAS1c fragment detected by the TAS1c long probe, but not by the 5′ probe (hereafter referred to as the 3′-fragment). This 3′-fragment migrated at a slightly higher molecular weight than the 3′-cleavage fragments detectable in rdr6-12 with wild-type AGO1 activity (Figure 4B). The size difference was particularly obvious in samples from the FLAG-AGO1D762A line in an rdr6-12/ago1-3 heterozygous background in which two distinct 3′-fragments were identified (Figure 4B). Consistent with RDR6-DCL4-mediated turnover of the TAS1c precursor, an increase in TAS1c full-length transcript abundance was seen in rdr6-12/ago1-3/FLAG-AGO1D762A compared with ago1-3/FLAG-AGO1D762A (Figure 4B). Such an increase was not observed upon inactivation of SGS3 (Figure 4B).
To analyze the 3′-fragment in more detail, we performed 5′-RACE analysis of 5′-phosphorylated RNA fragments with primers either close to the miR173 cleavage site in TAS1c (Primer 1) or further downstream 3′ to the TAS1c 3′D10(−) and TAS1c 3′D6(−) cleavage sites (Primer 2, Figure 4A). The 5′-RACE experiment was technically robust because cleavage fragments of AUXIN RESPONSE FACTOR10, which is targeted by the AGO1-associated miR160 (Llave et al., 2002; Kasschau et al., 2003), were undetectable in AGO1 slicer-deficient backgrounds, while cleavage fragments of PLANTACYANIN, targeted by the AGO1- and AGO2-associated miR408 (Maunoury and Vaucheret, 2011), were present in all genetic backgrounds tested (Figure 4C). For TAS1c, cleavage at the miR173 site was easily detectable in FLAG-AGO1WT/ago1-3, while RACE products corresponding to TAS1c 3′D10(−) or TAS1c 3′D6(−) cleavage sites were not identified (Figure 4C). In the ago1-3 null background, miR173-guided cleavage of TAS1c was also detected, although cleavage fragments were less abundant than in the wild type (Figure 4C). This result demonstrates that other AGO proteins can provide miR173-guided cleavage activity in the absence of AGO1 protein and is consistent with the presence of residual, phased tasiRNAs in ago1-3 mutants (Figures 3B and 3C). In striking contrast, virtually no cleavage was detected at the miR173 site in the slicer-deficient ago1 background (Figures 4C and 4D). Nonetheless, two longer 5′-RACE products were amplified, which is consistent with the results of RNA gel blot analysis (Figures 4B and 4C). Sequencing revealed that these products corresponded to distributions of 5′-ends around two sites 16 and 40 nucleotides upstream of the miR173 cleavage site (Figure 4D). A similar pattern of absence of miR173-guided cleavage, but presence of fragments with 5′-ends upstream of the miR173 binding site was also observed in TAS1a, TAS1b, and TAS2 (Figure 4C). These data indicate that no other AGO protein substitutes for AGO1 to guide cleavage by miR173 in ago1-3/AGO1D762A mutants. Indeed, the 3′ fragments detected in ago1-3/AGO1D762A are unlikely to be produced via small RNA-guided cleavage because siRNAs matching the TAS1c RNA 5′ to the miR173 site could not be found in rdr6-12/ago1-3/FLAG-AGO1D762A (Figure 5A) and because no TAS1c 5′-cleavage fragment was detectable in this background (Figure 4B).
Figure 5.
RDR6, RDR1, RDR2, and SGS3 Dependence of TasiRNAs Produced in the Absence of AGO1 Slicer Activity.
(A) Small RNA reads mapping to TAS1c, TAS2, and TAS3 in the indicated mutants and transgenic lines. The representation is done as in Figure 3C. Small RNA libraries were prepared from total RNAs from 13-d-old seedlings. Reads mapped to additional TAS transcripts can be found in Supplemental Figure 5, and counts of tasiRNAs and miRNAs are detailed in Supplemental Data Set 2.
(B) to (E) RNA gel blot analysis of TAS1c, TAS3 siRNAs, and miR173. Total RNA from 13-d-old seedlings was used. Probe positions are shown in (A). In all cases, consecutive hybridizations to the same membranes were done with the indicated probes. Ethidium bromide (EtBr) staining of the upper part of the acrylamide gels was used as loading control.
(B) Analysis of rdr6-12, sgs3-1, and sgs3-14 null alleles in the ago1 slicer-deficient background. RNA samples were prepared from independently grown tissue compared with the sRNA-seq experiment shown in (A).
(C) Analysis of the hypomorphic RDR6 allele sgs2-18 in the slicer-deficient ago1 background. Two biological replicates of sgs2-18/ago1-3/FLAG-AGO1D762A and rdr6-12/ago1-3/FLAG-AGO1D762A were analyzed.
(D) Analysis of the rdr1-1 null allele and of rdr1-1/rdr6-12 in the ago1 slicer-deficient background.
(E) Analysis of the rdr2-1 null allele in ago1-3 null and slicer-deficient backgrounds. Membranes 1 (M1) and 2 (M2) are blotted from gels 1 (G1) and 2 (G2), respectively. Asterisks in TAS3 5′D7(+) panels indicate a 22-nucleotide species whose origin and biological function are unclear. The white lines visible in (C) (TAS1c p2) and (E) (TAS1c p2, p3) are digital artifacts produced during phosphor imaging and do not indicate presence of additional lanes.
Dependence on RDRs and SGS3 of Unphased TasiRNAs
We next analyzed the effect of rdr6-12 and sgs3-1 null mutations on tasiRNA production in the ago1-3/AGO1D762A line. Sequencing of total small RNA libraries revealed a nearly complete loss of small RNA reads from TAS1abc and TAS2 in the rdr6-12 and sgs3-1 mutant backgrounds (Figure 5A; Supplemental Figure 6 and Supplemental Data Set 2), suggesting that tasiRNA production in the absence of AGO1 slicer activity proceeds via the known SGS3/RDR6-dependent pathway. Consistent with the sequencing data, RNA gel blots showed a loss of TAS1c siRNAs in ago1-3/AGO1D762A containing the rdr6-12, sgs3-1, and sgs3-14 mutations (Figure 5B). To further support the conclusion that the same molecular machinery generates tasiRNAs in the absence of AGO1 slicer activity as in the wild type, we also introduced rdr1-1, rdr1-1/rdr6-12, rdr2-1, and the rdr6 mutant allele sgs2-18 into ago1-3/AGO1D762A. sgs2-18 is defective in siRNA production from sense transgenes but makes normal levels of tasiRNAs (Adenot et al., 2006). TAS1c siRNA production was also unaffected by the sgs2-18 mutation in the absence of AGO1 slicer activity (Figure 5C), suggesting that the same type of RDR6 activity is involved in TAS1c siRNA production in the absence and presence of AGO1 slicer activity. RDR1 is involved in viral siRNA amplification (Garcia-Ruiz et al., 2010) and can participate in the production of siRNAs that require both RDR6 and RDR1 (Lam et al., 2015), demonstrating the existence of more than one RDR6-dependent siRNA biogenesis pathway. In the slicer-deficient ago1 background, TAS1c siRNAs remained unaffected by inactivation of RDR1 (Figure 5D). RDR2 is mainly involved in the biosynthesis of 24-nucleotide siRNAs that guide transcriptional gene silencing (Xie et al., 2004), but it can also influence the production of other small RNAs (Jauvion et al., 2012; Klein-Cosson et al., 2015). Mutation of RDR2 had only a small effect on TAS1c siRNA accumulation in the slicer-deficient ago1 background (Figure 5E). Taken together, our results show that siRNAs are generated from TAS precursor transcripts in the absence of AGO1 slicer activity by the same molecular machinery as in the wild type despite the fact that miR173-guided precursor cleavage does not occur. In addition, we note that secondary siRNAs generated 5′ to the miR173 site in the catalytic ago1 mutant exhibited several apparent size classes. While the TAS1/2 dsRNA produced in this mutant may be substrates for more than one Dicer-like enzyme, the high-resolution gels shown in Figures 5B to 5D suggest that the siRNAs are also covalently modified because a distinct signal is detectable between the 21- and 22-nucleotide marker bands. Hybridizations detecting siR255 and TAS1c 3′D6(−) showed similar siRNA species in both ago1 slicer-deficient and wild-type backgrounds.
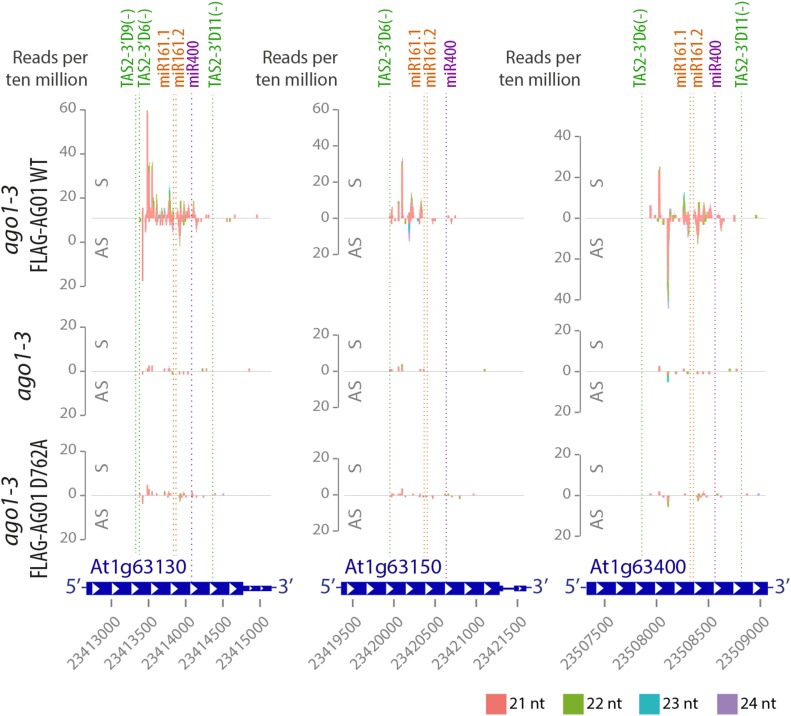
AGO1 Slicer Activity Is Required for siRNAs Spawned by TAS2/miR161 Targets
In addition to TAS transcripts, a number of pentatricopeptide repeat protein (PPR)-encoding mRNAs with multiple target sites for TAS2 tasiRNAs and the miRNAs miR161 and miR400 has been shown to produce RDR6-dependent secondary siRNAs (Howell et al., 2007). These siRNA populations were lost or strongly reduced in abundance in both ago1 null and in slicer-deficient AGO1 mutants (Figure 6; Supplemental Figure 7), indicating that slicer activity is required for their production. This observation does not necessarily mean that slicing of PPR mRNAs is required for secondary siRNA formation, however. Transcripts with more than one primary small RNA target site tend to produce secondary siRNAs particularly efficiently (Axtell et al., 2006). For the PPRs, miR400 may not contribute to secondary siRNA formation in the tissue analyzed due to its very low abundance (Arribas-Hernández et al., 2016), since secondary siRNAs were not found in proximity to the miR400 target site even in the wild type in our experiments (Figure 6; Supplemental Figure 7). On the other hand, the TAS2 target site appears to be of special importance because it defines the border of siRNA-spawning segments on all PPR mRNAs (Figure 6; Supplemental Figure 7). In slicer-deficient AGO1 mutants, passenger strand cleavage of TAS2 siRNA duplexes is defective (Figure 2C), causing effective loss of function of the TAS2 target site in PPR mRNAs. Therefore, the requirement for AGO1 slicer activity for the production of secondary PPR siRNAs is likely to be caused by defective passenger strand cleavage of TAS2 siRNA duplexes rather than defective PPR mRNA cleavage.
Figure 6.
AGO1 Slicer Activity Is Required for siRNAs Spawned by TAS2/miR161/miR400 Targets.
Small RNA reads of 13-d-old seedlings of Arabidopsis ago1-3 and stable transgenic lines expressing wild-type or slicer-deficient FLAG-AGO1 in the ago1-3 null background mapping to AT1G63130, AT1G63150, and AT1G63400 transcripts. Abscissae, TAIR9 coordinates; ordinates, reads per 10 million. S indicates sense siRNAs, and AS indicates antisense siRNAs, relative to mRNAs. Cleavage sites of TAS2 3′D6(−), TAS2 3′D9(−), TAS2 3′D11(−), miR161.1, miR161.2, and miR400 are indicated by dashed lines. The target sites of the couples miR161.1/miR161.2 and TAS2-3′D9(−)/TAS2-3′D6(−) have an overlap of 10 to 12 nucleotides and therefore cannot be considered as “double hits” on their own. Data for additional TAS2/miR161/miR400 target loci, 8-d-old wild-type control, and two other slicer-deficient mutants can be found in Supplemental Figure 7.
DISCUSSION
Mechanism of RDR6 Recruitment to TAS Transcripts
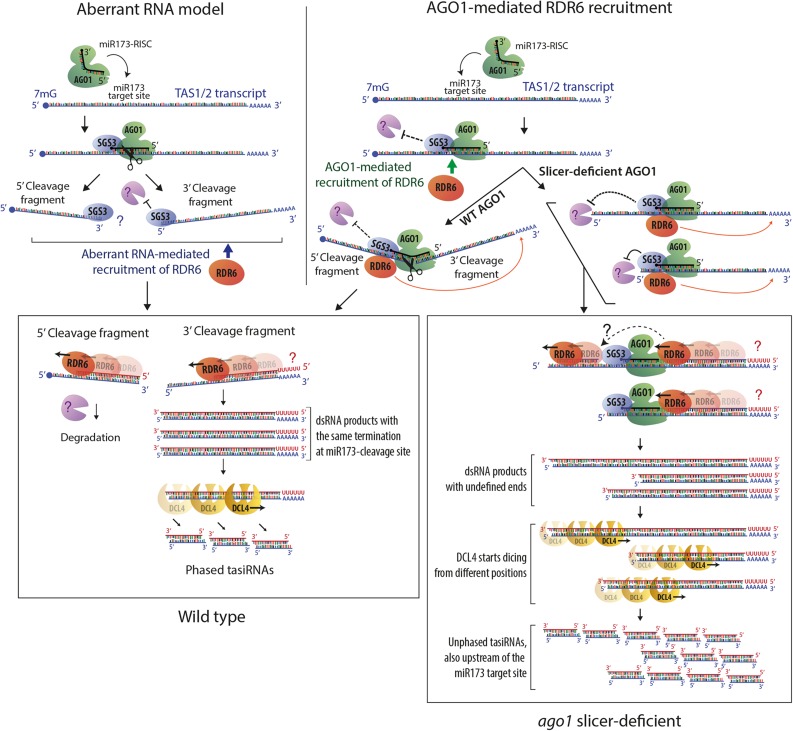
It is an important result of our study that the role of AGO1 slicer activity in tasiRNA biogenesis is limited to the definition of the phase of siRNAs rather than to the production of transitive siRNAs per se. In particular, since TAS1/2 precursor transcripts show no significant cleavage at the miR173 site in catalytic ago1 mutants, this result calls into question the model that the generation of cleavage fragments is a requisite step in secondary siRNA formation (Figure 7, left). Although a 3′-fragment of the TAS1c precursor transcript was detected in the slicer-deficient mutant in an rdr6 background, we do not consider it likely that this fragment arises by precursor cleavage. Rather, two observations suggest that it may be a degradation intermediate that accumulates because slicer-deficient AGO1 bound tightly to the miR173 target site acts as a road block to 5′-3′ exonucleolysis. First, a 5′-cleavage fragment was not detectable in ago1 slicer-deficient backgrounds, even in rdr6-12, where the 3′-fragment is clearly detectable by RNA gel blot analysis. Second, the 5′-ends constitute distributions around a preferred site rather than a distinct site (Figure 4D), which is consistent with the termination of exonucleolysis upon collision with bound AGO1 (Bagga et al., 2005). Nonetheless, regardless of their precise mechanism of generation, one may argue that these TAS1c aberrant RNAs constitute the source of RDR6-dependent siRNA formation. Two observations suggest that the 3′-fragments cannot be the exclusive source of TAS1c siRNAs. First, siRNAs extend beyond the 5′-end of the 3′-fragment. Second, the abundance of the full-length TAS1c precursor increases upon mutation of RDR6 in ago1-3/FLAG-AGO1D762A (Figure 4B), suggesting that RDR6 mediates turnover of the full-length transcript. Both observations could be explained if one assumes that the 3′-fragments merely trigger siRNA formation and that siRNAs are amplified subsequent to slicing of full-length TAS1c precursors by AGO proteins other than AGO1. While we cannot completely exclude such a scenario based on the present data, we do not consider it likely for two reasons. First, the sgs2-18 allele of RDR6 did not have any effect on the abundance of TAS1c siRNAs in the catalytic ago1 mutant. sgs2-18 was previously shown to be required for the production of amplified transgene siRNAs, but not for tasiRNAs (Adenot et al., 2006), whose phased pattern of accumulation in wild-type plants implies only a single round of RDR6 action on tasiRNA precursors. This suggests that despite their loss of phasing, tasiRNAs produced in ago1 slicer-deficient mutants may not involve reiterative siRNA amplification. Second, 5′-RACE analysis with the downstream TAS1c primer did not reveal any cleavage fragments that might be detectable if a few siRNAs guided cleavage of the TAS1c precursor. For these reasons, we favor the alternative model that recruitment of RDR6 by AGO1 underlies secondary siRNA production (Figure 7, right). Four other observations are in agreement with this model. First, in vitro-translated AGO1 loaded with the 22-nucleotide-long miR173 interacted with SGS3 in cell-free lysates (Yoshikawa et al., 2013). Second, a function of AGO1 in secondary siRNA production in addition to slicer activity was inferred from the observation that the miRNA duplex structure is a determinant of whether secondary siRNA formation is triggered (Manavella et al., 2012). Third, in contrast to AGO1, AGO2 loaded with miR173 failed to induce tasiRNA formation despite similar TAS1c precursor cleavage in transient expression assays, suggesting a more direct role of AGO1 in secondary siRNA formation (Carbonell et al., 2012). Fourth, two features of ectopic, miRNA-triggered secondary siRNAs in ski2 mutants pointed to a direct involvement of AGO1 in recruitment of RDR6. Although only RISC 5′-cleavage fragments were stable in mutants of the exosome cofactor SUPERKILLER2, examples of miRNA targets with secondary siRNAs exclusively 3′ to miRNA cleavage sites were identified (Branscheid et al., 2015). In addition, spreading of secondary siRNAs occurred on the cleavage fragment least stably base-paired to the miRNA, suggesting that RDR6 may be present at the AGO1-miRNA-target RNA complex prior to release of cleavage fragments and may use the cleavage fragment released first from RISC as template (Branscheid et al., 2015). Nonetheless, three observations have favored the model that slicing is required for secondary siRNA production. First, the 3′-cleavage fragment of TAS1 precursors can be observed by RNA gel blot analysis in rdr6, but not in sgs3/rdr6 mutants, supporting a model in which SGS3 stabilizes cleavage fragments to be used as template for RDR6 (Yoshikawa et al., 2005). However, this observation does not exclude direct recruitment of SGS3/RDR6 by AGO1 rather than by aberrant features of the cleaved RNA. Second, siR255 abundance as measured by RNA gel blot analysis was strongly reduced upon transient expression of TAS1 precursor and slicer-deficient AGO1 in N. benthamiana compared with coexpression with wild-type AGO1 (Carbonell et al., 2012). Since production of only a single siRNA species with phased accumulation pattern in the wild type was analyzed, this leaves open the possibility that loss of phasing leads to reduction of hybridization signal to this one probe. Third, AGO7 slicer activity was strictly required for production of a species of TAS3 siRNA in stable transgenic lines in Arabidopsis (Carbonell et al., 2012). Subsequent studies of miR390 loading onto AGO7 showed that cleavage of the miR390* strand is required for maturation of AGO7-miR390 RISC (Endo et al., 2013), suggesting that not only TAS3 precursor cleavage, but AGO7 recruitment to TAS3 precursors altogether, is abolished in slicer-deficient ago7 mutants as a consequence of defective miR390/miR390* duplex unwinding. We note that recruitment of RDRs via AGO proteins rather than cleavage fragments may also occur in the roundworm Caenorhabditis elegans whose RNAi response is effected by secondary RDR-dependent small RNAs (22G RNAs) (Yigit et al., 2006). Indeed, slicer-deficient mutants in the AGO protein RDE-1 generated a normal RDR-dependent RNAi response as long as the primary small RNA duplex loaded into RDE-1 contained enough mismatches to enable duplex unwinding without passenger strand cleavage (Steiner et al., 2009). In addition, in the RNAi pathway directing heterochromatin formation at outer centromeric repeats in fission yeast (Schizosaccharomyces pombe), the Ago-containing RITS complex and the RDR-containing RDRC physically interact (Motamedi et al., 2004). Therefore, direct recruitment of RDRs via AGO proteins may be a general principle underlying secondary siRNA production in animals, plants, and fungi.
Figure 7.
Models of RDR6 Recruitment to TAS Transcripts.
Schematic diagram highlighting main features of two models to explain RDR6 recruitment to TAS transcripts and the function of SGS3. In the aberrant RNA model (left), cleavage of TAS precursors is required to produce RNA fragments with aberrant features. SGS3 binds to the miR173/TAS duplex region protruding from RISC, and the role of SGS3 is to stabilize cleavage fragments to provide substrate for RDR6 (Fukunaga and Doudna, 2009; Yoshikawa et al., 2013). Protection of the 5′-cleavage fragment by SGS3, and RDR6-mediated conversion into dsRNA for degradation is postulated because steady state levels of the 5′-cleavage fragment increase substantially in rdr6 mutants and disappear in sgs3 mutants (Yoshikawa et al., 2005). In the model on the right, the AGO1 protein is required to recruit RDR6, and its miR173-guided precursor cleavage ensures phasing because it generates well-defined 5′-ends of the RDR6 substrate. This model readily explains how tasiRNAs are generated in catalytic AGO1 mutants. The model assumes that two mechanisms may contribute to preferential accumulation of siRNAs 3′ to the miR173 site: A road block function of bound AGO1/SGS3 limits RDR6 processivity to the miR173 site in most cases (Rajeswaran and Pooggin, 2012) and stops 5′-3′-exonucleolysis of those transcripts that escape protection against degradation provided by bound SGS3. On transcripts in which AGO1/SGS3 dissociate, RDR6 continues until the 5′-end, consistent with dsRNA mapping performed by Rajeswaran et al. (2012). Together, these processes give rise to a pool of DCL substrate dsRNAs with different ends, thus explaining the unphased nature of tasiRNAs in catalytic ago1 mutants. In both models, RDR6 is shown to initiate at the end of poly(A) tails, but the question marks indicate that it is unclear whether deadenylated or polyadenylated RNAs are used as templates in vivo.
Function of SGS3 in Secondary siRNA Formation
Previous studies suggested that SGS3 acts to stabilize cleavage fragments to be used as RDR6 templates (Yoshikawa et al., 2005, 2013). Our study suggests that SGS3 function is unlikely to be confined to specific stabilization of cleavage fragments. This is because of the full requirement of SGS3 for tasiRNA formation in the ago1-3/AGO1D762A line, in which miR173-guided cleavage fragments are not produced. SGS3 may mediate AGO1-dependent recruitment of RDR6 to template RNA, particularly in light of the AGO1-SGS3 and SGS3-RDR6 associations reported previously (Kumakura et al., 2009; Yoshikawa et al., 2013). Such a scenario would predict that mutation of SGS3 and RDR6 would lead to similar increases in TAS1 precursor transcript levels in the ago1 slicer-deficient background. However, this is not what we observe: In rdr6, TAS1c transcript levels show the expected increase, but not in sgs3-14 or sgs3-1, in which the levels are as low as in the presence of RDR6 and SGS3 (Figure 4B). This observation suggests that SGS3 associates with AGO1 and TAS precursor transcript and protects TAS RNAs from degradation regardless of whether they are cleaved or not (Figure 7). In AGO1 wild type, this would result in observable stabilization of cleavage fragments, but not full-length TAS RNA, if the cleavage rate is much higher than the rate of RISC dissociation. Such a result was indeed observed in a cell-free system by Yoshikawa et al. (2013). Conversely, in an AGO1 slicer-deficient background, SGS3 would protect TAS precursors from degradation, as observed in the stable transgenic lines used here. Immunodepletion of SGS3 in cell-free lysates failed to reveal such a protective effect of SGS3 on full-length TAS transcripts with bound slicer-deficient AGO1 (Yoshikawa et al., 2013). Despite this minor discrepancy that may well be attributed to the substantial differences in the two experimental systems, our observations on the effect of SGS3 inactivation support the model proposed by Yoshikawa et al. (2013) overall: Binding of a RISC/SGS3 complex to a target RNA stabilizes SGS3-bound RNA to allow its use as an RDR6 template. The observation that SGS3 localizes to distinct cytoplasmic foci (Kumakura et al., 2009; Jouannet et al., 2012) suggests that the basis for protection of TAS RNAs may be simple spatial sequestration away from RNA decay factors.
Importance of Two-Hit Triggers for Secondary siRNA Formation
In contrast to miR173-triggered tasiRNAs, RDR6-dependent siRNAs whose precursor mRNAs are targeted by miR161, miR400, and TAS2-derived siRNAs were nearly completely lost in AGO1 slicer-deficient mutants. At first glance, this result may suggest a requirement for mRNA cleavage for secondary siRNA production. Nonetheless, passenger strand cleavage and duplex unwinding of TAS2 tasiRNAs are defective in slicer-deficient AGO1, as evidenced by the enrichment of TAS2 passenger strands in immunopurified slicer-deficient AGO1 mutants compared with AGO1WT. Thus, in slicer-deficient AGO1, targeting by AGO1-TAS2 altogether is likely to be impaired, such that miR161/TAS2 two-hit targets are effectively converted into miR161 single hit targets. Thus, the loss of these siRNAs in slicer-deficient AGO1 mutants is also consistent with a requirement for AGO1 recruitment, but not necessarily target cleavage, for secondary siRNA production. It is possible that two closely spaced target sites on an RNA molecule may prolong RISC-target association, thus allowing RDR6 recruitment by AGO1 to occur particularly efficiently. Nonetheless, long RISC-target dwell times are unlikely to be sufficient for secondary siRNA generation via RDR6 because we did not observe ectopic secondary siRNAs on miRNA targets in the catalytic ago1 mutants.
METHODS
Plant Material and Growth Conditions
All lines used in this study are in the Arabidopsis thaliana Col-0 ecotype. The following mutants were described previously: ago1-3 (Bohmert et al., 1998), rdr6-12, sgs3-14 (SALK_001394) (Peragine et al., 2004), sgs2-18, sgs3-1 (Mourrain et al., 2000; Adenot et al., 2006), rdr1-1 (SAIL_672F11), and rdr2-1 (SAIL_1277H08) (Xie et al., 2004). T-DNA insertion lines were obtained from Nottingham Arabidopsis Stock Centre.
Seeds were surface-sterilized by immersion in 70% ethanol for 2 min followed by incubation in 1.5% hypochlorite 0.05% Tween 20 for 10 min and immediately rinsed with water. For molecular analyses of ago1 null or slicer-deficient seedlings, ∼500 seeds (10 mg) were germinated after 5 d of stratification and grown in 1-liter Erlenmeyer flasks containing 300 mL of liquid Murashige and Skoog (MS) medium (4.4 g/L MS salt mixture and 10 g/L sucrose) under 16-h-light (70 μmol m−2 s−1)/8-h-dark light cycle and mild agitation (140 rpm). The temperature was kept constant at 21°C, and Philips fluorescent tubes (TL-D 90 De Luxe 36W) were used as the light source. Whole seedlings including roots displaying unambiguous ago1 null or slicer-deficient phenotypes were manually selected and frozen in liquid nitrogen for analysis. When adult plants were required for genetic crosses or propagation, seedlings grown on MS-agar medium (4.4 g/L MS salt mixture, 10 g/L sucrose, and 8 g/L agar), pH 5.7, for 10 d were transferred to soil and maintained in a greenhouse with a 16-h-light/8-h-dark supplemental light cycle. Inflorescences for FLAG affinity purification were harvested from plants grown in incubators (Percival) at 60% relative humidity, under a 16-h-light (130 μmol m−2 s−1)/21°C 8-h-dark/16°C day-night cycle.
Plant Transformation and Selection of Transgenic Lines
Transgenic lines were generated by floral dip transformation (Clough and Bent, 1998) of ago1-3/+ plants with Agrobacterium tumefaciens GV3101 bearing pCAMBIA3300U AGO1P:FLAG-AGO1-AGO1T constructs. Selection of primary transformants (T1) was done on soil by spraying seedlings with 0.2 g/L BASTA 10 d after germination. Segregation studies of T2 and T3 populations were performed on MS-agar supplemented with glufosinate ammonium (Fluka; 7.5 mg/L). For every construct, two independent lines with a single T-DNA locus and comparable protein expression levels were selected in T2, and descendants homozygous for the insertion were identified in T3. Experiments comparing the four slicer-deficient mutants were performed by pooling T4 seeds of the two independent lines of every point mutant.
Generation of Double and Triple Mutant Lines
ago1-3/+ FLAG-AGO1D762A T3 plants from the same transgenic line were crossed to rdr6-12, rdr1-1, rdr6-12/rdr1-1, rdr2-1, sgs2-18, sgs3-1, and sgs3-14 mutants. F1 plants were allowed to self-pollinate, and ago1-3/+ mutants homozygous for the desired rdr1, rdr2, rdr6, and sgs3 alleles and containing the FLAG-AGO1D762A transgene were selected by genotyping and BASTA treatment among the corresponding F2 populations. F3 seedlings bearing the FLAG-AGO1D762A transgene in homozygosity were compared with the parental lines for molecular analysis.
Vectors
pCAMBIA3300U with a double PacI USER cassette (Supplemental Data Set 3) inserted between the PstI-XmaI sites at the multiple cloning site of the original pCAMBIA3300 plasmid was generously provided by Morten Nørholm (Nour-Eldin et al., 2006) and was used to introduce AGO1 constructs into ago1-3 plants.
Cloning of AGO1 Constructs
Two Arabidopsis AGO1 genomic DNA fragments were amplified from BAC T1N15 and introduced into pGEM-T Easy (Promega) to serve as templates for subsequent cloning. AGO1-AGO1T included the coding sequence of AGO1 and 670 bp downstream of the stop codon, and AGO1P contained the sequence located 2659 bp upstream of the start codon of AGO1. A combination of overlap extension PCR and USER cloning methodologies was applied to generate the AGO1P:FLAG-AGO1-AGO1T construct, as detailed in the Supplemental Methods. All primers used are listed in Supplemental Data Set 3.
Site-Directed Mutagenesis of FLAG-AGO1 Constructs
Mutagenesis of the four At-AGO1 catalytic residues (Asp-762, Glu-803, Asp-848, and His-988) in pCAMBIA3300U-AGO1P:FLAG-AGO1-AGO1T was performed by QuickChange site-directed mutagenesis (Agilent Technologies) following the manufacturer’s instructions. Phusion High-Fidelity DNA Polymerase (NEB) was used for PCR, and NEB DH 5-alpha Competent E. coli (High Efficiency) was used for chemical transformation. Primer sequences (LA100-105 and LA185-186) are listed in Supplemental Data Set 3. Markers designed to detect the mutations in bacterial colonies, as well as in plants, are described in the genotyping section below.
Genotyping
Genomic DNA from young leaves or inflorescences was extracted by homogenization of tissue in urea buffer (42% [w/v] urea, 312.5 mM NaCl, 50 mM Tris-HCl, pH8, 20 mM EDTA, and 1% N-lauroylsarcosine) followed by phenol:chloroform extraction and nucleic acid precipitation from the aqueous phase.
The ago1-3 allele was detected with a primer set in which the forward primer (LA298) is specific for the chromosomal AGO1 locus because it spans the junction between the promoter and the coding sequence of AGO1. Thus, primers LA298-299 were used to amplify a 364-bp fragment from the endogenous AGO1 locus. The PCR product was digested with BseGI, which yields two bands of 324 and 40 bp for the wild-type AGO1 allele and three bands of 224, 100, and 40 bp for the ago1-3 allele.
Point mutations in the four catalytic residues were detected in plants as well as in bacteria by PCR with primers LA106-111 and LA189-190, followed by digestion with the restriction enzyme indicated in the primer name and electrophoretic analysis in 3 to 4% agarose gels. Similarly, genotyping of the point mutants rdr6-12, sgs2-18, and sgs3-1 was performed using primers LA281-LA282, LA475-476, and LA477-478, respectively. Genotyping of the T-DNA lines sgs3-14, rdr1-1, and rdr2-1 was done using LP/RP primers LA481-482, LA545-546, and LA551-552, respectively, combined with left border primers SALK_LBb1 (sgs3-14) or SAIL_LB2 (rdr1-1 and rdr2-1).
The sequences of all primers are listed in Supplemental Data Set 3.
FLAG-AGO1 Affinity Purification
The tissue was flash-frozen in liquid nitrogen and manually ground to a fine powder using a mortar and pestle. The powder was vigorously mixed to homogeneity with four volumes (in relation to fresh weight) of ice-cold IP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, and 0.1% Nonidet P-40) freshly supplemented with EDTA-free protease inhibitor (Roche Complete tablets) and 5 mM DTT. The plant extracts were kept on ice or at 4°C for all subsequent steps. The lysates were incubated for 1 h under slow rotation and centrifuged to pellet plant debris until a totally transparent supernatant was obtained. The clear lysate was then incubated in batches with anti-FLAG M2 affinity resin (Sigma-Aldrich) at a ratio of 0.1 mL of beads per gram of tissue, for 2 h under slow rotation. The beads were stringently washed three times with IP buffer supplemented with 0.5 M NaCl, followed by one wash with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) prior to incubation with radiolabeled substrate or addition of TRI reagent for RNA extraction.
Slicer Assay
To assay the enzymatic activity of different AGO1 mutants purified from plants, we incubated affinity purified FLAG-AGO1 with radioactively cap-labeled RNA substrates consisting of 200 to 300 nucleotides in vitro-transcribed MYB65 and PHB RNA fragments containing the endogenous miR159 and miR165 binding sites. The 5′ cleavage products were quantified by autoradiography. Details of RNA template preparation, cap labeling, and incubation with FLAG-AGO1 are given in Supplemental Methods.
RNA Extraction
Total RNA was extracted using 1 mL of TRI reagent (Sigma-Aldrich) per 100 mg of finely ground tissue powder, as detailed in Supplemental Methods. Polysaccharides were removed from pellets redissolved in water by precipitation following the addition of 1/10 volume ethanol and 1/30 volume 3 M NaOAc (pH 5.2) and incubation on ice for 30 min. For extraction of AGO1-bound RNA, 1 mL of TRI reagent was added directly to the FLAG beads with immunoprecipitated FLAG-AGO1. We then proceeded in the same way as for total RNA extraction, except that RNA precipitation was performed in the presence of 20 μg of glycogen.
RNA Gel Blot Analysis
RNA gel blot analysis of small RNAs and of high molecular weight RNA was performed by standard methodology as detailed in Supplemental Methods.
Oligonucleotide DNA probes for detection of single miRNA and siRNA species (Supplemental Data Set 3) were 32P-end-labeled by T4-polynucleotide kinase (Fermentas). Unbound [γ-32P]ATP was removed by gel filtration in MicroSpin G-25 columns (GE Healthcare) prior to hybridization.
Long probes for detection of IR-71 and TAS1c siRNAs or cleavage fragments were generated using the Prime-a-Gene (Promega) kit. Fifty nanograms of DNA obtained by PCR from oligo(dT)-primed cDNA from a dcl1-11 dcl4-2 double mutant was used as template. Unincorporated [α-32P]dCTP was removed by gel filtration in Sephadex G-50 columns. The sequences of primers used for amplification of long probes are listed in Supplemental Data Set 3.
Isolation of Cleavage Fragments
A modified 5′-RACE protocol was used to isolate TAS1c cleavage fragments (Kasschau et al., 2003). Briefly, 3.5 µg of total RNA was used as input to ligate an RNA adaptor oligonucleotide onto RNA molecules with a free 5′-phosphate. Ligated RNAs were reverse transcribed with an oligo(dT) anchor primer, and 25 cycles of PCR were run with primers matching the RNA adaptor and the oligo(dT) anchor sequence to generate a pool of DNAs representing polyadenylated, 5′-phosphorylated RNAs. One microliter of these reactions was used as template for 29 cycles of PCR with a nested adaptor primer and a gene-specific primer. The GeneRacer kit (Invitrogen) was used for RNA manipulations except for gene-specific amplifications. All oligonucleotide sequences used are listed in Supplemental Data Set 3.
Library Construction for RNA-Seq
Libraries were constructed using 1 μg of TRI reagent-extracted total RNA from 100 mg of seedlings or TRI reagent-extracted AGO1-bound small RNAs (sRNAs) from the affinity purified FLAG-AGO1 contained in 1 g of inflorescences. The quality of the RNA was monitored using an Agilent 2100 bioanalyzer.
The sRNA libraries were prepared using the NEBNext Multiplex small RNA Library Prep Set for Illumina. Bioanalyzer traces of cDNA after 14 cycles of PCR amplification showed distinct peaks of ∼140 bp in size for total RNA libraries, corresponding to 21-nucleotide sRNAs plus adaptors. AGO1-IP RNA libraries had even sharper peaks centered around 147 nucleotides, confirming the presence of 21- to 24-nucleotide sRNAs in AGO1 RNPs. The sRNA libraries were pooled according to their relative cDNA concentration and loaded in a 4% MetaPhor Agarose gel (Lonza) for isolation and purification of the 140-bp band of interest. The correct size and quality of the purified cDNA was confirmed before sequencing using a bioanalyzer. The 50-bp single-end reads were obtained on an Illumina HiSeq2000 platform.
Bioinformatic Analysis of Small RNAs
The ncPRO pipeline (Chen et al., 2012) was used to process the sRNA-sequencing raw data, map the reads against the Arabidopsis genome (version TAIR9 from www.arabidopsis.org), and analyze the quality of deep sequencing globally.
The radar plots represent the phasing by displaying the abundance of reads falling into each of 21 possible registers. The register abundance calculations were computed as the frequency of the modulo-21 of the coordinate of each read mapping to the Arabidopsis genome. The registers and the histograms displaying the reads were generated using in-house R-cran scripts.
Accession Numbers
High-throughput sequencing data have been submitted to the European Nucleotide Archive under accession number E-MTAB-4529. Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers AT1G48410 (AGO1), AT5G23570 (SGS3), AT1G14790 (RDR1), AT4G11130 (RDR2), AT3G23125 (MIR173), AT2G27400 (TAS1a), AT1G50055 (TAS1b), AT2G39675 (TAS1c), AT2G39681 (TAS2), and AT3G17185 (TAS3).
Supplemental Data
Supplemental Figure 1. Different catalytic roles of the four Mg2+-coordinating active site residues (extended data supporting Figure 1).
Supplemental Figure 2. Trimming of siRNAs bound to slicer-deficient AGO1 does not occur in seedlings.
Supplemental Figures 3 and 4. AGO1 slicer activity is required for phasing but not for production of tasiRNAs (extended data supporting Figure 3).
Supplemental Figure 5. miR173 does not guide TAS1c precursor cleavage in the absence of AGO1 slicer activity (extended data supporting Figure 4).
Supplemental Figure 6. RDR6 and SGS3 dependence of tasiRNAs produced in the absence of slicer activity (extended data supporting Figure 5).
Supplemental Figure 7. AGO1 slicer activity is required for siRNAs spawned by TAS2/miR161/miR400 (extended data supporting Figure 6).
Supplemental Data Set 1. Read counts of tasiRNAs detected in slicer-deficient AGO1 mutants, ago1-3 null, and wild-type seedlings.
Supplemental Data Set 2. Read counts of tasiRNAs and miRNAs detected in slicer-deficient AGO1 seedlings on otherwise wild-type, sgs3, or rdr6 mutant backgrounds.
Supplemental Data Set 3. Sequences of primers and probes.
Supplemental Methods. Description of the AGO1 slicer assay, GuTR expression and purification, RNA extraction, RNA gel blotting, construction of AGO1P:FLAG-AGO1-AGO1T, and protein gel blotting.
Supplementary Material
Acknowledgments
We thank Theo Bølsterli and his team for plant care, Mathias Henning Hansen for invaluable help with plant and seed handling, Lena Bjørn Johansen for help with RNA gel blot hybridizations, and Maria L. Vigh, Nynne Nielsen, Barbara Tavares-Brandao, Andrea Barghetti, Maïna Floris, and Ida Andersson for help with manual selection of ago1 mutants. We thank Morten Nørholm for pCAMBIA3300U and Hervé Vaucheret for sgs3-1 and sgs2-18 seeds. Seed of rdr1-1/rdr6-12 was a kind gift from Christophe Himber. We thank Phillip Zamore and Stewart Shuman for providing the expression clone of the vaccinia virus capping enzyme and advice on how to purify it. We acknowledge the Nottingham Arabidopsis Stock Centre for providing seeds of T-DNA insertion lines and the ABRC for BAC clones. We thank Augustinus Fonden for supporting the purchase of a Typhoon FLA7000 phosphor imager. This work was supported by a Hallas Møller stipend from the Novo Nordisk Foundation, a starting grant from the European Research Council (Micromecca 282460), and a project grant (2011-7859) from the Lundbeck Foundation to P.B.
AUTHOR CONTRIBUTIONS
L.A.-H. performed most of the experiments in the article, including cloning and construction of transgenic lines, double mutant construction, affinity purifications, slicer assays, protein gel blot analyses, and library preparations. A.M. performed analysis of RNA sequencing data. C.P. purified GuTR for slicer assays and provided assistance during the study. V.B. and B.H. performed RNA sequencing. G.M. and J.H. provided support for setting up and analyzing the slicer assays. P.B. and L.A.-H. performed RNA gel blot and 5′-RACE analyses, wrote the manuscript, and designed the study.
Glossary
- miRNA
microRNAs
- siRNA
short interfering RNAs
- dsRNA
double-stranded RNA
- RISC
RNA-induced silencing complex
- tasiRNA
trans-acting siRNA
Footnotes
Articles can be viewed without a subscription.
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365. [DOI] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Arribas-Hernández L., Kielpinski L.J., Brodersen P. (2016). mRNA decay of most Arabidopsis miRNA targets requires slicer activity of AGO1. Plant Physiol. 171: 2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara M., Sarazin A., Thiébeauld O., Jay F., Voinnet O., Navarro L., Colot V. (2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10: e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N.G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Branscheid A., Marchais A., Schott G., Lange H., Gagliardi D., Andersen S.U., Voinnet O., Brodersen P. (2015). SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43: 10975–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24: 3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8: 884–896. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Servant N., Toedling J., Sarazin A., Marchais A., Duvernois-Berthet E., Cognat V., Colot V., Voinnet O., Heard E., Ciaudo C., Barillot E. (2012). ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics 28: 3147–3149. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Endo Y., Iwakawa H.O., Tomari Y. (2013). Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 14: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944. [DOI] [PubMed] [Google Scholar]
- Fukunaga R., Doudna J.A. (2009). dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 28: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 22: 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Hauptmann J., Dueck A., Harlander S., Pfaff J., Merkl R., Meister G. (2013). Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 20: 814–817. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39: 282–291. [DOI] [PubMed] [Google Scholar]
- Jauvion V., Rivard M., Bouteiller N., Elmayan T., Vaucheret H. (2012). RDR2 partially antagonizes the production of RDR6-dependent siRNA in sense transgene-mediated PTGS. PLoS One 7: e29785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V., Moreno A.B., Elmayan T., Vaucheret H., Crespi M.D., Maizel A. (2012). Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 31: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kawamata T., Tomari Y. (2010). Making RISC. Trends Biochem. Sci. 35: 368–376. [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84. [DOI] [PubMed] [Google Scholar]
- Klein-Cosson C., Chambrier P., Rogowsky P.M., Vernoud V. (2015). Regulation of a maize HD-ZIP IV transcription factor by a non-conventional RDR2-dependent small RNA. Plant J. 81: 747–758. [DOI] [PubMed] [Google Scholar]
- Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R., Watanabe Y. (2009). SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Lam P., Zhao L., Eveleigh N., Yu Y., Chen X., Kunst L. (2015). The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 167: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner P.J., Ameres S.L., Kueng S., Martinez J. (2006). Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Luo Q.J., Mittal A., Jia F., Rock C.D. (2012). An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620. [DOI] [PubMed] [Google Scholar]
- Maunoury N., Vaucheret H. (2011). AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation. PLoS One 6: e28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C. (2005). Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19: 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. (2004). Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Mourrain P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Ascano M., Gogakos T., Ishibe-Murakami S., Serganov A.A., Briskin D., Morozov P., Tuschl T., Patel D.J. (2013). Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Reports 3: 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. (2012). Structure of yeast Argonaute with guide RNA. Nature 486: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Shin C. (2015). Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res. 43: 9418–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C., Vaucheret H., Brodersen P. (2013). Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell 25: 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19: 421–428. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R., Pooggin M.M. (2012). RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 40: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R., Aregger M., Zvereva A.S., Borah B.K., Gubaeva E.G., Pooggin M.M. (2012). Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 40: 6241–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand T.A., Petersen S., Du F., Wang X. (2005). Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629. [DOI] [PubMed] [Google Scholar]
- Ren G., Xie M., Zhang S., Vinovskis C., Chen X., Yu B. (2014). Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. USA 111: 6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.-J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349. [DOI] [PubMed] [Google Scholar]
- Schirle N.T., Sheu-Gruttadauria J., MacRae I.J. (2014). Structural basis for microRNA targeting. Science 346: 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann N., Trabuco L.G., Bender C., Russell R.B., Grimm D. (2013). Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 20: 818–826. [DOI] [PubMed] [Google Scholar]
- Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y. (2014). Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 111: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-J., Smith S.K., Hannon G.J., Joshua-Tor L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Steiner F.A., Okihara K.L., Hoogstrate S.W., Sijen T., Ketting R.F. (2009). RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16: 207–211. [DOI] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49: 493–500. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.-L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. (2008). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Iki T., Tsutsui Y., Miyashita K., Poethig R.S., Habu Y., Ishikawa M. (2013). 3′ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. USA 110: 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2013). Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25: 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2015). A One Precursor One siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.