The tomato CLAUSA gene encodes a unique MYB transcription factor that promotes leaf differentiation by attenuating cytokinin response.
Abstract
Leaf morphogenesis and differentiation are highly flexible processes, resulting in a large diversity of leaf forms. The development of compound leaves involves an extended morphogenesis stage compared with that of simple leaves, and the tomato (Solanum lycopersicum) mutant clausa (clau) exposes a potential for extended morphogenesis in tomato leaves. Here, we report that the CLAU gene encodes a MYB transcription factor that has evolved a unique role in compound-leaf species to promote an exit from the morphogenetic phase of tomato leaf development. We show that CLAU attenuates cytokinin signaling, and that clau plants have increased cytokinin sensitivity. The results suggest that flexible leaf patterning involves a coordinated interplay between transcription factors and hormones.
INTRODUCTION
Leaves are determinate organs, which differentiate to become major providers for the plant following a limited growth period. However, a transient period of indeterminate growth, termed primary morphogenesis, occurs during early stages of leaf development. During this stage, the leaf attains its basic shape and generates lateral appendages such as leaflets (Dengler and Tsukaya, 2001; Hagemann and Gleissberg, 1996; Kaplan, 2001). The spatial and temporal extent of this morphogenetic phase responds flexibly to the genetic, developmental, and environmental context. This flexibility gives rise to a wide diversity of leaf sizes and shapes that vary within and between species. Compound leaves, such as those of tomato (Solanum lycopersicum), are composed of multiple leaflets and are characterized by a prolonged morphogenetic phase. This extended morphogenesis is enabled by the activity of a meristematic region at the leaf margin, termed marginal blastozone (Hagemann and Gleissberg, 1996). The morphogenetic stage is particularly long in tomato, resulting in a wide diversity of leaf forms and unique developmental flexibility (Bar and Ori, 2014; Bar et al., 2015; Burko and Ori, 2013). Recent studies have uncovered an even higher potential for prolonged morphogenesis in tomato leaf development, which is normally restricted by differentiation-promoting factors (Bar et al., 2015; Shani et al., 2010; Yanai et al., 2011). Characterization of mutants with extended or restricted morphogenesis has pointed to this potential and led to the identification of factors that balance morphogenesis and differentiation during leaf development (Sluis and Hake, 2015; Canales et al., 2010; Efroni et al., 2010; Bar and Ori, 2014; Blein et al., 2010).
Transcription factors from the class I knotted1-like homeobox (KNOXI) family were implicated in promoting the extended morphogenesis in leaves of many plant species, including tomato (Hareven et al., 1996; Janssen et al., 1998; Kimura et al., 2008; Bharathan et al., 2002; Hay and Tsiantis, 2006; Shani et al., 2009; Barth et al., 2009; Hake et al., 2004; Rast-Somssich et al., 2015). The plant hormone cytokinin acts downstream of KNOXI proteins to promote morphogenesis, mainly during relatively late stages of leaf development (Shani et al., 2010). By contrast, transcription factors from the CIN-TCP (CINNCINATA- TEOSINTE BRANCHED1-CYCLOIDEA-PCF) family, belonging to class II TCPs, restrict the morphogenetic window and promote differentiation (Burko et al., 2013; Efroni et al., 2008; Shleizer-Burko et al., 2011; Ori et al., 2007; Palatnik et al., 2003). The hormone gibberellin also promotes differentiation and partially mediates CIN-TCP function in this process (Yanai et al., 2011; Fleishon et al., 2011; Van Tuinen et al., 1999; Jasinski et al., 2008). The ratio of SINGLE FLOWER TRUSS and SELF PRUNING has also been shown to affect the balance between growth and differentiation in many developmental processes including leaf development, in addition to their role in regulating flowering time (Lifschitz et al., 2014; Shalit et al., 2009).
clausa (clau), first described in the 1960s (Khush and Rick, 1967), is a classical tomato mutant characterized by increased morphogenetic potential that leads to abnormal, excessively divided leaves, often carrying shoot-like structures on the rachis. CLAU is a potential negative regulator of KNOXI genes and of the boundary gene GOBLET (GOB) (Bar et al., 2015; Avivi et al., 2000; Jasinski et al., 2007, 2008; Naz et al., 2013).
CLAU therefore emerges as a factor that puts a hold on the morphogenetic potential of tomato leaves, enabling controlled maturation and morphogenesis. Here, we identify CLAU as a unique MYB transcription factor that promotes differentiation and attenuates CK responses.
RESULTS
CLAU Is a Unique MYB Transcription Factor
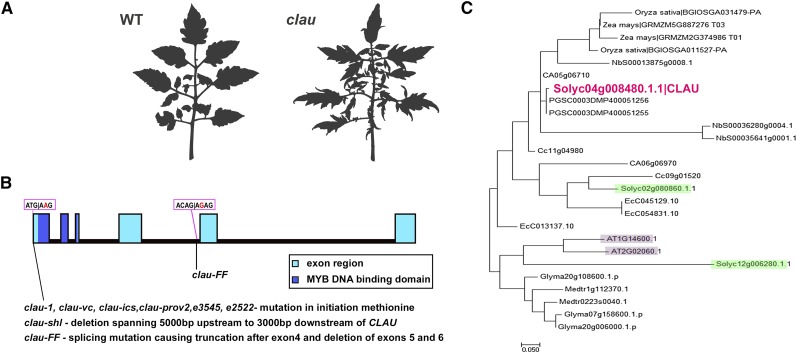
The classical tomato mutant clau possesses highly compound leaves (Figure 1A). We previously quantified the clau phenotype, demonstrating that clau leaves are on average 11 times more compound than wild-type leaves (Bar et al., 2015). The clau mutant phenotypes suggest that CLAU plays a central role in modulating the flexibility of leaf morphogenesis. We therefore wished to identify the CLAU gene to further understand this flexibility. We obtained eight clau alleles (Figure 1B) and, for the new alleles, confirmed allelism by complementation tests. Of these, clau:e3545 and clau:e2522 were identified in an EMS population (Menda et al., 2004), generated in the S. lycopersicum M82 background. Leaves of all eight alleles were similar, allowing for slight differences resulting from the different background cultivars (Supplemental Figure 1). CLAU was previously mapped to the short arm of chromosome 4 using radiation ablation mutagenesis (Khush and Rick, 1967). We created an F2 mapping population from a cross between clau:e3545 in the S. lycopersicum M82 background and Solanum pimpinellifolium and used this mapping population to pinpoint the physical location of CLAU between bases 2094866 and 2122247 on the short arm of chromosome 4, an area which contains two genes. RNA sequencing of shoot apices of the EMS allele clau-e3545, in comparison to wild-type M82 apices, revealed a mutation in the first methionine codon of a gene encoding a MYB transcription factor (Solyc04g008480, named CLAU hereafter), which is physically located within this interval. The mutation is predicted to prevent the formation of any protein from the CLAU open reading frame, which encodes a 297-amino acid SHAQKYF-class MYB transcription factor, possessing six exons (Figure 1B). clau and five additional alleles contain the exact same mutation, an ATG-to-AAG substitution in the codon for the initiation methionine. clau-shl is a deletion allele, where no other annotated genes are deleted, and clau-ff results from a splicing mutation leading to a truncated protein of 150 amino acids, lacking exons 5 and 6 (Figure 1B). We found CLAU homologs in several plant species (Figure 1C; Supplemental Data Set 1), including Arabidopsis thaliana (AT1G14600 and AT2G02060). Figure 1C depicts a phylogenetic analysis of CLAU homologs in a variety of different plant species that have published annotated genomes. CLAU is not highly conserved in Arabidopsis, as the Arabidopsis homologs, AT1G14600 and AT2G02060, possess only 35 and 32% global identity, respectively, to CLAU, with over 30% gaps in the alignment, and cluster in a different clade than CLAU, closer to its distant tomato homolog Solyc12g006280 (Figure 1C). To further examine possible CLAU homologs between different plant species, we examined the synteny of the genes within the tomato chromosomal region containing CLAU to Arabidopsis and several Solanaceae species. Although AT1G14600 is slightly more homologous to CLAU than AT2G02060, the genes within the respective region in Arabidopsis chromosome 2 that contains AT2G02060 show greater synteny with the CLAU region in tomato chromosome 4 (Supplemental Figure 2). Comparison of these syntenic regions suggests several rearrangements. The additional Solanaceae species presented have significant gene synteny in this region to tomato, in particular potato (Solanum tuberosum; chromosome 4) and pepper (Capsicum annuum; chromosome 5). The other species have only partially annotated published genomes, so the analysis may not be complete. Comparison between CLAU and other tomato MYB transcription factors shows that CLAU is relatively unique in tomato (Supplemental Figure 3 and Supplemental Data Set 2; see below), as the two closest tomato homologs, SolyC02g080860 and SolyC12g006280 (Supplemental Figures 3A and 3C; Figure 1C), share only 28 and 22% global identity, respectively, with CLAU. Local identities of 78 and 49%, respectively, are limited to the DBD region, where the SHAQKYF motif is highly conserved (amino acids 10 to 100; Supplemental Figures 3A and 3B). Overall, the 20 tomato proteins closest to CLAU all contain SHAQKYF domains (interpro IPR006447) and cluster on separate but close branches to CLAU, when compared with several tomato MYBs that are involved in developmental processes and contain R2R3 MYB domains (interpro IPR17930) (Supplemental Figure 3A).
Figure 1.
CLAU Is a MYB Transcription Factor.
(A) Leaf 5 of wild-type and clau:e3545 M82 tomato plants.
(B) Graphic representation of the CLAU ORF and the mutations found in the different clau alleles.
(C) Phylogenetic tree of CLAU homologs in different species. CLAU is in red. The two closest tomato homologs are marked in green, and the two closest Arabidopsis homologs are marked in purple. Rice, Oryza sativa; maize, Zea mays; Nicotiana benthamiana, Niben (Nb); pepper, Capsicum annum (CA); potato, Solanum tuberosum (Stub, PGSC); coffee, Coffea canefora (Cc); eucalyptus, Eucalyptus camaldulensis (Ec); soybean, Glycine max (Glyma); Medicago, Medicago truncatula (Medtr).
CLAU Is Expressed at the Leaf Margin to Restrict the Morphogenetic Window
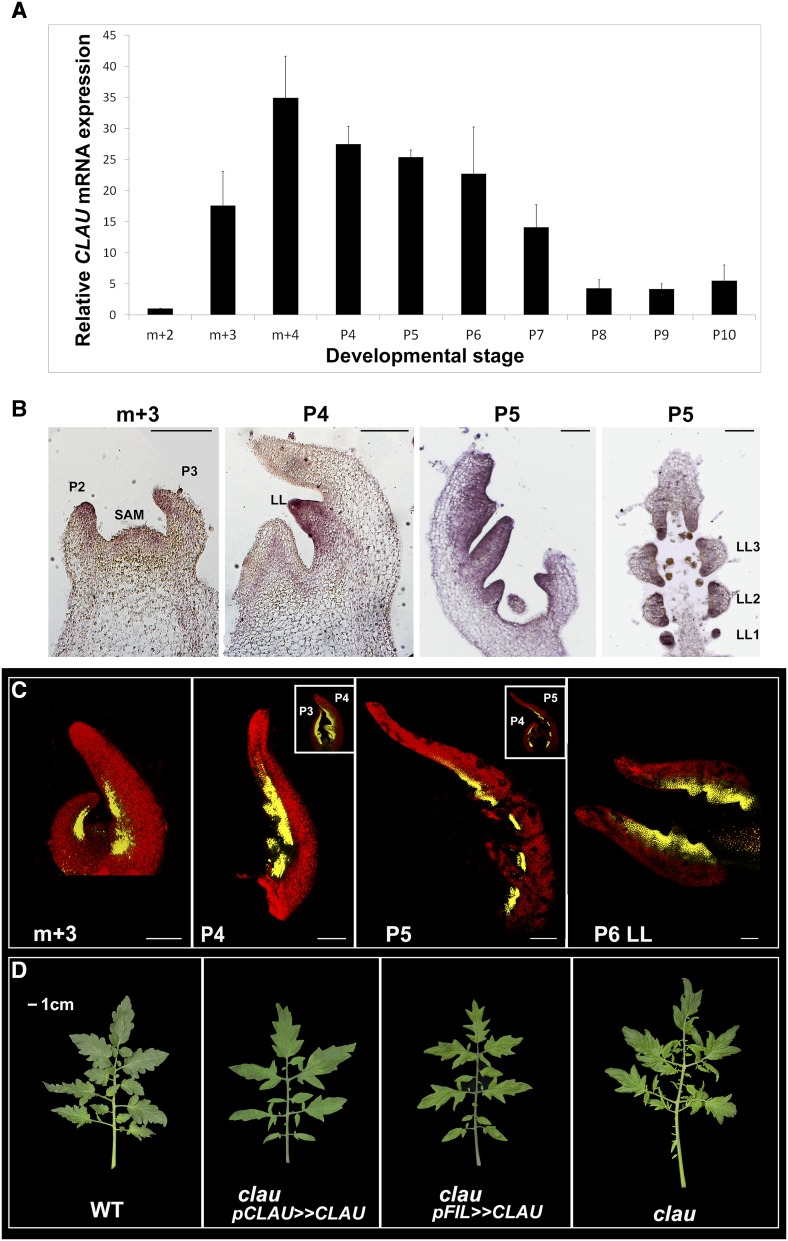
Pursuant to the identification of the CLAU gene, we proceeded to examine the expression patterns of the CLAU mRNA during leaf development using qRT-PCR (Figure 2). CLAU is expressed primarily during the P3-P7 stages of leaf development (Figure 2A), with expression peaking between P4 and P6. In situ mRNA hybridization localized CLAU mRNA expression to developing leaflets at the leaf margin (Figure 2B), correlating with the developmental stages during which the CLAU mRNA expression peaks. Expression of YFP under the control of the CLAU promoter, consisting of 2700 bp upstream of the CLAU translational start site, showed promoter activity at the leaf margin and in developing leaflets (Figure 2C), in a pattern similar to that observed for the CLAU mRNA in the in situ mRNA hybridization (compare Figures 2B and 2C). The 2700-bp CLAU promoter was able to rescue the clau mutant when driving CLAU cDNA expression (Figure 2D).
Figure 2.
CLAU Is Expressed during Leaflet Morphogenesis at the Leaf Margin.
Analysis of CLAU expression during development. Leaf development follows plastochrons, such that P1 is the most recently initiated leaf primordium, and it becomes P2, P3, etc., as new primordia emerge from the SAM, with leaflets initiating basipetally from the P3 stage on.
(A) qRT-PCR analyzing CLAU mRNA expression levels at successive stages of leaf development. Bars are the se of at least four biological replicates.
(B) In situ hybridization of CLAU mRNA in tissue sections of wild-type apices and leaf primordia, as indicated.
(C) Confocal micrographs of wild-type apices and leaf primordia, expressing YFP driven by the CLAU promoter. Insets show the plant shoot containing the depicted stage. Bars = 200 µm.
(D) Fifth leaf of the indicated genotypes. Expression of CLAU from the CLAU promoter rescues the clau mutant phenotype. m+2, m+3, m+4, SAM and 2, 3, or 4 youngest leaf primordia; LL, lateral leaflet.
Loss of CLAU function results in extended morphogenesis and extremely elaborate leaves. Identification of the CLAU gene allowed us to examine the effect of CLAU gain of function. Overexpression of CLAU under the control of its own promoter or the FIL promoter (Lifschitz et al., 2006; Shani et al., 2009), using a trans-activation system (Moore et al., 1998), led to a simplified leaf possessing primary leaflets and very few secondary leaflets (Figure 3). This confirms that CLAU is involved in curbing leaf morphogenesis, in line with the more elaborate leaf observed in the clau mutant. The simplified-leaf phenotype caused by expressing CLAU under the control of its own promoter suggests that CLAU affects morphogenesis in a dose-dependent manner. This is also supported by the observation that clau/+ heterozygotes occasionally showed a very mild clau phenotype. To further investigate the role of CLAU in leaf development, we overexpressed a fusion of CLAU with the SRDX repression motif (Hiratsu et al., 2003) in wild-type leaves, under the control of the FIL promoter, using the trans-activation system. The CLAU-SRDX fusion was expected to downregulate CLAU targets, affecting the downstream developmental pathways and providing insight into the usual mode of action of CLAU. Overexpression of CLAU-SRDX resulted in leaves very similar to those resulting from overexpression of CLAU alone (Figures 3E and 3D), suggesting that CLAU may act primarily as a repressor of its downstream targets.
Figure 3.
Overexpression of CLAU Promotes Differentiation.
Leaves of the wild type (A), clau (B), plants overexpressing CLAU under the control of the indicated promoters ([C] and [D]), and plants overexpressing CLAU fused to the SRDX repressor domain under the control of the FIL promoter (E). All leaves depicted are leaf 5. Bar = 1 cm.
CLAU Has a Unique Role in Compound-Leaf Development
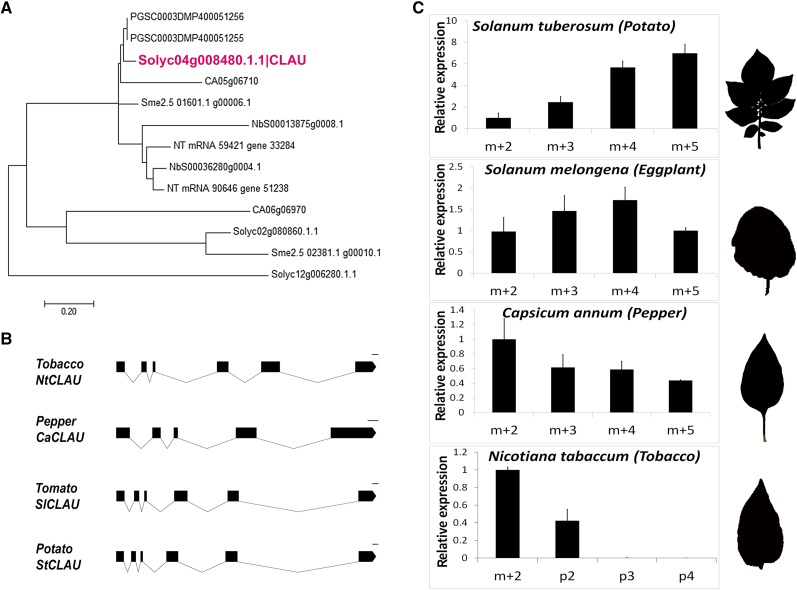
As detailed above, CLAU is relatively unique in tomato and its closest Arabidopsis homolog has low identity (32%) to CLAU. CLAU has close orthologs in additional Solanaceae species. To compare the nature of the CLAU genes in simple- and compound-leaf species, we analyzed the closest CLAU homologs in several additional Solanaceae species (Figure 4): potato, which has compound leaves, as well as eggplant (Solanum melongena), tobacco (Nicotiana tabacum), and pepper, with simple leaves. A phylogenetic tree of the two closest CLAU homologs in several Solanaceae species is presented in Figure 4A (sequence alignment in Supplemental Data Set 3). Potato CLAU (St-CLAU, PGSCDMP400051255) is the closest ortholog to CLAU; pepper (CA0506710) and eggplant (Sme2.501601) sequences are slightly more removed, and those of Nicotiana benthamiana (Nbs00013875) and tobacco (NT 33,284) are less similar (Figure 4A). St-CLAU bears 87% global identity to Sl-CLAU, spanning the entire protein. The St-CLAU mRNA had similar expression dynamics to Sl-CLAU during early leaf development (Figure 4C; compare with Figure 2A). The eggplant ortholog, Sm-CLAU, is 72% identical globally to CLAU. Its expression decreased at the P5 stage, when Sl-CLAU and St-CLAU were still highly expressed (Figure 4C). The pepper ortholog Ca-CLAU contains only five exons (Figure 4B) and encodes a shorter protein than Sl-CLAU. Ca-CLAU shares 54% global identity and 71% local identity with the N-terminal two-thirds of the Sl-CLAU protein. Ca-CLAU mRNA was expressed at earlier developmental stages and its expression decreased at the P3 stage, when Sl-CLAU and St-CLAU started to increase in expression, toward the formation of leaflets at the leaf margin (Figure 4C). The N. benthamiana Nb-CLAU and N. tabacum Nt-CLAU proteins share ∼55% global identity with Sl-CLAU. Similar to the observed expression pattern in pepper, the expression of Nt-CLAU decreased at the P3 stage (Figure 4C; developmental stages collected in tobacco were slightly different than in the other species due to technical constraints). Thus, the degree of similarity to Sl-CLAU and the expression dynamics and leaf shape were correlated in the tested species. This suggests that CLAU exerts part of its effect on leaf form during the formation of leaflets, in a developmental window that exists only in compound leaves. Simple leaves have a shorter morphogenetic window and thus lack the developmental window in which CLAU is active in compound leaves. In both leaf types, CLAU likely functions during early leaf development to attenuate meristematic activity and promote leaf fate.
Figure 4.
CLAU Has Unique Expression Dynamics during Compound Leaf Development.
(A) Phylogenetic analysis of CLAU in the Solanaceae. The Sl-CLAU protein is closest to the potato proteins (PGSC), followed by pepper and eggplant. The two closest tomato homologs (Solyc02g080860 and Solyc12g006280) cluster further away than the other Solanaceae homologs and have homologs of their own in the other Solanaceae species.
(B) Gene structure (bar = 100 bp) of CLAU in several Solanaceae species. The gene is composed of six exons and five introns, except in pepper, which possesses five exons and four introns. The genomic sequence of the eggplant CLAU is not available.
(C) qRT-PCR analyzing CLAU mRNA expression levels at early stages of leaf development in the indicated plastochrons in potato, eggplant, pepper, and tobacco. Final leaf shape is depicted to the right. Bars are the se of at least four biological replicates. m+2, m+3, m+4, SAM and 2, 3, or 4 youngest leaf primordia, respectively.
CLAU Attenuates Cytokinin Signaling
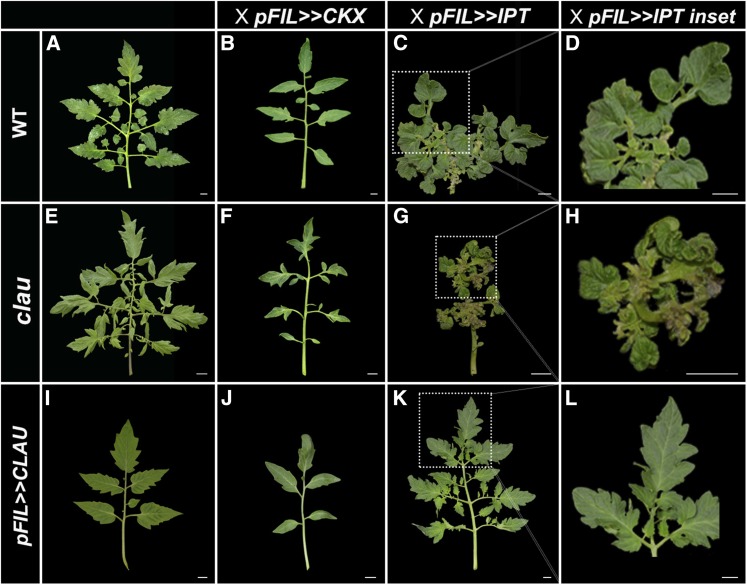
Overexpression of the Arabidopsis cytokinin (CK) biosynthesis gene ISOPENTENYLTRANSFERASE7 (IPT7) in tomato leaves led to the formation of highly compound leaves, whereas decreasing the CK levels via the expression of the Arabidopsis CK degradation gene CYTOKININ OXIDASE (CKX3) resulted in reduced leaf complexity (Shani et al., 2010) (Figure 5). Some aspects of the leaf phenotype caused by IPT7 overexpression are reminiscent of the clau phenotype, including a highly compound leaf, many ectopic meristems on the leaf, and an extension of the developmental window of leaflet morphogenesis (Bar et al., 2015). The simplified leaf phenotypes resulting from overexpression of CLAU and CKX were also similar. This prompted us to examine whether an altered CK response could be involved in the clau phenotype. Overexpressing pFIL>>IPT7 in a clau background (Figures 5G and 5H) enhanced the phenotype caused by elevated CK levels alone (Figures 5C and 5D). Conversely, reducing CK biosynthesis in the clau background was substantially epistatic to CKX overexpression (Figure 5F), as the leaf was only slightly more complex than the leaf overexpressing CKX alone (Figure 5B), suggesting that the clau phenotype depends at least in part on CK. Interestingly, overexpressing IPT7 and CLAU simultaneously under the control of the FIL promoter resulted in a significant rescue of the FIL>>IPT7 phenotype (Figures 5K and 5L), indicating that CLAU can affect the CK response. Taken together, these results suggest that CK dynamics may mediate CLAU function.
Figure 5.
CLAU Attenuates CK Response.
Genetic interactions between clau, pFIL>>CLAU, and pFIl>>CKX or pFIL>>IPT. Genotypes are indicated above each panel. All leaves depicted are fully expanded leaf 5. Bars = 1 cm.
Expression of the CK-Response Reporter TCS Is Upregulated in clau
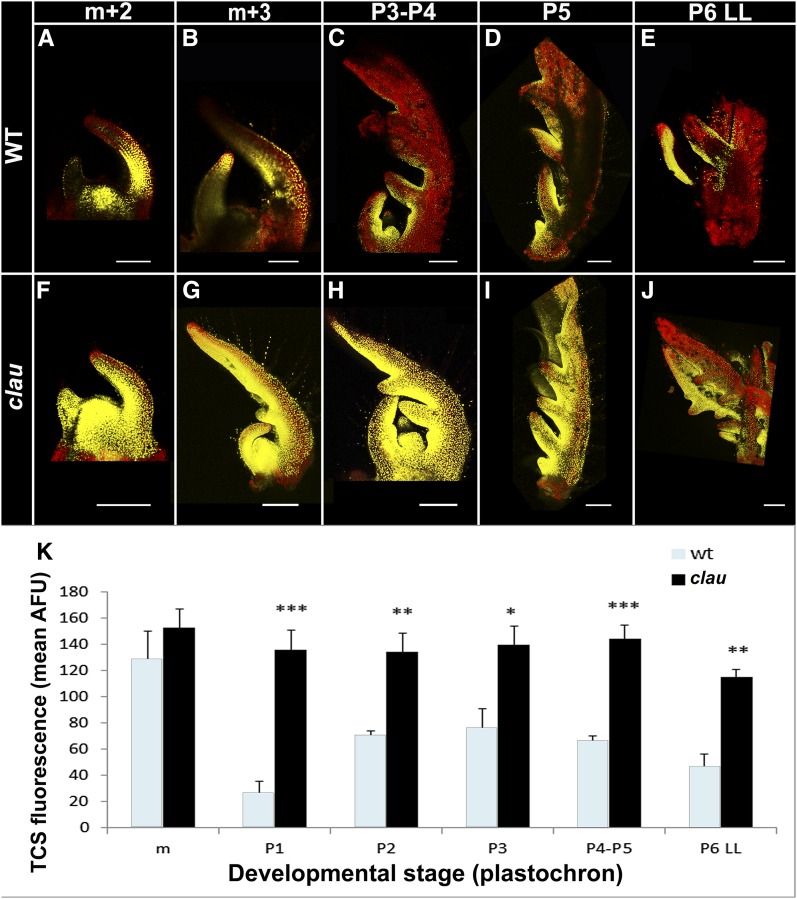
To further examine whether CK dynamics could be altered by CLAU, we compared the expression of a CK activity reporter TCS fused to VENUS (Zürcher et al., 2013; Steiner et al., 2016) between wild-type tomato plants and the clau mutant (Figure 6). We first confirmed that TCS is CK responsive in tomato as was reported in Arabidopsis (Supplemental Figure 4). To avoid positional effects, we compared a wild-type TCS-expressing line with the same line that was introgressed into clau by a cross. In wild-type shoot apices, TCS drove expression in the shoot apical meristem (SAM) and at the leaf margin of developing leaves (Figures 6A to 6E). TCS-driven expression was strongly upregulated in the clau background (Figures 6F to 6J). Quantification of TCS-driven fluorescence showed that TCS was significantly upregulated in clau at all developmental stages (Figure 6K), except in the SAM where its expression was already high in the wild-type background, and where CLAU expression was low (Figures 2A and 2B). In the wild type, TCS-driven expression was high in the SAM, decreased in P1, peaked again between P2-P5, and subsequently decreased (Figure 6K). In clau, however, all developmental stages showed similar, relatively high TCS-driven expression.
Figure 6.
clau Has an Upregulated CK Response.
The CK response reporter TCS is upregulated in clau. Confocal micrographs of wild-type ([A] to [E]) and clau ([F] to [J]) apices and/or leaves at the indicated developmental stages, expressing VENUS driven by the CK reporter TCS. VENUS quantification is presented in (K); three to seven meristems or primordia were analyzed for each developmental stage. Error bars represent se, and asterisks indicate statistical significance (two-tailed Student's t test) of upregulation in clau over the wild type. *P < 0.05, **P < 0.01, and ***P < 0.005. LL, lateral leaflet. Bars = 200 µm.
clau Has Increased CK Sensitivity
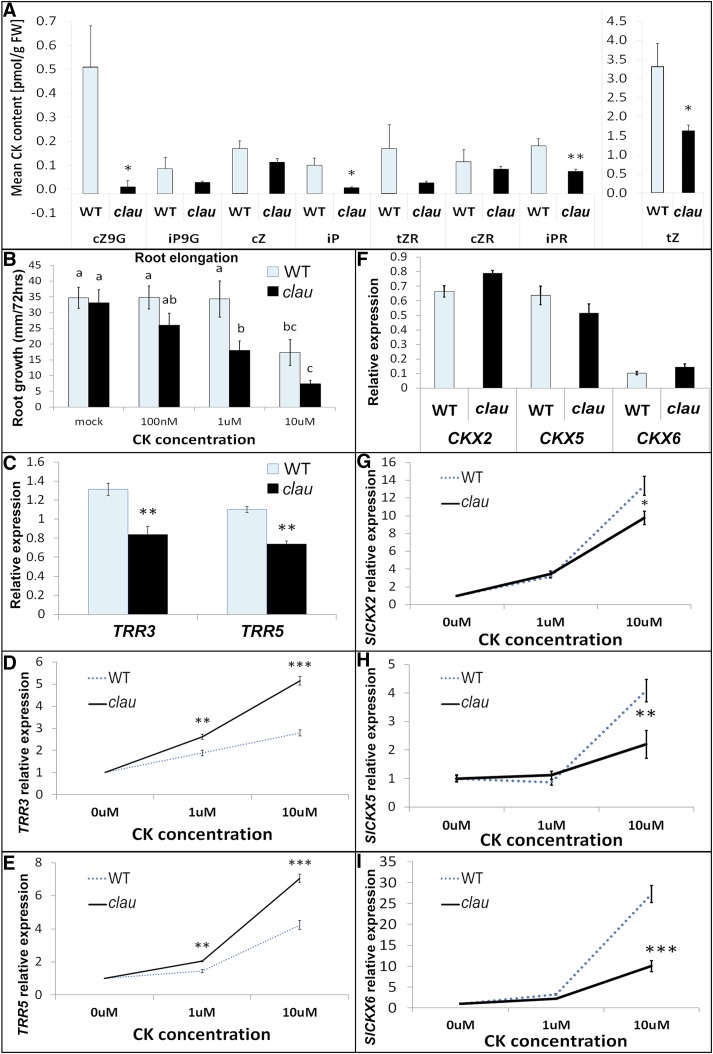
The genetic results suggesting an involvement of CK in the clau phenotype and the increase in TCS reporter-driven expression prompted us to examine whether clau could have increased CK content (Figure 7). Surprisingly, young clau primordia have a lower amount of CKs (Figure 7A), including trans-zeatin, which is the most abundant CK in these tissues. An increase in CK signaling coupled with a decrease in CK content led us to investigate whether clau affects CK sensitivity. In three independent biological assays (Coenen and Lomax, 1998; Fleishon et al., 2011), clau plants responded to CK concentrations a tenth (anthocyanin production; Supplemental Figure 5B) to a hundredth (root growth, Figure 7B; hypocotyl height, Supplemental Figure 5A) lower than the concentrations required to elicit a response in wild-type plants. Another hallmark of CK response is an increase in the transcripts of type A response regulator (RR) genes (Keshishian and Rashotte, 2015; Shi et al., 2013; D’Agostino and Kieber, 1999; Brandstatter and Kieber, 1998). The Arabidopsis type A RRs ARR3 and ARR5 respond strongly to CK and inhibit CK response (Shi et al., 2013; To et al., 2004). Interestingly, the baseline transcript level of the tomato type A RRs TRR3 and TRR5 was significantly lower in clau than in the wild type (Figure 7C), suggesting that TRRs may mediate the effect of CLAU on cytokinin sensitivity. Examining the type A TRR transcript response to CK treatment demonstrated that the response of TRR3 and TRR5 to CK was stronger in clau than in the wild type (Figures 7D and 7E). We also examined the response of TRR 8/9A, 8/9B, 8/9C, 16/17, and 16B to CK, and with the exception of TRR8/9C, all demonstrated a statistically significant higher transcriptional response following CK treatment in clau than in the wild type (Supplemental Figures 5C to 5H). We conclude that an absence of an active CLAU protein leads to increased CK sensitivity, suggesting that CLAU functions in attenuating the response to CK during development, possibly via positive regulation of TRR expression. A possible mechanism to counterbalance the increased CK sensitivity found in clau could be through CKX regulation. CKX2, 5, and 6 were all induced in tomato leaves following CK treatment (Shi et al., 2013). Interestingly, clau had similar levels of CKX2, 5, and 6 prior to CK treatment (Figure 7F). However, while CKX2 (Figure 7G), CKX5 (Figure 7H), and CKX6 (Figure 7I) were induced by 1 μM CK to a similar degree as in the wild type, they were induced to a lower degree in clau than in the wild type by 10 μM CK. This suggests that CKXs may feed back to the already low levels of CKs in clau rather than respond to the increased CK sensitivity.
Figure 7.
clau Has Decreased CK Content and Increased CK Sensitivity.
(A) Quantification various CK derivates in clau and wild-type young leaves (P5-6 developmental stage). cZ9G, cis-zeatin-9-glucoside; iP9G, iso-pentenyl-9-N-glucoside; cZ, cis-zeatin; iP, iso-pentenyl-adenine; tZR, trans-zeatin riboside; cZR, cis-zeatin riboside; iPR, iso-pentenyl riboside; tZ, trans-zeatin.
(B) Root growth response to CK in clau and wild-type seedlings.
(C) qRT-PCR analyzing TRR mRNA expression levels in untreated clau and wild-type shoot apices.
(D) and (E) qRT-PCR analyzing TRR mRNA expression levels in response to CK in clau and wild-type shoot apices.
(F) qRT-PCR analyzing CKX mRNA expression levels in untreated clau and wild-type shoot apices.
(G) to (I) qRT-PCR analyzing CKX mRNA expression levels in response to CK in clau and wild-type shoot apices.
In (D), (E), and (G) to (I), the expression in untreated apices is set to 1. In all cases, the SAM and five youngest leaf primordia were collected, and bars represent the mean ± se of at least five biological repeats. Asterisks indicate statistical significance (two-tailed Student's t test) of upregulation or downregulation in clau.*P < 0.05, **P < 0.01, and ***P < 0.005.
Overexpressing CLAU Decreases CK Sensitivity
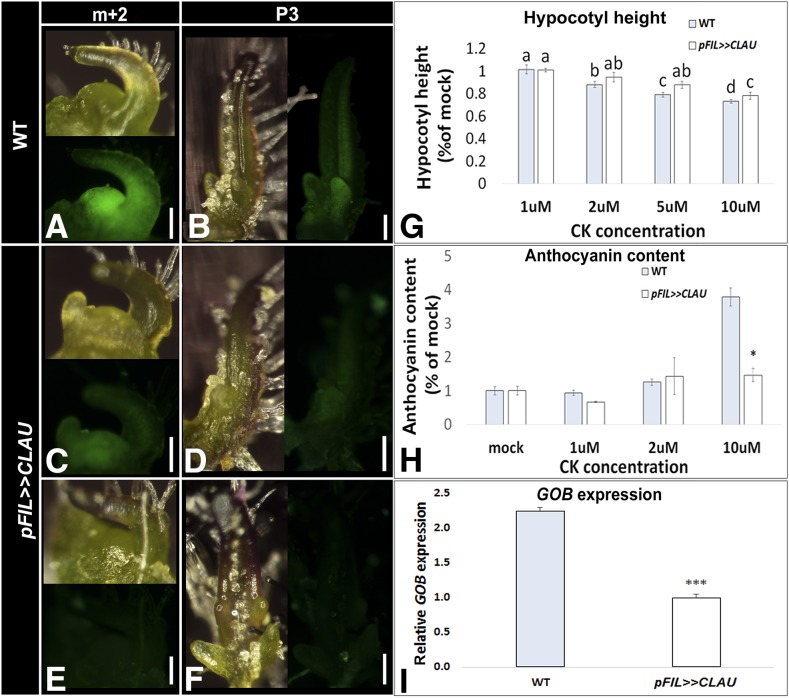
To complement the analysis presented in Figure 7, we examined whether overexpression of CLAU could decrease CK sensitivity. We first compared the expression of the TCSv2:3XVENUS reporter detailed above between wild-type tomato plants and several transgenic lines overexpressing pFIL>>CLAU (Figure 8). Once again, we compared a wild-type TCS-expressing line with the same line that was introgressed into pFIL>>CLAU. TCS-driven expression was downregulated in a pFIL>>CLAU background (Figures 8C to 8F). pFIL>>CLAU plants also showed a decrease in CK sensitivity, requiring higher CK concentrations to elicit a decrease in hypocotyl height (Figure 8G) or an increase in anthocyanin content (Figure 8H).
Figure 8.
pFIL>>CLAU Has Decreased CK Sensitivity and Decreased GOB Expression.
(A) to (F) The CK response reporter TCS is downregulated in pFIL>>CLAU. Stereomicroscope images of wild-type ([A] and [B]) and two lines of pFIL>>CLAU ([C] to [F]) apices and/or leaves at the indicated developmental stages, expressing VENUS driven by the CK reporter TCS. Bars = 100 µm.
(G) Hypocotyl height response to CK in pFIL>>CLAU and wild-type seedlings.
(H) Anthocyanin production response to CK in pFIL>>CLAU and wild-type leaves.
(I) qRT-PCR analyzing GOB mRNA expression levels in pFIL>>CLAU and wild-type shoot apices (m-P5).
Bars represent the mean ± se of 5 to 12 biological repeats, and asterisks indicate statistical significance (two-tailed Student's t test). *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
Overexpressing CLAU Leads to Reduced GOB Expression
In previous work (Bar et al., 2015), we showed that part of the CLAU mechanism of action in tomato involves negative regulation of the NAM/CUC gene GOB. We previously demonstrated that GOB is upregulated in a clau background and that downregulation of GOB via overexpression of its regulatory microRNA, miRNA164, results in a dramatic rescue of the clau phenotype (Bar et al., 2015). Upon identification of the CLAU gene, we wanted to examine whether upregulation of CLAU has an effect on GOB expression. Figure 8I shows that overexpression of CLAU caused downregulation of GOB, strengthening the assertion that CLAU negatively regulates GOB in leaf development.
CK Application Affects CLAU Expression
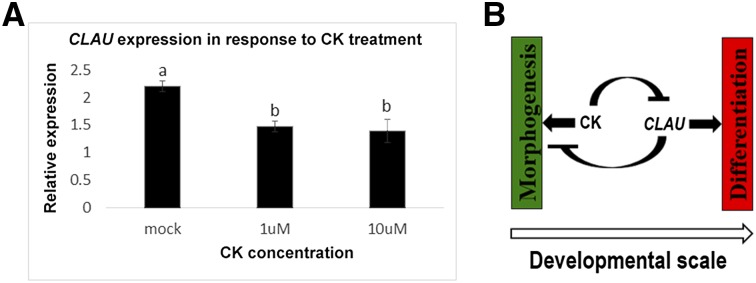
To further understand the interaction between CLAU and cytokinin, we examined whether CLAU expression is affected by CK (Figure 9). CK application led to a decrease in CLAU expression (Figure 9A), suggesting that CLAU promotes differentiation by inhibiting CK responses, and CK promotes morphogenesis by inhibiting CLAU expression (Figure 9B).
Figure 9.
CK Regulates CLAU Expression.
(A) qRT-PCR analyzing CLAU mRNA expression levels in mock and CK-treated wild-type shoot apices. Bars represent the mean ± se of 3 to 5 biological repeats, and letters indicate statistical significance (two-tailed Student's t test).
(B) Diagram indicating relationships between CK and CLAU during development. CK promotes morphogenesis and CLAU promotes differentiation. CLAU inhibits morphogenesis by inhibiting CK response, while CK in turn can inhibit differentiation by inhibiting CLAU expression.
DISCUSSION
Tomato leaves undergo extended morphogenesis, resulting in elaborate and variable leaf forms (Burko and Ori, 2013). Among tomato leaf phenotypes, the clau mutant is extreme in its leaf elaboration, exemplifying the potential of further extending the tomato leaf morphogenetic window and its possible results. Overexpressing CLAU curbs leaf morphogenesis, demonstrating that CLAU regulates the length of the leaf morphogenetic window.
How does CLAU regulate morphogenetic competence in the leaf? We recently reported that CLAU is involved in delimiting the expression of GOB, a central regulator of leaf morphogenesis (Bar et al., 2015; Ben-Gera et al., 2012; Berger et al., 2009; Brand et al., 2007; Busch et al., 2011; Rossmann et al., 2015). Here, GOB is shown to be downregulated by CLAU overexpression. In addition, clau has abnormally high expression of the KNOXI gene LeT6/Tkn2, which plays important role in promoting leaf morphogenesis (Avivi et al., 2000; Jasinski et al., 2007). Here, we show that a central mechanism via which CLAU regulates the leaf morphogenetic window is attenuation of CK response. Interestingly, KNOXI proteins positively regulate CK levels in the contexts of SAM maintenance and compound-leaf morphogenesis (Sakamoto et al., 2006; Scofield et al., 2013; Shani et al., 2010). It therefore appears that to promote differentiation and attenuate morphogenesis, CLAU interferes with the activity of several interconnected pathways that promote morphogenesis. It will be interesting to see which of these pathways are directly affected by CLAU and which are affected secondarily by the molecular and developmental context of differentiation.
How does CLAU affect CK response? clau mutants have decreased CK levels and increased CK sensitivity, and IPT overexpression phenotypes are enhanced by clau. These observations suggest that CLAU may negatively affect CK response, which in turn feeds back to CK levels. Notably, while TCS-driven reporter accumulation is increased and type A RRs respond to CK with higher sensitivity in clau, the steady state levels of these type A RRs are reduced in clau. These results suggest that CLAU is a positive regulator of type A RRs, which negatively regulate CK response (Kiba et al., 2004; To et al., 2004; Lee et al., 2008). In clau, their expression is reduced, leading to increased CK sensitivity. The effect of CLAU on type A RRs may be direct, or possibly, additional regulators are feeding back to downregulate type A RRs. In contrast to the increased CK sensitivity of TRR response in clau, CKX has reduced response to CK treatment in clau when compared with the wild type. These results suggest that the response of TRR and CKX expression to CK may be regulated by distinct pathways and that CK affects CKX expression in a pathway not affected by CLAU. In other words, some downstream components of the CK pathway respond to the elevated CK sensitivity mediated by CLAU, while other downstream components of the CK pathway respond to the decrease in CK content present in clau.
Our results show that CLAU is primarily expressed at the leaf margin and during the developmental stages where leaflets are formed, suggesting that CLAU functions to restrain morphogenesis. This is also evident from the fact that in wild-type leaves, CLAU expression decreases after leaflets have been determined (P8 onwards). Essentially, CLAU can be viewed as the gatekeeper for exiting morphogenesis, which can explain why its loss of function increases leaf complexity so drastically; CLAU functions as a repressor during the morphogenetic stage, and removing CLAU function causes an already elaborate leaf to continue elaboration indefinitely, subsequently being regulated only by the plant senescing (Bar et al., 2015).
TCP transcription factors, which also promote leaf differentiation and attenuate morphogenesis have been shown in Arabidopsis to decrease CK sensitivity by an interaction with the chromatin remodeling ATPase BRHAMA and direct binding to the ARR16 gene, which is rapidly induced by CK and inhibits CK response (Efroni et al., 2013). Therefore, transcription factors that antagonistically affect the balance between morphogenesis and differentiation appear to converge on modulating the CK response. As the tomato TCP factor LA was shown to also affect GA metabolism, the window of morphogenesis may be regulated through the balance between CK and GA (Shani et al., 2010; Bolduc and Hake, 2009; Steiner et al., 2012; Efroni et al., 2013; Yanai et al., 2011; Hay et al., 2002).
CLAU encodes a MYB transcription factor that possesses a DNA binding SHAQYF domain. Several MYB transcription factors have been previously reported to be involved in leaf development (Barkoulas et al., 2007; Luo et al., 2012; Busch et al., 2011; Naz et al., 2013), but most of them encode proteins containing R2R3- or R3-type DNA binding domains, which cluster distantly from CLAU (Supplemental Figure 3). CLAU homologs exist in simple-leaf species, where they are possibly involved in GOB regulation and restriction of the meristematic morphogenetic potential, but CLAU appears to have an expanded function in compound-leaf development. In tomato, and possibly additional compound-leaf species, CLAU is expressed and functions primarily at the stage when leaflets are already being formed, suggesting that it may have a more restricted role in the development of leaves that lack leaflets. In the Solanaceae, CLAU increases in expression at developmental stages where leaflets are being formed in the compound-leaf tomato and potato, decreasing after leaflets have been formed, while its expression decreases much earlier in the simple-leaf pepper and tobacco, possibly supporting a role in restricting lateral organ morphogenesis from the meristem. Interestingly, the pepper CLAU homolog is truncated, possessing five exons rather than the six in tomato and potato. The portion of the protein missing in Ca-CLAU is somewhat comparable to the clau-FF mutation (Ca-CLAU is ∼70% identical to amino acids 1 to 207 of CLAU, while the predicted protein in the clau-FF allele is composed of amino acids 1 to 150 of CLAU), which results in a loss-of-function clau phenotype. This suggests that the CLAU protein may not be functional or may have decreased or different function in pepper, demonstrating that the activity of CLAU in curbing leaf morphogenesis may not be required in the simple-leaf pepper. The function of CLAU in simple-leaf species remains to be investigated.
To summarize, we have shown here that CLAU is a MYB transcription factor that modulates leaf morphogenesis by curbing the morphogenetic potential, in part through attenuation of CK signaling. It appears that CLAU is of particular spatio-temporal relevance in compound leaves.
METHODS
Plant Material
Tomato seeds (Solanum lycopersicum cv M82 or as indicated) were sown in a commercial nursery and grown in the field or in a glasshouse under natural daylight with 25:18°C (day: night) temperatures and a maximum light intensity of ∼450 μmol m−2 s−1. For in situ hybridization and expression analyses, plants were grown in a controlled growth chamber under fluorescent bulbs providing ∼300 μmol m−2 s−1 at an 18 h/6-h light/dark regimen. The clau allele used throughout all experiments, clau:e3545, is from a tomato EMS population (Menda et al., 2004) and was previously characterized (Bar et al., 2015). Tomato (M82, sp) plants expressing pFIL>>IPT and pFIL>>CKX were generated by the LhG4 transactivation system (Moore et al., 1998) and described previously (Shani et al., 2010). Tomato plants overexpressing TCSv2:3XVENUS were obtained from the Muller, Eshed, and Weiss labs (Steiner et al., 2016; Zürcher et al., 2013), confirmed to be CK responsive in tomato, and backcrossed into a clau background.
Cloning and Plant Transformation
The CLAU gene was amplified from tomato M82 cDNA and cloned into the pENTR/d vector using a TOPO isomerase cloning system (Invitrogen). To generate op:CLAU and op:CLAU-SRDX, CLAU from pENTR was subcloned downstream to an Operator array (Moore et al., 1998). The SRDX fusion was generated by assembly PCR. At least five independent kanamycin-resistant transgenic lines from each transgene were crossed to the pFIL and pCLAU (see below) driver lines and examined, and a representative line was selected for further analysis. The CLAU promoter was generated by amplifying 2700 bp upstream of the CLAU ATG from genomic DNA (M82 background) and subcloned upstream to NLS-YFP generating pCLAU:nYFP or subcloned upstream to LhG4, generating a vector for driving expression in the trans-activation system (Moore et al., 1998).
CLAU Mapping and RNA-Seq
We created an F2 mapping population from a cross between clau:e3545 in the M82 background and Solanum pimpinellifolium. Markers used in the mapping of this population are presented in Supplemental Table 1. Using this population and these markers, we mapped CLAU between bases 2094866 and 2122247 on the short arm of chromosome 4. RNA was prepared from the shoot apices of 10-d-old clau:e3545 and wild-type M82 plants (m-P4) and subjected to RNA-seq. Two libraries per genotype were prepared according to the Illumina TruSeq RNA protocol and sequenced on an Illumina HiSeq2000 platform at the Genome Center of the Max Planck Institute for Plant Breeding Research. We obtained between 17.9 and 29.9 million 96-bp single-end reads per library (average of 25.8 million).
Reads were aligned to the S. lycopersicum reference sequence v2.40 (ftp://ftp.solgenomics.net/tomato_genome/wgs/assembly/build_2.40/) using TopHat v2.0.6 (Kim et al., 2013) with the following parameters: –max-insertion-length 12–max-deletion-length 12 -g 1–read-gap-length 12–read-edit-dist 20–read-mismatches 12–no-coverage-search–read-realign-edit-dist 0–segment-mismatches 3–splice-mismatches 1. An average of 93.2% of all reads were uniquely aligned to the reference genome.
To detect polymorphisms between clau:e3545 and wild-type M82, we first removed duplicated reads using default settings in Picard (http://broadinstitute.github.io/picard/), realigned indels, and called variants using default parameters in GATK v2.2-8 (DePristo et al., 2011). For all analyses, we retained only homozygous, biallelic variants covered in both genotypes with a phred-scaled variant quality higher than 30. This analysis returned 26,995 variants between the mutant and its M82 background. We estimated the effect of each variant in annotated transcripts (ITAG 2.3) using ANNOVAR (Wang et al., 2010). Deleterious variants in the candidate region in chromosome 4 were evaluated manually.
Phylogenetic Analysis
Protein sequence alignment was performed using Muscle (Edgar, 2004). The evolutionary history was inferred using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). In each case, the tree with the highest log likelihood is shown. Initial trees for the heuristic searches were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
Synteny
CLAU-like genes were chosen based on BLASTP to Sl-CLAU. Chromosomal regions were checked for synteny by seeking neighbor gene annotations and verified by global pairwise sequence alignment (Needle, EMBOSS). Genomic synteny was analyzed for chromosome 2 in Arabidopsis thaliana (TAIR10) and chromosome 4 in potato (Solanum tuberosum; ITAG1). Gene models for the synteny were drawn using published gff3 files. The following genomes were used for synteny analyses: Tomato ITAG2.4; Arabidopsis TAIR10; Potato ITAG1, PGSC3.4, and 4.03; Pepper CAv1.55; Eggplant SME2.5.1; Nicotiana benthamiana Niben101.
Tissue Collection and RNA Analysis
Tissue collection, RNA preparation, and qRT-PCR analysis were performed as previously described (Shleizer-Burko et al., 2011). Values are means of at least three biological repeats, each containing the shoot apices, including the SAM and P1-P5 leaf primordia, of 8 to 12 seedlings. Expression of all assayed genes was normalized relative to tomato EXPRESSED (EXP), except in the results presented in Figure 4, where each CLAU ortholog was normalized relative to the expression of TUBULIN within the same species. Primer sequences used for the qRT-PCR analyses are detailed in Supplemental Table 2. Student’s t test (two-tailed) was used for comparison of means, which were deemed significantly different at P ≤ 0.05. RNA in situ hybridization was performed as previously described (Shani et al., 2010). The CLAU mRNA antisense probe was generated as described previously (Berger et al., 2009) from a pENTR-CLAU construct (see above) containing the full-length CLAU cDNA, digested with HindIII.
Imaging
Intact leaves were photographed using a Nikon D5200 camera. For analysis of TCSv2:3XVENUS expression, dissected whole-leaf primordia were placed into drops of water on glass microscope slides and covered with cover slips. The pattern of VENUS expression was detected by a confocal laser scanning microscope (CLSMmodel SP8; Leica), with the solid-state laser set at 514 nm for excitation and 530 nm for emission. Chlorophyll expression was detected at 488 nm for excitation and 700 nm for emission. The pattern of VENUS expression was also observed with a Nikon SMZ1270 stereomicroscope equipped with a Nikon DS-RI2 camera and NIS elements software.
In situ hybridization slides were photographed with an Olympus 1X81 microscope using CellR software as described (Berger et al., 2009). LAS AF and ImageJ software were used for analysis, quantification, and measurements of captured images. Student’s t test (two-tailed) was used for comparison of means, which were deemed significantly different at P ≤ 0.05. Images were adjusted uniformly using Adobe Photoshop CS6.
CK Content Analysis
Cytokinins were isolated and purified as outlined by Novák et al. (2003) with some modifications. Frozen tomato tissue (30 mg) was homogenized using vibration mill MM 301 at a frequency of 30 Hz for 3 min (3-mm zirconium oxide beads; Retsch) with 1 mL ice-cold Bieleski solution (methanol-chloroform-formic acid-water; 12:5:1:2) as extraction solution. The cytokinins were then extracted overnight at 4°C using a benchtop laboratory rotator Stuart SB3 (Bibby Scientific) after addition of heavy labeled internal standards (OlChemIm). The samples were further purified by SPE (SCX), and the eluate was concentrated in vacuo at 37°C. For analysis, samples were redissolved in mobile phase and analyzed by ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS; Acquity UPLC System, Xevo TQ) according to Novák et al. (2008). Cytokinins were ionized by electrospray in positive mode and detected using multiple-reaction monitoring. Masslynx 4.1 software (Waters) was used to analyze the data and processed by the standard isotope dilution method (Rittenberg and Foster, 1940)
CK Treatment and Root Growth, Hypocotyl Height, and Anthocyanin Measurements
For anthocyanin measurement and hypocotyl elongation, plants were sprayed with the indicated CK concentrations three times a week for 3 weeks prior to analysis, starting upon emergence of the first leaf. Anthocyanins were extracted from the terminal leaflet of the third leaf by incubation overnight in methanol supplemented with a final concentration of 1% HCl. OD was measured in a plate spectrophotometer and anthocyanin content was calculated according to the following: (OD530-(0.25*OD660)), normalized to the starting tissue weight. Three technical replicates of 10 to 15 biological repeats were performed for each sample.
For root growth, seeds were germinated on sterile wetted paper filters and transferred to plates containing the indicated CK concentrations after 5 d, then grown upright on the CK concentrations for an additional 3 to 5 d in a growth chamber.
For qRT-PCR, 10-d-old seedlings were dipped in the indicated CK concentrations for 5 min and then incubated at room temperature for 3 h. Shoot apices containing the SAM and P1-P5 leaf primordia were then collected for RNA extraction. 6-Benzyl-aminopurine (Sigma-Aldrich) was used for all CK treatments.
Accession Numbers
Sequence data used in this study can be found in the Sol Genomic Network, The Arabidopsis Information Resource (TAIR), and the National Center for Biotechnology Information databases under the following accession numbers: CLAU, Solyc04g008480; CLAU homologs: tomato, Solyc02g080860.1.1 and Solyc12g006280.1.1; Arabidopsis, AT1G14600.1 and AT2G02060.1; coffee, Cc09g01520 and Cc11g04980; eucalyptus, EcC013137.10, EcC045129.10, and EcC054831.10; soybean, Glyma20g108600.1.p, Glyma07g158600.1.p, and Glyma20g006000.1.p; rice, Oryza_sativa|BGIOSGA011527-PA and Oryza_sativa|BGIOSGA031479-PA; Nicotiana benthamiana, NbS00036280g0004.1, NbS00013875g0008.1, and NbS00035641g0001.1; Nicotiana tabacum, NT_mRNA_59421_gene_33284 and NT_mRNA_90646_gene_51238; pepper, CA05g06710 and CA06g06970; potato, PGSC0003DMP400051256 and PGSC0003DMP400051255; eggplant, Sme2.5_01601.1_g00006.1 and Sme2.5_02381.1_g00010.1; maize, Zea_mays|GRMZM2G374986_T01 and Zea_mays|GRMZM5G887276_T03; and Medicago, Medtr0223s0040.1 and Medtr1g112370.1.
Supplemental Data
Supplemental Figure 1. Allelic representation of CLAU.
Supplemental Figure 2. Synteny of tomato CLAU chromosomal region with additional species.
Supplemental Figure 3. Analysis of various CLAU homologs and tomato MYB genes.
Supplemental Figure 4. TCSv2:3XVENUS is upregulated in response to CK treatment in tomato.
Supplemental Figure 5. Additional CK sensitivity experiments conducted on clau versus the wild type.
Supplemental Table 1. Markers used in CLAU mapping.
Supplemental Table 2. Primer pairs used in this work.
Supplemental Data Set 1. Text file of the CLAU alignment used for the phylogenetic analysis shown in Figure 1C.
Supplemental Data Set 2. Text file of the tomato MYB alignment used for the phylogenetic analysis shown in Supplemental Figure 3A.
Supplemental Data Set 3. Text file of the tomato MYB alignment used for the phylogenetic analysis shown in Figure 4A.
Supplementary Material
Acknowledgments
This research was supported by grants from the Israel Science foundation (539/14) and U.S.–Israel Binational Agricultural Research and Development Fund [IS 4531-12(c)] to N.O., the German-Israel Project Cooperation Foundation (OR309/1-1; FE552/12-1) to N.O. and J.M.J.-G., and by grant LO1204 (sustainable development of research in the Centre of Region Haná) from the National Program of Sustainability I, Ministry of Education, Youth, and Sports, Czech Republic, to P.T. We thank Bruno Muller for providing the TCSv2:3xVENUS construct, Yuval Eshed, Lior Tal Ziva Amsellem, and Evyatar Steiner for providing transgenic tomato lines expressing TCSv2:3xVENUS, Ori Ben Herzel for initial analysis of the clau alleles, Ori Ben Herzel and Hagai Kohay for generating the clau mapping population, Sharona Shleizer-Burko for tissue collection and RNA sample preparation, Matan Zeron and Mor Tsamir for technical assistance, David Weiss and Eilon Shani for critical reading of the manuscript, and members of the Ori group for continuous discussion and support.
AUTHOR CONTRIBUTIONS
M.B. and N.O. designed the research. M.B., A.I., H.B.G., M.L., J.M.J.-G., S.K., and P.T. performed research. M.B., A.I., M.L., H.B.G., N.O., J.M.J.-G., S.K., and P.T. analyzed data. M.B. and N.O. wrote the article.
References
- Avivi Y., Lev-Yadun S., Morozova N., Libs L., Williams L., Zhao J., Varghese G., Grafi G. (2000). Clausa, a tomato mutant with a wide range of phenotypic perturbations, displays a cell type-dependent expression of the homeobox gene LeT6/TKn2. Plant Physiol. 124: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Ben-Herzel O., Kohay H., Shtein I., Ori N. (2015). CLAUSA restricts tomato leaf morphogenesis and GOBLET expression. Plant J. 83: 888–902. [DOI] [PubMed] [Google Scholar]
- Bar M., Ori N. (2014). Leaf development and morphogenesis. Development 141: 4219–4230. [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Galinha C., Grigg S.P., Tsiantis M. (2007). From genes to shape: regulatory interactions in leaf development. Curr. Opin. Plant Biol. 10: 660–666. [DOI] [PubMed] [Google Scholar]
- Barth S., Geier T., Eimert K., Watillon B., Sangwan R.S., Gleissberg S. (2009). KNOX overexpression in transgenic Kohleria (Gesneriaceae) prolongs the activity of proximal leaf blastozones and drastically alters segment fate. Planta 230: 1081–1091. [DOI] [PubMed] [Google Scholar]
- Ben-Gera H., Shwartz I., Shao M.R., Shani E., Estelle M., Ori N. (2012). ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 70: 903–915. [DOI] [PubMed] [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J.P., Zinder M., Samach A., Eshed Y., Ori N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832. [DOI] [PubMed] [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Blein T., Hasson A., Laufs P. (2010). Leaf development: what it needs to be complex. Curr. Opin. Plant Biol. 13: 75–82. [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Shirding N., Shleizer S., Ori N. (2007). Meristem maintenance and compound-leaf patterning utilize common genetic mechanisms in tomato. Planta 226: 941–951. [DOI] [PubMed] [Google Scholar]
- Brandstatter I., Kieber J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y., Ori N. (2013). The tomato leaf as a model system for organogenesis. Methods Mol. Biol. 959: 1–19. [DOI] [PubMed] [Google Scholar]
- Burko Y., Shleizer-Burko S., Yanai O., Shwartz I., Zelnik I.D., Jacob-Hirsch J., Kela I., Eshed-Williams L., Ori N. (2013). A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell 25: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch B.L., Schmitz G., Rossmann S., Piron F., Ding J., Bendahmane A., Theres K. (2011). Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 23: 3595–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C., Barkoulas M., Galinha C., Tsiantis M. (2010). Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development. J. Plant Res. 123: 25–33. [DOI] [PubMed] [Google Scholar]
- Coenen C., Lomax T.L. (1998). The diageotropica gene differentially affects auxin and cytokinin responses throughout development in tomato. Plant Physiol. 117: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino I.B., Kieber J.J. (1999). Phosphorelay signal transduction: the emerging family of plant response regulators. Trends Biochem. Sci. 24: 452–456. [DOI] [PubMed] [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: Current issues. Int. J. Plant Sci. 162: 459–464. [Google Scholar]
- DePristo M.A., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishon S., Shani E., Ori N., Weiss D. (2011). Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 190: 609–617. [DOI] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: the significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152. [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151. [DOI] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744. [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Janssen B.J., Lund L., Sinha N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Kaur H., Tattersall A., Tsiantis M. (2007). Negative regulation of KNOX expression in tomato leaves. Planta 226: 1255–1263. [DOI] [PubMed] [Google Scholar]
- Jasinski S., Tattersall A., Piazza P., Hay A., Martinez-Garcia J.F., Schmitz G., Theres K., McCormick S., Tsiantis M. (2008). PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 56: 603–612. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kaplan D.R. (2001). Fundamental concepts of leaf morphology and morphogenesis: a contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162: 465–474. [Google Scholar]
- Keshishian E.A., Rashotte A.M. (2015). Plant cytokinin signalling. Essays Biochem. 58: 13–27. [DOI] [PubMed] [Google Scholar]
- Khush G., Rick C. (1967). Studies on the linkage map of chromosome 4 of the tomato and on the transmission of induced deficiencies. Genetica 38: 74–94. [Google Scholar]
- Kiba T., Aoki K., Sakakibara H., Mizuno T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45: 1063–1077. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.J., Kim S., Ha Y.-M., Kim J. (2008). Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta 227: 577–587. [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Ayre B.G., Eshed Y. (2014). Florigen and anti-florigen - a systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 5: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Yu C.W., Chen F.F., Zhao L., Tian G., Liu X., Cui Y., Yang J.Y., Wu K. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N., Semel Y., Peled D., Eshed Y., Zamir D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38: 861–872. [DOI] [PubMed] [Google Scholar]
- Moore I., Gälweiler L., Grosskopf D., Schell J., Palme K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz A.A., Raman S., Martinez C.C., Sinha N.R., Schmitz G., Theres K. (2013). Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc. Natl. Acad. Sci. USA 110: 2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Hauserová E., Amakorová P., Dolezal K., Strnad M. (2008). Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224. [DOI] [PubMed] [Google Scholar]
- Novák O., Tarkowski P., Tarkowská D., Doležal K., Lenobel R., Strnad M. (2003). Quantitative analysis of cytokinins in plants by liquid chromatography–single-quadrupole mass spectrometry. Anal. Chim. Acta 480: 207–218. [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Rast-Somssich M.I., et al. (2015). Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 29: 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg D., Foster G.L. (1940). A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. J. Biol. Chem. 133: 737–744. [Google Scholar]
- Rossmann S., Kohlen W., Hasson A., Theres K. (2015). Lateral suppressor and Goblet act in hierarchical order to regulate ectopic meristem formation at the base of tomato leaflets. Plant J. 81: 837–848. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Nieuwland J., Murray J.A. (2013). The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 75: 53–66. [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Gupta S., Lindquist I.E., Cameron C.T., Mudge J., Rashotte A.M. (2013). Transcriptome analysis of cytokinin response in tomato leaves. PLoS One 8: e55090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704. [DOI] [PubMed] [Google Scholar]
- Sluis A., Hake S. (2015). Organogenesis in plants: initiation and elaboration of leaves. Trends Genet. 31: 300–306. [DOI] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Livne S., Kobinson-Katz T., Tal L., Pri-Tal O., Mosquna A., Tarkowská D., Muller B., Tarkowski P., Weiss D. (2016). SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171: 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J.P., Haberer G., Ferreira F.J., Deruère J., Mason M.G., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuinen A., Peters A.H.L.J., Kendrick R.E., Zeevaart J.A.D., Koornneef M. (1999). Characterisation of the procera mutant of tomato and the interaction of gibberellins with end-of-day far-red light treatments. Physiol. Plant. 106: 121–128. [Google Scholar]
- Wang K., Li M., Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68: 571–582. [DOI] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P.T., Müller B. (2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.