Sequencing of 256 sorghum mutant families uncovered 1.8 million mutations, providing an efficient resource for causal gene/SNP variant to trait association with wide application to cereal crops.
Abstract
Sorghum (Sorghum bicolor) is a versatile C4 crop and a model for research in family Poaceae. High-quality genome sequence is available for the elite inbred line BTx623, but functional validation of genes remains challenging due to the limited genomic and germplasm resources available for comprehensive analysis of induced mutations. In this study, we generated 6400 pedigreed M4 mutant pools from EMS-mutagenized BTx623 seeds through single-seed descent. Whole-genome sequencing of 256 phenotyped mutant lines revealed >1.8 million canonical EMS-induced mutations, affecting >95% of genes in the sorghum genome. The vast majority (97.5%) of the induced mutations were distinct from natural variations. To demonstrate the utility of the sequenced sorghum mutant resource, we performed reverse genetics to identify eight genes potentially affecting drought tolerance, three of which had allelic mutations and two of which exhibited exact cosegregation with the phenotype of interest. Our results establish that a large-scale resource of sequenced pedigreed mutants provides an efficient platform for functional validation of genes in sorghum, thereby accelerating sorghum breeding. Moreover, findings made in sorghum could be readily translated to other members of the Poaceae via integrated genomics approaches.
INTRODUCTION
Sorghum (Sorghum bicolor) is a highly productive C4 crop that is well adapted to arid and semi-arid environments (Rooney et al., 2007). It is the fifth most important grain crop worldwide and a subsistence crop for millions of people in Africa and Southeast Asia. Moreover, because sorghum can be grown on marginal soil that is not suitable for food crops, it has also emerged as an important bioenergy crop. In addition to sorghum’s high grain yield, its stalk contains high levels of soluble sugars that can be easily converted to ethanol. The bagasse (residue after sugar extraction) is an excellent lignocellulosic source for biofuel and can also be used to generate electricity through direct burning. In addition, sorghum is an excellent forage crop with similar productivity to forage maize (Zea mays) but requiring much less water for irrigation (Rocateli et al., 2012).
The Saccharinae group in the Andropogoneae tribe of grasses includes the important grain crops sorghum and maize, as well as sugar crop sugarcane (Saccharum officinarum) and the highly productive biomass crop Miscanthus (de Siqueira Ferreira et al., 2013). As a diploid plant that has not experienced genome duplication for over 70 million years, the sorghum genome has minimal gene redundancy (Paterson et al., 2004, 2009). Consequently, sorghum exhibits a broad spectrum of phenotypes after EMS mutagenesis (Xin et al., 2008). In maize, by contrast, because of a recent whole-genome duplication and genome expansion (∼12 million years ago), many genes are buffered when mutated (Swigonová et al., 2004; Paterson et al., 2009; Schnable et al., 2009; Hughes et al., 2014). Genetic and genomic analysis of sugarcane and Miscanthus is even more challenging due to recent genome duplication, hybridization, and variability in polyploidy levels (Ming et al., 1998; Hodkinson et al., 2002; Jannoo et al., 2007; Premachandran et al., 2011). Fortunately, however, sorghum shares common ancestry with maize (∼12 to 15 million years ago) and sugarcane (5 to 9 million years ago) (Paterson et al., 2004, 2005). Due to the relatively small size of its genome (∼730 Mb), which has been completely sequenced, sorghum is an attractive model for functional genomics for maize, sugarcane, Miscanthus, and other C4 bioenergy crops with complex genomes (Swigonová et al., 2004; Paterson et al., 2009; Schnable et al., 2009; Hughes et al., 2014). This, along with its advantageous genomic features and suitability for forward genetics, makes sorghum a facile system in which to identify target genes that could be used to increase the quality and yield of grain and biomass. Once identified, such genes could then be modified via genome editing in crops with multiploid genomes (Bortesi and Fischer, 2015) by taking advantage of exciting progress on the RNA-guided CRISPR/CAS9 genomic editing system, which makes it possible to genetically improve organisms with complex genomes so long as the target genes are known (Jinek et al., 2012; Ran et al., 2013; Shalem et al., 2014). For example, brown midrib mutants increase the conversion efficiency of sorghum biomass to ethanol (Saballos et al., 2012; Sattler et al., 2012, 2014). As observed in sorghum, RNAi-mediated knockdown of the caffeic acid O-methyltransferase (bmr12) from sugarcane via RNAi increases fermentable sugar yield (Sattler et al., 2012; Jung et al., 2013). Therefore, discovery of gene mutations responsible for relevant traits in sorghum provides targets for genomic editing in other crops, thereby improving their utility for food and energy production.
EMS is a potent chemical mutagen that efficiently induces high-density mutations in genomes in many organisms (Greene et al., 2003). The development of TILLING (targeting induced local lesions in genomes) techniques, which can detect single-nucleotide mismatches in heteroduplexes of PCR amplicons, made EMS mutagenesis a workhorse technique for the study of recalcitrant metabolic and signaling processes in plants, animals, and microorganisms (Greene et al., 2003; Winkler et al., 2005; Gilchrist et al., 2006). In sorghum, a few mutated genes have been identified by TILLING based on individual amplicons (Xin et al., 2008; Blomstedt et al., 2012; Krothapalli et al., 2013). Recent advances in high-throughput next-generation sequencing have made it possible to catalog EMS-induced mutations on a large scale (Thompson et al., 2013; Henry et al., 2014). In rice (Oryza sativa) and wheat (Triticum aestivum), sequencing of captured exons has been used to identify large numbers of mutations from a limited number of mutant plants (Henry et al., 2014). However, this approach cannot capture mutations in regulatory regions that affect gene expression levels; moreover, it may not capture all exons with the same efficiency as whole-genome sequencing.
By linking large numbers of mutants with phenotypes of interest, crop plant mutation libraries have historically served as an important way to associate genes with their functions (Till et al., 2003; Settles et al., 2007; Krishnan et al., 2009). Previously, however, no comprehensive collection of sorghum mutant populations was available. To construct such a comprehensive mutant resource, we used whole-genome sequencing to discover mutations throughout the genome, as well as link mutations to phenotypes, in a pedigreed sorghum mutant library for which extensive phenotypic information is available (Xin et al., 2008, 2009, 2015). For this purpose, a mini-collection of 256 mutant lines randomly selected from the library was sequenced to an average coverage of 16x. Our results demonstrate that whole-genome sequencing of well-phenotyped mutant lines is an efficient way to identify causal mutations, as well as validate gene function by characterizing mutant phenotypes. Given sorghum’s relatively small diploid genome, low levels of gene duplication, and close phylogenetic relationship to maize and sugarcane, this resource should also facilitate the functional study of other grasses.
RESULTS
Pedigreed Mutant Library and Phenotypic Diversity
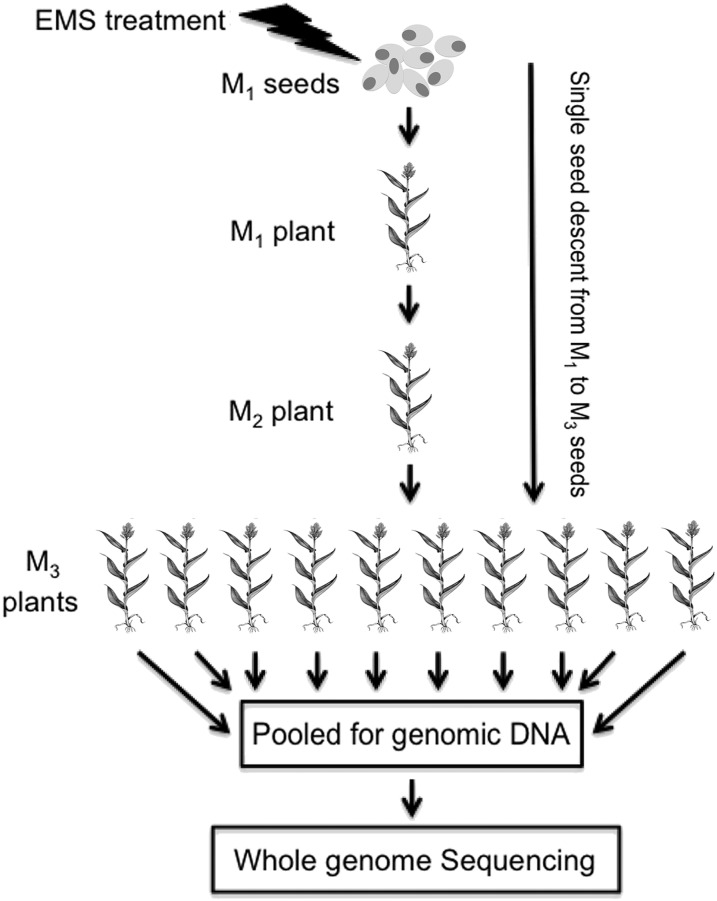
A pedigreed mutant library derived from a uniform genetic background is a powerful resource for linking genotypic variation to phenotypic diversity. We developed a mutant library in the sorghum inbred line BTx623, which was used to generate the sorghum reference genome. To generate the library, we introduced mutations using EMS and propagated individually mutagenized M1 seeds by pedigree to M3 seeds through single-seed descent (Figure 1). The mutants in the library exhibit a wide range of phenotypic variations, many of which segregate ∼1:3 (i.e., as Mendelian recessive traits) in M3 head rows. In addition to the variety of mutant phenotypes reported previously (Xin et al., 2008, 2009), we observed several traits in the mutant library that have the potential to be used for sorghum improvement (Table 1; Supplemental Data Set 1). For example, the multiseeded (msd) mutants, which double the seed number and seed weight per panicle, could be used to improve grain yield (Burow et al., 2014), and the brown midrib (bmr) mutants, mentioned above, could be used to improve biomass quality and ethanol conversion rate (Xin et al., 2009; Saballos et al., 2012; Sattler et al., 2012, 2014). Since the early 1930s (Duvick and Cassman, 1999), erect leaf architecture has contributed significantly to continuous yield gains in maize hybrids; however, sorghum architecture has not been improved in this manner due to the lack of erect leaf germplasm (Assefa and Staggenborg, 2011). From our library, we isolated several erect leaf mutants (erl) that have the potential to improve sorghum canopy architecture to maximize biomass yield and stress resilience (Xin et al., 2009, 2015).
Figure 1.
Construction of an EMS Mutant Population.
EMS-treated seeds (M1) were propagated by single-seed descent to M3 seeds, which were planted as a head row in the field for phenotypic evaluation. Leaf samples from 20 individual plants per mutant line were pooled for genomic DNA extraction. Ten panicles from M3 plants were pooled and deposited into the mutant library for distribution.
Table 1. Selection of Mutants That Could Be Used for Sorghum Improvement.
| Mutants | No. of Lines | Potential application of Trait |
|---|---|---|
| Erect leaf (erl) | 23 | Biomass and grain yield |
| Brown midrib (bmr) | 30 | Biomass conversion efficiency |
| Multiple tiller (mtl) | 120 | Biomass yield |
| Early flowering (efl) | 7 | Biomass and grain yield |
| Late flowering | 5 | Biomass yield |
| Stiff stem (stf) | 13 | Biomass and grain yield |
| Early senescence (esn) | 7 | Early dehydration of biomass |
| Multiple seed (msd) | 36 | Grain yield |
| Large seed (lsd) | 18 | Grain quality |
| Bloomless (blm) | 107 | Water-use efficiency |
| Heat sensitive (hs) | 120 | Tolerance to high temperature |
The mutants were isolated over the last several years based on visual inspection of individual M3 head plots from a mutant library of 6400 lines. Some mutants segregated 1:3 in the M3 head plots, indicating they were recessive mutations in single nuclear genes. Identified mutants were crossed to a nuclear male-sterile mutant also isolated from the mutant library. The F1 and F2 plants were used to determine the genetic mode of the mutations.
Population Sequencing, Quality Control, and Mutation Detection
The wide phenotypic variations in this library prompted us to develop a high-throughput platform for rapidly identifying the causal mutations underlying these traits. A total of 256 M3 families were randomly selected for whole-genome sequencing. Because many M2 plants were sterile, genomic DNA was prepared from leaf samples pooled from 20 individual M3 plants in order to capture most of the mutations present in the original M2 plant. We obtained a total of 3.1 terabytes raw sequence data, which covered each genome at an average depth of 16.4x (range, 11 to 60x). Over 98% of short reads in each line could be mapped to the sorghum v2.1 reference genome, covering 86.6% of the genome and 95.6% of gene space (Supplemental Data Set 2). Genome coverage did not improve with increased sequencing depth from 11x to 60x (Supplemental Figure 1), probably due to the presence of repetitive DNA sequences and the difficulty of generating unique mappings from short reads (Sims et al., 2014).
To determine the proportion of mutations that were captured at the average sequencing depth (16.4x), we selected three lines with a sequencing depth of ∼27x and compared the single-nucleotide polymorphism (SNP) discovery rate from 5x to 25x coverage. As shown in Supplemental Figure 2, 98% of homozygous SNPs and 85% of heterozygous SNPs that could be detected at 27x coverage were identified at 15x coverage. Thus, the vast majority of SNPs present in the mutant lines were captured at 16x genome coverage.
We developed a rigorous bioinformatics pipeline to identify most of the EMS-induced mutations while minimizing the false positives. First, we removed all SNPs with more than two alleles. Furthermore, only canonical EMS-induced GC-to-AT transition SNPs were included in the analysis (Greene et al., 2003). To exclude background SNPs already present in our parental line before mutagenesis, we sequenced the BTx623 inbred line used to generate the mutant library to 12.9x coverage. We identified 13,243 high-confidence variations in our own parental line BTx623 relative to the sorghum reference sequence v2.1 (Supplemental Data Set 3). These SNPs occurred at high frequencies in the 256 lines we sequenced (Supplemental Figure 3); therefore, they are very likely to represent differences between our BTx623 line and the reference genome. Due to limitations on sequencing depth, this approach is unlikely to capture all background SNPs. Therefore, we amended the background SNPs with 65,634 variations that had allele frequencies >0.05 in the sequenced mutant population or that were detected in five or more mutant lines.
It is technically difficult to eliminate all contamination and cross-pollination during development of a large pedigreed mutant library. Therefore, to ensure the quality of the mutation data, we first investigated whether any of the lines could have resulted from seed or pollen contamination. Two lines, ARS79 and ARS137, had extremely high apparent mutation rates (Supplemental Data Set 3 and Supplemental Figure 4A). Furthermore, the SNPs in these two lines overlapped extensively with natural variations (ARS79, 72%; ARS137, 77%; Supplemental Figure 4B). Therefore, it is likely that these two lines resulted from either seed contamination or pollination by unknown sorghum lines grown in the region; consequently, they were not included in subsequent analysis. We also compared the SNP overlap between each pair of lines to identify possible siblings or cross-pollination among the mutant lines. Among the 64,516 pairs, two (ARS80/ARS85 and ARS20/ARS26) exhibited >50% similarity (Supplemental Figure 5A); ARS85 and ARS20, which had higher sequencing depths, were retained for downstream analysis. Following removal of these four lines, the identity of mutations between any two lines was <5% (Supplemental Figure 5B). After filtering the mutation database using the criteria described above, we identified 1,862,560 bona fide EMS-induced SNPs. On average, each sample contained 7660 SNPs (1798 homozygous and 5862 heterozygous; Supplemental Data Set 3). The average mutation density in the population overall was 11 SNPs/Mb, with a range of 0.02 to 22.5 SNPs/Mb (Figure 2A). A recent study of rice using exome capture technology reported a similar large range of variation in the mutation rate (Henry et al., 2014).
Figure 2.
Characterization of EMS Mutations in 252 Sorghum Samples.
(A) Mutation density (SNP/Mb) in each sample.
(B) SIFT score distribution of the EMS mutations. Mutations with SIFT scores <0.05 were considered to be deleterious to protein structure or gene function.
(C) Distribution of large-effect mutations in each sample. Large-effect mutations include stop-gain mutations, nonsynonymous coding mutations with a SIFT score <0.05, and mutation at splice acceptor (two bases before exon start, except for the first exon) or donor sites (two bases after coding exon end, except for the last exon).
(D) Distribution of the number of large-effect mutations in each gene.
To validate the accuracy of the mutations identified using our bioinformatics pipeline, we randomly selected a total of 1024 SNPs (one heterozygous and three homozygous and SNPs from each mutant line) and subjected them to Sanger sequencing (Supplemental Data Set 4). Of the 1024 amplicons, 992 (96.8%) could be aligned to the targeted SNP regions, and 979 (95.6%) contained the predicted SNPs (Supplemental Table 1). The overall SNP accuracy was 98.7%/99.7% (767/769) for homozygous SNPs and 95.1% (212/223) for heterozygous SNPs. We divided the SNPs used for validation into five groups, according to the depth of sequencing coverage. SNP validation rates were similar (99%) for SNPs supported by 3 to 20 reads, whereas SNPs supported by 20 or more reads had a slightly lower validation rate, due to the heterozygous SNPs with high read depth. This slightly lower confirmation rate at high sequence depth might have been caused by paralogous alignment. The high SNP validation rate demonstrated that our variation call pipeline was robust, and the mutations discovered with our pipeline had high accuracy rate.
Functional Annotation of the EMS-Induced Mutations
Functional annotation of SNPs generated by EMS treatment was performed using the sorghum reference genome annotation v2.1. Based on the reference annotation, 22% of SNPs were located in gene space, covering ∼95% (31,363) of genes (Table 2). We identified 4652 SNPs that changed an amino acid codon to a stop codon; 862 and 636 SNPs at splice acceptor and donor sites, respectively; and 86,070 missense mutations (Cingolani et al., 2012). Among the nonsynonymous SNPs, 29,120 had SIFT (sort intolerant from tolerant) scores < 0.05 and were therefore predicted to be deleterious mutations (Ng and Henikoff, 2003; Kumar et al., 2009). Overall, these 35,817 disruptive mutations covered 18,684 (57%) of annotated genes in the sorghum genome. On average, each line contained 147 large-effect mutations (stop-gain mutations, nonsynonymous coding mutations with SIFT score < 0.05 [Figure 2B] or mutations at splice acceptor or donor sites), with a range of 2 to 336 mutations per line (Figure 2C). Each line harbored an average of 145 genes with disruptive mutations (Supplemental Data Set 5). Furthermore, 28% (9157) of genes had disruptive mutations in more than one line, whereas 96% (8836/9,157) of genes had unique mutations in two or more lines (Figure 2D; Supplemental Data Set 5). This finding provided a basis for identifying causal mutations defined by phenotypes with two or more independent alleles, allowing us to avoid the painstaking and costly approach of map-based cloning by classical forward genetics.
Table 2. Annotation of Mutations Detected in the Sequenced Mutant Population.
| Effect Type | SNPs | Genes |
|---|---|---|
| 3′ UTR | 44,120 | 18,772 |
| Start gained | 5,237 | 4,326 |
| 5′ UTR | 29,452 | 14,055 |
| Nonsynonymous coding | 86,070 | 25,311 |
| Start lost | 136 | 136 |
| Stop gained | 4,652 | 4,043 |
| Synonymous coding | 138,416 | 22,035 |
| Splice site acceptor | 862 | 834 |
| Splice site donor | 636 | 621 |
| Splice site region | 6,894 | 5,260 |
| Intron | 195,916 | 21,443 |
| Upstream 5 kb of genes | 480,763 | 31,358 |
| Downstream 5 kb of genes | 464,091 | 31,141 |
| Intergenic | 1,457,491 | – |
EMS-induced mutations (SNPs) were classified according to their location and potential effect on gene expression and protein structure. UTR, untranslated region.
Reverse Genetics Study Using Phenotype Data
To test the utility of our sequenced and phenotyped mutant lines, we focused on a frequently observed phenotype: bloomless (blm) mutants that lacked epicuticular wax deposition on their aerial surfaces. The blm mutants had well-defined shiny green sheaths that could be easily and unambiguously identified; moreover, this feature is potentially important in the context of drought tolerance and water-use efficiency (Burow et al., 2008).
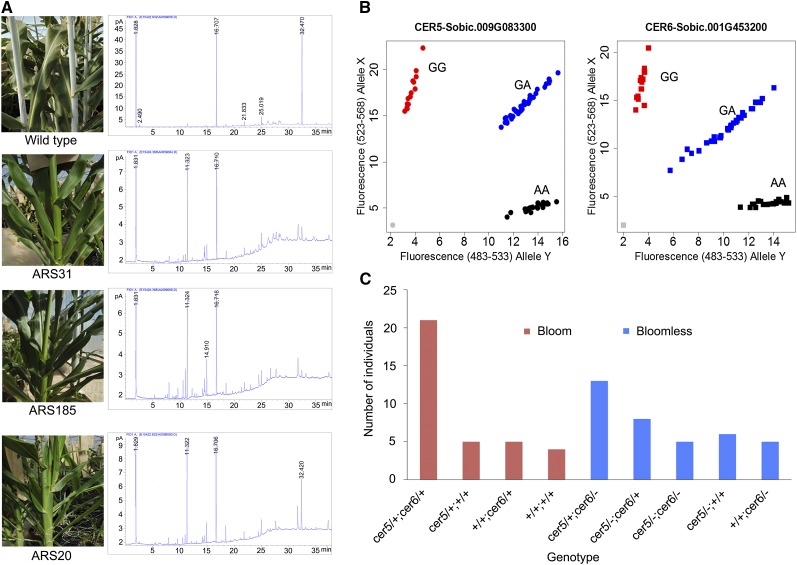
Compared with other crop plants, sorghum accumulates a very high level of epicuticular wax on its aerial surface, which is believed to be critical to its high water-use efficiency, and tolerance to drought and heat (Burow et al., 2008). To date, however, no gene that contributes to epicuticular wax metabolism and deposition has been identified in sorghum. Very-long-chain fatty acid (wax; VLCF) metabolism has been well studied in Arabidopsis thaliana (Pighin et al., 2004; Samuels et al., 2008; Haslam et al., 2012) and is proposed to underlie epicuticular wax production in that species. The biosynthesis of VLCFs in Arabidopsis involves ketoacyl-CoA synthase (KCS), β-ketoacyl-CoA reductase, enoyl-CoA reductase, and several other genes related to biosynthesis of wax, such as CER1-5 (Pighin et al., 2004; Samuels et al., 2008; Haslam et al., 2012). To determine whether these genes are related to the wax pathway in sorghum, we searched for mutations in their sorghum putative orthologs. Six lines with the bloomless phenotype harbored mutations in eight sorghum orthologs of genes involved in Arabidopsis VLCF metabolism (Table 3). The cer5 gene was represented by two allelic mutations in two different mutants. Fatty acid analysis of three blm lines (ARS20, ARS31, and ARS185) and the wild type confirmed that, as predicted, the three mutants lacked VLCFs. Moreover, the wax load was greatly reduced in blm mutants (Figure 3A).
Table 3. Mutants of Sorghum Putative Orthologs in Arabidopsis Very Long Fatty Acid Synthesis Pathway That Exhibited the Bloomless Phenotype and the Corresponding Allelic Mutations.
| Arabidopsis Gene | Sorghum Gene | Mutation Position | Amino Acid Change | Mutant | Independent Allele from Mutants Not in the Sequenced Population |
|---|---|---|---|---|---|
| AT1G68530 (CER6) | Sobic.001G453200 | Chr1:65789406 | E159K | ARS20 | Chr1:65789043;A38T |
| AT2G28630 (KCS12) | Sobic.004G341300 | Chr4:66583057 | R189C | ARS73 | |
| AT1G51500 (CER5) | Sobic.009G083300 | Chr9:12576446 | P581L | ARS73 | Chr9:12578091;R188H |
| AT1G51500 (CER5) | Sobic.009G083300 | Chr9:12577924 | L244F | ARS20 | Chr9:12578091;R188H |
| AT1G68530 (CER6) | Sobic.006G020600 | Chr6:3500267 | A133T | ARS205 | |
| AT1G71160 (KCS7) | Sobic.002G268300 | Chr2:65211661 | P449S | ARS31 | |
| AT1G19440 (KCS4) | Sobic.002G268500 | Chr2:65229433 | A49V | ARS31 | |
| AT5G43760 (KCS20) | Sobic.005G168700 | Chr5:55173639 | R303Q | ARS185 | Chr5:55173090;A486V |
| AT1G02205 (CER1) | Sobic.001G222700 | Chr1:21258089 | L100F | ARS185 |
Figure 3.
Phenotype of Bloomless Mutants and Genetic Evidence.
(A) Characterization of bloomless mutants. Phenotype shows the presence or absence of epicuticular wax in stems of wild-type and mutant lines. The chromatogram from fatty acid analyses confirmed that C30 fatty acid, which is predominant in the wild type, was significantly reduced in bloomless mutants.
(B) Scatterplots of endpoint genotyping results from an F2 population of ARS 20 for SbCER5 and SbCER6.
(C) Genotype and phenotype data of F2 individual plants from ARS20. +/+, Wild type; cer5/+ and cer6/+, heterozygous mutants; cer5/- and cer6/-, homozygous mutants.
We further validated the causal roles of these genes in the bloomless phenotype via two strategies: screening for independent allelic mutations and cosegregation test. A total of 30 blm mutants that were not included in the sequenced population were subjected to Sanger sequencing with overlap primers covering the eight sorghum orthologs of Arabidopsis VLCF genes. Six of the 30 mutants harbored distinct mutations in five of the eight genes. Two of the mutations were identical to those discovered by whole-genome sequencing of the 256 lines, whereas three were at different locations (Table 3). The detection of allelic mutations within genes with the same mutant phenotypes provides strong evidence for a causal link between gene and phenotype.
The ARS20 mutant line harbored heterozygous mutations in both the cer5 and cer6 genes. This unique line provided an ideal resource for cosegregation analysis between gene mutation and phenotype. Among 72 individual M4 plants obtained from a self-pollinated M3 plant (with heterozygous mutations in both genes), both cer5 and cer6 mutations followed an independent genotypic segregation ratio of 1 wild-type:2 heterozygote:1 mutant, whereas the bloom:bloomless phenotypic segregation was 3 bloom:1 bloomless. A goodness-of-fit test based on χ2 analyses of both genotypic and phenotypic segregation ratios revealed that the observed values were statistically similar to the expected values (Figure 3B). In the population, only individuals with homozygous mutations in either or both of these genes exhibited a bloomless phenotype, consistent with the cosegregation pattern of the two genes (Figure 3C). These results indicated that the bloomless phenotype is a recessive trait, as expected for EMS-induced mutations. Each of the SNP mutations followed a monogenic pattern of segregation, and when we analyzed them together, we observed a pattern consistent with two genes segregating independently. As a negative control, two other heterozygous mutations in ARS20 were randomly selected for genotyping; their genotypes exhibited no cosegregation with the bloomless phenotype. Furthermore, independent alleles were identified for both cer5 and cer6 in the 30 blm mutants sequenced by Sanger sequencing (Table 3). Taken together, these cosegregation results provided strong genetic evidence that we had identified two independent genes (Sobic.009G083300 and Sobic.001G453200) and that these specific mutations were responsible for the bloomless phenotype in line ARS20. This example demonstrates that the sequenced sorghum mutant lines provide a useful resource for efficiently identifying mutations in genes in order to deduce their function through reverse genetics.
In Silico Analysis of QTLs and Mutations Using the Sorghum Mutant Resource
Identification of genes that underlie complex traits is challenging in many organisms, particularly crop plants. Especially problematic are the relatively small-effect quantitative trait loci (QTLs), which account for a large share of agriculturally important variations, as well as fitness differences in natural populations (Ellegren and Sheldon, 2008). We investigated whether our sequenced mutant library could help validate genes located in QTL regions. To this end, we focused on 13 seed size QTL intervals reported in four mapping studies (Paterson et al., 1995; Feltus et al., 2006; Srinivas et al., 2009; Zhang et al., 2015). Four of these 13 genomic regions had been identified in both QTL and GWAS studies. Of the eight genes flanking the GWAS peaks, four candidates (Sobic.004G136600, Sobic.006G268800, Sobic.007G166600, and Sobic.010G144900) were significantly associated with seed size, and two of these genes contained mutations in the sequenced mutants. ARS110 and ARS118, which harbor mutations in Sobic.006G268800, had higher proportions of large seeds and thousand-kernel mass (TKM) (Supplemental Table 2). ARS235 and ARS253, which harbor homozygous mutations in Sobic.004G136600, also exhibited increases in the proportion of large seeds and TKM; a putative ortholog of this gene was recently characterized in maize and rice in regard to its role in grain filling (Sosso et al., 2015). ARS37, which harbors a stop-gain mutation in Sobic.010G144900, had reduced seed size, similar to the small seed phenotype observed in several accessions from the Kafir working group that have spontaneous mutations in this gene (Zhang et al., 2015). This result showed that our resources could be very useful in identifying candidate genes and thus greatly aid in advancing QTL cloning research in crop plants.
Mutation Landscape in Sorghum
Our large-scale sequencing efforts revealed characteristics of EMS-induced mutations in sorghum. Henry et al. (2014) sequenced the exomes captured from 72 rice mutant lines and demonstrated that EMS preferentially induces mutations in sequences matching the consensus motif gggrraR[G]CGrgg (Henry et al., 2014). We analyzed the frequencies of the 20 nucleotides up- and downstream of 1.8 million G/C-to-A/T mutation sites and found that the proportion of C residues was elevated at the −2 and +1 positions relative to the mutation. This result is consistent with observations made in Escherichia coli, in which EMS-alkylated O6-guanidine residues near A/T pairs are preferentially removed via excision repair (Burns et al., 1986). Whole-genome sequencing of sorghum revealed no other obvious pattern, suggesting that EMS-induced mutation is largely random in this species (Supplemental Figure 6). This conclusion is also supported by the nearly uniform distribution of mutations in all regions of the 10 sorghum chromosomes (Figure 4).
Figure 4.
Genome-Wide Distribution of EMS Mutations.
(A) Genomic coverage (sequencing depth) of the 252 sequenced lines.
(B) Mutation (SNP) density.
(C) Gene density.
(D) Karyotype.
DNA methylation in sorghum did not appear to reduce the frequency of EMS mutation, as it does in rice (Henry et al., 2014). DNA methylation is distributed in a characteristic pattern across the chromosome, with more methylation in sequences closer to the centromere; by contrast, our EMS-induced mutations were mostly uniformly distributed (Figure 4). We evaluated the methylation level at EMS-induced mutation sites using previously published sorghum shoot DNA bisulfite sequencing data (Olson et al., 2013). The levels of methylation at both CpG and CHG (where H is A, C, or T) around EMS-induced mutation sites were higher than the average methylation level in the whole genome (Supplemental Figure 7A). In exon regions, methylation levels at CpG and CHG at EMS mutation sites were almost double the average level in the exome (Supplemental Figure 7B). This result indicated either that EMS prefers to alkylate G residues in regions with higher methylation rates or that a high methylation rate hinders the mechanisms that repair EMS-induced mutations in high-methylation regions (Burns et al., 1986).
DISCUSSION
Genetic variation, due to either natural or induced mutations, is the raw material for plant breeding. Recent studies showed that genetic diversity within cultivated crop varieties has been reduced by artificial selection during plant breeding (Fu and Somers, 2009). Consequently, mutagenesis represents an important source of novel variations for use in breeding. For example, EMS mutagenesis has been successfully applied to breeding of salt-tolerant rice (Takagi et al., 2015). We identified novel traits from our mutant library, such as msd and erl mutants, that could also be applied to sorghum improvement (Burow et al., 2014; Xin et al., 2015). We compared the overlap of EMS-induced mutations with natural variations discovered by genotyping-by-sequencing and whole-genome sequencing in a total of 1016 sorghum lines (Mace et al., 2013; Morris et al., 2013). Only 6659 of 264,978 EMS-induced mutations in the corresponding region were present as natural variations (Supplemental Figure 4B). This observation implies that 97.5% of EMS-induced mutations from the sorghum mutant library are novel variations that could be explored to accelerate sorghum breeding to improve grain and biomass production.
A unique advantage of our mutant resource is that it provides the opportunity for swift integration of phenotypic data with the massive and detailed genomic sequence. As demonstrated here, functional genes and allelic series that control important agronomic traits (such as epicuticular wax production) can be identified and validated in mutant lines within a reasonably short period. Our cosegregation analyses of candidate genes (Sbcer5 and Sbcer6) for the bloomless trait in the M4-F2 population demonstrated that the mutant population could be used to resolve two independently segregating genes that affect a single trait. Thus, a translational genomics approach could be replicated with ease for candidate genes from model species such as Arabidopsis, especially for those involved in known biochemical pathways. The efficient validation of known genes in highly conserved pathways is an exciting and valuable application of the sorghum sequenced mutant resource.
In summary, we established a pedigreed mutant library in a sorghum inbred line, BTx623, that was used to generate the reference genome sequence. We sequenced a selection of 256 lines to an average of 16.4x coverage of the whole genome. The availability of this database will enable reverse genetics (in silico TILLING) to be conducted for a majority of the genes in sorghum. If additional alleles are needed for particular genes of interest, TILLING can be applied to an additional >6000 lines for which high-quality genomic DNA has been prepared. We demonstrated, through both simple genetics (blm) and in silico analysis of candidate genes for QTLs, that this mutation database can be used to efficiently identify causal mutations. We believe that other traits could be analyzed with similar levels of success. Fast and efficient discovery of causal genes for agronomically useful phenotypes will accelerate the improvements of not just sorghum, but for other grasses in which there are limited avenues of forward and genetic research techniques due to genome complexity.
METHODS
EMS Treatment
EMS treatment was performed as described (Xin et al., 2008). Briefly, dry BTx623 seeds in batches of 100 g (∼3300 seeds) were soaked with agitation for 16 h at 50 rpm on a rotary shaker in 200 mL of tap water containing 0.1 to 0.3% EMS (v/v). The treated seeds were thoroughly washed in ∼400 mL of tap water for 5 h at ambient temperature; wash water was changed every 30 min. Mutagenized seeds were air-dried and prepared for planting. The air-dried seeds were planted at a density of 120,000 seeds per hectare. Before anthesis, each panicle was covered with a 400-weight rainproof paper pollination bag (Lawson Bags) to prevent cross-pollination. Each bag was injected with 5 mL Chlorpyrifos (Dow AgroSciences) at 0.5 mL/L to control maize (Zea mays) earworms that might hatch within the bag and destroy the seeds. Sorghum (Sorghum bicolor) panicles were harvested manually and threshed individually, and M2 seeds were planted as one row per head. To ensure high mutagenesis efficiency, only panicles that set 10% or fewer seeds were allowed to propagate. Three panicles from each M2 head row were bagged before anthesis. Only one fertile panicle was used to produce the M3 seeds. For DNA extraction, duplicate leaf samples were collected from the same fertile plant, and both the leaf samples and the panicle were barcoded. Seeds from the barcoded M2 plants were harvested as M3 families of seeds. For phenotypic evaluation, each M3 family of seeds was planted as one row in the field. Many of the mutant lines exhibited diminished seed production during the M3 generation. Thus, 10 panicles were bagged for each M3 head row and pooled as M4 seeds, which will be distributed to end users upon request.
DNA Sample Preparation
Genomic DNA was prepared by a CTAB-based method (Xin and Chen, 2012). Because many lines failed to produce M3 seeds, and genomic DNA prepared from the original M2 plants was stored for several years, we extracted fresh genomic DNA from pooled M3 plants to obtain high-quality fresh DNA. To capture most of the mutations that existed in the original M2 plants, we pooled young leaf tissue samples from 20 individual plants for each M3 family. The pooled leaf samples were lyophilized for 2 d in a Labconco freeze dryer and used to prepare genomic DNA.
Variation Detection and Function Prediction
Sequencing was performed on Illumina HiSeq 2000 sequencing system by Beckman Genomic Services, acquired by Genewiz (https://www.genewiz.com). All reads were first aligned to the sorghum reference genome v2.1 (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Sbicolor) using Bowtie2 (Langmead and Salzberg, 2012). The alignment files were converted to BAM format and sorted using Samtools (Li et al., 2009). Picard (http://broadinstitute.github.io/picard/) was used to filter duplicated reads during library construction and sequencing. Variation calling was performed with Samtools and Bcftools (Li et al., 2009) using unique reads with both sequencing and alignment quality >20. Alleles present in more than two genotypes in the population were discarded. After removing all SNPs detected in BTx623, high-confidence SNPs were selected using the following criteria: (1) the SNP is supported by at least three reads or two complementary reads; (2) the mutation is detected in no more than five lines with an allele frequency ≤5%; and (3) the sequence change is GC→AT.
To validate a SNP, primers were designed based on the 2 kb of flanking genomic DNA sequence. The “eprimer3” module from EMBOSS (Rice et al., 2000) was used to find the primers with the following parameters: “-target 600,700 -psizeopt 600 -prang [500-700].” The copy number of primers was checked by searching the whole-genome sequence using BLAST (Altschul et al., 1990). Only single-copy primers were used for Sanger sequencing. The Sanger signals were converted to sequences using the SangerR (Poly Peak Parser) package (Hill et al., 2014). Heterozygous SNPs were identified based on ratio of peaks between two alternative nucleotides (Supplemental Figure 8): If the second-strongest peak signal was greater than one-third of the strongest peak signal, the SNP was scored as heterozygous (Hill et al., 2014). Amplicon sequences were aligned to the sorghum reference genome and then variations were called using the MUMMER package (Kurtz et al., 2004).
Functional annotation of SNPs was performed using SnpEff (Cingolani et al., 2012) based on the sorghum genome annotation v2.1. SIFT 4G was used to predict deleterious mutations (Vaser et al., 2016).
Characterization of Sorghum EMS-Induced SNPs
A sliding-window method (100 kb window; 10-kb step size) was used to determine the whole-genome distribution of EMS SNPs. For methylation analysis, published sorghum shoot bisulfite sequencing data were used (Olson et al., 2013). All reads were aligned to the sorghum reference genome V2.1 using Rmap (Smith et al., 2008), allowing a maximum of four mismatches. Only sites with more than five unique mapped reads were used to estimate the methylation level.
To determine the novelty of EMS-induced SNPs from natural variations annotated based on published literature (Mace et al., 2013; Morris et al., 2013), it is necessary to ensure that the EMS-induced SNPs and natural variations are at the same locations. Because all natural variations have been analyzed using the v1.4 sorghum genome annotation, the genes from the v1.4 annotation were first aligned to the v2.1 assembly using BLAST (Altschul et al., 1990). Only genes with a 100% match to the same chromosome from the v2.1 genome assembly were used for the SNP comparison. EMS-induced SNPs were compared with two natural variation sets available from the Gramene database (Monaco et al., 2014): The first population consists of 265,000 SNPs from 971 worldwide accessions, assayed by genotyping-by-sequencing (Morris et al., 2013); the second population consists of six million SNPs, assayed by whole-genome sequencing of 45 S. bicolor and two S. propinquum lines (Mace et al., 2013). The coordination of SNPs within the mapped genes was converted from the v1.4 assembly to v2.1 using a script written in house. Overlaps between VCF files were detected using BEDTools (Quinlan and Hall, 2010).
Phenotyping of Epicuticular Wax Mutants and Gas Chromatography
The 256 mutants were planted in the field for visual inspection in two consecutive years (2013 and 2014) and in a greenhouse (2015) at the experimental facilities of the Cropping Systems Research Laboratory, USDA-ARS. Mutant lines that were devoid of epicuticular wax (EW) were isolated, photographed, and subjected to EW analysis. EW from leaf and stem was extracted by dipping a cut sample of ∼6 cm2 in 6 mL of hexane for 60 s. Samples were dried under nitrogen, subjected to esterification using the MIDI protocol, dried again, and reconstituted in hexane containing internal standard (http://www.midi-inc.com). Wax analysis was performed on a gas chromatograph coupled with a flame ionization detector.
Cosegregation Test of the Bloomless Mutant Population
An M4 population of ARS20 obtained from M3 plants (with heterogynous mutation in both cer5 and cer6) was used as an F2 population to analyze the cosegregation between mutation and phenotype. Eighty M4 seeds of ARS20 were planted in the polyhouse under optimal conditions (12-h/12-h day-night light regime with temperatures of 24/20°C, day-night conditions; 50% RH) and provided with automatic irrigation. After 1 week, 72 seeds out of 80 germinated and were used for the study. Individual M4 (F2) plants were tagged and the bloom/bloomless phenotype was scored after 30 d for each of the lines in the population and continuously monitored throughout the plant cycle. Bloom plants produced profuse white EW in the stem, while bloomless plants are devoid of EW with shiny green stem color specifically at the four-leaf stage (30 d after planting).
Genomic DNA was extracted from each of the individual F2s using the CTAB method referenced above. Allele-specific genotyping was performed by endpoint analysis using Kompetitive Allele-Specific PCR (KASP) chemistry (LGC Genomics). In-house primers were designed and synthesized for the mutations in Sobic.009G083300 (SbCER5) and Sobic.001G453200 (SbCER6) and two negative control loci (Sobic.004G086300 and Sobic.006G127800) (Supplemental Table 3). The PCR conditions for KASP analysis of sorghum were adapted from Chopra et al. (2015). χ2 analyses were performed to assess the goodness of fit between the observed and expected values of the segregation ratio.
In Silico Analysis of QTLs and Mutations in Seed Size
Genes associated via bi-parental mapping or GWAS with QTLs for seed characteristics were identified in rice (Oryza sativa), maize, and sorghum. These genes were further assessed for the presence of mutations in whole-genome shotgun sequences of the sequenced lines. Seed samples from 63 selected mutant lines and BTx623 (wild type), grown in two successive years, were obtained based on whether a specific line harbored a mutation or SNPs in previously reported seed characteristic-related QTLs for sorghum, rice, or maize. To determine variations in seed diameter, the total harvested seed lot from each selected line was passed through a series of three sieves with 3.35 mm (#5), 2.8 mm (#6), and 2.45 mm (#7) screens. Percentages of total kernels retained in each screen were recorded, and average TKM was calculated for each of the selected mutant lines. To assess SNPs associated with seed size, different seed diameter classes and total kernel weight were statistically compared with those of the wild type.
Accession Numbers
All sequencing data have been deposited in the Sequence Read Archive (SRA) database under accession number SRP063947. The mutation data can be found at the Gramene database (http://ensembl.gramene.org/Sorghum_bicolor/Info/Index).
Supplemental Data
Supplemental Figure 1. Average sequencing depth and genome coverage in 256 samples.
Supplemental Figure 2. SNP discovery rate at various sequencing depths.
Supplemental Figure 3. Allele frequency of BTx623 SNPs and the NON-BTx623 SNPs in the 256 samples.
Supplemental Figure 4. Detection of two contaminating lines.
Supplemental Figure 5. Detection of sibling lines.
Supplemental Figure 6. Distribution of flanking nucleotides.
Supplemental Figure 7. Methylation profiling of EMS mutation.
Supplemental Figure 8. Validation of heterozygous mutations.
Supplemental Table 1. SNP validation rates at various sequencing depths.
Supplemental Table 2. Mutations associated with seed-size QTLs.
Supplemental Table 3. Primer sequences used for genotyping F2 population of ARS20.
Supplemental Data Set 1. Mutant phenotypes observed in each of the 256 sequenced lines.
Supplemental Data Set 2. Sequencing summary of 256 mutants.
Supplemental Data Set 3. Number of SNPs and mutation rate in each line.
Supplemental Data Set 4. Primers designed for SNP validation by Sanger sequencing.
Supplemental Data Set 5. Disruptive SNPs in each line.
Supplementary Material
Acknowledgments
We thank Lan Liu-Gitz and Halee Hughes (USDA-ARS) for technical support and Veronica Acosta-Martinez (USDA-ARS) for technical assistance with GC analysis of waxes. The project was funded by ARS in-house project 6208-21000-020-00D and the United Sorghum Checkoff Program. We also acknowledge the funding from National Science Foundation IOS 1127112 (Y.J. and B.W.) and USDA-ARS 1907-21000-030-00D (D.W.) for data analysis. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
AUTHOR CONTRIBUTIONS
Z.X., J.B., and D.W. conceived the idea. Y.J. and Z.X. drafted the manuscript. Z.X. selected the mutant lines and prepared the genomic DNA. Y.J. and B.W. analyzed the data. R.C. and G.B. conducted reverse genetics and integrated genomics analyses of the bloomless and seed size mutants and genes. J.C., G.B., C.H., and Y.E. assisted with phenotyping at various stages. All authors edited and agreed on the final manuscript.
Glossary
- SNP
single-nucleotide polymorphism
- SIFT
sort intolerant from tolerant
- VLCF
very-long-chain fatty acid
- QTL
quantitative trait locus
- TKM
thousand-kernel mass
- EW
epicuticular wax
Footnotes
Articles can be viewed without a subscription.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Assefa Y., Staggenborg S.A. (2011). Phenotypic changes in grain sorghum over the last five decades. J. Agron. Crop Sci. 197: 249–257. [Google Scholar]
- Blomstedt C.K., Gleadow R.M., O’Donnell N., Naur P., Jensen K., Laursen T., Olsen C.E., Stuart P., Hamill J.D., Møller B.L., Neale A.D. (2012). A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant Biotechnol. J. 10: 54–66. [DOI] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33: 41–52. [DOI] [PubMed] [Google Scholar]
- Burns P.A., Allen F.L., Glickman B.W. (1986). DNA sequence analysis of mutagenicity and site specificity of ethyl methanesulfonate in Uvr+ and UvrB- strains of Escherichia coli. Genetics 113: 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow G., Xin Z., Hayes C., Burke J. (2014). Characterization of a multiseeded mutant of Sorghum for increasing grain yield. Crop Sci. 54: 2030–2037. [Google Scholar]
- Burow G.B., Franks C.D., Xin Z. (2008). Genetic and physiological analysis of an irradiated bloomless mutant (epicuticular wax mutant) of sorghum. Crop Sci. 48: 41–48. [Google Scholar]
- Chopra R., Burow G., Hayes C., Emendack Y., Xin Z., Burke J. (2015). Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genomics 16: 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Siqueira Ferreira S., Nishiyama M.Y. Jr., Paterson A.H., Souza G.M. (2013). Biofuel and energy crops: high-yield Saccharinae take center stage in the post-genomics era. Genome Biol. 14: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D.N., Cassman K.G. (1999). Post-Green Revolution trends in yield potential of temperate maize in the North-Central United States. Crop Sci. 39: 1622–1630. [Google Scholar]
- Ellegren H., Sheldon B.C. (2008). Genetic basis of fitness differences in natural populations. Nature 452: 169–175. [DOI] [PubMed] [Google Scholar]
- Feltus F.A., Hart G.E., Schertz K.F., Casa A.M., Kresovich S., Abraham S., Klein P.E., Brown P.J., Paterson A.H. (2006). Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor. Appl. Genet. 112: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Fu Y.-B., Somers D.J. (2009). Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 49: 161–168. [Google Scholar]
- Gilchrist E.J., O’Neil N.J., Rose A.M., Zetka M.C., Haughn G.W. (2006). TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics 7: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Comai L., Henikoff S. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam T.M., Mañas-Fernández A., Zhao L., Kunst L. (2012). Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160: 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Nagalakshmi U., Lieberman M.C., Ngo K.J., Krasileva K.V., Vasquez-Gross H., Akhunova A., Akhunov E., Dubcovsky J., Tai T.H., Comai L. (2014). Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26: 1382–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.T., Demarest B.L., Bisgrove B.W., Su Y.C., Smith M., Yost H.J. (2014). Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 243: 1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson T.R., Chase M.W., Renvoize S.A. (2002). Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Ann. Bot. (Lond.) 89: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.E., Langdale J.A., Kelly S. (2014). The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 24: 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannoo N., Grivet L., Chantret N., Garsmeur O., Glaszmann J.C., Arruda P., D’Hont A. (2007). Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 50: 574–585. [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Vermerris W., Gallo M., Fedenko J.R., Erickson J.E., Altpeter F. (2013). RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol. J. 11: 709–716. [DOI] [PubMed] [Google Scholar]
- Krishnan A., et al. (2009). Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 149: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krothapalli K., Buescher E.M., Li X., Brown E., Chapple C., Dilkes B.P., Tuinstra M.R. (2013). Forward genetics by genome sequencing reveals that rapid cyanide release deters insect herbivory of Sorghum bicolor. Genetics 195: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace E.S., et al. (2013). Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4: 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., et al. (1998). Detailed alignment of saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150: 1663–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco M.K., et al. (2014). Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 42: D1193–D1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.P., et al. (2013). Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 110: 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A., Klein R.R., Dugas D.V., Lu Z., Regulski M., Klein P.E., Ware D. (2013). Expanding and vetting sorghum bicolor gene annotations through transcriptome and methylome sequencing. Plant Genome 10: 3835. [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Van de Peer Y., Vandepoele K. (2005). Ancient duplication of cereal genomes. New Phytol. 165: 658–661. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Lin Y.-R., Li Z., Schertz K.F., Doebley J.F., Pinson S.R., Liu S.-C., Stansel J.W., Irvine J.E. (1995). Convergent domestication of cereal crops by independent mutations at corresponding genetic Loci. Science 269: 1714–1718. [DOI] [PubMed] [Google Scholar]
- Pighin J.A., Zheng H., Balakshin L.J., Goodman I.P., Western T.L., Jetter R., Kunst L., Samuels A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704. [DOI] [PubMed] [Google Scholar]
- Premachandran M., Prathima P., Lekshmi M. (2011). Sugarcane and polyploidy: a review. J. Sugarcane Res. 1: 1–15. [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rocateli A.C., Raper R.L., Balkcom K.S., Arriaga F.J., Bransby D.I. (2012). Biomass sorghum production and components under different irrigation/tillage systems for the southeastern U.S. Ind. Crops Prod. 36: 589–598. [Google Scholar]
- Rooney W.L., Blumenthal J., Bean B., Mullet J.E. (2007). Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod. Biorefin. 1: 147–157. [Google Scholar]
- Saballos A., Sattler S.E., Sanchez E., Foster T.P., Xin Z., Kang C., Pedersen J.F., Vermerris W. (2012). Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70: 818–830. [DOI] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707. [DOI] [PubMed] [Google Scholar]
- Sattler S.E., Saballos A., Xin Z., Funnell-Harris D.L., Vermerris W., Pedersen J.F. (2014). Characterization of novel Sorghum brown midrib mutants from an EMS-mutagenized population. G3 (Bethesda) 4: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S.E., Palmer N.A., Saballos A., Greene A.M., Xin Z., Sarath G., Vermerris W., Pedersen J.F. (2012). Identification and characterization of four missense mutations in Brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. BioEnergy Res. 5: 855–865. [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Settles A.M., et al. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. (2014). Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Smith A.D., Xuan Z., Zhang M.Q. (2008). Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics 9: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47: 1489–1493. [DOI] [PubMed] [Google Scholar]
- Srinivas G., Satish K., Madhusudhana R., Reddy R.N., Mohan S.M., Seetharama N. (2009). Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor. Appl. Genet. 118: 1439–1454. [DOI] [PubMed] [Google Scholar]
- Swigonová Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., et al. (2015). MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 33: 445–449. [DOI] [PubMed] [Google Scholar]
- Thompson O., et al. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. (2016). SIFT missense predictions for genomes. Nat. Protoc. 11: 1–9. [DOI] [PubMed] [Google Scholar]
- Winkler S., Schwabedissen A., Backasch D., Bökel C., Seidel C., Bönisch S., Fürthauer M., Kuhrs A., Cobreros L., Brand M., González-Gaitán M. (2005). Target-selected mutant screen by TILLING in Drosophila. Genome Res. 15: 718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Chen J. (2012). A high throughput DNA extraction method with high yield and quality. Plant Methods 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Wang M., Burow G., Burke J. (2009). An induced sorghum mutant population suitable for bioenergy research. BioEnergy Res. 2: 10–16. [Google Scholar]
- Xin Z., Gitz D., Burow G., Hayes C., Burke J.J. (2015). Registration of two allelic erect leaf mutants of sorghum. J. Plant Regist. 9: 254–257. [Google Scholar]
- Xin Z., Wang M.L., Barkley N.A., Burow G., Franks C., Pederson G., Burke J. (2008). Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li J., Compton R.O., Robertson J., Goff V.H., Epps E., Kong W., Kim C., Paterson A.H. (2015). Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae). G3 (Bethesda) 5: 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.