Abstract
Cell-based, high-throughput screening (HTS) assays are increasingly important tools used in drug discovery, but frequently rely on readouts of gene expression or phenotypic changes and require development of specialized, labeled reporters. Here we introduce a cell-based, label-free assay compatible with HTS that can report quantitatively on enzyme activities by measuring mass changes of substrates with MALDI-mass spectrometry. The assay uses self-assembled monolayers to culture cells on arrays presenting substrates, which serve as reporters for a desired enzyme activity. Each spot of cells is treated with a compound, cultured and lysed, enabling endogenous enzymes to act on the immobilized peptide substrate. We demonstrate that the assay can measure protein tyrosine phosphatase (PTP) activity from as few as five cells and we describe a screen that identified a compound that reduces PTP activity in cell lysates. This approach offers a valuable addition to the methods available for cell-based screening.
Keywords: self-assembled monolayers, analytical methods, high-content screening, bioassays
Graphical Abstract
A cell-based, enzyme activity assay compatible with high-throughput screening is described in which cells are cultured on arrays of self-assembled monolayers presenting enzyme substrates. Cell lysis on the chip enables enzymes to modify the immobilized substrates, changing their mass, which is measured by MALDI mass spectrometry, detecting activity from as few as five cells.
1. Introduction
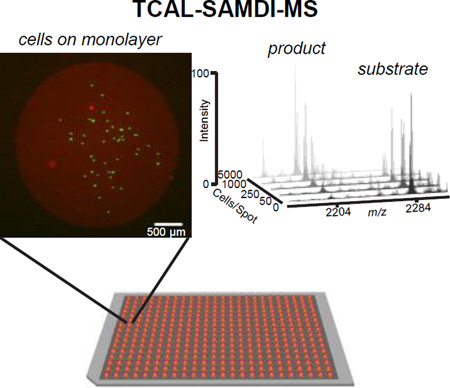
Cell-based assays are finding increasing use in modern drug discovery screens because they enable the concomitant evaluation of compound permeability, toxicity and activity within a more physiologically relevant cellular environment[1]. However, cell-based assays that measure the activities of specific enzymes can be substantially more difficult to implement than biochemical assays. The common strategies for measuring enzyme activities—including those based on absorbance, fluorescence and radioactivity—often require reagents that cannot be delivered to the appropriate cellular compartment or are not compatible with the cellular environment. In this paper, we describe a method that combines cell lysis with a label-free assay of enzyme activities in the lysate. The assay uses self-assembled monolayers for MALDI-mass spectrometry (SAMDI)[2], where the monolayers are engineered to present enzyme substrates together with a peptide that supports cell adhesion on the assay chip. In this way, lysis of a population of cells occurs in the presence of peptide substrates that record the activity of a defined enzyme. This approach, termed Tandem Culture and Lysis-SAMDI (TCAL-SAMDI) provides a general method for conducting cell-based, chemical screening with quantitative readouts of enzymatic activity, easily adaptable to a wide range of targets.
Most cell-based screens use gene expression or phenotypic changes as a readout and require a labeled reporter in addition to compatibility with automated data acquisition and analysis methods. Gene expression reporter systems, such as β-lactamase paired with fluorescence resonance energy transfer (FRET)[3], fluorogenic[4], or chromogenic[5] substrates, have been of significant value in cell-based screening[6]. Protein and other biomolecule labeling methods, including genetic encoding of fusion proteins incorporating fluorescent proteins[7], chromophoric, fluorescent, and immuno-labeling[8], have been used to visualize protein expression[9], localization[10], and translocation between cellular compartments[11]. High content screens (with automated image acquisition and analysis) using these methods have been used to identify compounds that produce desired molecular and phenotypic changes[12].
Our previous work has developed SAMDI mass spectrometry as a label-free assay for measuring enzyme activities[13]. In SAMDI, an enzyme substrate is immobilized to a self-assembled monolayer presenting tri(ethylene glycol) groups. The substrate can be immobilized through a variety of chemical reactions and the glycol groups serve the important role of preventing non-specific adsorption of proteins to the surface, giving a more quantitative measure of activity. Further, these monolayers are well-suited for analysis by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) because irradiation of the monolayer with a laser results in dissociation of the thiolate-gold bond and release of the intact alkanethiolates. In this way, treatment of the immobilized peptide with an enzyme that can modify its structure will result in a change in mass of the peptide-alkanethiolate conjugate, which can be directly observed in the SAMDI spectrum.
In this paper, we demonstrate a strategy for analyzing lysates from small numbers of cells, which relies on culturing cells on a monolayer that presents a peptide for cell adhesion together with a peptide substrate to report on a desired enzyme activity. In this way, cells can be cultured on the monolayer and lysed in place, where enzymes in the lysate can directly and immediately act on the immobilized substrates. The monolayer is then rinsed and analyzed by SAMDI mass spectrometry to quantitate the amount of product. We apply this method in a 384-array format for measuring both protein tyrosine phosphatase (PTP) and caspase-3 activities and we also show how this platform was to perform a cell-based screen to identify modulators of PTP activity.
2. Results
2.1. TCAL-SAMDI assay of phosphatase activity
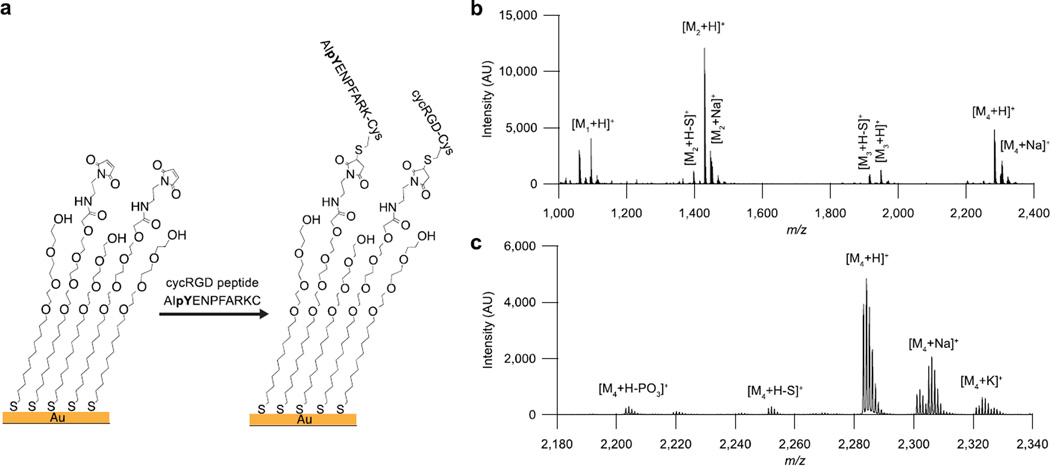
We first prepared an array of monolayers to measure PTP activity in HeLa cell cultures. Each monolayer was composed of alkanethiolates terminated with maleimide groups at a density of 10% relative to total alkanethiolates, against a background of tri(ethylene glycol) groups (Figure 1a). We applied a solution containing a cyclic peptide having the cell adhesion RGD motif together with a peptide having a phosphorylated tyrosine residue (AIpYENPFARKC)[14] to report on phosphatase activity, and the peptides were immobilized onto the monolayer. The RGD motif is found in fibronectin[15] and mediates cell adhesion and spreading by binding to integrin receptors[16]. The monolayers were formed on a stainless steel plate having an array of gold spots (2.8-mm diameter) positioned to match a 384-well plate format, as recently described[17]. The area surrounding each spot consisted of a thin layer of evaporated titanium with a monolayer of hexadecylphosphonic acid formed on the titanium dioxide to render the outer area hydrophobic, enabling droplets to be isolated on the monolayer-coated gold spots. HeLa or MDA-MB-231 cells were plated and cultured on the monolayers in individual volumes of media (3 µL) that were isolated on each spot (Figure 2a). After two hours in culture, the media was rapidly removed from all spots with a robotic liquid-handling instrument and immediately replaced with lysis buffer (1 µL). After incubating the plates with lysis buffer for 1 hr, the plates were rinsed with water and then ethanol and analyzed by mass spectrometry.
Figure 1.
SAMDI on monolayers with two peptides. (a) Two peptides with terminal cysteines (a cell adhesion ligand (cyclic RGD peptide – cycRGD) and an phosphatase substrate (AIpYENPFARKC)) are immobilized on alkanethiolate monolayers with 10% of the molecules presenting maleimides and 90% terminated with tri(ethylene glycol). (b) SAMDI spectrum of a monolayer with immobilized RGD peptide and phosphatase peptide. M1: alkanethiolate with RGD, M2: alkyldisulfide with RGD, M3: alkanethiolate with phosphatase peptide, M4: alkyldisulfide with phosphatase peptide (c) The same spectrum, showing the portion of the spectrum used for PTP activity analysis.
Figure 2.
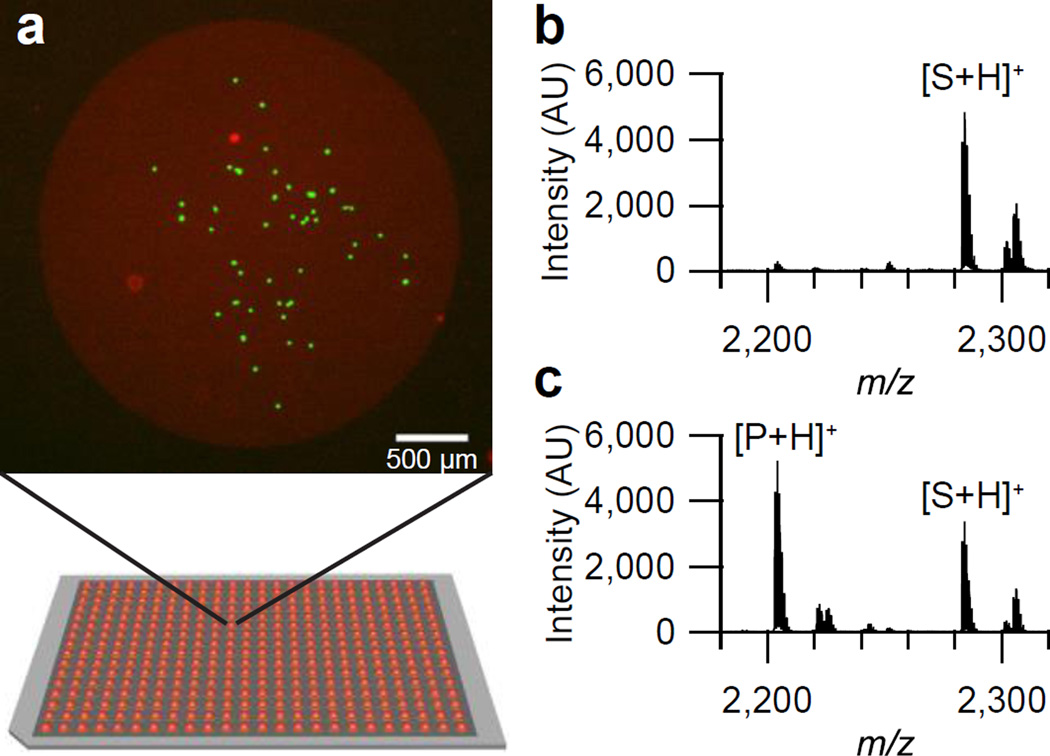
Tandem Culture and Lysis SAMDI (TCAL-SAMDI). (a) Cells (MDA-MB-231) are cultured on monolayers presenting both cell adhesion ligands and a phosphatase substrate, on a 384-spot plate. Green: live cells, Red: dead cells and gold spot. (b) A SAMDI spectrum from a spot without cells. (c) A SAMDI spectrum after lysis of 50 cells.
Prior to analysis, the plates were treated with 2,4,6-trihydroxyacetophenone (THAP) matrix and MALDI spectra were collected for each monolayer island. Performing MALDI on spots without cells produced spectra with peaks at mass-to-charge (m/z) values corresponding to the peptide-alkanethiolate conjugates as well as disulfides formed between a peptide-modified alkanethiolate and a background tri-ethylene glycol-presenting alkanethiolate, along with Na+ adduct peaks (Figure 1b,c). After lysis of cells on the spots, dephosphorylation of the peptide substrate resulted in the formation of a product peak with a mass shift of −80 Da (Figure 2b). We confirmed that the peak at −80 Da relative to the substrate peak is the product peak by performing the same assay with a substrate of a different mass, in which case no peak appeared at the mass corresponding to the original product peak, but rather, at −80 Da relative to the new substrate.
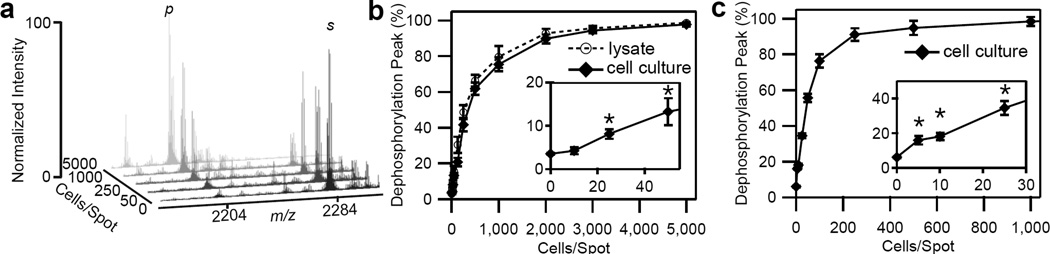
We observed that as the number of cells cultured on a spot increased, the relative intensity of the product peak grew larger while that for the substrate peak diminished (Figure 3a). The extent of phosphatase activity was determined by measuring the area under the curve for the product peak and dividing by the sum of areas for the substrate and product peaks. Because of differences in ionization efficiencies between the phosphorylated and dephosphorylated molecules, to calculate product yield, it would be necessary to scale the observed dephosphorylation peak fractions using a calibration curve as shown in Figure S1. We observed that the dephosphorylation peak fraction increased with HeLa cell number, before plateauing near 2,000 cells per spot. (Figure 3b). With this method, we were able to measure phosphatase activity from as few as 25 cells per spot (Figure 3b). With MDA-MB-231 cell cultures, we measured PTP activity from only 5 cells per spot (Figure 3c).
Figure 3.
Enzyme activity measurement with TCAL-SAMDI. (a) SAMDI-MS spectra showing the conversion of substrate (s) to product (p) as the number of HeLa cells cultured and lysed on monolayer-coated gold spots increases. Quantification of the dephosphorylation peak fraction, defined as the area under the curve of the product peak relative to that of the substrate and product peaks in the SAMDI spectra, resulting from culturing and lysing (b) HeLa cells and (c) MDA-MB-231 cells on monolayers presenting adhesion and substrate peptides. Insets in (b) and (c) are magnified regions of the graphs. (*: P < 0.01).
To determine if culturing cells on the monolayer would interfere with enzyme activity on immobilized substrates, we also measured phosphatase activity from previously prepared HeLa cell lysates. Here, HeLa cells were not cultured on the monolayers but rather lysed and then applied to the monolayers to measure PTP activity (Figure 3b, dashed line). Comparing these two methods revealed that the activity measured with TCAL-SAMDI (Figure 3b, black line) was not significantly different at most cell concentrations from the activity measured from cell lysates (Figure 3b, dashed line). This result demonstrates that culturing and lysing cells directly on the surface did not interfere with the ability of enzymes to act on the immobilized substrate or the ability to perform SAMDI on these surfaces.
2.2 Duplexing enzyme activity measurements with TCAL-SAMDI
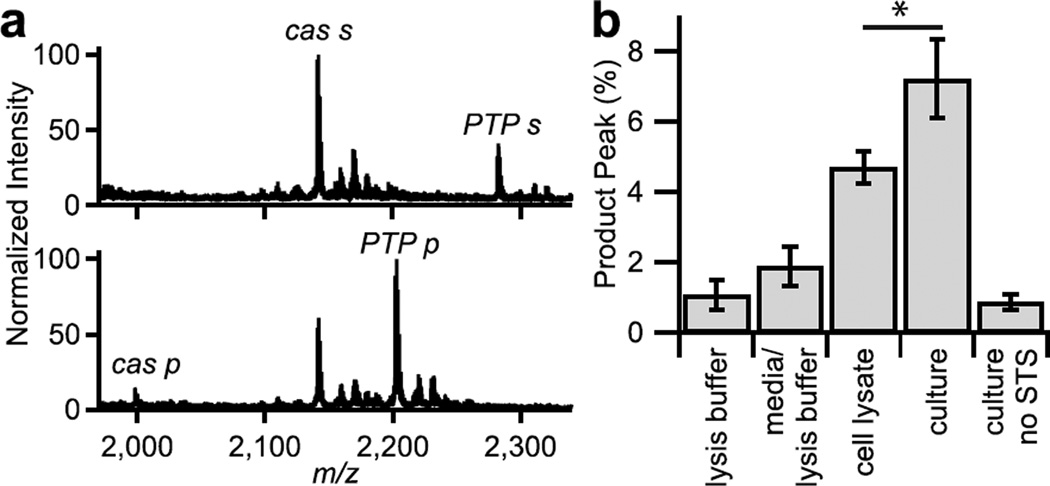
One significant benefit of mass spectrometric assays is that they are well suited to multiplexed formats[13e]. To illustrate this advantage, we prepared monolayers that had the adhesion peptide, the phosphatase substrate and a peptide substrate for the protease caspase-3 (CGKRKGDEVDSG)[13g]. We cultured HeLa cells on monolayers presenting these three peptides for one hour and then induced apoptosis by adding staurosporine to the medium[18]. After four hours of treatment with staurosporine, the cells were lysed as described above and the monolayers were similarly analyzed by SAMDI mass spectrometry. The mass spectra clearly show the conversion of both substrates to their corresponding products (Figure 4a), demonstrating the ability to duplex activity measurements with TCAL-SAMDI. In addition to peak for the dephosphorylated peptide, we also observed a peak corresponding to the caspase-3 product at 144 Da lower than the initial substrate mass (Figure 4a). Monolayers treated with lysis buffer and media, as well as monolayers with cells but without staurospaurine show a lack of the caspase-3 product (Figure 4b). Interestingly, the conversion of substrate to product was significantly greater when cells were lysed directly on the surface, compared to the conversion observed from applying the cell lysate to the monolayer. This benefit of the TCAL assay may reflect a loss of activity of the enzyme activity that accumulates in time.
Figure 4.
Duplexing enzyme activity measurements with TCAL-SAMDI. (a) SAMDI spectra of a spot with no cells (top) and with 10,000 cells (bottom) shows the conversion of two substrates (PTP s: PTP substrate; cas s: caspase-3 substrate) to their products when cells are cultured, treated with STS, and lysed on the surface. (b) Caspase-3 activity measured by SAMDI (*: P < 0.05).
2.3 Screening with TCAL-SAMDI
We next describe a screen where we evaluated 10,240 small molecules in MBD-231 cells to identify those that modulate phosphatase activity. We found that cultures having 100–150 cells/spot resulted in approximately equal-sized peaks for the substrate and product peptides in the SAMDI spectra. This small number of cells required in the assay means that the entire screen could be performed with just one million cells. We also measured the Z’-factor, a commonly used statistical measure of assay performance, and found a Z’-factor of 0.66 (see Experimental Section for additional details)[19].
A primary screen was carried out by applying one compound per spot, using approximately 30 plates for the entire screen. After culturing cells on the chips for 2 h, solutions of each compound (1 µL) were added to the media (3 µL) on each spot on the array so that the resulting compound concentration was 10 µM with 1% DMSO. The cells were incubated for 2 h, media was removed, lysis buffer was incubated on each spot for 1 h, and the plates were analyzed by SAMDI. The five compounds that produced the greatest inhibition of PTP activity on each plate were analyzed in a secondary screening step to verify the activity. This process identified four compounds of interest that were investigated further.
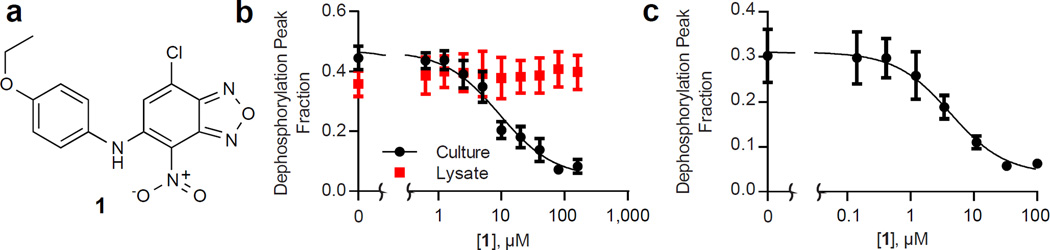
Compound 1 (Figure 5a) reduced PTP activity nearly completely with an IC50 of 9 µM (Figure 5b). However, we found that the compound did not directly inhibit the phosphatase because it was inactive when tested on cell lysates (Figure 5b). We also verified that the reduced activity was not an artifact stemming from the detachment of cells, by counting the number of cells per spot (Figure S2a). Addition of compound 1 did decrease cell viability (~30% at 100 µM of 1), but this decrease was small compared to the observed inhibition of PTP activity (Figure S2a,b). As an additional control to ensure that the reduction in PTP activity was not an artifact resulting from a loss of protein, lysates were prepared from cells treated with the compound while being cultured in standard 96-well tissue culture plates. Treatment of monolayers with these lysates confirmed that 1 reduced PTP activity, with a similar IC50 of 4.2 µM (Figure 5c), and this was not due to a decrease in protein concentration in the lysates (Figure S2c).
Figure 5.
Compound 1 identified by chemical screening via TCAL-SAMDI. (a) Chemical structure of 1. (b) SAMDI analysis of PTP activity measured from MDA-MB-231 cells cultured on the monolayer and from lysate applied to the monolayer, both treated with 1. (c) SAMDI analysis of lysate from cells cultured in 96-well plates and treated with 1.
3. Discussion
This paper demonstrates an efficient method for isolating lysates from cultured cells and assaying those lysates for enzyme activities. The efficiency of this process—which avoids the need to physically collect and manipulate the lysate—makes the method well-suited to high throughput applications comprising tens of thousands of distinct assay compositions. Here we illustrate the method with assays of phosphatase activity and we demonstrate a cell-based screen to identify molecules that regulate phosphatase activities by acting on upstream targets.
The method is enabled by two properties of the self-assembled monolayers. First, the monolayers give excellent control over the ligand-receptor interactions at the interface, allowing the surface to simultaneously mediate cell adhesion and to present a peptide that serves as a substrate to probe the desired enzyme activity. Were the surface not inert to non-specific protein adsorption—which is a common challenge with many substrates used in bioanalytical methods—the peptide substrate would be blocked from interacting with the enzymes by way of an adsorbed protein layer. Second, monolayers of alkanethiolates on gold are compatible for analysis by MALDI mass spectrometry and therefore the assays can be performed in a label-free format. This is particularly significant because it allows measurement of virtually any enzyme activity. No other surface chemistry—including common hydrogel polymer layers or alkylsiloxane monolayers—has been shown to combine these two benefits and therefore the TCAL-SAMDI method offers a new capability in bioassays.
The measurement of phosphatase activities in this work is also significant because these activities are extremely challenging to measure in cell lysates. The commonly used colorimetric assay based on p-nitrophenylphosphate (pNPP), which undergoes a shift in absorbance after dephosphorylation, is not able to discriminate between the activities of many phosphatases (acid, alkaline, protein tyrosine and serine/threonine). Assays that report on the generation of free phosphate ion that is released from a phosphopeptide of choice, such as the commonly used malachite green assay, can offer greater specificity, but are incompatible with lysate samples because of the difficulty involved in eliminating sources of phosphate present in the cell. The SAMDI assay can be used with any peptide substrate and therefore provide a more specific response on activity. Thus, there are no currently available phosphatase activity assays other than the TCAL assay that can be used to conduct a cell-based screen of protein tyrosine phosphatase activity specifically. While this alone would be valuable, this method could be applied to a broad range of enzyme activities, including glycosyltransferases, deacetylases, kinases, proteases, and others[13b–h].
An important advantage of the TCAL assay is that it does not require the lysate to be physically manipulated. At the time of the assay, the media is removed from the cell cultures and a lysis buffer is applied to the cells. No further manipulation is required because the lysate that is generated is in contact with the monolayer presenting the substrate for the relevant enzyme activity. For this reason, the TCAL method does not introduce any time delay between generating the lysate and assaying for activities; these time delays often lead to losses of enzyme activities, as does adsorption of proteins from the sample to the walls of pipettes and wells. Because the lysate is not manipulated in TCAL, it is possible to use smaller volumes of lysate and in this work we demonstrated the preparation of lysates from as few as five cells.
An important application of the TCAL method is in screening libraries of small molecules that modulate biochemical activities in cultured cells. Cell-based screens have been particularly important in those cases where a desired downstream biochemical activity is modulated—but where there exist many distinct targets that are relevant—or where a phenotypic response is desired. Further, cell-based assays have the advantage that they do not identify as hits those molecules that are cytotoxic or that are unable to cross the membrane. However, it can be challenging to develop the reagents required in a cell-based assay. For example, FRET-based reporters of kinase activities[20] have required a substantial effort to develop, and those assays have a limited quantitative resolution. In other cases, it can be difficult to deliver the reagent to the appropriate cellular compartment. The label-free SAMDI assay can be readily formatted to detect many enzyme activities. Further, the assay is performed after the cell has been lysed and therefore avoids limitations of getting a reagent into the cell.
The TCAL method provides a practical method for performing cell-based assays. Gold-coated glass substrates are already commercially available, and metal plates like those used in this work could similarly be made. All components of the monolayer are also commercially available and the required amount of chemicals needed to form the monolayer is minimal. Our 10,000 compound screen required 32 plates for the initial screen, which required approximately twenty hours to analyze by mass spectrometry. The same liquid handling robotic instruments were used for this screen as are used for typical high-throughput screens. Standard cell culture was used, though the number of cells required was very small compared to most cell-based assays. The volume of media per assay (4 µL) and amount of screening compound (0.25 nmol) was very small compared to most assays, minimizing reagent costs. A commercially available MALDI-TOF instrument was used to read the plates. Available software that analyzes the area under the peaks in the mass spectra was used to determine activity, with some post-processing in Microsoft Excel, requiring only a few hours to process the data from 32 plates. Hence, as a screening assay, the TCAL method is reasonably practical, cost-efficient and rapid.
4. Conclusion
The TCAL-SAMDI method enables a broad range of cell-based assays that have as an endpoint a biochemical activity. As such, this method removes the constraints stemming from the incompatibility of many enzyme activity assays with the cellular environment. By integrating the cell culture with the assay on the same spot, the TCAL method requires only tens of cells. Further, because the method uses 384-array plates, it can take advantage of available liquid handling and automation tools. These benefits make the TCAL assay an exciting addition to the current methods for cell-based assays.
5. Experimental Section
Reagents
PTP Inhibitor I (PTPI-I) was purchased from Santa Cruz Biotechnology and PHPS1 was obtained from Sigma Aldrich. Hexadecyl phosphonic acid and 2,4,6-trihodroxyacetophenone were also purchased from Sigma Aldrich. The 10,240-chemical library was purchased from Chembridge. Amino acids and peptide synthesis reagents were obtained from Anaspec. The phosphatase substrate (pY peptide; sequence: AIpYENPFARKC), caspase-3 substrate (CGKRKGDEVDSG), and cyclic RGD peptides were synthesized following standard solid phase peptide synthesis protocols as previously described[21]. The Presto Blue kit, calcein-AM and ethidium homodimer-1 were purchased from Life Technologies and the cell viability assays were performed following manufacturer’s instructions.
Preparation of SAMs
Custom fabricated stainless steel plates (8 × 12.3 cm) were first cleaned using hexanes, ethanol and DI water. An electron beam evaporator was used to first deposit 5 Ti (5 nm, 0.02 nm/s) onto the steel plates. The evaporator was vented to oxidize the Ti layer. Next, an aluminum mask having holes in a 384-well format was placed on top of the plate and an additional Ti (5 nm, 0.02 nm/s) were deposited followed by Au (35 nm, 0.05 nm/s). The Au-coated steel plates were soaked overnight at 4 °C in an ethanolic solution containing a 1:4 ratio of an asymmetric disulfide terminated with a maleimide group and a tri(ethylene glycol) group and a symmetric disulfide terminated with tri(ethylene glycol) groups, with a 0.5 mM total disulfide concentration. The plates were rinsed with ethanol and then immersed in a 10 mM ethanolic solution of hexadecyl phosphonic acid for 10 min. After rising with ethanol and drying under air, an automated reagent dispenser (Multidrop Combi, Thermo Scientific) was used to spot 3 µL of a peptide solution consisting of 32 µM pY peptide and 8 µM cyclic RGD in 1× PBS at pH 7.5 onto the arrayed plates. The peptide immobilization solution used for duplexed phosphatase and caspase-3 activity measurements consisted of 8 µM pY peptide, 8 µM cyclic RGD and 24 µM caspase-3 peptides. All peptide immobilization steps were carried out for 1 h at 37 °C in a humidity chamber.
TCAL-SAMDI assay for PTP and caspase-3 activity
HeLa cells and MDA-MB-231 cells were obtained from ATCC and cultured in αMEM (for HeLa cells) or high-glucose DMEM (for MDA-MB-231 cells) medium supplemented with 10% fetal bovine serum, glutamax, penicillin and streptomycin. All cells were cultured in a humidified incubator at 37 °C and CO2. Cells were trypsinized and suspended in media, and the average number of cells per µL was counted using a hemocytometer and Countess automated cell counter (Life Technologies), and cell concentrations were adjusted to seed the desired number of cells per spot in 3 µL media. Cells were cultured on the monolayers presenting RGD and peptide substrates on steel plates or glass slides for 2 h under standard growth conditions. For PTP activity assays, cells were cultured for 2 hours before addition of inhibitors, if any. For caspase-3 activity assays, 1 µL of 4 µM staurosporine (STS) was added to each spot (for a final concentration of 1 µM STS), and incubated for 4 hours. After culture, media was removed and lysis buffer (1 or 1.5 µL) was delivered manually or with an automated reagent dispenser to each spot and the lysate was allowed to react with the monolayer for 1 h at 37 °C in a humidity chamber. Lysis buffer was composed of 20 mM Tris, 136 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, pH 7.4. A protease inhibitor tablet obtained from Roche was added to the lysis buffer. For caspase activity assays, 10 mM dithiothreitol was added to the lysis buffer. The surfaces were then rinsed with DI water and ethanol, and dried with air. A 30 mg/mL solution of 2,4,6-trihydroxyacetophenone in acetone was delivered to each spot on the array and the surfaces were analyzed using an AB Sciex 5800 MALDI TOF/TOF instrument in positive reflector mode. The area under the curves for the [M + H]+ peaks of disulfides was measured with the Data Explorer software (AB Sciex). All experiments were repeated at least three times, with at least three spots per condition each time. Presented data represent the means and standard errors of all spots. For lysate experiments (see below for more detail), data represents the averages and standard errors from at least three independently prepared lysates. Statistical comparisons between mean activities were made using Student’s t-tests.
TCAL-SAMDI assay for chemical screen
A 10,240 chemical library was used to screen for phosphatase inhibitors. For the chemical screen, 100 or 150 MDA-MB-231 cells were seeded on each spot presenting cyclic RGD and pY peptide (phosphatase substrate) and cultured for 2 h. A stock solution of each compound was first prepared in DMSO then diluted in media. Each compound was delivered (1 µL) to each spot on the array to a final concentration equivalent to 10 µM and 1% DMSO. The cells were exposed to the compounds for 2 h under standard cell growth conditions. After media removal, the lysis buffer with protease inhibitor was applied to each spot independently and incubated for 1 h at 37 °C in a humidity chamber. The plates were then rinsed with water, ethanol, and dried under air. Matrix was applied prior to mass spectrometry analysis. As described above, the data was analyzed to quantify the levels of phosphatase activity and hits were ranked. The five compounds that produced the greatest inhibition of PTP activity on each plate were chosen for a secondary screen to confirm hits. The secondary screen was carried out following the same conditions stated above, except that each compound was tested on six spots instead of one. Additionally, some compounds tested in the secondary screen step were tested at 50 µM.
Evaluation of dose-dependent inhibition by TCAL-SAMDI
MDA-MB-231 cells were seeded at 75 cells per spot on monolayers presenting phosphatase peptide substrates and cyclic RGD as described above. Following cell attachment and culture for 2 h, inhibitors (1 µL solution in media) were delivered to each spot to achieve a range of final concentrations from 0 to 640 µM in media and incubated for 2 h. Following media removal, the lysis buffer with protease inhibitor was applied to each spot independently and incubated for 1 h at 37 °C in a humidified chamber. Plates were then rinsed with water, ethanol and dried. Matrix was applied prior to analysis by mass spectrometry. All experiments were carried out at least twice, with 6 spots per condition each time. Presented data represent the averages and standard errors of all spots. IC50 values and curves were determined using GraphPad prism software.
Evaluation of dose-dependent inhibition in cell lysates using SAMDI
MDA-MB-231 cells were lysed using the lysis buffer containing protease inhibitor described above to achieve the equivalent of 75 cells per 1.5 µL, after mixing with inhibitor solutions. Inhibitor solutions in lysis buffer at concentrations ranging from 0 to 640 µM were added to the lysate and 1.5 µL of the mixture was spotted on monolayers presenting cyclic RGD and phosphatase substrate. The reaction was carried out for 1 h at 37 °C in a humidified chamber. To evaluate dose-response inhibition under standard culture conditions, MDA-MB-231 cells were plated on a 96-well plate at a density of 6,400 cells per well and cultured for 2 h. Culture medium containing inhibitors at final concentrations ranging from 0 to 300 µM and 1% DMSO were delivered to each well and culture proceeded for another 2 h. The mixture containing media and inhibitor was removed from each well and centrifuged. Lysis buffer containing protease inhibitor was applied to each well in the plate and incubated for 10 min at room temperature. The lysate was collected and added to the corresponding cell pellet for each inhibitor concentration sample. The lysate (1.5 µL) was spotted on monolayers presenting RGD and phosphatase substrate. Protein concentrations were measured using a BCA assay (Santa Cruz Biotechnology), following manufacturer instructions. Sample analysis using SAMDI followed as described above. Presented data represents the averages and standard errors from at least three independently prepared lysates. IC50 values and curves were determined using GraphPad prism software.
Cell viability assays
The Presto Blue assay was performed using MDA-MB-231 cells seeded at 6,400 cells per well in a 96 well plate format. After a 2 h culture period, medium containing inhibitors at a range of concentrations from 0 to 300 µM was added to each well and incubated for 2 h. PrestoBlue reagent was added to the wells, incubated for 25 min, and fluorescence was measured using a Cytation 3 (BioTek) plate reader. For calcein-AM and ethidium homodimer-1 staining, MDA-MB-231 cells were seeded on glass slides presenting monolayers of RGD and phosphatase peptide substrate at 75 cells per spot. After 2 h of culture, 1 µL of media containing inhibitors for a final concentration of 10 µM and 80 µM was added to each spot and incubated for an additional 2 h. Media was removed and a solution of calcein-AM and ethidium homodimer-1 in PBS were delivered (3 µL) to each spot and incubated at 37 °C for 20 min. Each spot was imaged using an epifluorescent microscope and cells were counted using ImageJ Cell Counter plug-in.
Z’ factor determination
A gold-coated steel plate with monolayers arrayed in a 384-well format was used to seed 150 MDA-MB-231 cells per spot. After 2 hr culture, DMSO was added to a final concentration of 1% on 160 spots (negative controls) and PTPI-I was added to a final concentration of 300 µM, 1% DMSO, on 160 spots (positive controls). After 2 hr, the media was removed and lysis buffer applied for 1 hr at 37 °C. Sample analysis using SAMDI followed as described above. Z’ factor was calculated Equation 1,
| (1) |
where σc+ and σc− represent the standard deviations of the positive and negative controls, respectively, and µc+ and µc− represent the means of the positive and negative controls, respectively.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Adam H. Eisenberg and Michael D. Scholle for technical assistance and advice with the screen.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.a) Fox S, Farr-Jones S, Sopchak L, Boggs A, Nicely HW, Khoury R, Biros M. J. Biomol. Screen. 2006;11 doi: 10.1177/1087057106292473. [DOI] [PubMed] [Google Scholar]; b) Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Nat. Rev. Drug Discov. 2011;10 doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 2.Su J, Mrksich M. Angew. Chem. Int. Ed. Engl. 2002;41 doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]
- 3.Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng LX, Whitney M, Roemer K, Tsien RY. Science. 1998;279 doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 4.Whitney M, Rockenstein E, Cantin G, Knapp T, Zlokarnik G, Sanders P, Durick K, Craig FF, Negulescu PA. Nat. Biotechnol. 1998;16 doi: 10.1038/4302. [DOI] [PubMed] [Google Scholar]
- 5.Moore JT, Davis ST, Dev IK. Anal. Biochem. 1997;247 doi: 10.1006/abio.1997.2092. [DOI] [PubMed] [Google Scholar]
- 6.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat. Chem. Biol. 2007;3 doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 7.Tsien RY. Annu. Rev. Biochem. 1998;67 doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 8.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312 doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 9.Deshmukh VA, Tardif V, Lyssiotis CA, Green CC, Kerman B, Kim HJ, Padmanabhan K, Swoboda JG, Ahmad I, Kondo T, Gage FH, Theofilopoulos AN, Lawson BR, Schultz PG, Lairson LL. Nature. 2013;502 doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Rourke NA, Meyer T, Chandy G. Curr. Opin. Chem. Biol. 2005;9 doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Wehrman TS, Casipit CL, Gewertz NM, Blau HM. Nat. Methods. 2005;2 doi: 10.1038/nmeth771. [DOI] [PubMed] [Google Scholar]
- 12.a) Ding S, Schultz PG. Nat. Biotechnol. 2004;22 doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]; b) Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, Schultz PG. Science. 2012;336 doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]; c) Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Science. 2004;306 doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]; d) Lang P, Yeow K, Nichols A, Scheer A. Nat. Rev. Drug Discov. 2006;5 doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 13.a) Min DH, Yeo WS, Mrksich M. Anal. Chem. 2004;76 doi: 10.1021/ac049816z. [DOI] [PubMed] [Google Scholar]; b) Ban L, Pettit N, Li L, Stuparu AD, Cai L, Chen W, Guan W, Han W, Wang PG, Mrksich M. Nat. Chem. Biol. 2012;8 doi: 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gurard-Levin ZA, Kilian KA, Kim J, Bähr K, Mrksich M. ACS Chem. Biol. 2010;5 doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kuo HY, Deluca TA, Miller WM, Mrksich M. Anal. Chem. 2013;85 doi: 10.1021/ac402614x. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Min DH, Su J, Mrksich M. Angew. Chem. Int. Ed. Engl. 2004;43 doi: 10.1002/anie.200461061. [DOI] [PubMed] [Google Scholar]; f) Mrksich M. ACS Nano. 2008;2 doi: 10.1021/nn7004156. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Su J, Rajapaksha TW, Peter ME, Mrksich M. Anal. Chem. 2006;78 doi: 10.1021/ac051974i. [DOI] [PubMed] [Google Scholar]; h) Min DH, Tang WJ, Mrksich M. Nat. Biotechnol. 2004;22 doi: 10.1038/nbt973. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Liao X, Mrksich M. Langmuir. 2013;29 doi: 10.1021/la3034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruoslahti E. Annu. Rev. Cell. Dev. Biol. 1996;12 doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 16.a) Houseman BT, Mrksich M. Biomaterials. 2001;22 doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]; b) Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. J. Am. Chem. Soc. 1998;120 [Google Scholar]
- 17.a) Gurard-Levin ZA, Scholle MD, Eisenberg AH, Mrksich M. ACS Comb. Sci. 2011;13 doi: 10.1021/co2000373. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Patel K, Sherrill J, Mrksich M, Scholle MD. J. Biomol. Screen. 2015 doi: 10.1177/1087057115588512. [DOI] [PubMed] [Google Scholar]
- 18.Bernard B, Fest T, Pretet JL, Mougin C. Cell Death Differ. 2001;8 doi: 10.1038/sj.cdd.4400796. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TDY, Oldenburg KR. J. Biomol. Screen. 1999;4 doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.a) Ting AY, Kain KH, Klemke RL, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2001;98 doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang J, Ma Y, Taylor SS, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2001;98 doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilian KA, Mrksich M. Angew. Chem. Int. Ed. Engl. 2012;51 doi: 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.