Abstract
Human cancer genome studies have identified the SWI/SNF chromatin remodeling complex member ARID1A as one of the most frequently altered genes in several tumor types. Its role as an ovarian tumor suppressor has been supported in compound knockout mice. Here, we provide genetic and functional evidence that Arid1a is a bona fide mammary tumor suppressor, using the Chromosome aberrations occurring spontaneously 3 (Chaos3) mouse model of sporadic breast cancer. About 70% of mammary tumors that formed in these mice contained a spontaneous deletion removing all or part of one Arid1a allele. Restoration of Arid1a expression in a Chaos3 mammary tumor line with low Arid1a levels greatly impaired its ability to form tumors following injection into cleared mammary glands, indicating that ARID1A insufficiency is crucial for maintenance of these Trp53-proficient tumors. Transcriptome analysis of tumor cells before and after reintroduction of Arid1a expression revealed alterations in growth signaling and cell-cycle checkpoint pathways, in particular the activation of the TRP53 pathway. Consistent with the latter, Arid1a reexpression in tumor cells led to increased p21 (Cdkn1a) expression and dramatic accumulation of cells in G2 phase of the cell cycle. These results not only provide in vivo evidence for a tumor suppressive and/or maintenance role in breast cancer, but also indicate a potential opportunity for therapeutic intervention in ARID1A-deficient human breast cancer subtypes that retain one intact copy of the gene and also maintain wild-type TRP53 activity.
Keywords: Arid1a, tumor suppressor, breast cancer, cell cycle, senescence, mouse
THE identification of genetic drivers of specific types and subtypes of cancer continues to be an important goal of cancer biology research. Major efforts including The Cancer Genome Atlas (TCGA) project have cataloged mutations in diverse human tumors. This wealth of data has been instrumental in identifying genes that may be playing a direct or indirect role in carcinogenesis by virtue of their being commonly altered in a particular cancer type. However, proving causality of these candidate “driver” genes, and elucidating their roles in tumorigenesis, requires relevant experimental validation.
ARID1A (also called BAF250a), encoding an important component of the mammalian SWI/SNF complex, has emerged as one of the most commonly mutated or downregulated genes in diverse tumors, including gastrointestinal (Wang et al. 2011; Cajuso et al. 2014), endometrial (Liang et al. 2012; The Cancer Genome Atlas Research Network et al. 2013), ovarian clear cell (Jones et al. 2010; Wiegand et al. 2010), pancreatic (Waddell et al. 2015), lung (Imielinski et al. 2012), and breast (Cornen et al. 2012; Mamo et al. 2012). ARID1A impacts epigenetic gene regulation by altering chromatin structure around promoters of specific loci in conjunction with its associated SWI/SNF complex components (Inoue et al. 2011; Chandler et al. 2013). Therefore, its downregulation or mutation in somatic cells can have profound consequences, including inappropriate proliferation (Romero and Sanchez-Cespedes 2014). Despite the accumulating correlative data implicating ARID1A as a tumor suppressor, functional proof has been lacking in part due to the fact that knockout of Arid1a in mice causes embryonic lethality even in the heterozygous state (Gao et al. 2008). However, two recent reports have shown that conditional biallelic knockout of Arid1a in ovarian surface epithelial cells, in conjunction with either conditional expression of a mutant phosphoinositide 3-kinase catalytic subunit (PIK3CA) (Chandler et al. 2015), or conditional disruption of Pten (Guan et al. 2014), caused carcinomas resembling clear cell in the former, and endometriod/undifferentiated in the latter. In both studies, deletion of Arid1a alone, or deletion of only one Arid1a allele in the compound mutant situations, was insufficient to cause cancer. While these studies provided compelling evidence for the tumor suppressive role of Arid1a in ovarian cancer, they (and most other genetically engineered cancer models) do not model the process of sporadic cancer development. Furthermore, the dependency of biallelic Arid1a inactivation upon mutation of Pten or Pik3ca in driving tumor formation in these models seems to be specific to the pathogenesis of endometrium-related ovarian neoplasms (Maeda and Shih Ie 2013) and does not appear to apply to several of the other human cancers in which ARID1A is commonly mutated (Kandoth et al. 2013). Thus, it is important to validate cancer genes/pathways in the context of their tumor-type-specific environments, as the behavior of these genes and pathways may vary by tissue type. Sporadic breast cancer (i.e., not associated with inherited neoplasia-driving mutations) accounts for the vast majority of breast cancer cases in the U.S. (80–85%) (American Cancer Society 2014). Although ARID1A has not yet been widely recognized as a key suppressor of breast carcinogenesis, it is heterozygously deleted in a substantial fraction of tumors (Cornen et al. 2012; Mamo et al. 2012), and low ARID1A expression in tumors of patients with breast cancer correlates significantly with poorer prognosis and overall survival (Mamo et al. 2011; Zhao et al. 2014; Cho et al. 2015; Zhang et al. 2015). Here, we report functional evidence that Arid1a loss is critical for mammary tumorigenesis in a mouse model of spontaneous breast cancer and present data on how this leads to deregulated cancer cell growth.
Results and Discussion
The Chaos3 mouse, bearing a missense allele (Mcm4Chaos3) of the DNA replication gene Mcm4, exhibits high levels of genomic instability caused by the mutation’s destabilization of the MCM2–7 replicative helicase complex (Shima et al. 2007; Kawabata et al. 2011; Chuang et al. 2012). Most females homozygous for the Chaos3 mutation congenic in the C3HeB/FeJ strain background (C3H-Chaos3) develop spontaneous mammary tumors (MTs) with an average latency of 12 months (Shima et al. 2007). Array Comparative Genomic Hybridization (aCGH) analyses of nine C3H-Chaos3 MTs revealed interstitial deletions common to a small number of chromosomal regions (Wallace et al. 2012). Almost all tumors were missing both copies of Nf1, a tumor suppressor that functions as negative regulator of Ras, but positive for Trp53 (Wallace et al. 2014).
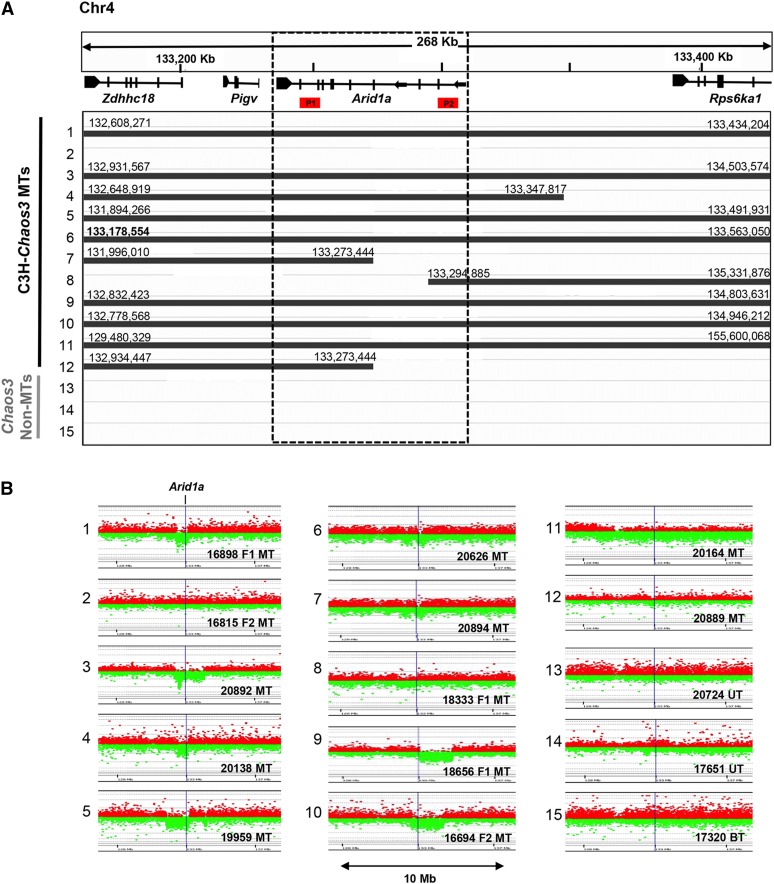
Those aCGH data, plus an additional 12 reported here, indicated that most (18/21) MTs also contained deletions involving part or all of an ∼100-kb region on chromosome 4 (Chr4) (Figure 1) containing Arid1a (Figure 1). To further validate the aCGH results, we performed digital droplet PCR (ddPCR) on DNA from the same 12 MTs plus three non-MTs using probes situated at both ends of Arid1a. All 15 calls for probe 2, and 13/15 for probe 1 (Figure 1A) were consonant. The two discrepancies were in MTs 7 and 12, which according to aCGH results, have an identical deletion breakpoint within Arid1a. It is possible that in these cases, the breakpoint is actually proximal to that called by the software. As an alternative confirmation of Arid1a hemizygosity in these tumors, we took advantage of genetic polymorphisms in two F1 (C3HeB/FeJ × C57BL/6J) MTs deleted for Arid1a (Figure 1, nos. 1 and 8) and an F2 MT having no deletion (Figure 1, no. 2), based on aCGH calls. Genotyping of SNPs at the 3′ end of Arid1a revealed agreement with the aCGH and ddPCR data (Figure 1A and Supplemental Material, Figure S1).
Figure 1.
Arid1a is recurrently deleted in C3H-Chaos3 mammary tumors. (A) Overview of aCGH data near the Arid1a locus from 15 tumor samples, adapted from an IGV depiction. Solid lines denote stretches of contiguous probes with reduced hybridization signal, thus representing deleted regions. Nucleotide coordinates of deletion endpoints are indicated and correspond to the last probe with reduced hybridization signal on the array. The control non-MTs consist of two uterine tumors and one bone tumor. (B) Plot of probe intensities near the Arid1a from aCGH hybridization. Each dot is a probe on the array, with the green and red representing control vs. tumors, respectively. Locations of primer pairs used for Arid1a CNV analyses by ddPCR are depicted as P1 and P2 (see Materials and Methods). MT, mammary tumor.
We next scored 33 additional C3H-Chaos3 MTs and five cell lines derived from C3H-Chaos3 MTs for deletions in the Arid1a coding region by ddPCR. In total, ∼70% of the Chaos3 MTs analyzed had monoallelic deletions for all or part of Arid1a (Table S1). Hemizygosity for ARID1A also appears to be common in human breast carcinomas at frequencies as high as ∼40% depending on the dataset (The Cancer Genome Atlas Network 2012; Ciriello et al. 2015; Eirew et al. 2015) (Figure S2).
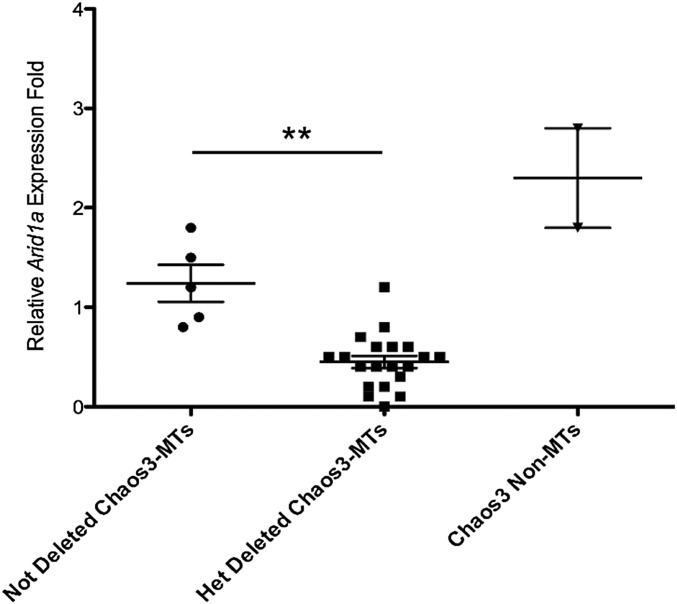
If hemizygosity of Arid1a contributes to tumorigenesis, then either it is haploinsufficient (i.e., 50% expression contributes to the transformed state) or the nondeleted allele is also altered in a genetic or epigenetic manner that reduces Arid1a expression to a level below that which is necessary to prevent transformation and/or tumor growth. To test this, we quantified Arid1a messenger RNA (mRNA) in hemizygous and nondeleted C3H-Chaos3 MTs. On average, transcript levels in 24 Arid1a-deleted tumors, but not nondeleted tumors, was about half that present in WT mammary tissue (Figure 2 and Table S2). The results suggest that ARID1A reduction, but not elimination, may contribute to tumorigenesis or tumor maintenance. Interestingly, the two Chaos3 non-MTs tested had approximately fivefold more Arid1a than the deleted MTs (Figure 2).
Figure 2.
Tumors hemizygous for Arid1a have less Arid1a mRNA. Plotted are qRT-PCR expression data from the Chaos3 non-MTs (two uterine tumors) and C3H-Chaos3 MTs that were either heterozygously deleted (n = 21) or not deleted (n = 5) for ARID1A, based on ddPCR data. Expression levels for each sample were calculated relative to an average of two WT RNA tissue samples. Significant differences were calculated using a two-tailed Student’s t-test (** P < 0.001).
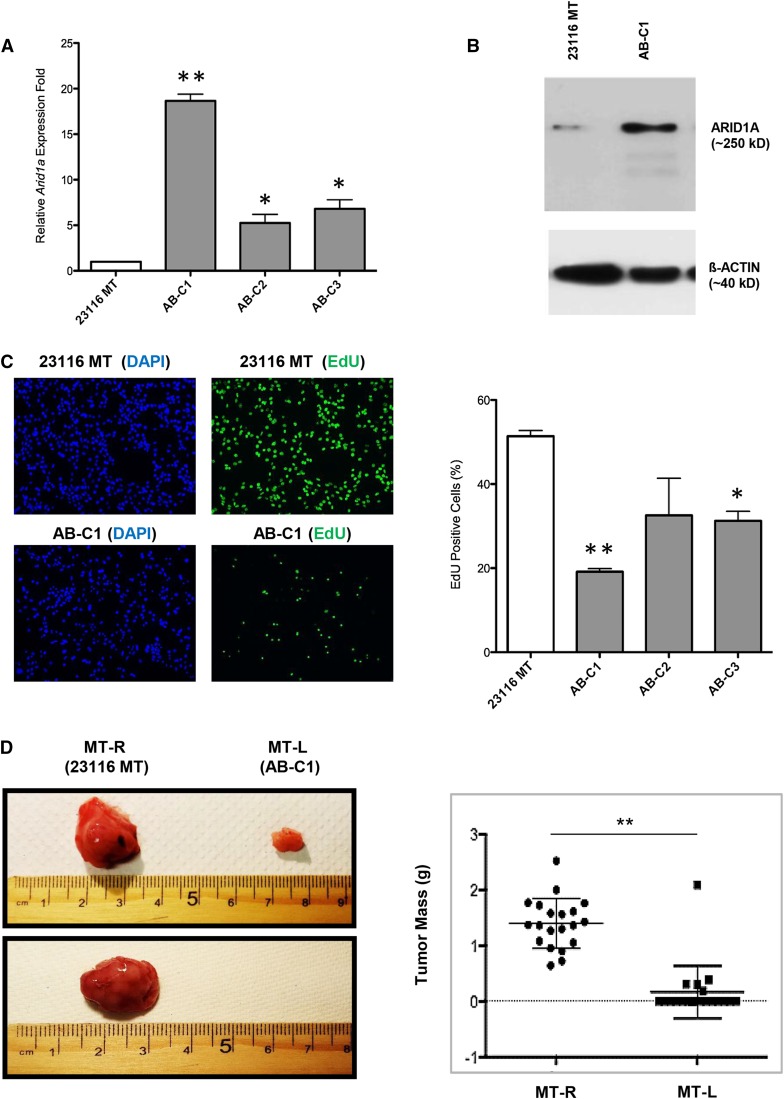
The genetic and molecular data described above imply, but do not prove, that decreased ARID1A expression is involved in either neoplastic transformation or maintenance of the transformed state. To address this question functionally, we conducted experiments to restore Arid1a expression in ARID1A-deficient C3H-Chaos3 MT cells, followed by analyses of the in vitro and in vivo consequences. First, we generated a Chaos3 MT cell line (23116 MT) that has one copy of Arid1a deleted (Table S1) and very low levels of Arid1a expression (Figure 3, A and B), and then stably introduced an Arid1a complementary DNA (cDNA) expression construct into these cells using lentivirus-mediated transduction. These transformed lines were termed “Addback” (AB) cells. We then assessed cell proliferation activity via 5-ethynyl-2′-deoxyuridine (EdU) incorporation in the parental vs. three transduced cell lines, and found that ectopic Arid1a expression caused a dramatic decrease (approximately threefold) in EdU incorporation in each of the lines tested (Figure 3C).
Figure 3.
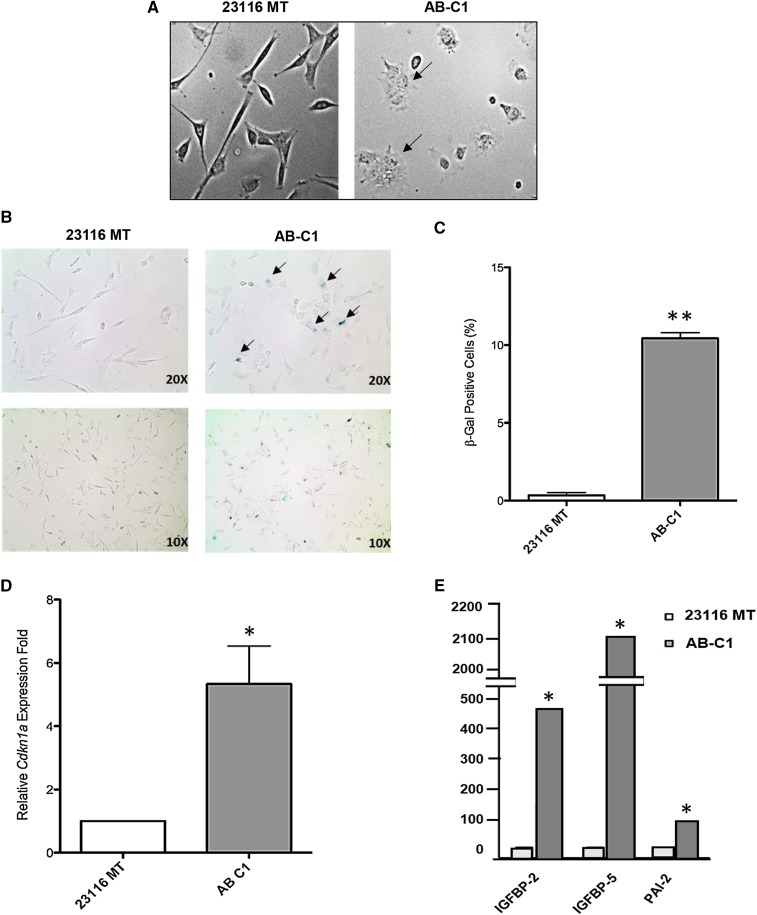
Overexpression of Arid1a in C3H-Chaos3 MT cell line reduces proliferation rate and prevents tumor growth. (A) Arid1a expression levels quantified by qRT-PCR in untransduced Chaos3 MT cell line (23116 MT) and three individual clonal lines (AB-C1, A-C2, and AB-C3) transduced with the Arid1a expression vector. Results are shown as the mean ± SEM of three technical replicates. Significant differences were calculated using a two-tailed Student’s t-test (* P < 0.05; ** P < 0.001). Values are relative to untransduced parental cell line 23116 MT. (B) Arid1a expression levels quantified by immunoblotting in indicated cell lines. (C) EdU incorporation assays of 23116 MT vs. AB clones (C1–C3). Error bars signify the mean ± SEM of three experimental replicates, with >1000 cells counted per sample for each replicate. Significant differences were calculated using a two-tailed Student’s t-test (* P < 0.05; ** P < 0.001). Values are relative to untransduced parental cell line 23116 MT. (D) Representative images of tumors (or lack thereof) arising from transplantation of 23116 MT (MT-R) and AB-C1 (MT-L) cells into cleared fad pads of recipient syngeneic C3H females. MT-R and MT-L refer to right and left sides of mouse, respectively (see Materials and Methods). Next to it is a plot depicting individual weights of tumors (n = 20 mice, 40 potential tumors). Significant differences between the control and experimental groups were calculated using a two-tailed Student’s t-test (** P < 0.001).
To determine if ARID1A is required for tumorigenicity, we tested whether one of the transduced clones (AB-C1) exhibiting elevated levels of mRNA (Figure 3A) and protein (Figure 3B) would reduce/abolish the ability of 23116 MT cells to form tumors when transplanted into host animals. The parental and AB-C1 cancer cells were injected into cleared mammary fat pads of WT C3H female mice (23116 MT on one side and AB-C1 on the other of each mouse; see Materials and Methods) and monitored for tumor formation. Overexpression of Arid1a significantly decreased MT formation frequency and size (Figure 3D). These results indicate that loss/reduction of Arid1a expression is crucial for the growth and/or formation of C3H-Chaos3 MTs. As shown below, ectopic Arid1a overexpression did not inhibit tumor formation in an unrelated non-Chaos3 MT cell line MCN1, indicating that excessive ARID1A itself is not cell toxic.
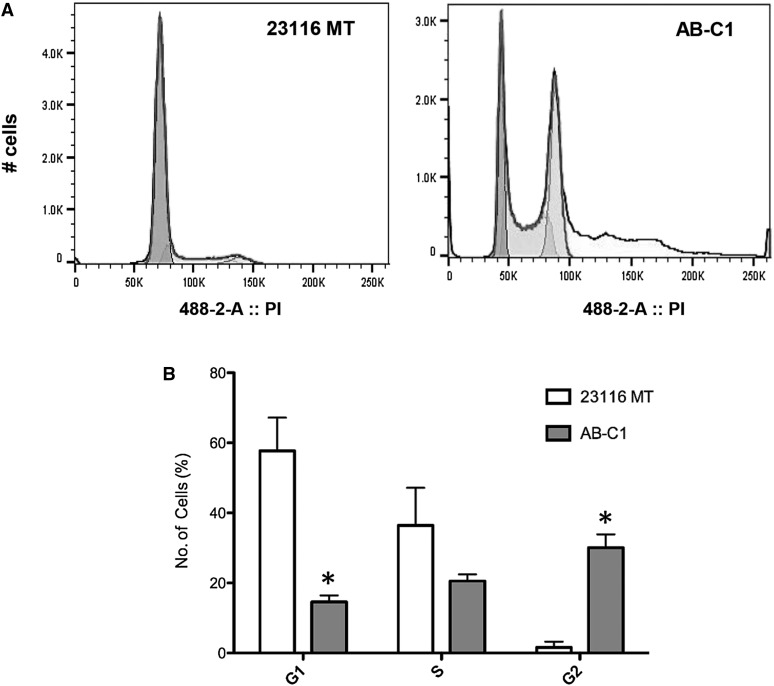
To gain insight into the mechanisms by which Arid1a loss promotes tumorigenesis in the Chaos3 model, we considered data showing that Arid1a is required for efficient functioning of the DNA damage response (DDR), specifically the G2/M cell-cycle checkpoint that helps suppress genomic instability (GIN) and tumorigenesis (Lobrich and Jeggo 2007; Shen et al. 2015). Since Chaos3 cells have chronic replication stress and GIN (Shima et al. 2007; Kawabata et al. 2011; Bai et al. 2016), it is possible that a loss or reduction of ARID1A in a cell allows escape from DDR-mediated growth arrest or apoptosis, thus promoting carcinogenesis. Therefore, we examined the cell cycle of AB-C1 cultures. This revealed an accumulation of cells in the G2 phase (Figure 4, A and B), suggesting that Arid1a overexpression might be inducing a checkpoint response and consequent growth arrest.
Figure 4.
Cell-cycle analysis of C3H-Chaos3 mammary tumor cell lines. (A) Cell-cycle profiles of 23116 and AB-C1 mammary tumor lines. (B) Percentage of cells in different phases of the cell cycle (G1, S, and G2) based on FloJo statistical analyses. Significant differences were calculated using a two-tailed Student’s t-test (* P < 0.05; ** P < 0.001). Values are relative to untransduced parental cell line 23116 MT.
Since ectopic Arid1a expression in ARID1A-deficient MT cells caused cell-cycle arrest, we assessed whether either senescence or apoptosis was triggered as a consequence. TUNEL assays did not reveal a significant increase in apoptosis, based on relative percentages of positively stained cells (23116 MT = 0.6%, ± 0.2% SEM; AB-C1 = 1.5%, ± 0.4%), raising the possibility that these cells were instead senescing. Another indication was the dramatic change in morphological features of the cells in which Arid1a was overexpressed. They appeared larger and flatter (Figure 5A), characteristic of cells undergoing senescence (Kuilman et al. 2010). Finally, we conducted senescence-associated β-galactosidase (SABG) assays, showing that the AB-C1 cell population had nearly 10-fold more SABG+ cells than the parental cultures (Figure 5, B and C).
Figure 5.
Senescence characteristics of AB-C1 cancer cells. (A) Morphological comparison of 23116 MT vs. AB-C1 cells (×20 magnification). (B) Representative images of indicated cells stained for senescence-associated β-galactosidase activity. (C) Average percentages of positively stained cells (blue color) calculated from four technical replicates. (D) qRT-PCR validation of senescence-associated genes. (E) qRT-PCR assay comparing relative transcript levels of p21 in AB-C1 vs. 23116 MT cells. Significant differences were calculated using a two-tailed Student’s t-test (* P < 0.05; ** P < 0.001). Values are relative to untransduced parental cell line 23116 MT.
Multiple studies have demonstrated that DNA damage-induced G2 arrest activates cellular senescence in a TRP53- and p21-dependent manner (Bunz et al. 1998; Mao et al. 2012; Krenning et al. 2014). mRNA levels of p21, which is transcriptionally regulated by TRP53, was approximately fivefold higher in AB-C1 MT cells compared to the 23116 parental line (Figure 5D). Taken together, these data indicate that restoring or overexpressing Arid1a in C3H-Chaos3 MT cells enables G2/M arrest and subsequent cellular senescence. This is consistent with data indicating that ARID1A functions as both a “gatekeeper” in its control of cell proliferation and a “caretaker” in its maintenance of genomic integrity (Wu et al. 2014).
To better understand the mechanism by which restoration/overexpression of Arid1a impairs growth and tumorigenesis, we conducted RNA sequencing (RNA-seq) comparing the transcriptomes of 23116 MT vs. AB-C1 and AB-C2 cells. A total of 554 genes were significantly differentially expressed (DE) between the parental stock and the Arid1a-transduced lines [fragments per kilobase of exon per million fragments mapped (FPKM) > 5; log2 > 1 or < −1 Table S3]. Ingenuity pathway analysis (IPA) of these DE genes revealed that the TRP53 pathway was activated in AB-C1/C2 cells, while the TGFβ pathway was repressed (Figure S3).
RNA-seq data also showed that the most highly upregulated genes within the TRP53 pathway in AB-C1 cells were Igfbp5, Igfbp2, and Serpinb2 (Pai-2). We validated these data by quantitative RT-PCR (qRT-PCR) (Figure 5E). All three genes have been implicated in tumor growth suppression (Andreasen et al. 1997; Butt et al. 2003; Pereira et al. 2004). IGFBP5 was found to activate cellular senescence through a TRP53-dependent mechanism in human endothelial cells (Kim et al. 2007). These data further support the idea of a TRP53-dependent senescence checkpoint response being activated when Arid1a is overexpressed in these cancer cells.
TRP53 was reported to interact physically with ARID1A and the rest of the SWI/SNF complex, enabling transcriptional regulation of downstream gene targets (Guan et al. 2011). Several human cancer studies have found that loss of ARID1A expression correlates with high amounts of potentially functionally inactive TRP53 (Wang et al. 2011; Zang et al. 2012; Bosse et al. 2013; Bitler et al. 2015), suggesting they have codependent tumor suppressive functions, and that ARID1A deficiency would have a similar effect as losing TRP53 activity. A similar mutually exclusive pattern of Trp53 and Arid1a expression seems to exist in C3H-Chaos3 MTs. They exhibit high levels of TRP53 (Wallace et al. 2014), but it does not seem to be effective or functional in the sense of its established role in suppressing uncontrolled cell growth or malignant transformation. Based on this hypothesis as well as the RNA-seq data, it is possible that reexpressing Arid1a in the C3H-Chaos3 tumor cells restores the ability of TRP53 to regulate downstream target genes.
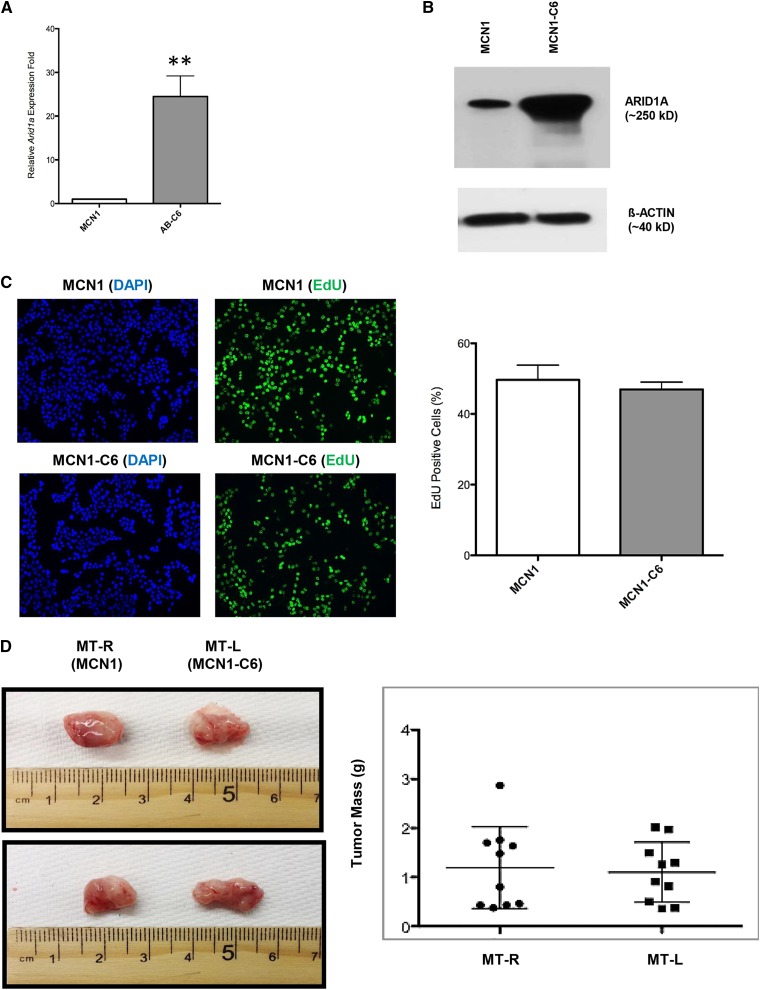
To test this hypothesis, we overexpressed Arid1a in a Trp53 null mouse MT cell line MCN1 (Cheng et al. 2010), which we found to not only have low Arid1a expression (Figure 6, A and B), but also apparent hemizygosity of Arid1a (Table S1). EdU incorporation assays comparing the untransduced and AB cells showed that unlike the TRP53-proficient Chaos3 cell line 23116, proliferation was unaltered upon overexpressing Arid1a in MCN1 (Figure 6C), and subsequent mammary fat pad growth assays revealed that tumorigenecity in vivo was also unaffected (Figure 6D). This is consistent with the hypothesis that active TRP53 is necessary for ARID1A to function in its tumor suppressive role.
Figure 6.
Overexpression of Arid1a in the TRP53-deficient MCN1 has no effect on proliferation rate or tumor growth. Arid1a expression levels were quantified by (A) qRT-PCR and (B) immunoblotting in MCN1 MT cell line and its transduced counterpart cell line (MCN1-C6). Significant differences were calculated using a two-tailed Student's t-test (** P < 0.001). Values are relative to untransduced parental cell line MCN1. (C) EdU incorporation assays of MCN1 vs. MCN1-C6. Error bars signify the mean ± SEM of three experimental replicates, with >1000 cells counted per sample for each replicate. (D) Representative images of tumors arising from transplantation of MCN1 (MT-R) and MCN1-C6 (MT-L) cells into cleared fad pads of recipient syngeneic FVB/N recipient females. MT-R and MT-L refer to right and left sides of mouse, respectively (see Materials and Methods). The right panel plots individual weights of tumors (n = 10 mice, 20 tumors).
C3H-Chaos3 tumors have a manageable number of recurring spontaneous alterations, making it feasible to unravel the molecular events leading to tumor formation—a crucial question in cancer genetics. The experiments here provide genetic and functional evidence that ARID1A is a suppressor of mammary tumorigenesis, and particularly, that it is required for maintenance of tumorigenic potential as revealed by transplantation assays. This role in tumor maintenance also appears to be the case in human ovarian cancer, where it was shown that reintroduction of the gene into tumor cells bearing ARID1A mutations inhibited xenograft growth (Guan et al. 2011). Our finding that Arid1a is almost exclusively monoallelically (not biallelically) deleted in the Chaos3 model of spontaneous breast cancer, apparently similar to the situation in human breast cancers, is important in terms of potential therapeutic intervention. We showed that overexpressing Arid1a ectopically in MT cells greatly suppresses tumor growth in a TRP53-dependent manner. Therefore, in breast cancer cases that retain an intact ARID1A allele in trans to a mutant/deleted allele, and also contain wild-type (WT) TRP53, it may be possible to employ methods for specific reactivation of the remaining allele, thus suppressing tumor growth and triggering cell-cycle arrest. Recent development of sequence-specific, chimeric transcriptional regulators offers one such potential avenue to accomplish this (Maeder et al. 2013; Gilbert et al. 2014; Chavez et al. 2015; Konermann et al. 2015; Zhang et al. 2015).
Materials and Methods
Cancer cell lines
The 23116 MT cell line was generated from a primary MT that arose in a C3H-Chaos3 mouse. The tumor was dissected and mechanical pulverized, then seeded on gelatin-coated culture dishes in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). “Add-back” (AB) cell lines were generated by lentiviral transduction of Arid1a expression vector pLenti-puro-ARID1A (Addgene plasmid no. 39478), followed by puromycin selection (2 µg/ml) and growth of clonal lines (AB-C1/C2/C3) expressing ectopic Arid1a. All qRT-PCR primers are shown in Table S4.
aCGH
Genomic DNA was isolated from primary tumors by solubilizing in lysis buffer (50 mM Tris-HCl pH 8.0; 100 mM EDTA pH 8.0; 100 mM NaCl; 1% SDS; 0.5 mg/ml Proteinase K) for 3 hr at 55°, phenol/chloroform extraction, precipitation of the DNA in 0.8 volumes isopropanol, followed by spooling and washes in 70%, then 100% ethanol. One microgram of genomic DNA was used for labeling and hybridization to the SurePrint G3 Mouse Genome Comparative Genomic Hybridization (CGH) Microarray, 1 × 1 M (Agilent; product no. G4838A). Two independent reference WT DNAs (from strain C3HeB/FeJ mammary tissue) were used as hybridization controls. This array consists of 60-mer probes with an overall median spacing of 1.8 kb [1.5 kb in Reference Sequence (RefSeq) genes]. Content for probe design was sourced from University of California Santa Cruz (UCSC) mm9 (National Center for Biotechnology Information Build 37). DNA labeling, hybridization, and posthybridization processing were performed according to the manufacturer’s protocol. Images were scanned using the Agilent’s SureScan Microarray Scanner. Agilent’s Cytogenomics software was used for spot identification and signal quantification, following normalization of test/reference ratios and background correction. Criteria for calling amplifications/deletions were as follows: minimum number of contiguous probes ≥3, minimum average absolute log2 ratio ≥0.25. Copy number alterations were visualized using the Integrative Genomics Viewer (IGV) software package (Robinson et al. 2011).
ddPCR
ddPCR was carried out using the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA). Approximately 60 ng genomic DNA extracted from 51 different tumor samples (Chaos3-MTs and Chaos3 non-MT controls) was used per reaction. Individual tumor samples were analyzed for copy number variations (CNVs) occurring in the target gene Arid1a, by probing two different genomic locations spanning the length of the entire gene. Primer and probe combinations used for the assay are shown in Table S4. Gapdh was used as a reference gene in CNV analyses. Droplet generation and droplet reading for ddPCR were carried out using Bio-Rad equipment and reagents, according to the manufacturer’s instructions. Results were analyzed using QuantaSoft Software (Bio-Rad) and represented as concentration of DNA (copies per microliter). Each DNA sample was run in duplicate. Results for all samples analyzed are shown in Table S1.
qRT-PCR
Total RNA was isolated from cells and tissues using the E.Z.N.A. Total RNA Kit I (Omega Biotek). A total of 500 ng of RNA was used for cDNA synthesis using the qScript cDNA Supermix Kit (Quantabio). qRT-PCR analyses was done using Fast SYBR Green Master Mix (Life Technologies) and custom designed primers (Table S4), using GAPDH as an endogenous reference. Assays were run on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each sample was run in triplicate wells, from which mean Ct values were obtained. Relative quantification of gene expression was calculated using the ΔΔCt method. At least two technical replicates were run for each experiment to obtain standard error values.
EdU proliferation assay
Cells were grown overnight on coverslips and pulse labeled with 10 μM EdU for 30 min. Cells were then fixed with formaldehyde (final concentration of 1%) for 10 min, followed by permeabilization (0.3% Triton X-100 in PBS) for 15 min. The “Click” reaction cocktail [10 mM (+)-sodium-L-ascorbate; 0.1 mM 6-caboxyfluorescein-TEG azide; 2 mM CuSO4] was added to cells and incubated for 30 min at room temperature. Nuclei were counterstained with DAPI and coverslips were mounted on slides for EdU+ cell counting by fluorescence microscopy. Experiments were conducted in triplicate, with >1000 cells counted per replicate.
Mammary fat pad injection surgeries
MT cell lines were injected into nos. 4 and 9 inguinal fat pads of 3-week-old nulliparous WT C3HeB/FeJ female mice, following clearance of the endogenous epithelium. Volume of cells injected per fat pad was 10 µl, at a concentration of 1 × 106 cells/ml. Tumors were allowed to develop until one grew to ∼2 cm in diameter, at which point the animals were killed and tissues were collected for analyses. The time to tumor formation following surgery varied from 3 to 12 weeks.
Cell-cycle analyses
One million cells were centrifuged and resuspended in 200 µl of a cold hypotonic solution of propidium iodide (PI) (50 µg/ml PI, 1 mg/ml sodium citrate, and 1 µl/ml Triton X-100). Cells were incubated at 4° overnight for complete lysis and staining of nuclei. Cell-cycle profiles were analyzed using a flow cytometer (BD LSR II), with 488-nm excitation and emission collected with a 576/26 band-pass filter. Using a PI signal-specific width vs. area plot, only single nuclei were included in the analyses of all profiles.
Senescence-associated β-galactosidase assay
AB-C1 and 23116 MT cells were seeded onto a six-well dish at a concentration of 0.2 × 106 cells/ml. The next day, cells were stained using a senescence detection kit (Abcam, ab65351) as per the manufacturer’s instructions. In brief, cells were fixed for 10 min at room temperature with the provided fixation solution and then stained for 16 hr at 37°. The next day, they were visualized using light microscopy for development of blue color. Images were taken at ×10 magnification and the number of positive cells were counted using ImageJ software. Experiments were carried out in triplicate to calculate the average percentage values.
RNA-seq
Total RNA was isolated from replicate samples of 23116 MT and AB clones (C1 and C2) cells using the E.Z.N.A. Total RNA Kit I. A total of 500 ng per sample was used to prepare cDNA libraries, with the NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (both from New England Biolabs). The libraries were then sequenced on Illumina’s Hi-Seq platform, generating single-end 100-bp reads. Reads were aligned to the mouse genome (UCSC mm10) using Tophat v2.0.13 (Trapnell et al. 2012). Significant DE genes between 23116 MT and AB clones were determined with the help of the TopHat tool CuffDiff (v2.2.1), which uses a Q-value cutoff of 0.05 (Q-value = P-value corrected for multiple hypothesis testing) (Trapnell et al. 2012). DE genes were additionally sorted based on more stringent criteria where at least one condition (average of replicates) must have FPKM ≥5 and the minimum log2 (fold change) between conditions is twofold (up or down).
Data and reagent availability
Cell lines and constructs are available upon request. RNA-seq data and aCGH data are available at the Gene Expression Omnibus (accession nos. GSE81575 and GSE81967, respectively).
Acknowledgments
The authors are grateful to Alex Nikitin for providing MCN1 cells, Jennifer Grenier for the RNA-seq analysis, Christa Heyward and Rodica Bunaciu for assistance with flow cytometry, and Robert Weiss and Adrian McNairn for feedback on the manuscript. This work was supported by National Institutes of Health grant R21-CA175961 to J.C.S.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184879/-/DC1.
Communicating editor: T. R. Magnuson
Literature Cited
- American Cancer Society, 2014 Breast Cancer Facts and Figures 2015–2016. Available at cancer.org. Accessed June 21, 2016. [Google Scholar]
- Andreasen P. A., Kjoller L., Christensen L., Duffy M. J., 1997. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int. J. Cancer 72: 1–22. [DOI] [PubMed] [Google Scholar]
- Bai G., Smolka M. B., Schimenti J. C., 2016. Chronic DNA replication stress reduces replicative lifespan of cells by TRP53-dependent, microRNA-assisted MCM2–7 downregulation. PLoS Genet. 12: e1005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler B. G., Fatkhutdinov N., Zhang R., 2015. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin. Ther. Targets 19: 1419–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse T., ter Haar N. T., Seeber L. M., v Diest P. J., Hes F. J., et al. , 2013. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod. Pathol. 26: 1525–1535. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., et al. , 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501. [DOI] [PubMed] [Google Scholar]
- Butt A. J., Dickson K. A., McDougall F., Baxter R. C., 2003. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J. Biol. Chem. 278: 29676–29685. [DOI] [PubMed] [Google Scholar]
- Cajuso T., Hanninen U. A., Kondelin J., Gylfe A. E., Tanskanen T., et al. , 2014. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer 135: 611–623. [DOI] [PubMed] [Google Scholar]
- Chandler R. L., Brennan J., Schisler J. C., Serber D., Patterson C., et al. , 2013. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33: 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler R. L., Damrauer J. S., Raab J. R., Schisler J. C., Wilkerson M. D., et al. , 2015. Coexistent ARID1A–PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 6: 6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., et al. , 2015. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Zhou Z., Flesken-Nikitin A., Toshkov I. A., Wang W., et al. , 2010. Rb inactivation accelerates neoplastic growth and substitutes for recurrent amplification of cIAP1, cIAP2 and Yap1 in sporadic mammary carcinoma associated with p53 deficiency. Oncogene 29: 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. D., Lee J. E., Jung H. Y., Oh M. H., Lee J. H., et al. , 2015. Loss of tumor suppressor ARID1A protein expression correlates with poor prognosis in patients with primary breast cancer. J. Breast Cancer 18: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. H., Yang D., Bai G., Freeland A., Pruitt S. C., et al. , 2012. Post-transcriptional homeostasis and regulation of MCM2–7 in mammalian cells. Nucleic Acids Res. 40: 4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., Gatza M. L., Beck A. H., Wilkerson M. D., Rhie S. K., et al. , 2015. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornen S., Adelaide J., Bertucci F., Finetti P., Guille A., et al. , 2012. Mutations and deletions of ARID1A in breast tumors. Oncogene 31: 4255–4256. [DOI] [PubMed] [Google Scholar]
- Eirew P., Steif A., Khattra J., Ha G., Yap D., et al. , 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Tate P., Hu P., Tjian R., Skarnes W. C., et al. , 2008. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA 105: 6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., et al. , 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B., Wang T. L., Shih Ie M., 2011. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71: 6718–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B., Rahmanto Y. S., Wu R. C., Wang Y., Wang Z., et al. , 2014. Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J. Natl. Cancer Inst. Avalable at: http://jnci.oxfordjournals.org/content/106/7/dju146.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M., Berger A. H., Hammerman P. S., Hernandez B., Pugh T. J., et al. , 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Giannakopoulos S., Parkhurst C. N., Matsumura T., Kono E. A., et al. , 2011. Target genes of the largest human SWI/SNF complex subunit control cell growth. Biochem. J. 434: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Wang T. L., Shih Ie M., Mao T. L., Nakayama K., et al. , 2010. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., et al. , 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Luebben S. W., Yamaguchi S., Ilves I., Matise I., et al. , 2011. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Seu Y. B., Baek S. H., Kim M. J., Kim K. J., et al. , 2007. Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol. Biol. Cell 18: 4543–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., et al. , 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenning L., Feringa F. M., Shaltiel I. A., van den Berg J., Medema R. H., 2014. Transient activation of p53 in G2 phase is sufficient to induce senescence. Mol. Cell 55: 59–72. [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S., 2010. The essence of senescence. Genes Dev. 24: 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Cheung L. W., Li J., Ju Z., Yu S., et al. , 2012. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 22: 2120–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M., Jeggo P. A., 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 7: 861–869. [DOI] [PubMed] [Google Scholar]
- Maeda D., Shih Ie M., 2013. Pathogenesis and the role of ARID1A mutation in endometriosis-related ovarian neoplasms. Adv. Anat. Pathol. 20: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Angstman J. F., Richardson M. E., Linder S. J., Cascio V. M., et al. , 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo A., Cavallone L., Tuzmen S., Chabot C., Ferrario C., et al. , 2012. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene 31: 2090–2100. [DOI] [PubMed] [Google Scholar]
- Mao Z., Ke Z., Gorbunova V., Seluanov A., 2012. Replicatively senescent cells are arrested in G1 and G2 phases. Aging (Albany, NY) 4: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. J., Meyer T., Docherty S. E., Reid H. H., Marshall J., et al. , 2004. Bimolecular interaction of insulin-like growth factor (IGF) binding protein-2 with alphavbeta3 negatively modulates IGF-I-mediated migration and tumor growth. Cancer Res. 64: 977–984. [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero O. A., Sanchez-Cespedes M., 2014. The SWI/SNF genetic blockade: effects in cell differentiation, cancer and developmental diseases. Oncogene 33: 2681–2689. [DOI] [PubMed] [Google Scholar]
- Shen J., Peng Y., Wei L., Zhang W., Yang L., et al. , 2015. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5: 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T. R., Andrews C. A., et al. , 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 39: 93–98. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network , 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network ; C. Kandoth, N. Schultz, A. D. Cherniack, R. Akbani et al, 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N., Pajic M., Patch A. M., Chang D. K., Kassahn K. S., et al. , 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. D., Pfefferle A. D., Shen L., McNairn A. J., Cerami E. G., et al. , 2012. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics 192: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. D., Southard T. L., Schimenti K. J., Schimenti J. C., 2014. Role of DNA damage response pathways in preventing carcinogenesis caused by intrinsic replication stress. Oncogene 33: 3688–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Kan J., Yuen S. T., Shi S. T., Chu K. M., et al. , 2011. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43: 1219–1223. [DOI] [PubMed] [Google Scholar]
- Wiegand K. C., Shah S. P., Al-Agha O. M., Zhao Y., Tse K., et al. , 2010. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. C., Wang T. L., Shih Ie M., 2014. The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 15: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Z. J., Cutcutache I., Poon S. L., Zhang S. L., McPherson J. R., et al. , 2012. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 44: 570–574. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yin C., Zhang T., Li F., Yang W., et al. , 2015. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 5: 16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu C., Zhao Z., 2014. ARID1A: a potential prognostic factor for breast cancer. Tumour Biol. 35: 4813–4819. [DOI] [PubMed] [Google Scholar]