Abstract
Environmental stress triggers multilevel adaptations in animal development that depend in part on epigenetic mechanisms. In response to harsh environmental conditions and pheromone signals, Caenorhabditis elegans larvae become the highly stress-resistant and long-lived dauer. Despite extensive studies of dauer formation pathways that integrate specific environmental cues and appear to depend on transcriptional reprogramming, the role of epigenetic regulation in dauer development has remained unclear. Here we report that BLMP-1, the BLIMP-1 ortholog, regulates dauer formation via epigenetic pathways; in the absence of TGF-β signaling (in daf-7 mutants), lack of blmp-1 caused lethality. Using this phenotype, we screened 283 epigenetic factors, and identified lin-40, a homolog of metastasis-associate protein 1 (MTA1) as an interactor of BLMP-1. The interaction between LIN-40 and BLMP-1 is conserved because mammalian homologs for both MTA1 and BLIMP-1 could also interact. From microarray studies, we identified several downstream target genes of blmp-1: npr-3, nhr-23, ptr-4, and sams-1. Among them S-adenosyl methionine synthase (SAMS-1), is the key enzyme for production of SAM used in histone methylation. Indeed, blmp-1 is necessary for controlling histone methylation level in daf-7 mutants, suggesting BLMP-1 regulates the expression of SAMS-1, which in turn may regulate histone methylation and dauer formation. Our results reveal a new interaction between BLMP-1/BLIMP-1 and LIN-40/MTA1, as well as potential epigenetic downstream pathways, whereby these proteins cooperate to regulate stress-specific developmental adaptations.
Keywords: stress resistant development, BLMP-1, epigenetics, TGF-β, dauer
DURING development, epigenetic changes in gene expression are passed on to the daughter cells to dictate cell fate without changing the DNA sequence itself (Hemberger et al. 2009). Epigenetic regulation is critical not only for normal development but also for tumor proliferation (Jones and Baylin 2007). The PRDM (PR domain-containing genes) family regulates many epigenetic events through interactions with histone modification and nucleosome remodeling factors (Hohenauer and Moore 2012; Pinheiro et al. 2012). Within this family, PRDM-1/BLIMP-1 regulates differentiation of various tissues and cell types including germ cells and B cells (Turner et al. 1994; Bikoff et al. 2009; John and Garrett-Sinha 2009). PRDM-1/BLIMP-1 is a transcriptional repressor, interacting with chromatin factors, such as the SET domain protein G9a (Yu et al. 2000), histone deacetylase HDAC1/2 (Gyory et al. 2004), and demethylase LSD1 (Su et al. 2009). Defects in the gene function are associated with certain type of lymphoma (Mandelbaum et al. 2010), demonstrating its critical role in B cell development. Termination of B cell differentiation is controlled by PRDM-1/BLIMP-1 (Kallies et al. 2004) and an abnormal downregulation of PRDM-1/BLIMP-1 may prevent the terminal differentiation process in diffuse large B-cell lymphoma (Nie et al. 2010).
Recently, Horn et al. (2014) and Huang et al. (2014) reported that BLMP-1, which encodes a homolog of PRDM-1/BLIMP-1, regulates the Caenorhabditis elegans developmental process. Both groups showed that blmp-1 is required for cell migration and the molting process via its interaction with DRE-1 (a C. elegans homolog of FBXO11). Their results show that BLMP-1 in C. elegans plays a significant role in development with several conserved features. Both BLIMP-1 of mammals and BLMP-1 of C. elegans interact with the conserved molecule FBXO11 or DRE-1, respectively. In addition, both BLIMP-1 and BLMP-1 regulate similar development processes in mammals and in C. elegans, such as germ cell migration. These results demonstrate that C. elegans blmp-1 has a conserved function and operates through similar molecular pathways as those of mammals.
The nematode C. elegans undergoes specialized development to become a stress-resistant larva called a dauer to survive harsh conditions such as starvation or high temperatures (Cassada and Russell 1975). Dauers are characterized by a distinct morphology and behavior: dauers are stress resistant, can survive for many months under adverse conditions, and do not eat (Cassada and Russell 1975). These differences indicate that dauer larvae employ a specific development program that nondauers do not execute in order to maximize their fitness under stress. Dauer formation is controlled by the nuclear hormone receptor DAF-12, a vitamin D and liver-X receptor homolog that functions as a ligand-regulated switch between dauer and nondauer programs (Fielenbach and Antebi 2008; Wang et al. 2015). Notably, worms that have been dauers have been shown to retain persistent histone modifications that change gene expression to affect life span and brood size of the postdauer adult animals, demonstrating that going through a different form of development leaves epigenetic marks (Hall et al. 2010). However, the mechanisms by which BLMP-1 regulates dauer formation in an epigenetic manner have not been investigated.
Here we report a new molecular pathway where BLMP-1 interacts with a MTA1 homolog of LIN-40 to specifically regulate dauer development in the absence of TGF-β signaling. Although both BLIMP-1 and MTA1 are known to interact with the TGF-β pathway, it is unknown whether MTA1 interacts with BLIMP-1. Through the study of the dauer development process, we have discovered a new and potentially conserved pathway whereby two tumorigenic and epigenetic factors (BLIMP-1 and MTA1/LIN-40) interact to enable an animal to be resistant to stress. Furthermore, our study reveals that the stress-resistant developmental process employs a distinct molecular pathway from that of a reproductive (nonstress resistant) development process. These results are consistent with prior reports for a role of BLIMP-1 in C. elegans development, but further extend those findings by showing a role for BLIMP-1 in dauer formation that is DRE-1 independent through a novel epigenetic mechanism (Horn et al. 2014). Our study suggests that in the absence of TGF-β signal, BLMP-1 interacts specifically with LIN-40 to differentially regulate the transcription profile to execute a dauer-specific development program.
Materials and Methods
Strains and culture conditions
Worms were maintained as described previously (Sulston and Hodgkin 1988) with the following modifications: worms were routinely grown on NGM containing streptomycin plates (Avery 1993) . Worms were maintained at 20° on Escherichia coli strain HB101 unless indicated differently. The wild-type strain was C. elegans variant Bristol, N2. Mutant strains used were DR40 daf-1(m40ts) IV, CB1393 daf-8(e1393ts) I, DR77 daf-14(m77ts) IV, CB1372 daf-7(e1372ts) III, YJ99 daf-7(m62ts) III, CB1376 daf-3(e1376) X, YJ55 blmp-1(tm548) I, YJ56 blmp-1 (tm548) I; daf-7(e1372ts) III, YJ57 daf-7(e1372ts) III; daf-3(e1376) X, MH1951 unc-119(ed3) III; Ex[lin-40::gfp unc-119(+)], YJ78 blmp-1(tm548) I; uyEx74[blmp-1p::blmp-1 rol-6p::GFP].
Cell culture, transfection, and Western blot
HEK 293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS. Flag/MTA1 expression plasmids were obtained from Dr. Paul Wade. His-BLIMP-1 expression plasmids were obtained from Dr. Adam Antebi. Cells were seeded at 50–70% confluence/six-well plate in DMEM media for 24 hr. Total plasmid DNA (5 μg) of Flag-MTA1 and BLIMP-1/His were cotransfected into 293T cells using FuGENE HD transfection reagent (Promega, Madison, WI; E2311). After 72 hr, cells were washed by 1× PBS (pH 7.4) and then harvested. The pellet was resuspended in lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol and protease inhibitor cocktail) and then the lysates were spinned down by centrifugation (10,000 × g for 10 min), precleared by protein G agarose beads (Millipore, 16-266) then incubated with either anti-BLIMP-1 (Abcam, Ab96479; 1:100 dilution) antibody or anti-Flag antibody (Sigma, F1804, 1:100) and mouse IgG (Cell Signaling, 5415) for 16 hr at 4°. A total of 20 μl of protein G agarose was added to each sample and incubated for 2 hr at 4°. The precipitate samples were washed and then analyzed by Western blot.
For Western blot, the following primary antibodies were used: anti-BLIMP-1 antibody (Abcam, Ab96479; 1:1,000 dilution) and anti-Flag antibody (Sigma, F1804; 1:1,000 dilution). Secondary antibody used was anti-mouse antibody conjugated with HRP (GE Healthcare, NA931V; 1: 5,000 dilution). The bands were detected using ECL Plus Kit (GE Healthcare, RPN2232).
Analysis of dauer formation
Mutants were grown at a permissive temperature (15°) until they became L4 larva (fourth larva stage). They were then transferred to and kept at a nonpermissive temperature (25°) throughout the test period. For dauer formation assays, 5–10 L4s were allowed to grow and lay eggs for ∼24 hr at 25°, a nonpermissive temperature, and then removed. The scoring time points were selected for each genotype (96 hr for daf-2 and 72 hr for daf-7 and daf-11, because daf-2 grows a lot slower than daf-7 or daf-11 at 25°) so that all animals had passed L2 stages at the time of scoring. Dauers were scored based on intestinal reorganization and radial shrinkage of the body and the pharynx. Dauer morphology was observed under the DIC setting of an optical magnification of ×100. To confirm dauer formation, worms were also tested for resistance to 1% SDS (Cassada and Russell 1975).
Growth assay
Worms were prepared and synchronized by egg preparation (Lewis and Fleming 1995). After each day of L1 starvation, ∼100 L1s were plated on each of three E. coli-seeded NGM plates to grow at 20°. Every hour from 42 hr after plating, worms were examined under a dissecting microscope at ×50 magnification to count worms that had molted into young adults (Lee et al. 2012).
RNA interference screen
The bacteria-mediated feeding RNA interference (RNAi) screen was performed as described (Fraser et al. 2000), with the following modifications. The wild-type and CB1372 strain were screened with the clones of nucleosome modification and chromatin remodeling factor genes from the Ahringer feeding library (Fraser et al. 2000; Kamath and Ahringer 2003). The plates containing NGM agar with 1 mM IPTG and 50 mg/ml carbenicillin were inoculated with bacterial cultures grown 16–18 hr for each targeted gene. L4 stage worms were transferred in the plates for each gene at 25°. Twenty-four hours later, adults were removed. Five days later, the number of progeny that had become dauers was counted.
Quantitative RT-PCR
Total RNA preparation:
C. elegans (from mixed and individual stages) were grown on NGM plates at 20° or 25°, washed with M9 buffer, and resuspended in TRIzol (Invitrogen). After vortexing for 60 sec, the mixture was frozen in liquid nitrogen and thawed at room temperature. After chloroform extraction, DNA was removed using DNase I. After ethanol precipitation, the air-dried pellet was dissolved in DEPC water.
Complimentary DNA preparation:
Approximately 1–2 µg of total RNA in a 20-µl reaction was used to synthesize the complimentary DNA (cDNA) (Biovision, Bio65043 synthesis kit). Quantitative RT-PCR (qPCR) was carried out in a C-1000 thermal cycler Real-Time PCR system (Bio-Rad, Hercules, CA, CFX96 optics module) and analyzed using the Ct method (Lee et al. 2009). The mRNA levels of ama-1 (RNA polymerase II) and inf-1 (Initiation factor 4A) were used for normalization as previously described (Potts et al. 2009). The average of at least three repeats was used for each data point. qPCR was performed using primers as described in the Supplemental Material, Table S11.
Western blot analysis and antibodies
Worms were washed from NGM plates (approximately one to two plates, 1000 worms) with M9 buffer. Worm pellets were resuspended in lysis buffer (1× PBS, pH 7.4, 10% glycerol, protease inhibitor cocktail tablet (Roche, 11836170001) and lysed by sonication (Misonix Sonicator 3000, 10 bursts at 10-sec intervals). Then Western blot analysis was performed as described (You et al. 2006). We used the following antibodies: anti-BLMP-1 antibody (Novus Biologicals, 42010002; 1:5000 dilution), anti-GFP (You et al. 2006), anti-di/trimethylhistone H3K9 (Cell Signaling, 5327; 1:1000 dilution), anti-trimethylhistone H3K4 (Cell Signaling, 9751; 1:1000 dilution), and anti-histone H3 (Cell Signaling, 9715; 1:5000 dilution) for primary antibodies. We used the following for secondary antibodies: anti-rabbit antibody conjugated with HRP (Santa Cruz Biotechnology, SC2030; 1:5000 dilution), anti-mouse antibody conjugated with HRP (GE Healthcare, NA931V; 1:5000 dilution). The bands were detected using ECL Plus Kit (GE Healthcare, RPN2232).
Photography
Dauer morphology was observed under DIC using a Zeiss Axio A2 Imager at either ×63 or ×100 magnifications. Images were acquired using Zeiss Axiovision software.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) assays were performed as described, with minor modification (Mukhopadhyay et al. 2008). L1 stage worms were grown on NGM plates at 25° and then harvested 24 hr later. The worms were cross-linked by PBS containing 1% formaldehyde at room temperature for 30 min. Formaldehyde was quenched with PBS/2.5 M glycine and washed five times with PBS. The pellets were suspended in lysis buffer (50 mM HEPES⋅KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, and protease inhibitor cocktail (Roche, 11836170001) and lysed by sonication (five times at 10-sec intervals). The lysates were precleared by incubating salmon sperm DNA/protein A agarose beads (Millipore, 16-157) and incubated overnight at 4° with either anti-BLMP-1 antibody or IgG. The precipitates were washed and the cross-links were reversed by heating at 65° with proteinase K. DNA was recovered by phenol–chloroform extraction, precipitation, and then eluted. PCR were performed using primers as described in Table S11.
Co-immunoprecipitation
Worms were harvested and washed from NGM plates with M9 buffer. A total of 1 mg of worm pellets (LIN-40::GFP) was resuspended in lysis buffer (50 mM HEPES·KOH, pH 7.5, 150 mM KCl, 1 mM EGTA, 0.05% NP-40, 10% glycerol and protease inhibitor cocktail (Roche, 11836170001) and lysed by sonication (five times at 10-sec intervals). After sonication, the lysates were spinned down by centrifugation, precleared by protein G agarose beads (Millipore, 16-266), and then incubated with either anti-BLMP-1 (1:100 dilution) antibody or anti-GFP antibody (1:100 dilution) and mouse IgG (Cell Signaling, no. 5415) for 16 hr at 4°. A total of 20 μl of protein G agarose was added to each sample, and incubated for 2 hr at 4°. The precipitate samples were washed five times with PBS plus 0.1% Tween-20 and then resolved on SDS/PAGE, transferred to nitrocellulose membrane, and analyzed by Western blot.
Microarray
RNA extraction:
Total RNA was extracted and the quality evaluated using a sample processing method previously established in our laboratory (Dumur et al. 2004). Total RNA was extracted from C. elegans (after 24 hr from L1 at 25°) using the MagMAX-96 for Microarrays Total RNA Isolation Kit (Invitrogen Life Technologies, Carlsbad, CA), in an automated fashion using the magnetic particle processors MagMAXTM Express. RNA purity was judged by spectrophotometry at 260, 270, and 280 nm. RNA integrity as well as cDNA and cRNA synthesis products were assessed by running 1 µl of every sample in RNA 6000 Nano LabChips on the 2100 Bioanalyzer (Agilent Technologies).
Gene expression microarray analyses:
The Affymetrix protocol utilized for our microarray analyses has been previously described (Dumur et al. 2004) and was used with the following modifications. Starting from 500 ng of total RNA, we performed a single-strand cDNA synthesis primed with a T7(dT24) oligonucleotide. Second-strand cDNA synthesis was performed with the E. coli DNA Polymerase I, and biotinylation of the cRNA was achieved by in vitro transcription (IVT) reaction using the GeneChip 3′ IVT Express Kit (Affymetrix, Santa Clara, CA). After a 37° incubation for 16 hr, the labeled cRNA was purified using the cRNA cleanup reagents from the GeneChip Sample Cleanup Module. As per the Affymetrix protocol, 10 μg of fragmented cRNA was hybridized on the GeneChip C. elegans genome array (Affymetrix) for 16 hr at 60 rpm in a 45° hybridization oven. The GeneChip C. elegans genome array provides comprehensive coverage of the transcribed C. elegans genome by analyzing the expression level of >22,500 well-characterized transcripts. The arrays were washed and stained with streptavidin phycoerythrin (SAPE) (Molecular Probes) in the Affymetrix Fluidics Workstation. Every chip was scanned at a high resolution, on the Affymetrix GeneChip Scanner 3000 7G according to the GeneChip Expression Analysis Technical Manual procedures (Affymetrix). After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip Operating Software v1.4 (Affymetrix). Overall quality of each array was assessed by monitoring the 3′/5′ ratios for the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (Gapdh), and the percentage of “present” genes (%P). Arrays exhibiting Gapdh 3′/5′ <3.0 and %P >40% were considered good-quality arrays.
Statistical analysis:
For the microarray data analysis, background correction, normalization, and estimation of probe set expression summaries were performed using the log-scale robust multiarray analysis method (Irizarry et al. 2003). Hierarchical cluster analyses were performed with the BRB-ArrayTools v3.1.0 (Biometric Research Branch, National Cancer Institute), an Excel add-in that collates microarray data with sample annotations. In order to identify differentially expressed genes between the different classes, we performed t-tests for each probe set from biological replicates in each class. Statistical significance for multivariate analysis to assess probe-set-specific false discovery rates (FDRs) was performed by estimating the q-values, using the Bioconductor q-value package (Storey 2002).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
blmp-1 is necessary for development processes
As previously reported, blmp-1 mutations or RNAi of blmp-1 in C. elegans causes gonadal migration defects and molting defect, along with a small body size (Figure S1, A–D) (Horn et al. 2014; Huang et al. 2014).
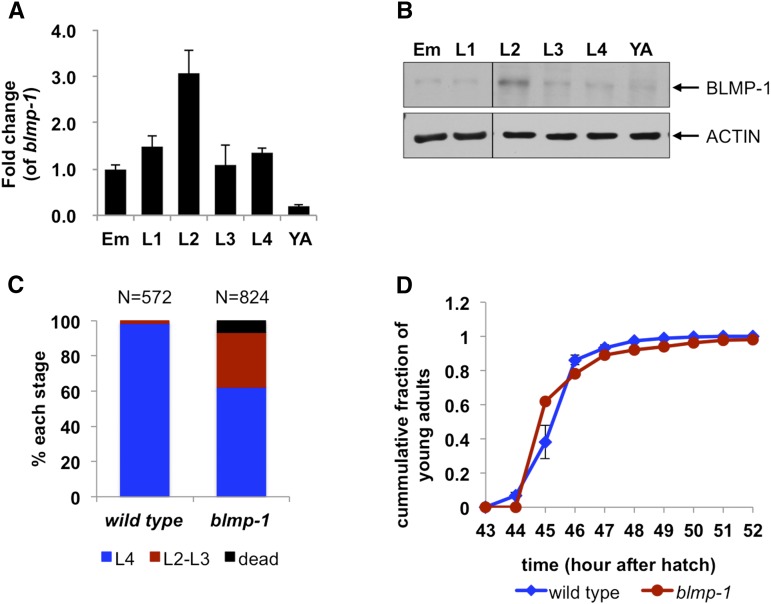
Upon examining the expression pattern of BLMP-1 during development by qPCR and Western blot analysis we noted that the messenger RNA (mRNA) and protein levels of BLMP-1 highly increased at the L2 (second larval) stage compared with other developmental stages (Figure 1, A and B). In addition, ∼6.7% of animals subjected to RNAi for blimp-1 died around L2 stage (Figure 1C), confirming the previous reports where it was suggested that blmp-1 plays an important role at the L2 stage. Most of blmp-1 mutants, however, are able to reach adulthood (Figure 1D), indicating that blmp-1 is not essential for survival during reproductive development.
Figure 1.
The levels of BLMP-1 during development and the growth rate and lethality of blmp-1 mutants. (A and B) The levels of blmp-1 mRNA measured by qPCR (A) and BLMP-1 protein measured by Western blot analysis (B) are highest at L2 stage compared to other stages in wild-type C. elegans. In A, the values are average ± SEM of three independent experiments. Em, embryos; YA, young adults. In B, actin is shown as a loading control. (C) The numbers of wild-type animals and blmp-1 mutants in different development stages (L2, L3, L4, and dead) were counted at 48 hr from egg hatching to yield the fraction of each stage and the death rate. blmp-1 mutants show more L2/L3 stages and dead worms compared to wild-type animals. (D) Growth rates of wild-type animals and blmp-1 mutants. Starting at 43 hr after hatching, the numbers of adult worms were counted every hour until 100% became adults (Lee et al. 2012; Wang et al. 2015). blmp-1 mutants showed grossly normal grow rate.
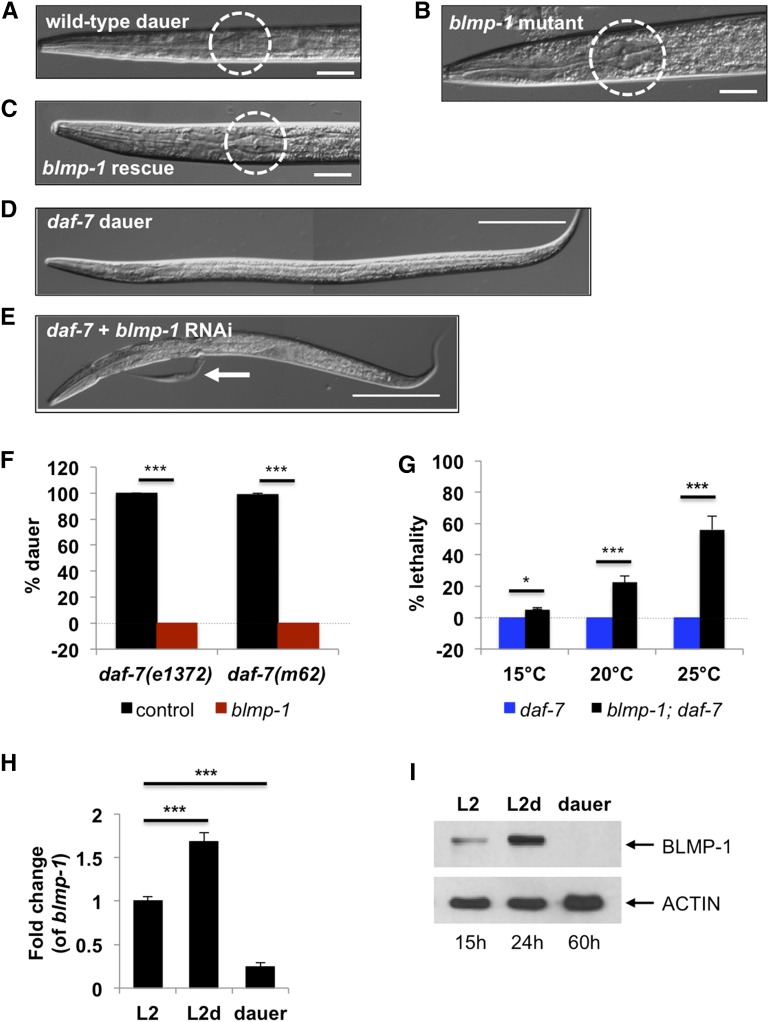
Horn et al. (2014) showed that when there is a lack of cholesterol, RNAi for blmp-1 prevents C. elegans from becoming a dauer. Interestingly, however, DRE-1, the interactor of BLMP-1 for reproductive (nondauer) development, was not necessary for dauer development. This result shows that blmp-1 could play a role in dauer development as well as reproductive (nondauer) development through distinct molecular partners that drive different cellular pathways. To investigate this further, the role of blmp-1 in dauer development was interrogated to examine how BLMP-1 may differentially regulate two distinct development programs. When we induced dauer formation using a synthetic dauer pheromone (a mixture of ascarosides (ascr) nos. 2, 3, and 5) and by limiting the amount of food (Butcher et al. 2007, 2008), blmp-1 mutants failed to become dauers under conditions where >90% of wild-type worms became dauers (Figure 2A and Figure S2A). The defective dauer formation phenotype (Daf-d) of blmp-1 mutants was rescued by extrachromosomal copies of the blmp-1 gene, confirming that the phenotype is caused by the mutation of blmp-1 (Figure 2, B and C).
Figure 2.
blmp-1 is essential for dauer development. (A) Representational photo of a wild-type dauer after treatment with the synthetic dauer pheromone (a mixture of ascarosides). Dotted circle shows a shrunken pharynx, an indication of a dauer. (B) A blmp-1 mutant after treatment with the synthetic dauer pheromone. Dotted circle shows that the pharynx was not shrunken, indicating the mutant does not develop into a dauer. (C) A transgenic animal carrying a wild-type copy of the blmp-1 gene in a blmp-1 mutation background (blmp-1 rescue) becomes a dauer. (D and E) A daf-7 mutant normally becomes a dauer (D) but fails and dies after treatment with blmp-1 RNAi (E). White arrow indicates a piece of cuticle separated from the body during dauer molt. Bar in A–C, 20 µm and in D and E, 100 µm. (F) Dauer formation of two different daf-7 mutants was counted after growth and treatment with blmp-1 RNAi from the mother generation at 25°. L4 worms (P0) were treated with RNAi and the dauer formation of the progeny (F1) was measured after 96 hr from the start of the treatment (see Materials and Methods). The values are from three independent experiments. The y-axis was lowered to start from −20 to visualize the 0% dauer. *** P < 0.001 by Student’s t-test. (G) The lethality of blmp-1; daf-7 mutants is temperature dependent, showing that lethality is specific for the dauer development process, whose incidence increases as temperature increases. The y-axis was lowered to start from −20 to visualize the 0% dauer. *** P < 0.001 by Student’s t-test. (H and I) The levels of blmp-1 mRNA (H) and BLMP-1 protein (I) increase at L2d stage compared to the preceding stage of L2 in daf-7 mutants. In H, the values are mean ± SEM of three independent experiments. *** P < 0.001 by Student’s t-test.
Dauer is induced mainly by the absence of one of three signals: insulin, TGF-β, and cGMP, all of which are necessary in worms to indicate a favorable environment (Riddle et al. 1981; Thomas et al. 1993). Mutants lacking any of these signals constitutively become dauers (Daf-c) regardless of food availability. When we treated three Daf-c mutants [daf-2 (insulin receptor mutant), daf-7 (TGF-β ligand mutant), and daf-11 (guanylate cyclase mutant)] with blmp-1 RNAi, none became dauers (Figure 2, D–F and Figure S2B). This confirms that blmp-1 is essential for dauer formation. Among three daf-7 mutants when treated with blmp-1 RNAi showed the most consistent and strongest phenotypes; after 4 days, ∼30% of daf-7 mutants treated with blmp-1 RNAi arrested before becoming dauers. Further, ∼70% died during dauer molting (Figure 2F and Figure S2, C and D). After 9 days, there were no viable blmp-1 RNAi-treated daf-7 worms (Figure 4B). Therefore we focused on daf-7 to further study the mechanisms of blmp-1 in dauer development.
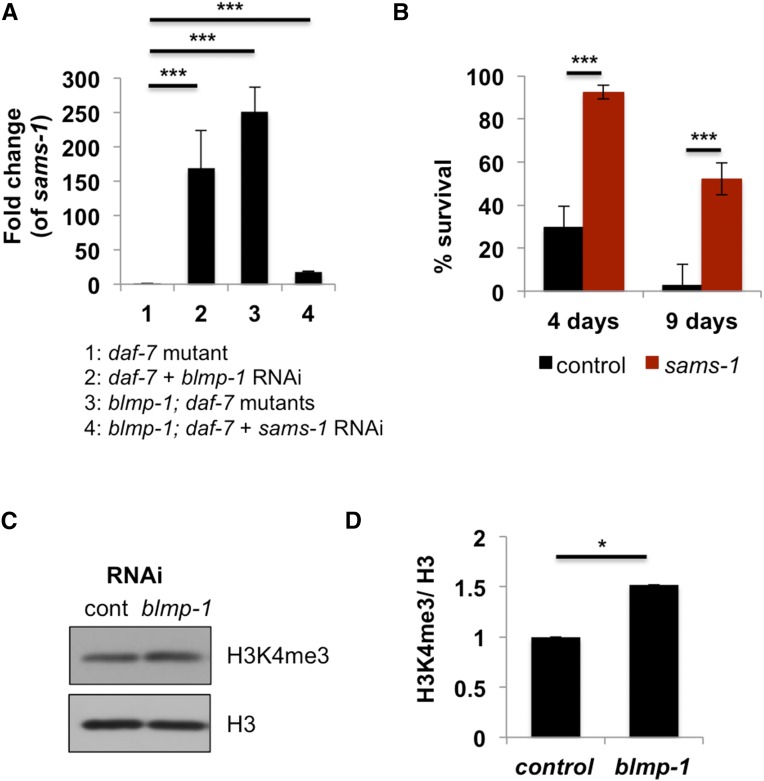
Figure 4.
BLMP-1 regulates dauer development by repressing SAMS-1 expression. (A) The mRNA level of sams-1 increases in the absence of BLMP-1 (either by RNAi, lane 2 or a mutation, lane 3) in the daf-7 mutant background. *** P < 0.001 by Student’s t-test. (B) Percentage of survival of blmp-1; daf-7 mutants with each RNAi was counted on the indicated days (4d and 9d). *** P < 0.001 by Student’s t-test. (C) The levels of histone trimethylation (H3K4me3) increase in knockdown of blmp-1 in daf-7 mutant compared to daf-7 treated with control RNAi. (D) Quantitation of the results in C. The values are normalized by total histone H3. The values are mean ± SEM of three independent experiments. *P < 0.05 by Student’s t-test.
To examine whether the arrest and lethality that blmp-1 RNAi causes in daf-7 mutants is due to BLMP-1’s role in general development in these mutant backgrounds or due to its specific role in dauer development, we performed two independent experiments. First, we treated daf-7 and daf-7; daf-3 double mutants with blmp-1 RNAi. Both mutants have a defect in the daf-7 gene. However, the double mutants cannot become dauers because of the missing downstream effector daf-3 (SMAD). If the arrest and the lethality induced by RNAi of blmp-1 in the daf-7 mutant background is simply because of lack of daf-7, both mutants should show the same arrest and lethality phenotypes. If the phenotypes are specifically due to the daf-7 role in dauer formation, however, only the daf-7 single mutant will show the phenotypes because the double mutants do not become dauers. The double mutants grown at 25° were not arrested or dead (Figure S2E). This confirms that the arrest and the lethality phenotypes observed in the absence of blimp-1 are due to BLMP-1’s specific role in dauer development, but not due to simple absence of daf-7 function. Second, to test whether downregulation of blmp-1 leads to lethality during dauer formation is dauer specific, we treated blmp-1; daf-7 double mutants with three different temperatures: 15°, 20°, and 25°. Most Daf-c mutants, including daf-7, grow to adult stage at 15° or 20° but they become dauers at a high temperature such as 25°. Under our conditions, ∼5% of daf-7 mutants became dauer at 15°, 20% at 20°, and 100% at 25°. In all three conditions, none of the daf-7 single mutants were dead. In contrast, blmp-1; daf-7 double mutants show increased lethality that is temperature dependent (Figure 2G), showing that the lethality is linked to the dauer formation process. When we measured the levels of mRNA and protein of BLMP-1, we noted that there was an increase at the L2d stage (Figure 2, H and I) in daf-7 mutants that then decreased upon entering dauer (Figure 2, H and I). These results also support a role for blmp-1 in dauer formation.
LIN-40 (MTA1) interacts with BLMP-1 to promote dauer formation in daf-7 mutants
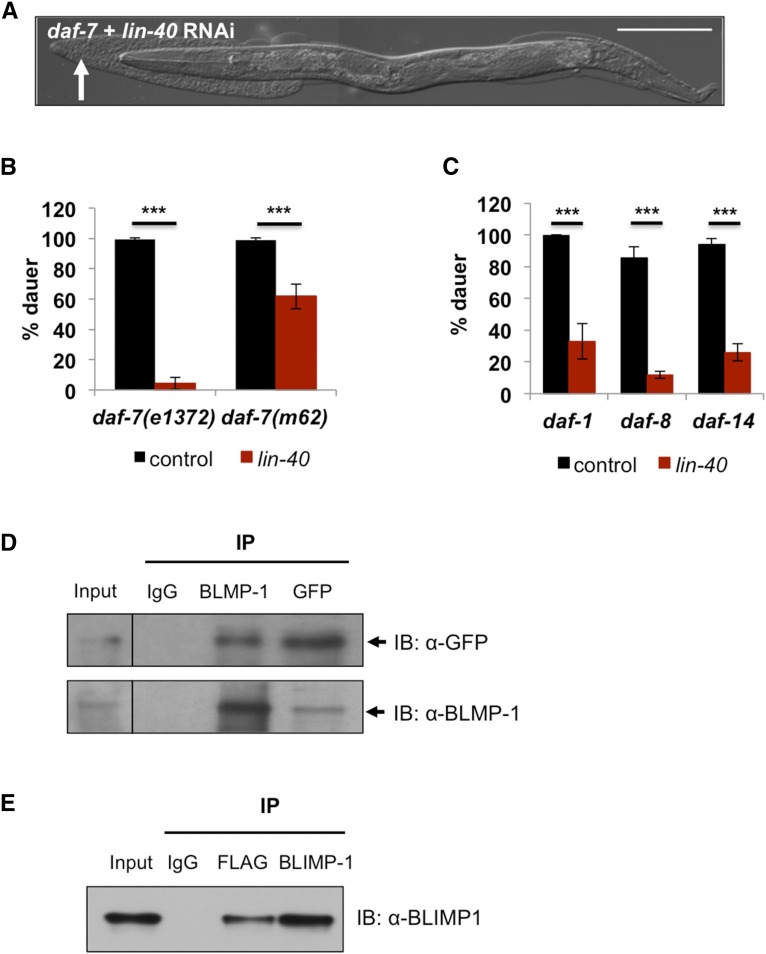
To determine the molecular mechanisms by which blmp-1 regulate dauer development in daf-7 mutants, we performed an RNAi screen of 283 histone modification and nucleosome remodeling genes from the Ahringer library (Fraser et al. 2000; Kamath and Ahringer 2003). We chose to screen those genes because of the known roles of the PRDM family in epigenetic regulation in mammals (Hohenauer and Moore 2012). Sixteen genes prevented dauer formation in daf-7 mutants when their expression was reduced by RNAi (Table S1). Among these, knockdown of 11 genes by RNAi caused lethality or larval arrest or delayed growth of reproductive (nondauer) wild-type animals (Table S2), indicating their essential roles in general development. This could suggest that knockdown of these genes prevents dauer formation in daf-7 mutants simply because the worms were unable to reach the developmental stage to become dauers. Among the remaining 5 genes, lin-40, a homolog of MTA1, which is implicated in tumor metastasis downstream of TGF-β in mammals (Thiery 2002), phenocopied RNAi of blmp-1 best (Figure 3A); >35% of the worms died during dauer molt and none of the survivors became dauers (Figure 3B).When we tested another mutant daf-7(m62) allele, although the percentage was reduced, lin-40 RNAi still prevented daf-7 mutants from becoming dauers (Figure 3B). The m62 allele of daf-7 shows a lot weaker dauer phenotype than the e1372 allele and the mutants have a high frequency of spontaneous recovery from dauers unlike e1372 allele. One possible explanation could be that BLMP-1 and LIN-40 interaction is weak in this mutant and somehow the weak commitment to dauer development could save the mutant from death in the absence of lin-40. Also, BLMP-1 could interact with a different partner in addition to LIN-40 and somehow the redundancy could selectively benefit the m62 allele. Nonetheless, when we examined all Daf-c mutants downstream of TGF-β daf-7 after treating them with blmp-1 RNAi or lin-40 RNAi, all reduced dauer formation, confirming that blmp-1 and lin-40 are required for the TGF-β pathway mutants to become dauers (Figure 3C and Figure S3). Like daf-7 mutants, mutants of the TGF-β receptor (daf-1) and two downstream SMADs (daf-8 and daf-14) are Daf-c. However, they too failed to become dauers when treated with RNAi for blmp-1 or lin-40. Under the screening conditions, lin-40 RNAi did not kill wild-type worms (Table S2).
Figure 3.
LIN-40 (MTA1) interacts with BLMP-1 to promote dauer formation. (A) RNAi of lin-40 phenocopies RNAi of blmp-1 in daf-7 mutants. lin-40 RNAi causes lethality during dauer molt. White arrow indicates a piece of cuticle separated from the body during dauer molt. Bar, 100 µm. (B) lin-40 RNAi prevents dauer formation in two different daf-7 alleles. The values are mean ± SEM, *** P < 0.001 by Student’s t-test. (C) lin-40 is essential for dauer formation in the TGF-β pathway mutants. L4 worms (P0) were treated with RNAi throughout the experiments and the dauer formation of the progeny (F1) was measured. All experiments were performed at 25° (see Materials and Methods). The numbers are mean ± SD, *** P < 0.001 by Student’s t-test. (D) BLMP-1 directly interacts with LIN-40 (see the fourth lane, GFP). A GFP antibody is used to pull down LIN-40 fused with GFP. Lane 1, input; lane 2, immunoprecipitated (IP) with rabbit IgG; lane 3, IP with α-BLMP-1 antibody; and lane 4, IP with α-GFP antibody. (E) HEK 293T cells were cotransfected with His-BLIMP-1 and Flag-MTA1. Immunoblots were developed with α-BLIMP-1 after immunoprecipitation using as a marked. Lane 1, input; lane 2, immunoprecipitated (IP) with mouse IgG; lane 3, IP with α-Flag antibody; and lane 4, IP with α-BLIMP-1 antibody.
Next we tested whether BLMP-1 and LIN-40 interact directly. BLMP-1 was co-immunoprecipitated (co-IP) with LIN-40, confirming the interaction between BLMP-1 and LIN-40 (Figure 3D). To test if this interaction is also conserved in mammals, we coexpressed BLIMP-1/PRDM-1 and MTA1 (the mammalian homolog of BLMP-1 and LIN-40, respectively) in HEK 293T cells and performed a co-IP experiment. BLIMP-1 co-immunoprecipitated with MTA1 (Figure 3E), showing that LIN-40/MTA1 directly associates with BLIMP-1/PRDM-1 and that this interaction could be conserved in mammals.
C. elegans has two MTA1 homologs: lin-40 and egl-27. However, egl-27 RNAi did not prevent daf-7 mutants from dauer formation (Figure S4). Because MTA1 proteins function as a part of the nucleosome remodeling and deacetylase (NuRD) complex (Xue et al. 1998; Zhang et al. 1998), we tested two of the known components of the NuRD complex in worms (Solari and Ahringer 2000; Passannante et al. 2010). Both wild type and daf-7 mutants treated with the RNAi of the NuRD genes (lin-53 and hda-1) became sick and arrested at L1 or L2 stages (or displayed embryonic lethality), suggesting that NuRD complex genes are essential for animals’ growth. Therefore we could not conclude if the NuRD components are required specifically for dauer formation in daf-7 mutants.
BLMP-1 regulates histone H3 trimethylation via SAMS-1
To find downstream targets of blmp-1 in daf-7 dauer development, we performed microarrays and compared the gene expression profiles between daf-7 mutants and blmp-1; daf-7 mutants after 24 hr from L1 when most daf-7 mutants enter the L2d stage. L2d stage is a prior stage of a dauer, when C. elegans is preparing to enter dauer development. L2ds are similar to L2s in size but differ slightly in age with a 9-hr developmental delay. This delay is believed to let them prepare to become stress-resistant larvae. They show signs of entering into the dauer development process, such as dark body color (Golden and Riddle 1984). We chose this stage to collect the samples for microarray, because this stage is the latest time point we could collect live animals and also because this stage would give us the most distinguished expression profiles relevant to dauer development but not to reproductive (nondauer) development. We identified that the expression levels of 117 genes (59 up, 58 down) were significantly changed in the absence of blmp-1 (Table S3, Table S4, Table S5, and Table S6) in the daf-7 mutant background. The 117 genes included targets relevant in signaling, metabolism, development, and nuclear hormone receptor regulation.
We also performed another set of microarrays to compare blmp-1 mutants to wild type collected at L2 stage to examine whether there are common genes that are regulated by blmp-1 in both developmental processes (Table S7, Table S8, Table S9, and Table S10). Most of them do not overlap with those regulated during dauer development (including the seven genes we tested below), demonstrating that BLMP-1 regulates different sets of genes depending on the developmental processes.
To test if the genes identified from the microarrays regulate dauer development as BLMP-1 does, we first tested the several upregulated genes from the list by individually knocking down their expression by RNAi in the blmp-1; daf-7 double mutant background. We reasoned that because knockdown of blmp-1 in daf-7 background causes lethality, if we remove the upregulated gene, then it would rescue the lethality. We found that haf-6, pept-1 (Figure S5A), and sams-1 RNAi rescued lethality in blmp-1; daf-7. haf-6 encodes a half-molecule ATP-binding cassette (ABC) transporter (Sundaram et al. 2006) and pept-1 encodes a low-affinity/high-capacity oligopeptide transporter whose activity is required for uptake of intact peptides from the intestine (Fei et al. 1998). At this point, we do not know how knockdown of these genes reduced the lethality of blmp-1; daf-7 mutants. Still, these results show that our microarray results successfully identified those genetic interactions that are relevant to this pathway. The rescue of blmp-1; daf-7 by knockdown of sams-1 was most interesting to us because of the known roles of SAMS-1. SAMS-1 encodes S-adenosyl methionine synthase to produce SAM (S-adenosyl methionine), which is a methyl group donor for histone methylation and plays a significant role in tumor suppression in mammals and life span in C. elegans. In mammals, overexpression of S-adenosyl methionine synthase isoform type 1 (MAT1A, the SAMS-1 homolog) increased the levels of DNA methylation and histone methylation (Reytor et al. 2009) and suppressed tumor growth rate and tumor weight (Li et al. 2010). In C. elegans, sams-1 is essential for lipid homoeostasis, which supports survival under harsh conditions (Li et al. 2011), and knockdown of sams-1 extends lifespan (Hansen et al. 2005). Because both homologs of BLMP-1 and LIN-40 in mammals function with histone modification machinery, and because BLMP-1 mainly functions as a repressor, SAMS-1 could be an appropriate target of BLMP-1 in C. elegans dauer development.
First, we confirmed the microarray result by qPCR; in blmp-1; daf-7 mutants sams-1 expression was increased compared to daf-7 mutant (Figure 4A). After 4 days at 25°, 26.6% of blmp-1; daf-7 mutants treated with control RNAi survived, whereas 92.4% of the blmp-1; daf-7 mutant treated with sams-1 RNAi survived (Figure 4B). To test whether the increase of SAMS-1 leads to changes in histone methylation, we tested two different histone modifications: H3K4me3 and H3K9me2/3 methylation. Generally, H3K4me3 are associated with active transcription and H3K9me2/3 are associated with transcriptional repression. Knockdown of blmp-1 in daf-7 mutants indeed increased histone H3K4me3 methylation (Figure 4, C and D); however, H3K9me2/3 methylation is not changed (Figure S5, B and C). These results suggest that blmp-1 regulates SAM expression that led to changes in histone modification and eventually to a differential development program specific for dauers.
LPR-3, NHR-23, and PTR-4 regulate dauer development acting downstream of BLMP-1
We then examined the genes that were most downregulated when blmp-1 was absent. We reasoned that if these genes are downstream targets of blmp-1, knockdown of these genes in the daf-7 mutant background will phenocopy blmp-1; daf-7 lethality or inability to develop into a dauer. Among several top-hit genes, knockdown of lpr-3 prevented daf-7 mutants from becoming dauers (Figure S6A). lpr-3 encodes a protein related to the lipocalin family that bind and transport lipophilic molecules. lpr-1, a member of the same family, is required for early larval development and normal growth rate (Stone et al. 2009). Despite the fact that the phenotype of lpr-3 RNAi is similar to that of blmp-1 RNAi in daf-7 mutants, and that the expression level of lpr-3 only significantly changed in dauer development (but not in reproductive nondauer development), lpr-3 seems necessary for both nondauer and dauer development programs (Figure S6B). We speculate that it is probable that lpr-3 is a common target for both dauer and nondauer development programs and different transcription machinery and transcription factors could regulate its expression. For instance, for the dauer development program, BLMP-1 could mediate its expression and for the nondauer development program, other factors do. Furthermore, this difference in transcriptional machinery could regulate the timing or the level of the gene expression.
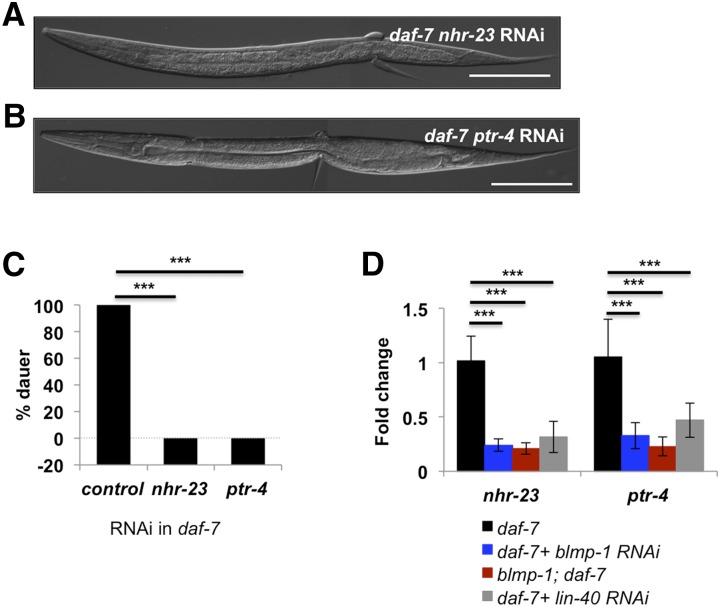
Because knockdown of blmp-1 or lin-40 in daf-7 mutant background causes lethality mostly during dauer molting, we next focused on molting-related genes among the list from the microarrays. We noted two such genes: nhr-23 and ptr-4 (Frand et al. 2005). nhr-23 is a nuclear hormone receptor known to function in all four molts during development (Kostrouchova et al. 2001), while ptr-4 is a distant homolog of Drosophila PATCHED and human PTCH (Zugasti et al. 2005; Burglin and Kuwabara 2006) and is required for normal molting in C. elegans from L4 to adult. Again, their gene expression depends on blmp-1 only during dauer development and not in reproductive nondauer development; yet these genes regulate molting progress for both development programs (Figure S6B). Nonetheless, RNAi of nhr-23 or ptr-4 in daf-7 mutants phenocopied the lethality during dauer molt in daf-7 mutants caused by blmp-1 RNAi (Figure 5, A–C). Moreover, ChIP assays of daf-7 mutant showed that BLMP-1 directly binds to the promoters of nhr-23 and ptr-4 through a consensus sequence (Kuo and Calame 2004) and did not bind in blmp-1; daf-7 mutant (Figure S7, A–C). Lastly, the mRNA levels of nhr-23 and ptr-4 were downregulated in lin-40 RNAi-treated worms as well as in blmp-1 RNAi or blmp-1 mutants (Figure 5D). Taken together, our results show that BLMP-1 differentially regulates expression of multiple genes to regulate a specific development process through an interaction with LIN-40 in the absence of TGF-β signaling.
Figure 5.
BLMP-1 and LIN-40 regulate the same downstream targets to regulate dauer development. (A and B) nhr-23 RNAi (A) or ptr-4 RNAi (B) causes lethality in daf-7 mutants during dauer molt. Bar, 100 µm. (C) Dauer formation of daf-7(e1372) were counted after treatment with nhr-23 RNAi or ptr-4 RNAi. The y-axis was lowered to start from −20 to visualize the 0% dauer. *** P < 0.001 by Student’s t-test. (D) The levels of mRNA of nhr-23 and ptr-4 decrease by RNAi of blmp-1 or lin-40. *** P < 0.001 by Student’s t-test.
Discussion
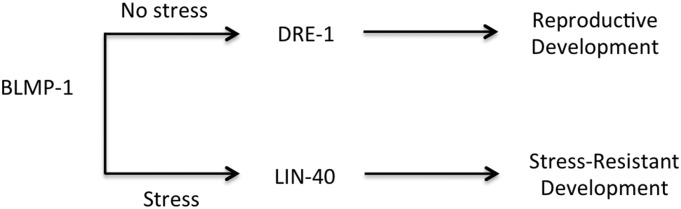
Recent studies suggest that chromatin regulators are required not for steady-state transcription but for normal transcriptional reprogramming in response to environmental cues (Weiner et al. 2012). C. elegans have conserved epigenetic regulation markers and genes to mediate epigenetic modification (Gerstein et al. 2010; Liu et al. 2011; Wenzel et al. 2011). Histone modifiers including SET domain-containing proteins play critical roles in developmental programming as well as reprogramming by environmental cues, including germ line differentiation and lifespan determination (Xu and Strome 2001; Yang et al. 2002; Bender et al. 2004; Agger et al. 2007; Andersen and Horvitz 2007; Christensen et al. 2007; Fisher et al. 2010; Greer et al. 2010, 2011). Yet, despite extensive studies of dauer formation pathways that integrate specific environmental cues and appear to depend on transcriptional reprogramming, the role of epigenetic regulation in dauer development has remained unclear. Here we propose that during dauer development, cells undergo transcriptional reprogramming via BLMP-1. BLMP-1 employs epigenetic processes recruiting a distinct partner such as LIN-40 in the absence of TGF-β signaling. In their recent studies, Horn et al. (2014) discovered that DRE-1 regulates various developmental processes in C. elegans interacting with BLMP-1. They showed that blmp-1 is required for dauer formation under cholesterol-deficient conditions. We also found that BLMP-1 is necessary for dauer formation in the absence of cGMP, insulin, and TGF-β signaling, the three signals whose absence leads to dauer formation regardless of the environment. Interestingly, their data showed that DRE-1 interacts with BLMP-1 for reproductive developmental processes but not for dauer formation, suggesting that BLMP-1 could play a unique role in dauer formation employing different pathways or partners. Overall, our and their work both show that BLMP-1 plays differential roles in the developmental processes depending on the environment (Figure 6).
Figure 6.
A model of BLMP-1 function in two distinct development processes. Under nonstressful conditions, BLMP-1 regulates reproductive growth interacting with DRE-1 (FBXO11). Under stressful conditions, however, the animals undergo transcriptional reprogramming via BLMP-1 specifically interacting with LIN-40.
The TGF-β pathway plays a central role in modulating cell proliferation in mammals, and, correspondingly, mutations of the TGF-β pathway contribute to cancer development and progression. Downstream of TGF-β signaling, MTA1 function has been shown to promote tumor metastasis in mammals (Thiery 2002; Li et al. 2012); however, MTA1 interaction with the BLIMP-1/PRDM-1 was unknown. Intriguingly, dysregulation of BLIMP-1 or MTA1 causes the same B cell neoplasm in the form of diffuse large B cell lymphomas (DLBCL) in mammals, pointing to their genetic interaction in B cell differentiation (Pasqualucci et al. 2006; Tam et al. 2006; Bagheri-Yarmand et al. 2007). Our finding that BLMP-1/PRDM-1 and LIN-40/MTA1 interact in C. elegans suggests that BLIMP-1 and MTA1 may also function together in tumorigenesis in mammals.
We found several potential downstream targets of blmp-1 that regulate dauer formation in daf-7 mutants. Knockdown of blmp-1 in daf-7 mutants increased sams-1 expression that leads to an increase in methylation of H3K4me3. It is possible that BLMP-1 regulates SAM levels via repressing the transcription of sams-1, and the low level methylation of H3K4me3 in certain genes could be critical for dauer formation. Additionally the role of ptr-4 as a downstream target of both BLMP-1 and LIN-40 is intriguing, given that mammalian PTCH, a distant homolog of PTR-4, serves as a negative regulator for the SHH/PTC pathway as well as for TGF-β-dependent tumorigenesis (Pearse et al. 2001).
LIN-40/MTA1 interacts with the NuRD complex (LET-418, CHD-3, and HAD-1) (Passannante et al. 2010) in C. elegans. Our findings support the idea that BLIMP-1/PRDM-1 may be recruited to the NuRD complex via LIN-40/MTA1 whereby it then mediates transcriptional repression. Our results suggest that BLIMP-1/BLMP-1 and MTA1/LIN-40 may act in a conserved epigenetic pathway that controls larval development in worms as well as cancer development and stress in mammals.
Acknowledgments
We thank Dr. Robert Horvitz, Dr. Min Han, Dr. Chantal Wicky, Dr. Chris Gissendanner, and Dr. Ann Sluder for RNAi constructs, Tana Blevins for technical help, Jeremy A. Meier for helpful discussions, Paul Wade (Flag-MTA1 plasmid) and Adam Antebi (His-BLIMP-1 plasmid) for providing plasmids, and the Caenorhabditis Genetics Center [National Institutes of Health (NIH) P40-OD010440] and National BioResource Project in Japan for strains. This work was supported by Virginia Commonwealth University School of Medicine (Y.-J.Y.), Nagoya Research Center for Brain and Neural Circuits (Y.-J.Y.), Inha University (J.K.), R01-DK080074-01 from NIH (C.D.), and R01-GM088290 from NIH (F.C.S.).
Author contributions: M.H., J.K., F.C.S., and Y.-J.Y. devised the experiments. C.D. performed microarray and analyzed the data. M.H. and J.K. performed all the other experiments. F.C.S. provided ascarocides. All authors contributed to writing the paper.
Footnotes
Communicating editor: B. Goldstein
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190793/-/DC1.
Literature Cited
- Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., et al. , 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Yarmand R., Balasenthil S., Gururaj A. E., Talukder A. H., Wang Y. H., et al. , 2007. Metastasis-associated protein 1 transgenic mice: a new model of spontaneous B-cell lymphomas. Cancer Res. 67: 7062–7067. [DOI] [PubMed] [Google Scholar]
- Bender L. B., Cao R., Zhang Y., Strome S., 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14: 1639–1643. [DOI] [PubMed] [Google Scholar]
- Bikoff E. K., Morgan M. A., Robertson E. J., 2009. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 19: 379–385. [DOI] [PubMed] [Google Scholar]
- Burglin T. R., Kuwabara P. E., 2006. Homologs of the Hh signalling network in C. elegans. WormBook 28: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R. A., Fujita M., Schroeder F. C., Clardy J., 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3: 420–422. [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Kim E., Clardy J., 2008. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc. Natl. Acad. Sci. USA 105: 14288–14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P. A., Pasini D., Rose S., et al. , 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Dumur C. I., Nasim S., Best A. M., Archer K. J., Ladd A. C., et al. , 2004. Evaluation of quality-control criteria for microarray gene expression analysis. Clin. Chem. 50: 1994–2002. [DOI] [PubMed] [Google Scholar]
- Fei Y. J., Fujita T., Lapp D. F., Ganapathy V., Leibach F. H., 1998. Two oligopeptide transporters from Caenorhabditis elegans: molecular cloning and functional expression. Biochem. J. 332(Pt 2): 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K., Southall S. M., Wilson J. R., Poulin G. B., 2010. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev. Biol. 341: 142–153. [DOI] [PubMed] [Google Scholar]
- Frand A. R., Russel S., Ruvkun G., 2005. Functional genomic analysis of C. elegans molting. PLoS Biol. 3: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., et al. , 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L., 1984. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102: 368–378. [DOI] [PubMed] [Google Scholar]
- Greer E. L., Maures T. J., Hauswirth A. G., Green E. M., Leeman D. S., et al. , 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Maures T. J., Ucar D., Hauswirth A. G., Mancini E., et al. , 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I., Wu J., Fejer G., Seto E., Wright K. L., 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5: 299–308. [DOI] [PubMed] [Google Scholar]
- Hall S. E., Beverly M., Russ C., Nusbaum C., Sengupta P., 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20: 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A. L., Dillin A., Kenyon C., 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M., Dean W., Reik W., 2009. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 10: 526–537. [DOI] [PubMed] [Google Scholar]
- Hohenauer T., Moore A. W., 2012. The Prdm family: expanding roles in stem cells and development. Development 139: 2267–2282. [DOI] [PubMed] [Google Scholar]
- Horn M., Geisen C., Cermak L., Becker B., Nakamura S., et al. , 2014. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev. Cell 28: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. F., Cho C. Y., Cheng Y. T., Huang J. W., Wu Y. Z., et al. , 2014. BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10: e1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., et al. , 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S. A., Garrett-Sinha L. A., 2009. Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp. Cell Res. 315: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Baylin S. B., 2007. The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Tarlinton D. M., Dietrich W., Corcoran L. M., et al. , 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z., Rall J. E., 2001. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98: 7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. C., Calame K. L., 2004. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 173: 5556–5563. [DOI] [PubMed] [Google Scholar]
- Lee I., Hendrix A., Kim J., Yoshimoto J., You Y. J., 2012. Metabolic rate regulates L1 longevity in C. elegans. PLoS One 7: e44720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Murphy C. T., Kenyon C., 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 10: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., Fleming J. T., 1995. Basic culture methods. Methods Cell Biol. 48: 3–29. [PubMed] [Google Scholar]
- Li D. Q., Pakala S. B., Nair S. S., Eswaran J., Kumar R., 2012. Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer Res. 72: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ramani K., Sun Z., Zee C., Grant E. G., et al. , 2010. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenicity in mice. Am. J. Pathol. 176: 2456–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Na K., Lee H. J., Lee E. Y., Paik Y. K., 2011. Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J. Biochem. 149: 529–538. [DOI] [PubMed] [Google Scholar]
- Liu T., Rechtsteiner A., Egelhofer T. A., Vielle A., Latorre I., et al. , 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbaum J., Bhagat G., Tang H., Mo T., Brahmachary M., et al. , 2010. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell 18: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A. J., Tissenbaum H. A., 2008. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3: 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K., Zhang T., Allawi H., Gomez M., Liu Y., et al. , 2010. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: a potential role of the microRNA let-7. Am. J. Pathol. 177: 1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Compagno M., Houldsworth J., Monti S., Grunn A., et al. , 2006. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J. Exp. Med. 203: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passannante M., Marti C. O., Pfefferli C., Moroni P. S., Kaeser-Pebernard S., et al. , 2010. Different Mi-2 complexes for various developmental functions in Caenorhabditis elegans. PLoS One 5: e13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse R. V., 2nd, Vogan K. J., Tabin C. J., 2001. Ptc1 and Ptc2 transcripts provide distinct readouts of Hedgehog signaling activity during chick embryogenesis. Dev. Biol. 239: 15–29. [DOI] [PubMed] [Google Scholar]
- Pinheiro I., Margueron R., Shukeir N., Eisold M., Fritzsch C., et al. , 2012. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150: 948–960. [DOI] [PubMed] [Google Scholar]
- Potts M. B., Wang D. P., Cameron S., 2009. Trithorax, Hox, and TALE-class homeodomain proteins ensure cell survival through repression of the BH3-only gene egl-1. Dev. Biol. 329: 374–385. [DOI] [PubMed] [Google Scholar]
- Reytor E., Perez-Miguelsanz J., Alvarez L., Perez-Sala D., Pajares M. A., 2009. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 23: 3347–3360. [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Swanson M. M., Albert P. S., 1981. Interacting genes in nematode dauer larva formation. Nature 290: 668–671. [DOI] [PubMed] [Google Scholar]
- Solari F., Ahringer J., 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10: 223–226. [DOI] [PubMed] [Google Scholar]
- Stone C. E., Hall D. H., Sundaram M. V., 2009. Lipocalin signaling controls unicellular tube development in the Caenorhabditis elegans excretory system. Dev. Biol. 329: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B. Stat. Methodol. 64: 479–498. [Google Scholar]
- Su S. T., Ying H. Y., Chiu Y. K., Lin F. R., Chen M. Y., et al. , 2009. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell. Biol. 29: 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988 Methods, pp. 587–606 in The Nematode Caenorhabditis Elegans, edited by W.B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sundaram P., Echalier B., Han W., Hull D., Timmons L., 2006. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol. Biol. Cell 17: 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W., Gomez M., Chadburn A., Lee J. W., Chan W. C., et al. , 2006. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 107: 4090–4100. [DOI] [PubMed] [Google Scholar]
- Thiery J. P., 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Thomas J. H., Birnby D. A., Vowels J. J., 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. A., Jr, Mack D. H., Davis M. M., 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77: 297–306. [DOI] [PubMed] [Google Scholar]
- Wang Z., Stoltzfus J., You Y. J., Ranjit N., Tang H., et al. , 2015. The nuclear receptor DAF-12 regulates nutrient metabolism and reproductive growth in nematodes. PLoS Genet. 11: e1005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Chen H. V., Liu C. L., Rahat A., Klien A., et al. , 2012. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10: e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D., Palladino F., Jedrusik-Bode M., 2011. Epigenetics in C. elegans: facts and challenges. Genesis 49: 647–661. [DOI] [PubMed] [Google Scholar]
- Xu L., Strome S., 2001. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics 159: 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G. T., Young M. K., Cote J., et al. , 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2: 851–861. [DOI] [PubMed] [Google Scholar]
- Yang L., Xia L., Wu D. Y., Wang H., Chansky H. A., et al. , 2002. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21: 148–152. [DOI] [PubMed] [Google Scholar]
- You Y. J., Kim J., Cobb M., Avery L., 2006. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K., 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20: 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D., 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289. [DOI] [PubMed] [Google Scholar]
- Zugasti O., Rajan J., Kuwabara P. E., 2005. The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans. Genome Res. 15: 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.