Abstract
Cell proliferation and tissue growth depend on the coordinated regulation of multiple signaling molecules and pathways during animal development. Previous studies have linked mitochondrial function and the Hippo signaling pathway in growth control. However, the underlying molecular mechanisms are not fully understood. Here we identify a Drosophila mitochondrial inner membrane protein ChChd3 as a novel regulator for tissue growth. Loss of ChChd3 leads to tissue undergrowth and cell proliferation defects. ChChd3 is required for mitochondrial fusion and removal of ChChd3 increases mitochondrial fragmentation. ChChd3 is another mitochondrial target of the Hippo pathway, although it is only partially required for Hippo pathway-mediated overgrowth. Interestingly, lack of ChChd3 leads to inactivation of Hippo activity under normal development, which is also dependent on the transcriptional coactivator Yorkie (Yki). Furthermore, loss of ChChd3 induces oxidative stress and activates the JNK pathway. In addition, depletion of other mitochondrial fusion components, Opa1 or Marf, inactivates the Hippo pathway as well. Taken together, we propose that there is a cross-talk between mitochondrial fusion and the Hippo pathway, which is essential in controlling cell proliferation and tissue homeostasis in Drosophila.
Keywords: ChChd3, mitochondria, Hippo pathway, cell proliferation
MITOCHONDRIA are highly dynamic organelles that continually move, fuse, and divide (Chan 2006; van der Bliek et al. 2013). Fusion and fission play an important role in shaping the complex tubular network and maintaining mitochondrial function during development (Chan 2012; Mishra and Chan 2014). Defective mitochondrial fusion and fission are often associated with aging, metabolic malfunction, neurodegenerative disorder, and cancer (Nunnari and Suomalainen 2012; Boland et al. 2013; Itoh et al. 2013). Although many nuclear signaling cascades that target mitochondrial dynamics have been discovered, it remains less clear how mitochondrial dynamics conversely influences different signaling pathways to regulate development and metabolism (Mitra 2013; Kasahara and Scorrano 2014; Mishra and Chan 2014).
The evolutionarily conserved Hippo pathway is a signaling cascade that controls tissue growth and regeneration through the regulation of cell proliferation and apoptosis (Pan 2010; Halder and Johnson 2011; Yu and Guan 2013; Irvine and Harvey 2015). Core components of the Hippo pathway in Drosophila include the Sterile 20-like kinase Hpo (MST1/2 in mammals) and the downstream NDR family kinase Wts (LAST1/2 in mammals), which inhibits the key transcriptional coactivator Yki (YAP/TAZ in mammals) through phosphorylation at S168 (Xu et al. 1995; Harvey et al. 2003; Pantalacci et al. 2003; Udan et al. 2003; Wu et al. 2003; Huang et al. 2005). This phosphorylation leads to the sequestration of Yki in the cytoplasm by interactions with 14-3-3 proteins and prevents the activation of Yki target genes, such as Diap-1, expanded, Cyclin E, and Bantam, which are responsible for cell proliferation and suppression of apoptosis (Ren et al. 2010). Inactivation of most genes of the Hippo pathway causes tissue overgrowth in flies and cancer in mammals (Harvey et al. 2013; Plouffe et al. 2015). In recent years, many upstream signals for the Hippo pathway have been identified through genetic and biochemical studies. These signals include apical–basal polarity, planer cell polarity, mechanical forces, and G-protein-coupled receptor signaling (Schroeder and Halder 2012; Yu and Guan 2013). One key mediator for integrating these signals to the Hippo pathway is the actin cytoskeleton, although the underlying mechanism is largely unknown (Gaspar and Tapon 2014).
The connection between mitochondrial function and the Hippo pathway has been recently discovered in both flies and mammalian cells (Nagaraj et al. 2012; Ohsawa et al. 2012; Sing et al. 2014). Overexpression of Yki or YAP2 leads to the expansion of mitochondria due to increased mitochondrial fusion, in addition to cell overproliferation (Nagaraj et al. 2012). Further genome-wide microarray analysis reveals that many genes associated with mitochondrial function are upregulated by Yki overexpression, including the two mitochondrial fusion genes optic atrophy 1-like (Opa1) and mitochondria assembly regulatory factor (Marf) (Nagaraj et al. 2012). RNA interference (RNAi) knockdown of Opa1 or Marf suppresses the mitochondrial fusion phenotype in Yki-overexpressing cells and also partially inhibits cell proliferation (Nagaraj et al. 2012). These data indicate that the Hippo pathway influences mitochondrial structure and function, which in turn may affect cell proliferation. On the other hand, mutations in components of the mitochondrial respiratory complexes cooperate with ectopic expression of oncogenic Ras to induce nonautonomous overgrowth in the Drosophila developing eye, and this involves the inactivation of the Hippo pathway (Ohsawa et al. 2012). A more recent study has shown that the Hippo pathway upstream component Fat binds to the core component of complex I, Ndufv2, with its cytoplasmic domain and regulates mitochondrial function, although it is independent on Fat’s role in Hippo signaling (Sing et al. 2014). These findings point to a complex relationship between mitochondria and the Hippo pathway.
In this study, we identify the coiled-coil-helix-coiled-coil-helix domain containing 3 (ChChd3) as a novel Drosophila mitochondrial fusion component and show that loss of function of ChChd3 leads to mitochondrial fragmentation and tissue undergrowth. We provide evidence that ChChd3 is an additional mitochondrial fusion target for the Hippo pathway. On the other hand, defects in mitochondrial fusion due to lack of ChChd3 or Opa1/Marf depletion cause inactivation of the Hippo pathway. Thus, our data support the notion that mitochondrial fusion can cross-talk with the Hippo pathway in controlling cell proliferation during Drosophila development.
Materials and Methods
Drosophila stocks and genetics
The following fly stocks were used: w1118, UAS-ChChd3 RNAi I (ChChd3-IR1, NIG-FLY stock center 1715R-1; used in Figure 1), UAS-ChChd3 RNAi II (ChChd3-IR2, Bloomington Drosophila Stock Center BL38984, used in Figure 1 and other figures), UAS-yki RNAi (Vienna Drosophila RNAi Center, V104523), UAS-Opa1 RNAi (BL32358), UAS-Marf RNAi (BL55189), P{PZ}l(3)03670[03670] (BL 11599), Df(3R)BSC749 (BL26847), UAS-yki.S168A.GFP.HA (BL28816), tubulin-Gal4, ey-Gal4/Cyo, GstD1-GFP/Cyo; FRT82B ChChd3D1/TM6B, Mhc-Gal4/TM3 Sb, ptc-Gal4 UAS-GFP/Cyo; puc-lacZ/TM6B, FRT82B, FRT82B ChChd3D1/TM6B, FRT82B wtsx1/TM6B (BL44251), FRT82B ChChd3D1 wtsx1/TM6B, en-Gal4 UAS-GFP/Cyo; Diap1-lacZ/TM6B, CycE-lacZ/Cyo; hh-Gal4 UAS-GFP/TM6B, ex-lacZ/Cyo; hh-Gal4 UAS-GFP/TM6B, hsFLP; FRT82B arm-lacZ/TM6B, hsFLP; Sp/Cyo; FRT82B ubi-mRFP.nls/TM6B, Diap1-lacZ FRT82B ChChd3D1/TM6B, CycE-lacZ/Cyo; FRT82B ChChd3D1/TM6B, ex-lacZ/Cyo; FRT82B ChChd3D1/TM6B, and Mef2-Gal4 UAS-Mito-GFP/TM2, MARCM82B (hsFLP; act-Gal4 UAS-GFP/Cyo; FRT82B tubulin-GAL80ts/TM6B).
Figure 1.
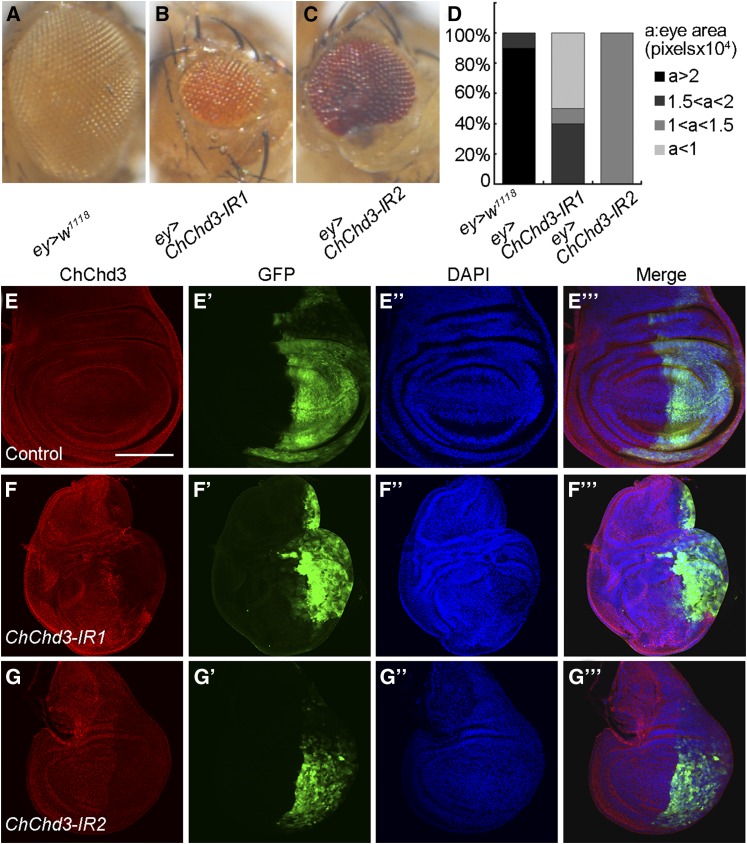
ChChd3 depletion causes tissue undergrowth. (A–C) Adult female eyes expressing the following transgenes under the control of ey-Gal4: (A) control, (B) UAS-ChChd3 RNAi I (ChChd3-IR1), and (C) UAS-ChChd3 RNAi II (ChChd3 IR2). (D) Quantification of eye size of the indicated genotypes. n = 10 for each genotype. (E–G′′′) ChChd3 expression in wing imaginal discs expressing UAS-GFP (E–E′′′), UAS-ChChd3 RNAi I (F–F′′′), and UAS-ChChd3 RNAi II (G–G′′′) under the control of en-Gal4. The en-expressing domain was marked by GFP. ChChd3 protein was effectively knocked down by RNAi. Bar, 100 μm.
To generate the pUAST-ChChd3 construct, the full-length ChChd3 cDNA was PCR amplified with the primers 5′-GAATTCATGGGAGCCCGACAGTCTCA-3′ and 5′-GCGGCCGCCCTAGGCCGCCTTGGCAGGAAC-3′ and cloned into the pUAST vector. This construct was then transformed into w1118 embryos using the standard P-element mediated transgenesis protocol. One line inserted on the second chromosome was used in this study.
The FLP/FRT system was used to induce mitotic clones in wing imaginal discs. Clones were labeled either negatively (absence of β-galactosidase or RFP) or positively [presence of GFP, mosaic analysis with a repressible cell marker (MARCM)]. Larvae were heat shocked for 1 hr at 36–42 hr after egg laying (AEL). Discs were dissected and fixed at 120 hr AEL.
Mutant generation
The ChChd3D1 mutat allele was generated by imprecise mobilization of a P-element insertion P{PZ}l(3)03670 with the standard procedure. Sequence analysis revealed that the deletion removes a 1069-bp genomic DNA fragment (from Ch3R: 31,052,786 to Ch3R: 31,053,854).
Immunostaining, 5-ethynyl-2′-deoxyuridine labeling, and microscopy
For S2 cell immunostaining, cells were resuspended and transferred to the concanavalin A-coated coverslip. Cells were then fixed for 20 min in PBS with 4% paraformaldehyde. For disc immunostaining, late third instar larval wing imaginal discs were dissected in ice-cold 1× PBS (10 mM NaH2PO4/Na2HPO4, 175 mM NaCl, pH 7.4) and fixed for 20 min in PBS with 4% paraformaldehyde. Then fixed cells or discs were washed three times with 0.1% Triton X-100 in PBS (PBT) and blocked in PBT with 3% BSA for 1 hr at room temperature. Next, samples were incubated with primary antibodies overnight at 4° and then washed three times before incubating with secondary antibodies for 2 hr. DAPI was added for the last 20 min. Samples were washed three times with PBT and mounted in Vectorshield. We used the following primary antibodies: chicken anti-GFP (1:2000; Abcam), mouse anti-RFP (1:2000; Abcam), mouse anti-β-galactosidase (1:2000; Abcam), mouse anti-armadillo (1:100; DHSB N2 7A1), rabbit anti-cleaved Caspase 3 (1:100; Cell Signaling), rabbit anti-phosphohistone 3 (Ser10) (1:500; Millipore, Bedford, MA), and mouse anti-ATP synthase-α (anti-ATP5A; 1:500; Mitosciences). Anti-ChChd3 was raised in rabbit against a GST–ChChd3 fusion protein harboring the C-terminal amino acid 77–224 of ChChd3 and used in 1:1000 dilution. Secondary antibodies (Alexa Fluor 488-, 555-, or 633-conjugated, anti-rabbit, anti-mouse, anti-chicken) were from Molecular Probes (1:250 or 1:500). DAPI (1 μg/ml; Sigma, St. Louis, MO) was used to stain for nuclei. For 5-ethynyl-2′-deoxyuridine (EdU) analysis, late third instar larvae were dissected in Schneider’s Drosophila medium, and tissues were incubated for 30 min in 5 μm EdU before fixation. Detection was performed according to the manufacturer’s protocol (C10338, Click-iT EdU Alexa Fluor 555 Imaging Kit; Life Technologies). To visualize mito-GFP-labeled mitochondria in larval body wall cells, the larval body wall was dissected in Schneider’s medium and observed under the confocal microscope. The images were taken on an Olympus FV1000 confocal microscope and processed using Adobe Photoshop. Measurement of clone area was performed using ImageJ software. For TEM analysis, samples were processed according to the standard protocol. Electron micrographs were taken on a Hitachi H-7650 TEM.
Western blotting
Protein extracts from larvae were prepared by grinding larvae in lysis buffer (1× RIPA buffer: 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS) containing the protease inhibitor cocktail (Roche). The lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°. Samples were subjected to SDS/PAGE and transferred to polyvinylidene fluoride membrane. Membranes were immunoblotted with the primary antibodies and then probed with the secondary antibodies. Rabbit anti-ChChd3 (1:2000) and mouse anti-tubulin (1:1000; Developmental Studies Hybridoma Bank, E7) were used as the primary antibodies. Blots were treated with the ChemiLucent ECL detection reagents (Millipore) and protein bands were visualized using the Chemiluminescence Imaging System (Clinx Science Instruments, Shanghai, China).
Quantitative PCR
Total RNA was extracted from 50 third instar larval wing imaginal discs with TRIzol (Invitrogen, Carlsbad, CA) reagent. Complementary DNA (cDNA) was synthesized using oligo-dT primers and PrimeScript RTase (TaKaRa, PrimeScript II 1st Strand cDNA Synthesis Kit). Quantitative PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and the ABI 7900HT Fast Real-Time PCR System with the following primers: ChChd3: 5′-CGACGATGTGGTCAAGCGACT-3′ and 5′-ACTTTCGGAGCAGGAGAAGC-3′; rp49: 5′-GCTAAGCTGTCGCACAAA-3′ and 5′-TCCGGTGGGCAGCATGTG-3′.
Mitochondrial fractionation
S2 cells were grown in Schneider’s medium supplemented with 10% FBS and harvested by centrifugation. Cytosolic and mitochondrial fractions were isolated by differential centrifugation using a commercial kit (Mitochondria Isolation Kit for Tissue, 89801; Thermo Scientific). Both fractions were analyzed by Western blotting using the following antibodies: rabbit anti-ChChd3 (1:2000), mouse anti-ATP5A (1:1000, as mitochondrial loading control), and mouse anti-tubulin (1:1000, as cytosolic loading control). For the assessment of submitochondrial protein localization, isolated mitochondrial pellet was suspended in isotonic buffer and treated with various concentrations of digitonin (0, 0.01, 0.02, and 0.04%). The samples were then subjected to proteolysis with Proteinase K. Proteins were then precipitated with 10% trichloroacetic acid (TCA) and analyzed by Western blotting using rabbit anti-ChChd3 (1:2000), mouse anti-total OXPHOS (1:4000; Abcam, ab110413) and rabbit anti-Tom20 (1:1000; Proteintech, 11802-1-AP) antibodies.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification of ChChd3 as a novel regulator for tissue growth
To identify novel regulators for tissue growth in Drosophila, we performed a genetic screen using ey-Gal4 to drive the expression of UAS-dsRNA (RNAi transgene) in the Drosophila eye and examined the eye size. We screened a small collection of RNAi lines from the National Institute of Genetics fly stock center (NIG-FLY), and found that knockdown of one gene (CG1715, hereafter called ChChd3, see below) caused a significant decrease in eye size (Figure 1, A and B, quantified in Figure 1D). Reduced eye size was also observed when using an independent RNAi line in which the short hairpin RNA (shRNA) was targeted to an additional region of the ChChd3 transcript (Figure 1C, quantified in Figure 1D). Furthermore, antibody staining in the wing imaginal disc coexpressing UAS-GFP and UAS-ChChd3 RNAi transgenes by en-Gal4 reveals that the level of ChChd3 protein was decreased in the posterior compartment of the wing disc for both RNAi lines, suggesting that knocking down of ChChd3 by RNAi was effective (Figure 1, E–G′′′). Thus, ChChd3 is a novel regulator to promote tissue growth in Drosophila eye development.
Generation and characterization of ChChd3 deletion mutant
To further explore the function of ChChd3 during Drosophila development, we generated the ChChd3D1 allele by imprecise excision of a P-element insertion line P{PZ}l(3)03670. ChChd3D1 is a deletion that removes the large portion of the ChChd3 coding region, including the translation start site and amino terminal 226 codons (Figure 2A). ChChd3D1 is homozygous lethal and expected to be a null allele of ChChd3. We confirmed this by performing Western blot analysis. In wild-type larval extracts, the antibody against ChChd3 specifically recognized one band of ∼26 kDa that was absent in ChChd3D1 homozygous mutant larval extracts (Figure 2B).
Figure 2.
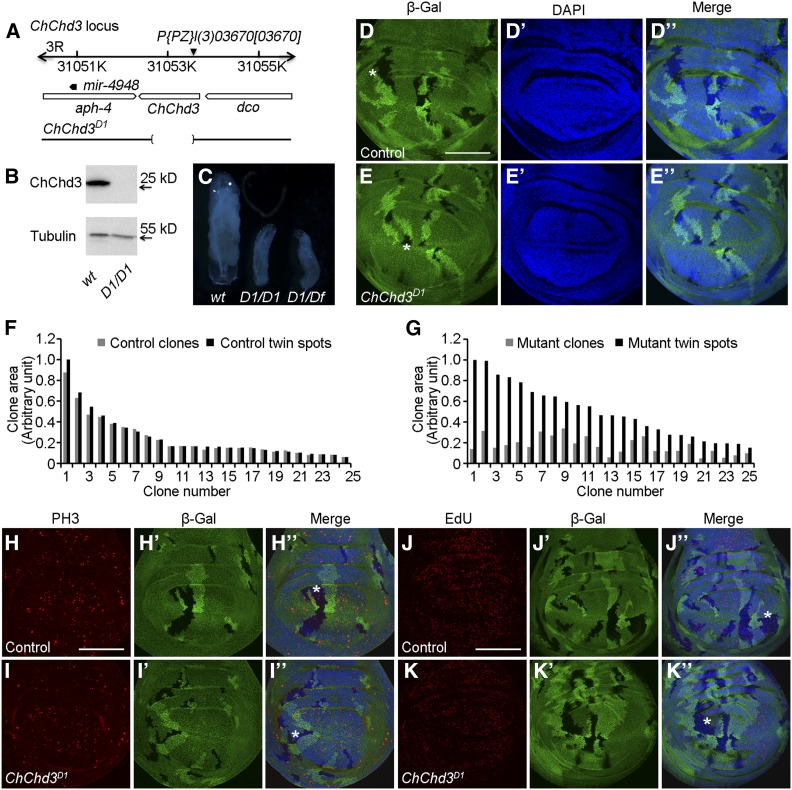
Loss of function of ChChd3 results in growth defects. (A) Schematic representation of the ChChd3 locus. The deletion in ChChd3D1 mutant allele is indicated by the bracketed area. (B) Western blot on second instar larval extracts from wild-type and ChChd3D1 mutant animals. Lysates were probed with anti-ChChd3 and anti-tubulin. (C) Wild-type control and mutant larvae homozygotic for ChChd3D1 or transheterozygous for ChChd3D1 and a deficiency after 5 days of growth. D1 denotes the ChChd3D1 mutant allele. Df denotes the Df(3R)BSC749 deficiency line removing the ChChd3 locus. (D–D′′) Wing imaginal disc with control mitotic clones (lack of β-Gal, black area) and their corresponding twin spots (two copies of β-Gal, brighter area) of similar size. (E–E′′) Wing imaginal disc with ChChd3D1 homozygous mutant clones (lack of β-Gal) that are smaller than their corresponding twin spots (two copies of β-Gal). (F) Measurements of clone area for 25 pairs of control clones and their sister twin spots. (G) Measurements of clone area for 25 pairs of ChChd3D1 homozygous mutant clones and their sister twin spots. (H–I′′) Wing imaginal discs containing control (H–H′′) or ChChd3D1 homozygous mutant (I-I′′) clones stained with anti-β-Gal and anti-PH3 antibodies. ChChd3 mutant clone had reduced number of cells positive for PH3 staining. (J–K′′) Wing imaginal discs containing control (J–J′′) or ChChd3D1 homozygous mutant (K–K′′) clones stained with anti-β-Gal and labeled with EdU. ChChd3 mutant clone had reduced number of cells positive for EdU labeling. Asterisks indicate the clone area. Bars, 100 μm.
Homozygous ChChd3D1 mutant animals displayed a notable growth defect as compared with the wild-type control. To analyze the growth defect in detail, we collected first instar larvae shortly after hatching and aged them for growth analysis. ChChd3D1 mutant larvae grew more slowly than wild-type animals. At 96 hr after larvae hatching, ChChd3D1 mutant larvae were much smaller than wild type and arrested at the second instar larval stage (Figure 2C). This phenotype was further confirmed using a transheterzygous combination for ChChd3D1 and a deficiency that uncovers the ChChd3 region (Figure 2C). In addition, we noticed that these mutant larvae had a protracted larval stage and were able to survive up to 15 days before they died (data not shown). ChChd3 expression from a UAS-ChChd3 transgene under the control of a ubiquitously expressed tubulin-Gal4 was able to rescue the larval growth and lethality defects in ChChd3D1 mutant, suggesting that the growth defects were specifically due to the loss of ChChd3 (Supplemental Material, Table S1).
To define the developmental basis for the undergrowth phenotype, we examined the ChChd3 loss-of-function effect on wing disc development. We recombined the ChChd3D1 onto an FRT82B chromosome and induced homozygous mutant cells in third larval instar wing imaginal discs. Homozygous mutant clones and their associated wild-type clones are recognized by the absence of lacZ and the presence of two copies of lacZ, respectively. ChChd3D1 mutant cells survived, but had a disadvantage for clone growth (Figure 2, D–E′′). Absence of ChChd3 protein in ChChd3D1 mutant clones was also confirmed with anti-ChChd3 antibody staining (Figure S1, A–A′′′). Measurement of the clone area showed that the size of mutant clones was smaller compared to the wild-type twin clones, suggesting that ChChd3D1 mutant cells proliferate less (Figure 2, F and G). The reduction of clone size was not due to the increase of apoptosis, since the Caspase 3 staining did not show detectable difference in both mutant and wild-type clones (Figure S1, B–C′′′). In addition, we did not observe the obvious difference of cell size using anti-Armadillo antibody to labeling the cell membrane in the mutant clone and its twin clone (Figure S1, D–E′′′). Instead, the number of EdU labeling and PH3+ cells was reduced in ChChd3 D1 mutant clones compared with the controls (Figure 2, H–K′′). Thus, we concluded that ChChd3 is required for cell proliferation during Drosophila wing development.
ChChd3 is a mitochondrial inner membrane protein and its mutation affects mitochondrial morphology
ChChd3 encodes a highly conserved protein predicted to be a mitochondrial inner membrane protein according to the role of its mammalian homolog (Schauble et al. 2007; Darshi et al. 2011). It contains a coiled-coil-helix-coiled-coil-helix (CHCH) domain at its C terminus (Figure 3A). To test whether Drosophila ChChd3 is localized to the mitochondrial inner membrane, we first performed immunofluorescence staining in S2 cells. ATP synthase (ATP5A) was used as a mitochondrial inner membrane marker. As shown in Figure 3, B–B′′′, we detected a colocalization of ChChd3 and ATP5A in the cytoplasm of S2 cells. The mitochondrial localization of ChChd3 was further demonstrated with cell fractionation experiments. S2 cell extracts were subjected to differential centrifugation, and each fraction was analyzed with Western blot to detect the presence of ChChd3. As shown in Figure 3C, the ChChd3 protein was highly enriched in the fraction containing the mitochondria, as determined by the mitochondrial marker ATP5A, and it was absent from the cytosolic fraction. Furthermore, we carried out proteolysis assay to determine ChChd3 submitochonrial localization (Figure 3D). ChChd3 was resistant to proteolysis after the outer membrane was disrupted by digitonin treatment, while the mitochondrial outer membrane protein (Tom20) was degraded under these conditions. In this assay, ChChd3 behaved similarly to ATP5A (detected by anti-Total OXPHOS), indicating ChChd3 is located inside the mitochondria. These experiments demonstrate that Drosophila ChChd3 is a mitochondrial inner membrane protein.
Figure 3.
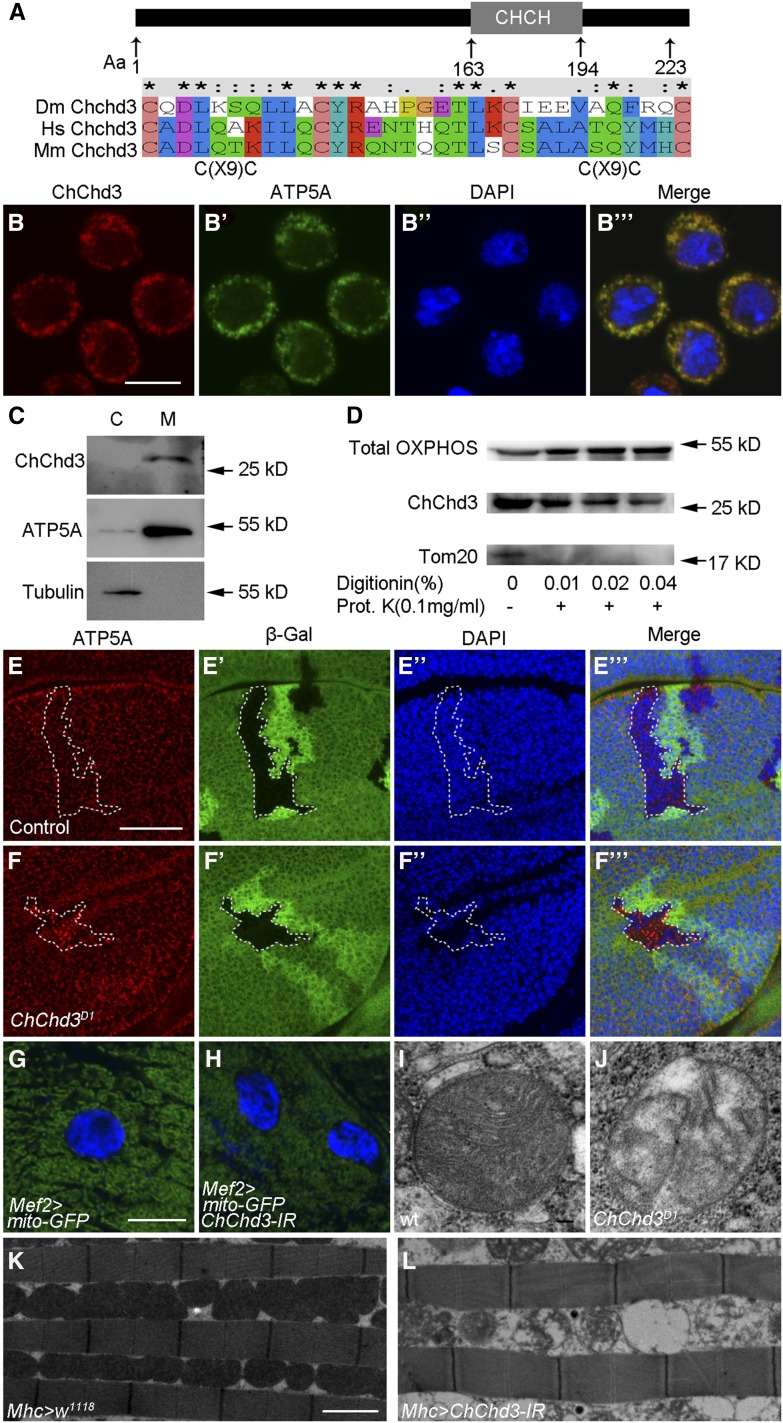
ChChd3 localizes to the inner mitochondrial membrane and is required for mitochondrial fusion and crista formation. (A) Schematic diagram of Drosophila ChChd3 protein domain structure and sequence comparison of Drosophila ChChd3 with human ChChd3 and mouse ChChd3 within the CHCH domain. (B–B′′′) S2 cells showing colocalization of ChChd3 and ATP5A. Samples were stained with anti-ChChd3 and anti-ATP5A antibodies. (C) Western blot analysis of cytosolic and mitochondrial fractions separated by centrifugation. Samples were probed with anti-ChChd3, anti-ATP5A (inner membrane), and anti-tubulin antibodies. ChChd3 is enriched in the mitochondrial fraction. (D) Western blot analysis of ATP5A (detected by anti-Total OXPHOS), ChChd3, and Tom20 proteins from S2 cell mitochondria. Isolated mitochondria were treated with the indicated concentrations of digitonin followed by Proteinase K digestion. (E–F′′′) Wing imaginal discs containing control (E–E′′′) or ChChd3D1 homozygous mutant (F–F′′′) clones stained with anti-β-Gal and anti-ATP5A antibodies. Clones are marked by the absence of β-Gal and the dashed white line. ChChd3 mutant clone cells display punctate ATP5A staining. (G) Larval body wall cells from the control larvae expressing UAS-mito-GFP with Mef2-Gal4 and showing mitochondria with tubular morphology. (H) Knockdown of ChChd3 in the larval body wall cells results in shorter mitochondria. (I and J) TEM images of mitochondria from wild-type (I) or ChChd3D1 mutant larvae. Crista content was reduced in ChChd3D1 mutant mitochondria. (K and L) TEM images of adult indirect flight muscle from the Mhc-Gal4 control (K) and ChChd3 knockdown flies (L). Knockdown of ChChd3 leads to fragmented mitochondria and reduced crista content. Bars, 10 μm in B; 40 μm in E; 10 μm in G; 0.1 μm in I; and 2 μm in K.
Given the fact that lacking ChChd3 leads to growth defects and ChChd3 localizes to the mitochondria, we examined the effect of ChChd3 mutation on mitochondrial morphology. First, we generated ChChd3D1 mutant clones in third instar larval wing imaginal discs and analyzed the cellular localization of ATP5A. The distribution of ATP5A in the ChChd3D1 mutant clone displayed a punctate and dispersed pattern while it is uniform in the twin wild-type clone (Figure 3, E–F′′′). Second, we made use of Mef2-Gal4 to drive the expression of UAS-mito-GFP to label the mitochondria in third instar larval body wall cells. The mitochondria in wild-type cells had a tubular shape; however, when ChChd3 activity was reduced using UAS-ChChd3 RNAi under the control of Mef2-Gal4 driver, the mitochondrial structure was found to be much shorter and fragmented compared with wild type (Figure 3, G and H). These results suggest that Drosophila ChChd3, similar to its mammalian homolog, is required for mitochondrial fusion (Darshi et al. 2011). Moreover, we analyzed the ultrastructure of mitochondria by TEM and found that loss of ChChd3 resulted in reduced crista content in larval cells (Figure 3, I and J). These results were further supported by examining the indirect flight muscle of ChChd3 RNAi flies using TEM analysis. Compared with the control, knockdown of ChChd3 with Mhc-Gal4 led to fragmented mitochondria and disintegration of cristae (Figure 3, K and L). Taken together, ChChd3 is essential for the maintenance of tubular mitochondria structure as well as crista architecture.
ChChd3 is partially required for Hippo pathway-mediated overgrowth
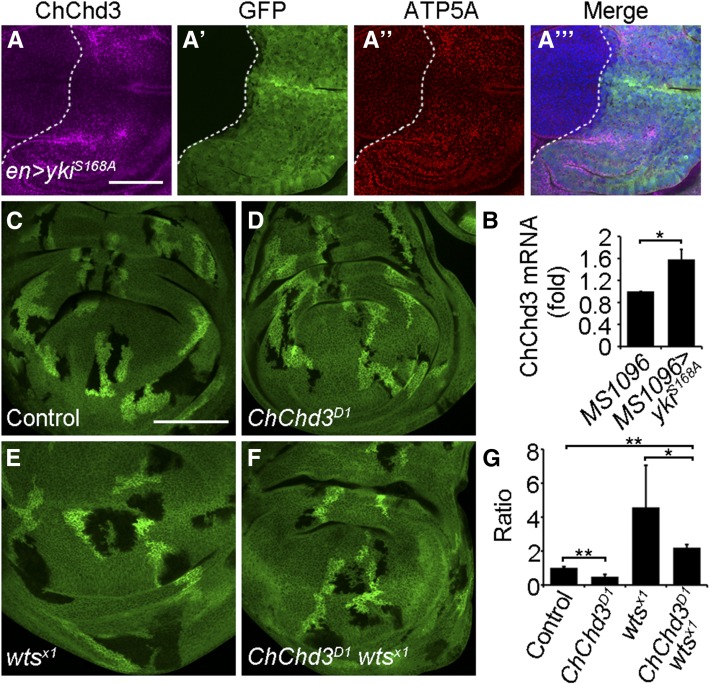
The Hippo pathway controls the structure and fusion of mitochondria through the regulation of expression of a subset of mitochondrial fusion genes, including Opa1 and Marf (Nagaraj et al. 2012). As ChChd3 is required for mitochondrial fusion and cell proliferation, we sought to investigate the relationship between ChChd3 and the Hippo pathway. To assess whether the Hippo pathway is able to regulate ChChd3 expression, we performed immunofluorescence staining with anti-ChChd3 antibody in wing disc tissues where yki was overexpressed. Overexpression of yki with a UAS-yki.S168A transgene under en-Gal4 control in the wing disc caused an increase of ChChd3 protein level in posterior cells (Figure 4, A–A′′′). Cells with increased expression of ChChd3 also show upregulation of ATP5A, which has been reported to be associated with mitochondrial expansion (Figure 4, A–A′′′) (Nagaraj et al. 2012). In addition, ChChd3 messenger RNA (mRNA) levels were also upregulated upon overexpression of yki by MS1096-Gal4 in the wing disc (Figure 4B). Thus, ChChd3 is an additional target of the Hippo pathway in controlling the mitochondrial fusion process. As the depletion of Opa1 or Marf can partially inhibit the overgrown phenotype caused by wts mutation or yki overexpression, we also tested whether removal of ChChd3 was able to suppress the overgrown phenotype induced by wts loss of function (Nagaraj et al. 2012). We used the FLP/FRT system to induce ChChd3, wts, or ChChd3 wts double mutant clones in wing discs. As expected, the size of the ChChd3 mutant clone was smaller than that of its wild-type clone, and the wts mutation produced larger clones and caused overgrowth (Figure 4, C–F, quantified in Figure 4G). Simultaneous removal of ChChd3 and wts led to the reduction of mutant clone size as compared with that of the wts single mutant clone (Figure 4, C–F, quantified in Figure 4G). However, the size of ChChd3 wts double mutant clones was still larger than their corresponding twin spot clones (Figure 4, C–F, quantified in Figure 4G). Taken together, these results demonstrate that the Hippo pathway regulates the expression of ChChd3 and ChChd3 is partially required for Hippo pathway-mediated overgrowth in Drosophila.
Figure 4.
ChChd3 is partially required for Hippo pathway-mediated overgrowth. (A–A′′′) Wing imaginal disc showing an increased ChChd3 protein level in posterior cells upon Yki overexpression by en-Gal4. (B) Quantitative PCR analysis of ChChd3 mRNA levels in wing discs expressing the UAS-yki.S168A transgene under MS1096-Gal4. Wing discs from MS1096-Gal4 flies served as a control. ChChd3 mRNA levels were normalized to rp49. (C–F) Wing imaginal discs with control (C), ChChd3D1 (D), wtsX1 (E), or ChChd3D1 wtsX1 (F) double mutant clones. Mutant clones are marked by lack of β-Gal and their corresponding twin spots are marked by two copies of β-Gal. (G) Quantification of the ratio between the mutant clone area and the twin spot area of wing imaginal discs with indicated genotype. n = 14 clones for each genotype. Bars, 50 μm in A and 100 μm in C.
Depletion of ChChd3 causes inactivation of Hippo activity
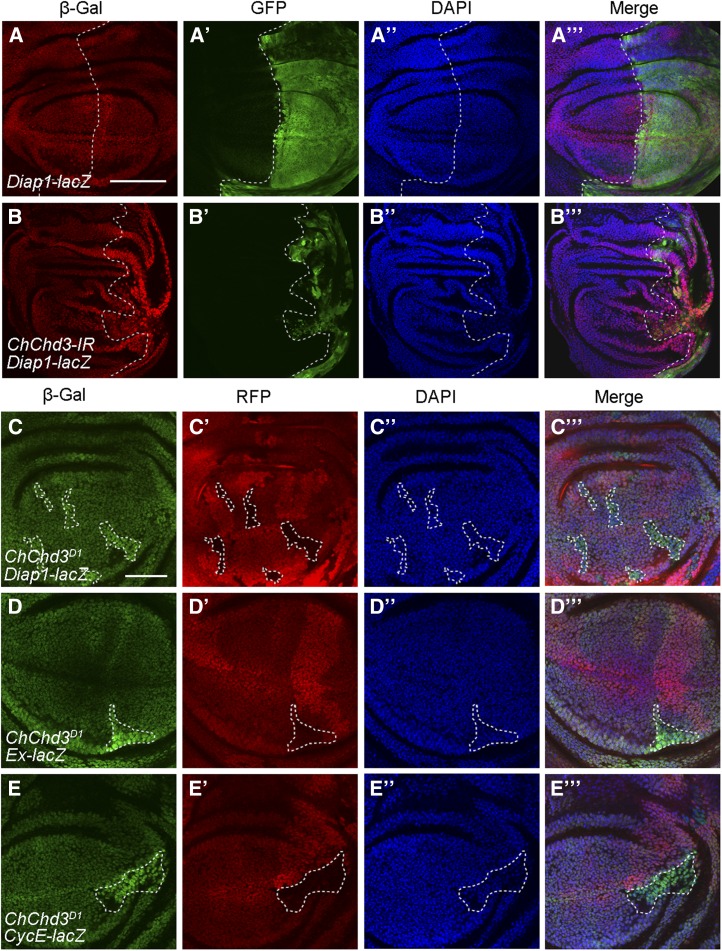
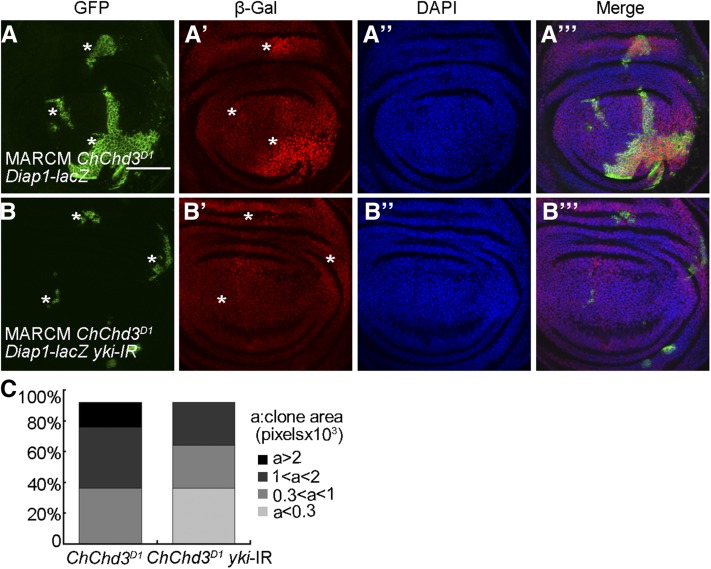
As loss of ChChd3 resulted in tissue undergrowth and slightly suppressed the overgrown phenotype mediated by wts loss of function, we speculated that the Hippo pathway might be hyperactivated in ChChd3 mutant tissues. To address this, we used a lacZ reporter for Diap1, a known Yki target gene, to monitor Hippo activity. In contrast to our expectation, wing discs expressing UAS-ChChd3-RNAi with en-Gal4 exhibited an increase level of Diap1-lacZ expression in the posterior compartment as compared to the control (Figure 5, A–B′′′), which was correlated to the inactivation of the Hippo pathway. Consistent with this finding, the expression of two other Yki target genes, expanded-lacZ (ex-lacZ) and CyclinE-lacZ (CycE-lacZ), was also elevated in the posterior compartments of wing discs upon ChChd3 knockdown by RNAi using hedgehog-Gal4 (hh-Gal4; Figure S2, A–D′′′). To further confirm the specificity of this effect, we examined the expression of these reporter genes in wing disc clones mutant for ChChd3. The expression levels of Diap1-lacZ, ex-lacZ, and CycE-lacZ were all increased in ChChd3 mutant clones compared to their levels in surrounding wild-type cells (Figure 5, C–E′′′). Next, we wanted to test whether Hippo target gene activation caused by ChChd3 mutation is dependent on yki. Using the MARCM system, we found that expression of UAS-yki-RNAi in the ChChd3 mutant background suppressed the upregulation of Diap1-lacZ and caused a significant reduction in the clone size compared to that of ChChd3 single mutant clones (Figure 6, A–B′′′, quantified in Figure 6C). Thus, depletion of ChChd3 causes undergrowth defects, but results in the inactivation of the Hippo pathway.
Figure 5.
Loss of ChChd3 increases Hippo pathway target gene expression. (A–A′′′) Expression pattern of the Diap1-lacZ reporter gene in a control wing imaginal disc expressing the UAS-GFP transgene with en-Gal4. (B–B′′′) Knockdown of ChChd3 by RNAi with en-Gal4 increases the level of Diap1-lacZ expression in posterior cells. Discs (in A–B′′′) were stained with anti-β-Gal and anti-GFP. DAPI was used to label DNA. Posterior cells are marked by GFP. Dashed lines indicate the anterior/posterior compartment boundary. (C–E′′′) Upregulation of Diap1-lacZ (C–C′′′), ex-lacZ (D–D′′′), or CycE-lacZ (E–E′′′) expression in ChChd3D1 homozygous mutant clones in wing imaginal discs. Discs (in C–E′′′) were stained with anti-β-Gal and anti-RFP. DAPI was used to label DNA. Mutant clones are marked by lack of RFP and their corresponding twin spots are marked by two copies of RFP. Dashed lines indicate the clone outline. Bars, 100 μm in A and D; 50 μm in C.
Figure 6.
Yki is required for Hippo target gene expression and cell proliferation in ChChd3 mutant clones. (A–A′′′) Wing imaginal disc containing ChChd3D1 mutant MARCM clones. (B–B′′′) Wing imaginal disc containing ChChd3D1 mutant clones with overexpression of UAS-yki-RNAi. Note overexpression of UAS-yki-RNAi suppresses the upregulation of Diap1-lacZ expression in the ChChd3D1 mutant background. Discs were stained with anti-GFP and anti-β-Gal. DAPI was used to label DNA. Mutant MARCM clones are marked by GFP and indicated by white asterisks. (C) Quantification of clone area of ChChd3D1 mutant and ChChd3D1 mutant with overexpression of UAS-yki-RNAi. n = 23 clones for each genotype. Bars, 50 μm.
Loss of ChChd3 causes oxidative stress and activates the JNK pathway
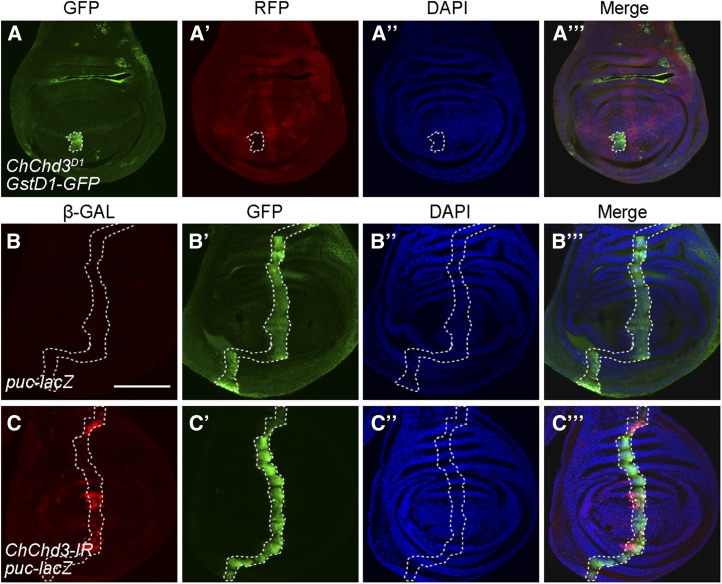
Mitochondrial dysfunction can lead to oxidative stress and subsequently activate JNK signaling, which contributes to Hippo inactivation (Ohsawa et al. 2012). To investigate how loss of ChChd3 leads to Hippo inactivation, we asked whether ChChd3 mutation affects oxidative stress and JNK signaling. To do this, we used the GstD1-GFP transgene as a marker for oxidative stress (Sykiotis and Bohmann 2008). Expression of GstD1-GFP was specifically induced in ChChd3 D1 mutant clones but not in surrounding wild-type cells in the wing disc (Figure 7, A–A′′′). To directly monitor JNK activity, we made use of an enhancer-trap line, puc-lacZ, as a reporter of JNK activation and examined its expression in third instar larval wing imaginal discs (Martin-Blanco et al. 1998). In control discs, puc-LacZ expression levels were low in ptc-expressing domain (Figure 7, B–B′′′). When ChChd3 was knocked down by RNAi using the ptc-Gal4 driver, we observed an increase of puc-lacZ expression in those cells expressing ptc-Gal4 (Figure 7, C–C′′′). Collectively, we concluded that loss of ChChd3 could induce oxidative stress and lead to the activation of JNK signaling.
Figure 7.
Loss of ChChd3 induces oxidative stress and activates JNK signaling. (A–A′′′) Upregulation of GstD1-GFP expression in ChChd3D1 homozygous mutant clones in a wing imaginal disc. Disc was stained with anti-GFP and anti-RFP. Mutant clones are marked by lack of RFP and outlined by the dashed line. (B–B′′′) Low expression of the puc-lacZ reporter gene in a control wing imaginal disc. (C–C′′′) Knockdown of ChChd3 by RNAi with ptc-Gal4 increases the level of puc-lacZ expression in a stripe of anterior compartment cells. ptc-expressing cells are marked by GFP and outlined by the dashed lines. Bars, 100 μm.
Disruption of the mitochondrial fusion pathway inactivates the Hippo pathway
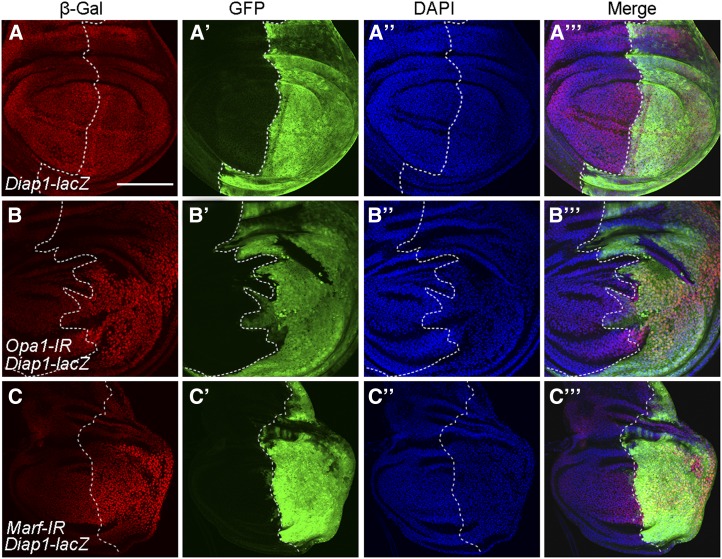
Since ChChd3 encodes a mitochondrial inner membrane protein and is required for mitochondrial fusion, we wonder whether other components of the mitochondrial fusion pathway have similar effects in regulating Hippo activity. To this purpose, we used RNAi to knock down two important mitochondrial fusion genes, Opa1 and Marf, and analyzed the effects on Hippo activity (Yarosh et al. 2008; Dorn et al. 2011; Sandoval et al. 2014). We first confirmed the specificity of RNAi lines by examining their effects on mitochondrial morphology in third instar larval body wall tissues. Indeed, RNAi of Opa1 or Marf with Mef2-Gal4 led to strong defects in mitochondrial fusion, as revealed by the presence of a short mitochondrial structure that was labeled with UAS-mito-GFP (Figure S3, A–C). Our TEM analysis also showed that the mitochondrial fragmentation defects were evident in adult indirect flight muscles when Opa1 or Marf was knocked down by RNAi using Mhc-Gal4 (Figure S3, D–F). These data suggest that both RNAi lines were efficient. Interestingly, knockdown of Opa1 by RNAi using en-Gal4 also led to a significantly increase of Diap1-lacZ expression levels in the posterior compartment of wing discs (Figure 8, A–B′′′). Similarly, we observed elevated expression of two other reporters, ex-lacZ and CycE-lacZ, in the posterior half of wing discs upon Opa1 knockdown by hh-Gal4 (Figure S4, A–A′′′ and C–C′′′). Knockdown of Marf produced the same effect on Hippo reporters (Figure 8, C–C′′′; Figure S4, B–B′′′ and D–D′′′). Together with our previous finding that ChChd3 mutation inactivates Hippo signaling, these results provide evidence that disruption of the mitochondrial fusion pathway can inactivate the Hippo pathway.
Figure 8.
Depletion of either Opa1 or Marf increases Hippo pathway target gene expression. (A–A′′′) Expression pattern of the Diap1-lacZ reporter gene in a control wing imaginal disc. (B–B′′′) RNAi knockdown of Opa1 increases the level of Diap1-lacZ expression. (C–C′′′) RNAi knockdown of Marf increases the level of Diap1-lacZ expression. Discs were stained with anti-β-Gal and anti-GFP. DAPI was used to label DNA. Posterior cells are marked by GFP. Dashed lines indicate the anterior/posterior compartment boundary. Bar, 100 μm.
Discussion
In the present study, we have shown that the mitochondrial inner membrane protein ChChd3 is required for tissue growth and uncovered a novel link between mitochondrial fusion and the Hippo pathway.
ChChd3 is a highly conserved protein located at the mitochondrial inner membrane and functions as a scaffolding protein to maintain crista integrity and protein import in mammalian cells (Darshi et al. 2011). In the same study, it is also shown that knockdown of ChChd3 results in increased fragmentation of the mitochondrial network (Darshi et al. 2011). Consistent with these findings in mammalian cells, we observed strong mitochondrial fusion defects in Drosophila tissues when ChChd3 was depleted. Furthermore, our TEM analysis also confirmed that the crista structure was altered in ChChd3 mutant mitochondria. These data demonstrate the conserved role of ChChd3 in maintaining the mitochondrial structure in both flies and mammals. What are the physiological and developmental functions of ChChd3 at an organismic level? Our genetic analysis reveals an essential role of ChChd3 during Drosophila development. Loss of ChChd3 affects tissue growth and causes the lethality at the second instar larval stage. Clone analyses in wing imaginal discs reveal that ChChd3 is required for epithelial cell proliferation. Staining with anti-Caspase 3 antibody is not evident in ChChd3 mutant clones in wing imaginal discs, suggesting that the reduced clone growth is not due to ectopic apoptosis. Previous studies have shown that mutations in several mitochondrial components cause a cell cycle arrest during the larval stage in Drosophila (Mandal et al. 2005; Owusu-Ansah et al. 2008). It is likely that the reduced cell proliferation in ChChd3 mutant tissues is caused by inefficient cell cycle progression or delayed cell cycle.
Mitochondrial dynamics, including fusion and fission, have a critical role in determining mitochondrial morphology and function (Chan 2012). Many signaling pathways have been shown to control mitochondrial dynamics (Kasahara and Scorrano 2014; Mishra and Chan 2014). Among these pathways, the Hippo pathway functions to promote mitochondrial fusion and biogenesis through the activation of mitochondrial fusion genes as well as other mitochondrial-related genes (Nagaraj et al. 2012). An increase of ChChd3 mRNA and protein levels in Yki-overexpressing cells suggests that the Hippo pathway is able to regulate ChChd3 expression to enhance mitochondrial function. Our studies reveal that ChChd3, in addition to Opa1 and Marf, is another target of the Hippo pathway in promoting mitochondrial fusion. Although the size of ChChd3 and wts double mutant clones in wing discs is reduced compared to that of wts single mutant clones, it is still larger than that of the wild-type clones. This is similar to the observation that Opa1 or Marf knockdown only partially reduced Yki-induced overgrowth defects (Nagaraj et al. 2012). Thus, the intact mitochondrion is required but not essential for Hippo pathway-mediated overgrowth.
Depletion of ChChd3 causes inactivation of the Hippo pathway. Similarly, Opa1 or Marf knockdown also leads to Hippo inactivation. These findings provide direct evidence that the cross-talk between mitochondria and Hippo signaling is bidirectional. The increased expression of mitochondria-associated genes, could partially compensate the mitochondrial fusion defects in the ChChd3 mutant background. Such mechanism might be part of a feedback loop that attenuates the effect of increased mitochondrial fragmentation and would benefit proliferative growth in tissues with mild mitochondrial defects.
Our data support the idea that mitochondrial fusion might be a key factor for maintaining Hippo activity. Previous studies have shown that defects in mitochondrial respiratory function in combination with Ras activation can drive nonautonomous tissue overproliferation in Drosophila, which is due to stimulated production of reactive oxygen species, activation of JNK signaling, and inactivation of Hippo signaling (Ohsawa et al. 2012). However, mutations in mitochondrial respiratory components alone are not able to inactivate Hippo pathway (Ohsawa et al. 2012). It has recently been reported that oncogenic Ras promotes mitochondrial fission through increased Drp1 phosphorylation, which leads to increased mitochondrial fragmentation (Kashatus et al. 2015; Serasinghe et al. 2015). It remains unknown whether Ras activation also enhances mitochondrial fission in the Drosophila developing eye. Visible upregulation of Diap1-lacZ levels was detected in Ras-overexpressing clones in eye discs, raising the possibility that Ras alone could inactivate the Hippo pathway due to increased mitochondrial fragmentation (Ohsawa et al. 2012). How does the defect in mitochondrial fusion inactivate the Hippo pathway? We have shown that loss of ChChd3 induces oxidative stress and activates JNK signaling. It is possible that the JNK pathway mediates the inactivation of Hippo signaling when mitochondrial fusion is defective.
Acknowledgments
We thank S.M. Cohen, P. Rørth, J.C. Pastor-Pareja, D. Bohmann, Y. Cai, Z.H. Li, and C. Tong for fly stocks and antibodies and J. Yang for help with mutant generation. We also thank the Bloomington Drosophila Stock Center, the Drosophila Genetic Resource Center, the National Institute of Genetics Fly Stock Center, the Tsinghua Fly Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies. This study was supported by the National Natural Science Foundation of China (grants 31371381 and 31371319) and the National Key Basic Research Program of the Ministry of Science and Technology of China (grants 2012CB966800 and 2013CB945600). The authors declare no conflict of interest.
Footnotes
Communicating editor: N. Perrimon
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.186445/-/DC1.
Literature Cited
- Boland M. L., Chourasia A. H., Macleod K. F., 2013. Mitochondrial dysfunction in cancer. Front. Oncol. 3: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. C., 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252. [DOI] [PubMed] [Google Scholar]
- Chan D. C., 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46: 265–287. [DOI] [PubMed] [Google Scholar]
- Darshi M., Mendiola V. L., Mackey M. R., Murphy A. N., Koller A., et al. , 2011. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286: 2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn G. W., 2nd, Clark C. F., Eschenbacher W. H., Kang M. Y., Engelhard J. T., et al. , 2011. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ. Res. 108: 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P., Tapon N., 2014. Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 31: 74–83. [DOI] [PubMed] [Google Scholar]
- Halder G., Johnson R. L., 2011. Hippo signaling: growth control and beyond. Development 138: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Pfleger C. M., Hariharan I. K., 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467. [DOI] [PubMed] [Google Scholar]
- Harvey K. F., Zhang X., Thomas D. M., 2013. The Hippo pathway and human cancer. Nat. Rev. Cancer 13: 246–257. [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D., 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434. [DOI] [PubMed] [Google Scholar]
- Irvine K. D., Harvey K. F., 2015. Control of Organ Growth by Patterning and Hippo Signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 7: pii: a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Nakamura K., Iijima M., Sesaki H., 2013. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A., Scorrano L., 2014. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 24: 761–770. [DOI] [PubMed] [Google Scholar]
- Kashatus J. A., Nascimento A., Myers L. J., Sher A., Byrne F. L., et al. , 2015. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Guptan P., Owusu-Ansah E., Banerjee U., 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9: 843–854. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E., Gampel A., Ring J., Virdee K., Kirov N., et al. , 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Chan D. C., 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., 2013. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. BioEssays 35: 955–964. [DOI] [PubMed] [Google Scholar]
- Nagaraj R., Gururaja-Rao S., Jones K. T., Slattery M., Negre N., et al. , 2012. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 26: 2027–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Suomalainen A., 2012. Mitochondria: in sickness and in health. Cell 148: 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S., Sato Y., Enomoto M., Nakamura M., Betsumiya A., et al. , 2012. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature 490: 547–551. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E., Yavari A., Mandal S., Banerjee U., 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 40: 356–361. [DOI] [PubMed] [Google Scholar]
- Pan D., 2010. The hippo signaling pathway in development and cancer. Dev. Cell 19: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S., Tapon N., Leopold P., 2003. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5: 921–927. [DOI] [PubMed] [Google Scholar]
- Plouffe S. W., Hong A. W., Guan K. L., 2015. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 21: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Zhang L., Jiang J., 2010. Hippo signaling regulates Yorkie nuclear localization and activity through 14–3-3 dependent and independent mechanisms. Dev. Biol. 337: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H., Yao C. K., Chen K., Jaiswal M., Donti T., et al. , 2014. Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. eLife 3: e03558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauble S., King C. C., Darshi M., Koller A., Shah K., et al. , 2007. Identification of ChChd3 as a novel substrate of the cAMP-dependent protein kinase (PKA) using an analog-sensitive catalytic subunit. J. Biol. Chem. 282: 14952–14959. [DOI] [PubMed] [Google Scholar]
- Schroeder M. C., Halder G., 2012. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin. Cell Dev. Biol. 23: 803–811. [DOI] [PubMed] [Google Scholar]
- Serasinghe M. N., Wieder S. Y., Renault T. T., Elkholi R., Asciolla J. J., et al. , 2015. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57: 521–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A., Tsatskis Y., Fabian L., Hester I., Rosenfeld R., et al. , 2014. The atypical cadherin fat directly regulates mitochondrial function and metabolic state. Cell 158: 1293–1308. [DOI] [PubMed] [Google Scholar]
- Sykiotis G. P., Bohmann D., 2008. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14: 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G., 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5: 914–920. [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., Shen Q., Kawajiri S., 2013. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5: pii: a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., Pan D., 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456. [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A., Yu W., 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yarosh W., Monserrate J., Tong J. J., Tse S., Le P. K., et al. , 2008. The molecular mechanisms of OPA1-mediated optic atrophy in Drosophila model and prospects for antioxidant treatment. PLoS Genet. 4: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. X., Guan K. L., 2013. The Hippo pathway: regulators and regulations. Genes Dev. 27: 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.