Abstract
Objective
Systemic lupus erythematosus (SLE) is a chronic autoimmune disorder whose etiology is incompletely understood, but likely involves environmental triggers in genetically susceptible individuals. We sought to identify the genetic loci associated with SLE in a Korean population by performing an unbiased genome-wide association scan.
Methods
A total of 1,174 Korean SLE cases and 4,248 population controls were genotyped with strict quality control measures and analyzed for association. For select variants, replication was tested in an independent set of 1,412 SLE cases and 1,163 population controls of Korean and Chinese ancestries.
Results
Eleven regions outside the HLA exceeded genome-wide significance (P<5×10−8). A novel SNP-SLE association was identified between FCHSD2 and P2RY2 peaking at rs11235667 (P = 1.0×10−8, odds ratio (OR) = 0.59) on a 33kb haplotype upstream to ATG16L2. Replication for rs11235667 resulted in Pmeta-rep=0.001 and Pmeta-overall=6.67×10−11 (OR=0.63). Within the HLA region, association peaked in the Class II region at rs116727542 with multiple independent effects. Classical HLA allele imputation identified HLA-DRB1*1501 and HLA-DQB1*0602, both highly correlated, as most strongly associated with SLE. We replicated ten previously established SLE risk loci: STAT1-STAT4, TNFSF4, TNFAIP3, IKZF1, HIP1, IRF5, BLK, WDFY4, ETS1 and IRAK1-MECP2. Of these loci, we identified previously unreported independent second effects in TNFAIP3 and TNFSF4 as well as differences in the association for a putative causal variant in the WDFY4 region.
Conclusions
Further studies are needed to identify true SLE risk effects in other suggestive loci and to identify the causal variant(s) in the regions of ATG16L2, FCHSD2, and P2RY2.
Systemic lupus erythematosus (SLE; [MIM152700]) is a chronic, heterogeneous autoimmune disease characterized by the loss of tolerance to self-antigens, dysregulated type I interferon responses, and inflammation, often resulting in systemic end-organ damage(1). Immune dysfunction of SLE involves both B and T lymphocytes of the adaptive immune system, together with elements of the innate immune system, including dendritic cells and the complement system(1). The clinical manifestations of SLE can be quite variable and can involve virtually any organ system. Although the precise etiology of SLE is largely unknown, the pathogenic mechanism likely involves environmental triggers in a genetically susceptible host(2). Few effective treatment options exist, largely due to an incomplete understanding of the pathophysiological basis of the disease.
Genetic predisposition leading to increased risk of SLE is supported by high heritability (>66%), increased risk among siblings of affected patients (λs≈30), and an ~25% monozygotic twin concordance(3). Today, associations of more than 50 loci with SLE susceptibility have been identified and confirmed(4). Many of these genes fall into known pathways that are key to innate and adaptive immune responses, lymphocyte activation and/or function, and immune complex clearance(4). However, a significant proportion of heritable risk to SLE has yet to be explained(5). The identification of SLE-associated genes and their pathogenic mechanisms will greatly enhance our understanding of lupus pathophysiology and facilitate the development of effective diagnostic, prognostic, and therapeutic tools. To date, large-scale genome-wide genetic studies of Asian SLE populations have focused on Han Chinese(6-8) and Japanese(9). Moreover, several reports have shown that transracial mapping of SLE loci can aid in the dissection of risk effects(4). In this study, we performed a genome-wide association (GWA) scan to identify genes associated with SLE in an East Asian population from Korea.
Methods
Subjects
A total of 1,174 patients with SLE were recruited from the Hanyang University Hospital for Rheumatic Diseases (HUHRD) and six other university hospitals in Korea(10). In addition, 552 ethnically matched healthy controls were recruited from HUHRD. The 3,700 ethnically matched out-of-study population controls were recruited from the Korean National Institutes of Health(10). In addition, an independent cohort of 1,412 SLE cases and 1,163 population controls were used for the replication studies(11, 12). This sample set consisted of 739 Korean SLE cases and 436 Korean controls as well as 677 Chinese SLE cases and 709 Chinese controls (Supplementary Table 1).
Written, informed consent from each participant was obtained by each participant following protocols approved by the Institutional Review Boards of participating institutes. All cases used in this study fulfilled at least 4 of the 11 American College of Rheumatology criteria for SLE(13), while healthy, population-based controls were without family history of SLE or any other autoimmune disease.
GWA scan Genotyping, Sample Quality Control, and Ascertainment of Populations Stratification
Samples were genotyped using the Illumina HumanOmni1-Quad or HumanOmniExpress arrays using Infinium chemistry at Oklahoma Medical Research Foundation (OMRF) following the manufacturer's protocol (Illumina, Inc., San Diego, CA). The out-of-study GWA controls were genotyped on the HumanOmni1-Quad arrays by the Korea National Institutes of Health. Strict quality control standards were implemented for SNPs retained in the association analysis, including requirements for well-defined cluster scatter plots. Samples were excluded if they had a SNP call rate <90%. SNPs were considered high quality SNPs if they had call rates >95%, no evidence of differential missingness between cases and controls (P < 0.05) and no evidence of a departure from expected Hardy-Weinberg proportions (controls P < 0.01, cases P < 0.000001). Inference is primarily based on those SNPs with minor allele frequency (MAF) greater than 1%.
Based on the SNPs that passed the above quality control thresholds, samples were removed if there were inconsistencies between recorded and genotype-inferred gender or excess heterozygosity on the autosomes. Duplicates and first- or second-degree relatives were removed based on identity-by-descent statistics computed by the program KING(14). Principal components (PCs) were computed with the samples and merged with HapMap phase 3 individuals (CEU, YRI, and CHB) as reference populations(15) using EIGENSOFT(16). Principal component analysis (PCA) was performed on a subset of autosomal SNPs that were selected by removing regions of known high linkage disequilibrium (LD), removing variants with MAF < 0.05, and pruning markers to reduce extended pairwise LD. The PCs were used to remove genetic outliers (Supplementary Figure 1). The dataset that passed laboratory and statistical quality control was composed of 1174 SLE cases (1096 females and 78 males) and 548 within-study controls (547 females and 1 male). In addition, 3698 out-of-study controls (2330 females and 1368 males) were merged into the within-study genotype data.
Statistical Analysis
To test for an association between a SNP and SLE status, a logistic regression analysis was computed including PC 3 as a covariate since no additional PC significantly changed the inflation factor (λ). Primary inference was based on the additive genetic model unless there was significant lack-of-fit (P < 0.05). If there was evidence of a departure from an additive model, then inference was based on the most significant value from the dominant, additive or recessive genetic models. The additive and recessive models were computed only if there were at least 10 and 30 individuals homozygous for the minor allele, respectively. The analyses were completed using the program SNPGWA version 4.0. For the analysis of chromosome X SNPs, the samples were stratified by gender and then meta-analyzed across gender using the program METAL(17).
To determine the number of independent associations within each SLE-risk locus exceeding the genome-wide significance threshold, a manual stepwise model or conditional analysis was computed. The stepwise modeling or conditional analysis was implemented using forward selection with backward elimination using the entry and exit criteria of P < 0.0001, which accounted for approximately 500 independent variants within a given genomic region. Specifically, for each region of interest, the top SNP was included as a covariate and the association statistics were re-calculated. SNPs were allowed to enter and exit models in this stepwise fashion until no additional SNPs met a significance threshold of P < 0.0001.
Replication Genotyping, Sample Quality Control, and Ascertainment of Populations Stratification
Genotypes were obtained using TaqMan assays (Life Technologies, Grand Island, NY) for four SNPs: rs2267828, rs10901656, rs11235667, and rs1048257. Analysis was conducted for these cohorts independently to allow for PC analysis using previously collected data. Ancestry adjustments for the Koreans were described previously in Lessard et al., (11). For the Chinese subjects, the PCA was done with slight modification from what was reported in Kaiser et al. (12). In this study, 7,918 randomly selected autosomal ImmunoChip SNPs with MAF>1%, low pairwise LD (r2 < 0.1), and no evidence of association with SLE (P > 0.01) were used to perform PC analysis using EIGENSOFT. PC analysis plots of the CHB and CHS subjects in the 1000 Genomes Project along with our subjects were used to select and remove genetic outliers. The first PC (Chinese cohort) and PCs 1, 2, and 3 (OMRF and UCLA Korean datasets) were included as covariates in the logistic regression models based on the variance explained in each dataset. These dataset were than meta-analyzed using the program METAL(17). To test for heterogeneity among the individual association results in the meta-analysis, we utilized both the Cochran's Q test statistic(18) and I2 index(19).
Imputation
To help localize the associations in the genome-wide significant regions, ungenotyped variants were imputed based on the reference panel from the 1000 Genomes Project(20). Specifically, the program SHAPEIT was used to pre-phase the genotype data(21). After phasing the data, IMPUTE2 was used for the imputation with the 1000 Genomes Phase I integrated reference panel(22). The imputed data was filtered using standard post-imputation quality control based on IMPUTE2 information scores >0.5 and confidence scores >0.9 for subsequent association tests. Post-association analysis required genotyped SNPs in LD with imputed variants to support the inferred alleles as true signals. The program SNPTESTv2 was used to test for association of the imputed variants(23).
Imputation of the HLA classical alleles in the genes HLA-A, -B, -C, -DPB1, -DQA1, -DQB1, and -DRB1 was done using the program HiBAG(24) and the Asian reference panel. In this sample, ~21% of the reference SNPs used by HiBAG were missing genotype data. To address this issue, HLA imputation was repeated after filling in the missing genotype data with the “best guess” imputed SNP data from the 1000 Genomes imputation described above. By using the “best guess” genotype data with a posterior probability >0.90, the percent of missing variants in the reference set was reduced to 0.36%.
Results
Summary of the genome-wide association phase
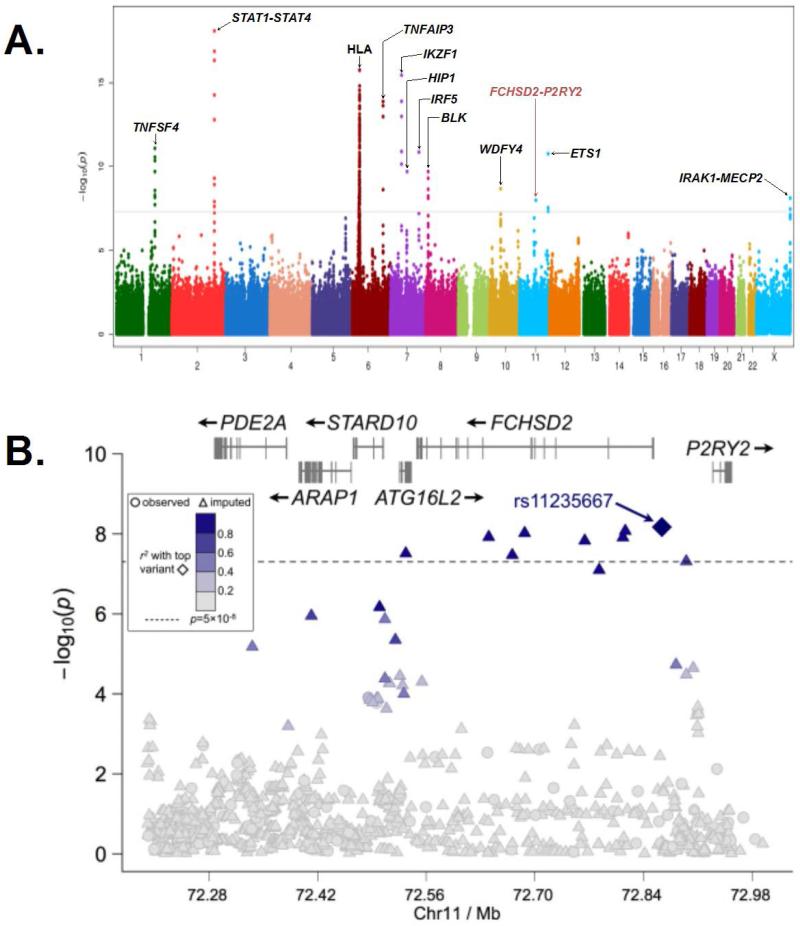
We observed modest inflation in the test statistic (λ=1.09) with only slight deviation from expected once the HLA and other known SLE loci were removed (Supplementary Figure 2). A total of eleven regions surpassed the genome-wide significance threshold of P < 5×10−8 with STAT4 (MIM600558) yielding the most significant genotyped association with SLE at rs11889341 (P = 8.02×10−19; Figure 1A and Table 1). Of the non-HLA regions, 10 risk loci had been previously identified and confirmed as risk loci for SLE, including STAT1 (MIM600555)-STAT4, IKZF1 (MIM603023), TNFAIP3 (MIM191163), TNFSF4 (MIM603594), HIP1 (MIM601767), IRF5 (MIM607218), ETS1 (MIM164740), BLK (MIM191305), WDFY4 (MIM613316), and IRAK1 (MIM300283)-MECP2 (MIM300005). In addition, association not previously described for SLE risk was observed at 11q14 (Figure 1A).
Figure 1. Summary of the genome-wide association results for 1174 SLE cases and 3698 controls of Korean ancestry and zoomed plot of the region associated with SLE at 11q14.
(A) The -log10(P-value) for each genotyped variant is plotted along the Y-axis with the chromosome and chromosomal position along the X-axis. The gray line indicates the genome-wide significance threshold of P = 5×10−8. (B) The −log10(P-value) is plotted for each genotyped (shown as circles) and imputed (shown as triangles) variants, and with the peak association, rs11235667, is plotted as a diamond. The linkage disequilibrium with rs11235667 is given by the scale on the figure. The genome-wide significance threshold is displayed as a dashed line at P = 5 ×10−8. Association exceeding this threshold was found extending from ATG16L2 through FCHSD2 to the shared promoter region with P2RY2.
Table 1.
Single locus analysis of previous SLE associations outside of HLA region
| Marker | ChrA | Position | Upstream Gene | Downstream Gene | Within Gene | MAFB | PGWAS | Model | Obs/ImpC | OR (95% CID) | Maj/MinE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1234314 | 1 | 173177392 | 1kb from TNFSF4 | 269kb from PRDX6 | - | 0.37 | 9.25×10−11 | Add | Imp | 1.37 (1.25-1.51) | C/G |
| rs2205960 | 1 | 173191475 | 15kb from TNFSF4 | 255kb from PRDX6 | - | 0.25 | 1.03×10−11 | Add | Imp | 1.44 (1.30-1.60) | G/T |
| rs76413021 | 1 | 173206297 | 30kb from TNFSF4 | 240kb from PRDX6 | - | 0.23 | 3.26×10−13 | Add | Imp | 1.52 (1.36-1.71) | G/A |
| rs844644 | 1 | 173209495 | 33kb from TNFSF4 | 237kb from PRDX6 | - | 0.4 | 4.47×10−11 | Add | Obs | 1.37 (1.25-1.50) | A/C |
| rs10489265 | 1 | 173236065 | 60kb from TNFSF4 | 211kb from PRDX6 | - | 0.25 | 8.19×10−12 | Add | Obs | 1.43 (1.29-1.58) | A/C |
| rs4916342 | 1 | 173347837 | 172kb from TNFSF4 | 99kb from PRDX6 | - | 0.3 | 8.22×10−9 | Add | Imp | 0.75 (0.67-0.82) | A/G |
| rs16833239 | 2 | 191940260 | - | - | STAT4 | 0.15 | 9.69×10−10 | Add | Imp | 0.67 (0.59-0.76) | G/A |
| rs11889341 | 2 | 191943742 | - | - | STAT4 | 0.32 | 8.02×10−19 | Add | Obs | 1.53 (1.40-1.69) | C/T |
| rs12612769 | 2 | 191953998 | - | - | STAT4 | 0.31 | 2.37×10−19 | Add | Imp | 1.59 (1.43-1.75) | A/C |
| rs13192841 | 6 | 137967214 | 152kb from OLIG3 | 221kb from TNFAIP3 | - | 0.13 | 7.50×10−3 | Dom | Obs | 1.23 (1.06-1.43) | G/A |
| rs5029937 | 6 | 138195151 | - | - | TNFAIP3 | 0.07 | 3.98×10−14 | Dom | Imp | 2.11 (1.74-2.55) | G/T |
| rs5029939 | 6 | 138195723 | - | - | TNFAIP3 | 0.07 | 4.09×10−14 | Dom | Imp | 2.10 (1.74-2.55) | C/G |
| rs2230926 | 6 | 138196066 | - | - | TNFAIP3 | 0.07 | 2.34×10−14 | Dom | Obs | 1.93 (1.63-2.28) | T/G |
| rs9373203 | 6 | 138289848 | 86kb from TNFAIP3 | 120kb from PERP | - | 0.37 | 4.89×10−6 | Add | Imp | 1.25 (1.14-1.37) | C/T |
| rs6922466 | 6 | 138444930 | 16kb from PERP | 38kb from KIAA1244 | - | 0.18 | 0.297 | Dom | Obs | 1.08 (0.94-1.24) | A/G |
| rs11185602 | 7 | 50299077 | 100kb from C7orf72 | 45kb from IKZF1 | - | 0.33 | 1.53×10−16 | Add | Imp | 0.66 (0.60-0.73) | A/G |
| rs17552904 | 7 | 50318308 | 120kb from C7of72 | 26kb from IKZF1 | - | 0.33 | 3.51×10−16 | Add | Obs | 0.65 (0.59-0.72) | G/T |
| rs6964720 | 7 | 75180344 | - | - | HIP1 | 0.24 | 2.00×10−10 | Add | Obs | 1.40 (1.26-1.56) | A/G |
| rs139110493 | 7 | 75209951 | - | - | HIP1 | 0.06 | 1.21×10−12 | Dom | Imp | 2.48 (1.93-3.19) | G/C |
| rs4728142 | 7 | 128573967 | 11kb from LOC392787 | 4kb from IRF5 | - | 0.14 | 1.38×10−11 | Add | Obs | 1.53 (1.35-1.73) | G/A |
| rs113478424 | 7 | 128575797 | 13kb from LOC392787 | 2kb from IRF5 | - | 0.14 | 3.97×10−12 | Add | Imp | 1.59 (1.39-1.81) | 15-mer*/T |
| rs922483 | 8 | 11351912 | - | - | BLK | 0.25 | 2.00×10−10 | Add | Obs | 0.71 (0.64-0.79) | T/C |
| rs2736345 | 8 | 11352485 | - | - | BLK | 0.24 | 7.88×10−11 | Add | Imp | 0.70 (0.63-0.78) | G/A |
| rs10857631 | 10 | 49955821 | - | - | WDFY4 | 0.13 | 1.67×10−4 | Add | Imp | 1.30 (1.13-1.48) | A/G |
| rs7097397 | 10 | 50025396 | - | - | WDFY4 | 0.37 | 2.10×10−9 | Add | Obs | 1.33 (1.21-1.46) | A/G |
| rs877819 | 10 | 50042951 | - | - | WDFY4 | 0.16 | 0.0558 | Add | Imp | 1.13 (1.00-1.28) | G/A |
| rs10776651 | 10 | 50084526 | - | - | WDFY4 | 0.34 | 1.54×10−7 | Add | Imp | 1.29 (1.18-1.43) | C/T |
| rs1913517 | 10 | 50119054 | - | - | WDFY4 | 0.31 | 2.54×10−5 | Add | Obs | 1.24 (1.12-1.37) | G/A |
| rs12576753 | 11 | 128304141 | None within 500kb | 25kb from ETS1 | - | 0.39 | 1.74×10−11 | Add | Obs | 1.37 (1.25-1.56) | C/A |
| rs1128334 | 11 | 128328959 | - | - | ETS1 | 0.38 | 7.17×10−12 | Add | Imp | 1.39 (1.26-1.52) | G/A |
| rs5986948 | X | 153266172 | 17.3kb from TMEM187 | 9.8kb from IRAK1 | - | 0.24 | 4.36 ×10−10 | Rec | Imp | 0.64 (0.56-0.74) | C/T |
| rs1059702 | X | 153284192 | - | - | IRAK1 | 0.26 | 5.14×10−10 | Rec | Imp | 0.65 (0.57-0.74) | A/G |
| rs2734647 | X | 153292180 | - | - | MECP2 | 0.25 | 7.54×10−9 | Dom | Obs | 0.62 (0.51-0.75) | T/C |
Chr = Chromosome
MAF = Minor allele frequency
Obs/Imp = observed/imputed
CI = Confidence interval
Maj/Min = Major/Minor allele
15-mer= CTTAGCTATTGCTC
Association at 11q14 with SLE
This SNP-SLE association was observed with a single variant located between FCHSD2 (MIM not available) and P2RY2 (MIM600041) (rs11235667; P=1.03×10−8; odds ratio (OR) = 0.59; 95% confidence interval (CI) = 0.50-0.71; Figure 1B, Table 2, and Supplementary Table 2). Moreover, additional support was observed with genotyped variants in the region (Supplementary Table 2). After imputation of the 11q14 region showing association with SLE, rs11235667 remained the most significant association (Figure 1B and Supplementary Table 2). However, a haplotype was identified with 8 variants exceeding the genome-wide significance threshold that spanned from ATG16L2 (MIM not available) through FCHSD2 to the shared promoter region with P2RY2. Stepwise logistic regression analysis adjusting for rs11235667 indicated the presence of only a single effect (Supplementary Figure 3).
Table 2.
Single locus analysis results for regions genotyped in the replication study.
| Marker | Region Name | Maj/MinA | MAFB Case / CtrlC | PGWAS | Model | OR (95% CID) | P Meta RepE | Q / I2 Meta RepE | OR (95% CID) Meta RepE | P Meta OverallF | Q / I2 Meta OverallF | OR (95%CID) Meta Overall |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2267828 | GTF2IRD1 | A/G | 0.40 / 0.45 | 7.02×10−7 | Add | 0.79 (0.72-0.87) | 0.02 | 0.56 / 0 | 0.87 (0.77-0.98) | 6.46×10−8 | 0.41 / 0 | 0.81 (0.76-0.88) |
| rs10901656 | DOCK1 | C/T | 0.27 / 0.23 | 6.91×10−7 | Dom | 1.39 (1.22-1.58) | 0.095 | 0.6 / 0 | 1.14 (0.98-1.31) | 9.56×10−6 | 0.23 / 28.72 | 1.21 (1.12-1.32) |
| rs11235667 | FCHSD2-P2RY2 | A/G | 0.07 / 0.11 | 1.03×10−8 | Add | 0.59 (0.50-0.71) | 0.0014 | 0.29 / 18.43 | 0.71 (0.57-0.87) | 6.67×10−11 | 0.14 / 44.37 | 0.63 (0.55-0.72) |
| rs1048257 | AHNAK2 | T/C | 0.34 / 0.39 | 1.67×10−6 | Add | 0.79 (0.72-0.87) | 0.086 | 0.29 / 17.06 | 0.90 (0.80-1.01) | 8.66×10−7 | 0.12 / 47.82 | 0.82 (0.76-0.89) |
Maj/Min = Major/Minor allele
MAF = Minor allele frequency
Case/Ctrl = Case/Control
CI = Confidence interval
Meta Rep = Meta-analysis for the replication
Replication analysis for the primary signal in the region of FCHSD2-P2RY2 was done using independent cohorts from Korea and China (Supplementary Table 1). The SNP rs11235667 between FCHSD2 and P2RY2 continued to show significant SLE association and similar effect size (Pmeta-rep = 0.001; OR = 0.71, 95% CI = 0.57-0.87). The overall meta-analysis between GWA and replication studies yielded a Pmeta-overall = 6.67×10−11 (OR = 0.63, 95% CI = 0.55-0.72; Table 2). No evidence of heterogeneity was observed in the meta-analysis (Table 2).
Bioinformatics analysis using Haploreg v2(25) revealed that the region around rs11235667 was hypersensitive to DNase 1 in B cells by the ENCODE project(26). This variant has been shown to be located within an enhancer element in multiple immunological cell types based on the Epigenetic Roadmap data (Supplementary Table 3)(27). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) experiments carried out by the ENCODE project found POL2 and YY1 proteins cross-linked to this region. Moreover, sequence prediction methods indicate that rs11235667 can alter the binding motif for the FOXa family of transcription factors using sequence prediction methods according to Haploreg v2(25). These data suggest the likely functional mechanism involves regulation of expression of ATG16L2, FCHSD2 and/or P2RY2. However, current eQTL databases do not suggest that rs11235667 influences the expression of these loci(25, 28). This could be due to the lack of data in the correct cell and/or tissue type and/or that some databases do not interrogate this SNP in their studies.
Of the other 8 variants that exceeded genome-wide significance on the haplotype, several findings make rs11235604 an intriguing potential causal variant (Supplemental Table 3). This variant is a missense allele (R220W) that resides in the coding region of ATG16L2. Although it is predicted to be benign by PolyPhen-2(29), it is possible this variant my still impact SLE. Haploreg v2(25) does report that rs11235604 alters 8 predicted regulatory motifs, and this variant is thought to be an active enhancer in several immunologically relevant cell types (Supplementary Table 3). Further work is needed to conclusively identify the polymorphism(s) responsible for this association signal. These studies would include evaluating the potential impact of rs11235667 on the expression of ATG16L2, FCHSD2, and/or P2RY2. In addition, experiments are needed toassess the impact on ATG16L2 of the missense allele arising from rs11235604 and/or any other variant(s) within this haplotype.
Association in the HLA region in Koreans with SLE
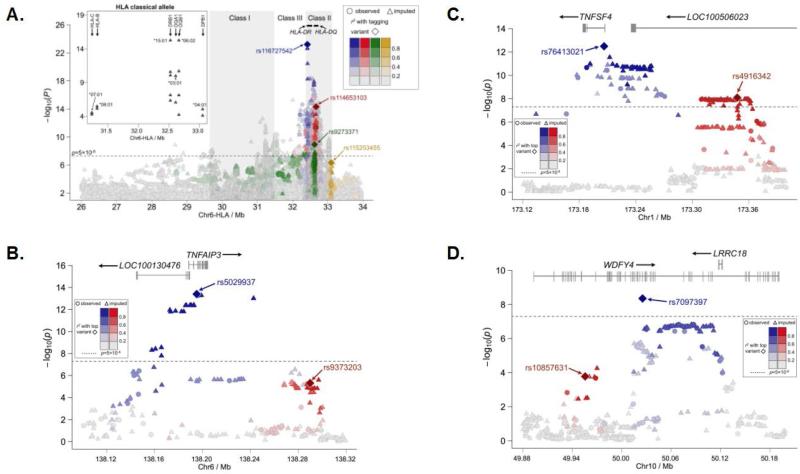
One of the most consistent associations with SLE has been with the human leukocyte antigen (HLA) region. Although the HLA was not the most statistically significant genotyped region, the SNP rs116727542 (P = 6.15×10−24; Table 3), which is located in a broad peak of association that was observed spanning HLA-DR (MIM142860) through -DQ (MIM146880), showed the strongest SNP-SLE association after imputation (Figure 2A, Table 3, and Supplementary Table 4). The interval between HLA-DR and -DQ has previously been implicated Koreans(30). In an attempt to identify the number of independent effects in this complex region, we used the stepwise approach described above and found ten independent effects (see Supplementary Figure 4A for results of the stepwise regression analysis). The first four variants identified in the stepwise regression analysis, rs116727542, rs9273371, rs114653103, and rs115253455, are all located in the HLA Class II region (Figure 2A, Supplementary Figure 4A, and Supplementary Table 4).
Table 3.
Single locus and stepwise results for top ten independent HLA associations
| MarkerA | Position | Upstream Gene | Downstream Gene | Within Gene | MAFB | P-value | OR (95%CIC) | Maj/MinD | Stepwise P-value | Stepwise OR (95%CIC) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs116727542 | 32421227 | 8.4kb from HLA-DRA | 64kb from HLA-DRB5 | - | 0.1700 | 6.15×10−24 | 0.53 (0.47-0.60) | G/A | 1.96×10−18 | 1.74 (1.54-1.97) |
| rs9273371 | 32626565 | 14kb from HLA-DQA1 | 675bp from HLA-DQB1 | - | 0.1000 | 1.18×10−9 | 1.61 (1.38-1.87) | C/T | 8.43×10−5 | 1.38 (1.18-1.63) |
| rs114653103 | 32668846 | 34kb from HLA-DQB1 | 40kb from HLA-DQA2 | - | 0.1200 | 7.31×10−15 | 0.57 (0.49-0.66) | G/T | 1.81×10−13 | 0.50 (0.41-0.60) |
| rs115253455 | 33100021 | 43kb from HLA-DPB1 | 30kb from COL11A2 | - | 0.1300 | 4.68×10−7 | 0.70 (0.61-0.80) | T/A | 4.51×10−7 | 0.66 (0.56-0.77) |
| chr6:31996524 | 31996524 | - | - | C4B | 0.1700 | 4.95×10−8 | 0.71 (0.63-0.80) | C/A | 4.87×10−10 | 0.64 (0.55-0.73) |
| rs113833333 | 32594898 | 37kb from HLA-DRB1 | 10kb from HLA-DQA1 | - | 0.4100 | 2.30×10−5 | 0.82 (0.74-0.90) | C/T | 3.09×10−8 | 0.74 (0.67-0.82) |
| rs116427960 | 31319226 | 79kb from HLA-C | 2.4kb from HLA-B | - | 0.0081 | 8.96×10−7 | 4.57 (2.49-8.38) | C/T | 1.69×10−5 | 3.17 (1.87-5.35) |
| rs114904515 | 29362756 | 20kb from OR12D3 | 1.7kb from OR12D2 | - | 0.1000 | 1.55×10−6 | 0.69 (0.60-0.80) | C/T | 5.70×10−6 | 0.67 (0.56-0.79) |
| rs118044183 | 30954150 | - | - | MUC21 | 0.1200 | 3.61×10−6 | 1.50 (1.27-1.79) | C/T | 1.39×10−4 | 1.39 (1.17-1.64) |
| rs2736191 | 31560910 | ~150bp from NCR3 | 22kb from AIF1 | - | 0.3800 | 7.53×10−7 | 0.79 (0.71-0.86) | C/G | 1.01×10−4 | 0.81 (0.73-0.90) |
All variants within this table have been imputed.
CI = Confidence interval
MAF = Minor allele frequency
Maj/Min = Major allele/Minor allele
Note: Tables are in the order they were identified in the stepwise model. The stepwise results presented in this table are for adjusting for all other variants in the table. For complete results in the HLA region, please refer to Supplementary Table 4.
Figure 2. Expanded view of the association between SLE and the HLA, TNFAIP3, TNFSF4, and WDFY4 regions.
The –log10(P-value) is plotted for each observed (shown as circles) and imputed (shown as triangles) variant in the MHC region according to base-pair position from 26Mb to 34Mb on chromosome 6 (A). Linkage disequilibrium with the first four variants included in the stepwise logistic regression analysis is shown with rs116727542 (blue diamond), rs114653103 (red diamond), rs9273371 (green diamond), and rs115253455 (gold diamond) all located within the HLA Class II region. The insert on the left shows the –log10(P-value) of the imputed classical alleles plotted according to base-pair position from 31Mb to 33.5Mb. The additional plots show results for Chromosome 6 in the region of TNFAIP3 (B), Chromosome 1 for the TNFSF4 (C) effects, and Chromosome 10 for WDFY4 (D). For each independent effect, the peak associations are represented by a diamond (blue for the first effect and red for the second, if applicable), and the correlation of variants accounted for by each effect is given in their respective color according the legends present in each plot. The genome-wide significance threshold is displayed as a dashed line on each plot at P = 5 ×10−8.
To better understand the relationship between the variants reported in this study and the classical HLA alleles, we imputed alleles at HLA A, B, C, DPB1, DQA1, DQB1 and DRB1. The peak statistical significance was observed at P = 5.55×10−16 for the two tightly linked alleles, HLA-DRB1*1501 (OR = 1.85; 95% CI = 1.59-2.14) and HLA-DQB1*0602 (OR = 1.90; 95% CI = 1.62-2.21; Figure 2A, Table 4, and Supplementary Table 5). Stepwise logistic regression modeling of the HiBAG-imputed HLA alleles identified 13 independent effects (Table 4). To better relate the classical alleles to the variants identified in this GWA scan, stepwise modeling was done with both SNPs and classical HLA alleles. The peak effect after 1000 Genomes imputation was rs116727542, which accounted for by HLA-DQB1*0602 and HLA-DRB1*0803 (see Supplementary Figure 4B for results of the additional 8 rounds of the stepwise regression analysis).
Table 4.
Multi-locus model of HIBAG-imputed HLA dosages, single locus and stepwise results
| Dosage Frequency | Best Guess Count | |||||||
|---|---|---|---|---|---|---|---|---|
| HLA Allele | Cases | Controls | Cases | Controls | OR (95% CIA) | Single Locus P-value | Stepwise P-value | Stepwise OR (95%CIA) |
| DQB1*0602B | 0.25 | 0.15 | 301 | 636 | 1.90 (1.62 - 2.21) | 5.55×10−16 | 1.93×10−23 | 2.35 (1.99 – 2.78) |
| DRB1*0803 | 0.20 | 0.13 | 251 | 613 | 1.59 (1.34 - 1.88) | 7.37×10−8 | 7.63×10−16 | 2.14 (1.78 - 2.58) |
| DQB1*0202 | 0.18 | 0.12 | 212 | 502 | 1.60 (1.35 - 1.90) | 7.57×10−8 | 2.19×10−18 | 2.50 (2.04 - 3.07) |
| DQA1*0302 | 0.19 | 0.16 | 286 | 847 | 1.41 (1.15 - 1.75) | 1.27×10−3 | 4.92×10−9 | 1.98 (1.58 - 2.50) |
| B*0801 | 0.02 | 0.004 | 20 | 14 | 5.43 (2.66 - 11.08) | 3.42×10−6 | 5.15×10−6 | 5.71 (2.70 -12.07) |
| DQA1*0401 | 0.04 | 0.03 | 54 | 133 | 1.73 (1.17 - 2.57) | 5.94×10−3 | 3.11×10−5 | 2.36 (1.58 - 3.54) |
| C*0702 | 0.22 | 0.17 | 261 | 711 | 1.37 (1.17 - 1.59) | 5.45×10−5 | 2.06×10−3 | 1.30 (1.10 - 1.54) |
| DRB1*0406 | 0.03 | 0.07 | 41 | 354 | 0.15 (0.09 - 0.26) | 2.63×10−11 | 1.11×10−4 | 0.32 (0.18 - 0.57) |
| DPB1*0501 | 0.79 | 0.72 | 930 | 3092 | 1.16 (1.05 - 1.28) | 2.87×10−3 | 1.06×10−4 | 1.23 (1.11 - 1.36) |
| DRB1*1602 | 0.03 | 0.02 | 38 | 87 | 1.78 (1.16 - 2.72) | 7.73×10−3 | 1.53×10−3 | 2.05 (1.32 - 3.20) |
| DPB1*1701 | 0.03 | 0.04 | 41 | 160 | 0.90 (0.62 - 1.30) | 5.66×10−1 | 1.07×10−3 | 0.5 (0.33 - 0.76) |
| C*0102 | 0.29 | 0.32 | 359 | 1429 | 0.88 (0.77 - 1.01) | 6.51×10−2 | 5.82×10−3 | 0.82 (0.71 - 0.94) |
| DRB1*1202 | 0.04 | 0.07 | 39 | 266 | 0.52 (0.37 - 0.74) | 2.98×10−4 | 7.07×10−3 | 0.61 (0.42 - 0.87) |
CI = Confidence interval
The allele HLA-DRB1*1501 had the same P-value as HLA-DQB1*0602, but the later was selected by the stepwise modeling procedure. For complete results, please refer to Supplementary Table 5.
Note: Tables are in the order they were identified in the stepwise model. The stepwise results presented in this table are for adjusting for all other variants in the table
Non-HLA SLE associations previously reported and identification of novel independent effects
Several previously identified non-HLA SLE loci were also replicated in this study, including: STAT1-STAT4, TNFSF4, TNFAIP3, IKZF1, HIP1, IRF5, BLK, WDFY4, ETS1, and IRAK1-MECP2 (Table 1, Figure 2B-D, Supplementary Figures 5 to 11, and Supplementary Tables 6 to 15. Of these loci, the associations in the region of TNFAIP3, TNFSF4, and WDFY4 have notable differences from previous studies.
After imputation of the TNFAIP3 region, the primary independent effect in the stepwise model was observed at rs5029937 located within the second intron of TNFAIP3 (Table 1 and Figure 2B). The second independent effect was identified at rs9373203 3’ of the TNFAIP3 coding region. A previous SLE study in Han Chinese(6) reported rs2230926 as associated with disease, and another SLE transracial mapping study in Koreans (with partial overlap of subjects with the current study) and Europeans(31) identified risk of SLE with rs7749323. Both variants (rs2230926 and rs7749323) are highly correlated with rs5029937, with D’ = 1.0 and r2 > 0.98, indicating consistency between our results and these previous reports (Supplementary Figure 12). In addition, the second effect tagged by rs9373203 was not identified in either the Han et al or Adrianto et al. studies (Figure 2B). Musone et al. (32) identified multiple effects, some of which spanned even further 3’ of TNFAIP3 than rs9373203. After their stepwise analysis, they identified rs6922466 as the tagging variant accounting for this association; however, this variant is not associated with SLE in the current study of Koreans. Moreover, the LD between these variants is very weak in Koreans (r2 = 0.00; D’ = 0.43), giving additional evidence that rs9373203 may be an independent effect warranting further study (Supplementary Figure 12).
In the region of TNFSF4 after imputation, two independent effects were observed in the stepwise model. The first effect, peaking at rs76413021, is located in the first intron of the TNFSF4 coding region (Table 1 and Figure 2C). This variant is in LD with rs2205960 (D’=0.98, r2=0.94) and rs1234315 (D’ = 0.97, r2 = 0.48) and was previously identified from the Han Chinese GWA scan(6) (Supplementary Figure 13). Moreover, this effect is consistent with the results reported in Europeans(33). The second independent effect, which is distinct from previous studies, peaks at rs4916342, which is located in an intron LOC100506023 just 5’ of TNFSF4 (Figure 2C). Neither Han et al. (6) nor Cunninghame Graham et al. (33) reported association signals as far 5’ of TNFSF4 as our observation of the second independent effect tagged by rs4916342. In a transracial mapping study of this region by Manku et al. (34) that included subjects from East Asia, a second independent effect (tagged by rs1234314) was identified that is. In our current study of Koreans, we found that the first effect, tagged by rs76413021, accounted for rs1234314 in the stepwise model. Although rs1234314 and rs4916342 are located in the same general genomic location, the LD structure further supports the observation that they are not the same genetic effect; however, all the risk variants are located on a single risk haplotype (Supplementary Figure 13). This suggests that risk alleles for both rs76413021 and rs4916342 are needed to confer susceptibility to disease.
In the region of WDFY4, the current study did not replicate rs877819, which has been previously reported to result in a downregulation of WDFY4 through modification of a YY1 binding site (Table 1)(35). However, the results for WDFY4 in Koreans are consistent with two previous studies. First, the most statistically significant association within this region, the coding variant rs7097397 leading to an amino acid substitution R1816Q (P = 2.10×10−9), was previously reported by Yang et al. (8). Second, we also demonstrate association for rs1913517, which was identified previously by Han et al. (6) (P = 2.54×10−5; Table 1 and Figure 2D). Our haplotype and stepwise regression analysis indicated that there were two independent effects in the region, with rs7097397 accounting for the association observed at rs1913517, and rs10857631 tagging the second independent effect (Figure 2D and Supplementary Figures 14 and 15).
Suggestive association with SLE identified in the genome-wide phase
In total, 15 genotyped variants surpassed the suggestive threshold of P < 2×10−6 and were considered for further replication (Supplementary Table 16). Replication was attempted for three additional variants located within GTF2IRD1, DOCK1, and AHNAK2, all of which have multiple genotyped variants showing suggestive significance and/or have been previously implicated in other related phenotypes (Table 2). Only rs2267828 near GTF2IRD1 yielded a Pmeta-rep < 0.05, but this variant did not surpass genome-wide significance after meta-analysis with the GWA scan (Table 2). The variant in the region of AHNAK2, rs1048257, was trending towards significance, while rs10901656 near DOCK1 showed association in one replication cohort with the opposite allele (Table 2). Outside of the 10 regions previously reported SLE loci described above, we observed only eight additional loci with 5×10−8 < P < 5×10−5 on the list of ~50 that have been described previously (Supplemental Table 17). This is likely due to the limited power of this study and/or results from population-specific differences from the studies in which these discoveries were originally identified.
Discussion
The association in this region peaks between three candidate genes, ATG16L2, FCHSD2, and P2RY2, all of which have the biological potential to impact SLE pathophysiology. While this locus has not been reported in other systemic autoimmune diseases, variants in this region are associated with Crohn's disease (MIM266600) in Korean subjects(36). Moreover, the peak variant identified in Crohn's disease, rs11235667, was also the variant discovered in this current study with SLE. The missense variant, rs11235604, was also reported to be associated in Crohn's disease(36). GWA studies conducted in SLE in Europeans did not identify this locus since it is monomorphic in that population. Moreover, GWA scans in Han Chinese both used the Illumina Human 610-Quad bead chip, which did not contain rs11235667(6-8).
ATG16L2 (autophagy related 16-like 2) is a ubiquitously expressed homologue of the gene ATG16L1 (MIM610767) that has been implicated as a risk locus for Crohn's disease in patients of European descent(37, 38). Both loci are involved in autophagy; however, little is known about the role ATG16L2 plays in the process. Interestingly, this pathway has been previously implicated in SLE. The gene ATG5 (MIM604261) has also been implicated as a risk locus for lupus(6, 39). Studies in the mouse have shown that Apg16l (the mouse equivalent of human ATG16L) interacts with Apg5 (the mouse equivalent of human ATG5) suggesting that it is possible that ATG16L2 and ATG5 may also interact humans(40). More studies are needed to understand the function ATG16L2 and if it is involved in the association with SLE.
FCHSD2 (FCH and double SH3 domains 2) has been described as regulator of F-actin assembly through interactions with WAS (also known as WASP) and WASL (also known as N-WASP)(41). FCHSD2 is primarily expressed in CD19+ B cells, dendritic cells, myeloid cells, CD4+ T cells, and CD8+ T cells(38). Previous studies have shown that WAS plays an important role in the migration of T cells through reorganization of the actin cytoskeleton subsequent to interactions with dendritic or B cells(42).
P2RY2 (purinergic receptor P2Y, G-protein coupled, 2) is known to be involved in many cellular functions and is expressed in myeloid cells including monocytes(38). P2RY2 is a receptor for ATP and UTP that acts as a sensor for the release of nucleotides by apoptotic cells(43). Mice null for P2RY2 showed a decreased ability to recruit monocytes and macrophages upon activation of nucleotides from apoptotic cells(43). P2RY2 is also known to induce CCL2 secretion in macrophages, and coding variants in the receptor have been shown to influence secretion of this proinflammatory chemokine(44).
Although the HLA region has been implicated in SLE susceptibility since the 1970s, the precise loci responsible for risk have not been fully characterized. A further cross comparison of populations will be beneficial to take advantage of differences in linkage disequilibrium and will likely help further refine association signals seen in the GWA studies. For the classical alleles, previous studies have identified associations with alleles in the HLA-DR locus in Europeans, Chinese, Japanese, and Koreans, but HLA-DQB1*0602 has not been implicated in Koreans before this current study(45-48). Two prominent Classical HLA alleles identified in Europeans with SLE showed differences in association in Koreans. While HLA-DRB1*1501 was among the most significantly associated with SLE, HLA-DRB1*0301 was found to be at low frequency in this population and is not associated with SLE (Supplementary Table 5). These results are consistent with a recent study in Koreans that evaluated the role of HLA in this population(49). Moreover, this study report that amino acid changes to HLA-DRB1 at positions 11, 13, and 26 account for the HLA association in SLE(49). However, it is possible that there are other amino acid changing variants, non-coding RNAs, and/or transcriptional changes for other HLA loci that are co-inherited with HLA-DRB1 on these haplotypes may also contributing to SLE risk.
This GWA scan replicated several loci that have been identified by prior studies, including STAT1-STAT4, TNFSF4, TNFAIP3, IKZF1, HIP1, IRF5, BLK, WDFY4, ETS1, and IRAK1-MECP2. It is important to note that a previous GWA scan of Korean women with SLE has also reported replication of STAT4 and BLK at a genome-wide significant level(30). Of these replicated regions, TNFSF4, TNFAIP3, IKZF1, HIP1, IRF5, BLK, and ETS1 have functional effects that have been previously described (see details in Supplementary Table 18). Although most of the signals in these loci are identical, we did describe notable differences with independent effects in TNFSF4 and TNFAIP3. Moreover, we did not observe association with rs877819, which had been proposed as putative causal variant leading to expression differences of WDFY4(35).
In conclusion, we performed a GWA scan of Korean SLE cases and population controls in which we identified 12 regions that surpassed genome-wide significance. The region from ATG16L2 through FCHSD2 to the promoter region of the P2RY2 locus was identified and confirmed as an SLE-associated region. Here, we also observed strong associations in the HLA region and showed the relationship between the classical HLA alleles and the variants reported within this GWA study in Koreans. The ten additional regions, STAT1-STAT4, TNFSF4, TNFAIP3, IKZF1, HIP1, IRF5, BLK, WDFY4, ETS1, and IRAK1-MECP2 have previously been implicated in SLE. Additional replication is needed for the suggestive loci identified in this study to determine their relationship with SLE. Although GWA approaches have been very successful in the identification of risk loci, continued efforts are need to narrow association signals to the causal variant(s) and to determine the functional causal mechanism(s) contributing to SLE pathogenesis.
Supplementary Material
Acknowledgments
We are grateful to all the individuals with SLE and those serving as healthy controls who participated in this study. Sample collection and phenotyping of the subjects utilized in this study was made possible by funds from the Korea Healthcare Technology R&D Project of the Ministry for Health & Welfare (HI13C2124, S.C.B.). This publication was made possible by grants 5P01 AI083194 (C.J.L., A.R., P.M.G., M.E.A.R., L.A.C., C.O.J., R.P.K., T.J.V., J.B.H., C.D.L., K.L.S, and B.P.T.), 5P01 AR049084 (R.P.K. and C.D.L.), R01 AR043814 (B.P.T.), R21 AR065626 (B.P.T.) by the NIH. The contents are the sole responsibility of the authors and do not necessarily represent the official views of the NIH. Additional support was obtained from Wake Forest School of Medicine Center for Public Health Genomics (C.D.L.), Oklahoma Medical Research Foundation (C.J.L., P.M.G., and K.L.S.), the Alliance for Lupus Research (L.A.C. and B.P.T.), and Kirkland Scholar Award (L.A.C.). The out-of-study Korean control data was provided from Korean Biobank Project supported by the Korea Center for Disease Control and Prevention at the Korea National Institute of Health.
Footnotes
Author Contributions: C.J.L., J.A.K., A.R., M.E.A.R., L.A.C., C.O.J., R.P.K., T.J.V., J.B.H., K.L.S., S.-C.B., C.D.L., and B.P.T were responsible for the study design. K.K, S.-Y.B., Y.B.J., J.C., H.-S.L., Y.M.K., C.-H.S., W.T.C., S.-K.L., J.-Y.C., S.C.S., J.H.O., Y.J.K., B.-G.H., N.S., H.S.H., Y.W.S., S.-C.B., and B.P.T. assisted in the collection and characterization of the SS cases and healthy controls. J.Z., J.A.I., E.K.W., Q.-Z.L., and P.M.G. performed the genotyping. C.J.L., S.S, H.A., performed all analyses under the guidance of K.L.S., C.D.L. and B.P.T. C.J.L., S.S., J.Z., K.K., H.L., J.A.I., K.L.S., S.-C.B., C.D.L., and B.P.T. prepared the manuscript, and all authors approved the final draft.
Competing Financial Interests:
The authors declare no competing financial interests.
Supplemental Data Summary
Supplemental data includes 16 tables and 14 figures.
Web-based Resources:
OMIM, www.omim.org/
SNPGWA version 4.0, www.phs.wfubmc.edu
SHAPEIT, www.shapeit.fr/
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 3.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes and immunity. 2009;10(5):373–9. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr Opin Rheumatol. 2014;26(5):482–92. doi: 10.1097/BOR.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai C, Deng Y, Quinlan A, Gaskin F, Tsao BP, Fu SM. Genetics of systemic lupus erythematosus: immune responses and end organ resistance to damage. Curr Opin Immunol. 2014;31C:87–96. doi: 10.1016/j.coi.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–7. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. American journal of human genetics. 2013;92(1):41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6(2):e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada Y, Shimane K, Kochi Y, Tahira T, Suzuki A, Higasa K, et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 2012;8(1):e1002455. doi: 10.1371/journal.pgen.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Annals of the rheumatic diseases. 2014 doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. American journal of human genetics. 2012;90(4):648–60. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser R, Taylor KE, Deng Y, Zhao J, Li Y, Nititham J, et al. Brief Report: Single-nucleotide polymorphisms in VKORC1 are risk factors for systemic lupus erythematosus in Asians. Arthritis and rheumatism. 2013;65(1):211–5. doi: 10.1002/art.37751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 14.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- 19.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 24.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, et al. HIBAG--HLA genotype imputation with attribute bagging. The pharmacogenomics journal. 2014;14(2):192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344(6183):519–23. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HS, Kim T, Bang SY, Na YJ, Kim I, Kim K, et al. Ethnic specificity of lupus-associated loci identified in a genome-wide association study in Korean women. Annals of the rheumatic diseases. 2014;73(6):1240–5. doi: 10.1136/annrheumdis-2012-202675. [DOI] [PubMed] [Google Scholar]
- 31.Adrianto I, Wen F, Templeton A, Wiley G, King JB, Lessard CJ, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nature genetics. 2011;43(3):253–8. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40(9):1062–4. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunninghame Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40(1):83–9. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manku H, Langefeld CD, Guerra SG, Malik TH, Alarcon-Riquelme M, Anaya JM, et al. Trans-ancestral studies fine map the SLE-susceptibility locus TNFSF4. PLoS Genet. 2013;9(7):e1003554. doi: 10.1371/journal.pgen.1003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Yang W, Qiu R, Li J, Xin Q, Wang X, et al. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes and immunity. 2012;13(7):536–42. doi: 10.1038/gene.2012.33. [DOI] [PubMed] [Google Scholar]
- 36.Yang SK, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, et al. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63(1):80–7. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 37.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 38.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. Journal of cell science. 2003;116(Pt 9):1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 41.Cao H, Yin X, Cao Y, Jin Y, Wang S, Kong Y, et al. FCHSD1 and FCHSD2 are expressed in hair cell stereocilia and cuticular plate and regulate actin polymerization in vitro. PLoS One. 2013;8(2):e56516. doi: 10.1371/journal.pone.0056516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafouresse F, Cotta-de-Almeida V, Malet-Engra G, Galy A, Valitutti S, Dupre L. Wiskott-Aldrich syndrome protein controls antigen-presenting cell-driven CD4+ T-cell motility by regulating adhesion to intercellular adhesion molecule-1. Immunology. 2012;137(2):183–96. doi: 10.1111/j.1365-2567.2012.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature. 2009;461(7261):282–6. doi: 10.1038/nature08296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins KR, Kovacevic W, Stokes L. Nucleotides regulate secretion of the inflammatory chemokine CCL2 from human macrophages and monocytes. Mediators of inflammation. 2014;2014:293925. doi: 10.1155/2014/293925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HS, Chung YH, Kim TG, Kim TH, Jun JB, Jung S, et al. Independent association of HLA-DR and FCgamma receptor polymorphisms in Korean patients with systemic lupus erythematosus. Rheumatology (Oxford) 2003;42(12):1501–7. doi: 10.1093/rheumatology/keg404. [DOI] [PubMed] [Google Scholar]
- 46.Graham RR, Ortmann W, Rodine P, Espe K, Langefeld C, Lange E, et al. Specific combinations of HLA-DR2 and DR3 class II haplotypes contribute graded risk for disease susceptibility and autoantibodies in human SLE. Eur J Hum Genet. 2007;15(8):823–30. doi: 10.1038/sj.ejhg.5201827. [DOI] [PubMed] [Google Scholar]
- 47.Furukawa H, Oka S, Shimada K, Sugii S, Hashimoto A, Komiya A, et al. Association of increased frequencies of HLA-DPB1*05:01 with the presence of anti-Ro/SS-A and anti-La/SS-B antibodies in Japanese rheumatoid arthritis and systemic lupus erythematosus patients. PLoS One. 2013;8(1):e53910. doi: 10.1371/journal.pone.0053910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furukawa H, Kawasaki A, Oka S, Ito I, Shimada K, Sugii S, et al. Human leukocyte antigens and systemic lupus erythematosus: a protective role for the HLA-DR6 alleles DRB1*13:02 and *14:03. PLoS One. 2014;9(2):e87792. doi: 10.1371/journal.pone.0087792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K, Bang SY, Lee HS, Okada Y, Han B, Saw WY, et al. The HLADRbeta1 amino acid positions 11-13-26 explain the majority of SLE-MHC associations. Nature communications. 2014;5:5902. doi: 10.1038/ncomms6902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.