Abstract
Pluripotent stem cells (PSC) may provide a potential source of haematopoietic stem/progenitor cells (HSPCs) for transplantation; however, unknown molecular barriers prevent the self-renewal of PSC-HSPCs. Using two-step differentiation, human embryonic stem cells (hESCs) differentiated in vitro into multipotent haematopoietic cells that had CD34+CD38−/loCD90+CD45+GPI-80+ foetal liver (FL) HSC immunophenotype, but displayed poor expansion potential and engraftment ability. Transcriptome analysis of immunophenotypic hESC-HSPCs revealed that, despite their molecular resemblance to FL-HSPCs, medial HOXA genes remained suppressed. Knockdown of HOXA7 disrupted FL-HSPC function and caused transcriptome dysregulation that resembled hESC-derived progenitors. Overexpression of medial HOXA genes prolonged FL-HSPC maintenance but was insufficient to confer self-renewal to hESC-HSPCs. Stimulation of retinoic acid signalling during endothelial-to-haematopoietic transition induced the HOXA cluster and other HSC/definitive haemogenic endothelium genes, and prolonged HSPC maintenance in culture. Thus, retinoic acid signalling-induced medial HOXA gene expression marks the establishment of the definitive HSC fate and controls HSC identity and function.
Haematopoietic stem cells (HSCs) regenerate the blood system upon transplantation, and can therefore cure inherited and acquired blood diseases. However, lack of HLA-matched bone marrow (BM) or cord blood (CB) donors limits their therapeutic use1. Generation of HSCs from human embryonic stem cells (hESCs) or induced pluripotent stem cells could provide alternative HSC sources. Recent studies used transcription factor reprogramming to convert fibroblasts or mature blood cells2–4 to haematopoietic cells possessing some properties of HSCs. Despite these promising approaches, clinical application of in vitro generated HSCs remains unachieved.
While hESCs can differentiate into most blood lineages5, efforts to produce engraftable HSCs have failed6. The molecular barriers preventing HSC generation in vitro are poorly understood due to lack of studies comparing candidate HSCs from PSC-cultures and human conceptus that match by immunophenotype and developmental stage. During embryogenesis, haematopoiesis starts in the yolk sac by the generation of two distinct waves of myelo-erythroid progenitors (primitive and transient definitive) that can be distinguished by the specific globins expressed in their progeny7. These progenitors lack self-renewal ability and robust lymphoid potential8,9. Definitive HSCs possessing these properties emerge in the third haematopoietic wave from specialized haemogenic endothelium in major arteries in the AGM (aorta-gonad-mesonephros) region, yolk sac, placenta and vitelline and umbilical vessels10. Human haemogenic endothelial cells express CD34 and CD3111 and up-regulate CD43 upon haematopoietic commitment12,13, whereas HSCs also co-express CD45 (pan-haematopoietic), CD90 (HSC, endothelium), GPI-80 (human foetal HSCs14), and typically have low CD38 expression (lineage commitment/HSC activation). Haematopoietic differentiation of mouse and human ESCs mirrors embryonic haematopoiesis8,15 and recapitulates mesoderm and haemato-vascular commitment16,17 followed by waves of primitive and definitive erythropoiesis18,19. However, hESC-derived haematopoietic cells lack reconstitution ability6,20,21 and full lymphoid and adult-type erythroid potential22,23, resembling yolk sac-derived lineage-restricted progenitors24. A long-standing goal has been to identify regulatory cues and molecular landmarks that distinguish the definitive HSC fate from the short-lived embryonic progenitors.
We used a two-step hESC differentiation to generate HSPCs with human foetal HSC surface phenotype (CD45+CD34+CD38−/loCD90+GPI-80+). Molecular profiling showed remarkable resemblance of hESC-HSPCs to FL-HSPCs, yet revealed distinct differences in HSC regulatory programs, including the HOXA genes. Knockdown and overexpression studies revealed that medial HOXA genes, in particular HOXA7, govern definitive HSPC identity and function. Rescue of retinoic acid (RA) signalling during endothelial-to-haematopoietic transition induced medial HOXA genes and definitive haematopoietic program in hESC-HSPCs. These studies uncover regulatory programs that distinguish human definitive HSC lineage from embryonic progenitors and offer a blueprint for identifying missing cues required for HSC generation.
Results
hESCs generate HSPCs that are unable to engraft
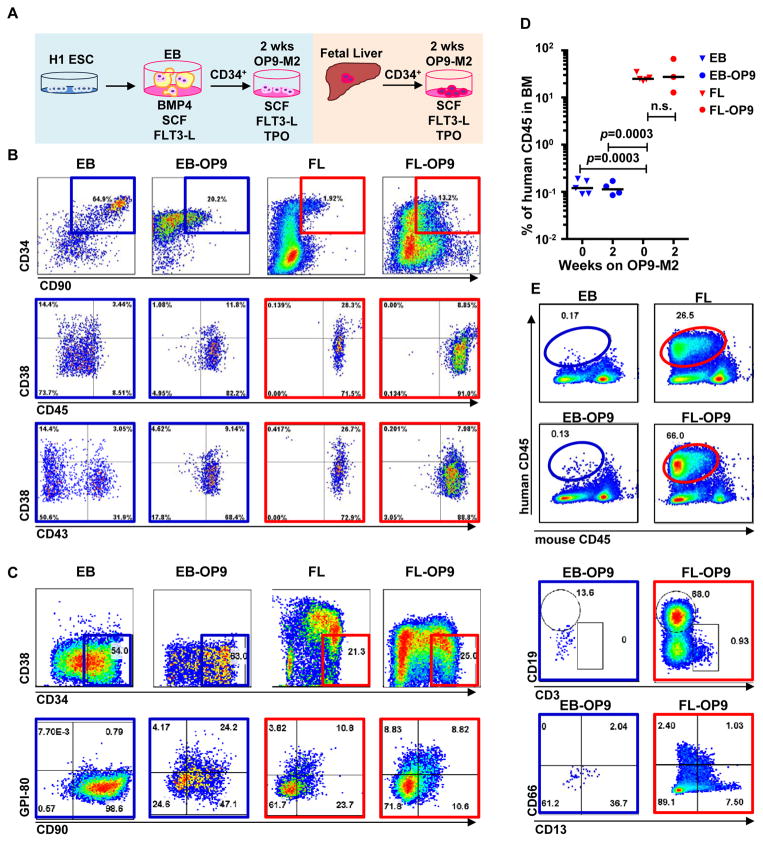
To identify barriers for generating HSCs from PSCs, we compared hESC-derived HSPCs and human foetal HSCs based on immunophenotypic, functional and molecular criteria. Second trimester FL-HSCs were used as controls as they are ontologically closer to PSC-derived cells than CB or BM. A two-step differentiation protocol was employed to generate haematopoietic cells from H1 hESCs (Figure 1A). To promote haemato-vascular differentiation, embryoid bodies (EB) were cultured with BMP4 (days 4–10), FLT-3L and SCF (days 4–14)25. Day 14 EBs had generated cells that co-expressed CD34, CD90 and CD43 (Figure 1B), but not CD45. To promote the development of hemato-vascular precursors toward HSCs, EB-derived CD34+ cells were plated with TPO, SCF and FLT3L on OP9-M2 stroma, which supports the expansion of multipotent human HSPCs26. Two-week OP9-M2 co-culture (EB-OP9) generated cells with CD34+CD38−/loCD90+CD45+ HSPC immunophenotype (Figure 1B) and some co-expressed the HSC marker GPI-80 (Figure 1C). These data showed that two-step differentiation generates cells with the immunophenotype of human foetal HSCs.
Figure 1. Two-step culture of hESCs generates immunophenotypic HSPCs that engraft poorly.
(A) Culture and isolation strategy for differentiating H1 hESCs to HSPCs. (B) Representative FACS plots from 11 experiments staining for CD34, CD90, CD38, CD45 and CD43 on hESC-derived CD34+ cells isolated from 2 week EBs (EB), and after 2 week maturation culture on OP9-M2 (EB-OP9), compared to cells from second trimester foetal liver that were isolated directly (FL) or cultured on OP9-M2 (FL-OP9). (C) Representative FACS plots from 9 experiments staining for CD38, CD34, CD90 and human foetal HSC self-renewal marker GPI-80 on hESC- and FL-derived cells. (D) Human engraftment in NSG mice with hESC-derived and FL-derived CD34+ cells, before and after OP9-M2 co-culture (individual values and mean are shown, n=5 EB, n=4 EB-OP9 and FL, and n=3 FL-OP9 transplanted mice, statistical significance was calculated using the Wilcoxon Rank Sum test, see Supplementary Table 7 for statistics source data). (E) Representative FACS plots of showing human CD45+ fraction in the mouse BM 12 weeks post-transplantation. Multi-lineage engraftment is assessed by CD19 and CD3 (B-and T-lymphoid), and CD66 and CD13 (myeloid) stainings.
To assess whether the hESC-HSPC maturation culture on OP9-M2 stroma confers functional properties of HSCs, CD34+ cells from EBs (isolated directly or after two-step differentiation), and FL (isolated directly or after 2 week culture) were transplanted into sublethally irradiated NOD-scid IL2Rγ-null (NSG) mice (Supplementary Figure 1A). Human CD45+ chimerism in BM was measured 12 weeks post-transplantation. While FL-HSPCs engrafted successfully before or after OP9-M2 culture, hESC-derived cells showed minimal engraftment (Figure 1D). Human CD45+ cells in the BM of mice transplanted with FL contained HSPCs (Supplemental Figure 1B), CD19+ B-cells, CD3+ T-cells and CD13+ or CD66+ myeloid cells, whereas the mice transplanted with hESC-derived cells only harboured rare human myeloid cells (Figure 1E). These data show that hESC-HSPCs are severely impaired functionally.
hESC-HSPCs have poor proliferative potential
To understand the functional defects in hESC-HSPCs, hESC- and cultured FL-HSPCs (CD34+CD38−/loCD90+CD45+) were sorted and re-plated on OP9-M2 co-culture to assess their expansion (Figure 2A). Both FL- and hESC-HSPC cultures maintained an immunophenotypic HSPC population one week later (Figure 2B, 2C), however, at three weeks, hESC-HSPCs had disappeared (Figure 2B, 2C). BrdU incorporation analysis did not reveal differences in cell cycle between FL- and hESC-HSPCs (Supplementary figure 2A), suggesting that loss of hESC-HSPCs was not due to inability to divide.
Figure 2. hESC-derived haematopoietic cells have limited proliferative potential in vitro.
(A) Strategy for comparing the expansion of hESC- and FL-HSPCs. (B) FACS staining for HSPC surface markers CD34+, CD38−/lo, CD45+ and CD90+ at various time points in OP9-M2 co-culture. (C) Expansion of FL and hESC-derived haematopoietic cells sorted for HSPC phenotype after two-step differentiation, and cultured for additional weeks on OP9-M2 (mean +/- SEM – upward bars – from n=4 experiments, statistical significance was assessed using the Wilcoxon Rank Sum test. (D) CFU-C expansions from 10,000 hESC-derived or FL-derived immunophenotypic HSPCs in methylcellulose following 0, 1 and 3 additional weeks on OP9-M2 co-culture (mean +/- SEM – upward bars –from n=4 experiments, statistical significance was assessed using the Wilcoxon Rank Sum test. (E) The morphology of myelo-erythroid colonies generated from hESC- or FL-HSPCs on methylcellulose as assessed by light microscopy (green scale bar = 1mm) and May-Grünwald-Giemsa (MGG) staining (black scale bar = 100 μm). Statistics source data used to generate graphs in C and D can be found in Supplementary Table 7.
hESC-HSPCs also produced fewer clonogenic progenitors on OP9-M2 than FL-HSPCs (Figure 2D), although both formed erythroid (BFU-E), granulocyte-macrophage (CFU-GM), macrophage (CFU-M) and mixed myelo-erythroid (CFU-Mixed) colonies (Figure 2E). CFU-C potential in EB-OP9 cells was higher in CD34+CD38−/loCD90+CD45+ fraction than total CD34+ cells (Supplementary Figure 2B), which also showed minimal CFU-C expansion (Supplementary Figure 2C). These data indicate that hESC-derived immunophenotypic HSPCs display poor proliferative potential.
Previous studies documented impaired differentiation of hESCs to adult-type erythroid cells23. Colonies from EB CD34+ cells generated erythroid cells that predominantly expressed embryonic ε-globin, whereas foetal γ-globin (HBG1) was low (16.3% of FL levels) and adult β-globin (HBB) nearly undetectable (1.8% of FL levels). However, colonies derived from EB CD34+ cells after 2 weeks on OP9-M2 induced some expression of γ- and β-globins (84.9% and 54.0% of FL-derived cells, respectively) (Supplementary Figure 2D). β-globin induction suggests that hESC-HSPCs can differentiate to adult-type erythroid cells if given adequate maturation time in culture.
T-cells generation is another hallmark of definitive haematopoiesis27. Both EB- and FL-derived CD34+ cells generated CD4 and CD8 single- and double-positive T-lymphoid cells in OP9-DL1 culture (Supplementary Figure 2E). These results imply that the main defect in hESC-derived haematopoietic cells is not differentiation, but self-renewal.
Microarray uncovers molecular defects in hESC-HSPCs
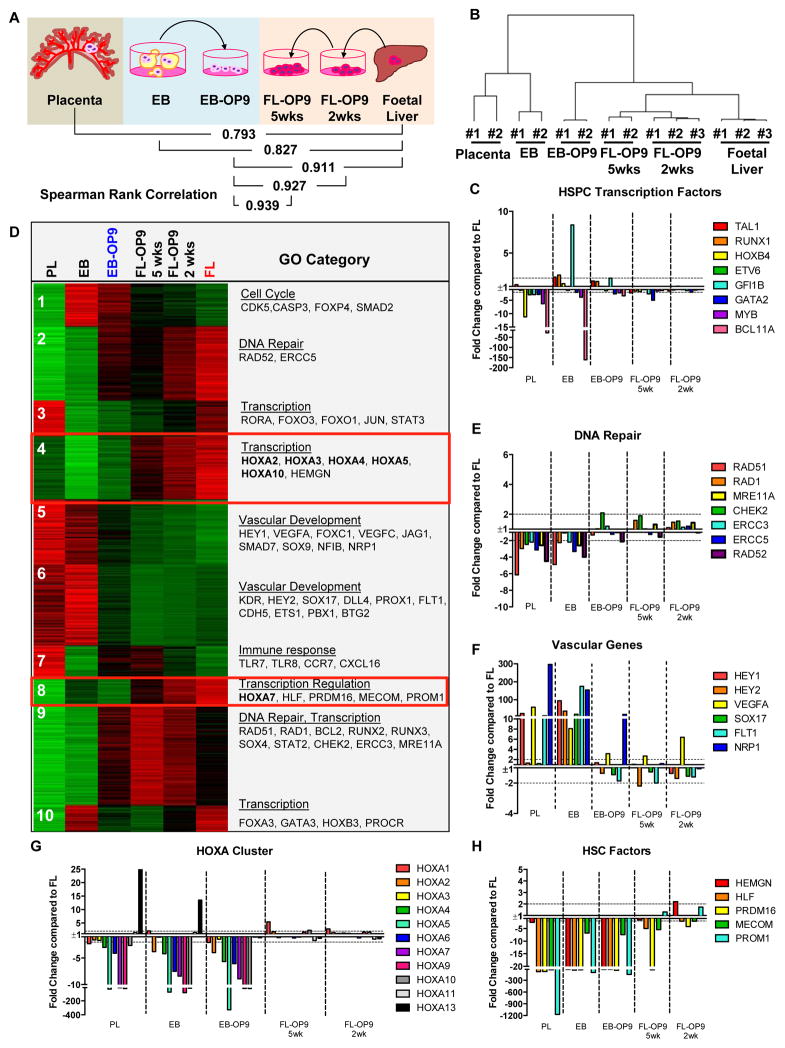
To understand the defective self-renewal of hESC-HSPCs, we assessed their relationship to primary human foetal HSPCs using microarray analysis. Gene expression differences were compared between immunophenotypic HSPCs (CD34+CD38−/loCD90+CD43+CD45+/−) derived from hESCs via EB or two-step differentiation (EB-OP9), and from FL. To assess culture-induced effects, FL-HSPCs were cultured on OP9-M2 for 2 or 5 weeks (FL-OP9). CD34+CD38−/loCD90+CD43+ cells from early human placenta (PL) were used as a reference for immature human HSPCs (Supplementary Table 1). Spearman-rank coefficient comparisons and hierarchical clustering of samples revealed that FL-HSPCs are more similar to EB-OP9-HSPCs than EB- or PL-HSPCs (Figure 3A and B). EB-OP9-HSPCs were most similar to cultured FL-HSPCs, implying that hESC-HSPC transcriptome was influenced by prolonged culture (Figure 3A). Many transcription factors governing the development of definitive HSPCs (SCL/TAL1, GATA2, RUNX1, MYB, ETV6, HOXB4, GFI1B, BCL11A etc.) were expressed in both EB-OP9-HSPCs and FL-HSPCs (Figure 3C). These data revealed that EB-OP9-HSPCs are remarkably similar to FL-HSPCs at the molecular level.
Figure 3. Identification of differentially expressed programs in hESC- and FL-HSPCs.
(A) Spearman rank correlation of HSPCs isolated at different stages of development: 3–5 week placenta (PL, CD34+CD38−/lo CD90+CD43+ n=2), hESC-HSPCs isolated from 2 week EBs (EB, CD34+CD38−/loCD90+CD43+ n=2) or after two-step differentiation (EB-OP9, CD34+CD38−/lo CD90+CD43+CD45+ n=2), and 2nd trimester FL isolated freshly (FL, CD34+CD38−/lo CD90+CD45+ n=3) or after 2 or 5 weeks on OP9-M2 (FL-OP9, CD34+CD38−/loCD90+CD45) (n=3 and n=2, respectively). n represents number of tissue samples collected from separate specimens per condition. Each replicate was collected from independent experiments and analysed together. (B) Dendrogram showing hierarchical clustering of microarray samples. (C) Relative levels of haematopoietic transcription factors in different samples compared to FL-HSPCs. (D) K-means clustering of differentially expressed genes in HSPCs from different stages of human haematopoietic development with representative examples of GO terms and genes in clusters (see Supplementary Tables 2 and 3 and GEO database GSE64865). (E) Levels of DNA repair genes compared to FL-HSPCs from Clusters 2 and 9, (F) vascular genes from Clusters 5 and 6,. (G) HOXA genes and (H) other HSC factors from clusters 4 and 8. See Supplementary Tables 1 and 3, and GEO database GSE64865 for values.
To identify co-regulated and differentially expressed programs between samples K-means clustering and DAVID GO (gene ontology) analysis was performed for all differentially expressed genes (> 2-fold, p value <0.05) (Supplementary Tables 2 and 3). K-means clustering showed that OP9-M2 co-culture of EB-derived cells induced many FL-HSPC associated genes (Figure 3D, clusters 2 and 9) that encoded DNA-repair factors and transcriptional regulators (Figure 3E), while vascular genes were downregulated (Figure 3D, clusters 5 and 6, Figure 3F).
K-means clustering also identified genes that remained dysregulated in hESC-HSPCs. The most striking differences were observed in clusters 4 and 8 that contained genes highly expressed in FL-HSPCs but suppressed in hESC-HSPCs and early PL. The most highly enriched GO category was “Transcription” (Figure 3D), which included many HOXA genes. All HOXA genes except for HOXA13 (a regulator of placental vascular labyrinth specification28) were severely suppressed in hESC-derived HSPCs (Figure 3G). Similar HOXA pattern was observed with early PL, raising the hypothesis that medial HOXA gene induction in FL-HSPCs reflects developmental maturation and acquisition of HSC properties. Clusters 4 and 8 also included HSC regulators HMGN, HLF, PRDM16 and MECOM/EVI129,30, and HSC surface markers PROM1/CD133, EMCN and ROBO4 (Figure 3H). These analyses demonstrated that the two-step conditioning fails to induce transcriptional regulators highly expressed in FL-HSCs, including HOXA genes.
Medial HOXA genes govern human HSPC function
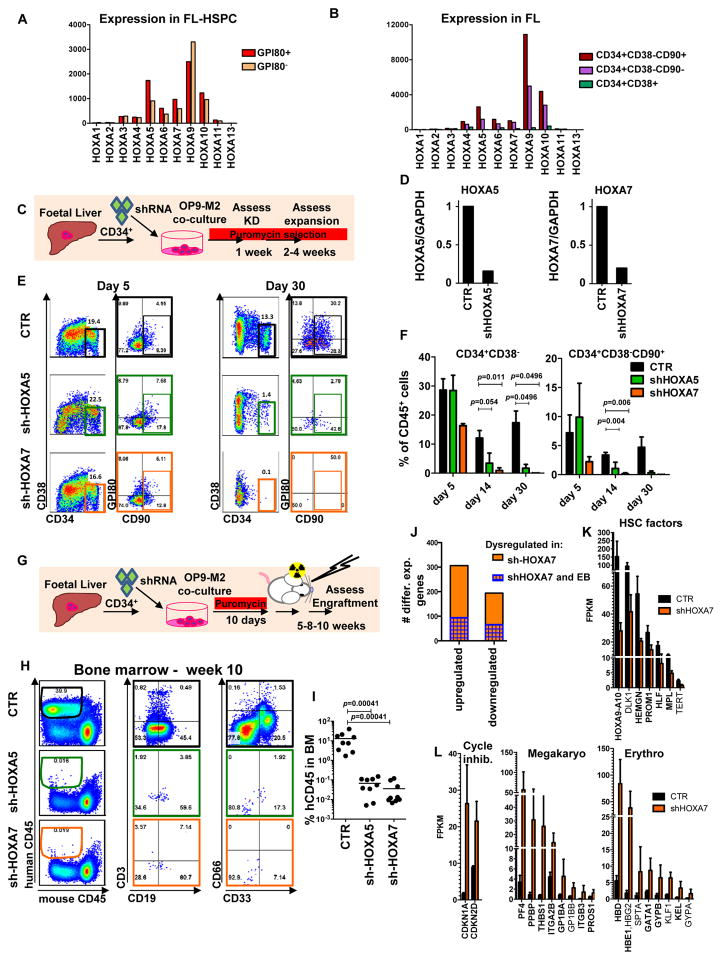
We next asked whether HOXA gene silencing contributes to the poor self-renewal of hESC-HSPCs. Microarray analysis of human FL-HSPC subsets14 documented expression of several medial HOXA genes in GPI-80+ HSCs and their immediate progeny (Figure 4A), and down-regulation upon differentiation (Figure 4B). As HOXA9 is a known regulator of mouse HSC proliferation31,32, FL-HSPCs were transduced with pLKO.1 shRNA lentiviral vectors targeting HOXA5 or HOXA7 (Figure 4C) to test if other medial HOXA genes regulate human HSCs. Knockdown was confirmed using q-RT-PCR one week post-transduction (Figure 4D, Supplementary Table 6). By 2–4 weeks, HOXA5 and HOXA7 shRNA-treated cells were depleted of HSPCs (Figure 4E, 4F). Cell cycle analysis 1 week post-transduction did not reveal significant differences in BrdU incorporation of HOXA5 or HOXA7 knockdown HSPCs, (Supplementary Figure 3A), implying that HOXA5- and HOXA7-deficient HSPCs can divide, but cannot self-renew. Transplantation of HOXA5 and HOXA7 shRNA-transduced FL-HSPCs into NSG mice (Figure 4G) showed minimal engraftment (Figure 4H, 4I and). These results suggest that both HOXA5 and HOXA7 are necessary for human FL-HSPC expansion in vitro and reconstitution in vivo. HOXA7 showed stronger phenotypes in both assays.
Figure 4. Medial HOXA genes govern the function and identity of human foetal HSPCs.
(A, B) Microarray analysis of HOXA gene expression in CD34+CD38−/lo CD90+GPI-80+ cells and their progeny (Mean values are shown, left, n=3 samples, GEO database GSE54316, and right, n=3 samples (CD34+CD38-CD90+) or 2 (CD34+CD38-CD90- and CD34+CD38-) GSE34974. (C) Schematic showing the strategy for lentiviral shRNA knockdown of HOXA5 or HOXA7 in FL-HSPCs. (D) Knockdown is confirmed using q-RT-PCR 1 week post-infection (mean +/- SD shown from n=3 different FL samples). (E) Representative FACS plots 30 days after HOXA5 or HOXA7 knockdown. (F) Quantification of HSPC subsets in empty-vector (CTR) and shRNA infected cells (shHOXA5 or shHOXA7) after 5, 14 and 30 days in culture (mean and SEM, n=6 independent experiments per condition for day 14 and n=3 for day 5 and 30). Statistical significance was assessed using Wilcoxon Signed Rank test. (G) Schematic showing the transplantation strategy with HOXA5 or HOXA7 knockdown FL-HSPCs. (H) Representative FACS plots from mouse BM 10 weeks post-transplantation assessing human CD45+ cells and multi-lineage engraftment (CD19 and CD3 for B-and T-lymphoid, and CD66 and CD33 for myeloid). (I) Quantification of human engraftment (n=9 mice per condition from 3 independent experiments Individual values and mean are shown.) Statistical significance was assessed using the Wilcoxon Rank Sum test (J) RNA-sequencing of HOXA7 knockdown FL-HSPCs at day 5 post-infection. Number of genes up- or down-regulated in shHOXA7 FL-HSPCs are shown. Genes dysregulated both in HOXA7 knockdown FL-HSPCs (RNA-seq 1.8-fold change, n=4 independent experiments, p-value < 0.05) and in EB-OP9-HSPCs compared to FL-HSPCs (microarray, 2-fold change, p-value < 0.05) are shown in blue pattern overlay. (K) Examples of HSC factors downregulated in HOXA7 knockdown FL-HSPCs and (L) differentiation associated genes upregulated in HOXA7 knockdown FL-HSPCs. Mean shown for n=4 independent specimens, values used to generate graphs can be found in Supplementary Table 4 and GEO database GSE76685). See Supplementary Table 7 for Statistics source data for 4D, F and I.
To investigate how HOXA7 regulates human HSCs, pLKO1-control- and HOXA7- shRNA-transduced FL-HSPCs from four different FL tissues were sorted 5 days post-infection and subjected to RNA sequencing (Figure 4E). 500 significantly differentially expressed genes (306 up-regulated, 194 downregulated, 1.8-fold, p-value <0.05) between HOXA7 knockdown and pLKO1-control HSPCs were identified (Figure 4J and Supplementary Table 4). Comparison to hESC-HSPC microarray data revealed that 30.1% of the genes upregulated upon HOXA7 knockdown were also significantly upregulated in EB-OP9-HSPCs compared to FL-HSPCs, and 34.0% of the downregulated HOXA7 dependent genes showed low expression in EB-OP9 HSPCs. The shared downregulated genes included HSC regulators HOXA9-10, HLF and HMGN, and HSC surface proteins PROM1/CD133 and MPL (Figure 4K). HOXA7 knockdown HSPCs and EB-OP9-HSPCs also upregulated genes associated with megakaryocytic and erythroid differentiation (Figure 4L). The shared upregulated genes also included cell cycle inhibitors CDKN1A (p21Cip1)33 and CDKN2D (p19Ink4d)34, while other cell cycle regulators and proliferation markers were unaffected at mRNA level (Supplementary Figure 3B, C). These results imply that HOXA7 activates factors regulating definitive HSC identity and suppresses programs associated with differentiation-primed embryonic progenitors.
HOXA gene overexpression expands FL-HSPCs
We next assessed if HOXA gene overexpression improves in vitro expansion of FL-HSPCs and hESC-HSPCs. Tetracyclin-inducible PNL lentiviral vector35 (Supplementary figure 4A), which induced 30-fold overexpression of HOXA5 or HOXA7 in FL-HSPCs (Supplementary figure 4B), prolonged FL-HSPC maintenance in culture with both HOXA5 and HOXA7 (Supplementary figure 4C–D). Constitutively active FUGW vector36, which resulted in 4–5-fold transgene overexpression, prolonged FL-HSPC maintenance with HOXA7 (Figure 5A–E). HSPCs overexpressing HOXA genes showed comparable differentiation ability in colony assays, implying that HOXA5 or HOXA7 expression does not prevent differentiation (Figure 5F).
Figure 5. Overexpression of medial HOXA genes enhances proliferative potential in FL-HSPCs but does not confer HSC properties to hESC-HSPCs.
(A) Schematic showing the strategy for constitutive lentiviral overexpression of HOXA5 or HOXA7 in FUGW vectors in FL-HSPCs. (B) Representative FACS plots of FUGW empty vector, HOXA5- or HOXA7- overexpressing FL-HSPCs. (C, D) Expansion of total FL cells (C) or HSPCs (D) transduced with HOXA5- or HOXA7- overexpression vectors or empty vector control (CTR), (mean and SEM values from n=3 independent experiments; statistical significance was assessed using the paired Student’s t-test. (E) q-RT-PCR confirming overexpression in transduced HSPCs sorted 1 week post-infection (n=1 experiment with 2 pooled donors). (F) CFU-Cs from 2000 HSPCs sorted after day 10 of infection with vectors overexpressing HOXA5 or HOXA7 or FUGW empty vector control (mean and SD values shown from n=4 transductions from 2 independent experiments, p-values shown correspond to CTR vs. OE-HOXA7). (G) Schematic showing the strategy for lentiviral overexpression of HOXA5 and/or HOXA7 and/or HOXA9 in FUGW vectors in EB CD34+ cells. (H) Representative examples of FACS plots of EB CD34+ cells overexpressing HOXA5 or HOXA7 or a combination of HOXA5, HOXA7 and HOXA9. Un-transduced FL is shown as a control. (I) Quantification of CD34+CD38−/loCD45+ haematopoietic cells from (H), mean from n=4 independent experiments for CTR and n=3 for HOXA5/7/9, HOXA5, and HOXA7 at days 0 and 24, and n=2 at all other time points. J) Representative FACS plots and (K) quantification of human CD45+ cells in the BM of NSG mice 12 weeks post-transplantation. Multi-lineage engraftment is assessed by CD19 and CD3 (B-and T-lymphoid) and CD66 and CD33 (myeloid) (mean from n=5 mice per condition (except for FL n=4) from two independent experiments). (L). Q-RT-PCR for HOXA7 from transduced EB-OP9-HSPCs 2 weeks post-infection from one representative experiment. (M) Graphs representing RNA-seq of EB-OP9 cells overexpressing HOXA7 for genes regulated by HOXA7 in FL-HSPCs (Figure 4K,L) (one representative experiment, GEO database GSE76685). See Supplementary Table 7 for statistics source data in D, E, F, I and K.
Although HOXA5 or HOXA7 overexpression promoted FL-HSPC expansion, their overexpression in EB-derived CD34+ cells did not improve hESC-HSPC expansion in vitro (Figure 5G–I and Supplementary figure 4E) or engraftment in vivo (Figure 5J–K). Similar results were observed overexpressing HOXA5, HOXA7 and HOXA9 simultaneously. RNA-sequencing of HOXA7 overexpressing hESC-HSPCs showed that despite the confirmed HOXA7 overexpression (Figure 5L), there was minimal change in putative HOXA7 target genes (Figure 5M). Altogether these studies showed that medial HOXA genes promote HSPC expansion when expressed in correct cellular context.
RA signalling induces HOXA genes in hESC-HSPCs
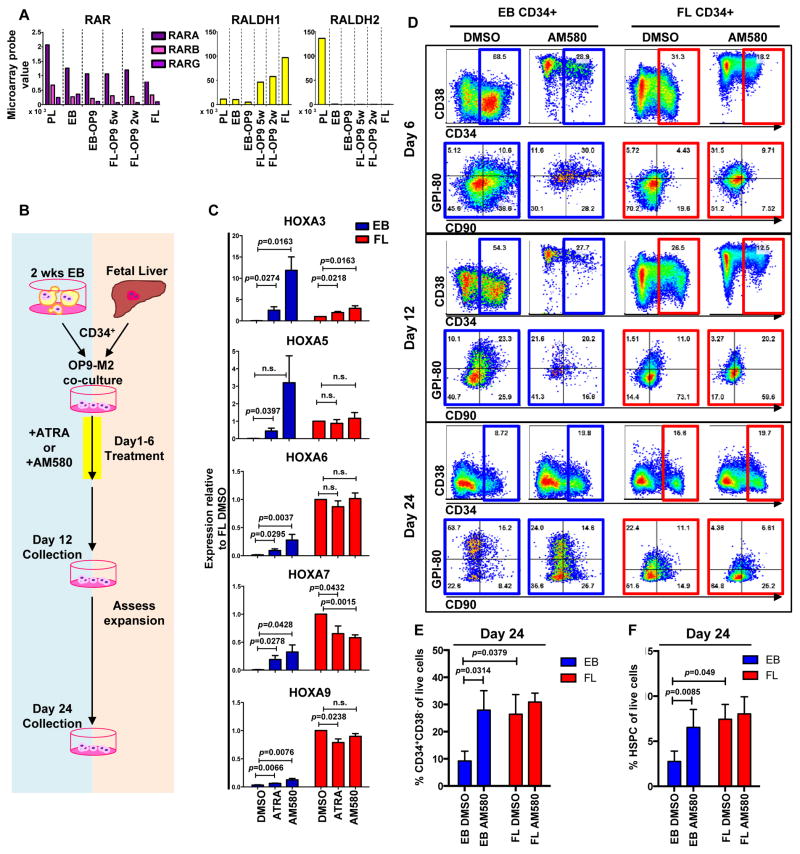
We next sought for upstream regulators that could specify the definitive HSPC fate and induce HOXA genes in a correct cellular context. Medial HOXA genes are developmentally regulated by retinoic acid (RA) signalling in other cell types37,38. RA generated by ALDH1A2 (RALDH2) in haemogenic endothelium and its binding to RA receptor alpha (RAR-α) is necessary for generating HSCs in mouse AGM39. Microarray analysis showed robust expression of RAR-α, and to lesser extent RAR-β and RAR-γ in HSPCs at different stages. Notably, RALDH2 was expressed in CD34+CD38−/loCD90+CD43+ cells in the early placenta, but not in EB. FL-HSPCs did not express RALDH2, consistent with its function in haemogenic endothelium (Figure 6A). RALDH1 was expressed in FL-HSPCs but low in both EB- and PL-derived cells. These results nominated defective RA signalling during hESC haematopoietic specification as a potential barrier for inducing HOXA genes.
Figure 6. Retinoic acid signalling activates medial HOXA genes during human haematopoietic development.
(A) Microarray analysis of gene expression of components of RA signalling pathway compared to FL-HSPCs (for n see Figure 4B); for mean values see Supplementary Table 1 and GEO database GSE64865). (B) Schematic showing 6-day treatment of CD34+ EB and FL cells by all-trans retinoic acid (ATRA) and RAR-α agonist AM580. Cells were re-seeded on OP9-M2 stroma after 12 days and analysed after additional 12±1 days (day 24±1). (C) q-RT-PCR of HOXA3, HOXA5, HOXA6, HOXA7 and HOXA9 expression in EB or FL cells treated with RA and AM580 (mean +/- SEM from n=4 independent experiments, see Supplementary Table 7 for statistics source data). (D) Representative FACS plots of CD45+ cells from AM580-treated EB and FL cells at 6, 12 and 24 days on OP9-M2 culture (n=8 independent experiments). (E) Quantification of CD34+CD38−/lo and (F) HSPC fraction of EB- and FL-derived haematopoietic cells at day 24±1 of OP9-M2 culture (mean +/− S.E.M from n=8 independent experiments). Statistical significance was assessed using the Student’s paired t-test for C, E, and F, one-tailed for C and two-tailed for E and F.
We next asked whether RALDH2 function could be bypassed by administering all-trans retinoic acid (ATRA) or the RAR-α agonist AM58040 during hESC differentiation. CD34+ cells were isolated from EBs at 2 week and cultured for 6 days on OP9-M2 with or without ATRA (1.0 μM) or AM580 (0.2 μM) (Figure 6B). Q-RT-PCR analysis showed that treatment of EB-derived CD34+ cells with AM580 or ATRA induced several medial HOXA genes (Figure 6C). AM580 had more robust effect on HOXA genes and was used for further studies.
FACS analysis at 6 days of AM580 treatment revealed strong induction of CD38, a known RA target41, in hESC- and FL-HSPCs (Figure 6D). Persistent CD90 expression suggested they had not differentiated. AM580-induced CD38 expression was reversible (Figure 6D), and AM580-treated EB CD34+ cells typically retained a higher fraction of CD34+CD38−/lo cells and undifferentiated HSPCs after 23–25 days than DMSO-treated controls (Figure 6E, F). A concomitant increase was observed with CFU-Cs, in particular with mixed myelo-erythroid colonies (Supplemental Figure 5A, B). These data show that inducing RA-signalling in haemogenic endothelium stage prolongs hESC-HSPC maintenance.
RA signalling promotes definitive haemogenic endothelium and HSC fate
To understand how RA signalling modulates hESC-derived haemato-vascular precursors, RNA-sequencing was performed to define AM580-induced genes in hESC-HSPCs (CD45+CD34+CD90+). Six days after AM580 treatment, RAR-β and RAR-γ, known RAR-α42, were upregulated (Figure 7A). HOXA cluster was also induced (Fig 7B–C), while RUNX1, which is expressed in both progenitors and HSCs, showed no difference. Altogether, AM580 induced 408 genes and repressed 562 (2-fold, p value <0.05) (Supplementary Table 5). AM580-induced genes showed highest enrichment in GO categories reflecting vasculature development, cell adhesion and migration (Fig 7D). These include factors implicated in definitive haemogenic endothelium and HSC generation such as CDH5, NOS3, and SOX17; Notch ligands and targets DLL4 and HEY2; and HSC surface markers ROBO4, EMCN and PROCR. Another highly enriched category was regulation of transcription, which included several HSC regulators (e.g. HOX co-factor PBX1, MECOM/EVI1, ERG, GFI1, GATA3 and HLF) (Figure 7E).
Figure 7. Retinoic acid signalling pulse in EB haemato-vascular cells induces transcriptional programs associated with definitive haemogenic endothelium and HSC fate.
(A) FPKM quantification from RNA-seq for RAR-α targets RAR-β and RARA-γ and (B) HOXA genes in sorted AM580-treated and DMSO control hESC-HSPCs (mean from 2 independent experiments). (C) RNA-seq genome browser screen shot for HOXA cluster and RUNX1 in hESC- and FL-HSPCs after 6 days of AM580 treatment. (D) GO categories of biological processes significantly upregulated in hESC-HSPCs by AM580 treatment at day 6. (E) FPKM quantification values from representative genes from vasculature development and transcription GO categories from genes significantly upregulated in hESC-HSPCs by 6 day AM580 treatment (2-fold or greater change, p-value <0.05, mean from 2 independent experiments). (F) ATAC-seq genome browser shot for HOXA cluster and RUNX1 assessing change in accessibility of regulatory regions in hESC- and FL-HSPCs upon AM580 treatment. (G) Peaks significantly induced by AM580-treatment grouped based on the distance from TSS. (H) GO categories enriched among genes showing significant difference in accessibility after AM580 treatment. (I) ATAC-seq signal proximal to the TSS of genes up- or downregulated by AM580 treatment. (ATAC-seq data shows one representative data set from two independent experiments that showed comparable results). See Supplementary Table 5 and GEO database GSE76685 for values used to generate graphs in 6 B, E, G, H and I.
Analysis of hESC-HSPCs at day 12 of OP9-M2 culture (6 days after AM580 removal) evidenced expression of HOXA, albeit at lower levels, and many vascular factors and HSC regulators (Supplementary figures 6A, B). Interestingly, many genes upregulated in HOXA7 knockdown FL-HSPCs and hESC-HSPCs, including cell cycle inhibitors CDKN1A and CDKN2D and erythroid and megakaryocytic genes, started to decline in AM580-treated hESC-HSPCs (Supplementary figure 6C). These data suggest that RAR-α signalling during endothelial-to-haematopoietic transition induces HOXA genes and other regulators that establish the definitive HSC fate, while suppressing embryonic progenitor programs.
To investigate if RA signalling induces genes by modulating chromatin accessibility, ATAC-sequencing was performed for CD45+CD34+CD90+ hESC-HSPCs (DMSO- or AM580-treated for 6 days) and FL-HSPCs. AM580 stimulation increased chromatin accessibility throughout the HOXA cluster (Figure 7F). Unbiased analysis of differentially accessible peaks (q-value <0.05) between DMSO- and AM580-treated hESC-HSPCs showed that most ATAC-sequencing signal was 50–500 kb from transcriptional start site, likely representing enhancers (Figure 7G). GREAT analysis identified vasculature-related categories as top differentially accessible gene groups (Figure 7H). The average ATAC-sequencing signal around the TSS of AM580-induced genes increased two-fold upon AM580 treatment (Figure 7I), whereas genes repressed upon AM580 treatment did not show difference. These data suggest that RA signalling facilitates chromatin opening in the regulatory regions of the genes it activates.
Discussion
Inability to replicate the microenvironment where HSCs develop has prevented the generation of clinically valuable HSCs from PSCs. However, the molecular barriers underlying the dysfunction of PSC-HSPCs are unknown. Here we document the generation of hESC-HSPCs that possess the immunophenotype of human foetal HSCs and differentiate into adult-type erythroid cells and T-lymphoid cells, but lack self-renewal ability. We pinpointed defective medial HOXA gene activation as a major developmental barrier preventing the establishment of self-renewing HSCs from hESCs, and identified RA signalling during endothelial-to-haematopoietic transition as a key inducer of HOXA genes and HSC fate.
Cells obtained from EBs possessed a surface phenotype of haemogenic endothelium and immature haematopoietic precursors comparable to the early placenta (CD34+CD38−/loCD90+CD43+CD45−GPI-80−)13,14. Upon culture on HSC-supportive stroma, hESC-HSPCs acquired surface expression of CD45 and GPI-8014 and a closer molecular resemblance to FL-HSPCs. However, specific developmentally regulated genes, including the HOXA genes, remained silenced in hESC-HSPCs. Low expression of HOXA genes can also be found in published transcriptome analysis of hESC-derived CD34+ cells2,43–45, implying that HOXA gene silencing is independent of the differentiation protocol or PSC-line used.
HOXA genes are dysregulated in leukemias46,47 and HOXA9 has been previously implicated in mouse foetal HSC self-renewal32,48. Knockout mice for other Hoxa genes have not revealed strong HSC phenotypes, presumably due to compensation by other Hox genes49,50. Deletion of the entire Hoxa cluster in adult mice severely reduced HSC activity and even Hoxa cluster haploinsufficiency compromised HSC function51,52. HOXA9 overexpression enhanced haematopoietic specification from hESCs, but did not confer HSC function44. Our finding that HOXA5 and HOXA7 are critical for in vitro expansion and engraftment of FL-HSPCs, and their overexpression promotes FL-HSPC maintenance, documents a broader requirement for medial HOXA genes in human HSC regulation.
As HOXA7 had strongest loss- and gain-of-function phenotype, it was chosen for molecular analysis. Many genes dysregulated in HOXA7-deficient FL-HSPCs were also dysregulated in EB-OP9-HSPCs. Similar to hESC-HSPCs, HOXA7 deficient FL-HSPCs upregulated embryonic and foetal globins and other erythroid and megakaryocyte differentiation genes, implying they acquire embryonic progenitor-like identity. Although cell cycle was unaffected in HOXA7 deficient FL-HSPCs and hESC-HSPCs, they upregulated cell cycle inhibitors CDKN1A (p21Cip1) and CDKN2D (p19Ink4d). These inhibitors are low in human FL-HSPCs, but become induced after prolonged culture26. They are also highly expressed in FL CD34+CD38+CD90− progenitors that proliferate, but cannot self-renew14. Thus, upregulation of these CDKNs in haematopoietic cells associates with poor proliferative potential and onset of differentiation.
While the overexpression of HOXA7, and to a lesser extent HOXA5, improved FL-HSPC maintenance in culture, their overexpression, even together with HOXA9, was insufficient to rescue hESC-HSPC function. HOXA7 overexpression in hESC-HSPCs did not rescue the genes regulated by HOXA7 in FL-HSPCs. The context-dependent regulation of HOXA target genes may depend on their epigenetic state and/or complementary regulatory factors present only in properly specified HSPCs.
We identified RA-signalling as an upstream regulator for HOXA genes and definitive HSPC transcriptional program. RA production by RALDH2 regulates HSC development during endothelial-to-haematopoietic transition in mouse AGM39. EB-derived haemato-vascular cells showed low RALDH2 expression compared to developmentally matched PL-HSPCs, suggesting that hESC-derived haemato-vascular precursors cannot produce RA. Supplementing RAR-α agonist to EB-derived haemato-endothelial cells for 6 days induced the expression of medial HOXA genes, and increased chromatin accessibility in HOXA cluster and other RA-induced genes. Although many RA-regulated vascular genes are not highly expressed in FL-HSPCs, they promote definitive haemogenic endothelium17 and HSC fate. RA stimulation also induced HSC regulators MECOM, HLF, GFI1, GATA3 and HSC surface markers ROBO4, EMCN and PROCR. Strikingly, RAR-α stimulation partially suppressed the HOXA7-controlled differentiation programs and cell cycle inhibitors in hESC-HSPCs. These results suggest that RAR-α-induced medial HOXA gene activation is critical for HSC fate.
The level of HOXA expression in hESC-HSPCs attenuated after RA stimulation ended, implying that other regulators contribute to maintaining high HOXA expression in HSCs. Moreover, prolonged RA signalling can be detrimental for HSC development and maintenance 53,54 and RA effects vary with experimental conditions and developmental stage54–56. Therefore, the timing and dosage of RA stimulation must be optimised. Although other genetic programs are needed to impart full self-renewal capacity in HSCs, RA signalling and medial HOXA genes emerge as key regulators governing the patterning of haemogenic endothelium to definitive HSC fate.
Methods
Human ESCs culture
The human ESC line H1, obtained from WiCell (WA01), was maintained on 50 Gy irradiated (CF1) murine embryonic fibroblasts feeders, in DMEM/F12 medium (Invitrogen) containing 20% knockout serum replacement (Invitrogen), 0.5% penicillin/streptomycin (50 U/ml, Invitrogen), 1mM L-glutamine, 0.5% MEM non-essential amino acids (NEA, Invitrogen), 10 ng/ml basic FGF (R&D Systems), and 0.11mM BME (Invitrogen). hESCs were passaged weekly using 1mg/ml collagenase IV (Invitrogen) in DMEM/F12 for 10 minutes, detached by gentle pipetting or cell scraper to maintain cells in small clumps, washed twice and split to new 6-well plates containing irradiated CF1 MEFs. Cells were mycoplasma-tested and authenticated by short-term repeat analysis. hESC work was approved by the UCLA Embryonic Stem Cell Research Oversight committee. WA01 is not listed in the ICLAC or NCBI Biosample as commonly misidentified cell line.
Differentiation of embryoid bodies
hESCs were treated with 0.5 mg/ml Dispase (Invitrogen) in DMEM/F12 for 5 minutes at 37°C. Clumps were mechanically detached and transferred to low attachment plates (Corning) in IMDM (Invitrogen), 15% FBS (Hyclone), 1% MEM-NEA, 0.3 μM BME (Invitrogen) and, 1% penicillin/streptomycin (100 U/ml, Invitrogen)/1mM L-glutamine (P/S/G). Medium was changed every 2 days and supplemented with 10 ng/ml BMP-4 (R&D Systems), 50 ng/ml FLT-3L (Peprotech) and 300 ng/ml SCF (Peprotech) at day 4 of EB differentiation. BMP-4 was removed from the culture medium at day 12. EBs were harvested at 2 weeks and dissociated with 0.25% trypsin-EDTA (Invitrogen) with 2% chick serum (Sigma-Aldrich) for 30 minutes at 37°C with periodic agitation and filtered with 70 μm cell strainer. A step-by-step protocol detailing EB generation and AM580/ATRA treatment can be found at Nature Protocol Exchange57.
Human tissue collection
Placenta and fetal liver were de-identified, discarded material obtained from elective termination of first and second trimester pregnancies following informed consent. As these tissues are discarded material with no personal identifiers, this research does not constitute research on Human Subjects. This protocol was reviewed by the UCLA IRB committee, who determined such studies can be performed without further IRB review. Specimen age for this study is denoted as developmental age, two weeks less than gestational age, and was determined by ultrasound or estimated by the date of the last menstrual period. Tissues were harvested into PBS 5% FBS (Hyclone), Ciprofloxacin HCl (10 ng/mL, Sigma), amphotericin B (2.50 μg/mL, Invitrogen), 1% penicillin/streptomycin, transported on ice and processed the same day.
Human tissue processing
Single cell suspensions were prepared from FL at 14–17 weeks of developmental age. Tissues were mechanically dissociated using scalpels and syringes. Mononuclear cells were enriched on a Ficoll layer according to manufacturer’s protocol (GE Healthcare Biosciences AB) and strained through a 70 μm mesh.
Single cell suspensions were prepared from placenta at 3–5 weeks of developmental age. Tissues were mechanically dissociated and digested in 2.5 U dispase (Gibco), 90 mg collagenase (Worthington), and 0.075 mg DNase I (Sigma) per gram of tissue for 90 minutes at 37°C with agitation. Cells were then filtered through 70 μm cell strainer.
Selection of CD34+ cells by magnetic beads
Single cell suspensions obtained from 2 week EB differentiation cultures, human fetal livers or human placentas were magnetically isolated with anti-CD34 microbeads (Miltenyi Biotech) according to manufacturer’s protocol.
OP9-M2 stromal co-culture for HSPC maturation and expansion
OP9-M2 cells26 were irradiated (20 Gy) and pre-plated (50,000 cells/cm2) onto tissue culture-treated wells 24 hours before start of co-culture in OP9-medium (α-MEM (Invitrogen), 20% FBS (Hyclone), P/S/G). Haematopoietic cells derived from hESCs or haematopoietic tissues were plated on stromal layer in OP9-medium supplemented with SCF (25 ng/ml, Peprotech), Flt-3 (25 ng/ml, Peprotech) and TPO (25 ng/ml, Peprotech) (HSC-medium). Cells were co-cultured at 37°C and 5% CO2 and re-plated or analyzed/sorted by flow cytometry every 7–14 days. Half of the HSC-medium was replaced every other day.
Flow cytometry and cell sorting
FACS analysis was performed using single cell suspensions prepared as described. Cells were stained with mouse anti-human monoclonal antibodies against human-CD45-PE cl.J.33 (IM2078U; Beckman coulter) -APC, -APC-H7 cl.2D1 (368512, 368516; Biolegend), -BV711 cl.HI30 (304050; Biolegend), mouse-CD45-APC-H7 cl. 30-F11 (557659; BD) (all diluted 1:100), CD34-APC cl. 581 (555824; BD, 1:20), CD90-FITC cl. 5E10 (555595; BD, 1:100), CD38-PE-Cy7 cl. HIT2 (560677; BD, 1:100), CD19-PE cl. 1D3 or HIB19 (12-0193, 12-0199; eBiosciences, 1:50), CD43-PE cl. MT1 (51727; SCB, used at 1:25) or cl.1G10 -FITC (BD 1:20), GPI-80-PE cl. 3H9 (D087-5; MBL International Corporation, used at 1:50), CD3-PE-Cy7 cl. SK7 (557851, BD) (eBiosciences, 1:50), CD4 APC (MHCD0405; Invitrogen, 1:50), CD8-PE cl. HIT8A (555635; BD, 1:50), CD13-APC cl. WM15 (557454; BD, 1:500), and CD66b FITC cl. G10F5 (555724; BD, 1:50) CD33-PE cl. WM53 (561816; BD, 1:100).
Dead cells were excluded with 7-amino-actinomycin D (7AAD) (BD Biosciences, used at 1:50). Cells were assayed on a BD-LSRII flow cytometer and data were analyzed with FlowJo software (Tree Star Inc.). Cell sorting was performed using a BD FACS Aria II.
Methylcellulose colony-forming assays
Myelo-erythroid progenitor potential was assessed on methylcellulose (MethoCult GF+ H4435, SCT) supplemented with TPO (10 ng/mL, Peprotech), 1% penicillin/streptomycin (100 U/ml, Invitrogen) and 1% amphotericin B (2.50 μg/mL, Invitrogen). Cultures were incubated at 37°C and 5% CO2 for 14–16 days and colonies scored based on morphological characteristics. Images were taken using an Olympus BX51 microscope with DP72 camera.
Cytospins
Representative myelo-erythroid colonies from methylcellulose assays were picked and resuspended in PBS with 40% FBS (Hyclone). The cell suspension was cytospun on slides using a Shandon Cytospin 4 (Thermo Electron Corporation) spun at 300 rpm under medium acceleration. Slides were air-dried overnight and stained using May-Grunwald-Giemsa stain (Sigma Aldrich).
Imaging
Bright field images of individual colonies and images of MGG stained colonies were taken using the Zeiss Axovert 40 CFL microscope under the 10X objective with attached Canon PC 1089 camera at 4X zoom for a total magnification of 40X.
T lymphoid differentiation
hESC- or FL-derived CD34+ cells were plated on non-irradiated OP9-DL1 stroma (25,000 cells/cm2) in OP9-medium supplemented with SCF (25 ng/ml, Peprotech), FLT3-L (10 ng/ml, Peprotech) and IL-7 (20 ng/ml, Peprotech). Cells were lifted and reseeded on OP9-DL1 every week until FACS analysis.
Cell cycle analysis
Cultured cells were pulse-labeled with 10μM BrdU for 35 min in culture. Cells were sorted for the indicated surface phenotypes and processed according to the FITC-BrdU flow kit (BD) instructions.
RNA isolation, cDNA synthesis and quantitative reverse transcriptase PCR
RNA isolation was performed using the RNeasy Mini kit (Qiagen) with additional DNase (Qiagen) step using manufacturer’s protocol. cDNAs were prepared using Quantitech reverse transcription kit (Qiagen), and quantitative polymerase chain reaction (qPCR) for GAPDH, glycophorin A (GYPA), haemoglobin subunit epsilon (HBE), haemoglobin subunit gamma (HBG), and beta globin (HBB) was performed with the LightCycler 480 SYBR Green I Master Mix (Roche) on the Lightcycler 480 (Roche); qPCR for GAPDH and HOXA7 with the SYBR Select Master Mix (Life Technology, LT) using the ViiA™ 7 Real-Time PCR System (LT); and qPCR for GAPDH, HOXA3, HOXA5, HOXA6, and HOXA9 was performed with the TaqMan%reg; Gene Expression Master Mix (LT) and analyzed using the ViiA™ 7 Real-Time PCR System (LT). SYBR Green-compatible primers were obtained from the PrimerBank database or literature58, 59. Taqman%reg; primers were purchased from LT. Primers were tested against OP9-M2 cDNA to rule out amplification of murine genes, and gDNA or water as negative controls. Primers are presented in Supplementary Table 6.
Microarray analysis
RNA isolation was performed using RNEasy Mini kit (>50,000 cells) or RNeasy Micro kit (<50,000 cells) (Qiagen) with DNase digestion using manufacturer’s protocol. RNA was amplified by the NuGen amplification kit and hybridized on Affymetrix arrays (Human U133plus2.0 Array). Samples were quantile normalized.
K-means clustering of differentially expressed genes
Pairwise differential expression analysis was performed for the various populations by independently comparing immunophenotypic HSPCs derived from hESCs (EB and EB-OP9), placenta (PL), cultured fetal liver (FL-OP9 2 weeks and FL-OP9 5 weeks), to freshly isolated FL-HSPC (FL). The following criteria were used to filter probes and identify differentially expressed genes: (i) fold change value of 2 or higher, (ii) a p-value less than 0.05, (iii) probes that are called “Absent” in all replicates in all samples were excluded and (iv) probes that have an absolute expression level of 50 or less were excluded. The union of all probes that met these criteria was chosen for further analysis, after which normalized expression values, which were replicate-averaged and standardized, were obtained. The standardized expression values were clustered using K-means method. GO enrichment analysis was performed for genes in the various clusters. Differential expression assessment was performed using Limma R package60 through Bioconductor61. To account for multiple comparisons, p-values were adjusted using the Benjamini-Hochberg correction method to control the False Discovery Rate (FDR). A 5% FDR was used as cutoff. K-means clustering of RMA normalized, replicate-averaged and standardized expression values was done using Cluster 3.0 software62 and the resulting clusters were viewed using Java Treeview software63.
RNA-sequencing analysis
Total RNA from 2000 to 20000 sorted cells was extracted using the RNeasy Mini kit (Qiagen) and library was constructed using Ovation Rna-seq system System v2 (Nugen), followed by KAPA LTP Library Preparation Kit. Libraries were sequenced using HIseq-2000 (Illumina) to obtain single end 50 bp long reads. Demultiplexing of the reads based on the barcoding was performed using in house Unix shell script. Splice junction mapping to the human genome (hg19) was performed using TopHat v2.0.9 or v2.0.1464 with the parameters --no-coverage-search -M -T -x 1. Coverage files were created with the Genomecov tool from Bedtools65 with the parameters -bg -split –ibam. For abundance estimations (FPKMs) the aligned read files were further processed with Cufflinks v2.2.166 --compatible-hits-norm -N -u --GTF gencode.v19.annotation.gtf. Assemblies for all samples were merged using Cuffmerge and differential expression was determined using Cuffdiff.
Production of lentiviral shRNA and overexpression vectors
shRNA experiments were performed with pLKO lentiviral vectors from the TRC library (TRCN000017529 for HOXA5, TRCN000015084 for HOXA7) containing puromycin resistance gene. Human HOXA5, HOXA7 and HOXA9 were cloned from human FL full-length cDNA, into either the constitutive pFUGW lentiviral vector, (Addgene plasmid # 14883, from David Baltimore), downstream and in frame with the GFP sequence with the synthetic addition a P2A sequence between the 2 ORFs, or the inducible lentiviral overexpression system pNL-EGFP/TREPittdU3, (Addgene plasmid # 18659, from Jakob Reiser), between the sites BamHI and NheI. pNL-TREpitt vectors were co-transduced with the constitutive pNL-EF1α-rTTA-M2 lentiviral vector to provide in trans the Tet transactivator. For lentiviral vectors production, 20 milions 293T cells were transfected with 12.5 ug deltaR8.2, 5 ug VSV-G, and 12.5 ug DNA/shRNA and 90 uL of lipofectamine-2000 (Invitrogen) in OPTI-MEM and incubated for 5–6 hours at 37°C. After incubation for 48 hours in complete medium, supernatant was filtered and concentrated using an ultracentrifuge (Beckman Coulter) at 20,200 rpm for 1.5 hours at 4°C. Following centrifugation, pelleted viruses were resuspended in 125 uL of SFEM and stored at −80°C.
Lentiviral transduction of CD34+ cells
FL-HSPCs cells were prestimulated 6–18 h in fresh SFEM culture medium supplemented with 100 ng/mL SCF, 100 ng/mL TPO and 100 ng/mL FLT3-L. Wells were treated with 40 ug/mL RetroNectin (Takara) and seeded with pre-stimulated FL-HSPC in 300 uL SFEM culture medium. Lentivirus (5 uL) was added twice during 24 hours incubation. Transduced cells were washed and seeded on OP9-M2 with HSC-medium. For shRNA-lentiviral vectors, puromycin (1.0 μg/ml) treatment was used for selection of transduced HSPCs and maintained throughout culture. EB-derived CD34+ cells were infected in SFEM for 2 hours on a rocking platform at 37°C and then moved to OP9-M2 co-culture on HSC medium. For overexpression experiments with PNL vectors, doxycycline was added immediately at the start of OP9-M2 culture and maintained at 1 μg/ml.
Transplantation into NOD-scid IL2Rγ-null mice
Female NOD-scid IL2Rγ-null (NSG, Jackson Laboratories) mice, 8–12 weeks old, were sublethally irradiated (325 rads) and intra-tibially injected with FL or hESC CD34+ cells in a volume of 35 μL. Mice were transplanted with either 1.2E6 EB-derived CD34+ enriched cells (60% CD34+) or with 10,000 FL CD34+ cells in 50 μL PBS. A second batch of mice was transplanted with 2.5E6 EB CD34+ derived cells (25% CD34+) or 3E6 FL CD34+ derived cells (35% CD34+) obtained after two weeks culture on OP9-M2,
For shRNA treated FL cells, 50,000 cells were infected with shRNA lentivirus and expanded on OP9-M2 for 9 days under puromycin selection. Transduced hematopoietic cells were retro-orbitally injected in sublethally irradiated NSG mice.
For engraftment of EB-derived cells overexpressing HOXA genes, 300,000 cells were infected with the indicated lentivirus. After infection cells were seeded on OP9-M2, expanded for 2 weeks, collected and injected in the right tibia of sublethally irradiated (275 rads) female NSG mice. FL CD34+ cells were injected as a positive control of engraftment.
Mice were sacrificed at 10–12 weeks to harvest bone marrow. Collected cells were FACS-analyzed to evaluate human engraftment (human CD45 and mouse CD45), differentiation into myelo-lymphoid lineages (CD13, CD33, CD66, CD3 or CD19) and preservation of the HSPC compartment (CD34+, CD38−, CD90+).
All studies and procedures involving mice were approved by the UCLA Animal Research Committee (Protocol 2005-109).
Induction of retinoic acid signaling
ATRA (All-trans retinoic acid, Sigma-Aldrich) was dissolved in DMSO at 25 mM and applied at final concentration of 1 μM; AM580 (Sigma-Aldrich) was dissolved in DMSO at 10 mM at a final concentration of 0.2 μM; DMSO was used at 1:25,000 final dilution. Treatments were performed on FL- or hESC-derived CD34+ cells in HSC-medium.. Cells were collected at day 6 for q-RT-PCR and RNA-sequencing analysis. For longer cultures, RA stimulation media were removed at day 6 from the wells avoiding disruption of the cell layer and replaced with HSC-medium with no treatment. Cells were then collected at day 12 reseeded on OP9M2 for another 12–13 days, and then assayed for HSPC expansion by FACS or colony-forming potential in methylcellulose. A step-by-step protocol detailing EB generation and AM580/ATRA treatment can be found at Nature Protocol Exchange57.
ATAC-sequencing
Cells (12,000 to 60,000) were sorted in PBS and processed according to the protocol indicated67, with minor adjustments. Nuclei were purified by the addition of 250μl of lysis buffer (10mM Tris-HCl, pH7.4, 10mM NaCl, 3mM MgCl2, 0.1%IGEPAL), pelleted and resuspended in the transposition reaction mix (Nextera DNA Library Prep Kit, Illumina) and incubated at 37°C for 30 minutes. Transposed DNA was column purified and used for library amplification with custom made adaptor primers (see primer table) using NEBNext High-Fidelity 2x PCR Master Mix (NEB). The amplification was interrupted after 5 cycles and a SyBR green qPCR was performed with 1/10 of the sample to estimate for each sample the additional number of cycles to perform, before saturation was achieved. Total amplification was between 10 and 15 cycles. Purified Libraries were sequenced using HIseq-2000 (Illumina) to obtain paired end 50 bp long reads. Demultiplexing of the reads and creation of the fastq files was performed using in house Unix shell script. Read mapping to the genome (hg19) was performed using Bowtie2 or v2.2.568 with parameters --local -X 2000 -N 1 --no-mixed. Bamcoverage tool from Deeptools was used to create the coverage .bw files for visualization69. Samtools v1.2 was used to remove duplicates and reads aligned to chrM. MACS270 was used to call the differentially accessible peaks between the AM580 treated and untreated EB-derived cells, using the parameters --broad --broad-cutoff 0.1.
Statistics and Reproducibility
Graphs were generated with GraphPad PRISM software. Statistical significance was calculated in R version.3.2.3. Statistical significance was assessed using the Wilcoxon rank sum test for unpaired data sets and the Student’s paired t-test for paired data sets. All tests are 2-tailed with the exception of Fig 6C, where a one-tailed distribution was used since it is an established fact that RA treatment enhances HOX gene expression. Data sets were considered paired when treatments (i.e., DMSO vs. AM580 or LKO vs. shRNA) were performed in parallel on the same batch of cells (EB) or from the same donor (FL) for the same time periods. Data were considered unpaired when cells from different batches, donors, or cell types were compared. The null hypothesis of the medians/means being equal was rejected at α = 0.05 and exact significant p-values are shown in each graph. The comparisons to be made were decided a priori with the intention of limiting the overall number of comparisons and are indicated by lines in the figures and adjustments for multiple comparisons would be applied to k > 4 comparisons. To account for multiple comparisons in microarray analysis, p-values were adjusted using the Benjamini-Hochberg correction method to control the False Discovery Rate (FDR). The non-parametric Wilcoxon Rank Sum test is robust to outliers. We tested for differences in sample dispersion using Ansari-Bradley and we could not reject the null hypothesis of equal dispersions. A Satterthwaite’s approximation was used to account for unequal variances when using the t-test. The investigators were not blinded to allocation during experiments and outcome assessment. For mouse engraftment studies, the experiments were not randomized, no statistical method was used to predetermine sample size and no animal was excluded from analysis.
Supplementary Material
Acknowledgments
We thank BSCRC FACS Core at UCLA; the UCLA Clinical Pathology Microarray Core; BSCRC Sequencing Core, UCLA Tissue and Pathology Core and CFAR Gene and Cell Therapy Core (NIH grant AI028697-21) and Novogenix LLC. We thank Hilary Coller for discussions, Tim Bolan for assistance with experiments, Yi Xing and Yu-Ting Tseng for consultation on RNA-sequencing analysis, and Tanya Stoyanova, Donny Johnson and Owen Witte for help with NSG mice. This work was supported by CIRM RN1-00557, NIH RO1 DK100959 and LLS Scholar award for H.K.A.M; Broad Stem Cell Center at UCLA and JCC Foundation; NIH P01 GM081621 for J.Z. and Z.G, and NIH PO1 HL073104 and CIRM RB3-05217 for G.M.C. D.R.D. was supported by the NSF GRFP and Ruth L. Kirschstein National Research Service Award GM007185. V.C. by LLS Special Fellow Award and BSCRC post-doctoral fellow award, M.I.S. by Ruth L. Kirschstein National Research Service Award HL086345, A.T.N by Beckman Scholarship, and P.S. by the Eugene V. Cota-Robles fellowship.
Footnotes
Authorship:
D.R.D., V.C., M.I.S., P.S., Z.G, J.Z, G.C and H.K.A.M. designed experiments and interpreted data. D.R.D., V.C., M.I.S., A.T.N, A.M and P.S., performed experiments. R.S. performed bioinformatics analysis of the microarray data and C.R assisted with statistical analysis, D.R.D., V.C. and H.K.A.M. wrote the manuscript, which all authors edited and approved.
The authors declare no competing financial interests.
References
- 1.Bordignon C. Stem-cell therapies for blood diseases. Nature. 2006;441:1100–1102. doi: 10.1038/nature04962. [DOI] [PubMed] [Google Scholar]
- 2.Doulatov S, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira CF, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riddell J, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen AL, Stachura DL, Weiss MJ. Designer blood: creating hematopoietic lineages from embryonic stem cells. Blood. 2006;107:1265–1275. doi: 10.1182/blood-2005-09-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slukvin II. Hematopoietic specification from human pluripotent stem cells: current advances and challenges toward de novo generation of hematopoietic stem cells. Blood. 2013;122:4035–4046. doi: 10.1182/blood-2013-07-474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrath KE, Palis J. Hematopoiesis in the yolk sac: more than meets the eye. Exp Hematol. 2005;33:1021–1028. doi: 10.1016/j.exphem.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Kyba M, Daley GQ. Hematopoiesis from embryonic stem cells: lessons from and for ontogeny. Exp Hematol. 2003;31:994–1006. doi: 10.1016/j.exphem.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 10.Swiers G, Rode C, Azzoni E, de Bruijn MF. A short history of hemogenic endothelium. Blood Cells Mol Dis. 2013;51:206–212. doi: 10.1016/j.bcmd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 12.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Handel B, et al. The first trimester human placenta is a site for terminal maturation of primitive erythroid cells. Blood. 2010;116:3321–3330. doi: 10.1182/blood-2010-04-279489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prashad SL, et al. GPI-80 Defines Self-Renewal Ability in Hematopoietic Stem Cells during Human Development. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 16.Pick M, Azzola L, Mossman A, Stanley EG, Elefanty AG. Differentiation of human embryonic stem cells in serum-free medium reveals distinct roles for bone morphogenetic protein 4, vascular endothelial growth factor, stem cell factor, and fibroblast growth factor 2 in hematopoiesis. Stem Cells. 2007;25:2206–2214. doi: 10.1634/stemcells.2006-0713. [DOI] [PubMed] [Google Scholar]
- 17.Ditadi A, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, et al. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Dravid G, Zhu Y, Scholes J, Evseenko D, Crooks GM. Dysregulated gene expression during hematopoietic differentiation from human embryonic stem cells. Mol Ther. 2011;19:768–781. doi: 10.1038/mt.2010.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shojaei F, Menendez P. Molecular profiling of candidate human hematopoietic stem cells derived from human embryonic stem cells. Exp Hematol. 2008;36:1436–1448. doi: 10.1016/j.exphem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Martin CH, Woll PS, Ni Z, Zuniga-Pflucker JC, Kaufman DS. Differences in lymphocyte developmental potential between human embryonic stem cell and umbilical cord blood-derived hematopoietic progenitor cells. Blood. 2008;112:2730–2737. doi: 10.1182/blood-2008-01-133801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu C, et al. Differentiation of human embryonic stem cells into hematopoietic cells by coculture with human fetal liver cells recapitulates the globin switch that occurs early in development. Exp Hematol. 2005;33:1450–1458. doi: 10.1016/j.exphem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Tian X, Kaufman DS. Differentiation of embryonic stem cells towards hematopoietic cells: progress and pitfalls. Curr Opin Hematol. 2008;15:312–318. doi: 10.1097/MOH.0b013e328302f429. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Cerdan C, Menendez P, Bhatia M. Derivation and characterization of hematopoietic cells from human embryonic stem cells. Methods Mol Biol. 2006;331:179–200. doi: 10.1385/1-59745-046-4:179. [DOI] [PubMed] [Google Scholar]
- 26.Magnusson M, et al. Expansion on stromal cells preserves the undifferentiated state of human hematopoietic stem cells despite compromised reconstitution ability. PLoS One. 2013;8:e53912. doi: 10.1371/journal.pone.0053912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy M, et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Shaut CA, Keene DR, Sorensen LK, Li DY, Stadler HS. HOXA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet. 2008;4:e1000073. doi: 10.1371/journal.pgen.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilo F, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117:5057–5066. doi: 10.1182/blood-2010-08-300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimmeck D, et al. Transcriptome-wide profiling and posttranscriptional analysis of hematopoietic stem/progenitor cell differentiation toward myeloid commitment. Stem Cell Reports. 2014;3:858–875. doi: 10.1016/j.stemcr.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsteinsdottir U, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence HJ, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilpert M, et al. p19 INK4d controls hematopoietic stem cells in a cell-autonomous manner during genotoxic stress and through the microenvironment during aging. Stem Cell Reports. 2014;3:1085–1102. doi: 10.1016/j.stemcr.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluta K, Luce MJ, Bao L, Agha-Mohammadi S, Reiser J. Tight control of transgene expression by lentivirus vectors containing second-generation tetracycline-responsive promoters. J Gene Med. 2005;7:803–817. doi: 10.1002/jgm.712. [DOI] [PubMed] [Google Scholar]
- 36.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 37.Marshall H, Morrison A, Studer M, Pöpperl H, Krumlauf R. Retinoids and Hox genes. FASEB J. 1996;10:969–978. [PubMed] [Google Scholar]
- 38.Gavalas A, Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev. 2000;10:380–386. doi: 10.1016/s0959-437x(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 39.Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155:215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 40.Delescluse C, et al. Selective high affinity retinoic acid receptor alpha or beta-gamma ligands. Mol Pharmacol. 1991;40:556–562. [PubMed] [Google Scholar]
- 41.Kishimoto H, et al. Molecular mechanism of human CD38 gene expression by retinoic acid. Identification of retinoic acid response element in the first intron. J Biol Chem. 1998;273:15429–15434. doi: 10.1074/jbc.273.25.15429. [DOI] [PubMed] [Google Scholar]
- 42.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 43.Salvagiotto G, et al. Molecular profiling reveals similarities and differences between primitive subsets of hematopoietic cells generated in vitro from human embryonic stem cells and in vivo during embryogenesis. Exp Hematol. 2008;36:1377–1389. doi: 10.1016/j.exphem.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos-Mejía V, et al. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124:3065–3075. doi: 10.1182/blood-2014-03-558825. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, et al. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. J Exp Med. 2005;201:1603–1614. doi: 10.1084/jem.20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beachy SH, et al. Isolated Hoxa9 overexpression predisposes to the development of lymphoid but not myeloid leukemia. Exp Hematol. 2013;41:518–529.e515. doi: 10.1016/j.exphem.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 48.McKinney-Freeman S, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen F, Greer J, Capecchi MR. Analysis of Hoxa7/Hoxb7 mutants suggests periodicity in the generation of the different sets of vertebrae. Mech Dev. 1998;77:49–57. doi: 10.1016/s0925-4773(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 50.Boucherat O, et al. Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2013;304:L817–830. doi: 10.1152/ajplung.00006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebert-Ghali CE, et al. HoxA cluster is haploinsufficient for activity of hematopoietic stem and progenitor cells. Exp Hematol. 2010;38:1074–1086. e1071–1075. doi: 10.1016/j.exphem.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Lebert-Ghali C, et al. Hoxa cluster genes determine the proliferative activity of adult mouse hematopoietic stem and progenitor cells. Blood. 2016;127:87–90. doi: 10.1182/blood-2015-02-626390. [DOI] [PubMed] [Google Scholar]
- 53.Muramoto GG, et al. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28:523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szatmari I, Iacovino M, Kyba M. The retinoid signaling pathway inhibits hematopoiesis and uncouples from the Hox genes during hematopoietic development. Stem Cells. 2010;28:1518–1529. doi: 10.1002/stem.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rönn RE, et al. Retinoic acid regulates hematopoietic development from human pluripotent stem cells. Stem Cell Reports. 2015;4:269–281. doi: 10.1016/j.stemcr.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cano E, Ariza L, Muñoz-Chápuli R, Carmona R. Signaling by Retinoic Acid in Embryonic and Adult Hematopoiesis. Journal of Developmental Biology. 2014;2:18. [Google Scholar]
- 57.Dou DR, Calvanese V, Saarikoski P, Galic Z, Mikkola HKA. Induction of HOXA genes in hESC-derived HSPC by two-step differentiation and RA signalling pulse. Nature Protocol Exchange. 2016 [Google Scholar]
- 58.Thoma SJ, Lamping CP, Ziegler BL. Phenotype analysis of hematopoietic CD34+ cell populations derived from human umbilical cord blood using flow cytometry and cDNA-polymerase chain reaction. Blood. 1994;83:2103–2114. [PubMed] [Google Scholar]
- 59.Bauchwitz R, Costantini F. Developmentally distinct effects on human epsilon-, gamma- and delta-globin levels caused by the absence or altered position of the human beta-globin gene in YAC transgenic mice. Hum Mol Genet. 2000;9:561–574. doi: 10.1093/hmg/9.4.561. [DOI] [PubMed] [Google Scholar]
- 60.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 63.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 64.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.