Abstract
Very long chain fatty acids are required for sphingolipid synthesis, lipid homeostasis, myelin formation, epidermal permeability, and retinal function. Seven different enzymes are known to be involved in the elongation cycle of fatty acids, with different chain-length specificities. Elovl1 is one of those enzymes whose function has been linked mainly to the synthesis of sphingolipids and the epidermal barrier. However, the role of Elovl1 in organogenesis is not clear. In zebrafish, 2 Elovl1 genes, elovl1a and elovl1b, are highly expressed in the swim bladder, and elovl1b is also expressed in the kidney. We found that both elovl1 knockdown embryos contain increased levels of long chain fatty acids from carbon number 14 to 20 as compared to control embryos. Oil-Red-O staining shows that yolk lipid consumption is greatly reduced, whereas lipid droplets accumulate within the swim bladder. Notably, knockdown of either elovl1a or elovl1b affects the expression of genes involved in swim bladder development and impairs inflation of the swim bladder. Consistent with its expression in the pronephros, knockdown of elovl1b alone affects the expression of genes required for kidney development and reduces renal clearance. Our findings strongly suggest that both elovl1 genes are a key determinant of swim bladder and kidney development in zebrafish, which may be comparatively applicable to lung and kidney development in humans.
KEYWORDS: fatty acid chain elongase, kidney development, swim bladder development, zebrafish
INTRODUCTION
Very long chain fatty acids (VLCFAs) are associated with several inherited disorders. They are also required in the development of the epidermal permeability barrier,1 retinal physiology and function,2-4 lipid homeostasis,5 myelin formation,6,7 spermatogenesis, and male fertility.8 They are predominantly synthesized in the endoplasmic reticulum (ER) by the action of various enzymes called elongases with different chain-length specificities.9,10 To date, 7 different elongases (Elovl) that catalyze the first step in the elongation cycle of fatty acids in the ER have been reported.10,11 It has been suggested that among them, Elovl 1, 3, and 6 are involved in the elongation of saturated and monounsaturated VLCFAs, whereas Elovl 2, 4, and 5 are involved in the elongation cycle of polyunsaturated fatty acids.11
Previous research has shown that production of C20 and C22 Coenzyme As (CoAs) by Elovl1 is essential for C24 sphingolipid synthesis. It has also been reported that Elovl1 activity is regulated by a ceramide synthase, CERS2, which is essential for synthesis of C24 sphingolipids.12 Elovl1 homozygous knockout mice have been observed to die shortly after birth, possibly because of an impaired epidermal permeability barrier.13 In these mice, the number of ceramides with fatty acids longer than C26 was reduced, whereas the number of those shorter than C24 increased. In contrast, the level of C24 sphingomyelin was reduced but was accompanied by an increase in C20 sphingomyelin levels. In humans, ELOVL1 knockdown lowers C26 fatty acid levels in X-linked adrenoleukodystrophy (X-ALD) fibroblasts by reducing the elongation of C22 fatty acids to C26, which highlights the therapeutic potential of ELOVL1 modulation for the treatment of X-ALD.14
Elovl1 is highly expressed in the stomach, lung, kidney, skin, and intestine of the mouse.15 Although its function has been linked largely to sphingolipid synthesis and the epidermal permeability barrier, the precise role of Elovl1 in vertebrate development or in different organs has not been studied extensively because of the premature death of elovl1 knockout mice during the early postnatal period.13 In the present study, we examined the role of elovl1 during zebrafish embryogenesis. Zebrafish possess 2 elovl1 genes, elovl1a and elovl1b; sequence analysis indicates that both elovl1 genes are evolutionarily conserved among vertebrates. Expression analysis showed that both genes are expressed in the swim bladder from 2 days post fertilization (dpf), and elovl1b is expressed in the kidney as well. We found that knockdown of 2 elovl1 genes affects yolk consumption, lipid metabolism, and swim bladder development. In addition, elovl1b knockdown affects kidney development. Developmental defects in both swim bladder and kidney are largely due to impaired expression of the genes that are necessary for proper formation of these organs. Notably, defective swim bladder development induced by elovl1 knockdown can be rescued by co-expression of elovl1 mRNA, confirming that the phenotype observed is specific to elovl1 knockdown. These results suggest that the function of Elovl1 is conserved among vertebrates, and to our knowledge, they represent the first report regarding the developmental role of elovl1 in zebrafish. Furthermore, our results also suggest that zebrafish can be used to characterize the roles of genes related to lipid metabolism during vertebrate development.
RESULTS
The zebrafish genome contains duplicated elovl1, the protein sequences of which are highly homologous to those of mammals
We found 2 zebrafish elovl genes, elovl1a and elovl1b, from the Ensembl genome database (www.ensembl.org). Sequence alignment suggests that Elovl1 is conserved in vertebrates, including humans, mice, and zebrafish. In particular, zebrafish Elovl1a and Elovl1b possess almost identical fatty acid desaturase motifs necessary for fatty acid elongation and thus conserved in Elovl family proteins in mammals (Fig. 1). While general sequence homology is around 62.5% between human Elovl1 and zebrafish Elovl1a and 67.9% between human Elovl1 and zebrafish Elovl1b, C-terminal regions of both Elovl1a and Elovl1b in zebrafish are composed of extra amino acids and significantly different from each other. We performed whole mount in situ hybridization to characterize the expression pattern of 2 zebrafish elovl1 genes during embryogenesis. Expression of both elovl1a and elovl1b was detected as early as 2 hpf, indicating that both genes are maternally derived (Fig. 2). Zygotic expression of elovl1a was observed at 6 hpf, while elovl1b was not expressed at this stage. At 12 hpf, expression of both elovl1a and elovl1b was ubiquitously observed, but persisted only in the head region at 1 dpf, after which expression of elovl1a was largely confined to the swim bladder region. In addition to the swim bladder, elovl1b was expressed in the pronephros, the first immature kidney. Weak expression of elovl1a and elovl1b was also detected in the pharyngeal area (Fig. 2). Consistent with these results, a previous study showed that elovl1a is expressed in the dorsal part of the swim bladder primordium.20 Based on the expression patterns, it is likely that elovl1b may have evolved to play a unique role in kidney development of zebrafish, whereas both elovl1a and elovl1b may act together in the development of tissues in which they are expressed.
FIGURE 1.
Elovl1 is highly conserved in zebrafish, mice, and humans. Protein sequence alignment of zebrafish, mice, and human elovl1. Zebrafish contains 2 elovl1 genes, elovl1a and elovl1b. Conserved amino acids are boxed in black. Similar amino acids are boxed in gray, and 5 conserved motifs in Elovl family members are underlined. Abbreviations: ZF, zebrafish; Hum, human; Mou, mouse.
FIGURE 2.
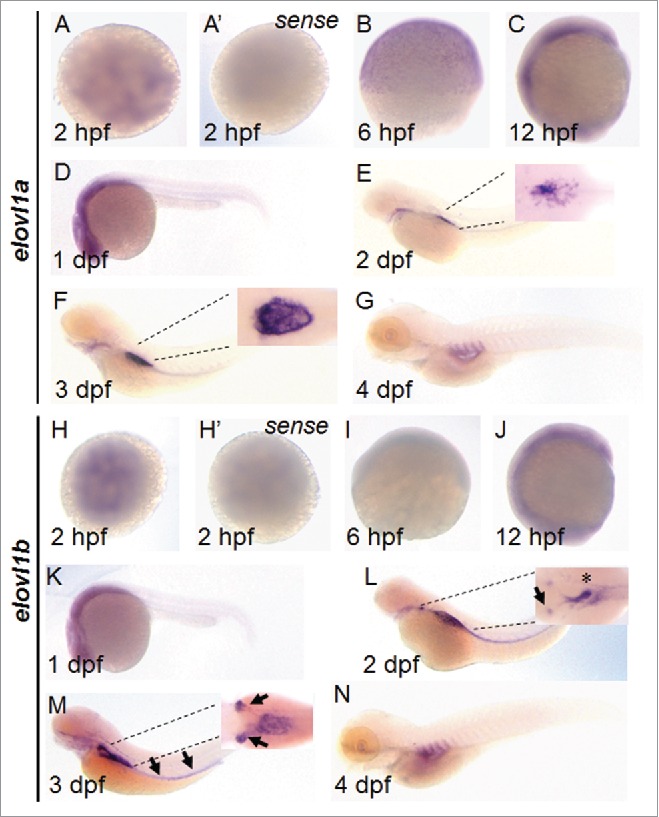
Expression pattern of elovl1a and elovl1b during early development. Expression of elovl1a and elovl1b during embryonic development was analyzed by in situ hybridization. Embryos are presented in animal views (A, A’, H, H’), dorsal views (insets in E, F, L, M) or lateral views (B, C, D, E, F, G, I, J, K, L, M, N). Sense probes for elovl1a (A’) and elovl1b (H’) were used to prove the specificity of antisense probes. Insets in E, F, N, and P are magnified images shown in dorsal views.
Elovl1 knockdown impairs swim bladder inflation
To examine the loss of elovl1a and elovl1b function, we designed morpholinos (MOs) to either interfere with protein synthesis (translation-blocking MO; trMO) or impair mRNA maturation (splice-blocking MO; spMO) of elovl1a or elovl1b. We injected the morpholino into the one cell stage of zebrafish embryos and examined the morphological phenotypes associated with knockdown of elovl1a or elovl1b. As shown in Fig. 3, embryos injected with either MOelovl1a or MOelovl1b display slightly curved body axes and deflated swim bladders. Since both elovl1 genes are maternally expressed, we expected that there might be a differential effect between trMOs and spMOs. We counted the number of embryos that display deflated swim bladders after injecting with a trMO or a spMO. We found that the percentages of embryos that display defective formation of the swim bladder in response to trMOs (43% for elovl1a and 27% for elovl1b: shown in Fig. 3F) are almost similar to those induced by spMOs (26% for elovl1a and 28% for elovl1b). Moreover, embryos injected with a trMO or a spMO are phenotypically indistinguishable. To determine whether the phenotype observed upon elovl1 knockdown is specific to the loss of elovl1, we cloned the open reading frame of zebrafish elovl1a and elovl1b and microinjected elovl1a or elovl1b mRNA together with the corresponding gene trMO. We reasoned that mRNA that contains only ORF should not be affected by a trMO, since trMOs are designed to contain a significant portion of 5′UTR (8/25 nucleotides for MOelovl1a and 6/25 for MOelovl1b). We found that overexpression of elovl1a mRNA efficiently rescues the swim bladder defect induced by elovl1a knockdown (16%, n = 184 for rescue in comparison with 43% for trMOelovl1a alone), and a similar result was obtained for elovl1b (10%, n = 201 for rescue in comparison with 27% for trMOelovl1b alone). In addition, we found that spMOs induce abnormal transcripts that are longer than those found in control embryos (Fig. S1), confirming that phenotypes induced by spMOs are also caused by knockdown of the corresponding gene. Interestingly, we found that the defective swim bladder phenotype that results from one evolvl1 gene knockdown could not be rescued by overexpression of the other elovl1 mRNA. Furthermore, a half dose of a single gene knockdown does not induce any particular phenotype, whereas a half dose of combined knockdown of elovl1 (MOelovl1a+MOelovl1b) induces the defective swim bladder phenotype as efficiently as a full dose of a single elovl1 knockdown. These results suggest that both elovl1a and elovl1b are required, but may not act redundantly, during vertebrate development.
FIGURE 3.
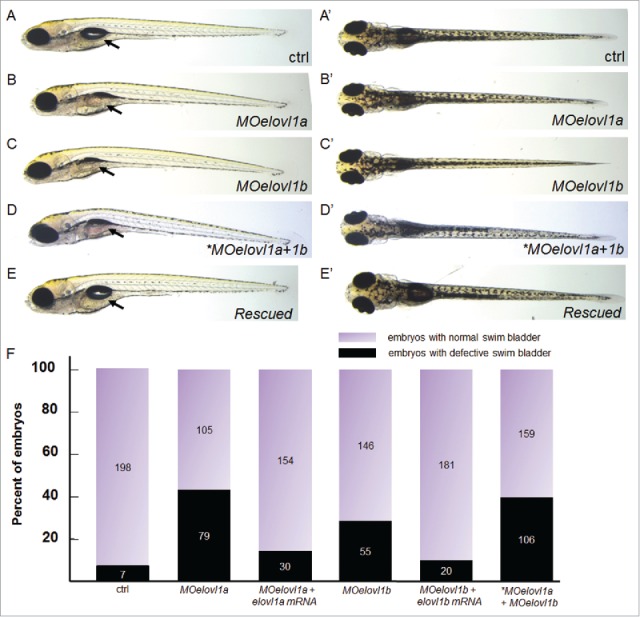
Elovl1 knockdown impairs swim bladder inflation. (A-E) 10 ng MOelovl1a or MOelovl1b alone, a half dose (5 ng each) of both MOs (*MOelovl1a+1b), or single MO + corresponding mRNA (50 pg elovl1a or 250 pg elovl1b) were injected at the one-cell stage of zebrafish embryos. Injection of either MOelovl1a or MOelovl1b, and a half dose of both MOs resulted in a deflated swim bladder, whereas co-injection of either MOelovl1a + elovl1a mRNA or MOelovl1b + elovl1b mRNA rescued the defective swim bladder phenotype. Embryos displaying the swim bladder inflation defect at 5 dpf are shown in lateral (B-D) or dorsal (B′-D′) views with the anterior aspect to the left. (F) The percentages of affected embryos (43% (n = 184) for MOelovl1a, 16% (n = 184) for MOelovl1a + elovl1a mRNA, 27% (n = 201) for MOelovl1b, 10% (n = 201) for MOelovl1b + elovl1b mRNA, and 40% (n = 265) for a half dose of combined *MOelovl1a+1b) with the number of embryos in each category are shown in a graph. The total number of embryos shown inside each bar was counted from at least 3 independent experiments for each condition. Arrow points the region where the swim bladder develop.
Elovl1 knockdown alters whole embryo lipid profiles
Previous studies have reported that elongases are required in the chain elongation process of saturated, monounsaturated, and polyunsaturated long chain fatty acids.11 We compared the levels of total fatty acids in control embryos at 5 dpf to those in embryos injected with MOelovl1a+MOelovl1b. We found that the levels of fatty acids composed of carbon numbers (C) 14–20 seem to be accumulated in MO-injected embryos as compared to those in control embryos (Fig. 4). Due to technical challenges associated with the very small size of zebrafish embryos and the paucity of very long chain fatty acids comprising their total cellular lipids, we were unable to detect any population of very long chain fatty acids (C > 22) under our assay conditions. Nevertheless, the increased levels of long chain fatty acids seem to be consistent with previous reports in which elovl1 was shown to act in chain-elongation for the production of very long chain fatty acids.12-15
FIGURE 4.

Elovl1 knockdown affects long chain fatty acid profiles in zebrafish embryos. Quantification of fatty acids from C14–C20 in control and embryos injected with a half dose (5 ng each) of both MOs at 5 dpf. Y-axis shows the amount of fatty acids per mg of tissue weight. Fatty acids from C14–C20 seem to be increased in MOelovl1-injected embryos as compared to those in control embryos. p values shown in each graph were determined by Student's t-test in Microsoft Excel.
Elovl1 knockdown suppresses lipid consumption from the yolk and lipid utilization in the swim bladder
A developing zebrafish embryo depends entirely on the yolk mass for its nutrient supply until approximately 5 dpf.16 Oil-Red-O (ORO) staining detects neutral lipids in the yolk, as well as in organs and tissues that require lipids as nutrients for growth. Several organs and tissues including head, heart, swim bladder, and vasculature are known to be ORO-positive,16,17 reflecting a high dependency on lipid metabolism. Among these organs, the swim bladder displays strong ORO staining following complete consumption of the yolk lipids 5 dpf and later, whereas the heart and vasculature are barely ORO-positive at the same developmental stage.18 To examine the potential link between deflated swim bladders and disturbed lipid profiles upon elovl1 knockdown, we performed ORO staining in zebrafish embryos at 4 dpf and 5 dpf. We found that embryos injected with MOelovl1a + MOelovl1b display a persistent ORO staining in the yolk at 5 dpf, whereas control embryos show no detectable ORO staining (Fig. 5), indicating impaired lipid consumption in response to elovl1a and elovl1b knockdown. In addition, MO-injected embryos show more condensed ORO staining in the swim bladder than control embryos, although both are ORO positive. We resected the swim bladders stained with ORO and examined the neutral lipids more closely. Control embryos showed lipid droplets on the surface of the swim bladder at 4 dpf, which dispersed homogeneously to completely cover the swim bladder at 5 dpf. In contrast, swim bladders of MO-injected embryos at 4 dpf look much smaller and contain a persistent cluster of lipid droplets in the central region at 5 dpf. The disparity of ORO staining between control and MO-injected embryos confirms a strong correlation between elovl1 activity and lipid metabolism during zebrafish embryogenesis.
FIGURE 5.
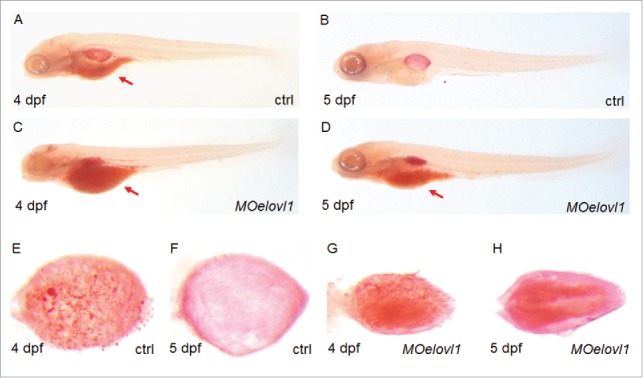
Elovl1 knockdown suppresses lipid consumption in the yolk and lipid utilization in the swim bladder. Oil-Red-O staining was performed to visualize the neutral lipids in developing embryos at 4 and 5 dpf. Note that neutral lipids accumulate in the yolk (compare B with D) as well as on the surface of swim bladder epithelium (compare F and H) in MOelovl1-injected embryos. Embryos are presented in lateral views (A–D) with the anterior aspect to the left. Dissected swim bladders from the wild type and knockdown embryos are presented in dorsal views (E–H) with the anterior aspect to the left.
Elovl1 knockdown affects genes required for swim bladder development
Defective inflation and lipid utilization in the swim bladder in response to elovl1a and elovl1b knockdown does not necessarily reflect abnormality from the early stages of swim bladder development. We therefore examined whether development of the swim bladder progresses normally, but its deflation becomes evident upon elovl1 and elovl1b knockdown. The zebrafish swim bladder is composed of 3 layers, i.e., epithelium, mesenchyme, and mesothelium and genes that mark each layer have been previously reported.19-21 We found that expression of genes in all layers of the swim bladder, including epithelial sox2 and hb9, mesenchymal acta2, and outer mesothelium hprt1l and anxa5, is severely reduced in MO-injected embryos as compared to that in the control (Fig. 6). In addition, we also examined expression of surfactant proteins (SP) in zebrafish swim bladder. Surfactant is composed of a mixture of lipids and SP and is essential for maintaining the integrity of the lung by reducing surface tension at the liquid-air interface22. According to a recent report in which homologs of surfactant genes in humans are expressed in the swim bladder,22 we examined expression of prosaposin (SP-B) and tenomodulin (SP-C) and found that expression of both genes is significantly decreased in MO-injected embryos compared to that in control. These results suggest that both elovl1 genes are important for the expression of genes in all 3 tissue layers of the swim bladder in addition to the expression of SP genes required for functional swim bladder.
FIGURE 6.
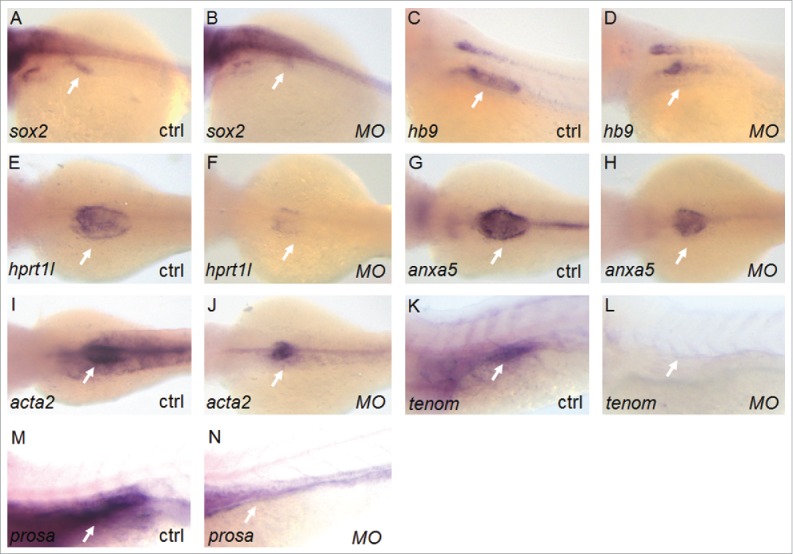
Elovl1 knockdown affects expression of genes involved in swim bladder development. Whole mount in situ hybridization of embryos at 3 or 4 dpf was performed to examine expression of genes involved in swim bladder development. Expression of genes in all 3 tissue layers of the swim bladder, for example, epithelial sox2 (28% of total injected embryos, n = 57) and hb9 (24%, n = 64), mesenchymal acta2 (24%, n = 54), and mesothelial hprt1l (28%, n = 53) and anxa5 (24%, n = 52), is significantly reduced in elovl1-knockdown embryos compared to the control at 3 dpf (n > 50 for each gene). Similarly, expression of surfactant genes (tenomodulin, tenom for short; prosaposin, prosa for short) is also decreased upon elovl1-knockdown embryos (25%, n = 61 for tenom and 27%, n = 55 for prosa) as compared to the control at 4 dpf. Arrows indicate the developing swim bladder region where each gene is expressed. Embryos are presented in dorsal views with the anterior aspect to the left.
Elovl1b is required for kidney development
We found a significant difference between the expression patterns of the 2 elovl1 genes (Fig. 1). Restricted expression of elovl1a was evident in the swim bladder at 2-3 dpf, whereas elovl1b was highly expressed in the pronephric kidney and the swim bladder region. Consistent with our findings, a previous study has reported Elovl1 expression in the kidney of mammals.15 Recent studies indicate that development of the kidney in zebrafish is quite similar to that in mammals.23-25 In particular, gene expression of the glomerulus and nephron tubule segments is conserved in both mammals and zebrafish, confirming the use of zebrafish as a suitable model to determine the mechanisms governing nephrogenesis. Based on the expression patterns observed, we hypothesized that elovl1b knockdown may affect kidney development in the zebrafish embryo. We found that expression of Wilms' tumor protein 1b (wt1b), which is a marker for renal precursors, was not changed upon elovl1b knockdown (Fig. 7), indicating that elovl1b might not play a role in the initial differentiation of renal progenitors. However, we found that expression of several marker genes in different regions of the developing kidney was significantly reduced upon elovl1b knockdown. Expression of cadherin 17 (cdh17), a pan tubule and duct marker, is significantly decreased in elovl1b-knockdown embryos at both 24 and 60 hpf. Expression of segment-specific markers, such as transient receptor potential melastatin 7 (trpm7) for the proximal straight tubule, solute carrier family 12 member 1 (slc12a1) for the early distal tubule, and solute carrier family 12 member 3 (slc12a3) for the late distal tubule, is slightly reduced at 24 hpf and significantly decreased at 60 hpf upon elovl1b knockdown.
FIGURE 7.
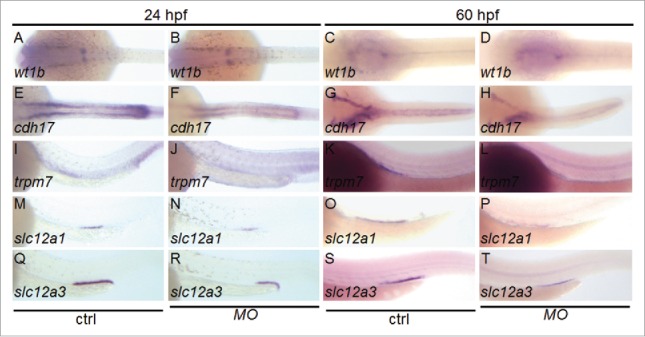
Elovl1b knockdown affects genes required for kidney development. Embryos were injected with MOelovl1b and whole mount in situ hybridization was performed to analyze expression of genes involved in kidney development. Genes expressed in the kidney tubule were significantly reduced in elovl1b knockdown embryos (for each gene at 24 hpf, wt1b, 0%, n = 51; cdh17, 24%, n = 50; trpm7, 32%, n = 51; slc12a1, 24%, n = 54; slc12a3, 40%, n = 49; and at 60 hpf, wt1b, 0%, n = 48; cdh17, 28%, n = 50; trpm7, 28%, n = 53; slc12a1, 32%, n = 51; slc12a3, 32%, n = 52) as compared to those in the control. Embryos are presented in dorsal views (A – H) or lateral views (I – T) with the anterior aspect to the left.
We performed a rescue experiment by overexpressing elovl1a or elovl1b mRNA. As shown in Fig. S2, overexpression of elovl1a cannot rescue decreased expression of genes in developing kidney while overexpression of elovl1b can. Consistent with the finding that overexpression of elovl1a cannot rescue the deflated swim bladder induced by MOelovl1b, and vice versa, this result further suggests a functional difference between elovl1a and elovl1b. Moreover, elovl1b might play an important role in maintaining a functional kidney in zebrafish, which has not been described in mammalian ELOVL1.
Elovl1b knockdown affects renal function
Previous studies have reported that rhodamine-labeled dextran can be used to assess renal function in zebrafish embryos.26,27 We injected 50 ng rhodamine-labeled dextran directly into the heart field at 3 dpf when the pronephros functions properly, and examined the passage of fluorescent dextran out of the embryos. Kidney function was normal in the wild type embryos, so the dextran was filtered through the glomerulus and was excreted in the urine, which results in the disappearance of fluorescence 24 hours post-injection. In contrast, in the case of the elovl1b-injected embryos, residual fluorescence remained in the heart, indicating compromised renal function, which may be due to reduced tubular secretion as a result of reduced expression of the genes involved in the development of kidney tubules (Fig. 8).
FIGURE 8.
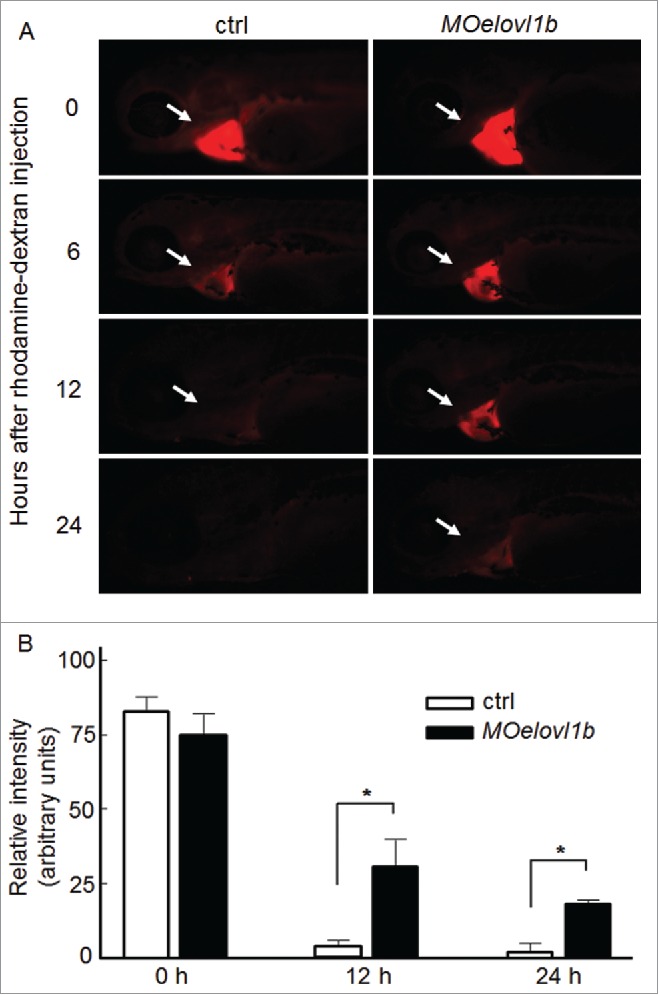
Elovl1b knockdown affects renal function. (A) Control and elovl1b-knockdown embryos were injected with rhodamine-labeled dextran in the pericardium of 3-day-old embryos and the fluorescence intensity was measured at 0, 6, 12, and 24 hours post-injection. The control embryo barely displays fluorescence after 24 hours of dextran injection (0%, n = 15), whereas evol1b-knockdown embryos show residual fluorescence in the heart region (30%, n = 27). Embryos are presented in lateral views with the anterior aspect to the left. (B) The relative intensity of fluorescent signals in Y-axis was quantified by making selections of the heart fields using ImageJ 1.47v (National Institute of Health). Intensity from each embryo was quantified and average intensities from each experimental group were calculated with the corresponding standard deviation. * indicates statistical significance with p values < 0.05 determined by Student's t test in Microsoft Excel.
Elovl1 knockdown does not affect neuronal myelination
A previous study has suggested that a reduction in the C24 lignoceric and nervonic acids of sphingolipids causes hypomyelination.7 We found accumulation of long chain fatty acids in response to elovl1 knockdown, possibly due to a defect in fatty acid chain elongation. Although we were unable to detect very long chain fatty acids even in control embryos (possibly because of the presence of very low amounts), we assume that the levels of very long chain fatty acids may have further been reduced in response to elovl knockdown. We used mbp:EGFP transgenic zebrafish28 to study the effects of elovl knockdown in neuronal myelination. In contrast to a previous report, the degree of neuronal myelination is indistinguishable between the control and MOelovl1-injected embryos (Fig. S3), suggesting that Elovl1 may be dispensable for myelination in zebrafish for which other elovl genes may be responsible. To examine whether knockdown of 2 elovl1 genes affect expression of other elovl genes, we performed RT-PCR from 1 to 3 dpf after injecting a half dose of MOelovl1a + MOelovl1b. We found that elovl2, elovl4a, elovl4b, elovl5, elovl6, elovl6l, elovl7 and elovl7b are expressed during zebrafish development but that expression of most genes are not affected by knockdown of elovl1a and elovl1b, except expression of elovl6l and elovl7b to be slightly increased (Fig. S4). Although a tissue-specific expression of elovl genes needs to be carefully examined for confirmation, these results suggest that an elovl gene in zebrafish may be independently expressed without being significantly affected by other elovl genes.
Discussion
The therapeutic importance of the fatty acid elongase enzyme ELOVL1 is highlighted in X-ALD fibroblasts, in which ELOVL1 knockdown reduces the elongation of C22 fatty acids to C26 thereby lowering C26 fatty acid levels and suggesting ELOVL1 modulation as an effective treatment for X-ALD.14 To date, to our knowledge, the role of the ELOVL1 in different organs has not been reported, with the exception of its role in the formation of the epidermal permeability barrier in mice.13 In this report, we describe that elovl1 deficiency causes defects in swim bladder and kidney development in zebrafish. We showed that ELOVL1 is highly conserved among vertebrates, and that the zebrafish genome contains 2 elovl1 genes, elovl1a and elovl1b. Gene expression patterns indicated that both elovl1 genes are maternally expressed during development. The maternal transcripts may be used to support rapid cell division during the cleavage stage at which various membrane lipids need to be synthesized to build plasma membrane of dividing cells. At 2 dpf and later during development, elovl1 is strongly expressed in the swim bladder, and elovl1b is expressed in the pronephros (Fig. 2). Consistent with strong expression of both elovl1 genes in the swim bladder, we found that one elovl1 gene knockdown results in a flat, deflated swim bladder. To confirm the link between the chain elongation function of elovl1 with the swim bladder defect, we measured lipid profiles in developing zebrafish embryos at 5 dpf. We found that long chain fatty acids from carbon number 14 to 20 seem to accumulate in elovl1 knockdown embryos compared to those in control embryos (Fig. 4). Abnormal lipid profiles may have an effect on developing swim bladders and tested the hypothesis by examining the swim bladders that were stained with Oil-Red-O. In agreement, we found that accumulated neutral lipids remain and persist within the epithelial surface of resected swim bladders of elovl1a or elovl1b knockdown embryos, whereas they disperse when the swim bladder becomes inflated at 5 dpf in the control (Fig. 5). In situ hybridization analysis revealed that genes involved in swim bladder development are largely downregulated in elovl1a or elovl1b knockdown embryos in comparison to those in the control (Fig. 6). One possible explanation for an organ-specific defect upon knockdown of an elovl1 gene is that a certain kind of fatty acids generated by the Elovl1 may be essential for proper development of the organ. Consistent with the hypothesis, knockdown of either elovl1a or elovl1b does not induce developmental abnormalities until 24 hpf although both genes are strongly expressed. It is conceivable that elovl genes including 2 elovl1 genes may act redundantly at least until 24 hpf, while development of some organs, the swim bladder as an example, that depend highly on lipid metabolism may require a specialized set of fatty acids for organogenesis. Indeed, we found that knockdown of elovl1a or elovl1b significantly decreased expression of surfactant genes required for the initial formation and maintenance of surfactant.
Regarding elovl1b expression in the pronephric kidney, we found that elovl1b knockdown impairs the expression of genes involved in normal kidney development (Fig. 7). By 24 hpf, the pronephros can be detected by expression of segment-specific genes, and we found no significant difference in wt1b expression between the control and elovl1b knockdown embryos, suggesting that elovl1b may not be required for early differentiation of the pronephros. Instead, Elovl1b may play a role in the maintenance of functional nephrons, since expression of the segment marker genes in the pronephros is markedly reduced upon elovl1b knockdown. Indeed, embryos injected with MOelovl1b are shown to be less efficient in excretion capability (Fig. 8), indicating a defect in the functional integrity of the kidney.
Several studies have reported previously that very long chain fatty acids are required for neuronal myelination.6,7 We tested the possibility of myelination defects on elovl1 knockdown in zebrafish, but did not observe any significant difference between control and elovl1 knockdown embryos. Since myelinated neurons can undergo demyelination under certain disease conditions, including a reduction in the levels of very long chain fatty acids, our analysis using MO-mediated transient knockdown could be limited by a relatively short period of the investigation. In addition, other Elovl proteins might be involved primarily in myelinating neurons, since we barely detected expression of elovl1a or elovl1b in developing neurons. In fact, our RT-PCR analysis indicates that expression of elovl6l and elovl7b is increased in response to knockdown of elovl1a and elovl1b.
Although highly similar to each other, Elovl1a and Elovl1b may not function redundantly. We showed that overexpression of elovl1a cannot compensate for the reduced function of elovl1b, and vice versa. This is clearly shown in the developing kidney where only elovl1b is expressed. Knockdown of elovl1b decreases expression of developing kidney marker genes, and overexpression of elovl1b, but not elovl1a, can restore expression of genes to a normal level. Although we do not currently understand apparently different roles of Elovl1a and Elovl1b, C-terminal regions of the proteins are quite different from each other and not even present in human ELOVL1, implying that the regions may play a important role in delineating one from the other.
In spite of some differences in Elovl1 between mammals and zebrafish, including elovl1 gene duplication in zebrafish, Elovl1 appears to evolve conservatively in both structure as well as function. First, zebrafish Elovl1 shows high sequence homology to that of humans and mice, all of which have 5 fatty acid desaturase motifs conserved in Elovl family members. Second, expression analysis in zebrafish indicates that both elovl1 transcripts are maternally derived and thus ubiquitously expressed during early development. Expression of elovl1a becomes restricted in the swim bladder, a structure homologous to the mammalian lung,19 whereas elovl1b is expressed in the pronephros and the swim bladder in later development. Previous studies have demonstrated Elovl1 expression in the lung and kidney of the mouse,15 suggesting that zebrafish may have evolved to possess organ-specific elovl1 expression. Third, total lipid profiles of elovl1 knockdown embryos show a strong trend toward accumulation of long chain fatty acids in comparison to those of the control, suggesting functional equivalence between ELOVL1 genes in mammals and zebrafish. The developmental roles of zebrafish elovl1 in the present study can possibly be applicable to those of mammals. In this regard, developmental defects in the kidney and lung of Elovl1 homozygous knockout mice could be another reason for early neonatal mortality,13 although further studies are necessary to confirm this assumption.
In conclusion, we report the previously uncharacterized roles of elovl1 in the development of vital organs such as the kidney and swim bladder during zebrafish embryogenesis, which may potentially be applicable to humans. Further detailed analysis will be necessary to understand the molecular mechanisms of elovl1 in the development of internal organs during vertebrate embryogenesis and to facilitate therapeutic applications in human diseases including X-ALD.
MATERIALS AND METHODS
Fish maintenance
The zebrafish and their embryos were collected from natural spawning and raised, staged, and handled according to standard protocols.29 Wild type and mbp:EGFP transgenic zebrafish28 were selected for the investigation. The experimental protocol was approved by the Committee for Ethics in Animal Experiments of the Wonkwang University (WKU15-126) and carried out under the guidelines for animal experiments.
Microinjection
MOs for elovl1a (translation: 5′-AGCACCTCTTGAAACATCTTGTCGT-3′, splicing: 5′-GCCACCCGCTGTAAATTTCACAAGA-3′); elovl1b (translation: 5′-CTTTCACAGTCTCCAGCATTTTTGC-3′, splicing: 5′-AGGAATCTGCATATTTACCTCTGGC-3′); and control MO (5′-AACATACATCAGTTTAATATATGTA-3′) were purchased from Gene Tools, USA. 10 ng trMOelovl1a or trMOelovl1b, 6 ng spMOelovl1a or 8 ng spMOelovl1b was microinjected into the one cell stage of zebrafish embryos as previously described.30 For rescue experiments, 50 or 250 pg of in vitro-synthesized elovl1a or elovl1b mRNA, respectively, was also microinjected with MOs.
Constructs and RNA synthesis
Zebrafish elovl1a and elovl1b open reading frames were individually amplified from total RNAs prepared from zebrafish embryos at 10 hours post fertilization (hpf), using a standard RT-PCR protocol and cloned in a pCRII-Topo vector (Invitrogen, USA). Antisense RNA probes of elovl1a and elovl1b were synthesized for in situ hybridization assays. For rescue experiments, each cDNA recovered from EcoRI and XhoI enzymes in pCRII-Topo was subcloned into EcoRI and XhoI sites in a pCS2+ vector and mRNAs were synthesized using the mMESSAGE mMACHINE kit (Ambion, Inc., USA).
Measurement of fatty acids from zebrafish larvae
~50 mg tissue mass from approximately 100 control and MO-injected zebrafish at 5 dpf was homogenized using TissueLyzer (Qiagen). Heicosanoic acid as an internal standard was added into the samples and fatty acids were extracted by Folch method.31 For fatty acid analysis, base hydrolysis using KOH was performed followed by neutralization with HCl. Methyl esterification was performed with BCl3-MeOH at 60 °C for 30 min, and fatty acids of the sample were analyzed by gas chromatography-mass spectrometry analysis using 7890A/5975A (Agilent). All standards including the internal standard for calibration were purchased from Avanti-Polar Lipids and Sigma-Aldrich. The detailed analytical conditions for fatty acids were described in our previous report.32
Whole mount Oil-Red-O staining
Control and MO-injected embryos at 96 hpf and 120 hpf were fixed with 4% paraformaldehyde overnight, washed 3 times with PBS, and depigmented with a bleaching solution (200 µL 1× SSC (saline sodium citrate), 33.8 mL water, 2 mL formamide, and 4 mL hydrogen peroxide). The embryos were then washed with PBS 3 times and pre-incubated in 60% isopropanol for 30 min. Embryos were then incubated in freshly prepared 0.3% solution of Oil-Red-O (Sigma) in 60% isopropanol for 3 hours, rinsed several times with PBS, and photographed.
Whole mount in situ hybridization and microscopy
Control and elovl1-knockdown embryos were collected at various developmental stages, fixed in 4% paraformaldehyde, and processed for in situ hybridization, as previously described30. Images were taken using a Leica M165FC microscope equipped with Leica DFC500.
Reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was performed as previously reported29. In brief, total RNA was prepared from 10 uninjected and MO-injected embryos from 1- to 3 dpf using Trizol (Ambion, USA) following manufacturer's instructions. First strand cDNA was synthesized (Roche, USA) and quantitative PCR was performed. Primer sequences used are listed in Tab. S1.
Functional assay for kidney function
Zebrafish embryos at 72 hpf were anesthetized in tricaine and 50 ng 10-kDa rhodamine-labeled dextran (Thermo Fischer, USA) was injected directly into the pericardium. Images of fluorescent embryos were taken at 0, 6, 12, and 24 hours post-injection using a Leica M165FC microscope equipped with Leica DFC500. The relative intensity of fluorescent signals was quantified by making selections of the heart fields using ImageJ 1.47v (National Institute of Health). Intensity from each embryo was quantified and average intensities from each experimental group were calculated with the corresponding standard deviation. Student's t-test in Microsoft Excel was used to determine statistical significance.
Abbreviations
- CoA
Coenzyme A
- dpf
day post fertilization
- Elovl
elongase
- ER
endoplasmic reticulum
- hpf
hour post fertilization
- ORO
Oil-Red-O
- spMO
splicing-blocking morpholino
- trMO
transcription-blocking morpholino
- VLCFAs
Very long chain fatty acids
- X-ALD
X-linked adrenoleukodystrophy
Supplementary Material
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the laboratories of Drs. Choe and Park for sharing materials and valuable comments and discussion on this project. We also thank Dr. Gong for providing invaluable constructs for making in situ probes (hb9, acta2 and anxa5).
AUTHOR CONTRIBUTIONS
Conceived and designed experiment: RP and S-KC. Performed experiments: SB, JNL, Y-IK, I-KN, S-JK, S-JK and HJY. Analyzed data: SB, JNL, SK, G-SO, H-JK, H-SS, RP and S-KC. Wrote the paper: SB, HJY, RP and S-KC.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Ministry of Education (2013R1A1A2010518) and Ministry of Science, ICT & Future Planning (2011-0030130 and 2014M3A9D8034463), and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [grant number : HI14C0384]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- [1].Vasireddy V, Uchida Y, Salem NJR, Kim SY, Mandal MN, Reddy GB, Bodepudi R, Alderson NL, Brown JC, Hama H, et al.. Loss of functional ELOVL4 depletes very long-chain fatty acids (>or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum Mol Genet 2007; 16: 471-82; PMID:17208947; http://dx.doi.org/ 10.1093/hmg/ddl480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harkewicz R, Du H, Tong Z, Alkuraya H, Bedell M, Sun W, Wang X, Hsu YH, Rudd JE, Hughes G, et al.. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem 2012; 287: 11469-80; PMID:22199362; http://dx.doi.org/ 10.1074/jbc.M111.256073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mcmahon A, Butovich IA, Mata NL, Klein M, Ritter R, Richardson J, Birch DG, Edwards AO, Kedzierski W. Retinal pathology and skin barrier defect in mice carrying a Stargardt disease-3 mutation in elongase of very long chain fatty acids-4. Mol Vis 2007; 13: 258-72; PMID:17356513 [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, LI Y, Metzker ML, Allikmets R, Zack DJ, Kakuk LE, Lagali PS, et al.. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet 2001; 27: 89-93; PMID:11138005 [DOI] [PubMed] [Google Scholar]
- [5].Pauter AM, Olsson P, Asadi A, Herslof B, Csikasz RI, Zadravec D, Jacobsson A. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J Lipid Res 2014; 55: 718-728; PMID:24489111; http://dx.doi.org/ 10.1194/jlr.M046151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sargent JR, Coupland K, Wilson R. Nervonic acid and demyelinating disease. Med Hypotheses 1994; 42: 237-42; PMID:8072429; http://dx.doi.org/ 10.1016/0306-9877(94)90122-8 [DOI] [PubMed] [Google Scholar]
- [7].Yeh YY. Long chain fatty acid deficits in brain myelin sphingolipids of undernourished rat pups. Lipids 1988; 23: 1114-8; PMID:3226227; http://dx.doi.org/ 10.1007/BF02535275 [DOI] [PubMed] [Google Scholar]
- [8].Zadravec D, Tvrdik P, Guillou H, Haslam R, Kobayashi T, Napier JA, Capecchi MR, Jacobsson A. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J Lipid Res 2011; 52: 245-55; PMID:21106902; http://dx.doi.org/ 10.1194/jlr.M011346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cinti DL, Cook L, Nagi MN, Suneja SK. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog Lipid Res 1992; 31: 1-51; PMID:1641395; http://dx.doi.org/ 10.1016/0163-7827(92)90014-A [DOI] [PubMed] [Google Scholar]
- [10].Leonard AE, Pereira SL, Sprecher H, Huang YS. Elongation of long-chain fatty acids. Prog Lipid Res 2004; 43: 36-54; PMID:14636670; http://dx.doi.org/ 10.1016/S0163-7827(03)00040-7 [DOI] [PubMed] [Google Scholar]
- [11].Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res 2006; 45: 237-49; PMID:16564093; http://dx.doi.org/ 10.1016/j.plipres.2006.01.004 [DOI] [PubMed] [Google Scholar]
- [12].Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci USA 2010; 107: 18439-44; PMID:20937905; http://dx.doi.org/ 10.1073/pnas.1005572107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sassa T, Ohno Y, Suzuki S, Nomura T, Nishioka C, Kashiwagi T, Hirayama T, Akiyama M, Taguchi R, Shimizu H, et al.. Impaired epidermal permeability barrier in mice lacking elovl1, the gene responsible for very-long-chain fatty acid production. Mol Cell Biol 2013; 33: 2787-96; PMID:23689133; http://dx.doi.org/ 10.1128/MCB.00192-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ofman R, Dijkstra IM, Van Roermund CW, Burger N, Turkenburg M, Van Cruchten A, Van Engen CE, Wanders RJ, Kemp S. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol Med 2010; 2: 90-7; PMID:20166112; http://dx.doi.org/ 10.1002/emmm.201000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol 2000; 149: 707-18; PMID:10791983; http://dx.doi.org/ 10.1083/jcb.149.3.707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Flynn EJ III, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J Lipid Res 2009; 50: 1641-52; PMID:19366995; http://dx.doi.org/ 10.1194/jlr.M800590-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim YI, Bhandari S, Lee JN, Yoo KW, Kim SJ, Oh GS, Kim HJ, Cho M, Kwak JY, So HS, Park R, Choe SK. Developmental roles of D-bifunctional protein-A zebrafish model of peroxisome dysfunction. Mol Cells 2014; 37: 74-80; PMID:24552713; http://dx.doi.org/ 10.14348/molcells.2014.2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry 2006; 45: 15179-87; PMID:17176039; http://dx.doi.org/ 10.1021/bi0619268 [DOI] [PubMed] [Google Scholar]
- [19].Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol 2009; 331: 222-36; PMID:19422819; http://dx.doi.org/ 10.1016/j.ydbio.2009.04.035 [DOI] [PubMed] [Google Scholar]
- [20].Yin A, Korzh S, Winata CL, Korzh V, Gong Z. Wnt signaling is required for early development of zebrafish swimbladder. PLoS One 2011; 6: e18431; PMID:21479192; http://dx.doi.org/ 10.1371/journal.pone.0018431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yin A, Winata CL, Korzh S, Korzh V, Gong Z. Expression of components of Wnt and Hedgehog pathways in different tissue layers during lung development in Xenopus laevis. Gene Expr Patterns 2010; 10: 338-44; PMID:20682360; http://dx.doi.org/ 10.1016/j.gep.2010.07.005 [DOI] [PubMed] [Google Scholar]
- [22].Zheng W, Wang Z, Collins JE, Andrews RM, Stemple D, G Z. Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalina lung. PLoS One 2011; 6:e24019; PMID:21887364; http://dx.doi.org/ 10.1371/journal.pone.0024019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, Mcmahon AP, Davidson AJ. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet 2007; 3: 1922-38; PMID:17953490; http://dx.doi.org/ 10.1371/journal.pgen.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cheng CN, Verdun VA, Wingert RA. Recent advances in elucidating the genetic mechanisms of nephrogenesis using zebrafish. Cells 2015; 4: 218-33; PMID:26024215; http://dx.doi.org/ 10.3390/cells4020218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Desgrange A, Cereghini S. Nephron Patterning: Lessons from Xenopus, Zebrafish, and Mouse Studies. Cells 2015; 4: 483-499; PMID:26378582; http://dx.doi.org/ 10.3390/cells4030483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hentschel DM, Park KM, Cilenti L, Zervos AS, Drummond I, Bonventre JV. Acute renal failure in zebrafish: a novel system to study a complex disease. Am J Physiol Renal Physiol 2005; 288: F923-929; PMID:15625083; http://dx.doi.org/ 10.1152/ajprenal.00386.2004 [DOI] [PubMed] [Google Scholar]
- [27].Tobin JL, Beales PL. Restoration of renal function in zebrafish models of ciliopathies. pediatr nephrol 2008; 23: 2095-9; PMID:18604564; http://dx.doi.org/ 10.1007/s00467-008-0898-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, Park HC, Kim CH. Visualization of myelination in GFP-transgenic zebrafish. Dev Dyn 2010; 239: 592-7; PMID:19918882; http://dx.doi.org/ 10.1002/dvdy.22166 [DOI] [PubMed] [Google Scholar]
- [29].Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203: 253-310; PMID: 8589427; http://dx.doi.org/ 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- [30].Choe SK, Ladam F, Sagerstrom CG. TALE factors poise promoters for activation by Hox proteins. Dev Cell 2014; 28: 203-11; PMID:24480644; http://dx.doi.org/ 10.1016/j.devcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957; 226: 497-509; PMID:13428781 [PubMed] [Google Scholar]
- [32].Kim HK, Shin MS, Youn BS, Kang GM, Gil SY, Lee CH, Choi JH, Lim HS, Yoo HJ, Kim MS. Regulation of energy balance by the hypothalamic lipoprotein lipase regulator Angptl3. Diabetes 2015; 64: 1142-53; PMID:25338813; http://dx.doi.org/ 10.2337/db14-0647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



