Abstract
We propose that sensory inputs are processed in terms of optimised predictions and prediction error signals within hierarchical neurocognitive models. The combination of non-invasive brain imaging and generative network models has provided support for hierarchical frontotemporal interactions in oddball tasks, including recent identification of a temporal expectancy signal acting on prefrontal cortex. However, these studies are limited by the need to invert magnetoencephalographic or electroencephalographic sensor signals to localise activity from cortical ‘nodes’ in the network, or to infer neural responses from indirect measures such as the fMRI BOLD signal. To overcome this limitation, we examined frontotemporal interactions estimated from direct cortical recordings from two human participants with cortical electrode grids (electrocorticography – ECoG). Their frontotemporal network dynamics were compared to those identified by magnetoencephalography (MEG) in forty healthy adults. All participants performed the same auditory oddball task with standard tones interspersed with five deviant tone types. We normalised post-operative electrode locations to standardised anatomic space, to compare across modalities, and inverted the MEG to cortical sources using the estimated lead field from subject-specific head models. A mismatch negativity signal in frontal and temporal cortex was identified in all subjects. Generative models of the electrocorticographic and magnetoencephalographic data were separately compared using the free-energy estimate of the model evidence. Model comparison confirmed the same critical features of hierarchical frontotemporal networks in each patient as in the group-wise MEG analysis. These features included bilateral, feedforward and feedback frontotemporal modulated connectivity, in addition to an asymmetric expectancy driving input on left frontal cortex. The invasive ECoG provides an important step in construct validation of the use of neural generative models of MEG, which in turn enables generalisation to larger populations. Together, they give convergent evidence for the hierarchical interactions in frontotemporal networks for expectation and processing of sensory inputs.
Keywords: Dynamic causal modelling, Mismatch negativity, Electrocorticography, Magnetoencephalography, Cognition
1. Introduction
The brain is proposed to efficiently process information from the world around us through optimising the feedback of predictions of sensory inputs and the feedforward signalling of prediction errors, in hierarchical information processing networks (Friston & Kiebel, 2009). Under this hypothesis, top-down predictions are compared to bottom-up sensory information and return a prediction error to update the prediction model when a mismatch occurs (Chennu et al., 2013, Friston, 2009b, Kiebel et al., 2008a, Lieder et al., 2013, Rao and Ballard, 1999). The information processing hierarchy may have multiple levels, with increasing abstraction of information and representation of complex arbitrary features (Carlin et al., 2011, Ewbank et al., 2011). To test this hypothesis, many studies have used auditory oddball paradigms which evoke a robust error signal in terms of the mismatch negativity response (MMN) to unexpected deviant stimuli that violate a learned regularity of standard stimuli (Chennu et al., 2013; Hughes et al., 2013, Näätänen et al., 1993, Phillips et al., 2015).
Evidence for the direction of influence in frontotemporal interactions underlying auditory prediction came initially from reduced MMN responses in patients with frontal cortical lesions (Alho, Woods, Algazi, Knight, & Näätänen, 1994). Evidence from generative models of electrophysiological responses in healthy humans also provides compelling support for hierarchical feedback and feedforward interactions, with prediction and error signals respectively, in frontotemporal connectivity (Garrido, Kilner, Kiebel, & Friston, 2009; Hughes & Rowe, 2013) and phase synchronisation (MacLean & Ward, 2014). We recently demonstrated the presence of high-order expectancy inputs driving the frontal cortex (Phillips et al., 2015) using an oddball task that alternated standard tones with deviants differing from the standard in one of five dimensions. In keeping with previous work (Boly et al., 2011, Garrido et al., 2009a, Garrido et al., 2008 Schmidt, Leventhal, Mallet, Chen, & Berke, 2013) we confirmed that frontotemporal connections were common to all deviant dimensions. Additionally, the frontal cortex was subject to an expectancy or pacemaker input, which was violated by temporal irregularities (duration and silent gap deviants) but not frequency, loudness or laterality. This provides a potential mechanism to explain MMN responses to unexpected absent stimuli (H. C. Hughes et al., 2001, Oceák et al., 2006, Raij et al., 1997, Wacongne et al., 2011).
To study these networks in humans, it has been necessary to invert generative models of neural interactions to fit the magneto-/electro-encephalography signal (Dietz et al., 2014, Garrido et al., 2008, Garrido et al., 2009a Hughes et al., 2013). This inversion can in principle be performed simultaneously with the optimisation of neural interactions in the model, using dynamic causal modelling (DCM, Friston, Harrison, & Penny, 2003). This combines the neural network optimisation and inversion of the estimated lead field, using neural mass models and mean field approximations. An important step in validation of DCM would be direct rather than indirect estimation of local field potentials generated by local neuronal ensembles. For example, David et al. (2008) provided face validation of the DCM method for functional magnetic resonance imaging, by comparing model parameters derived from fMRI to intracranial recordings of a known model of epileptic spiking and wave discharges in rats. Recently, Papadopoulou, Friston, and Marinazzo (in press) provided construct validation of steady-state DCM using simultaneous EEG and ECoG monkey recordings during wakefulness and sedation, finding the same winning models across imaging modalities.
We sought to provide construct validation of human magnetoencephalography (MEG) method for DCM using human electrocorticography (ECoG), which is sensitive to local field potentials. ECoG of frontal and temporal cortex in left or right hemisphere was undertaken in two patients undergoing pre-surgical assessment for intractable epilepsy. We used the same task and homologous generative model sets as for the analysis of MEG data from forty healthy adults (Fig. 1). The patient data enabled the comparison of generative models of hierarchical frontotemporal interactions without the need for inversion of the lead field inherent in MEG. Special procedures were required to normalise the patient data to standard anatomical space to enable fair comparison between methods, given the gross distortion of macroscopic anatomy following craniectomy.
Fig. 1.
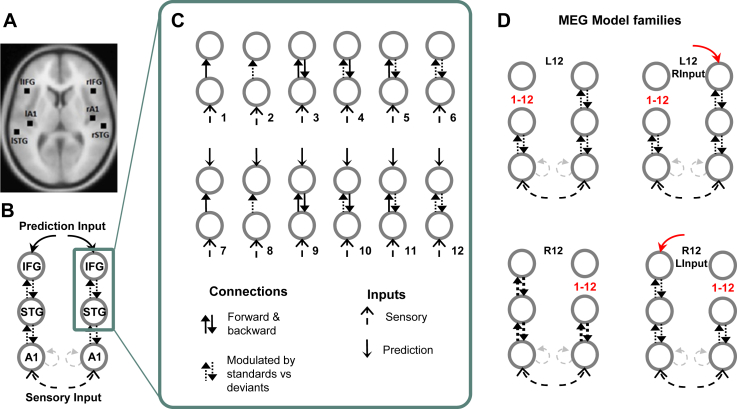
The connectivity model space to be used in the MEG and ECoG datasets. A) The source loci in bilateral primary auditory cortex (A1), STG and IFG, based on Garrido, Kilner, Kiebel, et al., (2009). B) The full model from Phillips et al. (2015) which provided an optimal parsimonious fit of the data. C) The subset of models studied here as intracranial electrodes have limited coverage of sources in the full model. The models are illustrated in order of complexity, beginning with a simple forward (bottom-up) connection between the two nodes and progress by adding modulated connections (dashed connections), backward (top-down) connections and the prediction inputs acting on the highest node. D) We defined four families of models, with variation across families shown in red. All MEG models include sensory input to A1 with intrinsic A1 connectivity and bidirectional connectivity between A1 and STG. Model family L12 includes the 12 models in C) for the left hemisphere whilst keeping full connectivity on the right hemisphere. Model family L12-RInput repeats L12 models and also includes a right frontal expectancy input. R12 and R12-LInput families include homologous models on the opposite hemispheres.
We predicted the identification of homologous networks across modalities, in terms of the principal features of hierarchical frontotemporal networks for sensory processing. Such homology would provide convergent evidence for the hierarchical interactions of frontotemporal networks in the MMN and an important construct validation of DCM methods.
2. Material and methods
2.1. Patients, surgery and intracranial recordings
Patients were selected for this study if their electrodes were located at two or more regions in the same hemisphere that have been associated with the MMN cortical sources in previous EEG and MEG studies: adjacent to primary auditory cortex (immediately superficial to A1 for surface electrodes); superior temporal gyrus (STG) and inferior frontal gyrus (IFG) (Alho et al., 1994, Opitz et al., 2002, Rinne et al., 2000, Rinne et al., 2005). Two adult patients met these criteria (female aged 20 and male aged 30, both right-handed). Both patients had drug-resistant epilepsy and were undergoing electrode implantation to localise epileptic foci and determine surrounding neural function prior to surgical resection. Electrode locations were determined by clinical criteria. Electrode grids and strips were comprised of platinum electrodes embedded in a 0.5 mm flexible silicon plate with 3 mm diameter contact area and 10 mm inter-electrode distance (AdTech, WI, USA).
Patient L1 was implanted with a 6 × 8 subdural electrode grid over the left hemisphere, extending over superior temporal cortex, covering prefrontal, motor and somatosensory cortex plus inferior parietal lobe. Patient R2 was implanted with subdural electrode grids and strips in the right hemisphere, including: 8 × 8 electrode grid covering prefrontal, motor and somatosensory cortex, and the posterior STG and two adjacent 8 × 2 electrode strips covering the temporal-parietal junction, posterior temporal pole, occipital pole and inferior parietal lobe. Electrode locations for both patients are shown in Fig. 2.
Fig. 2.
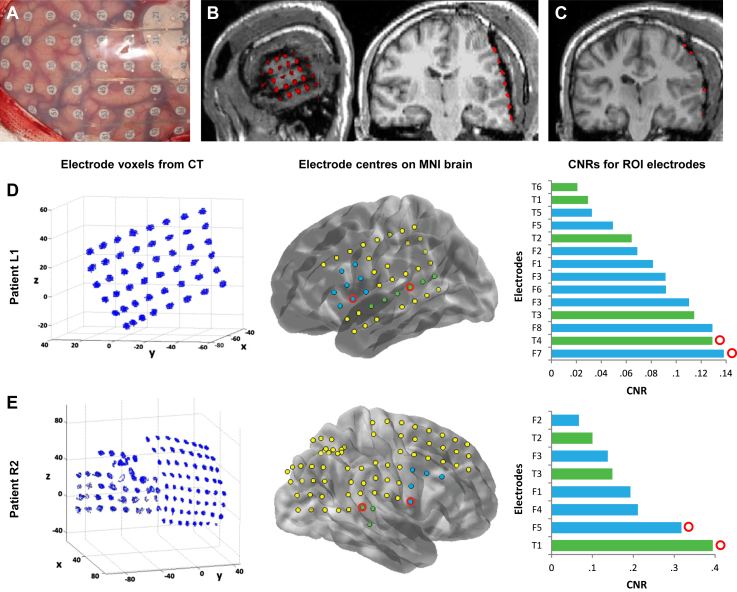
The localisation of ECoG electrodes in standardised image space. A) The patient electrode grid on cortical surface during surgical implantation. B) The lateral and coronal views showing the position of these electrodes in MRI (greyscale) overlaid with the coregistered CT image (red) in native space. The CT image is masked and thresholded to show just the densities corresponding to the electrodes. C) Coronal view of the patient's normalised MRI and coregistered CT. D & E) Electrode localisation and selection for the two patients, L1 and R2. Left column: CT image thresholded to show voxel densities corresponding to electrodes. The centre of each electrode voxel cluster is taken as the coordinate for that electrode and is shown overlaid on a MNI template brain in the centre column. Here electrodes are labelled according to the temporal (green) or frontal (blue) region of interest. The electrodes which disappear between the voxel clusters (left) and localised coordinates (centre) were either not recorded from or we removed them due to high epileptiform activity. Right column: Mean CNRs for each electrode in the regions of interest between 150 and 250 msec. The electrodes with highest mean CNR are circled in red and have the following MNI coordinates: Patient L1 STG [−69, −35, 7] and IFG [−63, 8, −1], patient R2 STG [71, −37, 14] and IFG [68, 0, 17].
Post-implantation structural MRI (Philips Achieva 1.5T, FFE sequence, TR = 15 msec, TE = 5.214 msec, field-of-view 256 × 256 for L1, 240 × 240 for R2, 1 mm slice thickness) and CT images (Siemens Emotion 16, .47 × 0.47 × 0.6 mm voxels, field-of-view 512 × 512) were acquired two-four days after implantation. Task related local field potentials were recorded four days after implantation using the ECoG electrode grids and strips. Patients gave written informed consent before testing and the study was approved by the Institutional Ethics Committee of Ramos Mejía Hospital.
2.2. Task
The task used in both MEG and ECoG has been described in detail previously (Hughes et al., 2013, Näätänen et al., 2004, Phillips et al., 2015). It is a time-efficient variant of the classic oddball task, in which standard tones alternate with deviant tones that deviate from the standard in one of five dimensions while holding other stimulus properties constant. This evokes MMN responses comparable to those seen in classic oddball tasks (Näätänen et al., 2004). In brief, standard tones were presented binaurally with 75 msec duration (including a 7 msec ramp up and ramp down), and contained three sinusoidal partials of 500, 100 and 1500 Hz. The deviant tones differed in one of the following: Frequency (550, 1100, 1650 Hz or 450, 900, 1350 Hz), intensity (±6 dB), location of sound source (right or left instead of binaural), shortened duration (25 msec) or a silent gap in the middle 25 msec. The task was presented using E-Prime® software (Psychology Software Tools, Inc, USA) via plastic tubes and earpieces and participant's hearing was checked before the task to assure tones were clearly audible.
Deviant tones were presented in a pseudo-random order such that a deviant type never appeared twice in a row and each deviant type would appear at least once in a sequence of ten tones. Tones were presented every 500 msec, in three blocks of five minutes. Fifteen standard tones were played at the beginning of each block and excluded from further analysis. In total, 900 standard and 900 deviant tones were then played over the three blocks. Therefore, the task includes many events in a finite study period, providing the potential advantages of efficiency in clinical populations as well as generalisation of inferences over multiple types of deviant.
2.3. Intracranial data collection, pre-processing and ERP analysis
ECoG local field potentials for patient L1 were recorded using Harmonie 5.2 software (Stellate Systems Inc., Canada) with a 64-channel amplifier Bioscience EEG64 (Bioscience SRL, Argentina) sampled at 200 Hz. Patient R2's data were recorded on a Blackrock Cervello Elite system (Blackrock NeuroMed, LLC, USA), sampled at 2000 Hz and downsampled to 250 Hz. Both patients' data were filtered between 1 and 40 Hz using high and low-pass Butterworth filters in forward and reverse directions to obtain zero-phase distortion. We extracted −100 to +400 msec epochs around stimulus tone onset and baseline corrected to the −100 to 0 msec period. Electrodes were rejected if they covered the clinically identified epileptic foci or were observed to contain high epileptic activity. We also used automatic analysis of trials data and rejected any epoch outside of a three standard deviation from the mean threshold (including many but not necessarily all epileptic spikes). Finally, we used visual inspection to remove any residual trials that contained epileptic spiking. Standard and deviant time-courses were compared for each electrode using a two-sample t-test with temporal cluster correction of 25 msec.
2.4. ECoG electrode localisation
We sought to use the same MMN sources in standardised MNI space to directly compare effective connectivity measures across modalities. ECoG electrodes were localised using patient CT images, which were normalised to standard space as described by Blenkmann, Phillips, Muravchik, and Kochen (2015). Briefly, we co-registered patient post-implantation T1-MRI and CT images using SPM8 software (Wellcome Trust Centre for Neuroimaging, UCL), segmented the T1-MRI pial surface using freesurfer (Dale and Sereno, 1993, Dale et al., 1999, Fischl and Dale, 2000, Princich et al., 2013) and used this segmentation to normalise the T1-MRI in SPM8. The normalisation transformation was used to normalise the CT image to MNI space. We used an in-skull mask and thresholded the CT image to just include voxel clusters corresponding to each electrode. The voxel clusters were grouped using k-means clustering and we took the mean centre of each cluster as the coordinate for each electrode (Ibáñez et al., 2013). This electrode localisation procedure is available as an open-source toolbox (http://sourceforge.net/projects/ielectrodes).
Once the ECoG electrode locations were in standardised space, it was necessary to select a single electrode from the region of interest, to be used in the connectivity analysis and to guide the specification of homologous coordinates in the MEG analysis. To identify the specific electrodes, we first selected a subset of electrodes residing in STG or IFG according to the gross anatomy (Cheng et al., 2013, Doeller et al., 2003, Molholm et al., 2005, Rinne et al., 2000). Within each subset, we calculated the contrast-to-noise ratio (CNR) between 150 and 250 msec for each electrode. This CNR was calculated from the pooled signal-to-noise ratio for standard and deviant conditions, as shown:
Where μs,d is the mean voltage for each condition, σs,d is the standard deviation and ns,d is the number of trials (Cui, Bray, & Reiss, 2010). The electrodes with highest CNR in each region were used in the ECoG connectivity analysis and their coordinates were used as sources for the MEG connectivity analysis.
2.5. MEG participants
Forty young healthy MEG participants completed the task (23 males, mean age 33.7, range 21–41, 4 left-handed) as part of the population-based sample collected by the Cambridge Centre for Ageing and Neuroscience (Cam-CAN, www.cam-can.com). Full protocols and exclusion criteria for this cohort are described by Shafto et al. (2014). Participants gave written informed consent and ethical approval for the Cam-CAN study was obtained from the Cambridgeshire 2 Research Ethics Committee.
2.6. MEG data acquisition and pre-processing
MEG data were collected using a 306-channel Vectorview system in a magnetically shielded room (ElektaNeuromag, Helsinki, Finland), including a magnetometer and two orthogonal planar gradiometers at each of the 102 positions. Five Head-Position Indicator (HPI) coils monitored head position plus paired EOG electrodes recorded vertical and horizontal eye movements. The three-dimensional locations of the coils and three anatomical fiducials (nasion and left and right pre-auricular points) were recorded using a 3D digitiser (Fastrak Polhemus Inc, Colchester, VA). Movement compensation and downsampling from 1 kHz to 250 Hz was completed using Maxfilter software (Elekta Neuromag). The remaining pre-processing steps were completed using SPM8 software (Wellcome Trust Centre for Neuroimaging, UCL). This included high-pass filtering at 1 Hz and low-pass filtering at 40 Hz using Butterworth filters in forward and reverse directions, and epoching −100 to 400 msec around each tone onset with baseline correction of the −100 to 0 msec period. We used automatic artefact rejection through thresholding of EOG electrodes at 200 μV. Trials were averaged using robust averaging (Wager, Keller, Lacey, & Jonides, 2005) followed by an additional low-pass filter at 40 Hz to remove high frequency noise that can be introduced by robust averaging.
2.7. MEG source space analysis
The sources of MMN responses (difference between standard and deviant trials) were reconstructed using the gradiometer data in SPM8. The forward leadfield model was estimated using a realistic single-shell head model, which was constructed from participant's individual structural MRI scan (T1-weighted, 3D MPRAGE sequence, TR = 2250 msec, TE = 2.99 msec, flip angle 9°, field-of-view 240 × 256 × 160, 1 mm slice thickness, collected on a 3T Siemens Tim Trio scanner, normalised to MNI space). The head model was co-registered to digitised fiducial markers and >60 scalp loci. The inverse source reconstruction was computed using the multiple sparse priors algorithm (MSP, Greedy Search; Friston et al., 2008) for the characteristic MMN time window of 150–250 msec after tone onset. The resulting source images were smoothed using an 8 mm FWHM Gaussian kernel. We identified significant sources of the MMN response using a one-sample t-test, comparing the MMN to zero mean (p < .01, family-wise error correction for multiple comparisons).
Additionally, we estimated the equivalent current dipoles in bilateral primary A1 (Garrido, Kilner, Kiebel, et al., 2009 MNI coordinates: [−42, −22, 7], [46, −14, 8]), the STG and the IFG (coordinates extracted from the ECoG datasets). These coordinates were used as informed priors to fix the dipole locations to these six sources. Dipole orientations and amplitudes were set with flat priors allowing them to be fitted to the data using the variational Bayes method of SPM8 (Kiebel, Daunizeau, Phillips, & Friston, 2008).
2.8. Network modelling
We used DCM to examine the hierarchical interactions in frontotemporal networks during the MMN task, with both ECoG and MEG data. DCM uses biophysically constrained neural mass models (David et al., 2006, Kiebel et al., 2006, Kiebel et al., 2009) to make inferences about the mechanisms behind observations of evoked electro- and magneto-encephalographic responses, in terms of the coupling between equivalent current dipole or local field potentials sources and how this coupling is changed by experimental stimuli.
We first repeated the methods of Phillips et al. (2015) to test the reliability of the findings with this larger MEG cohort and successfully reproduced their findings. This full model set could not be replicated in the ECoG dataset due to the limited electrode coverage, thus we were constrained to a sub-space of unilateral two-node models. We therefore inverted twelve generative models, representing alternative hypotheses behind frontotemporal MMN connections, as shown in Fig. 1. Based on previous studies (Garrido et al., 2007a, Garrido et al., 2007b), these models assessed the inclusion of forward and backward connections between MMN sources in the STG and IFG, and the modulation of these connections by the stimuli (models 1–6). Following this, models 7–12 included top-down internally-generated predictions acting on the frontal source, which we showed to be important for temporal expectations (Phillips et al., 2015). These models were based on anatomically motivated networks (Cheng et al., 2013, Doeller et al., 2003, Giard et al., 1995, Molholm et al., 2005, Rinne et al., 2000) and previous MMN studies using DCM (Garrido et al., 2007a, Phillips et al., 2015). The data were modelled across the post-stimulus period of 0–250 msec, for each patient using the biophysically constrained local field potential model. All deviant types were used together to maximise the number of deviant trials in the single subject analyses.
In the MEG dataset, all six MMN dipole sources were reconstructed to ensure a good dipole fit at each location and maximise variance explained. We used primary A1 sources as in previous studies (Boly et al., 2011, Cooray et al., 2014, Dietz et al., 2014, Garrido et al., 2007b, Moran et al., 2014) and patients' STG and IFG source coordinates. These sources were modelled as equivalent current dipoles. Standard and deviant tones were reconstructed separately using the forward modelling described above and models were inverted using SPM 8's DCM-10 standard algorithm.
We investigated the alternative models in Fig. 1C in each hemisphere separately and modelled the contralateral connectivity as fully connected (Garrido et al., 2007a, Phillips et al., 2015). These models also included sensory inputs into bilateral A1 and bidirectional connections between A1 and STG as shown in Fig. 1B. We repeated these models to investigate the presence and symmetry of the frontal expectancy inputs (Phillips et al., 2015), resulting in 48 models in total. We used a hierarchical model comparison approach to first compare model families and then compare the models within the winning family. The model families are shown in Fig. 1D. Model family L12 explores the 12 models in Fig. 1C for the left hemisphere whilst keeping full connectivity on the right hemisphere. Model family L12-RInput repeats L12 models and also includes a right frontal expectancy input. R12 and R12-LInput families explore these models again but in the opposite hemispheres.
Finally, we modelled all possible combinations of the twelve models in Fig. 1C across the two hemispheres in a post hoc analysis, resulting in 144 models (12 × 12). This accounts for the above family comparison always including full connectivity in one hemisphere.
2.9. Model comparison and selection
Bayesian model selection was used to compare the generative models (Penny, Stephan, Mechelli, & Friston, 2004). Bayesian Model Selection compares the free-energy estimate (F) of the bound on the log of model evidence of each model, ln p(y|m) (the probability of the data y given each model m). This measure of model evidence adjusts model fit for complexity to reduce over-fitting (Kiebel et al., 2009, Stephan et al., 2010). We used a fixed effects approach for both ECoG and MEG datasets, assuming our MEG population of healthy participants have the same network architecture underlying the observed data across the group with variation in connection strengths (Dietz et al., 2014, Stephan et al., 2010). It is also appropriate to use a fixed effects rather than random effects for single patient studies (Friston, Holmes, & Worsley, 1999).
The model with highest model evidence is referred to as the ‘winning’ model (implicitly, the winner from the inverted model set, not all possible models). A difference in model evidence between the winning and ‘second place’ models (ΔF) of five units or more is comparable to a Bayes factor of 150 and by convention this is regarded as strong evidence for one model over another (Kass & Raftery, 1995). We calculated the posterior probability of each model and model family, the probability of that model as the generator of the data, contingent upon the current model space. A posterior probability >.95 is regarded as informative (Stephan et al., 2010).
3. Results
3.1. ECoG localisation and analysis
ECoG electrode locations were extracted from normalised MRI and CT images (Fig. 2C). We masked and thresholded the CT image to contain voxels corresponding to the electrodes and clustered these to give electrode coordinates in standard anatomic space (Fig. 2D and E left). These are shown overlaid on a representative brain in standard anatomic space in Fig. 2D and E centre. Both patients had electrode coverage of STG and IFG regions close to the sources of the MMN. These electrodes are highlighted in green and blue respectively and form regions of interest for each area. CNRs for each electrode in these regions of interest are shown in Fig. 2D and E right, from which we chose the electrodes with highest mean CNR over the 150–250 msec time period. The MNI coordinates for each electrode are as follows: Patient L1 STG [−69, −35, 7] and IFG [−63, 8, −1], patient R2 STG [71, −37, 14] and IFG [68, 0, 17].
3.2. Source analysis
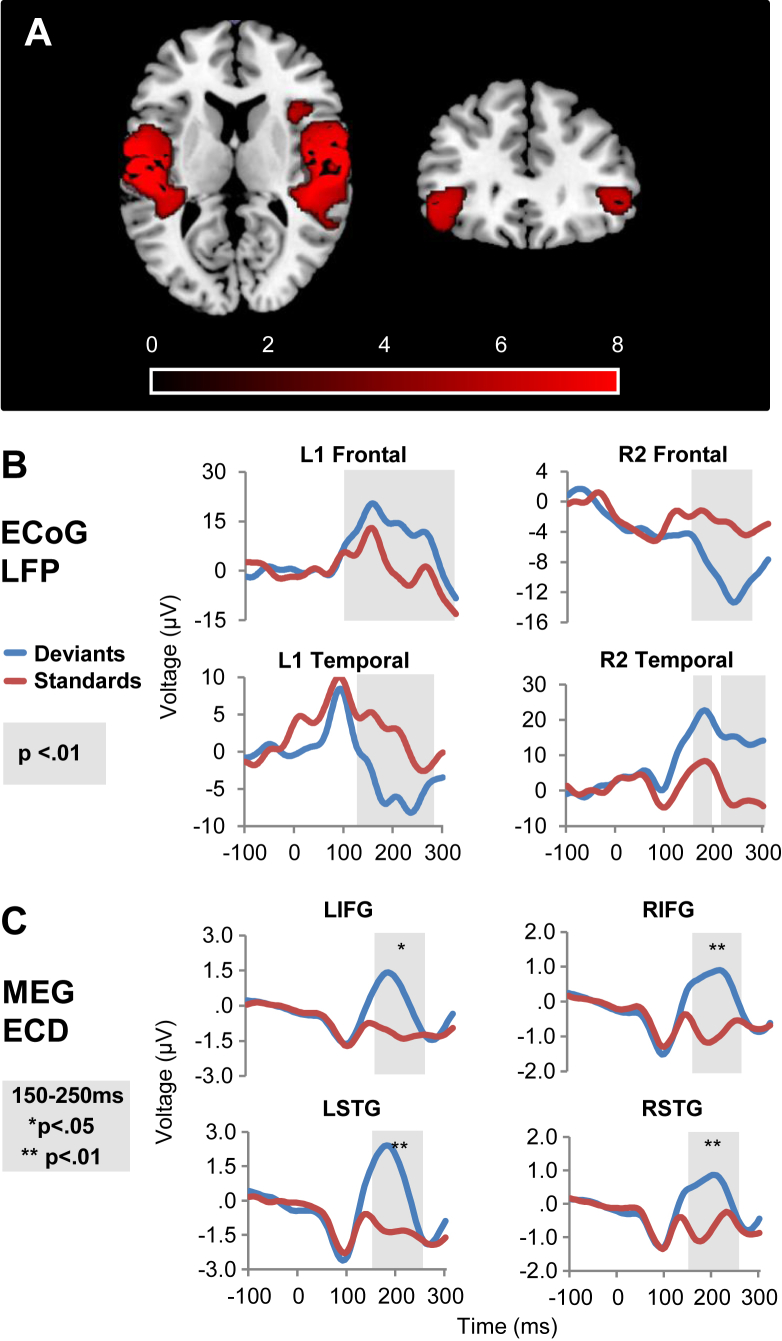
Distributed sources of the MMN from the MEG data were localised using participants' structural T1 MRI images for the forward model and the MSP algorithm for the inversion. Fig. 3A shows the group difference between standard and deviant tones using a one-sample t-test, p < .01 with FWE correction for multiple comparisons. There were significant differences between conditions (i.e., MMN) in primary A1, STG and IFG, as expected. The local field potentials for the standard and deviant conditions in the chosen ECoG electrodes are shown in Fig. 3B. The grey shaded areas show where there is a p < .01 significant difference between standard and deviant tones using a two-sample t-test across all trials, with 25 msec temporal cluster correction. All selected electrodes show significant differences during the characteristic MMN time-window of 150–250 msec. Additionally, Fig. 3C shows the group average standard and deviant dipoles at each of these electrode locations, from the healthy participants in the MEG study. The paired t-test results showed significant differences between standard and deviant group mean waveforms during the characteristic MMN time-window of 150–250 msec. Note, the differences in the sign of the waveforms between methods are likely caused by the differences between the methods of source reconstruction of dipoles using MEG and ECoG electrode recordings of local field potentials within the underlying cortex.
Fig. 3.
The source localisation of neural responses in MEG with local field potentials recorded in ECoG. A) Group MEG source localisation of MMN using multiple sparse priors inversion algorithm. There are significant MMN sources in bilateral primary auditory cortex, STG and IFG, shown in red with p < .01 and FWE correction. B) Average local field potential (LFP) for standard and deviant trials at each location for the patient with left (L1) and right (R2) electrodes respectively. Grey shading indicates significant (p < .01) differences between the conditions using a two-sample t-test with 25 msec temporal cluster threshold, comparing all trials at each time point. C) Group MEG reconstructed equivalent-current dipoles (ECD) waveforms at each of the source electrode locations used for ECoG in panel B). These coordinates are listed in text and in Fig. 2. Grey shading indicates 150–250 msec characteristic MMN time-window. *p < .05 and **p < .01 indicate significant differences between average standard and deviant conditions using a paired t-test across MEG participants.
3.3. Dynamic causal modelling
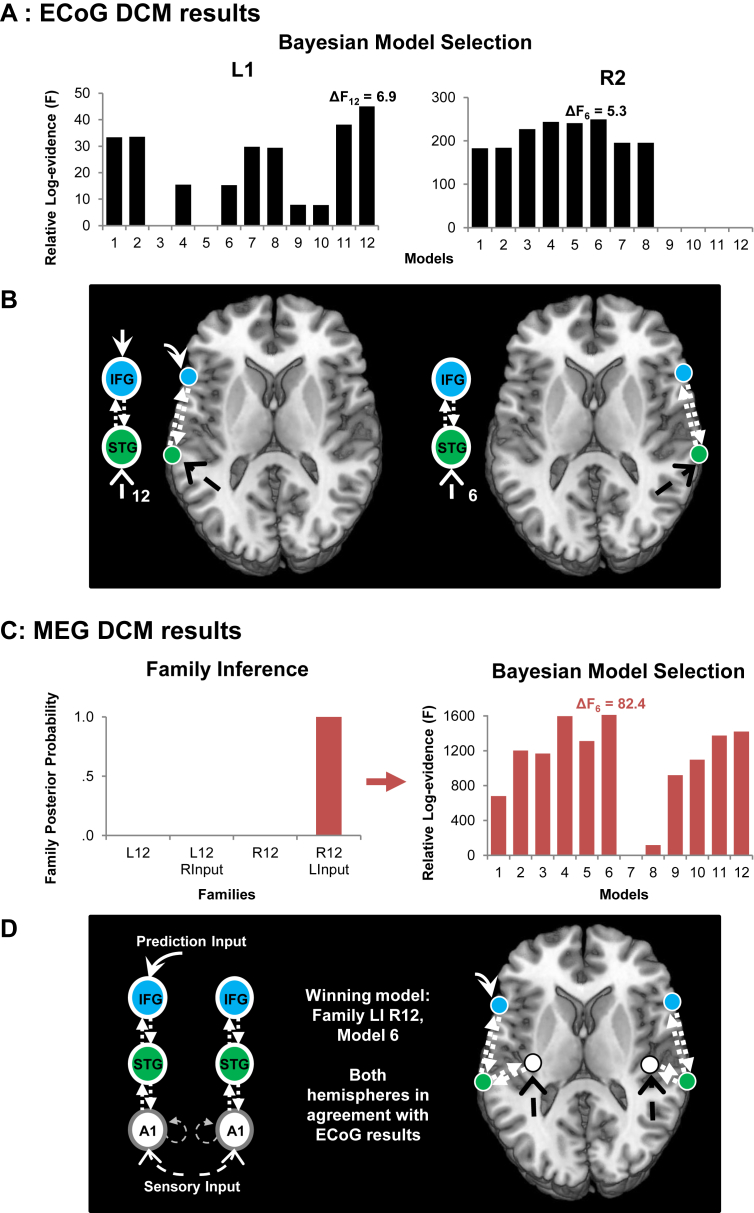
Fig. 4 shows the results of Bayesian model selection with model evidences for ECoG and MEG datasets. For all datasets, the winning model is the one with highest relative log-evidence compared to the other models tested. Fig. 4A shows model 12 is the winning model for patient L1 with Δf = 6.9, and has a posterior probability ≈ 1. Model 6 is the winning model for patient R2 with Δf = 5.3, and posterior probability ≈ 1.
Fig. 4.
Bayesian Model Selection for both patients and matched MEG. A) The model evidences for each model in the patient with let (L1) and right (R2) electrodes respectively. The model with highest relative log-evidence is the ‘winning’ model within the models used in the comparison. A difference in F between the first and second place models >5 is equivalent to a Bayes factor of 150. Model 12 ‘wins’ for patient L1 and model 6 wins for patient R2. B) These winning models are overlaid on a representative brain in MNI space. C) MEG model comparison. Taking a hierarchical approach, model families were first compared, grouping models investigating left or right changes in connection directions and modulation (L12 or R12) and grouping models where the contralateral side has inputs into the frontal sources (R/LInput) or does not. The winning family (R12 LInput) has a frontal input into the left hemisphere and variation in the direction and modulation of frontotemporal connections on the right side. Expanding this family to compare its twelve models, we find the overall winning model has modulated bidirectional connections between right frontotemporal sources but no frontal input. D) The MEG winning model. This matched the ECoG winning models for both hemispheres.
In the MEG group, we analysed the model set hierarchically, by firstly comparing model families: grouping models investigating left or right changes in connection directions and modulation (L12 or R12) and grouping models where the contralateral side has inputs into the frontal sources (L/R Input) or does not. The winning family (Fig. 4C) included models with frontal input into the left fully connected hemisphere and with variation in the direction and modulation of frontotemporal connections on the right side as shown in Fig. 1C.
We then compared the twelve models within this family (Fig. 4C). The overall winning model had modulated bidirectional connections between right frontotemporal sources but no frontal input (Δf = 82.4, posterior probability ≈1). Both sides of the MMN frontotemporal connections match the individual connectivity in each ECoG patient. All winning models have a Δf > 5, equivalent to a Bayes factor ∼150 against the second model, providing strong evidence for the winning model against all other tested models.
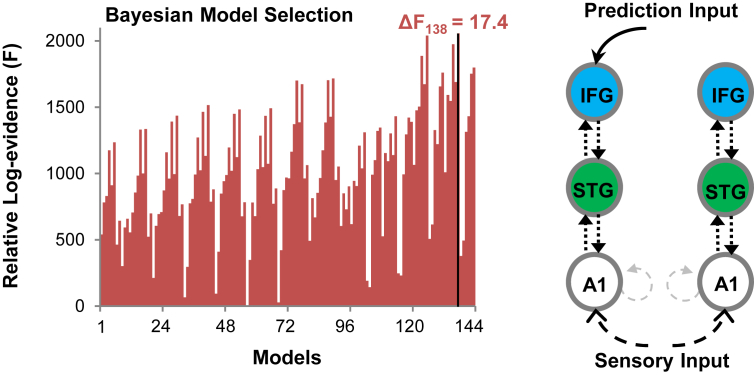
Finally, in a post hoc analysis, we varied all possible combinations of left and right connectivity resulting in 144 models to account for the family comparison always including full connectivity in one hemisphere (Fig. 5). We confirmed the same winning model as shown in Fig. 4, with Δf = 17.4 and posterior probability ≈ 1.
Fig. 5.
MEG Bayesian model selection for all combinations of left and right frontotemporal connectivity from Fig. 1. Left: Relative log-evidence for all 144 models. The corresponding posterior probability of this winning model ≈1. The winning model is number 138 indicated by the black bar, the same as illustrated in Fig. 4D, with a ΔF = 17.4 between the winning and second place model.
4. Discussion
The principal results of this study are that (i) DCM of invasive human ECoG supports the inferences derived from non-invasive MEG, providing construct validation; (ii) there is strong evidence for bilateral feedforward and feedback connections between frontal and superior temporal cortex, consistent with the predictive coding hypothesis and extending previous studies; and (iii) the expectancy input to prefrontal cortex is asymmetrical, being present on the left but not on the right according to the analysis of both modalities. The complementarity between ECoG and MEG balances the precise anatomical localisation and direct measurement of local field potentials against the ability to generalise to larger populations using inversion of safe non-invasive recordings. This is the first study to compare directly these two methods in the context of validation of DCM of task-based responses in humans, building on DCM of direct recordings in rodents and monkeys at rest.
Previous studies have identified a hierarchical network of primary A1, STG and IFG which are interpreted as supporting feedback sensory predictions and feedforward prediction errors (Chennu et al., 2013, Friston, 2005, Garrido et al., 2009b, Wacongne et al., 2011, Winkler, 2007). Several studies have examined networks that generate a MMN response, and differed slightly according to the inclusion of lateral connections and/or a left frontal source (Boly et al., 2011, Dietz et al., 2014, Garrido et al., 2009a, Garrido et al., 2008, Schmidt et al., 2013a, Schmidt et al., 2013b). These network features were brought together by Phillips et al. (2015), revealing evidence for bilateral frontotemporal feedforward and feedback connectivity across deviant dimensions, and variation in lateral connections across deviant dimensions. An important new feature was the expectancy input which can explain the activation of lower sensory areas in the partial or complete absence of an expected stimulus (Hughes et al., 2001, Raij et al., 1997, Wacongne et al., 2011). We successfully replicated the results of Phillips et al. (2015) in the current study using a larger and independent cohort of healthy adults. But, by broadening the model space we find that these expectancy signals act primarily on left prefrontal cortex.
Examination of the direct cortical recordings, with generative models using homologous nodes to the MEG dataset, confirmed modulation of bidirectional frontotemporal connections in both patients and across the group MEG participants. This is in agreement with previous studies using a singular deviant dimension such as classic and roving oddball paradigms (Dietz et al., 2014, Garrido et al., 2007a, Garrido et al., 2007b, Garrido et al., 2008). Roving paradigms use deviant and standard tones with identical physical features to discount response differences to differences in the stimuli and extract a ‘pure’ MMN response (Garrido et al., 2008). Despite the potential differences between the MMN responses in the multiple oddball paradigm used in this study and classic and roving paradigms, our results reinforce the presence of bidirectional, modulated frontotemporal connections, demonstrating the generalisation of these connections across MMN responses.
We also observed evidence for a frontal expectancy unit in the patient with left hemisphere electrodes but not in the patient with the right hemisphere electrodes. This was in agreement with the MEG group results. Previous studies suggest asymmetry of the frontal function in sensory processing (Downar, Crawley, Mikulis, & Davis, 2001), and there has been debate over the presence of a left prefrontal source for the mismatch negativity (Alho et al., 1994, Cheng et al., 2013, Giard et al., 1995, Jemel et al., 2002, Rinne et al., 2000, Rinne et al., 2005) and its inclusion in models of connectivity (Dietz et al., 2014, Garrido et al., 2009a, Garrido et al., 2008, Moran et al., 2014). However in this study as in Phillips et al. (2015) we confirmed a significant difference between standard and deviant evoked responses in the left IFG. The expectancy input asymmetry contrasts with the bilateral interactions lower in the frontotemporal hierarchy, but might be accounted by the subtle asymmetry in frontal source locations which were specified by patient electrode locations within inferior frontal and superior temporal regions of interest. Previous studies have tended to use unilateral sources (Boly et al., 2011, Cooray et al., 2014, Garrido et al., 2008, Schmidt et al., 2013a, Schmidt et al., 2013b), or impose symmetry of the expectancy input (Dietz et al., 2014, Garrido et al., 2009a, Moran et al., 2014, Phillips et al., 2015), and replication of the asymmetry of expectancy units would be helpful, from independent groups.
The location of sources is clearly critical in specifying the models. Here we used CNRs of the direct recordings to identify sources and applied these to the MEG data. Previous comparisons between ECoG and M/EEG have taken a similar approach, at least in the context of identifying the epileptogenic zone (Ding et al., 2009, Mikuni et al., 1997). There was good agreement of source location across modalities with additional validation by good outcomes after surgical resection.
Several studies have provided face validation of fMRI and electrophysiological DCM, showing the effective connectivity method correctly identifies the models behind simulated data (David et al., 2006, Friston et al., 2003, Razi et al., 2014), known connectivity and neural drivers in a rodent epilepsy model (David et al., 2008) and known connectivity changes due to anaesthesia (Moran et al., 2011). Other studies provide predictive validation by observing the same winning models across fMRI recordings in the same participants (Frassle et al., 2015, Friston et al., 2003, Rowe et al., 2010, Schuyler et al., 2010) and through the ability of stochastic fMRI DCM to predict frequency spectrum changes in simultaneous EEG recordings (Daunizeau, Lemieux, Vaudano, Friston, & Stephan, 2013).
Further studies provide construct validation of dynamical causal modelling through comparisons of winning models with other effective and functional connectivity measures (Friston et al., 2003, Papadopoulou et al., 2015) and comparisons across stochastic and spectral DCM methods (Razi et al., 2014). Additionally, multimodal imaging provides important construct validation of DCM (Daunizeau et al., 2011, Friston, 2009a) along with potential spatiotemporal resolution advantages (Riera et al., 2005, Sakkalis, 2011, Smith, 2012). For example, Papadopoulou et al. (in press) used simultaneous ECoG and EEG in a non-human primate for construct validation of steady-state DCM, showing the same winning models across modalities. Similar to our study they used reconstructed EEG sources at the same coordinates as their ECoG electrode sources.
Human multimodal effective connectivity studies have used EEG to identify epileptic seizure onset for fMRI based DCM (Murta, Leal, Garrido, & Figueiredo, 2012) and used EEG event related components to guide construction of fMRI based models (Nguyen, Breakspear, & Cunnington, 2014), but they do not compare models across modalities per se. This study is the first to provide construct validation of human evoked-response DCM through direct comparison of models across ECoG and MEG. As with Papadopoulou et al. (in press), we show good agreement across modalities for the critical features, which together with the replication of Phillips et al. (2015) indicates that DCM of human MEG is reliable.
The reliability of DCM supports its potential for clinical application. Connectivity measures complement functional and structural imaging, providing additional insights as well as greater sensitivity to disease presence, severity and treatment efficacy (Rowe, 2010). Using fMRI or M/EEG based methods, clinical applications include epilepsy (David et al., 2008), depression (de Almeida et al., 2009, Schlösser et al., 2008), Parkinson's disease (Herz et al., 2015, Michely et al., 2015, Rowe et al., 2010) and stroke rehabilitation (Grefkes & Fink, 2014). Additionally, DCM of the MMN has been used to study effective connectivity changes in neurodegenerative disease (Hughes et al., 2013), coma (Boly et al., 2011), drug effects (Schmidt et al., 2013a, Schmidt et al., 2013b) and changes in healthy ageing (Cooray et al., 2014, Moran et al., 2014, Tsvetanov et al., 2016).
Several limitations of this study arise from the nature of human invasive neuroimaging. Firstly, the two patients had drug resistant epilepsy, thus did not have healthy brains. However, we excluded electrodes that covered the epileptic foci as identified by the clinicians and electrodes that recorded epileptic spiking activity.
Secondly, electrode coverage for each patient was limited and did not provide full coverage of the six-node network modelled in previous studies (Dietz et al., 2014, Garrido et al., 2009a, Moran et al., 2014, Phillips et al., 2015). However, there is sufficient overlap, and replication of sites, to allow one to test the hypotheses related to DCM. The generative networks used for ECoG are nested within the generative models used for MEG, and we matched the coordinates in MNI space between ECoG and MEG analyses. The ECoG data cannot directly speak to the validation of the elements of the network that we do not have data for, thus we have not shown generalisation to all areas in the MMN network. This does not prevent the construct validation across modalities for the frontotemporal elements that are common to both ECoG and MEG and allow one to test the principal hypotheses related to DCM. We propose generalisation across modalities for frontotemporal feed-forward and feed-back influences in hierarchical models. Further, ECoG limitations include the electrode coverage which did not overlap across patients, thus we cannot directly compare models across patients. This is a common problem with human invasive studies. It was not practical for the patient participants to also undergo MEG.
There are also methodological considerations. We aimed to compare similar sources across methodologies, but the ECoG was from single subjects whereas MEG is a group wise analysis. Exact locations may vary. Moreover, the inversion of the lead field to a subject specific head model and the warping of this head model to standard anatomic space using coregistered MRI is common, with robust algorithms. In contrast, the gross distortion of anatomy due to the craniectomy and the presence of cortical surface electrodes may introduce normalisation difficulty. We therefore used a different method for normalisation of patient data (Blenkmann et al., 2015). We suggest however, that the spatial tolerance of source modelling is greater than the likely normalisation differences arising from the two methods.
Finally, DCM is intended for hypothesis testing and model comparison, not data driven searches amongst all possible models. Despite a large model set in comparison to many studies, other network configurations are possible. We took a structured and hierarchical approach, first identifying the most likely family of twelve models based on their shared critical features, and then the most likely model within this family. Post hoc examination across the whole set of 144 models confirmed this winning model, but this is not inevitable and future studies may also justify the preliminary identification of an optimal model family in a hierarchical approach to model selection (e.g., Boly et al., 2011, Ewbank et al., 2013, Goulden et al., 2012, Stephan et al., 2010).
In conclusion, we find strong agreement in the critical features of effective connectivity inferred from invasive and non-invasive neurophysiology, in a robust auditory oddball task. This bridges between invasive animal models and more common modes of non-invasive human neuroimaging. Both methods supported the presence of feedforward and feedback interactions in frontotemporal networks which we propose carry sensory errors and predictions respectively, in addition to left prefrontal expectancy signals.
Acknowledgements
Medical Research Council (MC-A060-5PQ30 and a Doctoral Training award to HNP), Wellcome Trust (103838 Senior Research Fellowship to JBR, LEH, Biomedical Research Fellowship WT093811MA to TAB), the James F. McDonnell Foundation 21st Century Science Initiative: Understanding Human Cognition. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) research was supported by the Biotechnology and Biological Sciences Research Council (grant number BB/H008217/1). PIDC 53/2012, PICT 0775/2012 and UNAJ investiga 2014 to AB and SK. We thank Robert Knight for his advice on processing steps in ECoG analysis.
Reviewed 6 February 2016
Action editor Emrah Duzel
Contributor Information
Holly N. Phillips, Email: holly.phillips@mrc-cbu.cam.ac.uk.
Alejandro Blenkmann, Email: ablenkmann@gmail.com.
Laura E. Hughes, Email: laura.hughes@mrc-cbu.cam.ac.uk.
Silvia Kochen, Email: skochen@retina.ar.
Tristan A. Bekinschtein, Email: tb419@cam.ac.uk.
Cam-CAN, Email: ccmanagement@cam-can.com.
James B. Rowe, Email: James.Rowe@mrc-cbu.cam.ac.uk.
References
- Alho K., Woods D.L., Algazi A., Knight R.T., Näätänen R. Lesions of frontal cortex diminish the auditory mismatch negativity. Electroencephalography and Clinical Neurophysiology. 1994;91(5):353–362. doi: 10.1016/0013-4694(94)00173-1. [DOI] [PubMed] [Google Scholar]
- de Almeida J.R.C., Versace A., Mechelli A., Hassel S., Quevedo K., Kupfer D.J. Abnormal amygdala-prefrontal effective connectivity to happy faces differentiates bipolar from major depression. Biological Psychiatry. 2009;66(5):451–459. doi: 10.1016/j.biopsych.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkmann A., Phillips H.N., Muravchik C.H., Kochen S. VI Latin American Congress on Biomedical Engineering CLAIB 2014. Springer International Publishing; 2015. Grid and depth intracranial electrodes localization in a normalized space using MRI and CT images; pp. 413–416. [Google Scholar]
- Boly M., Garrido M.I., Gosseries O., Bruno M.-A., Boveroux P., Schnakers C. Preserved feedforward but impaired top-down processes in the vegetative state. Science. 2011;332(6031):858–862. doi: 10.1126/science.1202043. [DOI] [PubMed] [Google Scholar]
- Carlin J.D., Calder A.J., Kriegeskorte N., Nili H., Rowe J.B. A head view-invariant representation of gaze direction in anterior superior temporal sulcus. Current Biology. 2011;21(21):1817–1821. doi: 10.1016/j.cub.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.H., Baillet S., Hsiao F.-J., Lin Y.-Y. Effects of aging on neuromagnetic mismatch responses to pitch changes. Neuroscience Letters. 2013;544:20–24. doi: 10.1016/j.neulet.2013.02.063. [DOI] [PubMed] [Google Scholar]
- Chennu S., Noreika V., Gueorguiev D., Blenkmann A., Kochen S., Ibáñez A. Expectation and attention in hierarchical auditory prediction. The Journal of Neuroscience. 2013;33(27):11194–11205. doi: 10.1523/JNEUROSCI.0114-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray G., Garrido M.I., Hyllienmark L., Brismar T. A mechanistic model of mismatch negativity in the ageing brain. Clinical Neurophysiology. 2014;125(9):1774–1782. doi: 10.1016/j.clinph.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Cui X., Bray S., Reiss A.L. Functional Near Infrared Spectroscopy (NIRS) signal improvement based on negative correlation between oxygenated and deoxygenated hemoglobin dynamics. NeuroImage. 2010;49(4):1199–1216. doi: 10.1016/j.neuroimage.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Sereno M.I. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: A linear approach. Journal of Cognitive Neuroscience. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Daunizeau J., David O., Stephan K.E. Dynamic causal modelling: A critical review of the biophysical and statistical foundations. NeuroImage. 2011;58(2):312–322. doi: 10.1016/j.neuroimage.2009.11.062. [DOI] [PubMed] [Google Scholar]
- Daunizeau J., Lemieux L., Vaudano A.E., Friston K.J., Stephan K.E. An electrophysiological validation of stochastic DCM for fMRI. Frontiers in Computational Neuroscience. 2013;6(January):103. doi: 10.3389/fncom.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O., Guillemain I., Saillet S., Reyt S., Deransart C., Segebarth C. Identifying neural drivers with functional MRI: An electrophysiological validation. PLoS Biology. 2008;6(12):2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O., Kiebel S.J., Harrison L.M., Mattout J., Kilner J.M., Friston K.J. Dynamic causal modeling of evoked responses in EEG and MEG. NeuroImage. 2006;30(4):1255–1272. doi: 10.1016/j.neuroimage.2005.10.045. [DOI] [PubMed] [Google Scholar]
- Dietz M.J., Friston K.J., Mattingley J.B., Roepstorff A., Garrido M.I. Effective connectivity reveals right-hemisphere dominance in audiospatial perception: Implications for models of spatial neglect. The Journal of Neuroscience. 2014;34(14):5003–5011. doi: 10.1523/JNEUROSCI.3765-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Wilke C., Xu B., Xu X., Drongelene W., He B. EEG source imaging: Correlate source locations and extents with ECoG and surgical resections in epilepsy patients. Journal of Clinical Neurophysiology. 2009;24(2):130–136. doi: 10.1097/WNP.0b013e318038fd52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeller C.F., Opitz B., Mecklinger A., Krick C., Reith W., Schröger E. Prefrontal cortex involvement in preattentive auditory deviance detection: Neuroimaging and electrophysiological evidence. NeuroImage. 2003;20(2):1270–1282. doi: 10.1016/S1053-8119(03)00389-6. [DOI] [PubMed] [Google Scholar]
- Downar J., Crawley A.P., Mikulis D.J., Davis K.D. The effect of task relevance on the cortical response to changes in visual and auditory stimuli: An event-related fMRI study. NeuroImage. 2001;14(6):1256–1267. doi: 10.1006/nimg.2001.0946. [DOI] [PubMed] [Google Scholar]
- Ewbank M.P., Henson R.N., Rowe J.B., Stoyanova R.S., Calder A.J. Different neural mechanisms within occipitotemporal cortex underlie repetition suppression across same and different-size faces. Cerebral Cortex. 2013;23(5):1073–1084. doi: 10.1093/cercor/bhs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank M.P., Lawson R.P., Henson R.N., Rowe J.B., Passamonti L., Calder A.J. Changes in “Top-Down” connectivity underlie repetition suppression in the ventral visual pathway. The Journal of Neuroscience. 2011;31(15):5635–5642. doi: 10.1523/JNEUROSCI.5013-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Dale A.M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frassle S., Stephan K.E., Friston K.J., Steup M., Krach S., Paulus F.M. Test-retest reliability of dynamic causal modeling for fMRI. NeuroImage. 2015;117:56–66. doi: 10.1016/j.neuroimage.2015.05.040. [DOI] [PubMed] [Google Scholar]
- Friston K.J. A theory of cortical responses. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2005;360(1456):815–836. doi: 10.1098/rstb.2005.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J. Causal modelling and brain connectivity in functional magnetic resonance imaging. PLoS Biology. 2009;7(2):0220–0225. doi: 10.1371/journal.pbio.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J. The free-energy principle: A rough guide to the brain? Trends in Cognitive Sciences. 2009;13(7):293–301. doi: 10.1016/j.tics.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Harrison L.M., Daunizeau J., Kiebel S.J., Phillips C., Trujillo-Barreto N. Multiple sparse priors for the M/EEG inverse problem. NeuroImage. 2008;39(3):1104–1120. doi: 10.1016/j.neuroimage.2007.09.048. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Harrison L.M., Penny W.D. Dynamic causal modelling. NeuroImage. 2003;19(4):1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Holmes A.P., Worsley K.J. How many subjects constitute a study? NeuroImage. 1999;10(1):1–5. doi: 10.1006/nimg.1999.0439. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Kiebel S.J. Predictive coding under the free-energy principle. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2009;364(1521):1211–1221. doi: 10.1098/rstb.2008.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M.I., Friston K.J., Kiebel S.J., Stephan K.E., Baldeweg T., Kilner J.M. The functional anatomy of the MMN: A DCM study of the roving paradigm. NeuroImage. 2008;42(2):936–944. doi: 10.1016/j.neuroimage.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M.I., Kilner J.M., Kiebel S.J., Friston K.J. Evoked brain responses are generated by feedback loops. Proceedings of the National Academy of Sciences. 2007;104(52):20961–20966. doi: 10.1073/pnas.0706274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M.I., Kilner J.M., Kiebel S.J., Friston K.J. Dynamic causal modeling of the response to frequency deviants. Journal of Neurophysiology. 2009;101(5):2620–2631. doi: 10.1152/jn.90291.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M.I., Kilner J.M., Kiebel S.J., Stephan K.E., Friston K.J. Dynamic causal modelling of evoked potentials: A reproducibility study. NeuroImage. 2007;36(3):571–580. doi: 10.1016/j.neuroimage.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M.I., Kilner J.M., Stephan K.E., Friston K.J. The mismatch negativity: A review of underlying mechanisms. Clinical Neurophysiology. 2009;120(3):453–463. doi: 10.1016/j.clinph.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giard M.H., Lavikahen J., Reinikainen K., Perrin F., Bertrand O., Pernier J. Separate representation of stimulus frequency, intensity, and duration in auditory sensory memory: An event-related potential and dipole-model analysis. Journal of Cognitive Neuroscience. 1995;7(2):133–143. doi: 10.1162/jocn.1995.7.2.133. [DOI] [PubMed] [Google Scholar]
- Goulden N., Elliott R., Suckling J., Williams S.R., Deakin J.F.W., McKie S. Sample size estimation for comparing parameters using dynamic causal modeling. Brain Connectivity. 2012;2(2):80–90. doi: 10.1089/brain.2011.0057. [DOI] [PubMed] [Google Scholar]
- Grefkes C., Fink G.R. Connectivity-based approaches in stroke and recovery of function. The Lancet Neurology. 2014;13(2):206–216. doi: 10.1016/S1474-4422(13)70264-3. [DOI] [PubMed] [Google Scholar]
- Herz D.M., Haagensen B.N., Christensen M.S., Madsen K.H., Rowe J.B., Løkkegaard A. Abnormal dopaminergic modulation of striato-cortical networks underlies levodopa-induced dyskinesias in humans. Brain: A Journal of Neurology. 2015;138(6):1658–1666. doi: 10.1093/brain/awv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H.C., Darcey T.M., Barkan H.I., Williamson P.D., Roberts D.W., Aslin C.H. Responses of human auditory association cortex to the omission of an expected acoustic event. NeuroImage. 2001;13(6 Pt 1):1073–1089. doi: 10.1006/nimg.2001.0766. [DOI] [PubMed] [Google Scholar]
- Hughes L., Ghosh B.C.P., Rowe J.B. Reorganisation of brain networks in frontotemporal dementia and progressive supranuclear palsy. NeuroImage: Clinical. 2013;2:459–468. doi: 10.1016/j.nicl.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L., Rowe J.B. The impact of neurodegeneration on network connectivity: A study of change detection in frontotemporal dementia. Journal of Cognitive Neuroscience. 2013;25(5):802–813. doi: 10.1162/jocn_a_00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez A., Cardona J.F., Dos Santos Y.V., Blenkmann A., Aravena P., Roca M. Motor-language coupling: Direct evidence from early Parkinson's disease and intracranial cortical recordings. Cortex. 2013;49(4):968–984. doi: 10.1016/j.cortex.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Jemel B., Achenbach C., Müller B.W., Röpcke B., Oades R.D. Mismatch negativity results from bilateral asymmetric dipole sources in the frontal and temporal lobes. Brain Topography. 2002;15(1):13–27. doi: 10.1023/a:1019944805499. [DOI] [PubMed] [Google Scholar]
- Kass R.E., Raftery A.E. Bayes factors. Journal of the American Statistical Association. 1995 [Google Scholar]
- Kiebel S.J., Daunizeau J., Friston K.J. A hierarchy of time-scales and the brain. PLoS Computational Biology. 2008;4(11):e1000209. doi: 10.1371/journal.pcbi.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebel S.J., Daunizeau J., Phillips C., Friston K.J. Variational Bayesian inversion of the equivalent current dipole model in EEG/MEG. NeuroImage. 2008;39(2):728–741. doi: 10.1016/j.neuroimage.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Kiebel S.J., David O., Friston K.J. Dynamic causal modelling of evoked responses in EEG/MEG with lead field parameterization. NeuroImage. 2006;30(4):1273–1284. doi: 10.1016/j.neuroimage.2005.12.055. [DOI] [PubMed] [Google Scholar]
- Kiebel S.J., Garrido M.I., Moran R.J., Chen C.-C., Friston K.J. Dynamic causal modeling for EEG and MEG. Human Brain Mapping. 2009;30(6):1866–1876. doi: 10.1002/hbm.20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieder F., Stephan K.E., Daunizeau J., Garrido M.I., Friston K.J. A neurocomputational model of the mismatch negativity. PLoS Computational Biology. 2013;9(11):e1003288. doi: 10.1371/journal.pcbi.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean S.E., Ward L.M. Temporo-frontal phase synchronization supports hierarchical network for mismatch negativity. Clinical Neurophysiology. 2014;125(8):1604–1617. doi: 10.1016/j.clinph.2013.12.109. [DOI] [PubMed] [Google Scholar]
- Michely J., Volz L.J., Barbe M.T., Hoffstaedter F., Viswanathan S., Timmermann L. Dopaminergic modulation of motor network dynamics in Parkinson's disease. Brain: A Journal of Neurology. 2015;138(3):664–678. doi: 10.1093/brain/awu381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni N., Nagamine T., Ikeda A., Terada K., Taki W., Kimura J. Simultaneous recording of epileptiform discharges by MEG and subdural electrodes in temporal lobe epilepsy. NeuroImage. 1997;5(4 Pt 1):298–306. doi: 10.1006/nimg.1997.0272. [DOI] [PubMed] [Google Scholar]
- Molholm S., Martinez A., Ritter W., Javitt D.C., Foxe J.J. The neural circuitry of pre-attentive auditory change-detection: An fMRI study of pitch and duration mismatch negativity generators. Cerebral Cortex. 2005;15(5):545–551. doi: 10.1093/cercor/bhh155. [DOI] [PubMed] [Google Scholar]
- Moran R.J., Jung F., Kumagai T., Endepols H., Graf R., Dolan R.J. Dynamic causal models and physiological inference: A validation study using isoflurane anaesthesia in rodents. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0022790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran R.J., Symmonds M., Dolan R.J., Friston K.J. The brain ages optimally to model its environment: Evidence from sensory learning over the adult lifespan. PLoS Computational Biology. 2014;10(1):e1003422. doi: 10.1371/journal.pcbi.1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murta T., Leal A., Garrido M.I., Figueiredo P. Dynamic Causal Modelling of epileptic seizure propagation pathways: A combined EEG-fMRI study. NeuroImage. 2012;62(3):1634–1642. doi: 10.1016/j.neuroimage.2012.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näätänen R., Paavilainen P., Tiitinen H., Jiang D., Alho K. Attention and mismatch negativity. Psychophysiology. 1993;30(5):436–450. doi: 10.1111/j.1469-8986.1993.tb02067.x. [DOI] [PubMed] [Google Scholar]
- Näätänen R., Pakarinen S., Rinne T., Takegata R. The mismatch negativity (MMN): Towards the optimal paradigm. Clinical Neurophysiology. 2004;115(1):140–144. doi: 10.1016/j.clinph.2003.04.001. [DOI] [PubMed] [Google Scholar]
- Nguyen V.T., Breakspear M., Cunnington R. Fusing concurrent EEG-fMRI with dynamic causal modeling: Application to effective connectivity during face perception. NeuroImage. 2014;102:60–70. doi: 10.1016/j.neuroimage.2013.06.083. [DOI] [PubMed] [Google Scholar]
- Oceák A., Winkler I., Sussman E., Alho K. Loudness summation and the mismatch negativity event-related brain potential in humans. Psychophysiology. 2006;43(1):13–20. doi: 10.1111/j.1469-8986.2006.00372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B., Rinne T., Mecklinger A., Von Cramon D.Y., Schröger E. Differential contribution of frontal and temporal cortices to auditory change detection: fMRI and ERP results. NeuroImage. 2002;15(1):167–174. doi: 10.1006/nimg.2001.0970. [DOI] [PubMed] [Google Scholar]
- Papadopoulou M., Friston K.J., Marinazzo D. Estimating directed connectivity from cortical recordings and reconstructed sources. Brain Topography. 2015:1–19. doi: 10.1007/s10548-015-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny W.D., Stephan K.E., Mechelli A., Friston K.J. Comparing dynamic causal models. NeuroImage. 2004;22(3):1157–1172. doi: 10.1016/j.neuroimage.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Phillips H.N., Blenkmann A., Hughes L., Bekinschtein T.A., Rowe J.B. Hierarchical Organization of frontotemporal networks for the prediction of stimuli across multiple dimensions. The Journal of Neuroscience. 2015;35(25):9255–9264. doi: 10.1523/JNEUROSCI.5095-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princich J.P., Wassermann D., Latini F., Oddo S., Blenkmann A., Seifer G. Rapid and efficient localization of depth electrodes and cortical labeling using free and open source medical software in epilepsy surgery candidates. Frontiers in Neuroscience. 2013;(7 DEC) doi: 10.3389/fnins.2013.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raij T., McEvoy L.K., Mäkelä J.P., Hari R. Human auditory cortex is activated by omissions of auditory stimuli. Brain Research. 1997;745(1–2):134–143. doi: 10.1016/s0006-8993(96)01140-7. [DOI] [PubMed] [Google Scholar]
- Rao R.P.N., Ballard D.H. Predictive coding in the visual cortex: A functional interpretation of some extra-classical receptive-field effects. Nature Neuroscience. 1999;2(1):79–87. doi: 10.1038/4580. [DOI] [PubMed] [Google Scholar]
- Razi A., Kahan J., Rees G., Friston K.J. Construct validation of a DCM for resting state fMRI. NeuroImage. 2014;106:1–14. doi: 10.1016/j.neuroimage.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera J., Aubert E., Iwata K., Kawashima R., Wan X., Ozaki T. Fusing EEG and fMRI based on a bottom-up model: Inferring activation and effective connectivity in neural masses. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2005;360(1457):1025–1041. doi: 10.1098/rstb.2005.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne T., Alho K., Ilmoniemi R.J., Virtanen J., Näätänen R. Separate time behaviors of the temporal and frontal mismatch negativity sources. NeuroImage. 2000;12(1):14–19. doi: 10.1006/nimg.2000.0591. [DOI] [PubMed] [Google Scholar]
- Rinne T., Degerman A., Alho K. Superior temporal and inferior frontal cortices are activated by infrequent sound duration decrements: An fMRI study. NeuroImage. 2005;26(1):66–72. doi: 10.1016/j.neuroimage.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Rowe J.B. Connectivity analysis is essential to understand neurological disorders. Frontiers in Systems Neuroscience. 2010;4(September):1–13. doi: 10.3389/fnsys.2010.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe J.B., Hughes L., Barker R.A., Owen A.M. Dynamic causal modelling of effective connectivity from fMRI: Are results reproducible and sensitive to Parkinson's disease and its treatment? NeuroImage. 2010;52(3):1015–1026. doi: 10.1016/j.neuroimage.2009.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkalis V. Review of advanced techniques for the estimation of brain connectivity measured with EEG/MEG. Computers in Biology and Medicine. 2011;41(12):1110–1117. doi: 10.1016/j.compbiomed.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Schlösser R.G.M., Wagner G., Koch K., Dahnke R., Reichenbach J.R., Sauer H. Fronto-cingulate effective connectivity in major depression: A study with fMRI and dynamic causal modeling. NeuroImage. 2008;43(3):645–655. doi: 10.1016/j.neuroimage.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Diaconescu A.O., Kometer M., Friston K.J., Stephan K.E., Vollenweider F.X. Modeling ketamine effects on synaptic plasticity during the mismatch negativity. Cerebral Cortex. 2013;23(10):2394–2406. doi: 10.1093/cercor/bhs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Leventhal D.K., Mallet N., Chen F., Berke J.D. Canceling actions involves a race between basal ganglia pathways. Nature Neuroscience. 2013;16(8):1118–1124. doi: 10.1038/nn.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler B., Ollinger J.M., Oakes T.R., Johnstone T., Davidson R.J. Dynamic Causal Modeling applied to fMRI data shows high reliability. NeuroImage. 2010;49(1):603–611. doi: 10.1016/j.neuroimage.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafto M.A., Tyler L.K., Dixon M., Taylor J.R., Rowe J.B., Cusack R. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) study protocol: A cross-sectional, lifespan, multidisciplinary examination of healthy cognitive ageing. BMC Neurology. 2014;14(204) doi: 10.1186/s12883-014-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M. The future of FMRI connectivity. NeuroImage. 2012;62(2):1257–1266. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Stephan K.E., Penny W.D., Moran R.J., den Ouden H.E.M., Daunizeau J., Friston K.J. Ten simple rules for dynamic causal modeling. NeuroImage. 2010;49(4):3099–3109. doi: 10.1016/j.neuroimage.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanov K.A., Henson R.N.A., Tyler L.K., Razi A., Geerligs L., Ham T. Extrinsic and intrinsic brain network connectivity maintains cognition across the lifespan despite accelerated decay of regional brain activation with age. The Journal of Neuroscience. 2016 doi: 10.1523/JNEUROSCI.2733-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacongne C., Labyt E., van Wassenhove V., Bekinschtein T.A., Naccache L., Dehaene S. Evidence for a hierarchy of predictions and prediction errors in human cortex. Proceedings of the National Academy of Sciences. 2011;108(51):20754–20759. doi: 10.1073/pnas.1117807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T.D., Keller M.C., Lacey S.C., Jonides J. Increased sensitivity in neuroimaging analyses using robust regression. NeuroImage. 2005;26(1):99–113. doi: 10.1016/j.neuroimage.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Winkler I. Interpreting the mismatch negativity. Journal of Psychophysiology. 2007;21(3):147–163. [Google Scholar]