Abstract
Angiostrongylus cantonensis is a zoonotic nematode parasite causing human eosinophilic meningitis (or meningoencephalitis) worldwide. A closely related species, Angiostrongylus malaysiensis, might also be a human pathogen. Larvae were obtained from land snails in Lao PDR, Cambodia, Myanmar and Thailand. We sequenced two nuclear gene regions (nuclear ribosomal ITS2 and SSU rRNA) and a portion of one mitochondrial gene (COI) from these larvae. Angiostrongylus cantonensis and A. malaysiensis were identified. This is the first report of the molecular identification of the two Angiostrongylus species in Lao PDR, Cambodia and Myanmar. The regional distributions of the two species broadly overlap. Phylogenetic relationships were inferred including data from Angiostrongylus species deposited in public databases. All the gene regions we sequenced have potential value in distinguishing between species of Angiostrongylus. The COI gene exhibited the greatest intraspecific variation in the study region (five haplotypes in A. cantonensis and four in A. malaysiensis) and might be suitable for more detailed phylogeographic studies.
Introduction
The genus Angiostrongylus contains nematodes parasitic in rodents and carnivores, in which they are found in the mesenteric or pulmonary arteries and lungs [1]. Angiostrongylus cantonensis is a primary cause of human eosinophilic meningitis or meningoencephalitis in many areas of the world: more than 2800 cases have been documented worldwide [2–6]. Humans are incidental hosts who become infected by eating infective larvae in snails, slugs, paratenic hosts or contaminated vegetables [4,7]. Additional species have been reported. Angiostrongylus costaricensis causes abdominal angiostrongyliasis in Latin American countries [8]. Angiostrongylus mackerrasae and Angiostrongylus malaysiensis may also be pathogenic in humans [9]. Identification of Angiostrongylus species based on morphological characters is difficult due to vague and similar descriptions of size and body shapes among different species [1,10,11]. Molecular methods, based on PCR-direct sequencing of the nuclear small subunit ribosomal RNA (SSU rRNA) gene, now provide a useful approach for identification of Angiostrongylus species [12]. The mitochondrial cytochrome c subunit I (COI) gene and the nuclear ribosomal internal transcribed spacer 2 (ITS2) region have also been used for constructing phylogenetic trees to differentiate geographical isolates of A. cantonensis, A. malaysiensis, A. costaricensis and Angiostrongylus vasorum [13–17]. Despite several reports of angiostrongyliasis in Thailand, confirmed using molecular methods [2], there have been no reports on molecular identification of Angiostrongylus species in Lao PDR, Myanmar and Cambodia, countries adjacent to Thailand in the Greater Mekong subregion. Therefore, in the present study, we characterized the DNA regions of SSU rRNA, ITS2 and COI of A. cantonensis and A. malaysiensis from these countries and their phylogenetic relationships with parasites recovered from Thailand elsewhere. Intra- and interspecific variation are discussed.
Materials and Methods
Angiostrongylus worms
Third-stage Angiostrongylus larvae (L3) recovered from land snails in Thailand came from the Northeast, Khon Kaen Province, the North, Phrae Province, and the South, Surat Thani and Patthalung Provinces. The L3 larvae from Myanmar were recovered from Achatina fulica collected from Mongla Township, Shan State, eastern Myanmar. The L3 larvae from Laos were recovered from Cryptozona siamensis from Vientiane, central Lao PDR. The L3 larvae from Cambodia were recovered from Ac. fulica from Siem Reap Province, northwestern Cambodia (kindly provided by Dr. Chivorn Leang, Phnom Penh, Cambodia). All snails were bought from daily food markets in each of these countries. The collection localities and host species of Angiostrongylus samples are presented in Fig 1. An adult A. cantonensis of the Hawaiian strain (kindly provided by Professor Yukifumi Nawa, Research Affairs, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand) was used as the reference sample from outside Asian countries. Information about samples is in Table 1. The parasite samples were kept in 75% ethanol at -70°C until used. These data do not report any studies with animals performed by any of the authors. We confirm that the process did not involve endangered or protected species.
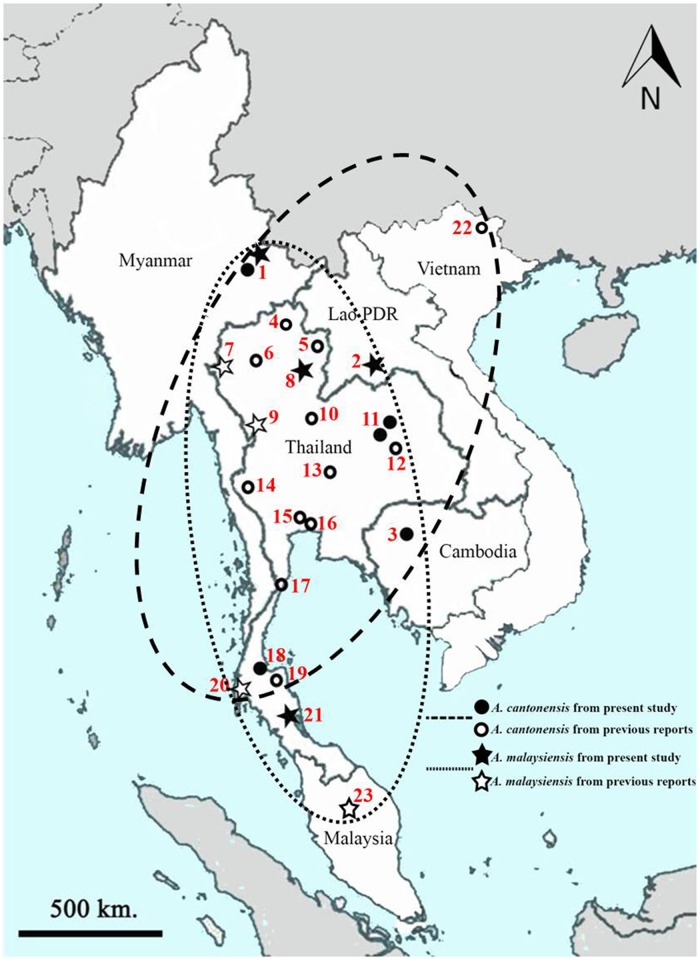
Fig 1. Map of sampling locations of Angiostrongylus cantonensis and Angiostrongylus malaysiensis showing their overlapping distributions in six countries.
Stars indicate A. malaysiensis; circles indicate A. cantonensis; the dotted line shows inferred range of A. malaysiensis; the dashed line shows range of A. cantonensis in the region. 1, Mongla Township in Myanmar (present study); 2, Vientiane capital city in Lao PDR (present study); 3, Siem Reap Province of Cambodia (present study); 4–21, Provinces in Thailand (4, Chiang Rai (KP721449); 5, Nan (KC995246); 6, Chiang Mai (KP721447); 7, Mae Hong Sorn (KP732099); 8, Phrae (present study); 9, Tak (KP732097); 10, Phitsanulok (KP721445); 11, Khon Kaen (present study); 12, Maha-Sarakham (KC995223); 13, Lopburi (KC995212); 14, Kanchanaburi (KC995206); 15, Bangkok (KC995188); 16, Samut Prakan (KP721442); 17, Prachuap Khiri Khan (KC995262); 18, Surat Thani (present study); 19, Nakhon Si Thammarat (KC995260); 20, Phang Nga (KP732100); 21, Phattalung (present study)); 22, Cao Bang Province of Vietnam (De et al [18]); 23, Peninsular Malaysia (JN663729).
Table 1. Angiostrongylus worms used in the present study and the mitochondrial COI haplotypes found.
| Locality | Worm species | No specimens * | Source host | Date | GenBank no. of COI gene | COI Haplotypes |
|---|---|---|---|---|---|---|
| Mancha Khiri, Khon Kaen, Thailand | A. cantonensis | 1 | Achatina fulica | 2013 | KU532143 | AC11 |
| Mueang, Khon Kaen, Thailand | A. cantonensis | 1 | Ac. fulica | 2010 | KU532147 | AC10 |
| Surat Thani, Thailand | A. cantonensis | 1 | Ac. fulica | 2014 | KU532146 | AC13 |
| Siem Reap, Cambodia | A. cantonensis | 10 | Ac. fulica | 2015 | KU532148 | AC12 |
| Mongla, Shan State, Myanmar | A. cantonensis | 7 | Ac. fulica | 2013 | KU532145 | AC2 |
| Hawaii, United States | A. cantonensis | 1 | Experimentally maintained rat | NA | KU532144 | AC5 |
| Phatthalung, Thailand | A. malaysiensis | 3 | Ac. fulica | 2014 | KU532152 | AM1 |
| Phrae, Thailand | A. malaysiensis | 2 | Cryptozona siamensis | 2014 | KU532151 | AM3 |
| Vientiane, Lao PDR | A. malaysiensis | 2 | C. siamensis | 2015 | KU532153 | AM3 |
| Vientiane, Lao PDR | A. malaysiensis | 8 | C. siamensis | 2015 | KU532154 | AM4 |
| Mongla, Shan State, Myanmar | A. malaysiensis | 1 | Ac. fulica | 2013 | KU532149 | AM3 |
| Mongla, Shan State, Myanmar | A. malaysiensis | 2 | Ac. fulica | 2013 | KU532150 | AM2 |
NA, not available.
*All specimens were third stage larvae, except for one adult (Hawaii isolate).
DNA extraction, PCR amplification and DNA sequencing
Extraction of genomic DNA from each Angiostrongylus sample was done using a Genomic DNA Mini Kit (Macherey-Nagel GmbH & Co., Duren, Germany) according to the manufacturer’s instructions. Specific primers and regions amplified are listed in Table 2. All polymerase chain reactions (PCRs) were done using a GeneAmp PCR System 9700 (Applied Biosystems, Singapore). The reaction mixture was prepared in a total volume of 25 μL containing 2.5 μL of 10x high fidelity PCR buffer, 2.0 μL MgCl2 (25 mM), 0.5μL of dNTP mix (10 mM), 0.5 μL of each primer (10 μM), 0.125 U of Taq high-fidelity PCR system (Roche Applied Science, Mannheim, Germany), adjusted to 25 μL with deionized water and 5 μL of DNA extracted from an individual worm. For the SSU rRNA and the COI amplifications, the PCR conditions were initial denaturation at 94°C for 2 min, followed by 10 cycles of denaturation at 94°C for 1 min, annealing at 40°C for 1 min and extension at 68°C for 2 min, then 30 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 2 min, extension at 68°C for 2 min and a final extension at 68°C for 7 min. For the ITS2 amplification, the PCR conditions were initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec and final extension at 72°C for 7 min. Amplified products were separated by electrophoresis on a 1.0% (w/v) agarose gel, stained with ethidium bromide, and visualized under ultraviolet light. DNA sequencing was conducted at First BASE Laboratories Sdn Bhd (Selangor, Malaysia) using the BigDye terminator v3.1 cycle-sequencing kit (ABI), and both strands were directly sequenced using the PCR primers as sequencing primers (Model 310 or 3100, Applied Biosystems).
Table 2. The specific primers used in the present study.
| Gene regions | Primers | Approximate amplicon size |
|---|---|---|
| SSU rRNA | Angi18S-1_forward 5’-AAAGTTAAGCCATGCATG-3’ Angi18S-2_reverse 5’-CATTCTTGGCAAATGCTTTCG-3’ [17] | 885 bp |
| COI | AngiCOI_forward 5’-TTTTTTGGGCATCCTGAGGTTTAT-3’ AngiCOI_reverse 5’-CGAGGATAACCATGTAAACCAGC-3’ (newly designed primers) | 605 bp |
| ITS2 | AngiITS2_ forward 5' ACATCTGGTTCAGGGTTGTT 3' AngiITS2_ reverse 5' AGCATACAAGCACATGATCAC 3' (newly designed primers) | 395 bp |
Sequence alignment and Phylogenetic analysis
The sequences were aligned and compared with those of Angiostrongylus species deposited in the GenBank database using the multiple sequence alignment program ClustalW available within BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) [19]. Phylogenetic trees were constructed using the maximum-likelihood (ML), maximum-parsimony (MP) and neighbour-joining (NJ) methods implemented in MEGA6 [20]. The substitution model for each dataset was chosen using the Bayesian Information Criterion (BIC) in MEGA6 software: the lowest BIC score is considered to best describe the substitution pattern [20, 21]. Models used were the Kimura two-parameter (K2) model (SSU rRNA alignment), Tamura three-parameter (T92) model (ITS2 alignment) and the General Time Reversible model allowing for a proportion of invariable sites (GTR+I) (COI alignment). Bootstrap percentages were computed using 1000 pseudoreplications. Bayesian analyses were performed using MrBayes v3.2. [22]. MrBayes implements a more limited number of substitution models than does MEGA6. The available model with the closest BIC score was therefore chosen. For the SSU rRNA alignment, this was the Hasegawa-Kishino-Yano model (HKY). The SSU analysis was run for three million generations (2 runs, each of four chains), by which time the standard deviation of split frequencies had fallen below 0.01, and sampled every 1000 generations. Examination of the output (using the “sump” command) indicated that the potential scale reduction factor was 1 for relevant parameters and that stationarity had been approached after 20% of generations. The first 20% of trees were therefore discarded as burnin (following recommendations in the MrBayes manual). For ITS2, the best model was the HKY model allowing for a proportion of invariable sites (HKY+I). Following the approach outlined above for the SSU alignment, the analysis was run for two million generations and the first 25% of trees discarded as burnin. For COI, given that this is a protein-coding gene and that each codon position might evolve in a different way, a different approach was used. Partition Finder v1 [23] indicated that the same substitution model should be applied to each codon position and that, for implementation in MrBayes, this model was the GTR+I. The analysis was run for five million generations and sampled every 1000 generations. The first 20% of trees were discarded as burnin. In all Bayesian analyses, consensus trees were generated using the command “contype = allcompat”, which adds all compatible groups to a 50% majority-rule tree. The COI haplotypes were numbered following Monte et al [16].
Results
A total of 39 Angiostrongylus specimens were used for PCR amplification and DNA sequencing (Table 1). A single representative of each unique nucleotide sequence from each collection site was retained for phylogenetic analysis (Fig 1). Ten new partial SSU rRNA and ITS2 sequences and 12 new partial COI sequences were used for phylogenetic analyses (GenBank accession numbers KU528678-KU528687, KU528688-KU528697 and KU532143-KU532154, respectively). The distributions of A. cantonensis and A. malaysiensis were found to overlap broadly in the region.
Small subunit ribosomal RNA gene
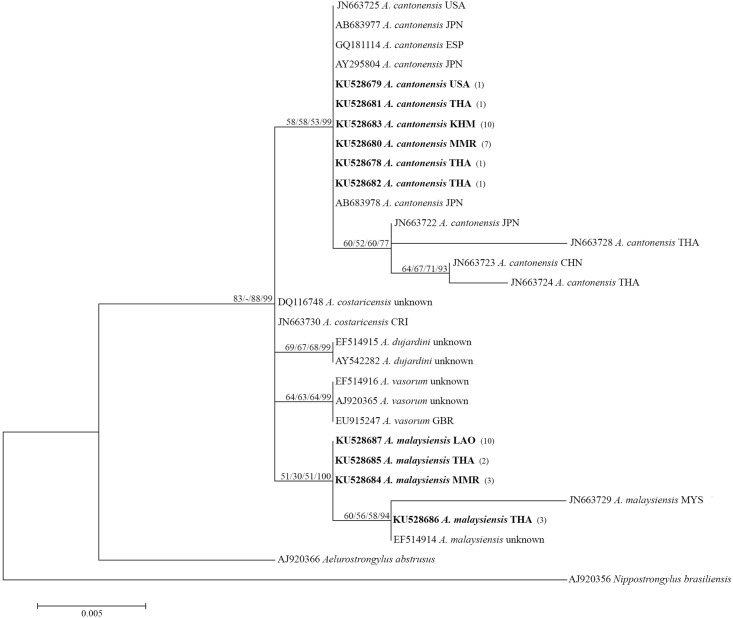
The two major species were clearly distinguished according to SSU rRNA sequences, as shown in Fig 2. Bootstrap/Bayesian posterior probability values of 58/58/53/99% and 51/30/51/100% supported A. cantonensis and A. malaysiensis, respectively. All new A. cantonensis SSU rRNA sequences were almost identical, regardless of geographical origin. However, there was slight variation among A. malaysiensis sequences. Based on analysis of 786 nucleotides between A. cantonensis and A. malaysiensis, a total of 8 variable sites were found (Table 3).
Fig 2. Maximum-likelihood phylogenetic tree of five species of Angiostrongylus (A. cantonensis, A. malaysiensis, A. dujardini, A. costaricensis and A. vasorum), Aelurostrogylus abstrusus and Nippostronglus braziliensis (outgroups) based on partial SSU rRNA sequences.
Support values (ML bootstrap/MP bootstrap/NJ bootstrap/Bayesian posterior probabilities) are shown above the branches. A dash (-) instead of a numerical support value indicates that a particular grouping was not found by that method of analysis. Bold letters indicate sequences obtained in the present study. Numbers in parentheses following each newly obtained sequence indicate the number of larvae possessing that sequence.
Table 3. Variable nucleotide positions within the SSU rRNA gene of A. cantonensis and A. malaysiensis.
| Nucleotide positions* | 29 | 181 | 227 | 480 | 656 | 662 | 665 | 689 |
|---|---|---|---|---|---|---|---|---|
| A. cantonensis (AY295804) | T | T | C | C | T | G | A | A |
| A.cantonensis THA (KU528678) | T | T | C | C | T | G | A | A |
| A.cantonensis THA (KU528682) | T | T | C | C | T | G | A | A |
| A.cantonensis THA (KU528681) | T | T | C | C | T | G | A | A |
| A.cantonensis KHM (KU528683) | T | T | C | C | T | G | A | G |
| A.cantonensis MMR (KU528680) | T | T | C | C | T | G | A | G |
| A.cantonensis USA (KU528679) | T | T | C | C | T | G | A | A |
| A. malaysiensis (EF514914) | C | C | T | A | C | A | G | G |
| A.malaysiensis THA (KU528686) | C | C | T | A | C | A | G | G |
| A.malaysiensis THA (KU528685) | C | T | T | A | C | G | G | G |
| A.malaysiensis LAO (KU528687) | C | T | T | A | C | G | G | G |
| A.malaysiensis MMR (KU528684) | C | T | T | A | C | G | G | G |
THA, Thailand; KHM, Cambodia; MMR, Myanmar; USA, United States of America; LAO, Lao PDR.
* Based on nucleotide positions in the A. cantonensis SSU rRNA sequence (AY295804). Bold letters indicated the present results.
Ribosomal DNA internal transcribed spacer 2 gene
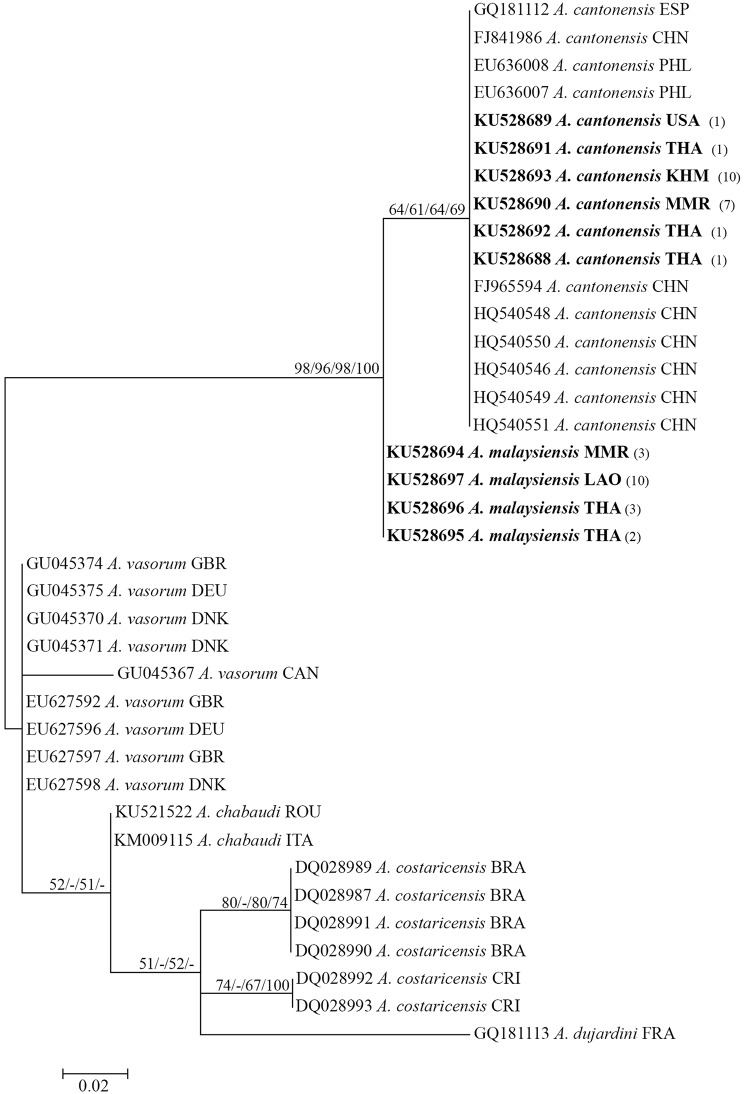
The phylogenetic tree based on the Angiostrongylus ITS2 sequences is shown in Fig 3. No outgroup was specified because of the difficulty in aligning species from other genera or families–this is a midpoint-rooted tree. Even within the genus Angiostrongylus, each species exhibited many unique repeats and indels. All A. cantonensis sequences fell into a well-supported group (bootstrap/Bayesian posterior probability values 64/61/64/69%). All A. malaysiensis sequences from Lao PDR, Myanmar and two provinces in Thailand (Phrae and Phatthalung) were identical, forming a group with 100% Bayesian posterior probability (Bayesian Inference, figure not shown).
Fig 3. Maximum-likelihood phylogenetic tree of Angiostrongylus cantonensis, A. malaysiensis, A. vasorum, A. chabaudi, A. costaricensis and A. dujardini based on the partial ITS2 sequences.
Support values (ML bootstrap/MP bootstrap/NJ bootstrap/Bayesian posterior probabilities) are shown above the branches. A dash (-) instead of a numerical support value indicates that a particular grouping was not found by that method of analysis. Bold letters indicate sequences obtained in the present study. Numbers in parentheses following each newly obtained sequence indicate the number of larvae possessing that sequence.
Mitochondrial cytochrome c oxidase subunit I gene
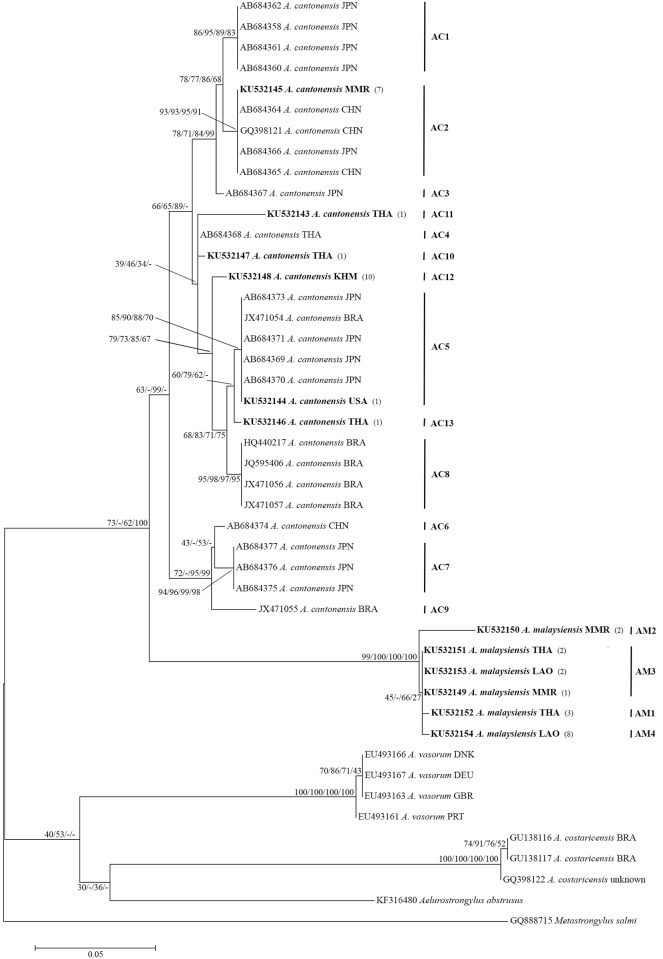
In the phylogenetic analysis based on the COI sequences, all A. cantonensis formed a monophyletic cluster with support values of 63/-/99/-%. The A. cantonensis sequences fell into 13 distinct haplotypes (four of them, AC10-AC13, not previously observed–Table 1) when all previously published data were included (Fig 4). The A. malaysiensis sequences formed a second monophyletic cluster with support values of 99/100/100/100%. This cluster contained four distinct haplotypes (AM1-AM4). Interspecific distances between A. cantonensis and A. malaysiensis sequences ranged from 9% to 13%. Intraspecific distances within A. cantonensis ranged from <1% to 4%, and among A. malaysiensis from <1 to 2% (Table 4).
Fig 4. Maximum-likelihood phylogenetic tree of Angiostrongylus cantonensis, A. malaysiensis, A. vasorum, A. costaricensis, Aelurostrogylus abstrusus and Metastrongylus salmi (the last two as outgroups) based on partial COI sequences.
Support values (ML bootstrap/MP bootstrap/NJ bootstrap/Bayesian posterior probabilities) are shown above the branches. A dash (-) instead of a numerical support value indicates that a particular grouping was not found by that method of analysis. Bold letters indicate sequences obtained in the present study. AC1-AC13 are the 13 distinct haplotypes of A. cantonensis; AM1-AM4 are the 4 distinct haplotypes of A. malaysiensis. Numbers in parentheses following each newly obtained sequence indicate the number of larvae possessing that sequence.
Table 4. Pairwise p-distances between COI sequences from A. cantonensis and A. malaysiensis samples.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. GQ398121 A.cantonensis CHN | - | |||||||||||||
| 2. AB684368 A.cantonensis THA | 0.02 | - | ||||||||||||
| 3. AB684370 A.cantonensis JPN | 0.03 | 0.01 | - | |||||||||||
| 4. KU532143 A.cantonensis THA | 0.03 | 0.02 | 0.03 | - | ||||||||||
| 5. KU532147 A.cantonensis THA | 0.02 | 0.00 | 0.02 | 0.03 | - | |||||||||
| 6. KU532145 A.cantonensis MMR | 0.00 | 0.02 | 0.03 | 0.03 | 0.02 | - | ||||||||
| 7. KU532146 A.cantonensis THA | 0.04 | 0.02 | 0.01 | 0.04 | 0.02 | 0.04 | - | |||||||
| 8. KU532144 A.cantonensis USA | 0.03 | 0.01 | 0.00 | 0.03 | 0.02 | 0.03 | 0.01 | - | ||||||
| 9. KU532148 A.cantonensis KHM | 0.03 | 0.01 | 0.01 | 0.03 | 0.01 | 0.03 | 0.02 | 0.01 | - | |||||
| 10. KU532149 A.malaysiensis MMR | 0.10 | 0.10 | 0.11 | 0.11 | 0.10 | 0.10 | 0.11 | 0.11 | 0.11 | - | ||||
| 11. KU532153 A.malaysiensis LAO | 0.10 | 0.10 | 0.11 | 0.11 | 0.10 | 0.10 | 0.11 | 0.11 | 0.11 | 0.00 | - | |||
| 12. KU532154 A.malaysiensis LAO | 0.10 | 0.09 | 0.10 | 0.10 | 0.10 | 0.10 | 0.11 | 0.10 | 0.10 | 0.00 | 0.00 | - | ||
| 13. KU532151 A.malaysiensis THA | 0.10 | 0.10 | 0.11 | 0.11 | 0.10 | 0.10 | 0.11 | 0.11 | 0.11 | 0.00 | 0.00 | 0.00 | - | |
| 14. KU532152 A.malaysiensis THA | 0.10 | 0.10 | 0.11 | 0.11 | 0.11 | 0.10 | 0.11 | 0.11 | 0.11 | 0.00 | 0.00 | 0.01 | 0.01 | - |
| 15. KU532150 A.malaysiensis MMR | 0.12 | 0.12 | 0.13 | 0.13 | 0.12 | 0.12 | 0.13 | 0.13 | 0.13 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 |
CHN, China; THA, Thailand; JPN, Japan; MMR, Myanmar; USA, United States of America; KHM, Cambodia; LAO, Lao PDR.
Bold letters indicated the present results.
Discussion
This study investigated the distribution and genetic relationships (based on SSU rRNA, ITS2 and COI nucleotide sequences) of A. cantonensis and A. malaysiensis specimens originating from land snails from different locations in Lao PDR, Cambodia, Myanmar and Thailand. To our knowledge, this is the first report of A. cantonensis in Cambodia and Myanmar, and also the first report of A. malaysiensis in Lao PDR and Myanmar. Distributions of these two species in the four countries broadly overlap (Fig 1). A recent molecular study in Thailand using mitochondrial cytochrome b (Cytb) nucleotide sequences reported A. cantonensis in two central provinces (Samut Prakan and Bangkok), two northern provinces (Phitsanulok and Chiang Mai), a northeastern province (Khon Kaen) and in southern province (Surat Thani) [24]. The same study reported A. malaysiensis from the north (Tak and Mae Hong Sorn) and south (Phang Nga) of Thailand. Our study found A. cantonensis in northeastern Thailand (Khon Kaen Province) and in the south (Surat Thani Province). We found A. malaysiensis in Phrae Province (northern Thailand) and Phatthalung Province (in the south). Interestingly, we also found A. cantonensis in eastern Myanmar and northwestern Cambodia and A. malaysiensis in central Lao PDR and eastern Myanmar.
There is little intraspecific variation of the SSU rRNA nucleotide sequences in A. cantonensis and A. malaysiensis (Table 3). Consequently, the 786-bp fragment of the SSU rRNA gene is a suitable marker to identify A. cantonensis and distinguish it from other Angiostrongylus species [12,17].
ITS2 sequences revealed distinct differences among A. cantonensis, A. malaysiensis, A. vasorum, A. chabaudi and A. costaricensis as reported previously [14,15], but minimal or no differences within each species. This conserved region could therefore be a useful genetic marker for the specific identification and genetic characterization of Angiostrongylus species.
The COI gene is one of the most commonly used sources of sequence data for studying geographical populations of Angiostrongylus species [13,14,16,17]. The present study supports previous findings, based on COI and Cytb sequences, that A. cantonensis is more closely related to A. malaysiensis than to A. costaricensis and A. vasorum [13,16,17,24]. Within A. cantonensis, we identified 13 haplotypes, an increase on the 9 haplotypes reported by Monte et al. [16]. Two haplotypes (AC2 and AC5) (Fig 4) have previously been reported from A. cantonensis from Brazil and Asia [16]. One sample (n = 7 larvae) from Myanmar exhibited the AC2 haplotype. In addition, we found four new haplotypes (AC10-AC13) from Thailand and Cambodia (Fig 4). In A. malaysiensis, one haplotype (AM3) occurred in worms from Laos, Myanmar and Thailand. A separate, distinct haplotype was also found in some specimens from each of those countries. The analysis also indicates a p-distance between A. cantonensis and A. malaysiensis ranging from 9% to 13% (Table 4), similar to previously reported values [13]. Pair-wise intraspecific distances among A. cantonensis haplotypes ranged from <1% to 4%, and among A. malaysiensis from <1% to 2% (Table 4). This suggests that COI sequences might be useful for differentiation of geographical isolates [16].
Conclusions
In summary, the present study has reported for the first time the phylogenetic relationships of A. cantonensis and A. malaysiensis in parts of the Greater Mekong subregion, based on sequences from three gene regions. The two species have broadly overlapping distributions in Lao PDR, Cambodia, Myanmar and Thailand. The techniques employed in this study can clearly be used in a molecular approach to identify Angiostrongylus species in the future. This provides a reliable alternative to morphological identification of parasite samples, especially in cases where morphological features are obscure in larval stages.
Acknowledgments
We wish to acknowledge David Blair (Khon Kaen University Publication Clinic, the Faculty of Medicine, Khon Kaen University) for valuable suggestions for improving the quality of this manuscript. We also thank the Academic Editor, Dr. Don Colgan, the Australian Museum and anonymous reviewers for the useful comments.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by: 1. The Higher Education Research Promotion and the National Research University Project of Thailand, Office of the Higher Education Commission, Thailand, through the Health Cluster (SHeP-GMS) Award Number: NRU592014, Recipient: Wanchai Maleewong, PhD. 2. The Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program Award Number: grant no. PHD/0053/2556, Recipient: Rutchanee Rodpai. 3. The Postdoctoral Training Program Graduate School and Khon Kaen University Award Number: grant no. 58101, Recipient: Oranuch Sanpool. 4. The TRF Senior Research Scholar Grant, Thailand Research Fund Award Number: grant number RTA5880001, Recipient: Wanchai Maleewong, PhD, and Pewpan M. Intapan. We certified that all the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript and the revised manuscript has not and will not be submitted for publication elsewhere, have reviewed and approved by all authors, and concurred in the revised submission by the corresponding author.
References
- 1.Robles Mdel R, Navone GT, Kinsella JM. A new angiostrongylid (Nematoda) species from the pulmonary arteries of Akodon azarae (Rodentia: Cricetidae) in Argentina. J Parasitol. 2008; 94: 515–519. 10.1645/GE-1340.1 [DOI] [PubMed] [Google Scholar]
- 2.Eamsobhana P, Lim PE, Yong HS. Genetic diversity of the rat lungworm, Angiostrongylus cantonensis, the major cause of eosinophilic meningitis. Hawaii J Med Public Health. 2013; 72(6 Suppl 2): 15–17. [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai HC, Liu YC, Kunin CM, Lee SS, Chen YS, Lin HH, et al. Eosinophilic meningitis caused by Angiostrongylus cantonensis: report of 17 cases. Am J Med. 2001; 111: 109–114. [DOI] [PubMed] [Google Scholar]
- 4.Tsai HC, Chen YS, Yen CM. Human parasitic meningitis caused by Angiostrongylus cantonensis infection in Taiwan. Hawaii J Med Public Health. 2013; 72(6 Suppl 2): 26–27. [PMC free article] [PubMed] [Google Scholar]
- 5.Thiengo SC, Simoes Rde O, Fernandez MA, Maldonado A. Angiostrongylus cantonensis and rat lungworm disease in Brazil. Hawaii J Med Public Health. 2013; 72(6 Suppl 2): 18–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Wang QP, Lai DH, Zhu XQ, Chen XG, Lun ZR. Human angiostrongyliasis. Lancet Infect Dis. 2008; 8: 621–630. 10.1016/S1473-3099(08)70229-9 [DOI] [PubMed] [Google Scholar]
- 7.Wan KS, Weng WC. Eosinophilic meningitis in a child raising snails as pets. Acta Trop. 2004; 90: 51–53. [DOI] [PubMed] [Google Scholar]
- 8.Kramer MH, Greer GJ, Quinonez JF, Padilla NR, Hernandez B, Arana BA, et al. First reported outbreak of abdominal angiostrongyliasis. Clin Infect Dis. 1998; 26: 365–372. [DOI] [PubMed] [Google Scholar]
- 9.Prociv P, Spratt DM, Carlisle MS. Neuro-angiostrongyliasis: unresolved issues. Int J Parasitol. 2000; 30: 1295–1303. [DOI] [PubMed] [Google Scholar]
- 10.Monte TC, Gentile R, Garcia J, Mota E, Santos JN, Maldonado A Junior. Brazilian Angiostrongylus cantonensis haplotypes, ac8 and ac9, have two different biological and morphological profiles. Mem Inst Oswaldo Cruz. 2014; 109: 1057–1063. 10.1590/0074-0276130378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton LA, Chilton NB, Beveridge I, Hoste H, Nansen P, Gasser RB. Genetic markers for strongylid nematodes of livestock defined by PCR-based restriction analysis of spacer rDNA. Acta Trop. 1998; 69: 1–15. [DOI] [PubMed] [Google Scholar]
- 12.Fontanilla IK, Wade CM. The small subunit (SSU) ribosomal (r) RNA gene as a genetic marker for identifying infective 3rd juvenile stage Angiostrongylus cantonensis. Acta Trop. 2008; 105: 181–186. [DOI] [PubMed] [Google Scholar]
- 13.Eamsobhana P, Lim PE, Solano G, Zhang H, Gan X, Yong HS. Molecular differentiation of Angiostrongylus taxa (Nematoda: Angiostrongylidae) by cytochrome c oxidase subunit I (COI) gene sequences. Acta Trop. 2010; 116: 152–156. 10.1016/j.actatropica.2010.07.005 [DOI] [PubMed] [Google Scholar]
- 14.Jefferies R, Shaw SE, Viney ME, Morgan ER. Angiostrongylus vasorum from South America and Europe represent distinct lineages. Parasitology. 2009; 136: 107–115. 10.1017/S0031182008005258 [DOI] [PubMed] [Google Scholar]
- 15.Liu CY, Zhang RL, Chen MX, Li J, Ai L, Wu CY, et al. Characterisation of Angiostrongylus cantonensis isolates from China by sequences of internal transcribed spacers of nuclear ribosomal DNA. J Anim Vet Adv. 2011; 10: 593–596. [Google Scholar]
- 16.Monte TC, Simoes RO, Oliveira AP, Novaes CF, Thiengo SC, Silva AJ, et al. Phylogenetic relationship of the Brazilian isolates of the rat lungworm Angiostrongylus cantonensis (Nematoda: Metastrongylidae) employing mitochondrial COI gene sequence data. Parasit Vectors. 2012; 5: 248 10.1186/1756-3305-5-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokiwa T, Harunari T, Tanikawa T, Komatsu N, Koizumi N, Tung KC, et al. Phylogenetic relationships of rat lungworm, Angiostrongylus cantonensis, isolated from different geographical regions revealed widespread multiple lineages. Parasitol Int. 2012; 61: 431–436. 10.1016/j.parint.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 18.De NV, Duyet le V, Chai JY. A case of ocular angiostrongyliasis with molecular identification of the species in Vietnam. Korean J Parasitol. 2015; 53: 713–717. 10.3347/kjp.2015.53.6.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95–98. [Google Scholar]
- 20.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; 2000. [Google Scholar]
- 22.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012; 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 24.Yong HS, Eamsobhana P, Song SL, Prasartvit A, Lim PE. Molecular phylogeography of Angiostrongylus cantonensis (Nematoda: Angiostrongylidae) and genetic relationships with congeners using cytochrome b gene marker. Acta Trop. 2015; 148: 66–71. 10.1016/j.actatropica.2015.04.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.