Abstract
Several bacterial species from the Burkholderia cepacia complex (Bcc) are feared opportunistic pathogens that lead to debilitating lung infections with a high risk of developing fatal septicemia in cystic fibrosis (CF) patients. However, the pathogenic potential of other Bcc species is yet unknown. To elucidate clinical relevance of Burkholderia contaminans, a species frequently isolated from CF respiratory samples in Ibero-American countries, we aimed to identify its key virulence factors possibly linked with an unfavorable clinical outcome. We performed a genome-wide comparative analysis of two isolates of B. contaminans ST872 from sputum and blood culture of a female CF patient in Argentina. RNA-seq data showed significant changes in expression for quorum sensing-regulated virulence factors and motility and chemotaxis. Furthermore, we detected expression changes in a recently described low-oxygen-activated (lxa) locus which encodes stress-related proteins, and for two clusters responsible for the biosynthesis of antifungal and hemolytic compounds pyrrolnitrin and occidiofungin. Based on phenotypic assays that confirmed changes in motility and in proteolytic, hemolytic and antifungal activities, we were able to distinguish two phenotypes of B. contaminans that coexisted in the host and entered her bloodstream. Whole genome sequencing revealed that the sputum and bloodstream isolates (each representing a distinct phenotype) differed by over 1,400 mutations as a result of a mismatch repair-deficient hypermutable state of the sputum isolate. The inferred lack of purifying selection against nonsynonymous mutations and the high rate of pseudogenization in the derived isolate indicated limited evolutionary pressure during evolution in the nutrient-rich, stable CF sputum environment. The present study is the first to examine the genomic and transcriptomic differences between longitudinal isolates of B. contaminans. Detected activity of a number of putative virulence factors implies a genuine pathogenic nature of this novel Bcc species.
Introduction
Bacteria from the Burkholderia cepacia complex (Bcc) cause serious respiratory infections in cystic fibrosis (CF) patients and thus significantly contribute to the reduction of life expectancy in CF [1]. Although the Bcc currently comprises 20 different bacterial species, only three of them have been under the spotlight of CF researchers until recently: Burkholderia cenocepacia (former genomovar III), which was recognized as a dominant Bcc species with arguably the highest potential for inter-patient transmission (evident from past large epidemic outbreaks caused by B. cenocepacia strains ET12, PHDC or ST32 [2]); Burkholderia multivorans (former genomovar II), which within the last 15 years replaced B. cenocepacia as the primary species of Bcc infection in many countries [3]; and Burkholderia dolosa (former genomovar VI), which is a far less common Bcc species than the former two but is equally clinically important [4]. The most worrisome feature of the three Bcc species in the CF care is the possibility of developing a fatal septic condition called cepacia syndrome [4–6].
Burkholderia contaminans was described taxonomically in 2009 as one of the latest Bcc members [7]. Its history related to the CF lung disease started in the early 2000s, when the species was recovered from respiratory samples from Portuguese and Argentinean patients [8, 9]. At that time, the bacteria were incorrectly or indefinitely classified, sometimes as taxon K, based on the restriction fragment length polymorphism (RFLP) pattern of the recA gene. Later, a study aiming to unravel the controversial identification of the B. contaminans metagenome in sea water (designated as Burkholderia SAR-1 at that time) indicated that B. contaminans was the species epidemiologically linked to the contamination of medical devices and products [10]. Today, B. contaminans is reported to be the most frequent Bcc species in Spanish [11] and Argentinean CF patients [12].
B. contaminans infections in CF are often of transient nature [13] and thus can be perceived as respiratory tract colonization rather than true infection. However, some patients develop chronic infection with long-term culture positivity [12, 13]. Furthermore, some of the cases can flare up and result in fatalities. Based on this unfavorable clinical manifestation, B. contaminans is likely an emerging CF pathogen. The circumstances leading to the establishment of true B. contaminans infections are unknown.
Recently, we performed a thorough transcriptomic analysis on B. cenocepacia isolates that were recovered from the bloodstream of CF patients with end-stage disease to identify the bacterial factors possibly associated with cepacia syndrome [14]. In this study, we applied an analogous approach. To gain insights into B. contaminans evolution during chronic infection, we compared transcriptomes and genomes of B. contaminans isolates recovered from different stages of CF infection from a single patient.
Results/Discussion
We analyzed seven B. contaminans isolates that had been recovered on four separate occasions over the course of 6-year long chronic infection (Table 1). The patient died at the age of 10 upon the developing septic state with B. contaminans detected in her blood culture. All isolates were assigned to the multilocus sequence type (ST) 872. ST872 shares the recA gene-ST-71 allele with the predominating B. contaminans population in Argentina [12].
Table 1. B. contaminans isolates used in the study.
| Isolate ID | Collection date | Source | Multilocus sequence type | Analyses performed |
|---|---|---|---|---|
| FFH2055 | 25-Jul-2005 | Sputum | 872 | WGS [15], phenotypic testing |
| i_S | 14-Oct-2009 | Sputum | 872 | phenotypic testing |
| L_S | 14-Oct-2009 | Sputum | 872 | phenotypic testing |
| 466_S | 5-Jan-2011 | Sputum | 872 | phenotypic testing |
| 467_S | 5-Jan-2011 | Sputum | 872 | RNA-Seq, WGS, phenotypic testing |
| MF16_B | 11-Oct-2011 | Blood | 872 | RNA-Seq, WGS, phenotypic testing |
| MF17_B | 11-Oct-2011 | Blood | 872 | phenotypic testing |
Transcriptomic differences between ST872 isolates
Initially, we performed comparative transcriptomics between isolate 467_S from sputum and isolate MF16_B from blood. Due to their different sites of origin and dates of collection, we expected to identify changes in gene expression that would be linked to the increasing severity of the infection. The experimental layout included three growth media (CF sputum, CF serum and control medium) and three replicates per isolate and condition. Among the 7,109 protein-coding genes, we observed a greater than 3-fold change in expression in more than 18% of the genes (1,308) under at least one of the three cultivation conditions assessed (for a complete set of genes, see S1, S2, S3 and S4 Tables, Table 2). Altered expression was detected for genes with a well established function in Burkholderia virulence as well as for more recently discovered genetic clusters with only a putative role in the Bcc pathogenicity.
Table 2. The lxa locus, putative regulators and co-regulated genes as derived from B. contaminans genomic and transcriptomic data.
| Gene product | Accession No: B. contaminans FFH2055 | Accession No: B. cenocepacia J2315 | Accession No: B. multivorans ATCC 17616 | Fold change of expression (MF16_B/467_S) | ||
|---|---|---|---|---|---|---|
| Serum | Sputum | BSM | ||||
| lxa locus | ||||||
| pyruvate, water dikinase PpsA | WR30_RS16490 | ortholog absent | BMULJ_05585 | 14.1 | 3.0 | 10.4 |
| esterase / lipase | WR30_RS16495 | ortholog absent | BMULJ_05586 | 16.4 | 2.1 | 12.6 |
| hypothetical protein | WR30_RS16500 | ortholog absent | BMULJ_05590 | 48.8 | 14.9 | 99.7 |
| hypothetical protein | WR30_RS16505 | ortholog absent | BMULJ_05591 | 18.8 | 4.0 | 9.3 |
| hypothetical protein | WR30_RS16510 | ortholog absent | BMULJ_05592 | 58.1 | 15.2 | 87.4 |
| hypothetical protein | WR30_RS16515 | ortholog absent | BMULJ_05594 | 149.1 | 8.7 | 62.2 |
| nicotinate phosphoribosyltransferase PncB | WR30_RS16520 | ortholog absent | BMULJ_05598 | 139.1 | 7.0 | 71.5 |
| sulfonate/nitrate/taurine transport system permease SsuC | WR30_RS16525 | ortholog absent | BMULJ_05602 | 50.6 | 5.7 | 37.5 |
| sulfonate/nitrate/taurine transport system ATP-binding protein SsuB | WR30_RS16530 | ortholog absent | BMULJ_05603 | 16.4 | 2.6 | 8.1 |
| hypothetical protein | WR30_RS16540 | ortholog absent | BMULJ_05604 | 104.7 | 12.1 | 70.5 |
| putative signal transduction protein | WR30_RS16545 | ortholog absent | BMULJ_05606 | 48.8 | 6.6 | 20.8 |
| hypothetical protein | WR30_RS16550 | ortholog absent | BMULJ_05607 | 15.2 | 9.9 | 22.5 |
| hypothetical protein | WR30_RS16570 | BCAM0275A | BMULJ_05608 | 46.2 | 10.9 | 39.7 |
| putative universal stress protein | WR30_RS16575 | BCAM0276 | BMULJ_05609 | 40.2 | 3.3 | 38.3 |
| protein of unknown function | WR30_RS16580 | BCAM0277 | BMULJ_05610 | 40.5 | 3.8 | 32.9 |
| putative heat shock protein | WR30_RS16585 | BCAM0278 | BMULJ_05611 | 230.7 | 33.1 | 328.6 |
| hypothetical protein | WR30_RS16590 | BCAM0279 | BMULJ_05612 | 81.6 | 3.9 | 50.2 |
| putative phospholipid-binding protein | WR30_RS16595 | BCAM0280 | BMULJ_05613 | 101.8 | 18.4 | 85.6 |
| hypothetical protein | WR30_RS16600 | BCAM0280A | BMULJ_05614 | 136.2 | 6.9 | 48.5 |
| putative sulfate transporter family protein | WR30_RS16605 | BCAM0281 | BMULJ_05615 | 67.2 | 2.8 | 18.0 |
| hypothetical protein | WR30_RS16610 | BCAM0282 | BMULJ_05616 | 4.2 | 4.1 | 8.5 |
| putative lysine decarboxylase | WR30_RS16615 | BCAM0283 | BMULJ_05617 | 8.7 | 2.8 | 22.2 |
| putative cytochrome c | WR30_RS16620 | BCAM0284 | BMULJ_05618 | 3.7 | 1.2 | 4.2 |
| hypothetical protein | WR30_RS16625 | BCAM0285 | BMULJ_05619 | 5.2 | 2.8 | 6.5 |
| putative alcohol dehydrogenase | WR30_RS16630 | BCAM0286 | BMULJ_05620 | 12.9 | 3.0 | 9.2 |
| two-component regulatory system response regulator protein | WR30_RS16645 | BCAM0288 | BMULJ_05622 | 1.9 | 1.8 | 1.7 |
| putative universal stress protein | WR30_RS16650 | BCAM0290 | BMULJ_05624 | 44.3 | 2.7 | 26.9 |
| putative universal stress protein | WR30_RS16655 | BCAM0291 | BMULJ_05625 | 75.6 | 9.1 | 28.4 |
| putative universal stress protein | WR30_RS16660 | BCAM0292 | BMULJ_05626 | 51.6 | 11.6 | 124.5 |
| putative acetate kinase | WR30_RS16665 | BCAM0293 | BMULJ_05627 | 37.3 | 13.1 | 30.7 |
| putative universal stress protein | WR30_RS16670 | BCAM0294 | BMULJ_05628 | 119.4 | 22.2 | 135.3 |
| hypothetical protein | WR30_RS16675 | BCAM0295 | BMULJ_05629 | 58.1 | 23.3 | 41.9 |
| acetoacetyl-CoA reductase PhbB | WR30_RS16680 | BCAM0296 | BMULJ_05630 | 168.9 | 15.6 | 82.1 |
| putative poly(3-hydroxyalkanoate) polymerase | WR30_RS16685 | BCAM0297 | BMULJ_05631 | 54.2 | 15.3 | 49.2 |
| putative phosphate acetyl/butyryl transferase | WR30_RS16690 | BCAM0298 | BMULJ_05632 | 132.5 | 15.9 | 34.5 |
| putative zinc-binding alcoholdehydrogenase | WR30_RS16695 | BCAM0299 | BMULJ_05633 | 108.4 | 9.5 | 21.9 |
| metallo-beta-lactamase superfamily protein | WR30_RS16700 | BCAM0300 | BMULJ_05634 | 52.0 | 7.3 | 25.8 |
| hypothetical protein | WR30_RS16705 | ortholog absent | BMULJ_05635 | 196.7 | 4.9 | 33.6 |
| RND efflux system outer membrane lipoprotein | WR30_RS16710 | ortholog absent | BMULJ_05636 | 102.5 | 5.0 | 9.8 |
| HlyD family secretion protein | WR30_RS16715 | ortholog absent | BMULJ_05637 | 17.6 | 4.5 | 16.8 |
| ABC-type antimicrobial peptide transport system ATPase component | WR30_RS16720 | ortholog absent | BMULJ_05638 | 14.9 | 3.2 | 12.0 |
| putative ABC-type antimicrobial peptide transport system permease | WR30_RS16725 | ortholog absent | BMULJ_05639 | 29.2 | 6.6 | 31.8 |
| hypothetical protein | WR30_RS16730 | BCAM0301 | BMULJ_05640 | 43.7 | 1.2 | 5.4 |
| putative ABC transporter protein | WR30_RS16735 | BCAM0302 | BMULJ_05641 | 20.4 | 7.2 | 16.2 |
| ABC transporter ATP-binding membrane protein | WR30_RS16740 | BCAM0303 | BMULJ_05642 | 34.1 | 4.0 | 21.4 |
| transporter system transport protein | WR30_RS16745 | BCAM0304 | BMULJ_05643 | 51.3 | 3.8 | 49.5 |
| hypothetical protein | WR30_RS16755 | BCAM0306 | BMULJ_05645 | 16.2 | 7.8 | 34.5 |
| hypothetical protein | WR30_RS16760 | BCAM0307 | BMULJ_05646 | 148.7 | 24.0 | 88.1 |
| hypothetical protein | WR30_RS16765 | BCAM0308 | BMULJ_05647 | 89.9 | 11.6 | 61.8 |
| putative cell division-related metallo peptidase | WR30_RS16770 | BCAM0309 | BMULJ_05648 | 118.6 | 11.0 | 63.6 |
| ribonucleotide reductase-like protein | WR30_RS16775 | BCAM0310 | BMULJ_05649 | 128.9 | 1.0 | 16.9 |
| putative 6-phosphofructokinase | WR30_RS16780 | BCAM0311 | BMULJ_05650 | 61.0 | 3.9 | 17.5 |
| putative polysaccharide deacetylase | WR30_RS16785 | BCAM0312 | BMULJ_05651 | 17.8 | 4.1 | 23.9 |
| hypothetical protein | WR30_RS16805 | BCAM0313 | BMULJ_05652 | 38.1 | 3.4 | 23.1 |
| hypothetical protein | WR30_RS16795 | BCAM0314 | BMULJ_05653 | 23.3 | 5.8 | 16.8 |
| hypothetical protein | WR30_RS16800 | BCAM0315 | BMULJ_05654 | 15.5 | 3.9 | 33.6 |
| cytochrome c553 | WR30_RS29415 | ortholog absent | BMULJ_05656 | 5.5 | 1.4 | 4.3 |
| putative cytochrome b | WR30_RS29420 | ortholog absent | BMULJ_05658 | 3.7 | -1.4 | 4.2 |
| hypothetical protein | WR30_RS16555 | BCAM0317 | BMULJ_05663 | 173.6 | 32.9 | 102.5 |
| putative cation-transporting ATPase | WR30_RS16560 | BCAM0318 | BMULJ_05664 | 26.4 | 3.1 | 23.8 |
| putative universal stress protein | WR30_RS16565 | BCAM0319 | BMULJ_05665 | 36.3 | 12.2 | 43.1 |
| putative cytochrome b561 | WR30_RS29470 | BCAM0320 | ortholog absent | 6.4 | 1.7 | 17.3 |
| hypothetical protein | WR30_RS29475 | BCAM0321 | ortholog absent | 46.2 | -1.1 | 58.1 |
| two-component regulatory system, response regulator protein | WR30_RS29480 | BCAM0322 | ortholog absent | 7.3 | 1.3 | 8.9 |
| two-component regulatory system, sensor kinase protein | WR30_RS29485 | BCAM0323 | ortholog absent | 5.9 | 1.2 | 5.8 |
| Putative regulators of lxa locus | ||||||
| CRP family regulatory protein | WR30_RS16635 | BCAM0287 | BMULJ_05621 | 7.4 | 7.2 | 8.8 |
| CRP family regulatory protein | WR30_RS29560 | BCAM0049 | BMULJ_03224 | 6.9 | -1.7 | 2.7 |
| cyclic nucleotide-binding transcriptional regulator | WR30_RS23390 | BCAM1483 | BMULJ_04241 | 18.8 | 9.3 | 10.9 |
| Co-regulated genes | ||||||
| protein of unknown function (DUF1488) | WR30_RS23375 | BCAM1480 | ortholog absent | 44.3 | 5.1 | 20.1 |
| hypothetical protein | WR30_RS23380 | BCAM1481 | BMULJ_04240 | 22.2 | 2.5 | 12.2 |
| hypothetical protein | WR30_RS23385 | BCAM1482 | ortholog absent | 32.7 | 3.1 | 3.6 |
| putative universal stress protein | WR30_RS23455 | BCAM1495 | BMULJ_04252 | 38.5 | 7.3 | 23.6 |
| hypothetical protein | WR30_RS23460 | BCAM1496 | BMULJ_04253 | 3.6 | 1.5 | 1.4 |
| alcohol dehydrogenase AdhA | WR30_RS24145 | BCAM1570 | ortholog absent | 106.2 | 2.4 | 21.7 |
| TonB-dependent receptor | WR30_RS24150 | BCAM1571 | BMULJ_04320 | 3.7 | -2.5 | 3.7 |
Quorum sensing (QS) systems
QS is known to regulate a multitude of functions that are involved in Bcc virulence in various laboratory models (reviewed in [16]). The genes encoding signal molecule synthases as well as virtually all QS-regulated virulence-related functions were expressed at higher levels in MF16_B compared with 467_S. They included the extracellular metalloproteases ZmpA and ZmpB [17, 18], siderophore ornibactin [19], lectins BclA and BclC [20–22], the nematocidal protein AidA [23], Flp-type pili and type III (T3SS) and type VI secretion systems [24] (S2 Table). However, the extent of expression differences varied considerably among particular genes and among cultivation conditions, with no obvious trend between the expression levels of QS signaling molecule synthases and the transcription of QS-regulated genes.
Bcc QS systems represent an intricate network of pleiotropic, mutually agonistic or antagonistic and often redundant regulators [16, 25], which together leads to a complex output response. A study that examined changes in AHL production by a large Bcc collection during the course of the CF infection found QS signaling to be functionally preserved [26]. Comparison of the gene expression between two B. cenocepacia isolates from different stages of the CF pulmonary infection revealed both upregulation (ornibactin) and downregulation (secretion systems, extracellular proteases) of QS-dependent functions [27]. Our recent study, similar in design, but involving a bloodstream B. cenocepacia isolate, demonstrated a significant increase in expression of the entire T3SS cluster [14], a finding consistent with T3SS expression in MF16_B.
Motility and chemotaxis
Over 70 genes encoding proteins with a motility-related function and spanning all previously described motility clusters [24] exhibited increased expression in the blood isolate MF16_B (S3 Table). This was in marked contrast with our previous observation on B. cenocepacia strain ST32 where all tested blood isolates (obtained at the time of cepacia syndrome) showed a uniform loss of motility or rapid decrease in motility as measured by swimming activity of the isolates [14]. Another study on a wide range of clinical isolates that belonged to various Bcc species found no apparent association between changes in motility and the clinical outcome of the Bcc infection [28].
Biosynthesis of antifungal compounds
Two gene clusters responsible for the biosynthesis of antifungal compounds showed significantly higher expression in MF16_B than in 467_S (S4 Table). One of them, pyrrolnitrin, which is produced by some Bcc and several other Gram-negative bacteria, inhibits growth of a wide range of fungi and of some Gram-positive bacteria [29]. Interestingly, a study by Schmidt et al. [30] noted the absence of the pyrrolnitrin biosynthetic operon in all of their examined strains of B. contaminans. However, in our studied B. contaminans strain ST872, all four genes of the pyrrolnitrin biosynthetic pathway were present, and these genes were also found in every other B. contaminans sequenced up to now. Pyrrolnitrin production depends entirely on QS [30]; the detected expression changes are thus in accordance with those observed for other QS-regulated functions whose expression was upregulated in the bloodstream isolate MF16_B.
Another antifungal compound, occidiofungin, was first described in B. contaminans MS14 [31], displaying its activity against a wide range of fungi [32]. The biosynthetic cluster, consisting of 18 genes [33], is cis-regulated by two regulators [34]. This cluster is absent in the genomes of some Bcc species, including B. cenocepacia and B. multivorans [35]. The occidiofungin biosynthetic cluster, including genes encoding both regulators (AmbR1 and AmbR2), showed increased expression in MF16_B, most markedly in sputum medium (S4 Table). Similar to pyrrolnitrin, occidiofungin production was found to be positively regulated by QS [36].
lxa locus
In 2013, Sass et al. described a gene cluster in B. cenocepacia strain J2315 (BCAM0275A - BCAM0323) whose expression was induced specifically under low oxygen concentration (low-oxygen-activated or lxa locus) [37]. The locus exhibits an irregular pattern of presence in Bcc bacteria, e.g., being absent in some strains of B. cenocepacia or B. multivorans while being present in the others [37]. Notably, the lxa locus (as delineated in B. cenocepacia J2315) can be found in genomes of all sequenced strains of B. contaminans, either as a complete set of genes (i.e., B. contaminans FFH2055 or B. contaminans MS14; see Table 2) or truncated (missing approximately one quarter of genes from the 3’ end of the locus, i.e., B. contaminans LMG 23361), whereas it is absent in Burkholderia lata strain 383, the closest relative of B. contaminans [37]. We detected a rapid increase in expression of the entire cluster in MF16_B vs. 467_S. The expression was most pronounced when the bacteria were cultivated in serum and a control medium (Table 2). Importantly, all three genes that were predicted to regulate lxa expression (BCAM0049, BCAM0287 and BCAM1483) [37] also displayed increased expression in MF16_B. Moreover, several genes neighboring the lxa cluster (as delineated in B. cenocepacia J2315 annotation) exhibited a similar pattern of differential expression. These genes were homologous to lxa-adjacent genes in the annotated genome of B. multivorans ATCC17616 (Table 2), suggesting that additional genes form extended, strain-specific lxa clusters. Although low-oxygen level was the only condition capable of inducing lxa transcription in the study of Sass et al. [37], we detected expression of the entire locus under normal aerobic conditions in B. contaminans. We believe that there is another eminent functional significance of this enigmatic part of multiple Bcc genomes.
Phenotypic differences between B. contaminans ST872 isolates
The transcriptomic differences between B. contaminans isolates 467_S and MF16_B allowed predicting changes in their phenotypic features. To verify the transcriptomic results, we assessed four phenotypes: swimming motility (a proxy of flagellar gene activity), proteolysis (zmpA and zmpB genes), antifungal activity (pyrrolnitrin and occidiofungin biosynthetic genes) and hemolysis. We assumed that occidiofungin had hemolytic properties because molecules synthesized by pathways highly homologous to that of occidiofungin were tested hemolytic in B. vietnamiensis [35] and B. ambifaria [38].
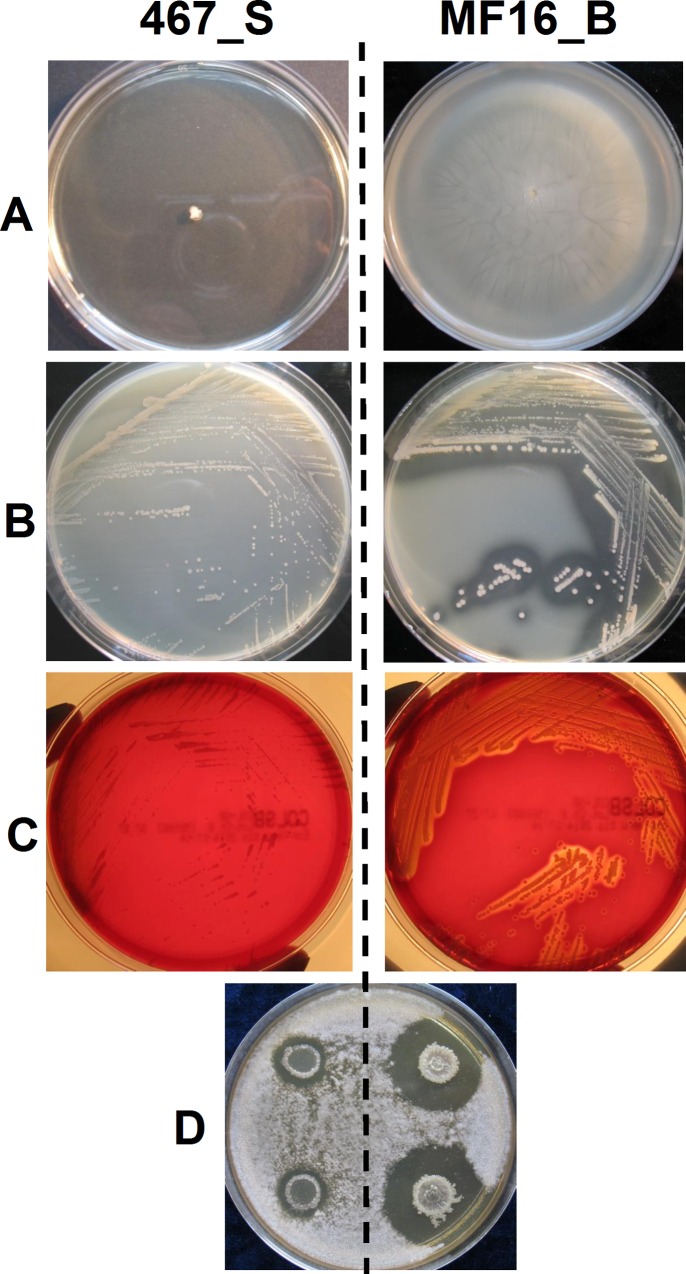
The phenotypic differences that had been revealed in agar plate assays were in agreement with changes observed at the transcriptional level (Fig 1): isolate 467_S was nonmotile and did not show any proteolytic or hemolytic activity after 48 hours of incubation; isolate MF16_B exhibited high motility, proteolysis and hemolysis. Antagonism against the fungus Geotrichum candidum was very pronounced in MF16_B, not in 467_S. Although 467_S exhibited slower growth on all agar plates, the differences were equally apparent after prolonged incubation, which gave rise to colonies of 467_S of similar size to that of faster-growing MF16_B (data not shown).
Fig 1. Agar plate phenotypic assays.
A) swimming motility, B) proteolysis, C) hemolysis and D) antifungal activity against Geotrichum candidum. Left–isolate 467_S, right–isolate MF16_B.
These phenotypic tests were used to screen additional five B. contaminans isolates that had been collected from the patient at various time points. Strikingly, this simple screening indicated coexistence of two distinct phenotypic groups in the subject’s lung as well as in blood, termed group A (represented by MF16_B) and group B (represented by 467_S) (Table 3). It is noteworthy that the phenotype of the blood isolate MF16_B matched the sputum isolate FFH2055 (with the exception of swimming motility), which was retrieved six years earlier. The sputum isolate 467_S had its phenotypic counterpart, isolate MF17, which was obtained from the same blood culture as the MF16_B.
Table 3. Phenotypic properties of B. contaminans ST872 isolates.
| Isolate ID | Swimming | Hemolysis | Proteolysis | Activity against G. candidum | Mutation frequency§ (x 10−8) | Phenotypic class |
|---|---|---|---|---|---|---|
| FFH2055 | - | +++ | +++ | +++ | 11 | A |
| i_S | ++ | - | - | - | 1,560 | B |
| L_S | + | - | + | - | 5,300 | B |
| 466_S | - | - | - | - | 2,400 | B |
| 467_S | - | - | - | - | 1,200 | B |
| MF16_B | +++ | +++ | +++ | +++ | 9 | A |
| MF17_B | - | - | - | - | 1,250 | B |
§ Median frequency of mutations towards rifampicin resistance from three independent experiments
Genomic differences between B. contaminans isolates
Genome sequencing of 467_S and MF16_B was performed (Illumina, 2 x 150 bp paired-end sequencing) in order to identify the extent of genetic differences between the phenotypic groups A and B and to complement information from their transcriptomes. Genomic sequences were compared with the reference genome of B. contaminans FFH2055, the initial isolate obtained from the studied patient and sequenced earlier in another study (8 contigs, PacBio sequencing) [15]. Of note, the reference genome was 8.21 Mb in size, encoded 7,109 proteins, and had a GC content of 66.40%.
When the sequencing reads covering MF16_B genome were mapped to the reference assembly, 44 variants were detected between both genomes. These were exclusively single-nucleotide insertions at tandem poly-GC repeats in MF16_B, indicating possible sequencing bias. To resolve this, we remapped Illumina sequencing reads for FFH2055 on the PacBio-assembled reference of the same isolate (S5 Table). This approach revealed that the called differences between FFH2055 and MF16_B genomes come from different sequencing technologies. The omission of nucleotides on poly-GC runs is in line with other observations for PacBio assemblies [39]. After this correction, the genomic sequences of isolates MF16_B and FFH2055 proved to be identical.
Contrary to genome uniformity observed within the phenotypic group A, a substantial number of genetic differences were detected when groups A and B were compared between each other. In total, 1,433 mutations separated the isolates 467_S and MF16_B, representing 0.017% of the total genome size and affecting 1,002 genes (14%) (Table 4, S6 Table).
Table 4. Summary of the genetic differences between B. contaminans isolates 467_S and MF16_B.
| Type of mutations | Particular mutations§ (class: number) | Total number | ||||
|---|---|---|---|---|---|---|
| single-nucleotide substitutions | AT→GC: 855 | GC→AT: 327 | transversions (all types): 43 | 1,225 | ||
| insertions (tandem repeats) | +1 GC: 105 | +1 AT: 6 | + two or more nucleotides: 39 | 150 | ||
| deletions (tandem repeats) | -1 GC: 15 | -1 AT: 12 | - two or more nucleotides: 19 | 46 | ||
| other indel mutations | 12 | |||||
| Total | 1,433 | |||||
| intragenic mutations | synonymous: 230 | missense: 796 | nonsense: 13 | frameshifts: 94 | other: 12 | 1,145 |
| intergenic mutations | 288 | |||||
| Total | 1,433 | |||||
§ Mutations in 467_S with respect to the genomic sequence of MF16_B
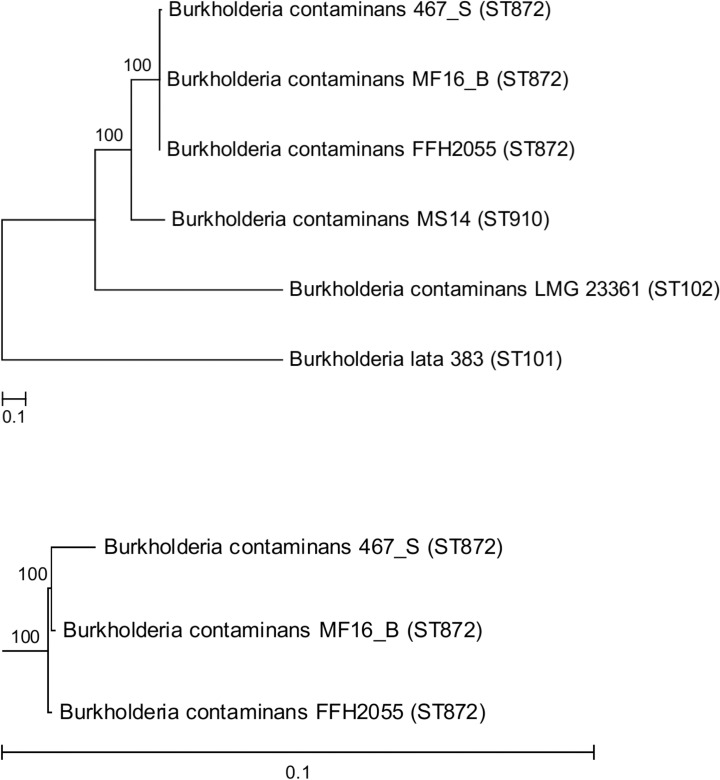
Over 200 indel mutations were detected among the two ST872 genomes representing the two phenotypic groups. Most indels occurred as single-nucleotide insertions or deletions at tandem repeats with a strong bias towards GC repeats. The indel mutations caused 94 frameshifts in coding sequences (S6 Table). All affected genes coded for full-length proteins in the reference genome FFH2055 (and in MF16_B), whereas frameshift mutations resulted in their pseudogenization in 467_S. This suggests that isolates from phenotypic group A represent a state ancestral to the derived isolate 467_S, regardless the opposite chronology of collection of respective clinical samples. The notion about the evolutionary order of 467_S vs. MF16_B and FFH2055 was further supported by the basal position of MF16_B (and its genotypically identical early isolate FFH2055) in the phylogenetic tree of B. contaminans ST872 (Fig 2), where isolate 467_S appeared as their genomic derivative. Therefore, the longitudinal position of MF16_B, which was isolated from blood culture, indicates that it might have been re-introduced into the patient from an unknown source (such as some disinfectants, cosmetic products, ultrasound gels or stopcocks for i.v. administration that were all found contaminated with B. contaminans in different Argentinian hospitals in the past; JD unpublished data). It is unlikely for a bacterium to remain genetically unchanged for 6 years of chronic infection; a study on B. dolosa reported average genomic substitution rate of approximately 2 SNPs per year of infection [40]. Nevertheless, we cannot definitely rule out an alternative that some members of the clonal phenotypic group A have persisted genetically unchanged, probably in a dormant state intracellularly. The coexistence of diverse lineages as a result of independent transfer of multiple clones into the patient’s bloodstream has been documented in B. dolosa chronic infection [41].
Fig 2. Genetic relationship of sequenced B. contaminans.
The overall tree (top) depicts whole-genome phylogeny inferred from SNPs. Multilocus sequence types of the isolates are denoted in parentheses. Bottom–the subtree containing only ST872 isolates.
80% of the detected mutations in 467_S were present in protein-coding genomic regions that occupied 85% of the genome. Among the intragenic single-base substitution mutations, only 230 (22%) were synonymous. This proportion was even lower than a value of 27.8%, which was calculated by Dillon et al. as the theoretical probability of synonymous coding substitutions in B. cenocepacia HI2424 [42]. Taken together with the high number of frameshift mutations, this proportion of synonymous/nonsynonymous mutations indicated absence of strong selection against mutations that would affect a protein function. We can speculate that most of the gene-inactivating mutations (such as frameshifts, nonsense and missense mutations) are tolerable by bacterial cells because they do not disturb vital processes needed for survival of bacteria in CF sputum. A recent study demonstrated that less than 500 genes were essential for P. aeruginosa growth in CF sputum [43], which is a complex environment very rich in organic nutrients that can be utilized by bacteria. Furthermore, it is known that mutations that negatively affect nutrient uptake or their utilization in a CF sputum are not eliminated by natural selection as rapidly as in more selective environments, as manifested by the frequently observed auxotrophy of Bcc and P. aeruginosa CF isolates [44, 45].
Regarding the differences in transcriptomes of the two lineages, we found no mutations in genes encoding signal molecule synthases or in their upstream sequences, implicating physiological rather than genetically-based causes of QS expression differences between MF16_B and 467_S. A considerable number of missense and frameshift mutations were identified within motility and chemotaxis genes (7 and 4, respectively–S3 Table). In combination with the very low expression of motility genes and a lack of swimming motility in 467_S and other isolates from phenotypic group B, it indicates that these genes are dispensable during long-term B. contaminans persistence in CF sputum. Decreased production of occidiofungin in isolate 467_S can be, in addition to its diminished transcription, explained by frameshift mutations in two biosynthetic genes (S4 Table).
Hypermutability in ST872
The spectrum of mutations that distinguished 467_S from MF16_B demonstrated a highly uneven distribution in particular mutation types (Table 4). Notably, a large excess (approximately 30-fold) of transitions compared with transversion nucleotide substitutions was observed. AT→GC mutations prevailed among transitions and accounted for 70% of all nucleotide substitutions. This pattern of mutations was in perfect accordance with the values obtained in a mutation-accumulation experiment with mismatch repair (MMR)-deficient Escherichia coli [46], which is believed to provide the best estimation of the spontaneous mutation rate in bacteria [47]. Therefore, we looked closely at mutations in the genes encoding MMR enzymes, mutS and mutL. Indeed, one amino acid change was identified in each of the translated proteins in 467_S isolate: 628N→D (1882A→G) in MutS and 317S→P (949A→G) in MutL (S6 Table). Both mutations were localized in conserved domains of the MMR proteins and may have therefore abolished their proper function in DNA repair. Notably, both mutations were unique for 467_S and have not been detected in the study that examined many hypermutable Argentinean B. contaminans isolates [48]. Additional analysis of homologous protein sequences of other Bcc clearly showed that corresponding amino acids in isolates FFH2055 and MF16_B represented wild-type state (not shown). These findings suggest that both mutations in MutS and MutL detected in isolate 467_S have arisen during the evolution of ST872 within the patient´s lungs.
Calculation of the mutation frequency confirmed that isolate 467_S, which carried both MMR mutations, was a hypermutator. The mutation frequency of this isolate was over two orders of magnitude higher than that of MF16_B. FFH2055, the first isolate from the patient, exhibited a normal mutation frequency. All other ST872 isolates, collected after FFH2055 and belonging to the phenotypic group B, were hypermutable (Table 3). Thus, the mutator phenotype likely arose early after patient´s colonization and has persisted since then. We assume that during the chronic infection, mutations that escaped correction by MMR have accumulated. The emergence of hypermutators during evolution in CF lungs is a widespread phenomenon that was reported in P. aeruginosa [49], Staphylococcus aureus and Haemophilus influenzae [50, 51]. Recently, Martina et al. reported a high prevalence of hypermutable B. contaminans among Argentinean CF patients [48]. Our findings are the first to demonstrate the effect of MMR deficiency on the genomic evolution of B. contaminans during infection in CF hosts.
Conclusions
The presented work is the first study that focuses on the genomic and transcriptomic changes that underlie the progression of chronic B. contaminans infection in CF. Unlike other Bcc species, only little attention has been paid to the mechanisms of pathogenicity of B. contaminans. This lack of information is worrisome in light of increasing proportion and even predominance of B. contaminans infections in Argentina [12], Spain [11] and other Ibero-American countries [8], indicating a large and characteristic geographic distribution of considerable concern.
Our results demonstrate that B. contaminans sequential isolates can hold high genomic diversity despite belonging to the same ST type. This diversity results in differential gene expression patterns, which in turn modify bacterial virulence. We demonstrate the profound effect of DNA repair deficiency on the evolution of the B. contaminans genome in the stable environment of CF sputum. Observed mutational and transcriptional inactivation of Bcc virulence factors might help to unravel the mechanisms that drive the adaptation of this primarily environmental bacterium to the CF lung during chronic infection.
Materials and Methods
Bacterial strains
Seven isolates of B. contaminans ST872 were obtained from the research Burkholderia strain collection of the University of Buenos Aires, Argentina. All isolates originated from a female CF patient (homozygous for the F508del mutation; born in January 2001) who attended Hospital de Niños Ricardo Gutierrez (Buenos Aires) and provided clinical specimens for microbiological investigation as part of her standard CF care. B. contaminans was cultivated from the patient´s sputum for the first time in July 2005; the lung remained infected since that time. S. aureus was isolated for the first time in January 2002 (episodes of pulmonary exacerbations were treated with amoxicilin-clavulanic acid); chronic infection with P. aeruginosa was first observed in March 2007 (inhaled tobramycin was administered in on/off cycles). Between 2008 and 2009, pulmonary function began to decrease (forced expiratory volume in 1 second [FEV1] 45–48% predicted). The patient started to be on frequent courses of outpatient anti-infective therapy (oral minocycline or trimethoprim-sulfamethoxazole), and was also admitted at the hospital every three months for i.v. antibiotic treatment, combining two or more of the following drugs: piperacillin-tazobactam, ceftazidime, amikacin, colistin and trimethoprim-sulfamethoxazole. The patient died in the intensive care unit in October 2011. The death was attributed to B. contaminans septicemia.
Bacterial cultivation and RNA extraction
All bacterial cultures were incubated at 37°C with shaking at 230 rpm in 50 ml Falcon conical centrifugation tubes. The bacteria were cultivated in synthetic basal salts medium (BSM) containing 14.3 mM glucose and 0.05% casamino acids [52], CF serum (pooled serum samples from 5 CF patients in various stages of infection, heat-inactivated at 56°C for 30 minutes) and CF sputum (pooled sputum samples from 5 CF patients of various stages of infection, diluted with BSM to a final concentration of 10% w/vol). We used CF serum and sputum samples that were collected and archived for the purpose of this study at the Motol University Hospital, Prague.
Starter cultures were grown overnight in Luria-Bertani broth and diluted to an OD600 of 0.5. The bacteria were harvested by centrifugation and resuspended in the same volume of BSM, serum or sputum. In total, 4 ml of BSM and serum and 6.25 ml of sputum were used. The cultures were incubated for 270 minutes (37°C, 230 rpm) into the mid-log growth phase. Then, the cultures were immediately snap-cooled in liquid nitrogen and centrifuged (5 minutes, 160 x g, 4°C). The pellets were resuspended in 1 ml of Trizol (Ambion), vortexed thoroughly and incubated for 5 minutes at room temperature. Chloroform (0.2 ml) was added; the samples were shaken for 30 seconds and incubated for 3 minutes at room temperature. The samples were centrifuged (15 minutes, 160 x g, 4°C), and the upper water phase was transferred to 0.5 ml of ice-cold 70% ethanol. The RNA samples were further processed using the RiboPure-Bacteria kit (Ambion), and the MICROBExpress kit (Invitrogen) was subsequently used to remove rRNA. The quality of the mRNA was assessed using Total RNA Nano chips on a Bioanalyzer 2100 (Agilent).
The complete procedure (bacterial cultivation, RNA extraction) was repeated three times to obtain three biological replicates per condition.
Whole genome sequencing and comparative genomics
To extract the genomic DNA, overnight cultures were grown in LB broth and DNA was extracted from collected cells using the DNAeasy kit (Qiagen). The genomic DNA was converted into a sequencing-ready library using the Nextera XT DNA sample Preparation Kit (Illumina). Illumina sequencing was performed in a MiSeq personal sequencing system with TrueSeqTM sequencing by synthesis (SBS) reversible terminator chemistry at the Next Generation Sequencing Core Platform at the Manitoba Institute for Children’s Health (MICH). Data output was assembled by Velvet [53]. Sequencing reads were deposited at the ArrayExpress Archive of Functional Genomics Data (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-4649.
Sequencing reads were subsequently mapped onto B. contaminans FFH2055, a reference genome of the multilocus sequence type ST872 [15]. Read mapping was performed with Geneious software 7.1.9 [54] using the Geneious mapper, with a maximum allowed mismatches set to 5%. Variant calling was performed (minimum coverage: 5 reads; minimum variant frequency: 0.75), and the effects of the mutations on the translated proteins were determined.
Complete and draft bacterial genomes for comparison were downloaded from GenBank [55]. A phylogenetic tree was inferred from single nucleotide polymorphisms (SNPs) among the compared genomes using CSI Phylogeny 1.1 [56]. The phylogram was visualized from the tree file using MEGA6 [57].
RNA-Seq and comparative transcriptomics
RNA samples were processed at the vertis Biotechnologie AG (Freising, Germany) service laboratory for cDNA preparation, library preparation (TruSeq, Illumina) and high-throughput sequencing (Illumina MiSeq, 100 bp reads). Raw RNA-Seq sequencing data were deposited at the ArrayExpress Archive of Functional Genomics Data (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-4645.
The read sequences were trimmed of low-quality portions and Illumina-specific adapters using Trimmomatic v 0.32 [58] with the following parameters: ILLUMINACLIP:./adapter.fa:2:30:8, LEADING:13, TRAILING:13, SLIDINGWINDOW:4:19, MINLEN:40, and HEADCROP:10. Any reads matching rRNA or tRNA sequences were removed using SortMeRna v 1.99beta [59] utilizing rRNA sequences from the SILVA [60] and Rfam [61] databases as well as rRNA and tRNA sequences from genome assembly of the reference B. contaminans genome. At each preprocessing step, the technical quality of the reads was assessed with FastQC v0.10.1 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Subsequently, the reads were mapped to the reference genome (this study). Mapping was performed using GSNAP version 2014-02-28 [62] with the default parameters.
Overlaps of the mapped reads with annotated genes were counted within the Bioconductor environment [63] using the packages GenomicFeatures and GenomicAlignments [64]. Generally, 60% of the raw reads mapped to the reference genome, which is typically 5.5 million mapped reads per sample (range 3.1–8.9, median 5.5 million reads). Quality control for the count data was performed utilizing principal component analysis (PCA), hierarchical clustering, and correlation analysis. All samples were considered adequate for further analyses. The count data were fitted with a generalized linear model (~ source + medium), and differences in the expression intensity between B. contaminans isolates 467_S and MF16_B in different cultivation media (serum, sputum, and BSM) were evaluated using DESeq2 [65]. Differentially expressed genes were selected with a 3-fold change and a p-value of less than 0.05 as the default cutoff. This choice was made due to the high fluctuation of levels of expression among replicates and to select the genes with most pronounced expression changes between the lung and bloodstream isolates.
The nucleotide sequences of differentially expressed genes were subjected to BLAST search [66] against complete Burkholderia genomes. Homologous genes from the first sequenced and best-studied B. cenocepacia strain J2315 [24] were assigned when possible. In cases where no homologs were identified in B. cenocepacia J2315, homologs from other completely sequenced Burkholderia strains with the highest similarity were chosen. Predicted functions were assigned to genes according to the Burkholderia Genome Database [67].
Phenotypic assays
Hemolysis was determined on Columbia sheep blood agar plates (Oxoid) after 48 hours of incubation at 37°C. Protease production was determined on D-BHI agar plates [68] after 48 hours of incubation at 37°C. Production of antifungal substances was determined by the modified method of Chen et al. [69]. Five microliters of B. contaminans suspension (OD600 = 1) were spot-inoculated on a malt extract agar plate (Oxoid) and incubated at 37°C for 48 hours. The indicator fungus Geotrichum candidum was then replica-plated from a completely covered agar plate (spread-inoculated and incubated for 3 days at room temperature) onto the B. contaminans plate. The inhibition zones were recorded after 24 hours of incubation at 30°C. Swimming motility was determined on Luria-Bertani (LB) plates containing 0.3% (w/v) Bacto agar (Sigma-Aldrich). The plates were inoculated by injecting 2 μl of bacteria in LB broth (OD600 = 1) under the surface of the center of the soft agar plate. The swimming zones were recorded after 48 hours of incubation at 37°C. The mutation frequency was determined as described in [48] with modifications. One microliter of an overnight culture grown from a single colony was diluted in 1 ml of LB broth. One microliter of the diluted culture was then inoculated into 5 ml of LB broth. The cultures were incubated at 37°C with shaking for 24 hours. The cell concentration was then determined by serial dilution of the cultures, followed by plating 100 μl aliquots in parallel onto LB agar and LB agar + rifampicin (100 μg/ml). Colonies were counted after incubation at 37°C for 48 hours, and the mutation frequency was calculated as the fraction of rifampicin-resistant cells in the population. The procedure was repeated three times per isolate.
Ethics statement
Sputum and serum samples used for bacterial cultivation were taken with the written informed consent of the CF adult subjects. The study was approved by the Ethics Committee for Multi-centric clinical trials of the University Hospital Motol, Prague on September 22, 2010, No.4.2.6.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Nucleotide sequence and reading frame of affected genes was assessed by comparison with homologues in B. contaminans MS14 genome.
(XLSX)
Differences were detected by mapping Illumina reads of 467_S on the FFH2055 reference assembly and subtracting variants present also in MF16_B. The mutations are denoted in the direction MF16_B → 467_S.
(XLSX)
Acknowledgments
We are grateful to Dr. Blanka Zikanova for providing Geotrichum candidum and to Dr. Laura Galanternik for providing the B. contaminans isolates from primary cultures.
Data Availability
All files are available from the ArrayExpress Archive of Functional Genomics Data under accession number E-MTAB-4645 and E-MTAB-4649.
Funding Statement
Ministry of Health of the Czech Republic, NT12405-5, www.iga.mzcr.cz; LK, PD, Ministry of Health of the Czech Republic, 15-28017A, www.azvcr.cz; PD, JN, Grant Agency of the Charles University, GAUK640214, www.cuni.cz/UK-33.html; LK, PD, and Academy of Sciences of the Czech Republic, RVO68378050, www.avcr.cz; MK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3(2):144–56. 10.1038/nrmicro1085 . [DOI] [PubMed] [Google Scholar]
- 2.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16(7):821–30. 10.1111/j.1469-0691.2010.03237.x . [DOI] [PubMed] [Google Scholar]
- 3.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23(2):299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalish LA, Waltz DA, Dovey M, Potter-Bynoe G, McAdam AJ, Lipuma JJ, et al. Impact of Burkholderia dolosa on lung function and survival in cystic fibrosis. Am J Respir Crit Care Med. 2006;173(4):421–5. 10.1164/rccm.200503-344OC . [DOI] [PubMed] [Google Scholar]
- 5.Blackburn L, Brownlee K, Conway S, Denton M. 'Cepacia syndrome' with Burkholderia multivorans, 9 years after initial colonization. J Cyst Fibros. 2004;3(2):133–4. 10.1016/j.jcf.2004.03.007 . [DOI] [PubMed] [Google Scholar]
- 6.Jones AM, Dodd ME, Govan JR, Barcus V, Doherty CJ, Morris J, et al. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax. 2004;59(11):948–51. 10.1136/thx.2003.017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, et al. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59(Pt 1):102–11. 10.1099/ijs.0.001123-0 . [DOI] [PubMed] [Google Scholar]
- 8.Cunha MV, Leitão JH, Mahenthiralingam E, Vandamme P, Lito L, Barreto C, et al. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a 7-year study. J Clin Microbiol. 2003;41(9):4113–20. 10.1128/JCM.41.9.4113-4120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordá-Vargas L, Degrossi J, Castañeda NC, D'Aquino M, Valvano MA, Procopio A, et al. Prevalence of indeterminate genetic species of Burkholderia cepacia complex in a cystic fibrosis center in Argentina. J Clin Microbiol. 2008;46(3):1151–2. 10.1128/JCM.01595-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahenthiralingam E, Baldwin A, Drevinek P, Vanlaere E, Vandamme P, LiPuma JJ, et al. Multilocus sequence typing breathes life into a microbial metagenome. PLoS One. 2006;1:e17 10.1371/journal.pone.0000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina-Pascual MJ, Valdezate S, Carrasco G, Villalón P, Garrido N, Saéz-Nieto JA. Increase in isolation of Burkholderia contaminans from Spanish patients with cystic fibrosis. Clin Microbiol Infect. 2015;21(2):150–6. 10.1016/j.cmi.2014.07.014 . [DOI] [PubMed] [Google Scholar]
- 12.Martina P, Bettiol M, Vescina C, Montanaro P, Mannino MC, Prieto CI, et al. Genetic diversity of Burkholderia contaminans isolates from cystic fibrosis patients in Argentina. J Clin Microbiol. 2013;51(1):339–44. 10.1128/JCM.02500-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho CP, Barreto C, Pereira L, Lito L, Melo Cristino J, Sá-Correia I. Incidence of Burkholderia contaminans at a cystic fibrosis center with an unusually high representation of Burkholderia cepacia during 15 years of epidemiological surveillance. J Med Microbiol. 2015;64:927–35. 10.1099/jmm.0.000094 . [DOI] [PubMed] [Google Scholar]
- 14.Kalferstova L, Kolar M, Fila L, Vavrova J, Drevinek P. Gene expression profiling of Burkholderia cenocepacia at the time of cepacia syndrome: loss of motility as a marker of poor prognosis? J Clin Microbiol. 2015;53(5):1515–22. 10.1128/JCM.03605-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloodworth RA, Selin C, López De Volder MA, Drevinek P, Galanternik L, Degrossi J, et al. Draft genome sequences of Burkholderia contaminans, a Burkholderia cepacia complex species that is increasingly recovered from cystic fibrosis patients. Genome Announc. 2015;3(4). 10.1128/genomeA.00766-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramoni S, Sokol PA. Quorum sensing systems influence Burkholderia cenocepacia virulence. Future Microbiol. 2012;7(12):1373–87. 10.2217/fmb.12.118 . [DOI] [PubMed] [Google Scholar]
- 17.Gingues S, Kooi C, Visser MB, Subsin B, Sokol PA. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J Bacteriol. 2005;187(24):8247–55. 10.1128/JB.187.24.8247-8255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect Immun. 2006;74(7):4083–93. 10.1128/IAI.00297-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding L-ornithine N(5)-oxygenase. Infect Immun. 1999;67(9):4443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lameignere E, Malinovská L, Sláviková M, Duchaud E, Mitchell EP, Varrot A, et al. Structural basis for mannose recognition by a lectin from opportunistic bacteria Burkholderia cenocepacia. Biochem J. 2008;411(2):307–18. 10.1042/bj20071276 . [DOI] [PubMed] [Google Scholar]
- 21.Lameignere E, Shiao TC, Roy R, Wimmerova M, Dubreuil F, Varrot A, et al. Structural basis of the affinity for oligomannosides and analogs displayed by BC2L-A, a Burkholderia cenocepacia soluble lectin. Glycobiology. 2010;20(1):87–98. 10.1093/glycob/cwp151 . [DOI] [PubMed] [Google Scholar]
- 22.Sulák O, Cioci G, Lameignère E, Balloy V, Round A, Gutsche I, et al. Burkholderia cenocepacia BC2L-C is a super lectin with dual specificity and proinflammatory activity. PLoS Pathog. 2011;7(9):e1002238 10.1371/journal.ppat.1002238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber B, Feldmann F, Köthe M, Vandamme P, Wopperer J, Riedel K, et al. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect Immun. 2004;72(12):7220–30. 10.1128/IAI.72.12.7220-7230.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeño-Tárraga AM, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;191(1):261–77. 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Grady EP, Viteri DF, Malott RJ, Sokol PA. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics. 2009;10:441 10.1186/1471-2164-10-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKeon SA, Nguyen DT, Viteri DF, Zlosnik JE, Sokol PA. Functional quorum sensing systems are maintained during chronic Burkholderia cepacia complex infections in patients with cystic fibrosis. J Infect Dis. 2011;203(3):383–92. 10.1093/infdis/jiq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mira NP, Madeira A, Moreira AS, Coutinho CP, Sá-Correia I. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients' airways and antimicrobial therapy. PLoS One. 2011;6(12):e28831 10.1371/journal.pone.0028831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zlosnik JE, Mori PY, To D, Leung J, Hird TJ, Speert DP. Swimming motility in a longitudinal collection of clinical isolates of Burkholderia cepacia complex bacteria from people with cystic fibrosis. PLoS One. 2014;9(9):e106428 10.1371/journal.pone.0106428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.el-Banna N, Winkelmann G. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol. 1998;85(1):69–78. 10.1046/j.1365-2672.1998.00473.x . [DOI] [PubMed] [Google Scholar]
- 30.Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, et al. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ Microbiol. 2009;11(6):1422–37. 10.1111/j.1462-2920.2009.01870.x . [DOI] [PubMed] [Google Scholar]
- 31.Gu G, Smith L, Wang N, Wang H, Lu SE. Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem Biophys Res Commun. 2009;380(2):328–32. 10.1016/j.bbrc.2009.01.073 . [DOI] [PubMed] [Google Scholar]
- 32.Lu SE, Novak J, Austin FW, Gu G, Ellis D, Kirk M, et al. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry. 2009;48(35):8312–21. 10.1021/bi900814c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu G, Smith L, Liu A, Lu SE. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl Environ Microbiol. 2011;77(17):6189–98. 10.1128/AEM.00377-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu G, Wang N, Chaney N, Smith L, Lu SE. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol Lett. 2009;297(1):54–60. 10.1111/j.1574-6968.2009.01653.x . [DOI] [PubMed] [Google Scholar]
- 35.Thomson EL, Dennis JJ. A Burkholderia cepacia complex non-ribosomal peptide-synthesized toxin is hemolytic and required for full virulence. Virulence. 2012;3(3):286–98. 10.4161/viru.19355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapalain A, Vial L, Laprade N, Dekimpe V, Perreault J, Déziel E. Identification of quorum sensing-controlled genes in Burkholderia ambifaria. Microbiologyopen. 2013;2(2):226–42. 10.1002/mbo3.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sass AM, Schmerk C, Agnoli K, Norville PJ, Eberl L, Valvano MA, et al. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J. 2013;7(8):1568–81. 10.1038/ismej.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z, Falkinham JO, Tawfik KA, Jeffs P, Bray B, Dubay G, et al. Burkholdines from Burkholderia ambifaria: antifungal agents and possible virulence factors. J Nat Prod. 2012;75(9):1518–23. 10.1021/np300108u . [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Escribano JP, Castro JF, Razmilic V, Chandra G, Andrews B, Asenjo JA, et al. The Streptomyces leeuwenhoekii genome: de novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genomics. 2015;16:485 10.1186/s12864-015-1652-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–80. 10.1038/ng.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46(1):82–7. 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillon MM, Sung W, Lynch M, Cooper VS. The rate and molecular spectrum of spontaneous mutations in the GC-rich multichromosome genome of Burkholderia cenocepacia. Genetics. 2015;200(3):935–46. 10.1534/genetics.115.176834 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 2015;112(13):4110–5. 10.1073/pnas.1419677112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barth AL, Pitt TL. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J Med Microbiol. 1996;45(2):110–9. . [DOI] [PubMed] [Google Scholar]
- 45.Barth AL, Pitt TL. Auxotrophy of Burkholderia (Pseudomonas) cepacia from cystic fibrosis patients. J Clin Microbiol. 1995;33(8):2192–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 2012;109(41):E2774–83. 10.1073/pnas.1210309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams AB. Spontaneous mutation rates come into focus in Escherichia coli. DNA Repair (Amst). 2014;24:73–9. 10.1016/j.dnarep.2014.09.009 . [DOI] [PubMed] [Google Scholar]
- 48.Martina P, Feliziani S, Juan C, Bettiol M, Gatti B, Yantorno O, et al. Hypermutation in Burkholderia cepacia complex is mediated by DNA mismatch repair inactivation and is highly prevalent in cystic fibrosis chronic respiratory infection. Int J Med Microbiol. 2014;304(8):1182–91. 10.1016/j.ijmm.2014.08.011 . [DOI] [PubMed] [Google Scholar]
- 49.Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs-a review. Pathogens. 2014;3(3):680–703. 10.3390/pathogens3030680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect. 2010;16(7):798–808. 10.1111/j.1469-0691.2010.03250.x . [DOI] [PubMed] [Google Scholar]
- 51.Oliver A. Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. Int J Med Microbiol. 2010;300(8):563–72. 10.1016/j.ijmm.2010.08.009 . [DOI] [PubMed] [Google Scholar]
- 52.O'Sullivan LA, Weightman AJ, Jones TH, Marchbank AM, Tiedje JM, Mahenthiralingam E. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ Microbiol. 2007;9(4):1017–34. 10.1111/j.1462-2920.2006.01228.x . [DOI] [PubMed] [Google Scholar]
- 53.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–9. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37(Database issue):D26–31. 10.1093/nar/gkn723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9(8):e104984 10.1371/journal.pone.0104984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–7. 10.1093/bioinformatics/bts611 . [DOI] [PubMed] [Google Scholar]
- 60.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(Database issue):D226–32. 10.1093/nar/gks1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–81. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 10.1186/PREACCEPT-8897612761307401 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41(Web Server issue):W29–33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24(23):2803–4. 10.1093/bioinformatics/btn524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sokol PA, Ohman DE, Iglewski BH. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J Clin Microbiol. 1979;9(4):538–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen KC, Ravichandran A, Guerrero A, Deng P, Baird SM, Smith L, et al. The Burkholderia contaminans MS14 ocfC gene encodes a xylosyltransferase for production of the antifungal occidiofungin. Appl Environ Microbiol. 2013;79(9):2899–905. 10.1128/AEM.00263-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Nucleotide sequence and reading frame of affected genes was assessed by comparison with homologues in B. contaminans MS14 genome.
(XLSX)
Differences were detected by mapping Illumina reads of 467_S on the FFH2055 reference assembly and subtracting variants present also in MF16_B. The mutations are denoted in the direction MF16_B → 467_S.
(XLSX)
Data Availability Statement
All files are available from the ArrayExpress Archive of Functional Genomics Data under accession number E-MTAB-4645 and E-MTAB-4649.