Abstract
We present a new method called DAISY (Dual Acquisition orIented ssNMR spectroScopY) for the simultaneous acquisition of 2D and 3D oriented solid-state NMR experiments for membrane proteins aligned in mechanically or magnetically lipid bilayers. DAISY utilizes dual acquisition of sine and cosine dipolar or chemical shift coherences and long living 15N longitudinal polarization to obtain two multi-dimensional spectra, simultaneously. In these new experiments, the first acquisition gives the polarization inversion spin exchange at the magic angle (PISEMA) or heteronuclear correlation (HETCOR) spectra, the second acquisition gives PISEMA-mixing or HETCOR-mixing spectra, where the mixing element enables inter-residue correlations through 15N-15N homonuclear polarization transfer. The analysis of the two 2D spectra (first and second acquisitions) enables one to distinguish 15N-15N inter-residue correlations for sequential assignment of membrane proteins. DAISY can be implemented in 3D experiments that include the polarization inversion spin exchange at magic angle via I spin coherence (PISEMAI) sequence, as we show for the simultaneous acquisition of 3D PISEMAI-HETCOR and 3D PISEMAI-HETCOR-mixing experiments.
Keywords: DAISY, POE, PISEMA, HETCOR, simultaneous acquisition, oriented solid-state NMR, membrane proteins, bicelles, aligned lipid bilayers
INTRODUCTION
The folding and function of membrane proteins are shaped by lipid membranes (Phillips et al. 2009). Therefore, a complete understanding of their structure-function relationships is possible only by characterizing the structures and dynamics of these proteins in native-like lipid bilayer environments. To this extent, oriented sample solid-state NMR (OS ssNMR) represents a powerful technique that enables the measurement of anisotropic NMR parameters for membrane proteins embedded in lipid membranes in the liquid crystalline phase (Veglia et al. 2012). Unlike the more popular magic angle spinning (MAS) technique, OS ssNMR necessitates the careful preparation of membrane protein samples in magnetically or mechanically aligned lipid membranes (Das et al. 2013). Although labor intensive, these preparations make it possible to more accurately control the parameters that contribute to the formation of well-oriented samples, such as hydration, lipid composition and phase, as well as membrane architecture. A key experiment for measuring anisotropic NMR parameters for aligned membrane proteins is the separated local fields (SLF) experiment developed by John Waugh (Waugh 1976). SLF experiments enable one to separate anisotropic NMR parameters such as 15N chemical shift (CS) and 15N-1H dipolar coupling (DC) in two dimensions. The most common versions of SLF experiments are PISEMA (Wu et al. 1995; Wu et al. 1994), SAMPI4 (Nevzorov et al. 2007), and HIMSELF (Dvinskikh et al. 2006), which are routinely used to analyze the topology of membrane bound proteins and peptides (Miao and Cross 2013; Murray et al. 2013). In addition, HETCOR experiments are also used to measure 1H and 15N anisotropic chemical shifts (Marassi et al. 2000), offering additional restraints to define the structure and architecture of membrane proteins (Fu et al. 2007). Historically, these experiments have suffered from poor sensitivity and resolution. In recent years, we significantly improved the performance of these experiments by implementing a sensitivity-enhanced (SE) element that enabled us to acquire both 2D and 3D spectra of membrane proteins aligned in lipid bicelles or bilayers (Gopinath et al. 2013). However, resonance assignment remains the Achilles’ hill of OS ssNMR. Although SLF and HETCOR spectra of U-15N labeled proteins give rise to regular patterns that are used to fingerprint membrane protein backbone topology (Marassi and Opella 2000; Mascioni et al. 2004; Mascioni and Veglia 2003; Mesleh et al. 2002; Wang et al. 2000), atomic-resolution structure determination requires residue-specific assignments of the ssNMR spectra (Mote et al. 2013; Traaseth et al. 2009; Verardi et al. 2011; Vostrikov et al. 2013). For single-pass membrane proteins, the regular patterns of the SLF spectra together with selective labeling of amino acids are often sufficient to assign the backbone resonances. However, this approach fails when deviations from ideal helices occur or when multispan membrane proteins are analyzed. In these cases, the amide resonance chemical shifts and dipolar correlations can be severely overlapped. A possible strategy to obtain backbone sequential assignments is to combine SLF experiments with through-space correlations of backbone amide nitrogens via 15N-15N polarization transfer schemes (Knox et al. 2010; Tang et al. 2012). There are three different strategies to accomplish polarization transfer: homonuclear 15N-15N polarization transfer via proton-driven spin diffusion (PDSD) (Suter and Enrnst 1985; Szeverenyi et al. 1982), cross-relaxation driven spin diffusion (CRDSD) (Xu et al. 2008), or mismatch Hartmann-Hahn (MMHH) (Nevzorov 2008). While PDSD uses longitudinal polarization transfer and provides sequential 15N-15N correlations, CRDSD and MMHH are both based on transverse polarization transfer and provide long-range inter-residue correlations (up to 10 Ǻ). A systematic study of these three polarization transfer techniques has been reported by our group, where we noted that the relative sensitivity of these experiments plays an important role in the design of the experiment to be carried out (Traaseth et al. 2010). In fact, high lipid-to-protein ratios are often necessary to obtain well-ordered bicellar phases (Veglia et al. 2012). As a result, ordered bicelle preparations contain only a few milligrams of U-15N labeled proteins, making their spectroscopic analysis quite challenging. In addition, the current probe technology, although significantly improved with the low-E coils (Gor'kov et al. 2007), requires long relaxation delays to prevent sample overheating. For these reasons, we propose a new method called DAISY (Dual Acquisition orIented ssNMR spectroScopY) that enables the acquisition of two OS ssNMR spectra, simultaneously. The overall philosophy of this approach impinges on our recent developments in the field of MAS NMR sequences for the dual and multiple acquisitions of 2D and 3D experiments (Gopinath and Veglia 2012a; Gopinath and Veglia 2012b; Gopinath and Veglia 2013). Specifically, we make use of the long living 15N polarization to acquire two experiments with only one pulse sequence and without the addition of a second receiver.
We demonstrate the performance of DAISY for the simultaneous acquisition of 2D PISEMA and 2D PISEMA-mixing experiments as well as 2D HETCOR and 2D HETCOR-mixing experiments. Finally, we also implemented a 3D experiment to simultaneously acquire PISEMAI-HETCOR and PISEMAI-HETCOR-mixing spectra. DAISY can be used with other variants of SLF, HETCOR and homonuclear mixing sequences and will speed up the acquisition of OS experiments without additional hardware.
MATERIAL AND METHODS
Sample preparation
U-15N labeled sarcolipin (SLN) was expressed in E. coli bacteria and purified as reported previously (Buck et al. 2003). Bicelles were prepared by drying 37.2 mg of DMPC and 7.6 mg of DHPC in chloroform into separate glass vials under a stream of N2 gas (DMPC/DHPC molar ratio of 3.2/1). Approximately 2 mg of SLN were reconstituted in a DHPC micelle solution, which was then added to DMPC lipids. Bicelles were formed after 3–5 freeze/thaw cycles, which resulted in a non-viscous solution between 0 and 15 °C and a viscous and clear solution above 30 °C. To align the bicelles with the bilayer normal parallel to the static magnetic fields, we doped our preparations with 5 mM YbCl3. The final volume was adjusted to 160 µL by addition of NMR buffer, giving a final lipid concentration of 28% (w/v). The NAL crystal was prepared as reported by Carroll et al. (Carroll et al. 1990).
NMR experiments
All of the NMR experiments were performed on an Agilent VNMRS spectrometer operating at a 1H frequency of 700 MHz and equipped with a low-E bicelle probe built by the RF program at the National High Magnetic Field Laboratory (NHMFL) in Florida (Gor'kov et al. 2007). A cross-polarization (CP) time of 1 ms was applied with 1H and 15N RF amplitudes set to 50 kHz. 5 µs 90° pulses were used for 1H and 15N, and a 50 kHz SPINAL decoupling (Manning et al. 2002) was used on 1H during the 15N acquisition period. A recycle delay of 3 s and an acquisition time of 15 ms were used with identical parameters for both acquisition periods (t2’ and t2” or t3’ and t3”). All the spectra were acquired with a τ1 value set to 1 ms to dephase any residual transverse magnetization. For SLN, the 15N-15N transfer was obtained using a 3 s PDSD mixing time (τmix), whereas for the NAL single crystal 6 ms MMHH mixing was used. For the PISEMA experiments, the FSLG sequence was used on the 1H channel during the t1 period with an effective RF amplitude of 62.5 kHz. A phase-switched spin-lock was applied on the 15N channel with a 62.5 kHz RF amplitude. The τ delay was set to 100 µs with an FSLG effective field corresponding to 80 kHz. For the HETCOR experiments, the FSLG homonuclear decoupling was used during t1 with 1H and 15N RF amplitudes of 62.5 kHz and 30 kHz, respectively. During the WIM24 heteronuclear polarization transfer, 90° pulses of 5 µs duration were used on both 1H and 15N with a τ delay set to 192 µs. The 2D spectra of the NAL crystal were acquired with 16 scans and 64 increments for 2D PISEMA and 32 increments for 2D HETCOR experiments. The 3D PISEMAI-HETCOR experiment was acquired with 8 scans with 16 t1 and t2 increments. Note that in the PISEMAI block the polarization inversion spin exchange at magic angle occurs via I spin dipolar coherence, dramatically increasing the signal (Gopinath et al. 2010a). For SLN, a total of 5000 scans and 20 t1 increments were used for simultaneous acquisition of the 2D PISEMA and 2D PISEMA-mixing experiments.
RESULTS
Simultaneous acquisition of PISEMA and PISEMA-mixing experiments
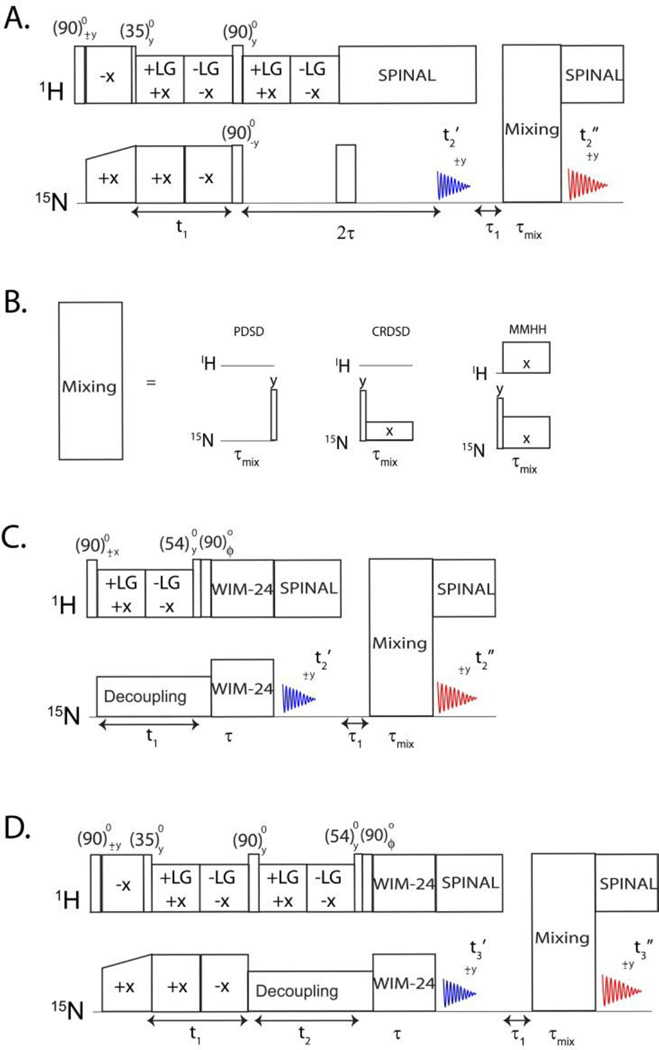
In a typical rotating frame SLF experiment (Ramamoorthy and Yamamoto 2006), polarization is transferred from the abundant I spin (1H) bath to the S spins (15N or 13C) to generate Sx polarization that evolves for a t1 period under I-S DC. The heteronuclear DC Hamiltonian is isolated from homonuclear DC interactions by means of homonuclear dipolar decoupling pulse scheme during t1. The S spins will then evolve under the CS Hamiltonian during the t2 acquisition period, resulting in a two-dimensional spectrum that correlates the S spin chemical shift with I-S DC. In the SE version, a spin-echo is utilized to recover both sine and cosine modulated dipolar coherences, thereby enhancing the signal by up to 40% (Gopinath et al. 2013; Gopinath et al. 2010a; Gopinath and Veglia 2009; Gopinath et al. 2010b). In the new experiment reported in Figure 1A, we utilize the dual acquisition method to separate sine and cosine coherences to give two separate spectra: 2D PISEMA and PISEMA-mixing. In this pulse scheme (Figure 1A), the polarization is transferred from 1H to 15N (I to S) via Hartmann-Hahn CP, a 35° pulse on 1H creates the polarization inversion state (Iz-Sz) in the doubly tilted rotating frame. During the t1 period, the Iz-Sz term evolves under zero quantum dipolar Hamiltonian, thereby creating cosine and sine dipolar coherences. After t1 evolution, a 90° pulse is applied to store the cosine dipolar coherence along the z-direction, whereas the two-spin order sine dipolar coherence is converted into a single spin term in the following 2τ period and detected during the first acquisition period to give a PISEMA spectrum associated with sine dipolar coherence. The evolution of the polarization is summarized as follows:
| (1) |
Figure 1.
(A). Pulse sequence for simultaneous acquisition of PISEMA and PISEMA-mixing experiments corresponding to the first and second acquisitions, respectively. One of the mixing sequences shown in (B) is used prior to the second acquisition. (C) Pulse sequence for the simultaneous acquisition of HETCOR and HETCOR-mixing experiments. (D) Pulse sequence for the simultaneous acquisition of two 3D experiments (PISEMAI-HETCOR and PISEMAI-HETCOR-mixing experiments).
In equation 1, U’ and U” refer to the rotating frame transformations and θm is the magic angle; SPISEMA, and SFSLG are dipolar scaling factors during t1 and τ periods, respectively; DIS and ωs are the I-S DC and S spin CS. The cosine dipolar term evolves through the mixing period followed by the second acquisition, giving a 2D PISEMA-mixing spectrum. Depending on the proximity of the 15N spin pairs as well as the length and type of mixing sequence, each Sx spin operator creates multiple cross peak terms () resulting from 15N-15N polarization transfer:
| (2) |
where . In the latter expression, H is the rotating frame Hamiltonian for PDSD, MMHH, or CRDSD (Traaseth et al. 2010) and τmix is the length of the mixing period. Note that the 15N nuclei have long T1 relaxation times, hence the cosine dipolar term can be stored without loss of polarization while recording the first FID. For the PDSD sequence, a 90° pulse is applied after the longitudinal mixing, while for the MMHH and CRDSD sequences the 90° pulse is applied prior to transverse mixing. The mixing times (τ) are typically 3s, 10 ms, and 6 ms for PDSD, CRDSD, and MMHH respectively. As we have shown previously, the PDSD sequence gives optimal cross peak intensities for sequential (i, i+1) correlations; on the other hand, the CRDSD and MMHH are more efficient for long-range distances of 6 to 10 Å.
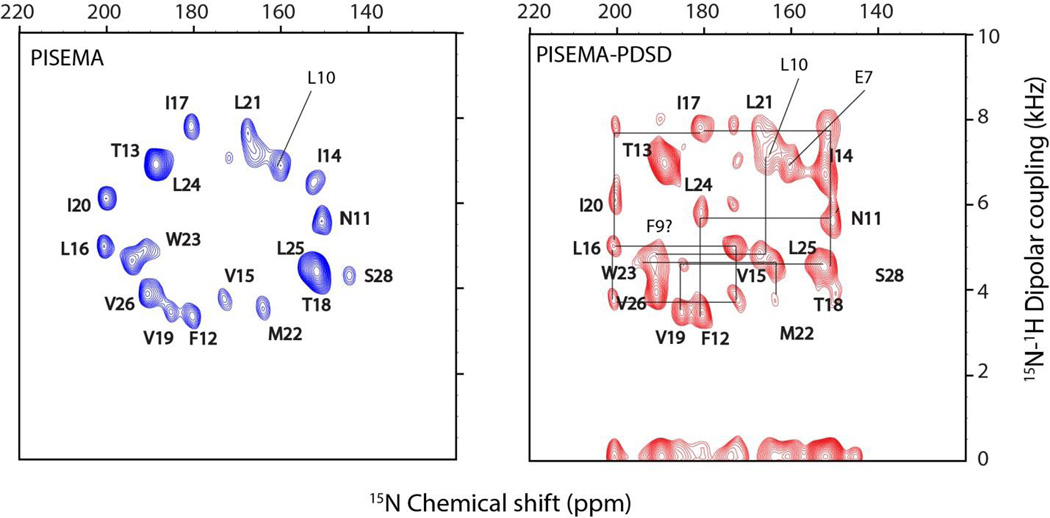
To demonstrate the performance of DAISY, we used the pulse sequence in Figure 1A to acquire 2D PISEMA and PISEMA-mixing experiments both on a single crystal of N-acetyl-leucine (NAL) and SLN in oriented bicelles. Figure 2A shows PISEMA and PISEMA-mixing experiments on the NAL crystal using the MMHH polarization transfer scheme with a 6ms mixing period. The four fold symmetry of the crystal unit generates a set of four distinct resonances with different values of the CS and DC. For SLN, we obtained intense cross peaks between sequential residues using a 3 s PDSD transfer (Figure 3). The sequential (i,i+1) inter-residue cross peak positions are in close agreement with the previous assignment of this protein obtained from a 3D version of PDSD experiment.
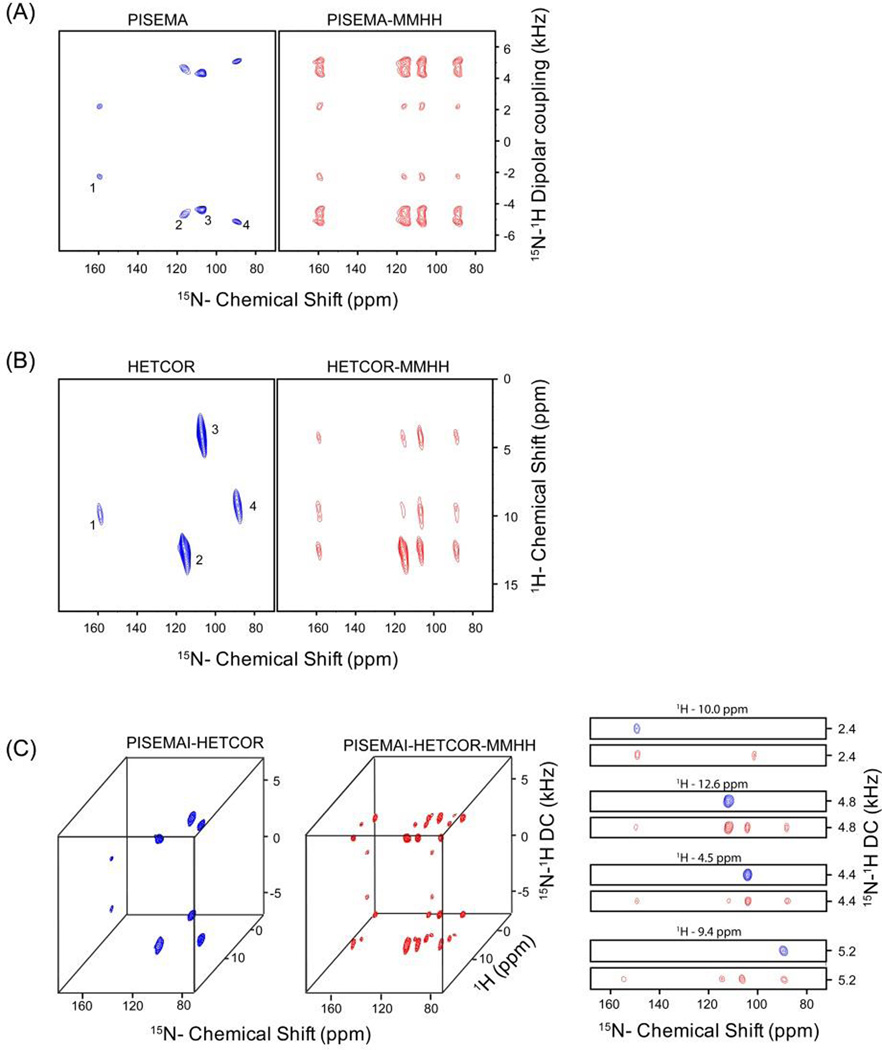
Figure 2.
Simultaneous acquisition of two and three-dimensional experiments on a NAL single crystal using the pulse sequences shown in Figure 1. (A) PISEMA and PISEMA-mixing, (B) HETCOR and HETCOR-mixing, (C) 3D PISEMAI-HETCOR and PISEMAI-HETCOR-mixing spectra obtained from respective sequences shown in Figure 1. MMHH (Figure 1B) was used with a 6 ms mixing period prior to the second acquisition. The spectra obtained in the first and second acquisitions (Figure 1) are shown in blue and red colors respectively; where the cross peaks resulting from 15N-15N mixing can be identified in the red spectra (along 15N dimension) by comparing the spectral positions with respect to reference spectrum shown in blue. Peaks from left to right in the 15N dimension are labeled as 1, 2, 3, and 4. The 2D strip plots shown in (C) are taken along the 1H chemical shift dimension of the 3D spectra.
Figure 3.
PISEMA and PISEMA-PDSD spectra of sarcolipin (SLN), acquired simultaneously using the pulse sequence shown in Figure 1A. PDSD mixing time of 3 s was used prior to the second acquisition. The cross peaks in the PISEMA-PDSD spectrum are identified by comparing with the reference PISEMA spectrum shown in blue. The cross peak positions are in agreement with previous assignment taken from reference (K. R. Mote et al, J. Biomol. NMR 2011, 51, 3, 339–346).
Simultaneous acquisition of 2D HETCOR and HETCOR-mixing experiments
The heteronuclear correlation (HETCOR) experiment maps 1H and 15N chemical shifts of 15N-1H spin systems. Sequential assignment can also be achieved via proton dimension by incorporating 15N-15N mixing in a 2D HETCOR experiment. For this purpose, we designed a new pulse scheme to simultaneously acquire two 2D experiments, HETCOR and HETCOR-mixing as reported in Figure 1C. A 90° pulse creates 1H single quantum coherence followed by chemical shift evolution during t1 under FSLG homonuclear decoupling (Bielecki et al. 1990). 15N spins are decoupled during t1 evolution using continuous wave (CW) decoupling. Cosine and sine modulated 1H chemical shift spin operators (Ix and Iz) are then transferred to 15N using WIM24 (Windowless Isotropic Mixing) cross polarization (Caravatti et al. 1983). The transverse polarization (Ix) is acquired during the first acquisition (t2’), whereas the longitudinal polarization undergoes homonuclear 15N-15N longitudinal/transverse mixing followed by the second acquisition (t2”). The phase ϕ of the 90° pulse prior to WIM transfer is alternated between×and y for the States mode of acquisition of the t1 dimension. A two-dimensional Fourier transform of the first (t1, t2’) and second (t1, t2”) acquisitions gives the 2D HETCOR and HETCOR-mixing spectra, respectively, where the cross peaks associated with each 15N-1H spin system are displayed along the 15N dimension. The evolution of the polarization is described as it follows:
| (3) |
In equation 3, ωI and ωs are the chemical shifts for spin I and S, respectively, and SWIM24 is the scaling factor (~0.66) for the WIM24 sequence. indicates the cross peaks as for equation 2. Figure 2B shows the HETCOR and HETCOR-mixing spectra of NAL single crystal using the pulse sequence of Figure 1B, with a 6 ms MMHH mixing period prior to the second acquisition.
Simultaneous acquisition of 3D experiments
The pulse sequence of Figure 1B can be easily extended to a 3D experiment by incorporating 15N-1H DC evolution prior to the 1H CS evolution period. This modification results in the pulse sequence reported in Figure 1D, which enables the acquisition of 3D PISEMAI-HETCOR and PISEMAI-HETCOR-mixing experiments in the first and second acquisitions, respectively. The spin operator formalism is similar to equations 1–3, where the 1H dipolar coherences are evolved during the t1 evolution period, and the chemical shift evolution takes place during t2, which is followed by WIM24 CP. As for the HETCOR experiment, two polarization transfer pathways result from the WIM24-CP sequence, which are then used to record 3D PISEMAI-HETCOR and PISEMAI-HETCOR-mixing experiments. To test this pulse sequence, we used an NAL single crystal; the resulting 3D PISEMAI-HETCOR and 3D PISEMAI-HETCOR-mixing spectra are shown in Figure 2C.
DISCUSSION
Oriented solid-state NMR is the most direct method to obtain the tilt and azimuthal angles of membrane proteins and peptides with respect to the lipid bilayer (Aisenbrey et al. 2013; Marassi and Opella 2000; Wang et al. 2000). A significant advantage of this approach is that the orientational restraints can be derived from the analytical expression of DC and CS values, or alternatively, obtained by refining protein structures using these values as harmonic restraints in simulated annealing calculations (De Simone et al. 2014; Shi et al. 2009). While the assignment procedures of microcrystalline and membrane proteins under magic angle spinning (MAS) conditions are now established (Hong et al. 2012; McDermott 2009; Siegal et al. 1999; Wang and Ladizhansky 2014), assignment continues to be a major bottleneck for OS ssNMR (Veglia et al. 2012) and in many cases site-specific resonance assignment can only be obtained using selectively labeled peptides and proteins (Michalek et al. 2013; Resende et al. 2014). A first example of the sequential assignment of a membrane protein was demonstrated for M2 domain of the influenza virus by Opella and co-workers (Marassi et al. 2000). Also, Nevzorov and co-workers proposed a 2D SLF experiment in which a 15N-15N MMHH scheme was used during the mixing period after DC evolution to sequentially assign the fd coat protein, with the assignment of the cross peaks of the SLF spectrum by comparison with the classical SLF spectrum (Knox et al. 2010; Tang et al. 2012). Our group recently applied a similar approach for the sequential resonance assignments of SLN in lipid bicelles using both 2D and 3D oriented solid-state NMR experiments (Mote et al. 2013). The new method DAISY presented here takes one step forward in the assignment protocol of oriented membrane proteins by integrating PISEMA and HETCOR experiments with 15N-15N polarization transfer. Sequential assignment of 15N-1H spin pairs can be obtained by comparing two spectra acquired simultaneously, thus avoiding the discrepancies in resonance positions due to different sample preparations and/or spectrometer instability. For single pass membrane proteins, the 2D version of DAISY shown in Figure 1A and B can used as a quick method for confirming the topology together with other experimental data and/or along with pattern recognition such as PISA wheels (Marassi and Opella 2000; Mascioni et al. 2004; Mascioni and Veglia 2003; Mesleh et al. 2002; Wang et al. 2000). As shown in Figure 2 for a NAL single crystal in arbitrary orientation, peaks 1 and 4 are well resolved in the dipolar dimension (Figure 2A), whereas peaks 2 and 3 are resolved in the 1H dimension (Figure 2B). This leads to unambiguous cross peak intensities for overlapped resonances in the F1 dimension of the 2D spectra. On the other hand, the 3D spectra shown in Figure 2C resolves all four peaks in either the F1 (DC) or F2 (1H CS) indirect dimensions.
Figure 3 shows the application of DAISY to SLN reconstituted in oriented in lipid bicelles. While the spectra of the single-span membrane protein SLN are relatively well-resolved, multispan membrane proteins display fairly overlapped spectra; in these cases, the 3D experiments will be crucial to determine the assignments of the spectra. A shortcoming of this approach is the low sensitivity of the second FID. This is due to weak polarization transfer efficiency among 15N nuclei as well as T1 or T1ρ relaxation effects during the 15N-15N mixing period. In other words, the first acquisition does not require as many scans as the second acquisition. However, the first acquisition that gives a reference spectrum may be counted a bonus if one decides to acquire 2D or 3D sequential correlation experiments. Moreover, the simultaneous acquisition of reference and the cross peaks spectra avoids the possibility of misinterpreting the peak positions caused by sample or spectrometer instability. Although this method is demonstrated for membrane proteins embedded in bicelle preparations, it can also be applied to mechanically aligned membrane proteins. Of course, larger line widths of mechanically aligned samples will limit the cross peak resolution, as bicelles give rise to sharper 15N resonances (Durr et al. 2012) However, based on the improvements in sample preparation, spectroscopy, and probe technology, we believe that DAISY represents a promising avenue by which to expand the boundaries of OS ssNMR.
CONCLUSIONS
Although selective labeling and computational approaches will continue to be important, sequential assignment is the only approach that allows full assignment of large membrane proteins. The new method presented in this paper (DAISY) enables one to acquire two PISEMA spectra simultaneously, with and without 15N-15N homonuclear polarization transfer. These two spectra together can be used for the sequential assignment of the 15N-1H spin systems of membrane proteins. Furthermore, DAISY was implemented for 2D HETCOR as well as 3D PISEMAI-HETCOR experiments. Together with recent developments in sample preparations and probe technology, the application of sensitivity enhanced methods (Gopinath et al. 2013) and dual acquisition methods such as DAISY will lead to the high-throughput determination of the structural topology of membrane proteins.
Acknowledgments
This work is supported by the National Institute of Health (GM 64742 and GM 72701). The NMR experiments were carried out at the Minnesota NMR Center.
REFERENCES
- Aisenbrey C, Michalek M, Salnikov ES, Bechinger B. Solid-state NMR approaches to study protein structure and protein-lipid interactions. Methods in molecular biology. 2013;974:357–387. doi: 10.1007/978-1-62703-275-9_16. [DOI] [PubMed] [Google Scholar]
- Bielecki A, Kolbert AC, de Groot HJM, Griffin RG, Levitt MH. Frequency-Switched Lee-Goldburg Sequences in Solids. Advances in Magnetic Resonance. 1990;14:111–124. [Google Scholar]
- Buck B, Zamoon J, Kirby TL, DeSilva TM, Karim C, Thomas D, Veglia G. Overexpression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein expression and purification. 2003;30:253–261. doi: 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- Caravatti P, Braunschweiler L, Ernst RR. Heteronuclear correlation spectroscopy in rotating solids. Chemical Physics Letters. 1983;100:305–310. [Google Scholar]
- Carroll PJ, Stewart PL, Opella SJ. Structures of Two Model Peptides: N-Acetyl-D, L-Valine and N-Acetyl-L-Valyl-L-Leucine. Acta Crystallogr, Sect C. 1990;46:243–246. [Google Scholar]
- Das N, Murray DT, Cross TA. Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. Nature protocols. 2013;8:2256–2270. doi: 10.1038/nprot.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A, Mote KR, Veglia G. Structural dynamics and conformational equilibria of SERCA regulatory proteins in membranes by solid-state NMR restrained simulations. Biophysical journal. 2014;106:2566–2576. doi: 10.1016/j.bpj.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr UH, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chemical reviews. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinskikh SV, Yamamoto K, Ramamoorthy A. Heteronuclear isotropic mixing separated local field NMR spectroscopy. Journal of Chemical Physics. 2006;125 doi: 10.1063/1.2212939. [DOI] [PubMed] [Google Scholar]
- Fu R, Truong M, Saager RJ, Cotten M, Cross TA. High-resolution heteronuclear correlation spectroscopy in solid state NMR of aligned samples. Journal of magnetic resonance. 2007;188:41–48. doi: 10.1016/j.jmr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Gopinath T, Mote KR, Veglia G. Sensitivity and resolution enhancement of oriented solid-state NMR: application to membrane proteins. Progress in nuclear magnetic resonance spectroscopy. 2013;75:50–68. doi: 10.1016/j.pnmrs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Traaseth NJ, Mote K, Veglia G. Sensitivity enhanced heteronuclear correlation spectroscopy in multidimensional solid-state NMR of oriented systems via chemical shift coherences. Journal of the American Chemical Society. 2010a;132:5357–5363. doi: 10.1021/ja905991s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Sensitivity enhancement in static solid-state NMR experiments via single- and multiple-quantum dipolar coherences. Journal of the American Chemical Society. 2009;131:5754–5756. doi: 10.1021/ja900096d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. 3D DUMAS: simultaneous acquisition of three-dimensional magic angle spinning solid-state NMR experiments of proteins. Journal of magnetic resonance. 2012a;220:79–84. doi: 10.1016/j.jmr.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angewandte Chemie. 2012b;51:2731–2735. doi: 10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Orphan spin operators enable the acquisition of multiple 2D and 3D magic angle spinning solid-state NMR spectra. The Journal of chemical physics. 2013;138:184201. doi: 10.1063/1.4803126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Verardi R, Traaseth NJ, Veglia G. Sensitivity enhancement of separated local field experiments: application to membrane proteins. The journal of physical chemistry B. 2010b;114:5089–5095. doi: 10.1021/jp909778a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gor'kov PL, et al. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. Journal of magnetic resonance. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annual review of physical chemistry. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox RW, Lu GJ, Opella SJ, Nevzorov AA. A resonance assignment method for oriented-sample solid-state NMR of proteins. Journal of the American Chemical Society. 2010;132:8255–8257. doi: 10.1021/ja102932n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marassi FM, Ma C, Gesell JJ, Opella SJ. Three-dimensional solid-state NMR spectroscopy is essential for resolution of resonances from in-plane residues in uniformly (15)N-labeled helical membrane proteins in oriented lipid bilayers. Journal of magnetic resonance. 2000;144:156–161. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. Journal of magnetic resonance. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascioni A, Eggimann BL, Veglia G. Determination of helical membrane protein topology using residual dipolar couplings and exhaustive search algorithm: application to phospholamban. Chemistry and physics of lipids. 2004;132:133–144. doi: 10.1016/j.chemphyslip.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Mascioni A, Veglia G. Theoretical analysis of residual dipolar coupling patterns in regular secondary structures of proteins. Journal of the American Chemical Society. 2003;125:12520–12526. doi: 10.1021/ja0354824. [DOI] [PubMed] [Google Scholar]
- McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annual Review of Biophysics, vol 38. Annual Review of Biophysics. 2009:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. Dipolar waves as NMR maps of protein structure. Journal of the American Chemical Society. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Cross TA. Solid state NMR and protein-protein interactions in membranes. Current opinion in structural biology. 2013;23:919–928. doi: 10.1016/j.sbi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M, Salnikov ES, Bechinger B. Structure and topology of the huntingtin 1–17 membrane anchor by a combined solution and solid-state NMR approach. Biophysical journal. 2013;105:699–710. doi: 10.1016/j.bpj.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mote KR, Gopinath T, Veglia G. Determination of structural topology of a membrane protein in lipid bilayers using polarization optimized experiments (POE) for static and MAS solid state NMR spectroscopy. Journal of biomolecular NMR. 2013;57:91–102. doi: 10.1007/s10858-013-9766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Das N, Cross TA. Solid state NMR strategy for characterizing native membrane protein structures. Accounts of chemical research. 2013;46:2172–2181. doi: 10.1021/ar3003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov AA. Mismatched Hartmann-Hahn conditions cause proton-mediated intermolecular magnetization transfer between dilute low-spin nuclei in NMR of static solids. Journal of the American Chemical Society. 2008;130:11282-+. doi: 10.1021/ja804326b. [DOI] [PubMed] [Google Scholar]
- Nevzorov AA, Park SH, Opella SJ. Three-dimensional experiment for solid-state NMR of aligned protein samples in high field magnets. Journal of Biomolecular Nmr. 2007;37:113–116. doi: 10.1007/s10858-006-9121-y. [DOI] [PubMed] [Google Scholar]
- Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A, Yamamoto K. A Family of PISEMA Experiments for Structural Studies of Biological Solids. In: Webb G, editor. Modern Magnetic Resonance. Netherlands: Springer; 2006. pp. 703–709. [Google Scholar]
- Resende JM, Verly RM, Aisenbrey C, Cesar A, Bertani P, Pilo-Veloso D, Bechinger B. Membrane interactions of phylloseptin-1, 2, and -3 peptides by oriented solid-state NMR spectroscopy. Biophysical journal. 2014;107:901–911. doi: 10.1016/j.bpj.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Traaseth NJ, Verardi R, Cembran A, Gao J, Veglia G. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. Journal of biomolecular NMR. 2009;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal G, van Duynhoven J, Baldus M. Biomolecular NMR: recent advances in liquids, solids and screening. Current Opinion in Chemical Biology. 1999;3:530–536. doi: 10.1016/s1367-5931(99)00004-6. [DOI] [PubMed] [Google Scholar]
- Suter D, Enrnst RR. Spin Diffusion in Resolved Solid-State. NMR Spectra Phys Rev B. 1985;32:5608–5627. doi: 10.1103/physrevb.32.5608. [DOI] [PubMed] [Google Scholar]
- Szeverenyi NM, Sullivan MJ, Maciel GE. Observation of spin exchange by two-dimensional fourier transform 13C cross polarization-magic-angle spinning. Journal of Magnetic Resonance (1969) 1982;47:462–475. doi: http://dx.doi.org/10.1016/0022-2364(82)90213-X. [Google Scholar]
- Tang W, Knox RW, Nevzorov AA. A spectroscopic assignment technique for membrane proteins reconstituted in magnetically aligned bicelles. Journal of biomolecular NMR. 2012;54:307–316. doi: 10.1007/s10858-012-9673-y. [DOI] [PubMed] [Google Scholar]
- Traaseth NJ, Gopinath T, Veglia G. On the performance of spin diffusion NMR techniques in oriented solids: prospects for resonance assignments and distance measurements from separated local field experiments. The journal of physical chemistry B. 2010;114:13872–13880. doi: 10.1021/jp105718r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia G, Traaseth NJ, Shi L, Verardi R, Gopinath T, Gustavsson M. 1.11 The Hybrid Solution/Solid-State NMR Method for Membrane Protein Structure Determination. In: Egelman EH, editor. Comprehensive Biophysics. Amsterdam: Elsevier; 2012. pp. 182–198. doi: http://dx.doi.org/10.1016/B978-0-12-374920-8.00115-6. [Google Scholar]
- Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov VV, Mote KR, Verardi R, Veglia G. Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure. 2013 doi: 10.1016/j.str.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. Journal of Magnetic Resonance. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- Wang S, Ladizhansky V. Recent advances in magic angle spinning solid state NMR of membrane proteins. Progress in nuclear magnetic resonance spectroscopy. 2014;82C:1–26. doi: 10.1016/j.pnmrs.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Waugh JS. Uncoupling of local field spectra in nuclear magnetic-resonance - determination of atomic positions in solids. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:1394–1397. doi: 10.1073/pnas.73.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Ramamoorthy A, Gierasch LM, Opella SJ. Simultaneous characterization of the amide H1 chemical shift, H1-N15 dipolar, and N15 chemical-shift interaction tensors in a peptide-bond by 3-dimensional solid-state NMR Spectroscopy. Journal of the American Chemical Society. 1995;117:6148–6149. [Google Scholar]
- Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolar solid-state NMR-spectroscopy. Journal of Magnetic Resonance Series A. 1994;109:270–272. [Google Scholar]
- Xu J, Struppe J, Ramamoorthy A. Two-dimensional homonuclear chemical shift correlation established by the cross-relaxation driven spin diffusion in solids. Journal of Chemical Physics. 2008;128 doi: 10.1063/1.2826323. [DOI] [PubMed] [Google Scholar]