Abstract
Background
Optical coherence tomography (OCT) is a non-invasive optical imaging method that has proven useful in various fields such as ophthalmology, dermatology and neuroscience. In ophthalmology, significant progress has been made in retinal layer segmentation and enhancement of OCT images. There are also segmentation algorithms to separate epidermal and dermal layers in OCT-acquired images of human skin.
New Method
We describe simple image processing methods that allow automatic segmentation and enhancement of OCT images of rodent brain.
Results
We demonstrate the effectiveness of the proposed methods for OCT-based microangiography (OMAG) and tissue injury mapping (TIM) of mouse cerebral cortex. The results show significant improvement in image contrast, delineation of tissue injury, allowing visualization of different layers of capillary beds.
Comparison with existing methods
Previously reported methods for other applications are yet to be used in neuroscience due to the complexity of tissue anatomy, unique physiology and technical challenges.
Conclusions
OCT is a promising tool that provides high resolution in vivo microvascular and structural images of rodent brain. By automatically segmenting and enhancing OCT images, structural and microvascular changes in mouse cerebral cortex after stroke can be monitored in vivo with high contrast.
Keywords: Optical coherence tomography, optical microangiography, Stroke, Image reconstruction techniques
1. INTRODUCTION
Accurate visualization and quantification of microvascular networks and blood perfusion are critical for the understanding cerebrovascular pathophysiology, and for evaluating therapies for neurovascular diseases, such as stroke, traumatic brain injury, vascular dementia, neuroinflammation, and cancer.
Optical coherence tomography (OCT) is a non-invasive method that allows volumetric imaging of living biological tissues at high resolution (<10μm), great depth (several millimeters) and high speed (up to 1.5 MHz line scan rate), without a need for contrast agents (Tomlins and Wang 2005; Wieser et al. 2014)
OCT-based microangiography (OMAG) analyzes the temporal fluctuations in OCT signals to produce a volumetric blood perfusion map, down to the capillary level (An et al. 2010). OMAG has been used to study the microvasculature of a variety of tissues in vivo, including healthy and diseased human skin (Baran et al. 2015a), human retina (Zhang et al. 2015), and mouse cerebral microvasculature (Baran et al. 2015c; Baran and Wang 2016). Moreover, OCT-based tissue injury mapping (TIM) depicts the changes in tissue properties by characterizing the attenuation of light inside the tissue (Baran et al. 2015b). TIM has been used to detect tissue injury development in mouse cerebral cortex after stroke and in human facial skin during acne development.
Recent progress has led to highly efficient methods to process retinal OCT images through enhancement and segmentation of its various layers (Yazdanpanah et al. 2009; Tian et al. 2015). Similarly, in dermatology, several studies have described algorithms to reduce speckle of OCT images, which allows segmentation of skin layers such as the stratum corneum and epidermal-dermal junction (Hori et al. 2006; Hojjatoleslami and Avanaki 2012).
Although these methods work well for ophthalmology and dermatology applications, they are not easily transferable to OCT images of brain due to the differences in tissue anatomy, physiology and experimental settings. The cerebral cortex consists of six layers, each exhibiting distinct functional properties (Merkle and Srinivasan 2016). Moreover, a cranial window is typically used to increase the penetration depth of light and overall quality of the OCT image (Li et al. 2014). There is a need for the development of image processing methods to separate the window and the skull from the cortex and accurately segment different cortical layers. Here, we propose simple, yet efficient algorithms to automatically segment cortical layers in OCT structural images. The results demonstrate improved contrast and details over standard en face OMAG and TIM images of the in vivo mouse cerebral cortex.
2. SYSTEM AND METHODS
2.1 OCT System Setup
A fiber-based SD-OCT system was used for these experiments (Reif et al. 2014). Briefly, a superluminescent diode (Thorlabs Inc., Newton, NJ, USA) was used as the light source, with central wavelength of 1340 nm, bandwidth of 110 nm, and axial resolution of ~7 μm in air. A 10X scan lens (Thorlabs Inc., Newton, NJ, USA) was used in the sample arm to achieve ~7 μm lateral resolution with 0.12 mm depth of field. The output light from the interferometer was routed to a custom-built spectrometer, which had a designed spectral resolution of ~0.141 nm that provided a detectable depth range of ~3 mm on each side of the zero delay line. The line rate of the line scan camera (1024 pixel detector-array, Goodrich Inc., Princeton, NJ, USA) employed in the spectrometer was 92 kHz. The system had a measured dynamic range of 105 dB with a light power of 3.5 mW at the sample surface. The operations for probe beam scanning, data acquisition, and data storage are controlled by a custom software package written in LabVIEW®.
2.2 Animal Preparation
All experimental procedures performed on animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Washington (Protocol number: 4262-01) and Oregon Health & Science University. During imaging, animals were deeply anesthetized using 1.5–2% isoflurane (0.2 L/min O2, 0.8 L/min air) and euthanized at the end of the experiments. Body temperature was maintained at 36.8 C with the homeothermic blanket system (507220-F, Harvard Apparatus, MA, USA).
A thinned-skull or open-skull cranial window with 3 – 4 mm diameter was created in the right parietal cortex 1 mm lateral and 1 mm posterior to bregma (Li et al. 2014). Stroke was induced using the intraluminal filament middle cerebral artery (MCA) occlusion method (Longa et al. 1989).
2.3 Tissue Injury Mapping
Tissue characterization techniques often rely on changes in the physical characteristics of disease-affected tissue, which alters the received signal, primarily through changes in tissue absorption and/or scattering. Changes in signal attenuation within tissue, measured as an attenuation coefficient, can be used to differentiate various disease-affected tissue types. Measuring the optical attenuation coefficient (OAC) using OCT signals has been used for in vivo characterization of various tissue injury or disease types, e.g., atherosclerosis (Xu et al. 2008), burn scar (Gong et al. 2013), and cerebral ischemia (Srinivasan et al. 2013).
We have recently developed a TIM method by using an algorithm called sorted average intensity projection (sAIP) for en face mapping of the OACs belonging to different tissue types (Baran et al. 2015b). The method works by selecting the tissue volume from the 3D OAC data, then sorting the values of OACs in each A-line in ascending order. The following equation is then used to calculate the average OAC at a specific en face location, defined as OACaverage[y]:
| (1) |
where OACsorted[i] is the value of the ith OAC and N is the total pixel number in the selected portion of each A-line. M is an adjustable parameter that determines the number of pixels to be averaged. Averaging several OACs after sorting removes potential fluctuations in individual OACs and produces a smooth en face map as in Fig. 5d–f. The M parameter should be chosen based on the size of the region of interest. Averaging excessive number of pixels makes it difficult to detect small changes.
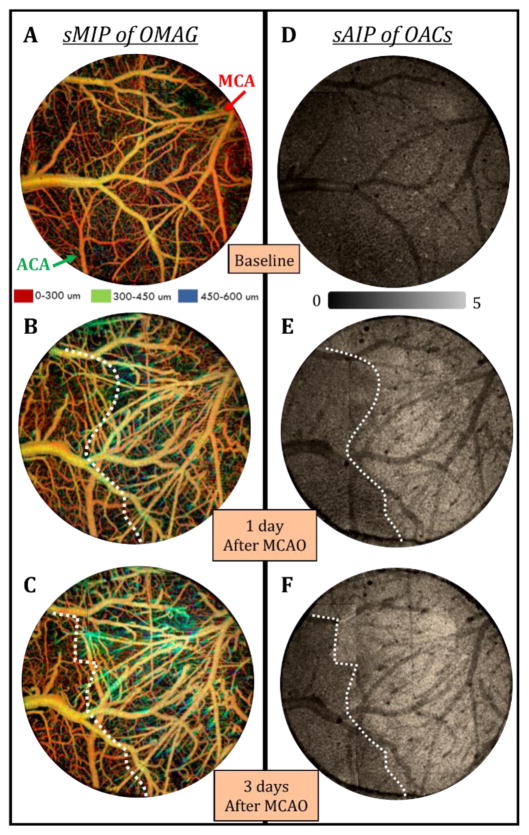
Fig. 5.
TIM images of mouse cerebral cortex after stroke using a thinned-skull cranial window. (a–c) En face depth-resolved (color coded) sMIP images of OMAG before (a) and 1 day (b) and 3 days after MCA occlusion (c). MCA and ACA branches are highlighted with arrows. (d–f) En face sAIP images of OACs from the 4th layer of the cortex before (d) and 1 day (e) and 3 days after MCA occlusion (f). Dotted white lines represent the border between normal and ischemic (infarcted) tissue. Field of view of the images is 3 mm x 3 mm.
OACs are calculated locally by using a recently developed method (Vermeer et al. 2014), where every pixel in the OCT data set is converted into a corresponding OAC pixel using the following relationship:
| (2) |
Here, I[i] is the OCT signal at a certain pixel, Δ is the pixel size and OAC[i] is the OAC at that pixel.
2.4 OCT-based Angiography
The OMAG technique is utilized to visualize the volumetric microvasculature down to the capillary level (An et al. 2011). For mouse cerebral cortex imaging, the data is acquired with 180 frames per second where every B-frame is made of 400 A-lines covering a total distance of approximately 2 mm. The slow axis (C-scan), consists of 3200 B-frames, covers the same distance by repeatedly scanning eight times at each location, and results in the construction of 3D images in about 18 seconds.
2.5 Automatic Segmentation of Cerebral Cortex Layers
Most automatic segmentation methods rely on the boundary detection of the first tissue layer in the OCT image. For example, these methods are used to detect the layers of the retina (Yazdanpanah et al. 2009; Tian et al. 2015) or the epidermis of skin (Hori et al. 2006; Hojjatoleslami and Avanaki 2012). There are many different types of boundary detection methods, a few of the more common and recent ones include peak detection (Koozekanani et al. 2001; Cabrera Fernández et al. 2005; Shahidi et al. 2005; Chan et al. 2006), active contours (Yazdanpanah et al. 2009), and shortest-path search image (Tian et al. 2015). In the peak detection algorithm, which has been often used in ophthalmology, the intensity peak within each A-line is used to create the location of the retinal layer boundary in the corresponding B scan (Koozekanani et al. 2001; Cabrera Fernández et al. 2005; Shahidi et al. 2005; Chan et al. 2006). The other two methods have been used mostly for retinal applications. The active contour method generates a spline, which gravitates towards edges to minimize energy (Yazdanpanah et al. 2009). The shortest-path based graph search finds the shortest distance between nodes using Dijkstra’s algorithm to form the segmented image (Tian et al. 2015).
However, these methods might not work well for OCT-based cerebral cortex imaging, due to the irregular surface reflections from the material covering the cortex (e.g. cover glass or plastic wrap). As can be seen in Fig. 1(a, b), the existing segmentation software may be deceived into viewing the surface reflection from the cover glass (highlighted with a yellow arrow in Fig. 1) as the first layer of the tissue. Since the presence of these reflections is at random locations of the image, it is not trivial to differentiate them from real surface boundary of the brain tissue.
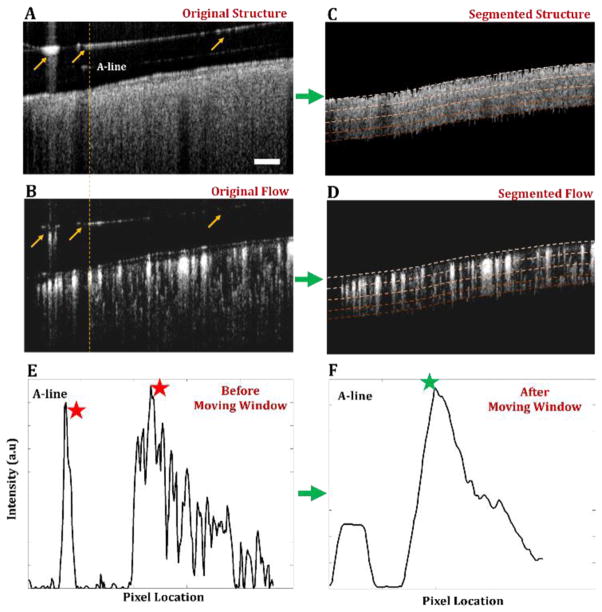
Fig. 1.
Implementation of automatic segmentation method using moving summation window. (a) Original OCT cross-sectional image of mouse cerebral cortex with a thinned-skull cranial window. A representative A-line with hyper-reflection from the cover glass is highlighted. (b) Original OMAG cross-sectional image. (c, d) Segmented OCT and OMAG cross-sectional images, respectively. Four segmented layers are highlighted with dashed lines. (e) Intensity plot of the A-line with hyper-reflection selected from (a, b). (f) Intensity plot of moving summation window on the A-line from (e). Red stars represent the peaks detected before moving summation window and green star represents the boundary location acquired using the peak detection after moving summation window. Scale bar represents 0.1 mm.
Here, instead of selecting the first peak intensity of the OCT structural A-lines, we detect the first peak intensity of the summation of a moving window as below:
| (3) |
| (4) |
where W is the moving window array along the OCT structural A-line starting from the top pixel, Wsize is the size of the window, and B is the boundary pixel location. Accordingly, random surface reflections above the cortex do not appear as a peak in the moving window array, since most of the signal coming from the non-tissue window would be background noise. Moreover, the highest peak of the moving window array would be the top surface pixel of the cortex due to the fact that light attenuation results in weaker pixel intensities in deeper compared to surface pixels.
Although this detection method is effective in eliminating surface reflections, it does not work as well when the size of the reflection is comparable to the size of the moving window. To overcome this issue, we add another mechanism to determine if the detected boundary location is accurate, by comparing the location of the peak pixel location with the neighboring peak pixel locations as below:
| (5) |
| (6) |
where Bmean[x] is the mean value of the neighboring peak pixel locations acquired from previous frame and T is the threshold value for the deviation of B[x] from Bmean[x]. Wsize and T are adjustable parameters, depending on the experimental settings. In our experiments, we use 30 pixels for Wsize and 10 pixels for T.
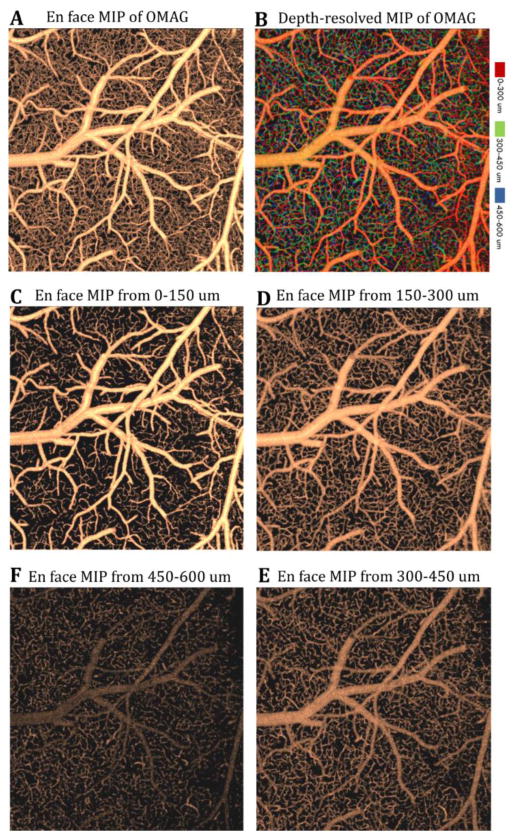
After detecting the boundary of the brain from OCT structural data, we select ~600 μm within the cortex and divide it into 4 layers, each with ~150 μm thickness on OMAG and TIM data, which have the identical volume size and shape with OCT structural data. For OMAG images, we apply sorted maximum intensity projection (sMIP) algorithm (Baran et al. 2015b) for each layer of every A-line where the maximum intensity of flow is selected and mapped to an en face plane. We finally apply a Gaussian filter (with 2 x 2 window) to the en face projection maps. Figure 2 shows the resulting OMAG images after the proposed method. For TIM images, we use sAIP (Baran et al. 2015b) for each layer of every A-line, where the average intensity of OAC is calculated and mapped to an en face plane and results are presented in Fig. 4.
Fig. 2.
En face sorted maximum intensity projection (sMIP) images of segmented OMAG data. (a) En face sMIP image of OMAG at 0 – 600μm depth. (b) En face depth-resolved (color-coded) sMIP image of OMAG at 0 – 600μm depth. (c–f) En face sMIP images of OMAG at various depths. Field of view of the images is 2 mm x 2 mm.
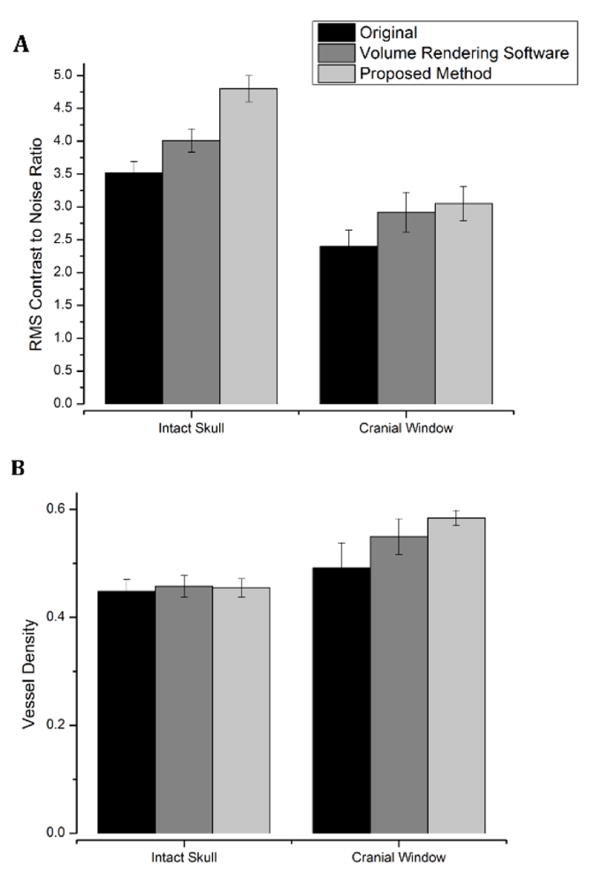
Fig. 4.
RMS contrast to noise ratio (a) and vessel density (b) comparison between image process results using the original, 3D volume rendering software and automatically segmented en face image projection. OMAG data of mouse cerebral cortex were acquired through an intact skull or open- cranial window.
3. RESULTS
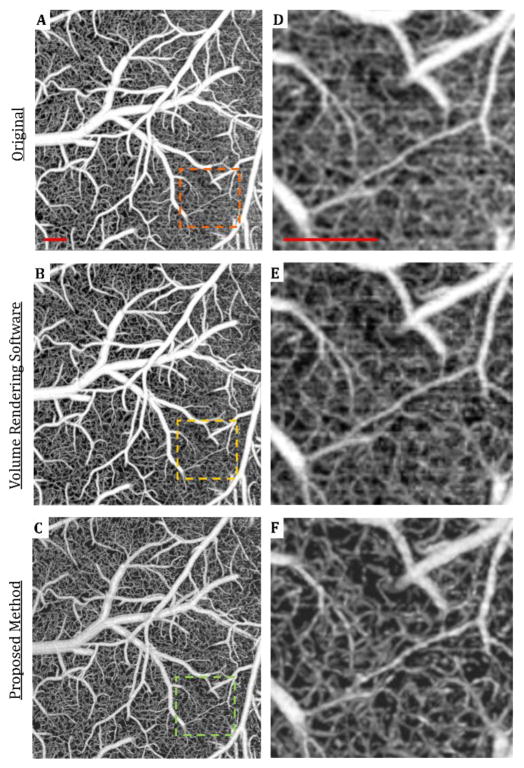
To demonstrate the effectiveness of the proposed methods, we compared the en face projections of OMAG images of mouse cerebral microvasculature acquired by three different methods in Figure 3(a–c). The first method is the maximum intensity projection (MIP) with the application of Gaussian filter (with 2 x 2 window) from volumetric OMAG data without segmentation. The second method is the Gaussian filtered (with 2 x 2 window) MIP using a common 3D volume rendering software for life sciences, Amira®. This software enables rendering and manipulation of a 3D volumetric image using the input of cross-sectional images. The third method is the Gaussian filtered (with 2 x 2 window) sMIP after the proposed automatic segmentation method. Figure 3(d–f) shows magnified views of OMAG images acquired using these methods compared to an image acquired by the proposed method (Fig. 3f). The image with the reduced background noise allowed for the differentiation of capillary segments.
Fig. 3.
Comparison between image processing results using the original, 3D volume rendering software and automatically segmented OMAG data of mouse cerebral cortex acquired through an open-skull cranial window. (a–c) En face maximum intensity projection (MIP) images of OMAG data using various methods. (d–f) Zoomed in views of the images in (a–c). Scale bar represents 0.2 mm.
The steps of accurate segmentation and separate processing of each layer reduces noise in the final en face image and eliminates the need to use a stronger smoothing filter (such as median filter) that often leads to blurry images. To explain it further, accurate segmentation provides the separation of vessels in in-focus layers from the capillaries in out-of-focus layers which might look like a “noisy background” in standard non-segmented MIP OMAG images. By mapping them separately, this “noisy background” effect on the in-focus layers are removed. Secondly, sMIP algorithm sorts the pixel intensities in segmented layers of A-lines and takes the average of the top values for each layer. This also helps reducing the noise in final en face images, because the possible noisy pixels are averaged out using a sMIP, versus using a standard MIP.
In order to evaluate the changes in image quality, we used RMS contrast and vessel density as metrics. RMS was favored by Bex and Makous (Bex and Makous 2002) over alternative contrast measurement methods. This method involves calculating the standard deviation of pixel intensities in an image. We compared the contrast of OMAG images generated by different projection methods using the RMS contrast value of each image, as defined in Eq. 7:
| (7) |
where M is the image pixel width, N is the image pixel height, Ī is the average intensity of the entire image, and Iij represents the intensity of the pixel at that location. Intensity values are normalized to values between 0 and 1 by dividing by the maximum possible intensity value. Finally, we divided the RMS contrast values with background noise values to acquire contrast to noise ratio. The mean contrast to noise ratio values are calculated from 9 images that represent different cases and shown in Fig. 4a. Accordingly, highest contrast to noise ratio is acquired with the proposed method.
In addition, vessel density in OMAG image is a commonly used metric to compare different methods. To calculate vessel density, vessel segmentation algorithm (Reif et al. 2012) is applied to each en face MIP blood flow image. In this algorithm, MIP images of microvasculature are binarized using an adaptive threshold to distinguish vessels from the background. The adaptive threshold of a moving window on the image is determined by the mean intensity value of the pixels in that window. Then, the vessel density is calculated by dividing the number of ones in the binary image by total pixel number. The mean vessel densities for three cases, each represented by 9 images, are shown in Fig. 4b. Higher vessel density in the images obtained by the proposed method is attributed to the less noisy images, which allows for more accurate vessel segmentation by distinguishing capillary segments from background.
We also multiplied the RMS contrast to noise ratio and the vessel density values for each case to have a more comprehensive single metric of quality for OMAG images. Results show that with the implementation of a cranial window, the proposed method provides 10% better image quality than OMAG images acquired using volume rendering software and 51% better contrast compared to original OMAG. For OMAG images with intact skull, the proposed method provides 19% higher image quality than the OMAG images acquired using volume rendering software and 38% better contrast compared to original OMAG.
Moreover, using the proposed method, we were able to obtain TIM images of ischemic mouse cerebral cortex after stroke with higher contrast. The area under the cranial window (parietal cortex) falls within the watershed region, where blood supply is carried by the distal branches of both the MCA and the anterior cerebral artery (ACA). During MCA occlusion (MCAO), lack of blood supply from the MCA induces ischemia, resulting in energy failure, anoxic depolarization and changes in tissue morphology and light scattering properties (Kawauchi et al. 2011). Using TIM, we were able to resolve the transition in light scattering properties between healthy and ischemic tissue.
Fig. 5 compares TIM images taken before and 1 day and 3 days after MCA occlusion in mouse. Fig. 5(a–c) shows the en face depth-resolved (color coded) sMIP images of OMAG within 0–600 um depth of the cerebral cortex. The OMAG images were obtained by merging 4 OMAG images into a mosaic. Lastly, the en face sAIP of OAC images are shown in Fig. 5(d–f). As can be seen in Fig. 5(b–c), blood vessels seem dilated in response to brain injury, and the vessel density is increased about 20% compared to baseline. However, this information is not sufficient to map the damaged area. The progression of infarction causes changes in tissue light scattering properties that result in an increase in OACs. The en face sAIP images of the OAC data from the 4th layer of the cortex in Fig. 5(d–f) show a clear delineation between the MCA (ischemic core) and ACA (watershed) territories after MCAO. Accordingly, on day 1 after MCAO, average OAC in the ischemic region (MCA territory) increases about 70% relative to baseline, revealing the infarcted region. On day 3 after the MCAO, the average OAC in the MCA territory continues to rise about 20%, likely reflecting infarct expansion. On the other hand, the average OAC increase in the ACA side might be due to an increased activity in this area or a slight change in imaging settings. A new study with larger number of animals is required to have a statistical assessment. Here, automatically segmented TIM proves to be useful in providing complimentary information about both the microvascular and structural tissue responses to stroke.
4. CONCLUSION
High quality and detailed visualization of the microvascular networks in the rodent cerebral cortex is important for evaluating therapies aimed at treating neurovascular diseases, such as stroke, traumatic brain injury, vascular dementia, neuroinflammation, and cancer. In particular, experimental stroke research can benefit from a non-invasive microscopic in vivo imaging capability to investigate recovery mechanisms in brain. Given its high spatial resolution and sensitivity, OMAG can provide useful information regarding highly intricate microcirculatory dynamics in brain. Moreover, in vivo non-invasive TIM can help delineate structural changes in brain tissue after stroke.
OCT imaging of mouse cerebral cortex undergoing ischemia (Fig. 5) indicate that OCT with improved image quality may serve as an indicator of tissue injury, potentially providing critical information regarding the therapeutic window of opportunity for tissue salvage after stroke. As illustrated by the dotted lines in Fig. 4, OCT can delineate the boundaries between infarcted and salvageable tissue at different stages of stroke injury progression. It is important to note that the presented boundary is not quantitatively detected, but provided as a visual aid. The accuracy of this boundary in TIM images should be confirmed using histology techniques, which is left as a future work. In this paper we only presented TIM results from layer 4, which provided the most pronounced changes in tissue properties compared to other layers. It is reasonable to hypothesize that there might be differences in response to MCAO between layers of cortex (Merkle and Srinivasan 2016). However, assessment of these differences requires additional animal studies.
In summary, OCT is a promising tool that provides high resolution in vivo microvascular and structural images of rodent brain. By automatically segmenting OCT images, structural and microvascular changes in mouse cerebral cortex after stroke can be monitored with high contrast. As a result, we foresee OCT as an indispensable tool in monitoring changes in tissue morphology and hemodynamics following pharmacological interventions in small animal models of tissue injury. Simple and effective enhancement techniques will help OCT to be widely adopted by the neuroscience community.
Highlights.
Automated algorithms are proposed for segmentation and enhancement of OCT images.
These algorithms are shown to be effective on OCT-acquired rodent brain images.
In vivo dynamics in mouse cerebral cortex is imaged after stroke with high contrast.
Acknowledgments
The authors would like to thank Tracy Petrie, Wei Wei and Kirsti Golgotiu for their help with the system setup and useful discussions.
Funding Information
National Institutes of Health R01HL093140 and R01EB009682 (RKW), and the Knight Cardiovascular Institute at Oregon Health & Science University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An L, Qin J, Wang RK. Ultrahigh sensitive optical microangiography for in vivo imaging of microcirculations within human skin tissue beds. Opt Express. 2010;18:8220–8. doi: 10.1364/OE.18.008220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An L, Shen TT, Wang RK. Using ultrahigh sensitive optical microangiography to achieve comprehensive depth resolved microvasculature mapping for human retina. J Biomed Opt. 2011;16:106013. doi: 10.1117/1.3642638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran U, Li Y, Choi WJ, Kalkan G, Wang RK. High resolution imaging of acne lesion development and scarring in human facial skin using OCT-based microangiography. Lasers Surg Med. 2015a;47:231–8. doi: 10.1002/lsm.22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran U, Li Y, Wang RK. In vivo tissue injury mapping using optical coherence tomography based methods. Appl Opt. 2015b;54:6448–53. doi: 10.1364/AO.54.006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran U, Li Y, Wang RK. Vasodynamics of pial and penetrating arterioles in relation to arteriolo-arteriolar anastomosis after focal stroke. Neurophotonics. 2015c;2:025006. doi: 10.1117/1.NPh.2.2.025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran U, Wang RK. Review of optical coherence tomography based angiography in neuroscience. Neurophotonics. 2016;3:010902. doi: 10.1117/1.NPh.3.1.010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bex PJ, Makous W. Spatial frequency, phase, and the contrast of natural images. J Opt Soc Am A. 2002;19:1096–106. doi: 10.1364/josaa.19.001096. [DOI] [PubMed] [Google Scholar]
- Cabrera Fernández D, Salinas HM, Puliafito CA. Automated detection of retinal layer structures on optical coherence tomography images. Opt Express. 2005;13:10200. doi: 10.1364/opex.13.010200. [DOI] [PubMed] [Google Scholar]
- Chan A, Duker JS, Ishikawa H, Ko TH, Schuman JS, Fujimoto JG. Quantification of photoreceptor layer thickness in normal eyes using optical coherence tomography. Retina. 2006;26:655–60. doi: 10.1097/01.iae.0000236468.33325.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, McLaughlin RA, Liew YM, Munro PRT, Wood FM, Sampson DD. Assessment of human burn scars with optical coherence tomography by imaging the attenuation coefficient of tissue after vascular masking. J Biomed Opt. 2013;19:21111. doi: 10.1117/1.JBO.19.2.021111. [DOI] [PubMed] [Google Scholar]
- Hojjatoleslami A, Avanaki MRN. OCT skin image enhancement through attenuation compensation. Appl Opt. 2012;51:4927–35. doi: 10.1364/AO.51.004927. [DOI] [PubMed] [Google Scholar]
- Hori Y, Yasuno Y, Sakai S, Matsumoto M, Sugawara T, Madjarova V, et al. Automatic characterization and segmentation of human skin using three-dimensional optical coherence tomography. Opt Express. 2006;14:1862–77. doi: 10.1364/oe.14.001862. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Sato S, Uozumi Y, Nawashiro H, Ishihara M, Kikuchi M. Light-scattering signal may indicate critical time zone to rescue brain tissue after hypoxia. J Biomed Opt. 2011;16:027002. doi: 10.1117/1.3542046. [DOI] [PubMed] [Google Scholar]
- Koozekanani D, Boyer K, Roberts C. Retinal thickness measurements from optical coherence tomography using a Markov boundary model. IEEE Trans Med Imaging. 2001;20:900–16. doi: 10.1109/42.952728. [DOI] [PubMed] [Google Scholar]
- Li Y, Baran U, Wang RK. Application of thinned-skull cranial window to mouse cerebral blood flow imaging using optical microangiography. PLoS One. 2014:9. doi: 10.1371/journal.pone.0113658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longa EZ, Weinstein PR, Carlson S, Cummins R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke. 1989;20:84–91. doi: 10.1161/01.str.20.1.84. [DOI] [PubMed] [Google Scholar]
- Merkle CW, Srinivasan VJ. Laminar microvascular transit time distribution in the mouse somatosensory cortex revealed by Dynamic Contrast Optical Coherence Tomography. Neuroimage. 2016;125:350–62. doi: 10.1016/j.neuroimage.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif R, Baran U, Wang RK. Motion artifact and background noise suppression on optical microangiography frames using a naïve Bayes mask. Appl Opt. 2014;53:4164. doi: 10.1364/AO.53.004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif R, Qin J, An L, Zhi Z, Dziennis S, Wang R. Quantifying Optical Microangiography Images Obtained from a Spectral Domain Optical Coherence Tomography System. Int J Biomed Imaging. 2012;2012:509783. doi: 10.1155/2012/509783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi M, Wang Z, Zelkha R. Quantitative thickness measurement of retinal layers imaged by optical coherence tomography. Am J Ophthalmol. 2005:139. doi: 10.1016/j.ajo.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Srinivasan VJ, Mandeville ET, Can A, Blasi F, Climov M, Daneshmand A, et al. Multiparametric, Longitudinal Optical Coherence Tomography Imaging Reveals Acute Injury and Chronic Recovery in Experimental Ischemic Stroke. PLoS One. 2013:8. doi: 10.1371/journal.pone.0071478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Varga B, Somfai GM, Lee WH, Smiddy WE, DeBuc DC. Real-time automatic segmentation of optical coherence tomography volume data of the macular region. PLoS One. 2015:10. doi: 10.1371/journal.pone.0133908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins PH, Wang RK. Theory, developments and applications of optical coherence tomography. J Phys D Appl Phys. 2005;38:2519–35. [Google Scholar]
- Vermeer KA, Mo J, Weda JJA, Lemij HG, de Boer JF. Depth-resolved model-based reconstruction of attenuation coefficients in optical coherence tomography. Biomed Opt Express. 2014;5:322. doi: 10.1364/BOE.5.000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser W, Draxinger W, Klein T, Karpf S, Pfeiffer T, Huber R. High definition live 3D-OCT in vivo: design and evaluation of a 4D OCT engine with 1 GVoxel/s. Biomed Opt Express. 2014;5:2963. doi: 10.1364/BOE.5.002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Schmitt JM, Carlier SG, Virmani R. Characterization of atherosclerosis plaques by measuring both backscattering and attenuation coefficients in optical coherence tomography. J Biomed Opt. 2008;13:034003. doi: 10.1117/1.2927464. [DOI] [PubMed] [Google Scholar]
- Yazdanpanah A, Hamarneh G, Smith B, Sarunic M. Intra-retinal layer segmentation in optical coherence tomography using an active contour approach. Med Image Comput Comput Assist Interv. 2009;12:649–56. doi: 10.1007/978-3-642-04271-3_79. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Huang Y, Zhang T, Kubach S, An L, Laron M, et al. Wide-field imaging of retinal vasculature using optical coherence tomography-based microangiography provided by motion tracking. J Biomed Opt. 2015;20:066008. doi: 10.1117/1.JBO.20.6.066008. [DOI] [PMC free article] [PubMed] [Google Scholar]