SUMMARY
Amino acid availability activates signaling by the mammalian target of rapamycin (mTOR) complex 1, mTORC1, a master regulator of cell growth. The class III PI-3-kinase Vps34 mediates amino acid signaling to mTORC1 by regulating lysosomal translocation and activation of the phospholipase PLD1. Here we identify leucyl-tRNA synthetase (LRS) as a leucine sensor for the activation of Vps34-PLD1 upstream of mTORC1. LRS is necessary for amino acid-induced Vps34 activation, cellular PI(3)P level increase, PLD1 activation, and PLD1 lysosomal translocation. Leucine binding but not tRNA charging activity of LRS is required for this regulation. Moreover, LRS physically interacts with Vps34 in amino acid-stimulatable non-autophagic complexes. Finally, purified LRS protein activates Vps34 kinase in vitro in a leucine-dependent manner. Collectively, our findings provide compelling evidence for a direct role of LRS in amino acid activation of Vps34 via a non-canonical mechanism, and fill a gap in the amino acid-sensing mTORC1 signaling network.
INTRODUCTION
Mammalian target of rapamycin (mTOR) is a Ser/Thr kinase that controls a wide spectrum of cellular processes including cell growth, differentiation and metabolism. mTOR complex 1 (mTORC1), characterized by the presence of raptor, regulates cell growth by integrating several extracellular and intracellular signals including mitogens, cellular energy status, oxygen levels and amino acid availability (Sarbassov et al., 2005; Wullschleger et al., 2006). Most signals upstream of mTORC1 merge at the tumor suppressor tuberous sclerosis complex TSC1-TSC2 and the target of its GTPase activity, Rheb (Li et al., 2004; Manning and Cantley, 2003). Once activated by Rheb, mTORC1 can phosphorylate its immediate targets, ribosomal S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E binding protein 1 (4EBP1), both of which regulate protein synthesis at the translational initiation level (Ma and Blenis, 2009).
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine (PC) to phosphatidic acid (PA), which binds to the FKBP12-rapamycin-binding domain of mTOR (Fang et al., 2001). We have reported that PLD1 and PA mediate mTORC1 activation by mitogens (Fang et al., 2003; Fang et al., 2001), and that PLD1 is also a critical mediator of amino acid-induced mTORC1 activation via vacuolar protein sorting 34 (Vps34) (Yoon et al., 2011). Vps34 is the only class III PI-3-kinase in mammals, responsible for producing phosphatidylinositol-3-phosphate (PI(3)P) from phosphatidylinositol. Vps34 exists in distinct complexes that contribute to a variety of cellular functions, including vesicular trafficking and autophagy (Backer, 2008; Russell et al., 2014). Notably, PI(3)P production by Vps34 is stimulated by amino acids (Byfield et al., 2005; Nobukuni et al., 2005). Upon amino acid stimulation, interaction between PI(3)P and the PX domain of PLD1 activates PLD1 and induces its subcellular translocation to the lysosome (Yoon et al., 2011), where mTOR is also recruited via regulation by the small GTPases Rag (Sancak et al., 2008).
mTORC1 lysosomal translocation and activation in response to amino acids requires the GTP-bound form of RagA or B as well as the GDP-bound form of RagC or D. The Ragulator complex and the GATOR1 complex act as GEF (guanine nucleotide exchange factor) and GAP (GTPase activating protein) for RagA/B, respectively (Bar-Peled et al., 2013; Bar-Peled et al., 2012). Sestrins have been reported to negatively regulate GATOR2, an inhibitor of GATOR1 and consequently activator of mTORC1 (Chantranupong et al., 2014; Parmigiani et al., 2014; Peng et al., 2014), and a most recent report from the Sabatini group suggests that sestrins directly sense leucine in the mTORC1 pathway (Wolfson et al., 2016). A critical regulator acting in parallel to the sestrin-GATOR pathway has been reported to be leucyl tRNA synthetase (LRS), which senses leucine and has GAP activity for RagD (Han et al., 2012). Although the GAP activity of LRS is under debate (Tsun et al., 2013), the role of LRS as a leucine sensor upstream of TORC1 has also been independently demonstrated in yeast (Bonfils et al., 2012).
What senses amino acids upstream of Vps34-PLD1 has remained an unanswered question. Here, we report that Vps34 is a downstream target of LRS in amino acid signaling. LRS directly interacts with Vps34 in a non-autophagic complex, and activates Vps34 in an amino acid-dependent manner. Vps34 and PLD1 are required to mediate LRS activation of mTORC1. Our findings reveal LRS as an amino acid sensor for the Vps34-PLD1-mTORC1 pathway.
RESULTS
LRS is required for amino acid-induced Vps34 signaling
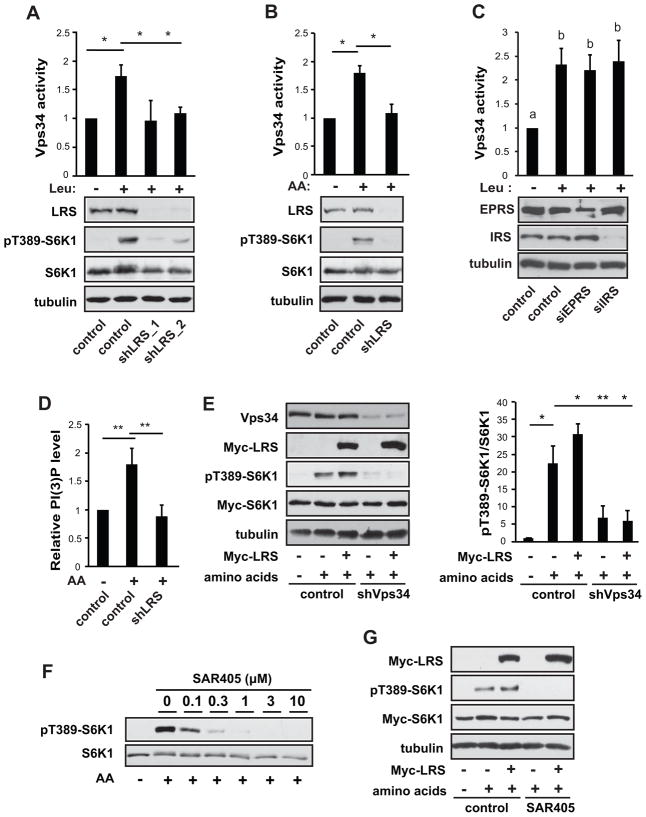
To validate the reported role of LRS in amino acid activation of mTORC1, we knocked down LRS in HEK293 cells and observed impaired leucine-stimulated S6K1 phosphorylation (Figure 1A). Total amino acid stimulation of pS6K1 was also significantly dampened (Figure 1B). Since there was no known sensor of amino acids upstream of the Vps34-PLD1-mTORC1 pathway, we set out to test whether LRS may fulfill that role by examining the effect of LRS knockdown on Vps34 lipid kinase activity. Co-expression of Vps15 has been reported to be necessary to ensure recombinant Vps34 stability and activity (Yan et al., 2009). Hence, we transfected bicistronic Myc-Vps34/V5-Vps15 into the cells, and immunoprecipitated Myc-Vps34 for in vitro kinase assays. Interestingly, knockdown of LRS decreased Vps34 activity induced by leucine (Figure 1A) or total amino acids (Figure 1B). As an important control, knockdown of two other tRNA synthetases, IRS (isoleucyl-tRNA synthetase) and EPRS (glutamyl-prolyl-tRNA synthetase), both in the multi-tRNA synthetase complex together with LRS (Park et al., 2008), had no effect on leucine-stimulated Vps34 activity (Figure 1C). This is consistent with the previous report that knockdown of these aaRS’s did not affect mTORC1 activation by amino acids (Han et al., 2012), suggesting that LRS has a unique function in mediating mTORC1 activation that is not shared by other aaRS’s. The change in Vps34 activity was paralleled by a reduction in the total cellular PI(3)P level in LRS knockdown cells (Figure 1D). Knockdown of LRS did not affect the protein levels of mTOR, raptor, Vps34, and Rag GTPases (Figure S1A). Hence, LRS appears to be necessary for amino acid activation of Vps34.
Figure 1. LRS is required for amino acid activation of Vps34.
(A) HEK293 cells were transduced with lentiviruses expressing shRNAs against LRS (shLRS) or a scrambled sequence as control, and selected with puromycin for 3 days. The cells were then transfected with bicistronic Myc-Vps34/V5-Vps15, serum starved overnight, and leucine (Leu) deprived for 2 hr, followed by leucine stimulation for 30 min. Cell lysates were analyzed by western blotting. Anti-Myc IP immunocomplexes were subjected to in vitro Vps34 kinase assays. (B) Cells were treated as in (A) except that amino acid (AA) deprivation and stimulation were performed. (C) Cells were treated as in (A) except that siRNA against IRS or EPRS were introduced by transfection. (D) Cells were treated as in (B) and PI(3)P levels were determined by quantitative PI(3)P ELISA assay. (E) Cells were transduced with shVps34 or control lentiviruses, and then co-transfected with Myc-LRS and Myc-S6K1, followed by AA stimulation as in (B). (F) AA stimulation was performed in the presence of varying concentrations of SAR405. (G) Cells were co-transfected with Myc-LRS and Myc-S6K1, stimulated with AA as in (B) in the absence or presence of 3 μM SAR405. All data shown are mean ± SD or representative blots from 3 to 5 independent experiments. *P<0.05; **P<0.01. Data denoted by different letters (a, b) are significantly different (P ≤ 0.05). See also Figure S1.
Next, we asked whether Vps34 mediates LRS activation of mTORC1. Overexpression of LRS enhanced amino acid activation of S6K1 as previously reported (Han et al., 2012), and this effect of LRS was abolished by Vps34 knockdown (Figure 1E). To further validate the link between LRS and Vps34, we utilized the Vps34-specific inhibitor SAR405, which had been reported to potently inhibit Vps34 with high selectivity at micromolar concentrations (Ronan et al., 2014). Amino acid activation of pS6K1 was inhibited by SAR405 in a dose-dependent manner, with complete inhibition occurring at 3 μM (Figure 1F). Furthermore, SAR405 blocked amino acid-stimulated PLD1 activation (Figure S1B) and translocation to the lysosome (Figure S1C), a process that requires Vps34 (Yoon et al., 2011). Amino acid-induced lysosomal translation of mTOR was not affected by the inhibitor (Figure S1D). Importantly, SAR405 also abolished pS6K1 under LRS overexpression (Figure 1G), confirming the requirement of Vps34 for LRS activation of mTORC1 in response to amino acid signals. Taken together, our results suggest that LRS is an upstream regulator of amino acid activation of Vps34.
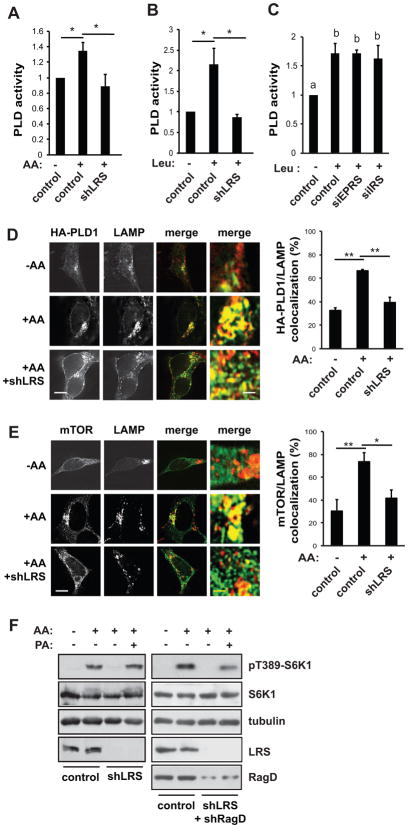
LRS is necessary for amino acid activation of PLD1
Since Vps34 activates PLD1 in amino acid signaling (Yoon et al., 2011), we proceeded to test the effect of LRS on PLD activity. LRS knockdown inhibited PLD activity induced by amino acids (Figure 2A) or leucine alone (Figure 2B), whereas knockdown of IRS or EPRS had no effect (Figure 2C). Furthermore, LRS knockdown blocked amino acid-stimulated PLD1 translocation to the lysosome (Figure 2D). Lysosomal translocation of mTOR was also suppressed by LRS knockdown (Figure 2E), as previously reported (Han et al., 2012). Collectively, these results indicate that LRS is required for amino acid regulation of PLD1.
Figure 2. LRS is necessary for amino acid activation of PLD1.
HEK293 cells were transduced with shLRS or control lentiviruses as described in Figure 1 legend. (A) Cells were stimulated with AA as described in Figure 1 legend, followed by in vivo PLD assays. (B) Cells were stimulated with Leu as described in Figure 1 legend. (C) Cells were treated as in (A) except that siRNA for IRS and EPRS were introduced by transfection. (D) Cells were transduced with shLRS lentivirus, and then transfected with HA-PLD1, followed by AA stimulation and immunostaining with anti-HA and anti-LAMP1/2 antibodies. (E) Cells were treated as in (D) without transfection, and immunostained with anti-mTOR and anti-LAMP1/2 antibodies. For D and E, representative images and quantification of co-localization (overlap coefficient) are shown. (F) Cells were transduced with shRNA lentiviruses as indicated, and stimulated with amino acids with or without 100 μM PA. All data shown are mean ± SD or representative blots from 3 to 5 independent experiments. *P<0.05; **P<0.01. Data denoted by different letters (a, b) are significantly different (P ≤ 0.05). Scale bars, 5 μm; (enlarged images) 0.5 μm.
If mTORC1 were downstream of this LRS-PLD pathway, one would expect exogenous PA (the product of PLD1) to compensate for signaling to mTORC1 when LRS was knocked down. This was indeed the case. As shown in Figure 2F (left panels), PA fully rescued amino acid-stimulated pS6K1 from LRS knockdown, suggesting that PLD is a major mediator of LRS signaling to mTORC1. PA partially restored pS6K1 when both LRS and RagD were knocked down (Figure 2F, right panels), which is consistent with Rag and PLD acting in parallel to activate mTORC1 (Yoon et al., 2011).
LRS is an amino acid sensor in the Vps34-PLD pathway
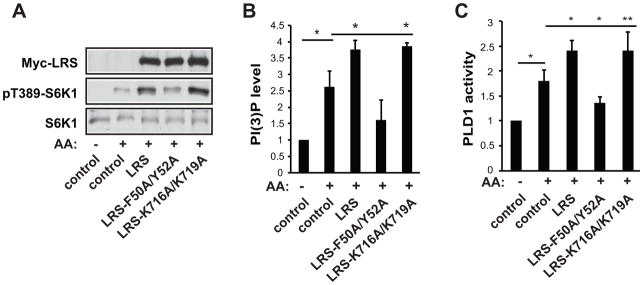
To investigate whether leucine binding or catalytic activity of LRS was involved in the regulation of Vps34-PLD1 signaling, we made use of two LRS mutants: a leucine binding deficient mutant (F50A/Y52A) and a tRNA charging deficient mutant (K716A/K719A) (Han et al., 2012). Overexpression of LRS or LRS-K716A/K719A, but not LRS-F50A/Y52A, increased amino acid stimulated S6K1 activity (Figure 3A), as reported previously (Han et al., 2012). Importantly, LRS, as well as the K716A/K719A mutant, augmented the total cellular PI(3)P level (i.e., Vps34 activity) and PLD1 activity, whereas the F50A/Y52A mutant did not have this effect (Figure 3B&C). These results indicate that leucine binding but not tRNA charging activity of LRS is required for activation of the Vps34-PLD1 pathway. This is consistent with the notion that LRS acts as a sensor for amino acids independently of its canonical function in translation regulation.
Figure 3. LRS is an amino acid sensor in Vps34/PLD1 signaling.
(A) HEK293 cells were transfected with LRS constructs as indicated and stimulated with AA as described in Figure 1 legend, followed by western analysis. (B) Cells were treated as in (A) and subjected to PI(3)P ELISA assays. (C) Cells were co-transfected with HA-PLD1 and LRS constructs, treated as in (A), and then subjected to in vivo PLD assays. All data shown are mean ± SD or representative blots from 3 to 5 independent experiments. *P<0.05; **P<0.01.
LRS physically interacts with Vps34 and the UNE-L domain is necessary for LRS regulation of Vps34
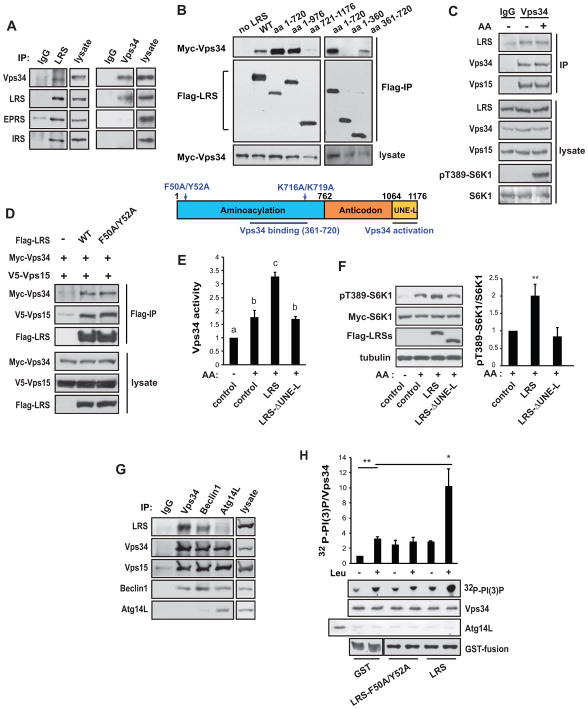
In order to uncover the mechanism underlying LRS regulation of Vps34, we evaluated a potential physical interaction between LRS and Vps34. We found that endogenous LRS co-immunoprecipitated with endogenous Vps34 (Figure 4A). Interestingly, while LRS immunoprecipitates contained Vps34 as well as EPRS and IRS (Figure 4A left panels), Vps34 immunoprecipitation brought down only LRS and not the other aaRS’s (Figure 4A right panels). This suggests that Vps34 interacts with LRS outside of the multi-tRNA synthetase complex. Recombinant Myc-Vps34 was also pulled down by Flag-LRS (Figure S2A). Co-expression of Vps15 did not increase the amount of recombinant Vps34 associated with LRS, suggesting that the Vps34-LRS interaction is not dependent on Vps15 (Figure S2B). As shown in Figure 4B, the minimum Vps34-binding site resided within amino acids 361–720 of LRS, although the presence of the rest of the N-terminus appeared to strengthen the interaction. We also found that the interaction between LRS and Vps34 was not affected by amino acid withdrawal or stimulation (Figure 4C), nor by the F50A/Y52A mutation (Figure 4D), suggesting that leucine binding is not necessary for the LRS-Vps34 interaction.
Figure 4. LRS interacts with and activates Vps34.
(A) LRS or Vps34 was immunoprecipitated from MEF cells, followed by western analysis. (B) HEK293 cells were transfected with Myc-Vps34 and Flag-LRS wild-type (WT) or fragments as indicated. Anti-Flag IP was performed, followed by western analysis. A schematic diagram of LRS is shown, in which the point mutations and regions required for Vps34 binding/activation are indicated (based on data in B and E). (C) MEF cells were stimulated with AA, followed by IP of Vps34 and western analysis. (D) HEK293 cells were transfected with Myc-Vps34 and Flag-LRS WT or F50A/Y52A. Anti-Flag IP was performed, followed by western analysis. (E) HEK293 cells were co-transfected with Myc-Vps34 and WT or mutant LRS, and stimulated with AA. Myc-Vps34 was immunoprecipitated and subjected to in vitro lipid kinase assay. (F) HEK293 cells were co-transfected with Myc-S6K1 and WT or mutant LRS, followed by AA stimulation and western analysis. (G) MEF cells were subjected to immunoprecipitation by antibodies against Vps34, Becline1, or Atg14L, followed by western analysis. (H) MEF cells were serum starved and amino acid deprived, and then subjected to Vps34 IP after pre-clearing by anti-Atg14L IP. The immunocomplexes were assayed for lipid kinase activity with or without 0.8 mM leucine. The first lane of the Atg14L blot shows Atg14L IP as a control. Purified GST, GST-LRS, and GST-LRS-F50A/Y52A proteins were included as indicated. *P<0.05; **P<0.01. Data denoted by different letters (a, b, c) are significantly different (P ≤ 0.05). See also Figures S2–4.
Despite their capacity to bind Vps34, LRS fragments containing up to N-terminal 976 amino acids did not activate Vps34, PLD1, or S6K1 (Figure S3A–C). This suggests that Vps34 binding alone is not sufficient for LRS function in the Vps34-PLD pathway, and that the C-terminus of LRS may be functionally important. Indeed, deletion of the C-terminal UNE-L domain (~110 amino acids), present only in higher eukaryotes and speculated to confer non-translational functions to LRS (Guo et al., 2010), abolished LRS activation of Vps34 (Figure 4E) and S6K1 (Figure 4F). Hence, the UNE-L domain appears to be required for the non-canonical function of LRS in activating Vps34-PLD-mTORC1 signaling.
LRS interacts with Vps34 in non-autophagic complexes
Vps34 is an important autophagy regulator in addition to its role in amino acid signaling, and it is believed that Vps34 functions in distinct protein complexes, some regulating autophagy and others non-autophagic (Backer, 2008; Russell et al., 2014). Following amino acid starvation, a Vps34 complex containing Atg14L is involved in autophagy, whereas Vps34 complexes lacking Atg14L are not (Yuan et al., 2013). Beclin1 is present in all the complexes. As shown in Figure S4A (left panels), when endogenous Vps34 was pulled down from mouse embryonic fibroblast (MEF) cells after preclearing the cell lysate with an anti-Atg14L antibody, this Vps34 was activated by amino acids. MEF cells are commonly used to study Vps34 complexes (Kim et al., 2013; Russell et al., 2013); we found non-autophagic Vps34 complexes to be more prevalent in these cells than in HEK293. The activity of Vps34 associated with Atg14L immunoprecipitates, however, was inhibited by amino acid stimulation (Figure S4A, right panels), similar to previous reports (Russell et al., 2013; Yuan et al., 2013). Hence, only non-autophagic Vps34 complexes are involved in amino acid signaling to mTOR. Next, we asked which Vps34 complex binds to LRS. As shown in Figure 4G, when non-autophagic Vps34 complexes were isolated by immunoprecipitation with Vps34 or Beclin1, LRS was found in both complexes. The autophagic Vps34 complex isolated by Atg14L immunoprecipitation, however, did not contain LRS. Therefore, our data suggest that LRS binds specifically to amino acid-activatable Vps34 in non-autophagic complexes.
LRS directly activates Vps34 kinase activity in a leucine-dependent manner
To further probe a mechanism by which LRS may regulate Vps34, we performed in vitro kinase assays on Atg14L-depleted Vps34 complex isolated from amino acid-deprived cells. As shown in Figure S4B, the addition of leucine to the Vps34 immunocomplex activated the kinase. Endogenous LRS present in the Vps34 complex most likely contributed to this activation. Importantly, as shown in Figure 4H, the addition of purified GST-LRS protein to the immunocomplex enhanced Vps34 kinase activity by ~3-fold when compared to the GST control, and this was dependent on leucine. The leucine-binding deficient LRS mutant (F50A/Y52A) also activated Vps34, but to a lesser degree and in a leucine-independent manner. These results strongly suggest that LRS directly regulates Vps34 activity. Even though the physical interaction between these two proteins is not dependent on amino acid or leucine binding (Figure 4C & D), leucine is critical for LRS regulation of Vps34 activity.
DISCUSSION
Although several regulators mediating amino acid signals in mTORC1 signaling had been reported, the mechanism by which amino acids activate Vps34 remained unclear. In the present study, we have identified LRS as a direct amino acid sensor upstream of the Vps34-PLD-mTORC1 pathway. LRS interacts with and activates Vps34, and this activation is dependent on leucine binding but not tRNA charging activity of LRS. Furthermore, we show that this regulatory event involves LRS outside of the multi-tRNA synthetase complex and Vps34 in a non-autophagic complex. Our structure-function analysis has led to the identification of a minimum Vps34-binding site within the N-terminus of LRS and the C-terminal UNE-L domain to be critical for the activity of LRS toward Vps34.
The multifaceted cellular roles of Vps34, especially in nutrient signaling, may appear paradoxical. Nutrient starvation has been shown to both inhibit and activate Vps34 kinase activity (Byfield et al., 2005; Kim et al., 2013; Nobukuni et al., 2005; Russell et al., 2013). Characterization of various protein complexes involving Vps34 has led to insights into its diverse function and regulation. Amino acid starvation activates Ulk1 following Beclin1 phosphorylation, thereby enhancing the activity of Atg14L-containing Vps34 complex that regulates autophagy, whereas Vps34 in Atg14L-free complexes is inhibited by amino acid starvation (Russell et al., 2013). Our observation that LRS associates specifically with Atg14L-free Vps34 complexes is consistent with the involvement of LRS in amino acid-activation of Vps34, and it suggests that LRS differentiates the role of Vps34 in mTORC1 activation from that of regulating autophagy.
It is not clear how LRS regulation of Vps34-PLD can be reconciled with the reported LRS regulation of Rag (Bonfils et al., 2012; Han et al., 2012). Curiously, the product of PLD, PA, fully rescues amino acid-stimulated mTORC1 signaling from LRS knockdown (Figure 2F), suggesting that the Vps34-PLD pathway is a major, if not only, mediator of LRS signaling to mTORC1. When RagD is knocked down in addition to LRS, PA only partially restores mTORC1 signaling (Figure 2F), consistent with Vps34-PLD and Rag functioning in parallel to regulate mTORC1 activation (Yoon et al., 2011). One would also expect the LRS-Vps34-PLD pathway to work in parallel with the sestrin-GATOR pathway upstream of mTORC1 (Chantranupong et al., 2014; Parmigiani et al., 2014; Peng et al., 2014; Wolfson et al., 2016).
Since Vps34 must sense amino acids prior to PLD1 translocation to the lysosome (Yoon et al., 2011), one would speculate that a cytosolic amino acid sensing event is required for the activation of the Vps34-PLD1 pathway. In support of this idea, we observed that neither P62 nor V-ATPase, two previously reported regulators that mediate amino acid-sensing on the lysosome (Duran et al., 2011; Zoncu et al., 2011), was involved in PLD1 translocation to the lysosome, even though V-ATPase inhibition decreased amino acid-induced PLD activity (M.-S. Yoon & J. Chen, unpublished data). LRS interacts with other aminoacyl tRNA synthetases and nonenzymatic components to form a multi-tRNA synthetase complex in the cytosol, which is thought to be involved in diverse non-canonical functions of the aaRS’s (Park et al., 2008). However, the LRS-Vps34 interaction appears to occur outside of this complex (Figure 4A). Nevertheless, it is possible that other factors may be involved in amino acid-sensing and/or signal transduction by LRS in the cytosol, which warrant future investigations.
EXPERIMENTAL PROCEDURES
Detailed information regarding reagents, cell lysis, immunoprecipitation, western blotting, and immunofluorescence imaging appear in the Supplemental Experimental Procedures.
Cell culture and transfection
HEK293 and MEF cells were grown in DMEM containing 10% fetal bovine serum (FBS) at 37 °C with 5% CO2. Transient transfections were performed with PolyFect (Qiagen) following manufacturers’ recommendations. Serum or amino acid starvation, and various stimulations were performed as previously described and in figure legends (Sun et al., 2008). Amino acid stimulation was performed by switching cells from amino acid-free to regular DMEM. For leucine stimulation, cells were incubated with leucine-free DMEM for 2 hrs, and stimulated with 0.8 mM leucine for 30 min.
RNAi
All shRNAs were in the pLKO.1-puro lentiviral vector from The RNAi Consortium (TRC; Sigma-Aldrich). The shRNA clones for human Leucyl tRNA synthetase are shLRS-1 (TRCN0000290440) and shLRS-2 (TRCN0000290511). The shRNA clones for Vps34 and scrambled sequence control were previously reported (Yoon et al., 2011). Lentivirus packaging and transduction were performed as previously described (Sun et al., 2008). The sequences of siEPRS and siIRS were also previously reported (Han et al., 2012; Han et al., 2006).
In vivo PLD assay
Cellular PLD activity was measured in a transphosphatidylation assay previously described (Sun et al., 2008). Detailed procedures can be found in Supplemental Experimental Procedures.
In vitro Vps34 lipid kinase assay
GST proteins were expressed in the E. coli strain BL21, and purified using glutathione Sepharose 4B (GE Healthcare) following the manufacturer’s protocols. Lipid kinase assays were performed with Vps34 immune complexes with the addition of various GST proteins. Detailed procedures are described in Supplemental Experimental Procedures.
Determination of cellular PI(3)P levels by ELISA
Lipids were extracted from cells and subjected to a competitive ELISA-format assay (Echelon Biosciences) according to the manufacturer’s manual. PI(3)P quantities were calculated from nonlinear fitting of PI(3)P standards.
Statistical analysis
All quantitative data are presented as mean ± standard deviation (SD) of at least three independent experiments. Whenever necessary, statistical significance of the data was analyzed by performing Student’s t tests. *P ≤ 0.05. **P ≤ 0.01. Data denoted with different letters (e.g., a, b, c) are significantly different (P ≤ 0.05).
Supplementary Material
Figure S1 – related to Figure 1. SAR405 inhibits PLD activation and lysosomal translocation.
(A) HEK293 cells were transduced with shLRS or control lentiviruses and selected with puromycin for 3 days, followed by western blot analysis.
(B) HEK293 cells were serum- and amino acid-starved, and then stimulated with amino acids in the absence or presence of 5 μM SAR405, followed by in vivo PLD assays.
(C) HEK293 cells were transfected with HA-PLD1, treated as in (B), and then immunostained with anti-HA and anti-LAMP1 antibodies. Quantification of colocalization was as described in Figure 2D and 2E legends.
(D) HEK293 cells were treated as in (B) and immunostained with anti-mTOR and anti-LAMP1 antibodies. Quantification of colocalization was as described in Figure 2D and 2E legends.
Figure S2 – related to Figure 4. Vps34-LRS interaction is independent of Vps15.
(A) HEK293 cells were transfected with Myc-Vps34 and Flag-LRS. Anti-Flag IP was performed, followed by western analysis.
(B) HEK293 cells were transfected as in (A) with or without V5-tagged Vps15, followed by anti-Flag IP and western analysis.
Figure S3 – related to Figure 4. Vps34-binding LRS fragments do not activate Vps34, PLD1, or mTORC1.
(A) HEK293 cells were co-transfected with bicistronic Myc-Vps34/V5-Vps15 and LRS constructs as indicated, serum starved overnight, and amino acid deprived for 2 hr, followed by amino acid stimulation for 30 min. Anti-Myc IP immunocomplexes were subjected to in vitro Vps34 kinase assays.
(B) HEK293 cells were co-transfected with HA-PLD1 and LRS constructs as indicated and then treated as above, followed by in vivo PLD assays.
(C) HEK293 cells co-transfected with HA-S6K1 and LRS constructs were treated as above and analyzed by western blotting.
Figure S4 – related to Figure 4. Non-autophagic Vps34 is activated by amino acids.
(A) MEF cells were serum starved overnight, amino acid deprived for 2 hr, and then stimulated with amino acids for 30 min. IP was performed with anti-Atg14L (right panels) or anti-Vps34 (left panels) after preclearing the cell lysate with an anti-Atg14L antibody, followed by in vitro Vps34 kinase assay.
(B) MEF cells were treated as in (A) without stimulation, and subjected to IP with anti-Vps34 after preclearing with anti-Atg14L, followed by in vitro Vps34 kinase assay with or without 0.8 mM leucine.
Acknowledgments
This paper is dedicated to the occasion of Professor Stuart L. Schreiber’s 60th birthday. We are most grateful to Professor Schreiber for his scientific vision and mentoring. We also thank Christina L. Rosenberger for critical reading and editing of the manuscript. This work was supported by grants from the National Institutes of Health (R01 AR048914 & GM089771 to JC), the Keck Foundation (to JC), the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A1A01058313 to MSY), and the Gachon University Gil Medical Center (2015–15 to MSY).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, M.S.Y., K.S., and J.C.; Methodology, M.S.Y., K.S., and E.A.; Investigation, M.S.Y., K.S., and E.A.; Resources, J.M.H. and S.H.K.; Writing–original draft, M.S.Y. and J.C.; Writing–review & editing, M.S.Y., K.S., J.M.H., and J.C.; Funding acquisition, M.S.Y. and J.C.; Supervision, M.S.Y. and J.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor Suppressor Complex with GAP Activity for the Rag GTPases That Signal Amino Acid Sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. doi:1110.1016/j.cell.2012.1107.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. Epub 32005 Jul 33027. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, Walker SJ, Brown HA, Chen J. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr Biol. 2003;13:2037–2044. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Han JM, Lee MJ, Park SG, Lee SH, Razin E, Choi EC, Kim S. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci U S A. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9:1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159:122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1. Mol Cell. 2013;2:00686–00682. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417:747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195:435–447. doi: 10.1083/jcb.201107033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–1995. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – related to Figure 1. SAR405 inhibits PLD activation and lysosomal translocation.
(A) HEK293 cells were transduced with shLRS or control lentiviruses and selected with puromycin for 3 days, followed by western blot analysis.
(B) HEK293 cells were serum- and amino acid-starved, and then stimulated with amino acids in the absence or presence of 5 μM SAR405, followed by in vivo PLD assays.
(C) HEK293 cells were transfected with HA-PLD1, treated as in (B), and then immunostained with anti-HA and anti-LAMP1 antibodies. Quantification of colocalization was as described in Figure 2D and 2E legends.
(D) HEK293 cells were treated as in (B) and immunostained with anti-mTOR and anti-LAMP1 antibodies. Quantification of colocalization was as described in Figure 2D and 2E legends.
Figure S2 – related to Figure 4. Vps34-LRS interaction is independent of Vps15.
(A) HEK293 cells were transfected with Myc-Vps34 and Flag-LRS. Anti-Flag IP was performed, followed by western analysis.
(B) HEK293 cells were transfected as in (A) with or without V5-tagged Vps15, followed by anti-Flag IP and western analysis.
Figure S3 – related to Figure 4. Vps34-binding LRS fragments do not activate Vps34, PLD1, or mTORC1.
(A) HEK293 cells were co-transfected with bicistronic Myc-Vps34/V5-Vps15 and LRS constructs as indicated, serum starved overnight, and amino acid deprived for 2 hr, followed by amino acid stimulation for 30 min. Anti-Myc IP immunocomplexes were subjected to in vitro Vps34 kinase assays.
(B) HEK293 cells were co-transfected with HA-PLD1 and LRS constructs as indicated and then treated as above, followed by in vivo PLD assays.
(C) HEK293 cells co-transfected with HA-S6K1 and LRS constructs were treated as above and analyzed by western blotting.
Figure S4 – related to Figure 4. Non-autophagic Vps34 is activated by amino acids.
(A) MEF cells were serum starved overnight, amino acid deprived for 2 hr, and then stimulated with amino acids for 30 min. IP was performed with anti-Atg14L (right panels) or anti-Vps34 (left panels) after preclearing the cell lysate with an anti-Atg14L antibody, followed by in vitro Vps34 kinase assay.
(B) MEF cells were treated as in (A) without stimulation, and subjected to IP with anti-Vps34 after preclearing with anti-Atg14L, followed by in vitro Vps34 kinase assay with or without 0.8 mM leucine.