Abstract
We recently optimized a procedure that directly yields aptameric sensors for small molecules in so-called structure-switching format. The protocol has a high success rate, short time, and is sufficiently simple to be readily implemented in a non-specialist laboratory. We provide a stepwise guide to this selection protocol.
Keywords: aptamer, biosensor, SELEX, small molecules, structure-switching
1. Introduction
Aptamers or oligonucleotide-based receptors are isolated through a process of in vitro selection or amplification, with a large number of protocols for aptamer isolation being reported over recent years [1–3]. We describe here a protocol that seems, at least in our hands, particularly suitable for isolation of aptamers for metal ions and small molecules [4–5]. All protocols for selection of aptamers are based on some type of affinity separation of binding and non-binding oligonucleotides, and when it comes to small molecules that often mean chemically modifying them and then attaching them to the solid-state matrix. In this protocol, we use small molecules in their native states, that is, unmodified and in solution-phase. The protocol is a streamlined version of previous reports by the groups of Ellington [6] and Li [7]. It is based on attaching an oligonucleotide library, via an oligonucleotide complementary to their constant region (part of PCR primer), to a column, and then passing over the column the target in solution, thus, isolating oligonucleotides in which the target impacts displacement of this complement.
The protocol has important advantages, but also some disadvantages. The advantages include: (1) no need to modify a small molecule (i.e., no synthetic skills are needed); (2) relatedly, using unmodified small molecules maximizes interactions with oligonucleotides, likely increasing the affinities of resulting receptors (e.g., we obtained with stringent selections aptamers for steroids and aromatic amines with Kd <100 nM); (3) direct screening for sensors via a structure-switching mechanism that comes straight from the principles on which the selection is based, (4) the ability to easily isolate and characterize even low-affinity aptamers (indeed, searching for high-affinity receptors is sometimes futile and by high-stringency of selections we loose on ‘good-enough’ binders; glucose is an example of this situation); and (5) the ability to finely tune counter-selection protocols. The biggest disadvantage is the need to have large quantities of small molecules, because the selection is done, at least initially, at high concentrations to minimize alternative mechanisms of release from the column. Another disadvantage is that we are limiting the structures of aptamers by the requirement that ligand binding conflicts with binding to the complementary oligonucleotide; this confines the diversity of aptameric structures. In our hands, the protocol did not work with several proteins that we tested, probably for the combination of these two reasons mentioned above. The protocol works excellently for metal ions, and in the case of small molecules [4–5], in our hands, it had at least 70% success rate. For example, it worked for (1) 14 amino acids we tried, with methionine and cysteine completely failing, (2) for all tested aromatic amine neurotransmitters, and (3) for all steroids that we could get in solution at concentrations above 50 µM (with deoxycholic acids only giving three-way junction structures).
2. Experimental section: Materials, Methods, and Tips
2.1. Basic preparation
2.1.1. Reagents and material preparations
We state the brand names and manufacturers of the materials used to help those who are new to SELEX experiments; other brands should work equally well. The storage condition of reagents and materials is at room temperature unless otherwise noted.
Molecular biology graded water: DNase/RNase free-water (Corning, cat. no. 46-000-CM). Nuclease-free water is used for all purposes unless otherwise noted.
Electrophoresis buffer: 0.5X Tris-Borate-EDTA (TBE) buffer diluted from 10X TBE (Corning, cat. no. 46-011-CM) or home-made TBE. For this dilution, distilled water may be use in lieu of DNAase/RNAase free water. Use of TBE is recommended rather than TAE (Tris-acetate-EDTA), as the oligonucleotides strands used in SELEX are usually small (e.g. 80 ~ 100 bp), and electrophoresis with TBE results in better image resolution.
SELEX reaction buffer: A variety of buffers were used. The basis of buffer selection was dependent on the final aptamer application conditions. One of the conditions in our previous research [5] is 20 mM HEPES, 1M NaCl, 10 mM MgCl2, 5 mM KCl, pH 7.5. After adjusting the pH, the buffer solution is filtered under vacuum with 0.22 µm stericup (EMD Milipore Corp. cat. no. SCGVU01RE).
2X SELEX reaction buffer: all components of the SELEX reaction buffer are doubled, vacuum filtering was also applied for this buffer. The 2X SELEX buffer is for use during the strand separation (see protocol listed below for this step).
Strand separation buffer: 20 mM HEPES, 300 mM NaCl (pH 7.5). Absence of MgCl2 from the buffer composition is essential, as Mg2+ stabilizes the DNA double strand and prevents complete denaturation of the DNA during the strand separation step.
Sodium hydroxide (NaOH): the diluted NaOH (0.25N) solution was prepared from 10N sodium hydroxide solution (Fisher Chemical, cat. no. SS255-1)
Hydrochloric acid (HCl): the diluted HCl (0.2N) solution was prepared from 36.5 to 38% w/w hydrochloric acid (Fisher Scientific, cat. no. A144-212)
Agarose gel: Depending on the size of the oligonucleotide used for the SELEX experiment, the appropriate agarose concentration is chosen. For up to ~ 100 bp length, 3% agarose gel is suitable. Freshly made gels can be kept in 0.5X TBE buffer for a long period of time (a few months) in room temperature, optionally, the gels may be re-used repeatedly, as the bands from the previously loaded samples in the gel will not overlap or interfere with the freshly loaded ones. This gel recycling will considerably save time and materials.
Agarose powder: Agarose LE (BioExcell, cat. no. 61133054)
4% commercial agarose gel (E-Gel EX Agarose Gels, 4%, Invitrogen, cat. no. G4010-04): This gel is needed to purify the desired PCR product if over-amplification occurs.
DNA ladder: O’RangeRuler 10 bp DNA Ladder (Thermo Sci Fermantas INC (FERM), cat. no. SM1313)
dNTP mixture: 10 mM premixed solution containing sodium salts of dATP, dCTP, dGTP and dTTP, (Promega, cat. no. U1515).
Taq polymerase: GoTaq Flexi DNA polymerase (Promega, cat. no. M829) or Choice-Taq DNA polymerase (Denville Scientific, cat. no. CB4050). Use the PCR buffer provided by the manufacturer.
PCR tubes: 0.2 ml strip tubes with individual cap.
Streptavidin resins: Streptavidin agarose resin (Thermo Sci Pierce Biotech, cat. no. PI20353).
Mini gravity column: Micro bio-spin chromatography columns (Bio-rad, cat. no. 732-6204)
Centrifugal filters for concentrating samples: 3K filter (EMD Millipore Amico Ultra-0.5 Centrifugal Filter Units, Milipore, cat. no. UFC UFC5003) for concentration of the single strands of library, 10K filter (Milipore, cat. no. UFC UFC5010) for the double strands of PCR products.
Cloning kit: TOPO TA cloning kit for Subcloning, with One Shot TOP10 chemically competent E. coli cells (Invitrogen, cat. no. K4500-01)
Solid E.coli growth media: ImMedia Amp Blue (Invitrogen, cat. no. 45-0038)
Liquid E.coli growth media: LB Medium (Powder), (MP biomedicals, cat. no. 113002-032)
Culture petri-dish: Falcon bacteriological petri dishes with lid (Corning Life Sciences, cat. no. 351029)
Liquid culture tubes: 12×75 mm polypropylene tubes, dual position cap, sterile (USA scientific, cat. no. 1450-0810)
Antibiotic: Ampicillin sodium salt (Sigma, cat. no. A9518).
DNA staining agent: Ethidium bromide (Sigma, cat. no. E7637). Non-toxic alternative dyes may be used.
3M NaOAC: 3M Sodium Acetate, pH 5.2 (MediaTech, INC. cat. no. 46033CI)
Absolute ethanol: Ethyl alcohol (Sigma, cat. no. E7023)
70% (v/v) ethanol: Reagent alcohol (Sigma, cat. no. 793213)
For most plastic-wares, we purchased free-DNAse, RNase, DNA & pyrogens levels, and for pipette tips, we prefer to use low-retention.
Microplate for fluorescent assay: Corning 384 Well Low Flange Black Flat Bottom Polystyrene NBS™ Microplate (Corning, cat. no. 3573)
Thermometer
Cell spreaders
Timer
Oligonucleotides: All oligonucleotides were purchased from Integrated DNA technologies (IDT DNA). We applied HPLC level purification if needed (see text and protocol).
2.1.2. Instrument/Equipment
Electrophoresis box (Mupid-exU, TaKaRa Clontech)
Gel document system: Alpha Innotech 3400 (Alpha Innotech)
UV/VIS spectrophotometer: NanoPhotometer P300 UV/Vis spectrophotometer (Implen)
Thermal cycler (Biometra)
Fluorescence plate reader: Victor II microplate reader (PerkinElmer)
Bench top centrifuge (Eppendorf)
Microwave
Analytical balance
Incubator for cell culture: shaker included.
Water bath
Beaker
2.1.3. Design of library, primers and capture
Choosing the size of the library: N8 ~ N60, wherein N indicates number of nucleotide positions generated by randomly mixing the A, C, G, and T bases (we do not order hand-mixed monomers, which is sometimes done to account for different synthetic yields). While it is certainly correct that, e.g., N60 library will encompass all possible subsets of, e.g., N20 library, we often considered it an advantage to use smaller sizes in our selections. For example, we used N8 [4], and in this case all different receptors of this size can be represented during selection, when we wanted to compare head-on structure-switching aptamers against similar targets and needed fully reproducible selections with multiple copies of each member; further, we considered implementing the selection on a microarray. Or another consideration: the same N30 sequences that are directly presented attached to the stem in N30 library, are (1) likely to be represented in larger libraries only in the context of additional secondary structures (due to folding in the presence of additional bases), thus yielding different receptors, and (2) may be removed from the stems, therefore not impacting release from the column upon binding to its ligand by a simple equilibrium (instead, perhaps yielding more complex mechanisms. In our experience, high-quality aptamers (e.g., with affinity of 100 nM and below), often result from multiple selections with different libraries, which maximizes the chances of success. That said, N30 ~ N40 seem to yield adequate results for the majority of the intended applications.
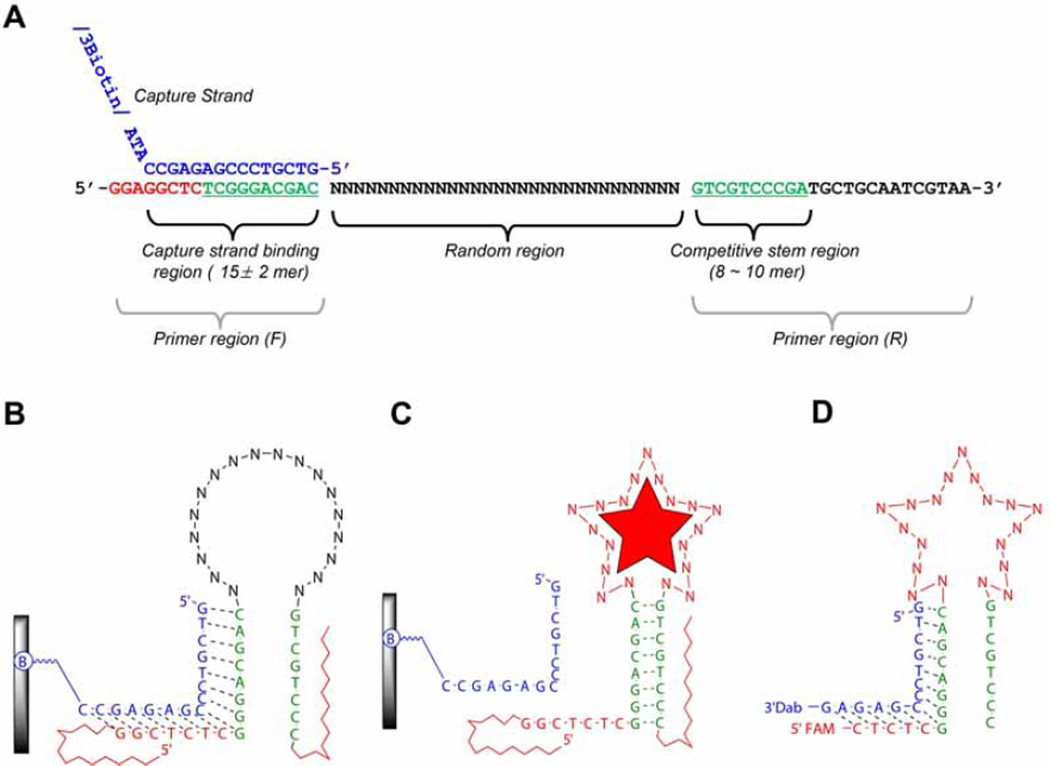
Design of the flanking regions of the library: The flanking regions of the library include the primer sequences with the capture region. When designing the primer sequences two things are required to ensure a high PCR efficiency. First, it is important to design primer regions that don’t self-dimerize and neither hetero-dimerize. Second, the primers must have a similar melting temperature (Tm). Good primer design ensures a clear single band after PCR. The primers can be analyzed through oligonucleotide analysis software, e.g. OligoAnalyzer 3.1, a free software from Integrated DNA technology (https://www.idtdna.com). Using the software analysis we discard primers expected to form hetero- or homo-dimers at ΔGs lower than -8 kcal/mole (usually this corresponds to dimerizations between four base-pairs or more). Shown in Fig. 1, the library design forms a short stem (8 ~ 10 bp, green letters) by the part of two primer regions. The capture strand which can partially hybridize (15 ± 2 mer in length), can compete with the stem by the structure-switching upon the target binding. Here is an example from our previous publication. (1) Library (72-mer): 5’–GGAGGCTCTCGGGACGAC(N30)GTCGTCCCGATGCTGCAATCGTAA-3’, (2) Forward-primer (18-mer): 5’-GGAGGCTCTCGGGACGAC-3’,(3) Reverse-primer (22-mer): 5’-TTACGATTGCAGCATCGGGACG-3’,(4) biotinylated reverse-primer (22-mer): 5’-biotin-TTACGATTGCAGCATCGGGACG-3’, (5) biotinylated column immobilizing capture (18-mer): 5’- GTCGTCCCGAGAGCCATA-BioTEG-3’. Note: HPLC purification is not mandatory for all these oligonucleotides. In case of the 72-mer library, without purification, incorporation of the shorter sequences cannot be avoided, but, we allow them as shorter type of library elements.
Fig. 1.
An example of SELEX library from Yang et al. [5]. (A) Library structure. This figure shows the relationship between a capture binding region and competitive stem region (green letters in both primer regions) which can induce the structural-switching upon target binding for the partitioning step. (B) An illustration of 2D structure of library binding on the streptavidin coated agarose column. A library element is attached to a column via a capture strand (blue letter in Fig. 1A). The capture strand is attached to an agarose-streptavidin column via a biotinylated oligonucleotide. “B” indicates biotinylation. (C) A library element partitioning mechanism. Upon binding the target, appropriate folding in the random region consequently induces the structural changes, which subsequently leads to dissociation from the capture stand. (D) An example of sensor design. After completion of SELEX, the final sensor is derived from trimming the flanking region of library element.
2.2. Selection Procedure
2.2.1. Choosing the concentration of target solution and concentration for negative selection
Generally, we start with a small molecule target concentration of 100 µM, and depending on the eventual intended purpose of the aptamer, we may reduce the concentration in the subsequent rounds. For negative/counter selection, we start with 100 µM of the negative selection molecules, and increase the concentration in the subsequent rounds. Negative selection is optional, and negative target molecules are dependent on the target molecules and purpose. For the most part, molecules which have similar chemical structures to the target are applied first, and any molecules which should not react with the selected aptamers for the purpose.
2.2.2. Selection step & elution profile
Oligonucleotide sample preparation: For the first round, depending on the library size, we can apply 100 ~ 1000 pmoles of library. The bigger the random region size, the larger the amount needed to cover the increased diversity. After deciding the library concentration, a 5x mole ratio of capture strand is used. From the third round of SELEX on, regardless of the library random size; we use only a portion of the product from the previous round, (e.g. ~100 pmoles) while reserving the rest for backup until the final round of SELEX. The SELEX steps are: (i) Heat water in a beaker to boiling e.g. microwave. (ii) Mix the library and capture sequence in 1:5 mole ratio, respectively, in 250 µL of SELEX buffer. (iii) Incubate the mixture of library and capture using the hot water for ~ 5 min to unfold any secondary forms of the library. (iv) Take the tube out of the beaker, and let it cool down to room temperature (> 10 min) to hybridize the capture strand to the library strands.
Bead column preparation: (i) Add 0.25 ml (or 0.2 ml minimum) of streptavidin agarose to a mini gravity column. (ii) Wash and equilibrate the column with the SELEX reaction buffer 5–6 times, each time with the same volume of the resin (e.g. 250 µL of buffer wash each time if 250 µL of resin is used), (iii) Add the mixture of library and capture, prepared in the previous step, to the column, collect the eluent, and re-apply this eluent two more times. This is to ensure the maximum amount of library is bound to the resin. (iv) Wash the column with SELEX reaction buffer to remove any unbound library molecules. This may be done as many times as desired. In general, we carried out the buffer wash 10 times, each time with the same volume of buffer as the resin used. It is important to not let the resin dry and to not apply pressure to elute the buffer. Gravity flow is sufficient. If the flow rate is extremely slow, more buffer may be added in addition to the previously added portion to help elute the solution. (v) Negative/counter selection is optional. The total number of negative selection rounds carried out and the concentration of the negative selection molecules used are adjusted based on the elution profile. You may increase the number of rounds of negative selection, or increase the concentration, or a combination of both to achieve the most stringent negative selection conditions. (vi) Positive selection: If negative selection is applied, additional buffer washes (~ 10 times) after the negative selection elution is required, prior to adding 250 µL of 100 µM positive target solution. The positive target solution is applied three times, and all three fractions should be collected. See Fig. 2A.
Small scale PCR for elution profile: (i) Prepare a master mix solution (e.g. for 500 µL of master mix solution: 50 µL of 10X PCR buffer, 10 µL of dNTP mixture (10 mM), 5 µL of each primer (100 pmole/ µL each), 402.5 µL of DNase/RNase-free water and 2.5 µL of Taq polymerase (5U/ µL) and mix thoroughly, (ii) pipet 47.5 µL into 10 individual PCR tubes, (iii) add 2.5 µL of template solution from the collected fractions, and (iv) perform the PCR reaction. PCR is run with 1 cycle of 95 °C, 2 min, N cycles of [92°C, 15 s; 59°C, 30 s; 72°C, 45 s], and 1 cycle of 72°C, 2 min. Annealing temperature (e.g. 59°C used here) can be adjusted depending on the melting temperature of primers. (v) Perform electrophoresis with the PCR product at 100 V for 20 min. (vi) Stain with EtBr and observe the bands under UV light and take an image of the gel. Note: Start with just 10 ± 1 cycle, check the elution profiles, if no bands are observed, add 2 more cycles, and perform the electrophoresis again. Keep repeating this until bands are eventually observed in the gel (Fig. 2B-D).
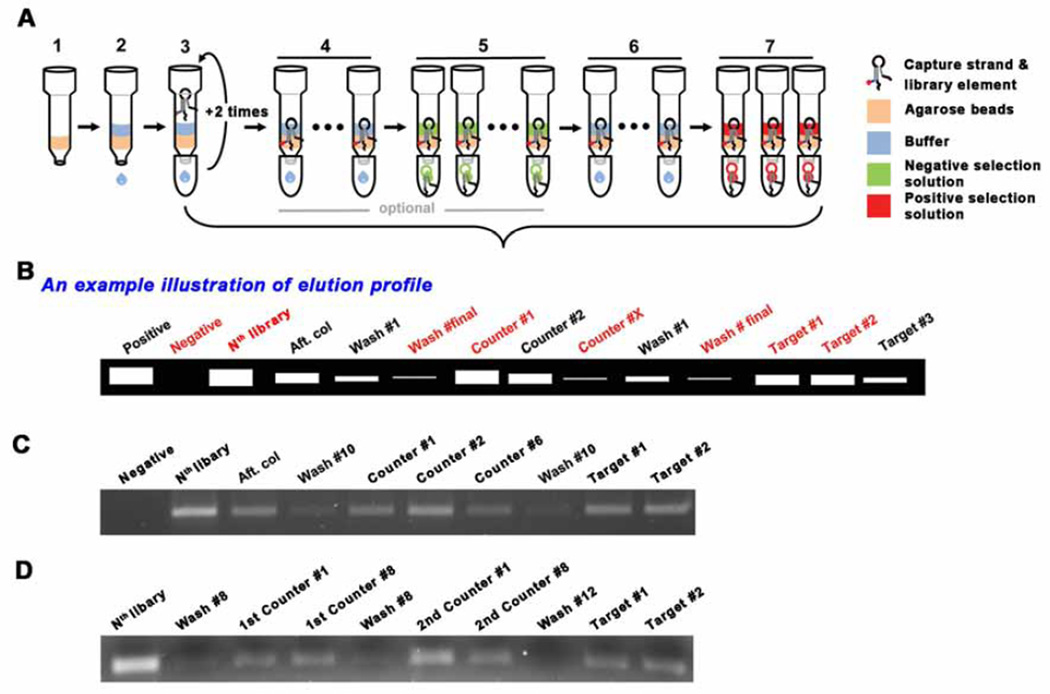
Fig. 2.
Schematic drawing for selection procedure. (A) (1) Agarose bead addition, (2) Wash the beads with SELEX buffer for equilbration, (3) Addition of the mixture of library and capture. After adding the mixture, the eluent is collected and re-applied into the column. This is to maximize the binding of the library-capture hybrid to the column. (4) Wash the column with the SELEX buffer ~10 times to remove uncaptured library elements. The number of washes can be adjusted based on the desired selection stringency. (5) Negative/Counter selection (optional). This elution step is to remove the less specific library elements that can bind to molecules which may compete with the desired target. (6) Additional wash - to remove residual solution from the negative selection, if applied. (7) Positive selection. During selection, several fractions need to be collected to monitor the elution profile. The choice of eluents to collect may be flexible, but the red labelled fractions should be collected, especially, the Nth library solution, the final buffer wash solution and the target solutions (#1 ~ #2). (B) An example illustration of elution profile. The collected eluents are compared though small scale PCR. The initial library templated PCR product can be utilized as a positive control for the PCR as well as the molecular weight marker. The commercial DNA ladder is used only when it is needed. The gel image of elution profile when one type of counter target is introduced (C), when two counter targets are applied (D).
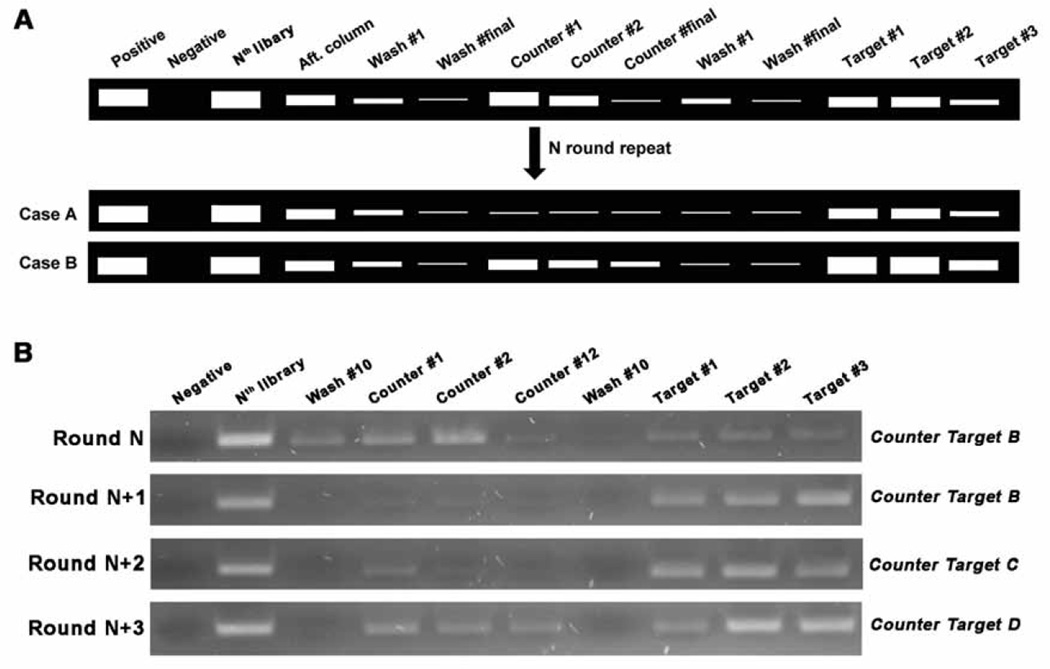
There are no firm rules on how may rounds (cycles) of SELEX for a target or how many rounds of negative selection, should be performed. This number will depend on the target molecules and the specificity and selectivity criteria the experimenters wish to impose. Therefore, the elution profiles, especially, the comparison of the differences between the last buffer wash and the target elution, is instrumental in helping determine when to stop the selection rounds. Fig. 3 shows examples.
Fig. 3.
The stop point of negative/counter selection. (A) After introducing negative selections, more rounds are carried out until the elution bands are dramatically decreased (Case A) or the target elution bands are brighter than the counter target elution bands (Case B). (B) Gel image example from “Target A” SELEX. At Round N (e.g. Round 9), ‘B’ was introduced as the counter target molecule, which is one of the analogs of target A, and showed similar elution bands in the counter and target lanes. At Round N+1, this counter target no longer bound to the library elements as observed by the lack of elution bands in the counter lanes. In the following round (Round N+2), we used another analog, ‘C,’ as the counter target, and observed a faint band in the counter target lanes of much weaker fluorescent intensity than in the target elution lanes. We then applied a third analog, ‘D,’ as the counter target in the next round (Round N+3) and we again observed some bands of much weaker fluorescent intensity than when the target was used. For the next two rounds, ‘D’ was used as a counter target and a similar elution profile was observed to Round (N + 3). This prompted us to stop the SELEX process and begin cloning.
2.2.3. Large scale PCR and strand separation
Large scale PCR: (i) Reduce the volume of all eluates to 50 ~ 100 µL using a centrifugal 3K MWCO concentrator (Fig. 4A), (ii) prepare a 1 mL volume of PCR mixture, concentrations of all reagents are the same as those in small scale PCR, except that for this reaction, we need to use the biotinylated reverse primer, instead of the non-biotinylated primer. After adding the template, (iii) take a 50 µL sample of the solution first, and perform small scale PCR to estimate the number of PCR cycles needed to amplify the template on the large scale. Apply 10 ± 1 cycles, take 10 µL of PCR product for gel loading, then put the tube back in the PCR machine and add 2 cycles more, and repeat it. Keep adding for ~15–18 cycles. Load all the samples on a gel, run the gel using the same conditions as before, and image the gel (Fig. 4B).
Strand separation: (i) Reduce the volume of all the PCR product to 50 ~ 100 µL using a centrifugal 10K MWCO concentrator - no washing with water is needed in this step. (ii) Prepare a streptavidin agarose column. Add 0.2 ml of streptavidin agarose resin to the column, and wash the column 5–6 times with the same volume of strand separation buffer as the resin to equilibrate. (iii) Add the concentrated PCR product, collect the eluent and apply this to the column two more times. This is to maximize the amount of library bound to the resin. (iv) Wash the column ten times with same volume as the resin using strand separation buffer to remove the PCR reaction buffer, unused dNTP, primers, etc. (v) add 300 µL of 0.2M NaOH, incubate for 10 min, collect the eluent, and add an additional 100 µL of 0.2M NaOH to collect the residual amount left in the column. (vi) Adjust the pH with HCl to approximately the same pH as the SELEX reaction buffer (e.g. pH 7.5). Use pH strips to check the pH. (vii) Add the same volume of 2X SELEX reaction buffer to adjust the ionic strength of the product for the next round of SELEX. (viii) Concentrate the solution with a 3K MWCO filter to 50 ~ 100 µL. This is the library solution for the next round of SELEX. (ix) Measure the library concentration using the nanophotometer, take ~ 100 pmoles for the next round, and store the rest of the solution at −20°C as a backup.
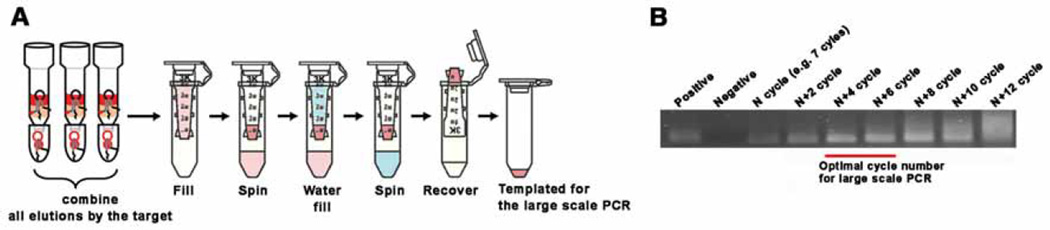
Fig. 4.
Large scale PCR. (A) Template preparation for the large scale PCR. Collect all target elutions, concentrate using a 3K MWCO centrifugal filter until it reaches 50 ~ 100 µL, then wash this concentrated solution with 400 ~ 500 µL of DNase/RNase-free water. This water washing is critical for high efficiency of large scale PCR. Use of all collected solution is recommended up until the ~5th round to allow amplification of all the oligonucleotide strands. After the 5th round, we recommend using only ~50% of template or less. The advantage of doing this is to keep a backup should the following rounds of SELEX fail, and after the 6th round, there are already multiple copies for many of the strands. (B) An example of small scale PCR (50 µL) for deciding the number of cycles before carrying out the large scale PCR (1 mL). Collect the PCR product after adding 2 cycles each time, and choose the number of cycles that give the highest yield, but not over-amplified. In this case, 11 ~ 13 cycles is recommended.
2.3. Post-selection step
2.3.1. Cloning & sequencing
Cloning: After obtaining a satisfactory elution profile (marked by a large increase in the PCR band intensity when the target is added, as compared to the buffer wash and negative selection), cloning is performed following the manufacturer’s instruction. (i) Prepare the insert DNA for cloning, PCR amplification is carried out under the same PCR conditions as the aforementioned, except for the final extension step where it was increased to 15 min at 72°C to add A-tail sufficiently for the T/A cloning system, and a minimum number of PCR cycles is applied (e.g. 9–10 cycles). Make sure to also use the non-biotinylated reverse primer for this process. In the manufacturer’s manual, fresh PCR product is directly used for the cloning, however, we purify and concentrate the PCR product though an ethanol precipitation method to get a higher concentration of DNA. The desirable DNA concentration for cloning is 500 ~ 1000 ng/ul, to maximize the positive colony number per plate. Sufficient numbers of colonies are necessary in order for us to be able to randomly choose several for sequencing. (ii) Incorporate directly the purified PCR to plasmid vectors using a cloning kit. The plasmids are subsequently transformed into competent cells, and positive colonies containing recombinant plasmid DNA is screened via blue/white screening. (iii) Some 20 to 30 colonies are picked at random in each cloning for liquid culture in 2 mL of LB media for > 16 hours. (iv) Plasmids are isolated from the cultured cell using Plasmid Miniprep Kit following the manufacturer’s manual.
Sequencing: PCR reaction is carried out with the isolated plasmid using the primers of the SELEX library, and then the plasmid samples are sent to DNA sequencing service facilities. For the sequencing, using a universal primer in the vector is recommended rather than using the library primer; the former will allow us to obtain the full library sequences without missing any in the primer regions.
2.3.2. Sequence analysis
DNA sequencing service facilities give two formats of sequencing results. One is in plain text files and the other is in native electropherograms files (e.g. AB1files). (i) Find the insert region, which starts from the forward primer until the end of the reverse primer binding region. If necessary, use the reverse complementary strands. (ii) Complete error correction or determine ‘N’ which is derived from failure of precise reading by the sequence analysis program manually using native electropherograms files. (iii) Count the frequency of sequences manually, and also try multiple sequence alignment using alignment programs such as CLUSTALW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). From these analyses, sequences are grouped.
Trimming sequences: the library contains the flanking region beyond the stem which is presumed not to be necessary for the functioning aptameric sensor (cf., protocols for testing clones below). After obtaining the whole sequences, trim the outside of the capture region in the forward primer region, and trim the outside of the capture competitive region (~ 8 mer next to the random region) (Fig. 1D).
Additional note on trimming: If we assume that the mechanism of selection is a direct stabilization of a stem, this protocol will provide a direct approach to characterize aptamers in their structure-switching sensor forms (cf., aptameric sensor validation below). We have, however, on rare occasions (<5% of sequences), encountered the situation in which a sequence would have an excellent elution profile but also a very poor response in its sensor form. In this case, there is a possibility of trimmed region participating in structure-switching in the presence of ligand, and such sequence should not be trimmed.
Secondary structure analysis: analyze 2D structure of sequences using Mfold site (http://mfold.rna.albany.edu/) or NUPACK (http://www.nupack.org/). From the 2D analysis, the sequences are grouped again based on the pocket region, as some sequences with different compositions may share similarities in the pocket regions, and they are assumed to have similar sensing capabilities for the target. In addition, through the homology analyses such as multiple sequence alignment and common motif finding, the candidate pool for the clone SELEX can be reduced
2.3.3. Clone SELEX protocol for testing affinity
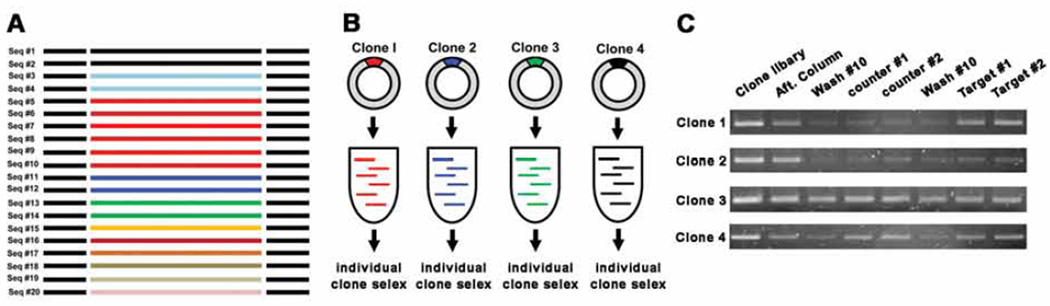
Individual clone SELEX: (i) Select the representative clones from the sequence analysis, e.g. one from each group. (ii) Perform a 500 µL large scale PCR with each selected clone. PCR conditions are 15 cycles when the template amount is about 500 ng, if the clone concentration is low, increase the number of PCR cycles or vice versa. (iii) Perform the strand separation and prepare the library which consists of strands of just one sequence, then (iv) perform regular SELEX. (v) Choose the clones which show the best and the second best elution profiles (Fig. 5C). These are the candidates for sensors.
Fig. 5.
A schematic for clone SELEX. (A) After sequencing, count repeats and choose ~ 4 clones based on their abundance. (B) Large scale PCR and strand separation for the selected clones. Strands of one particular sequence is amplified through large scale PCR and library is prepared. (C) Representative cases of clone SELEX elution profile. The elution profile of Clone 1 is the best, clone 4 is second. Cases 2 and 3 are not recommended for adaptation into sensors.
2.4. Aptameric sensor validation
2.4.1. Fluorescent sensor ordering
After completion of SELEX, the final sensor form is derived by trimming the flanking region of the library element (see Fig. 1D). The base pairing between capture and sensor is 13 or 14 bp, the main stem base pairing is 8 ~ 9 bp based on the original library structure. If the capturing efficiency is not sufficient with 13-mer, the length of the quencher may be extended (see Fig. 6B). The fluorophore choice is flexible, and the quencher should be compatible with the fluorophore. We use mostly fluorescein (FAM) conjugated to the 5’-end of sensor, and the quencher (dabcyl) should then be conjugated at 3’-end.
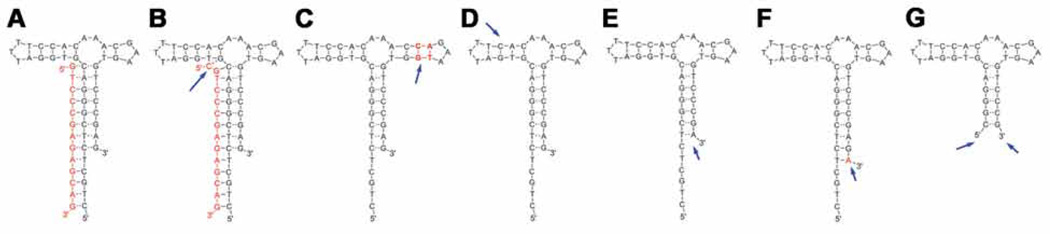
Fig. 6.
An example of sensor optimization. This is a steroid aptamer for deoxycorticosterone 21-glucoside [4]. Based on the original sensor (A), extended capture length (B), extended side stem (C), shortened side stem (D), shortened main stem length (E), or extended main stem base paring (F). (G) is the shortened sensor form for utilizing without the capture. Arrows indicate the modification sites.
2.4.2. Analysis of sensor reactivity
Quenching test: (i) Dissolve the sensor strand and the quencher strand separately in RNase/DNase-free water. (ii) Measure the concentrations. (iii) Prepare 8 different concentrations of quencher solution by two-fold serial dilution, and 2X concentrated sensor solution, e.g. we prepared 90 µL at a starting concentration of 1 µM for the quencher solution, and 7 more solutions by two-fold serial dilution. 350 µL of 100 nM sensor strand solution was prepared. (iv) Mix 40 µL of sensor solution and 40 µL of each quencher strand solution. (v) Incubate for 5 min in a hot water bath and let it cool to room temperature in the dark. Transfer 75 µL of each solution into a well in the plate. (vi) Read on a fluorescence plate reader (ex/em = FAM 485 nm /FAM 535 nm). (vii) Calculate the % quenching and plot the curve, (viii) Decide the ratio of quencher strand for the sensor, around 90% quenching is recommended, however, if 80% quenching is at 1:5 ratio, we choose this condition over using a higher quencher amount.
Sensor test: (i) Prepare 2X concentration of the sensor and quencher mixture using the quencher ratio determined from the quenching test above (ii) Incubate for 5 min in hot water, let it cool to room temperature and keep it protected from light (> 30 min). (iii) Prepare the 2X concentrated target solution and dilute serially. (iii) Combine 40 µL of the sensor-quencher mixture solution and 40 µL of target solution. (iv) Incubate it in the dark for 40 min. (v) Use 75 µL for measurement on the fluorescence plate reader, and (vi) plot the response curve.
2.4.3. Affinity measurement
The Kd is determined following Hu and Easley’s methods [8]. There are two separate binding affinity measurements for each sensor, Kd, eff1 and Kd, eff2. Kd, eff1 is the dissociation constants between sensor and capture oligonucleotide; it is calculated based on the decreasing fluorescent intensity (quenching) of sensor (50 nM fixed) in the presence of 0 ~ 500 nM range of the quencher strand concentrations (quencher strand contains DABCYL). The binding curve is plotted using ‘one phase decay’. Kd, eff2 is a unitless constant between the sensor-capture complex and its target (Kd, eff2 = [capture strand] [sensor-target] /[sensor-capture] [target]). Kd is derived from Kd, eff1/Kd, eff2, each binding curve is plotted using software (e.g. GraphPad Software).
2.4.4. Further optimization tips
Optimization of sensor or capture length: To improve the sensor reactivity, the length of sensor or quencher can be adjusted. At this stage, we cannot generalize any rules, as this needs to be optimized through trial and error by simple extensions and base-pair substitutions (Fig. 6), but in our experience, best results are achieved when Kd of competitor oligonucleotide is between 150–350 nM. We consider as good sensors those that show a ~10-fold increase in signal.
3. Conclusion
There are many approaches to the selection of aptamers and we suggest that the method we described herein is probably the most straightforward to be implemented in laboratories otherwise not specializing in nucleic acids. Together with the approach to derivatize in situ and in solution small molecules that are traditionally thought to interact poorly with nucleic acids, it has high probability to yield good aptamers for most molecules that could be of interest to an analyst.
Table 1. Overview of SELEX procedure.
The procedure consists of four main steps; the numbering for each sub-step is related in the following section.
| Main Steps | Sub-Steps | Time | |
|---|---|---|---|
| 2.1. | Basic preparations | 2.1.1. Reagents and material preparations | |
| 2.1.2. Instruments/Equipment | |||
| 2.1.3. Design of library, primers and capture | |||
| 2.2. | Selection procedure | 2.2.1. Choosing the concentration of target solution and compounds for negative selection |
Iterative step. 1 ~ 1.5 round of SELEX per day. Approximately, ~ 3 weeks |
| 2.2.2. Selection step & elution profile | |||
| 2.2.3. Large scale PCR & strand separation | |||
| 2.3. | Post-selection step | 2.3.1. Cloning & sequencing | 4 ~ 7 days |
| 2.3.2. Sequence analysis | |||
| 2.3.3. Clone SELEX | |||
| 2.4. |
Aptameric sensor validation |
2.4.1. Fluorescent sensor ordering | 4 ~ 7 days |
| 2.4.2. Analysis of sensor reactivity | > 1 day | ||
| 2.4.3. Affinity measurement | ~ 1 day | ||
| 2.4.4. Further optimization tips | Case by case | ||
Highlights.
-
▶
Highly useful method for the selection of aptamers or small-molecule targets.
-
▶
Provides a very detail step-wise guide to a selection protocol.
-
▶
Simple enough to be readily implemented in a non-specialist laboratory.
Acknowledgments
We are grateful to the National Science Foundation (CBET-1033288 and CBET-1026592), and National Institutes of Health (RGM104960). We thank Dr. Steven Taylor and Dr. Claudia Tapia-Alveal for comments that improved the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Biotechnol. Adv. 2015;33:1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. http://dx.doi.org/10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Zu Y. Molecules. 2015;20:11959–11980. doi: 10.3390/molecules200711959. http://dx.doi.org/10.3390/molecules200711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blind M, Blank M. Mol. Ther. Nucleic Acids. 2015:e233. doi: 10.1038/mtna.2014.74. http://dx.doi.org/10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K-A, Pei R, Stefanovic D, Stojanovic MN. J. Am. Chem. Soc. 2012;134:1642–1647. doi: 10.1021/ja2084256. http://dx.doi.org/10.1021/ja2084256. [DOI] [PubMed] [Google Scholar]
- 5.Yang K-A, Barbu MB, Halim M, Pallavi P, Kim B, Kolpashchikov DM, Pecic S, Taylor S, Worgall TS, Stojanovic MN. Nat. Chem. 2014;6:1003–1008. doi: 10.1038/nchem.2058. http://dx.doi.org/10.1038/nchem.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajendran M, Ellington AD. Anal. Bioanal. Chem. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. http://dx.doi.org/10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 7.Nutiu R, Li Y. Angew. Chem. Int. Ed. 2005;44:1061–1065. doi: 10.1002/anie.200461848. http://dx.doi.org/10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Easley CJ. Analyst. 2011;136:3461–3468. doi: 10.1039/c0an00842g. http://dx.doi.org/10.1039/c0an00842g. [DOI] [PubMed] [Google Scholar]