Abstract
Exclusively breast-fed infants can exhibit clear signs of IgE or non IgE-mediated cow's milk allergy. However, the definite characterization of dietary cow's milk proteins (CMP) that survive the maternal digestive tract to be absorbed into the bloodstream and secreted into breast milk remains missing. Herein, we aimed at assessing possible CMP-derived peptides in breast milk. Using high performance liquid chromatography (HPLC)-high resolution mass spectrometry (MS), we compared the peptide fraction of breast milk from 12 donors, among which 6 drank a cup of milk daily and 6 were on strict diary-free diet.
We identified two bovine β-lactoglobulin (β-Lg, 2 out 6 samples) and one αs1-casein (1 out 6 samples) fragments in breast milk from mothers receiving a cup of bovine milk daily. These CMP-derived fragments, namely β-Lg (f42-54), (f42-57) and αs1-casein (f180-197), were absent in milk from mothers on dairy-free diet. In contrast, neither intact nor hydrolyzed β-Lg was detected by Western blot and competitive ELISA in any breast milk sample. Eight additional bovine milk-derived peptides identified by software-assisted MS were most likely false positive. The results of this study demonstrate that CMP-derived peptides rather than intact CMP may sensitize or elicit allergic responses in the neonate through mother's milk. Immunologically active peptides from the maternal diet could be involved in priming the newborn's immune system, driving tolerogenic response.
Keywords: Breast milk, Cow's milk proteins, Protein digestion, Food allergens, Mass spectrometry
Graphical abstract
For the first time, peptides deriving from the hydrolysis of bovine milk allergens in the maternal gastrointestinal tract were identified in breast milk. Using HPLC-mass spectrometry, we identified fragments of bovine β-lactoglobulin and αs1-casein in the breast milk of mothers who had consumed bovine milk.
INTRODUCTION
Cow's milk proteins (CMP) deriving from maternal diet can elicit IgE- or non IgE-mediated allergic response in exclusively breastfed infants.1,2 In spite of the clinical evidence, a definite proof on a molecular basis that CMP flow into breast milk remains missing.
In addition to the potential primary (direct) and secondary (indirect) involvement in inducing adverse reactions for a selected cohort of allergic newborns, the exposure to allergens through breast milk has relevant physiological implications, as these allergens might be involved in priming the newborn's immune system to tolerate harmless food and foreign proteins.3
The passage of low amounts of food-derived allergens into breast milk as a possible route of sensitization has been suggested.4-7 Using immunochemical assays, Vadas et al. detected Ara h 1 and Ara h 2 allergens in breast milk of lactating women who had consumed roasted peanut.8 More recently, other authors identified a different peanut allergen in breast milk, i. e. Ara h 6, less than ten minutes after peanut ingestion.9 A direct correlation has been described between dose of egg intake and amount of intact ovalbumin in breast milk.10 On the contrary, wheat intake and levels of non-degraded gliadins in blood serum or breast milk are not correlated.11 Breast milk has been described to contain a basal level of immunoreactive gliadins even before intake of wheat, suggesting a certain degree of cross-reactivity between anti-gliadin antibodies and endogenous human milk proteins, provided that the mothers strictly complied with the avoidance diet during trial.12 Thus, in general the definition of the immunological determinants at this stage appears quite controversial, mainly due to the lack of validation of the immunochemical methods of detection, which can be biased by cross-reactivity or nonspecific antibody recognition.13,14
Whether β-lactoglobulin (β-Lg) occurs in human milk has been debated for many years. The scientific community now commonly accepts that humans do not express β-Lg. However, variable amounts of β-Lg have been detected in human milk by immunochemical techniques, supposedly being bovine β-Lg of dietary origin.15 β-Lg is highly resistant to pepsin, the main gastric protease, while it is extensively degraded by pancreatic proteases, before possible absorption in the small intestine.16 In contrast, caseins are quickly proteolyzed in both gastric and intestinal conditions.17 Interestingly, authors who had previously attributed the detection of β-Lg to antibody nonspecific recognition18,19 have later described intact bovine αs1-casein in human milk20 and colostrum.21 Although the identification of αs1-casein was based on mass spectrometry (MS)-based proteomics, these findings seem to conflict with the prompt digestion of caseins in the gastro-intestinal tract of the lactating mother. Indeed, the likelihood that CMP-derived peptides is transferred into milk through the mammary epithelium is much higher than for intact parent proteins. Nevertheless, no published research has attempted to detect immunologically active fragments of food allergens in human breast milk.
The aim of the current study was to assess the occurrence of CMP-derived peptides in breast milk using an antibody-independent method, such as high performance liquid chromatography (HPLC) coupled to high-resolution MS. Via HPLC-MS, the peptide fractions of mature (after colostrum-to milk transition) breast milk samples from mothers regularly consuming bovine milk and other dairy products were compared to those from mothers on a strict dairy-free diet.
MATERIALS AND METHODS
Samples of mature breast milk (40 mL) were obtained through a breast pump equipped with plastic disposable pump sets from 12 non-atopic healthy donors (median age: 32 years old) who delivered at term, 2–4 weeks after parturition. All subjects were from across Campania region in Italy. A written informed consent was provided by the mothers before the milk collection.
Six of the samples were collected from women who drank one cup of pasteurized bovine milk daily and regularly consumed other dairy products. Six control samples were collected from women on a strict milk- and dairy products-free diet since at least one week, opportunely instructed to avoid any possible sources of CMP. Breast milk of milk-drinking mothers was collected 2 hours after a regular milk load (200 mL). A skilled operator at the Department of Translational Medicine of the University of Naples “Federico II” (Italy) supervised the administration of bovine milk, sample collection and code labelling. Within less than 3 min after milk expression, the operator added a serine-protease inhibitor (Pefabloc®, Sigma, St. Louis, MI, USA) to each sample (1mM final concentration) and immediately froze milk at -20°C. High purity grade solvents and chemicals were from Sigma.
Peptide extraction
Aliquots of each milk sample were skimmed by centrifugation (3,000 × g for 15 min at 4°C) and the upper fat layer removed with a spatula (repeated twice). Proteins of skimmed milk were precipitated (at 4°C) with 12% (w/v) trichloroacetic acid (TCA) final concentration and soluble peptides were solid-phase extracted using Sep-pak C18 pre-packed columns (Waters, Milford, USA). After loading, peptides were extensively washed with 0.1% trifluoroacetic acid (TFA) and eluted with 70% acetonitrile containing 0.1% TFA. Organic solvent was evaporated in a vacuum centrifuge (Speed-vac, Savant) and peptides were finally lyophilized.
Competitive ELISA for β-Lg fragments
The presence of β-Lg-derived peptides in the breast milk samples was assayed using the RIDASCREEN® competitive ELISA kit, specifically developed for the detection of hydrolyzed fragments of β-Lg (R4901, R-Biopharm AG, Darmstadt, Germany). The assay was carried out according to the manufacturer's instruction, using intact bovine β-Lg as a standard. Samples were assayed in triplicate. Standard points fit a cubic spline curve that was built using the Ridasoft Win software vers. 1.91 (R-Biopharm AG). Breast milk was assayed at several dilutions, including 1/500 (recommended by the manufacturer), 1/200, 1/100, and 1/50 (v/v). To exclude matrix effects, control breast milk samples spiked with variable amounts of β-Lg were used as the positive control. The manufacturer's declared limit of detection and limit of quantification were 0.115 ppm and 5 ppm, respectively, with <1% cross-reactivity with caseins or other whey proteins.
Western blotting detection of intact β-Lg
The 12% TCA insoluble breast milk proteins were three-fold washed with diethyl ether and assayed for intact β-Lg by Western blot analysis. Proteins were dissolved in Laemmli sample buffer at 1 mg/mL and 10 μg were separated by SDS-PAGE using 12% acrylamide precast gels (Bio-Rad Laboratories, Hercules, CA, USA). Gels were run in duplicate; after separation, one gel was stained using G-250 Coomassie Blue Silver staining while the other was electroblotted onto nitrocellulose membranes using a Trans-Blot Cell (BioRad) at 400 mA at 4 °C for 1 h. Membranes were blocked for 1 h at room temperature with 5% (w/v) bovine serum albumin (Sigma) in Tris-buffered saline solution with 0.05% Tween 20 (TBS-T) and incubated overnight at 4 °C, with immunoaffinity purified anti-β-Lg IgG polyclonal antibody developed in rabbit (Abcam Ltd, Cambridge, UK) previously diluted 1/10000 in TBS-T. Monoclonal peroxidase-conjugated mouse anti-rabbit IgG antibody (Sigma) diluted in TBS-T (1/10000) was applied to the membrane for 1 h at room temperature. The membrane was extensively rinsed with TBS-T (3 × 10 min) and finally with TBS (1 × 10 min) before development. Chemiluminescence reagents (ECL Plus WB reagent, GE Healthcare) and X-ray film (Kodak, Chalons/Saône, France) were used to visualize the immunoreactive protein bands at various exposure times ranging from 0.5 to 5 min. A sample of bovine whey was used as the positive control. The positive control contained 2 μg or 1-100 pg of β-Lg for the SDS-PAGE and Western blot analyses, respectively.
HPLC-Orbitrap MS/MS
HPLC separation was performed on a Waters Nano Acquity UHPLC (Waters Corporation) with a Proxeon nanospray source. The digested peptides were reconstituted in 3% acetonitrile/0.1% formic acid and roughly 3 μg of each sample was loaded onto a 100 μm × 25 mm Magic C18 100Å 5U reverse phase trap, where they were desalted online before being separated on a 75 micron × 150 mm Magic C18 200Å 3U reverse phase column. Peptides were eluted using a gradient of 0.1% formic acid (A) and 100% acetonitrile (B) with a flow rate of 300 nL/min. A 60 min gradient was run with 5% to 35% B over 50 min, 35% to 80% B over 3 min, 80% B for 1 min, 80% to 5% B over 1 min, and finally held at 5% B for 5 minutes. Each sample run was followed by a 60 min column wash. Mass spectra were collected on a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) in a data-dependent mode with one MS precursor scan followed by 15 MS/MS scans. A dynamic exclusion of 20 s was used. MS spectra were acquired with a resolution of 70000 and a target of 1 × 106 ions or a maximum injection time of 30 ms. MS/MS spectra were acquired with a resolution of 17500 and a target of 5 × 104 ions or a maximum injection time of 50 ms. Peptide fragmentation was performed using high-energy collision dissociation with a normalized collision energy value of 27. Unassigned charge states as well as ions >+6 were excluded from MS/MS fragmentation.
Database search and peptide identification
LC-MS/MS raw data was converted to .mgf format and analyzed by the offline search engine X!Tandem22 as described previously,23 with some modifications. Briefly, mass spectral data were searched against the bovine proteome (downloaded from uniprot.org) to identify naturally-occurring peptides which could have originated from a bovine protein. An error tolerance of 10 and 20 ppm was specified for precursor and fragment m/z values, respectively. A minimum e value of 0.01 was required of all peptide matches, corresponding to a 99% confidence score. Oxidation of methionine was allowed as a potential modification, and phosphorylation of serine and threonine were allowed as potential modifications in a single refinement step. The output of the software was refined to exclude peptides which could have originated from a human protein. Since there is significant overlap between human and bovine milk protein sequences, it was necessary to refine the data further by identifying peptides that matched both proteomes. The output of the software was entered into the Uniprot BLAST search engine (uniprot.org/blast) and searched against mammalian proteomes, with a maximum allowable e value of 10. Peptides that were an exact match to a human sequence under these conditions were removed from the final results list. Next, a list of bovine milk proteins was assembled from prior proteomic studies.24-26 Peptides not originating from one of these proteins were also excluded from the final results.
To further characterize the samples and assess the performance of the peptide extraction, mass spectral data were also searched for naturally-occurring human milk peptides. The search employed the same parameters used to identify bovine milk-derived peptides and referenced a library of proteins previously identified in proteomic studies of human milk.27-30
3D Molecular Modelling
The molecular structure prediction of β-Lg was carried out using the on-line resource I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), provided by the Department of Computational Medicine and Bioinformatics University of Michigan, USA.
RESULTS AND DISCUSSION
LC-High Resolution Tandem Mass Spectrometry
Human milk is intrinsically rich in oligopeptides31 which are released from milk protein by endogenous proteases.32-33 These peptides are present at an even higher amount in pre-term than in term breast milk.34 The tandem MS-based identification of foreign peptides (if any) occurring at trace levels in breast milk challenges the dynamic range of MS alone. Therefore, prior to MS analysis, peptides must be separated with high resolution chromatography. Although the identification of possible bovine milk-derived peptides is restricted to a relatively low number of abundant proteins (casein and major whey proteins), it requires an untargeted approach, due to the lack of any predictable specificity of protein cleavage. The complexity of such a combination of “deep” and “untargeted” peptidomics is probably the reason why food allergen-derived peptides have not been detected in human milk so far.
In the current case, the identification of bovine milk-derived peptides in breast milk could be further complicated by the homology among the corresponding milk specific proteins. However, as it concerns the most abundant protein components of human milk, β-casein, αs1-casein and α-lactalbumin share 53, 31 and 74% homology, respectively, with their bovine counterparts, thereby offering a fair margin to discriminate peptides according to their origin. As the intent of the research is to identify peptides unique to bovine milk occurring in human milk (that may induce sensitization or allergic reaction to bovine milk), peptides completely homologous between the two species were irrelevant to our search.
On average, each milk sampled contained more than 1,100 endogenous human milk protein-origin peptides (e ≤ 0.01). These human milk peptides derived from mostly β-casein, osteopontin, and toll-like receptor 9. Overall, 11 peptides were identified as matching bovine milk protein-derived peptides, while they did not match protein sequences of human milk (Table 1). All raw data and X!Tandem peptide results are available in the ProteomeXchange database with identifier PXD003657 (provisional access for the Reviewers: ID: reviewer86363@ebi.ac.uk; password: JQQpzjK3).
Table 1.
Bovine milk-derived peptides identified in breast milk samples. M*= oxidized methionine
| Peptide sequence | Protein of origin (position) | Uniprot protein ID | Number of times identified | Mothers on milk-free diet (N=6) | Milk-drinking mothers (N=6) |
|---|---|---|---|---|---|
| GLKSRSKKF | Osteopontin (159-167) | P31096 | 5 | 1 | 4 |
| YGLKSRSKKF | Osteopontin (158-167) | P31096 | 1 | - | 1 |
| VAPGALLGLGNLTHLSLKYNNLTEVPRRLPPSLDTL | Toll-like receptor 9 (189-224) | Q5I2M5 | 3 | 1 | 2 |
| VAPGALLGLGNLTHLSLKYNNLTEVPRRLPPSLDT | Toll-like receptor 9 (189-223) | Q5I2M5 | 1 | - | 1 |
| YVEELKPTPEGDL | β-lactoglobulin (42-54) | P02754 | 2 | - | 2 |
| YVEELKPTPEGDLEIL | β-lactoglobulin (42-57) | P02754 | 1 | - | 1 |
| RCLQATLTQDSTYGNEDCLYLNIWVPQGR | Bile salt-activated lipase (81-109) | P30122 | 1 | 1 | - |
| SDIPNPIGSENSEKTTM*PLW | αs1-casein (180-197) | P02662 | 1 | - | 1 |
| ASVAVEPQLSVVTRVANLPLVS | Perilipin-2 (2-23) | Q9TUM6 | 1 | 1 | - |
| AKDTVTCVGHIIGA | Xanthine dehydrogenase/oxidase (656-669) | P80457 | 1 | - | 1 |
| YPNLCQLCKGEGENQ | Lactotransferrin (185-199) | P24627 | 1 | - | 1 |
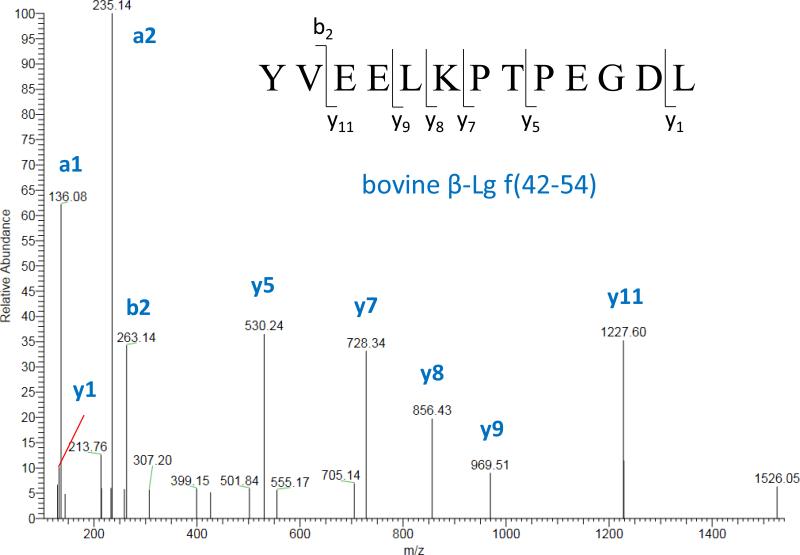
The most significant finding was the identification of a αs1-casein fragment and two peptides from β-Lg in the milk of lactating mothers who drank bovine milk. These peptides were not detected in any control samples. In relation to the β-Lg-derived peptides, one breast milk sample contained β-Lg (f42-54) and β-Lg (f42-57), while an additional sample contained only β-Lg (f42-54). The αs1-casein (f180-197) was found in an independent sample and it probably survived digestion, because it belongs to a relatively proteolysis-resistant region of the protein.35 No additional casein peptides were detected, most likely due to their prompt and extensive degradation by the gastro-intestinal system of the mother. The MS/MS spectrum of β-Lg (f42-57) is shown in Figure 1 with the assignment of the most intense fragments, also manually validated. Notably, this spectrum did not match any protein sequence in the human database. After identification, the CMP-derived peptides were unsuccessfully searched also in the remaining samples using a targeted approach (peptide ion extraction).
Figure 1.
Identification of β-Lg (f42-54) through the MS/MS spectrum of the peptide precursor [M+2H]2+ = 745.37. The most intense y- and b-/a- fragments are assigned.
The most frequently identified bovine milk protein peptides were those from osteopontin, a well-described milk protein, which also occurs in human milk.29,36 The peptide derived from bovine osteopontin has a closely related sequence in human osteopontin (GLKSRSKKF in bovine versus GLRSKSKKF in human), exhibiting the same molecular weight and differing only because of a positional K/R inversion. Hydrolytic events also release a significantly high number of osteopontin peptides in human milk.34 The manual inspection of the spectra confirmed that the lack of the fragmentation ions at level of the K or R residues resulted in software mismatch, thereby justifying the identification of this peptide as of bovine origin rather than human and its detection even in the control samples (milk from mothers on milk-free diet). Toll-like receptor 9 is a very minor protein of milk and it seems quite improbable that some specific domains could survive maternal digestion and flow into breast milk at a relatively high amount. Indeed, a peptide from Toll-like receptor 9 was also detected in a control sample. The peptides from xanthine dehydrogenase and lactoferrin (1 peptide each) identified in samples from milk-drinking mothers are probable candidate fragments of bovine origin. However, the confidence of their identification was low because they both contain free C residues, which in not-reducing conditions are expected to be engaged in disulfide bonds. A similar observation might hold for a peptide from bile salt-activated lipase, which contains two C residues and was identified in one control sample. The bovine and human sequences of bile salt-activated lipase in that region only differ by a N(bovine)→D(human) substitution inducing a shift of 1 mass unit besides an isobaric L→I. For the reasons above, the peptides from minor milk proteins, differently from those from β-Lg and αs1-casein, are most likely false positive identifications, thereby emphasizing that software-assisted characterizations should be carefully and critically evaluated. In the current case, the presence or absence of these peptides has only technical relevance, because they should not be involved in immunological mechanisms.
Competitive ELISA for β-Lg fragments
A competitive ELISA was applied to detect β-Lg peptides in breast milk samples. The ELISA kit is specifically designed to detect peptides arising from processed β-Lg in food matrices. Standard β-Lg solutions fit a cubic spline curve. Only one sample gave a positive response, with an apparent β-Lg concentration of 2.1 ppm; however, this concentration was below the limit of quantification of the assay (5 ppm). The ELISA β-Lg-positive sample was a control and therefore the presence of β-Lg peptides should be ruled out. In all the remaining samples, assayed at varying sample dilutions, β-Lg peptides were undetectable, at a 0.115 ppm limit of detection. The inconsistency of the ELISA results demonstrates that immunological methods suffer from bias, most likely due to the specificity of the antibody reactivity. The pitfalls of the ELISA-based methods in detecting and quantifying allergens in complex food matrices have been recently reviewed.14
Western blotting detection of intact β-Lg
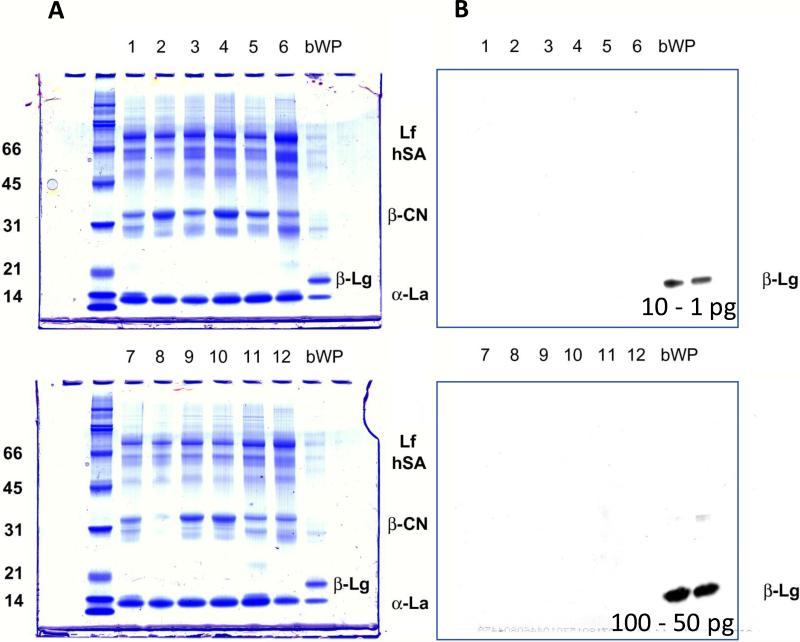
The SDS-PAGE and parallel Western blot analysis of the breast milk samples is shown in Figure 2. Bovine whey proteins were used in both cases as a positive control for β-Lg. Estimated amounts of β-Lg were 2 μg for the SDS-PAGE analysis and 1-100 pg for the ECL reagent based detection, respectively. The estimated limit of detection of β-Lg by Western blot was well below 0.1 ppm (1 pg with respect to 10 μg total protein). β-Lg was clearly detected in the whey protein extract with both the analyses, while no band ascribable to it was detected in any of the breast milk samples. These results confirmed the absence of intact bovine β-Lg or derived large immunoreactive fragments at detectable amount.
Figure 2.
SDS-PAGE electrophoresis (A) and anti-β-Lg Western blotting (B) of the 12 samples of human milk analyzed. Bovine whey proteins (bWP) were loaded as the β-Lg positive control. For the Western blotting analysis, the estimated amount of β-Lg in bWP control was 10 and 1 pg (upper panel) and 100 and 50 pg (lower panel).
Significance of the presence of bovine milk derived peptides in breast milk
β-Lg and αs1-casein are major allergens of bovine milk.37 For the first time, fragments released from allergenic proteins have been identified in human milk. The detection of allergen-derived peptides in breast milk opens a new perspective to monitor potential offending components arising from the maternal diet. Milk proteins are extensively processed along the gastro-intestinal tract of the mother, before intestinal absorption and systemic distribution. Although caseins are more abundant than whey proteins in bovine milk (roughly 80:20 w/w), they undergo a very early degradation in the stomach and then further in the intestine.17
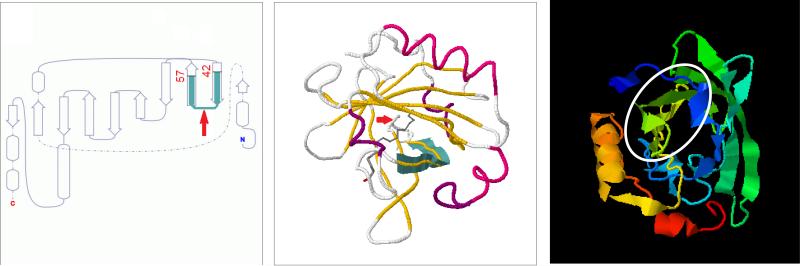
In contrast, β-Lg is highly resistant to gastric pepsin digestion and large peptides derived from it could escape duodenal degradation. The β-Lg (f42-54) and β-Lg (f42-57) peptides belong to a two β-sheets domain that is located in the lipid binding calix of the native protein (Fig. 3). In milk, β-Lg is most likely complexed with some small hydrophobic ligands, such as fatty acids or retinoids that could limit the access of proteases to the sensitive bonds. As assessed with I-TASSER simulations, β-Lg conformation should not considerably vary between free and fatty acid-complexed forms. The tertiary structure is also stable at the gastric acidic pH.38 Thus, even in the non-complexed protein the calix is buried inside the hydrophobic core of the protein and should be spared by proteolysis at least in part. To this purpose, the peptides of the region 40-60 of β-Lg are relatively resistant to the simulated gastric-duodenal-intestinal digestion of milk proteins, also including peptidases from Caco-2 cells39 and jejunal brush border membrane.35 The high local number of acidic residues also could explain the resistance of this protein domain to digestion.17
Figure 3.
3D structure of non-complexed β-Lg. The β-Lg 42-57 domain is highlighted in the unrolled model (left panel) and indicated in the X-ray crystal structure of monomer β-Lg (middle panel, PDB data bank). The molecular structure prediction (right panel) performed with I-TASSER confirms that β-Lg 42-57 belongs to the lipid binding calix of the protein.
Through breast milk, peptides from maternal diet can induce early and occult sensitization to food. Alternatively, the early exposure to low amounts of immunogenic epitopes could results in tolerogenic and protective response.40,41 The immunological mechanisms that might underlie these events are still unclear, further complicated by the variability in the frequency and concentration of foreign peptides/proteins in breast milk. Indeed, in our study β-Lg peptides were detected only in two milk samples, thus confirming that the kinetics of dietary components is affected by high individual variability,15 most likely depending on varying gastrointestinal digestion capacity or intestinal permeability as well as integrity of the mammary tissue. The secretion of β-Lg peptides into breast milk could also be related to long-term consumption of cow's milk, as higher levels of the protein have been reported in the milk of subjects that are used to consume milk in large amount.42
CONCLUSIONS
Immunologically active peptides derived from CMP in the maternal diet, rather than intact allergens, could be responsible of adverse responses in exclusively breast-fed newborns. These findings also indirectly prove that dietary protein fragments, albeit at low amounts, can survive gastro-intestinal digestion to be absorbed into the bloodstream and distributed to peripheral organs. Data herein are in line with the very recent detection of gluten-derived immunogenic peptides in the urine of celiac subjects.43
We did not accurately quantify the β-Lg peptides in breast milk samples. However, we can infer from the MS spectral count that they occur at a very low relative abundance. Whether trace amounts of these peptides are able to sensitize and elicit allergic responses or contribute to priming the immune system, promoting the acquisition of the oral tolerance, remains unknown. From this perspective, the time course of dietary peptide appearance in breast milk and the minimum amount of a dietary protein inducing a detectable presence of peptides in breast milk are important next steps for this research. In consideration of the very low amounts (if any) of potentially immunogenic protein fragments in breast milk, our data supports the current guidelines according to which maternal elimination diets during pregnancy and lactation are not recommended as a strategy to prevent childhood allergies of ‘at risk’ infants.7,44
ACKNOWLEDGEMENTS
G.P. acknowledges Regione Campania, Italy, which in part supported his research (L.R. 05/2002 grant). The authors thank the University of California Genome Center Proteomics Core (especially Brett Phinney and Michelle Salemi) for the mass spectrometry work, the Bioinformatics Core (especially Nikhil Joshi) for bioinformatics support. The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository45 with the dataset identifier PXD003657. D.C. Dallas acknowledges the K99/R00 Pathway to Independence Career Award, Eunice Kennedy Shriver Institute of Child Health & Development of the National Institutes of Health (K99HD079561) and D.B. acknowledges the National Institutes of Health (award R01 AT008759).
Footnotes
The authors have declared no conflict of interest.
REFERENCES
- 1.Järvinen KM, Mäkinen-Kiljunen S, Suomalainen H. Cow's milk challenge through human milk evokes immune responses in infants with cow's milk allergy. J. Pediatr. 1999;135:506. doi: 10.1016/s0022-3476(99)70175-7. [DOI] [PubMed] [Google Scholar]
- 2.Academy of Breastfeeding Medicine ABM Clinical Protocol #24: Allergic Proctocolitis in the Exclusively Breastfed Infant. Breastfeed Med. 2011;6:435. doi: 10.1089/bfm.2011.9977. [DOI] [PubMed] [Google Scholar]
- 3.Katz Y, Rajuan N, Goldberg MR, Eisenberg E, et al. Early exposure to cow's milk protein is protective against IgE-mediated cow's milk protein allergy. J. Allergy Clin. Immunol. 2010;126:77. doi: 10.1016/j.jaci.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Cant A, Marsden RA, Kilshaw PJ. Egg and cows' milk hypersensitivity in exclusively breast fed infants with eczema, and detection of egg protein in breast milk. Br. Med. J. (Clin. Res. Ed.) 1985;291:932. doi: 10.1136/bmj.291.6500.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Høst A, Husby S, Osterballe O. A prospective study of cow's milk allergy in exclusively breast-fed infants. Incidence, pathogenetic role of early inadvertent exposure to cow's milk formula, and characterization of bovine milk protein in human milk. Acta Paediatr. Scand. 1988;77:663. doi: 10.1111/j.1651-2227.1988.tb10727.x. [DOI] [PubMed] [Google Scholar]
- 6.Kilshaw PJ, Cant AJ. The passage of maternal dietary proteins into human breast milk. Int. Arch Allergy Appl. Immunol. 1984;75:8. doi: 10.1159/000233582. [DOI] [PubMed] [Google Scholar]
- 7.Denis M, Loras-Duclaux I, Lachaux A. Cow's milk protein allergy through human milk. Arch. Pediatr. 2012;19:305. doi: 10.1016/j.arcped.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Vadas P, Wai Y, Burks W, Perelman B. Detection of peanut allergens in breast milk of lactating women. JAMA. 2001;285:1746. doi: 10.1001/jama.285.13.1746. [DOI] [PubMed] [Google Scholar]
- 9.Bernard H, Ah-Leung S, Drumare MF, Feraudet-Tarisse C, et al. Peanut allergens are rapidly transferred in human breast milk and can prevent sensitization in mice. Allergy. 2014;69:888. doi: 10.1111/all.12411. [DOI] [PubMed] [Google Scholar]
- 10.Palmer DJ, Gold MS, Makrides M. Effect of cooked and raw egg consumption on ovalbumin content of human milk: a randomized, double-blind, cross-over trial. Clin. Exp. Allergy. 2005;35:173. doi: 10.1111/j.1365-2222.2005.02170.x. [DOI] [PubMed] [Google Scholar]
- 11.Chirdo FG, Rumbo M, Añón MC, Fossati CA. Presence of high levels of non-degraded gliadin in breast milk from healthy mothers. Scand. J. Gastroenterol. 1998;33:1186. doi: 10.1080/00365529850172557. [DOI] [PubMed] [Google Scholar]
- 12.Hopman E, Dekking L, Beelen F, Smoltsak N, et al. Presence of gluten proteins in breast milk: implications for the development of celiac disease. In: Hopman E, editor. Gluten intake and gluten-free diet in the Netherlands. Open access Doctoral thesis. Leiden University Medical Center; Netherland: 2008. pp. 17–29. Chapter 2. [Google Scholar]
- 13.Restani P, Gaiaschi A, Plebani A, Beretta A,B, et al. Evaluation of the presence of bovine proteins in human milk as a possible cause of allergic symptoms in breast-fed children. Ann. Allergy Asthma Immunol. 2000;84:353. doi: 10.1016/S1081-1206(10)62786-X. [DOI] [PubMed] [Google Scholar]
- 14.Koeberl M, Clarke D, Lopata AL. Next generation of food allergen quantification using mass spectrometric systems. J. Proteome Res. 2014;13:3499. doi: 10.1021/pr500247r. [DOI] [PubMed] [Google Scholar]
- 15.Sorva R, Mäkinen-Kiljunen S, Juntunen-Backman K. Beta-lactoglobulin secretion in human milk varies widely after cow's milk ingestion in mothers of infants with cow's milk allergy. J. Allergy Clin. Immunol. 1994;93:787. doi: 10.1016/0091-6749(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 16.Reddy IM, Kella NKD, Kinsella JE. Structural and conformational basis of the resistance of beta-lactoglobulin to peptic and chymotryptic digestion. J. Agric. Food Chem. 1988;36:737. [Google Scholar]
- 17.Picariello G, Ferranti P, Fierro O, Mamone G, et al. Peptides surviving the simulated gastrointestinal digestion of milk proteins: biological and toxicological implications. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010;878:295. doi: 10.1016/j.jchromb.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Bertino E, Prandi GM, Fabris C, Cavaletto M, et al. Human milk proteins may interfere in ELISA measurements of bovine beta-lactoglobulin in human milk. Acta Paediatr. 1996;85:543. doi: 10.1111/j.1651-2227.1996.tb14083.x. [DOI] [PubMed] [Google Scholar]
- 19.Bertino E, Coscia A, Costa S, Farinasso D, et al. Absence in human milk of bovine beta-lactoglobulin ingested by the mother. Unreliability of ELISA measurements. Acta Biomed. Ateneo Parmense. 1997;68:S15. [PubMed] [Google Scholar]
- 20.Coscia A, Orrù S, Di Nicola P, Giuliani F, et al. Detection of cow's milk proteins and minor components in human milk using proteomics techniques. J. Matern. Fetal Neonatal. Med. 2012;25:S54. doi: 10.3109/14767058.2012.715015. [DOI] [PubMed] [Google Scholar]
- 21.Orrù S, Di Nicola P, Giuliani F, Fabris C, et al. Detection of bovine alpha-S1-casein in term and preterm human colostrum with proteomic techniques. Int. J. Immunopathol. Pharmacol. 2013;26:435. doi: 10.1177/039463201302600216. [DOI] [PubMed] [Google Scholar]
- 22.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 23.Dallas DC, Guerrero A, Khaldi N, Castillo PA, et al. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J. Proteome Res. 2013;12:2295. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhardt TA, Lippolis JD. Bovine milk fat globule membrane proteome. J. Dairy Res. 2006;73:406. doi: 10.1017/S0022029906001889. [DOI] [PubMed] [Google Scholar]
- 25.Reinhardt TA, Lippolis JD. Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. J. Dairy Sci. 2008;91:2307. doi: 10.3168/jds.2007-0952. [DOI] [PubMed] [Google Scholar]
- 26.Wilson NL, Robinson LJ, Donnet A, Bovetto L, et al. Glycoproteomics of milk: differences in sugar epitopes on human and bovine milk fat globule membranes. J. Proteome Res. 2008;7:3687. doi: 10.1021/pr700793k. [DOI] [PubMed] [Google Scholar]
- 27.Mange A, Bellet V, Tuaillon E, Van de Perre P, Solassol J. Comprehensive proteomic analysis of the human milk proteome: contribution of protein fractionation. J. Chromatogr. B. 2008;876:252. doi: 10.1016/j.jchromb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Liao Y, Alvarado R, Phinney B, Lönnerdal B. Proteomic characterization of human milk fat globule membrane proteins during a 12 month lactation period. J. Proteome Res. 2011;10:3530. doi: 10.1021/pr200149t. [DOI] [PubMed] [Google Scholar]
- 29.Picariello G, Ferranti P, Mamone G, Klouckova I, et al. Gel-free shotgun proteomic analysis of human milk. J. Chromatogr. A. 2012;1227:219. doi: 10.1016/j.chroma.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Molinari CE, Casadio YS, Hartmann BT, Livk A, et al. Proteome mapping of human skim milk proteins in term and preterm milk. J. Proteome Res. 2012;11:1696. doi: 10.1021/pr2008797. [DOI] [PubMed] [Google Scholar]
- 31.Ferranti P, Traisci MV, Picariello G, Nasi A, et al. Casein proteolysis in human milk: tracing the pattern of casein breakdown and the formation of potential bioactive peptides. J. Dairy Res. 2004;71:74. doi: 10.1017/s0022029903006599. [DOI] [PubMed] [Google Scholar]
- 32.Guerrero A, Dallas DC, Contreras S, Chee S, et al. Mechanistic peptidomics: factors that dictate specificity in the formation of endogenous peptides in human milk. Mol. Cell. Proteomics. 2014;13:3343. doi: 10.1074/mcp.M113.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khaldi N, Vijayakumar V, Dallas DC, Guerrero A, et al. Predicting the important enzymes in human breast milk digestion. J. Agric. Food Chem. 2014;62:7225. doi: 10.1021/jf405601e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dallas DC, Smink CJ, Robinson RC, Tian T, et al. Endogenous human milk peptide release is greater after preterm birth than term birth. J. Nutr. 2015;145:425. doi: 10.3945/jn.114.203646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picariello G, Miralles B, Mamone G, Sánchez-Rivera L, et al. Role of intestinal brush border peptidases in the simulated digestion of milk proteins. Mol. Nutr. Food Res. 2015;59:948. doi: 10.1002/mnfr.201400856. [DOI] [PubMed] [Google Scholar]
- 36.Palmer DJ, Kelly VC, Smit AM, Kuy S, et al. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics. 2006;6:2208. doi: 10.1002/pmic.200500558. [DOI] [PubMed] [Google Scholar]
- 37.Wal JM. Structure and function of milk allergens. Allergy. 2001;56:S35. [PubMed] [Google Scholar]
- 38.Apenten RKO, Khokhar S, Galani D. Stability parameters for β-lactoglobulin thermal dissociation and unfolding in phosphate buffer at pH 7.0. Food Hydrocoll. 2002;16:95. [Google Scholar]
- 39.Picariello G, Iacomino G, Mamone G, Ferranti P, et al. Transport across Caco-2 monolayers of peptides arising from in vitro digestion of bovine milk proteins. Food Chem. 2013;139:203. doi: 10.1016/j.foodchem.2013.01.063. [DOI] [PubMed] [Google Scholar]
- 40.DuToit G, Katz Y, Sasieni P, Mesher D, et al. Early consumption of peanuts in infancy is associated with a low prevalence of peanut allergy. J. Allergy Clin. Immunol. 2008;122:984. doi: 10.1016/j.jaci.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Verhasselt V, Milcent V, Cazareth J, Kanda A, et al. Breast milk-mediated transfer of an antigen induces tolerance and protection from allergic asthma. Nat. Med. 2008;14:170. doi: 10.1038/nm1718. [DOI] [PubMed] [Google Scholar]
- 42.Fukushima Y, Kawata Y, Onda T, Kitagawa M. Consumption of cow milk and egg by lactating women and the presence of beta-lactoglobulin and ovalbumin in breast milk. Am. J. Clin. Nutr. 1997;65:30. doi: 10.1093/ajcn/65.1.30. [DOI] [PubMed] [Google Scholar]
- 43.Moreno ML, Cebolla Á, Muñoz-Suano A, Carrillo-Carrion C, et al. Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing. Gut. 2015 doi: 10.1136/gutjnl-2015-310148. In press: doi: 10.1136/gutjnl-2015-310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer DJ, Makrides M. Diet of lactating women and allergic reactions in their infants. Curr. Opin. Clin. Nutr. Metab. Care. 2006;9:284. doi: 10.1097/01.mco.0000222113.46042.50. [DOI] [PubMed] [Google Scholar]
- 45.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]