Abstract
Purpose
While targeted sequencing improves outcomes for many cancer patients, how somatic and germline whole-exome sequencing (WES) will integrate into care remains uncertain.
Methods
We conducted surveys and interviews, within a study of WES integration at an academic center, to determine oncologists' attitudes about WES and to identify lung and colorectal cancer patients' preferences for learning WES findings.
Results
167 patients (85% white, 58% female, mean age 60) and 27 oncologists (22% female) participated. Although oncologists had extensive experience ordering somatic tests (median 100/year), they had little experience ordering germline tests. Oncologists intended to disclose most WES results to patients but anticipated numerous challenges in using WES. Patients had moderately low levels of genetic knowledge (mean 4 correct of 7). Most patients chose to learn results that could help select a clinical trial, pharmacogenetic and positive prognostic results, and results suggesting inherited predisposition to cancer and treatable non-cancer conditions (all ≥95%). Fewer chose to receive negative prognostic results (84%) and results suggesting predisposition to untreatable non-cancer conditions (85%).
Conclusion
The majority of patients want most cancer-related and incidental WES results. Patients' low levels of genetic knowledge and oncologists' inexperience with large-scale sequencing presage challenges to implementing paired WES in practice.
Keywords: sequencing, cancer, incidental findings, return of results
Introduction
Exome and genome sequencing are disruptive technologies that may transform clinical practice. Physicians and patients will confront vast amounts of complex and uncertain data, including incidental findings unrelated to the testing indication. If recommendations regarding obligations to return incidental genomic results1,2 are adopted, patients will also face decisions about the types of genomic information that they would like to receive.
Given substantial uncertainty as to how to best deliver exome- and genome-guided medical care, it is imperative that we understand how whole-exome sequencing (WES) will alter clinical practice and anticipate the challenges that providers and patients will face. Oncology is an ideal setting in which to explore clinical sequencing because cancer is often driven by genomic changes. Targeted germline (normal tissue) and somatic (tumor) DNA sequencing, used separately, have dramatically improved outcomes in some high-risk 3-6 and cancer patient sub-populations,7-10 and larger gene panels are already used in practice.11,12 Sequencing's power increases when somatic and germline DNA are sequenced in parallel, as paired sequencing unequivocally distinguishes somatic from germline alterations13,14 and can uncover previously unsuspected inherited cancer risk.15
To inform the debate about how best to implement cancer-related WES, we initiated a prospective study to explore how introducing WES into care might affect cancer patients and oncologists. This manuscript reports findings from baseline surveys and interviews with patients and physicians. We hypothesized that patients would want to receive information about all potentially informative somatic and germline genomic alterations, and that oncologists would anticipate numerous clinical, psychosocial and ethical challenges as they prepare to evaluate and disclose WES results.
Participants and Methods
Study Setting, Dates, And Participants
The Dana-Farber Cancer Institute (DFCI) CanSeq study, launched in February 2013, is a single-arm prospective study of the integration of paired WES into clinical care. The eligible population for the CanSeq clinician study included all medical oncologists in the Thoracic and Gastrointestinal (GI) Oncology Centers who care for lung or colorectal adenocarcinoma patients. Two oncologists are study co-investigators and were excluded from baseline survey and interview participation; one oncologist declined to participate in the CanSeq study. Because CanSeq's goal is to understand both patients' and physicians' experiences with WES, only patients of participating oncologists were eligible.
The eligible population for the CanSeq patient study includes patients who 1) have stage IV lung or colorectal adenocarcinoma, 2) consent to companion genotyping protocols (to allow variant confirmation by a complementary technology), 3) have a life expectancy of at least 6 months, 4) have an Eastern Cooperative Oncology Group (ECOG) performance status of zero or one, 5) have sufficient tumor DNA for WES, 6) have a treating oncologist who is participating in the study, 7) speak English, 8) consent to participation and 9) receive ongoing care at DFCI. All study activities were approved by the DFCI Institutional Review Board, and both patient and physician participants gave written informed consent.
Study Procedures
Oncologist Survey And Interview Procedures
The oncologist survey was offered enrollment. Electronic reminders were sent out at one-week intervals until survey completion. After 3 contacts, a study investigator contacted non-responders to encourage participation. A sub-sample of oncologists, stratified by gender and academic rank, was invited to participate in individual interviews. Interviews were conducted in person or by telephone within approximately one month of physician enrollment. Interviews were audiorecorded and transcribed for analysis.
Patient Consent And Survey Procedures
After confirming patient eligibility with the treating physician, clinical research assistants (CRAs) approached patients during routine clinic visits to offer participation. At the time of consent, patients were asked to report their preferences for the receipt of cancer-related and non-cancer-related WES findings. Immediately following consent, patients were asked to complete the baseline survey on a computer tablet or on paper. Patients who did not complete the survey in clinic could complete it at home. Reminder letters/emails were sent to non-responders 2, 4 and 6 weeks after consent. The CRAs also approached patients during subsequent clinic appointments to facilitate survey completion. We paused reminders if medical record review or provider message identified an acute illness or hospitalization. Participants were considered non-responders if they had not completed the baseline survey within 2 months of enrollment or by the time of result reporting.
Measures And Domains
Physician Survey Measures And Interview Domains
The physician survey contained questions related to experience with somatic and germline testing in the prior year, attitudes about the return of sequencing results, confidence in the ability to perform relevant tasks (e.g., interpret data, explain concepts to patients, make treatment recommendations, provide psychosocial support, obtain informed consent), and socio-demographic and practice characteristics (Physician Baseline Survey Instrument, Supplementary Materials and Methods 1).
We created a measure to assess oncologists' attitudes about the return of genome results. The three-item measure asked how strongly oncologists agreed or disagreed with limiting return of results to those with clinical utility (evidence demonstrates that actions based on the results can change patient management decisions and improve net health outcomes), returning results with clinical validity (evidence of an established relationship between genotype and phenotype) but not utility, and returning all genomic sequencing results. The somatic genomic confidence measure was adapted from our prior work16 to include 2 additional items related to the oncologist's ability to identify consultants with expertise in integrating somatic genomic information into patient care and to provide psychosocial support related to coping with somatic information with adverse prognostic information. We adapted Nippert's germline confidence scale17 for the cancer context. We elicited intentions to disclose WES information using short vignettes describing an adult patient with a metastatic solid tumor. The hypothetical patient had undergone somatic and germline WES performed in a clinically certified lab, was on first-line chemotherapy, had an ECOG performance status of 0 or 1, had indicated that s/he would like to be told about all clinically valid results, and had biological children. We asked about intentions to disclose somatic predictive alterations (i.e., that could be targeted with a drug available through a phase II clinical trial or that is approved by the Food and Drug Administration (FDA) for a different cancer type) and somatic prognostic alterations (positive and negative). We also asked about intentions to disclose germline cancer risk alterations (with and without available risk-reduction strategies), pharmacogenetic polymorphisms (cancer and non-cancer related), alterations that conferred increased risk of developing a non-cancer condition (with and without available risk-reduction strategies), and carrier status. We conducted cognitive testing (structured survey review and feedback elicitation) of the draft survey instrument with 5 oncologists in disease centers other than GI and Thoracic, then revised and finalized the survey. The survey was administered on paper and took ≤10 minutes to complete.
The oncologist interviews were developed based on the researchers' prior experiences and the published literature (Physician Baseline Interview Guide, Supplementary Materials and Methods 2). We pilot tested the interview guide with 2 oncologists, then revised and finalized it. The interview took approximately 30 minutes and covered expectations related to WES; anticipated benefits, risks and challenges of using WES in clinical practice; and intentions to disclose results to patients. Data were collected until thematic saturation was achieved. Interviews were transcribed, reviewed for accuracy and completeness, and uploaded into NVivo 10 (QSR, Doncaster, Victoria, Australia).
Patient Measures
The patient consent form included 9 questions to elicit patients' preferences for the disclosure of somatic WES results (results that could be used to identify possible clinical trials; positive/negative prognostic results) and germline WES results (cancer risk; cancer and non-cancer pharmacogenetic; risk of developing treatable and non-treatable conditions other than cancer; and carrier status) (Patient Consent Preferences, Supplementary Materials and Methods 3). The patient baseline survey instrument (Supplementary Materials and Methods 4) included validated measures to assess patients' attitudes about getting a genetic test,18 experience with genetic testing,19 genetic knowledge,20,21 subjective numeracy,22 health literacy,23 self-reported ECOG performance status,24,25 quality of life,26 decision-making preferences,27 and socio-demographic characteristics. We cognitively tested the draft patient consent questions and survey instrument with 5 patients with advanced lung and colorectal cancer, then revised and finalized the instruments.
Analyses
Surveys
The aims of the oncologist survey were to describe oncologists' attitudes about WES disclosure, intentions to disclose WES results, somatic and germline genomic confidence, as well as the frequency of baseline genomic testing. The aims of the patient consent items and baseline survey were to describe patients' preferences for the return of WES results and their genetic knowledge. In exploratory analyses, we evaluated the associations between patient characteristics and 1) attitudes about getting a genetic test and 2) preferences to receive somatic and germline sequencing results. Attitudes about getting a genetic test were evaluated as a dichotomous outcome (<2/5 very positive vs. ≥2/5). Subjects who indicated a preference for return of all 3 types of somatic results and all 6 types of germline results were counted as having high preference for somatic results and germline results respectively (high preference vs. other). We explored the associations between attitudes/preferences and age, gender, cancer type, education, genetic knowledge, and attitudes about getting a genetic test (preferences only) in univariate analyses. Independent variables with univariate p-values less than 0.20 were included in a multivariable model. Relative risks estimates and 95% confidence intervals (CI) were determined using a modified Poisson regression with robust variance estimates.28
Interviews
Two team members independently coded qualitative data using NVivo 10 to develop a coding framework, guided by the interview guide domains. An iterative process of structured coding ensued, with discrepancies resolved through discussion and comparison to the raw data; a final kappa of 0.88 was achieved.
Results
Participant Characteristics
All 27 participating oncologists completed the baseline physician survey. One hundred and sixty-seven patients indicated preferences at the time of consent, and 153 patients completed the patient baseline survey (response rates 100% and 92% respectively, Figure S1). Patient and oncologist characteristics are shown in Table 1.
Table 1. Physician and Patient Characteristics.
| Physician Characteristics | Frequency (%) (n=27) |
|
|---|---|---|
| Program | ||
| GI | 52 | |
| Thoracic | 48 | |
| Principal Investigator | ||
| Clinical trials | 59 | |
| Translational research | 44 | |
| Basic science | 15 | |
| Outcomes/health services or cancer epidemiology | 22 | |
| Gender | ||
| Male | 78 | |
| Female | 22 | |
|
| ||
| Median | IQR | |
|
| ||
| Year completed fellowship | 2007 | 1999-2011 |
| # Unique patients seen per month | 50 | 28-70 |
| Percent of professional time spent in: | ||
| Patient care | 40 | 30-75 |
| Research | 40 | 19-60 |
| Teaching | 5 | 5-10 |
| Administration | 5 | 0-15 |
|
| ||
| Patient Characteristics |
Frequency (%) (n=167)* |
|
|
| ||
| Age at consent, mean (SD) | 59.8 (12.0) | |
| Gender | ||
| Female | 97 (58) | |
| Cancer | ||
| Lung | 89 (53) | |
| Colorectal | 78 (47) | |
| Race** | ||
| White | 130 (85) | |
| Non-white | 19 (13) | |
| Hispanic/Latino** | 3 (2) | |
| Education** | ||
| ≥ College graduate | 68 (44) | |
| Overall health, mean (SD) † ** | 5.2 (1.1) | |
| Prior genetic testing ‡ ** | ||
| Yes | 20 (13) | |
| No | 122 (80) | |
| Don't Know | 11 (7) | |
| Attitude toward genetic testing, mean (SD) §** | 1.3 (0.7) | |
Percentages may not add to 100% due to missing responses and/or rounding.
n=153 who completed baseline survey
Seven point scale from very poor (1) to excellent (7)1
Self reported
Attitude score range 1-5, lower numbers correspond to more positive attitudes2
Aaronson, N. K., Ahmedzai, S., Bergman, B., Bullinger, M., Cull, A., Duez, N. J., et al. (1993). The European Organization for Research and Treatment of Cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology. Journal of the National Cancer Institute, 85(5), 365–376.
Michie, S., di Lorenzo, E., Lane, R., Armstrong, K., & Sanderson, S. (2004). Genetic information leaflets: Influencing attitudes towards genetic testing. Genetics in Medicine, 6(4), 219–225.
Oncologists
Oncologists spent a majority of time in patient care and research. They reported ordering or interpreting a median of 100 somatic tests per year (interquartile range (IQR) 40-100) and 2 germline cancer predisposition tests per year (IQR 0-10, Table S1). Few oncologists had ordered or interpreted germline tests unrelated to cancer (n=4), cancer-related pharmacogenetic tests (n=9), non-cancer pharmacogenetic (n=2) or tests to identify carrier status (n=2).
Patients
Thirteen percent of patients reported having had genetic testing. Patients had positive attitudes about having a genetic test (mean 1.3 on a 1-5 point scale, with 1=most positive). No patient characteristics were associated with positive attitudes about getting a genetic test.
Physicians' Genomic Confidence, Disclosure Philosophy, And Intentions To Disclose WES Results
Most oncologists were very or moderately confident in their ability to carry out many somatic and germline genomic tasks (Table S2). Oncologists were less confident in their abilities to provide psychosocial support related to negative prognostic results and to perform activities related to cancer risk testing (i.e., provide pre-test counseling, obtain informed consent, and provide psychosocial support). Oncologists' attitudes about the return of genomic test results varied (Figure 1). Seventy-eight percent supported disclosure if WES results have established clinical validity; 67% did not support limiting return of results with to those with established clinical utility. Fifty-two percent agreed that patients should be offered as many sequencing results as they want, including raw sequencing data. When asked about disclosure intentions, most oncologists said that they would disclose somatic and germline WES information (Table 2). Some oncologists reported a reluctance to disclose somatic results that were in a pathway targeted by an agent that is FDA-approved for another tumor type, negative prognostic results, pharmacogenetic results, results related to non-treatable conditions, and carrier status.
Figure 1.
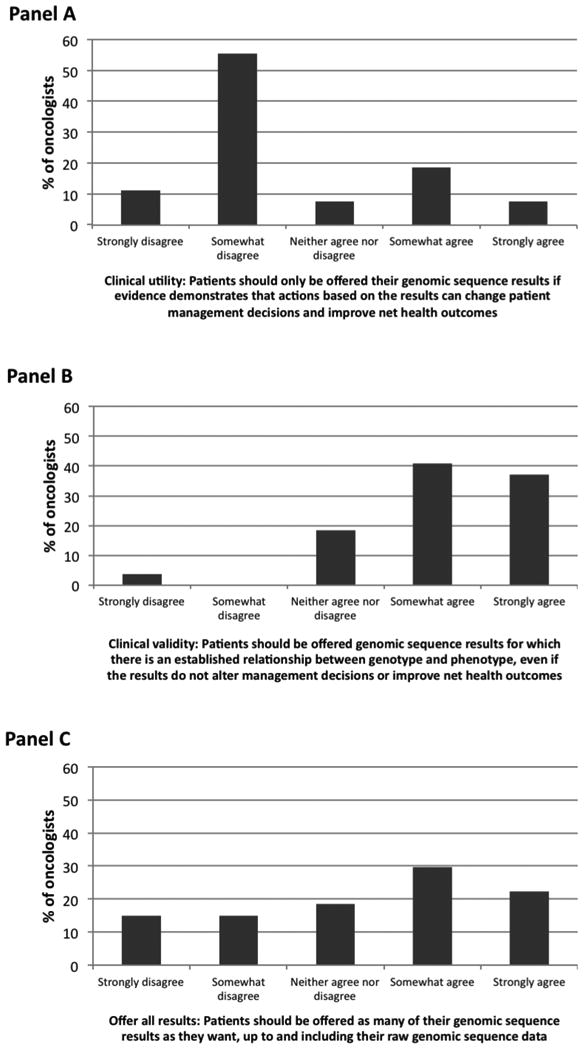
Oncologists' attitudes regarding return of genomic test results (n=27, %). Return based on clinical utility (Panel A), clinical validity (Panel B), and all results (Panel C).
Table 2.
Oncologists' intentions to disclose somatic and germline WES findings to patient (n=27)
| Definitely disclose % | Probably disclose % | Probably not disclose % | Definitely not disclose % | Unsure % | |
|---|---|---|---|---|---|
|
|
|||||
| Sequencing of tumor DNA identifies a somatic alteration that is… | |||||
| Predictive: targeted in a phase II clinical trial | 81 | 19 | -- | -- | -- |
| Predictive: targeted by an agent that is FDA-approved for a different cancer | 59 | 30 | 7 | -- | 4 |
| Prognostic: favorable | 56 | 44 | -- | -- | -- |
| Prognostic: unfavorable | 30 | 52 | 19 | -- | 4 |
| Sequencing of germline DNA identifies a(n)… | |||||
| Cancer risk alteration: treatable | 81 | 19 | -- | -- | -- |
| Cancer risk alteration: not treatable | 30 | 44 | 15 | -- | 11 |
| Pharmacogenetic polymorphism: cancer-related | 74 | 15 | 11 | -- | -- |
| Pharmacogenetic polymorphism: non-cancer-related | 56 | 0 | 15 | -- | -- |
| Non-cancer alteration: treatable | 78 | 22 | -- | -- | -- |
| Non-cancer alteration: not treatable | 11 | 67 | 19 | -- | 4 |
| Alteration associated with carrier status | 48 | 44 | 4 | -- | 4 |
Predictive: The alteration/pathway may be targeted by a therapeutic agent
Prognostic: The alteration confers a favorable or unfavorable prognosis
Physicians' Anticipated Challenges In Integrating WES Into Practice And Disclosure Intention Themes
Qualitative interviews generally reinforced the survey findings (Results S1). Oncologists anticipated various challenges in using WES (Table 3). Physicians reported that they generally intended to disclose somatic and germline findings (Interview Themes: Table S3). All physicians reported an intention to disclose predictive somatic findings; many also intended to disclose prognostic information. Some oncologists expressed reluctance to disclose germline information related to the risk of developing cancer and non-cancer conditions and non-cancer pharmacogenetic alterations. Several oncologists mentioned that such disclosures should involve another physician or a genetic counselor. Determinants of oncologists' intentions to disclose WES findings included patients' preferences and performance status, the physician's knowledge and the “actionability” of findings (e.g., the availability of relevant targeted therapies for somatic alterations or relevant risk reduction interventions for germline alterations). Some oncologists believed that patients have a right to know all information learned.
Table 3. Oncologists' anticipated challenges when using WES in clinical practice (n=19).
| Themes | Examples |
|---|---|
| Ability to distilling data and identify actionable findings |
|
| Large data volume and few actionable results |
|
| Determining what to disclose to patients |
|
| Managing patient expectations |
|
| Need for physician education |
|
| Managing and disclosing uncertain or incidental findings |
|
| Managing patient and family emotional response to WES disclosure |
|
| Disclosing non-cancer information |
|
Patients' Genetic Knowledge And Preference For Learning WES Results
Patients had moderately low genetic knowledge with a mean score of 4 correct out of 7 (Figure S2). A sizable minority did not know that genetic testing can be used to evaluate cancer risk, that fathers can pass on genetic conditions, and that people who have mutations do not always develop disease.
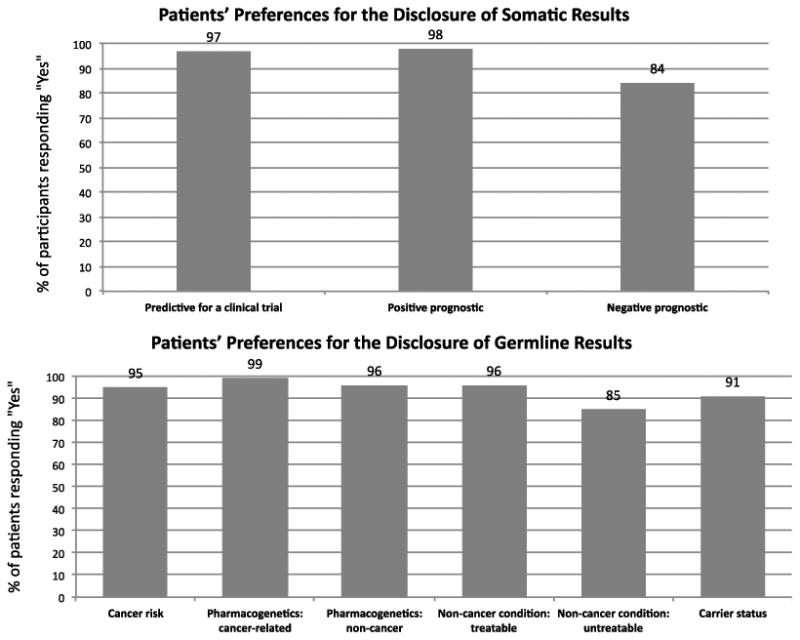
Almost all patients chose to learn most cancer-related, pharmacogenetic, and carrier status findings (Figure 2). Slightly fewer patients opted to receive negative prognostic results (84%) and information about the risk of developing an untreatable non-cancer condition (85%). After adjusting for gender, patients with less positive attitudes about getting a genetic test were less likely than those with very positive attitudes to indicate a high preference for the return of somatic (65% vs. 86%, adjusted RR 0.89, 95% CI 0.80-0.98, p=0.02) and germline (65% vs. 86%, adjusted RR 0.89, 95% CI 0.79-1.0, p=0.05) results. Additionally, men were more likely to indicate a high preference for the return of germline sequencing results than women (92% vs. 76%, adjusted RR 1.06, 95% CI 1.00-1.12, p=0.04).
Figure 2. Patients' preferences for the return of somatic and germline WES results.
Discussion
We examined the implementation of paired somatic and germline WES at a comprehensive cancer center. We found that although most oncologists have ample experience using and interpreting somatic genomic tests, they have little experience with germline testing. Nevertheless, respondents intended to disclose most WES results from both somatic and germline testing to patients. Oncologists also expressed concerns about data interpretation, disclosing non-cancer findings, and determining the “actionability” of alterations. We also found that patients with advanced lung and colorectal cancer have favorable attitudes towards having a genetic test but moderately low levels of genetic knowledge, and that most want to learn all WES results. Our findings advance the field by demonstrating that although physicians anticipate many challenges to delivering care involving large-scale sequencing, patients with incurable cancer express a strong desire to learn about genomic findings whether or not they have relevance to their immediate medical care.
When queried about somatic genomic testing, most oncologists were moderately or very confident in their ability to interpret somatic test results in their disease area, explain somatic genomic concepts to patients, make treatment recommendations based on somatic genomic information, and identify appropriate consultants. The high levels of confidence may relate to the fact that the oncologists in our study order and interpret large numbers of somatic tests and that lung and colorectal adenocarcinoma are malignancies in which genomic testing is part of guideline-based cancer care.29,30 Nevertheless, oncologists anticipated a number of challenges to delivering somatic WES care, including dealing with and interpreting large volumes of data and determining the actionability of somatic findings.31 Some oncologists expressed concerns about how to determine how much somatic data should be shared with patients and how to manage patients' expectations. In addition, oncologists expressed concerns about their ability to keep up with the literature in this rapidly evolving field.
Most oncologists planned to disclose all types of somatic WES findings to their patients with metastatic disease, assuming that patients desired WES information, that they had good performance status, and that a clinically certified lab performed the sequencing. Oncologists' responses during qualitative interviews help explain these findings. All interviewees planned to disclose somatic findings if there was an approved targeted therapy for another cancer type or if a clinical trial was available. Liberal attitudes to somatic disclosure were tied to oncologists' desire to explore different treatment options and to be able to offer enrollment in clinical trials. Several oncologists also stated that patients have a “right to know” somatic information.32,33
Ninety-two percent of our GI oncologists had ordered or interpreted germline tests in the prior year as compared to 38% of thoracic oncologists. Because genetic testing for familial colorectal cancer syndromes is integrated into standard practice, whereas standards for testing for hereditary lung cancer syndromes are just emerging, GI oncologists may be more accustomed to ordering germline tests than their thoracic counterparts. In addition, 35% of all oncologists had ordered or interpreted cancer-related pharmacogenetic tests in the prior year. This use of pharmacogenetic testing is notable because the reported uptake of pharmacogenetic testing generally is estimated to be low34-36 despite the fact that pharmacogenetic tests (e.g., DPYD, UGT1A1) are available, polymorphisms in these genes are associated with drug metabolism, and the FDA includes pharmacogenetic information in the label of drugs including 5-fluorouracil and irinotecan.37 Although oncologists have some experience with ordering and interpreting cancer-related germline genomic tests, they have less experience with tests that are unrelated to cancer. 38
Whereas most oncologists in our study intended to disclose most germline WES findings to patients, some expressed reluctance to disclose information about untreatable conditions, pharmacogenetic alterations, and information that would require further counseling and psychosocial support. Some oncologists noted that disclosure of a negative prognosis might facilitate patients' prognostic awareness, however, others reported a hesitance to share this information, particularly if the patient was at the end of life. Based on our interview data, oncologists' reluctance to disclose information seems less related to a desire to ‘protect’ patients from bad news than to a desire to give them information that is relevant to their situation and that accords with their preferences. Many oncologists noted that the “actionabiblity” of the information would be a key determinant of disclosure, and that they would be less willing to disclose information if it did not have implications for cancer therapy or prevention. For germline disclosure decisions, a few oncologists said that they may not disclose the information if the patient did not have or intend to have children. One might hypothesize that in the setting of advanced cancer, providers' and patients' risk-benefit calculus as they weigh the value of germline data and disclosure may differ substantially from that in other settings where the clinical utility of germline information for patients may be more clear. Additionally, several oncologists noted a desire to involve a genetic counselor or another provider when disclosing germline results. Given the complexity of the results and the familial implications of germline findings, institutions that offer WES or WGS may need to make relevant clinical and counseling expertise available to oncologists, patients, and patients' family members.
One of the most pressing questions in oncology is how to best support cancer providers as they integrate large amounts of genomic data into routine cancer care. Efforts to support providers must start with efforts to improve the quality of the data in reference genomic databases and with efforts to optimize bioinformatics algorithms and resources for variant calling and interpretation. In addition, sophisticated approaches to physician education and decision support will be essential. Given the rate at which information about genomic variants changes, dynamic genomic reports and point-of-care physician support will help providers to understand the potential implications of somatic and germline variants and to better personalize recommendations. In addition to systems interventions, physicians might also be supported through programs developed by their local institutions. For example, several institutions, including the DFCI, have developed multidisciplinary “Genomic Tumor Boards” where providers can discuss patients in a case-based format, highlighting their genomic or proteomic data, in order to get feedback or input from colleagues with expertise in medical oncology, molecular biology, clinical trials, pathology, and medical ethics. 14,39
The strong preference of most patients for return of all categories of somatic and germline genomic results, together with the relatively low levels of genetic knowledge, suggests that patients will also need assistance in understanding and making informed decisions based on genomic tests. Educational resources and decision aids will be needed at the time of initial consent and sample acquisition, so that patients can provide informed preferences about the types of results they desire. It will be important for patients undergoing cancer-related WES or WGS to understand basic genetic concepts such as the difference between somatic and germline testing, that men can pass germline genetic mutations onto their children, and that not all germline mutation carriers will develop disease. Computer-based education interventions have been shown to improve knowledge in the setting of germline cancer genetic counseling and may prove effective in this setting. 40 In addition, given that providers do not routinely elicit patients' preferences for the return of specific results when they order laboratory tests, more work needs to be done to determine how to best identify and respect patients' preferences for sequencing findings. One possible solution is to capture patients' preferences for sequencing results on the test requisition, therefore allowing the laboratory to customize result reporting. Finally, resources will be needed at the time of return of results to minimize misunderstanding and ensure that the actions patients and their family members take are based on evidence and consistent with their values. Given the speed with which genomic testing is entering oncology practice, the development of such resources for patients and families is an urgent priority. 2,41
The present study has several limitations. First, we conducted our study at a single academic center and restricted enrollment to adult patients with advanced lung and colorectal cancer and to their oncologists, suggesting the need for caution in generalizing our findings to other settings and cancer populations. Given that DFCI has an enterprise-wide, multiplex gene sequencing study underway, the oncologists in our study may use genomic testing more frequently than most oncologists. Second, we assessed oncologists' intentions to disclose WES results, which may or may not correspond to their actual behavior. Third, patients had high levels of education and included few members of racial/ethnic minority groups. Fourth, given the novelty of sequencing in the cancer context, several measures used with both patients and physicians were developed specifically for this study and had not previously been validated. Finally, alternative approaches to preference elicitation or pre-test education and counseling might have led patients to make different choices about return of results.
In sum, patients with advanced solid tumors express a strong desire for the return of genomic results, including incidental findings. However, these preferences may not be based upon a robust understanding of genetics or of the implications of the findings for patients' or their family members' health and medical care. Furthermore, oncologists who work with these patients express concern about their ability to evaluate, communicate and make decisions about the broad range of somatic findings that WES will produce, as well as about their ability to address germline findings that may result from parallel sequencing. Resources to assist physicians and patients in addressing these concerns represent a pressing priority for the cancer community.
Supplementary Material
Acknowledgments
We would like to acknowledge all members of the CanSeq team for their hard work and dedication. We also thank all of the oncologists and patients who have participated in the study.
Drs. Gray and Joffe had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Research Support: National Human Genome Research Institute U01HG006492 and U01 HG007303. Dr. Gray is also supported by the American Cancer Society (120529-MRSG-11-006-01-CPPB).
Footnotes
National Institutes of Health Clinical Trials.Gov Identifier: NCT02127359
Presented in part at the: American Society of Clinical Oncology Annual Meeting 2014 (Abstract 1535) and at the American Society of Human Genetics Annual Meeting 2014 (Session 11)
References
- 1.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol. 2015 Aug; doi: 10.1200/JCO.2015.63.0996. JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 3.Kauff ND, Satagopan JM, Robson ME, et al. Risk-reducing salpingo-oophorectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med. 2002;346(21):1609–1615. doi: 10.1056/NEJMoa020119. [DOI] [PubMed] [Google Scholar]
- 4.Domchek SM. Association of Risk-Reducing Surgery in BRCA1 or BRCA2 Mutation Carriers With Cancer Risk and Mortality. JAMA. 2010;304(9):967. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machens A, Niccoli-Sire P, Hoegel J, et al. Early malignant progression of hereditary medullary thyroid cancer. N Engl J Med. 2003;349(16):1517–1525. doi: 10.1056/NEJMoa012915. [DOI] [PubMed] [Google Scholar]
- 6.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118(5):829–834. doi: 10.1016/S0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 7.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.MacConaill LE, Garcia E, Shivdasani P, et al. Prospective enterprise-level molecular genotyping of a cohort of cancer patients. J Mol Diagn. 2014;16(6):660–672. doi: 10.1016/j.jmoldx.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol. 2013;31(15):1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Science Translational Medicine. 2011;3(111):111ra121–111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014;32(7):687–698. doi: 10.1200/JCO.2013.49.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray SW, Hicks-Courant K, Cronin A, Rollins BJ, Weeks JC. Physicians' attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32(13):1317–1323. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nippert I, Harris HJ, Julian-Reynier C, et al. Confidence of primary care physicians in their ability to carry out basic medical genetic tasks—a European survey in five countries—Part 1. J Community Genet. 2010;2(1):1–11. doi: 10.1007/s12687-010-0030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michie S, di Lorenzo E, Lane R, Armstrong K, Sanderson S. Genetic information leaflets: Influencing attitudes towards genetic testing. Genet Med. 2004;6(4):219–225. doi: 10.1097/01.GIM.0000132685.60259.EA. [DOI] [PubMed] [Google Scholar]
- 19.Sanderson SC. The effects of a genetic information leaflet on public attitudes towards genetic testing. Public Understanding of Science. 2005;14(2):213–224. doi: 10.1177/0963662505050993. [DOI] [PubMed] [Google Scholar]
- 20.Singer E, Antonucci T, Van Hoewyk J. Racial and ethnic variations in knowledge and attitudes about genetic testing. Genet Test. 2004;8(1):31–43. doi: 10.1089/109065704323016012. [DOI] [PubMed] [Google Scholar]
- 21.Furr LA, Kelly SE. The Genetic Knowledge Index: developing a standard measure of genetic knowledge. Genet Test. 1999;3(2):193–199. doi: 10.1089/gte.1999.3.193. [DOI] [PubMed] [Google Scholar]
- 22.Fagerlin A, Zikmund-Fisher BJ, Ubel PA, Jankovic A, Derry HA, Smith DM. Measuring numeracy without a math test: development of the Subjective Numeracy Scale. Med Decis Making. 2007;27(5):672–680. doi: 10.1177/0272989X07304449. [DOI] [PubMed] [Google Scholar]
- 23.Chew LD, Griffin JM, Partin MR, et al. Validation of screening questions for limited health literacy in a large VA outpatient population. J Gen Intern Med. 2008;23(5):561–566. doi: 10.1007/s11606-008-0520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5(6):649–655. [PubMed] [Google Scholar]
- 25.Basch E, Artz D, Dulko D, et al. Patient online self-reporting of toxicity symptoms during chemotherapy. J Clin Oncol. 2005;23(15):3552–3561. doi: 10.1200/JCO.2005.04.275. [DOI] [PubMed] [Google Scholar]
- 26.Aaronson NK, Ahmedzai S, Bergman B, et al. The European Organization for Research and Treatment of Cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology. J Natl Cancer Inst. 1993;85(5):365–376. doi: 10.1093/jnci/85.5.365. [DOI] [PubMed] [Google Scholar]
- 27.Apex CoVantage LLC. Degner_info needs decisional preferences women breast cancer_JAMA_1997. 2008 Jul;:1–8. [Google Scholar]
- 28.Zou G. A modified poisson regression approach to prospective studies with binary data. American Journal of Epidemiology. 2004;159(7):702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 29.NCCN. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology (NCCN Guidelines®)Non-Small Cell LungCancer. [Accessed February 1, 2014]; Version 3.2014.nccn.org. http://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Published January 24, 2014.
- 30.National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Colon Cancer Version 2.2015. J Natl Compr Canc Netw. 2014 Oct;:1–143. doi: 10.6004/jnccn.2018.0078. http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. [DOI] [PMC free article] [PubMed]
- 31.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramoni RB, McGuire AL, Robinson JO, Morley DS, Plon SE, Joffe S. Experiences and attitudes of genome investigators regarding return of individual genetic test results. Genet Med. 2013;15(11):882–887. doi: 10.1038/gim.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehan M. The right to know and genetic testing. Journal of Medical Ethics. 2015;41(4):287–288. doi: 10.1136/medethics-2015-102767. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JA. Pharmacogenetics in clinical practice: how far have we come and where are we going? Pharmacogenomics. 2013;14(7):835–843. doi: 10.2217/pgs.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13(12):987–995. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing. Clin Pharmacol Ther. 2011;89(3):387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Center for Drug Evaluation, Research. Genomics - Table of Pharmacogenomic Biomarkers in Drug Labeling. [Accessed January 23, 2015]; http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- 38.Parsons DW, Roy A, Plon SE, Roychowdhury S, Chinnaiyan AM. Clinical tumor sequencing: an incidental casualty of the American College of Medical Genetics and Genomics recommendations for reporting of incidental findings. J Clin Oncol. 2014;32(21):2203–2205. doi: 10.1200/JCO.2013.54.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker BA, Schwaederle M, Scur MD, et al. J Oncol Pract. University of California, San Diego; Moores Cancer Center: Aug, 2015. Breast Cancer Experience of the Molecular Tumor Board at the. [DOI] [PubMed] [Google Scholar]
- 40.Green MJ, Peterson SK, Baker MW, et al. Effect of a computer-based decision aid on knowledge, perceptions, and intentions about genetic testing for breast cancer susceptibility. JAMA. 2004;292(4):442–452. doi: 10.1001/jama.292.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robson ME, Storm CD, Weitzel J, Wollins DS, Offit K. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol. 2010;28(5):893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


