Summary
Ligation of the CD1d antigen‐presenting molecule by monoclonal antibodies (mAbs) can trigger important biological functions. For therapeutic purposes camelid‐derived variable domain of heavy‐chain‐only antibodies (VHH) have multiple advantages over mAbs because they are small, stable and have low immunogenicity. Here, we generated 21 human CD1d‐specific VHH by immunizing Lama glama and subsequent phage display. Two clones induced maturation of dendritic cells, one clone induced early apoptosis in CD1d‐expressing B lymphoblasts and multiple myeloma cells, and another clone blocked recognition of glycolipid‐loaded CD1d by CD1d‐restricted invariant natural killer T (iNKT) cells. In contrast to reported CD1d‐specific mAbs, these CD1d‐specific VHH have the unique characteristic that they induce specific and well‐defined biological effects. This feature, combined with the above‐indicated general advantages of VHH, make the CD1d‐specific VHH generated here unique and useful tools to exploit both CD1d ligation as well as disruption of CD1d–iNKT interactions in the treatment of cancer or inflammatory disorders.
Keywords: cancer, CD1d, dendritic cell, invariant natural killer T‐cell, variable domain of heavy‐chain‐only antibody
Abbreviations
- APC
allophycocyanin
- ATRA
all‐trans retinoic acid
- CFU
colony‐forming unit
- DC
dendritic cells
- FITC
fluorescein isothiocyanate
- IFN‐γ
interferon‐γ
- IL‐2
interleukin‐2
- iNKT
invariant natural killer T
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- MM
multiple myeloma
- moDC
monocyte derived DC
- PE
phycoerythrin
- PI
propidium iodide
- TCR
T‐cell receptor
- TNF‐α
tumour necrosis factor‐α
- VHH
variable domain of heavy‐chain‐only antibodies
- WT
wild‐type
- α‐GalCer
α‐galactosylceramide
Introduction
CD1d is a non‐polymorphic MHC‐like molecule that presents endogenous and exogenous glycolipid antigens to CD1d‐restricted T cells of which type 1 natural killer T cells, also known as invariant NKT (iNKT) cells, comprise the most extensively studied subset.1, 2 The iNKT cells are a specialized T‐cell subset characterized by the expression of a (semi‐)invariant T‐cell receptor (TCR) (Vα24‐Jα18 paired with Vβ11 in humans)3, 4 that rapidly produces large amounts of cytokines upon stimulation [e.g. interleukin‐2 (IL‐2), IL‐4, IL‐10, tumour necrosis factor‐α (TNF‐α) and interferon‐γ (IFN‐γ)].5, 6 Cognate interaction between iNKT cells and CD1d‐expressing dendritic cells (DC) has proven important for reciprocal activation. Ligation of glycolipid‐loaded CD1d molecules by iNKT cells, iNKT‐derived IFN‐γ and CD40–CD40 ligand interactions amplify DC IL‐12 production and enhance co‐stimulatory receptor expression by DC, thereby in turn boosting iNKT cytokine production and promoting T‐cell activation and NK cell transactivation.1, 7, 8 Moreover, bidirectional iNKT‐cell–DC interactions licence DC to cross‐present extracellular antigens to cytotoxic T cells, promoting the development of an adaptive immune response.9 Similarly, iNKT cells can provide cognate (via CD1d) and non‐cognate (via DC) help to B cells and induce and/or enhance humoral immune responses to various antigens.1, 10 As CD1d is also expressed on certain epithelial cells, biologically relevant interactions between iNKT and epithelial cells have been proposed.11, 12 Hence iNKT cells have been recognized for their ability to orchestrate microbial immunity as well as auto‐ and antitumour immunity.1, 10, 13
Mouse studies have provided important evidence regarding the role of iNKT cells in antitumour immunity. Models in iNKT‐deficient mice indicated a central role in tumour immunosurveillance, and activation of iNKT cells by the strong agonistic glycolipid‐ligand α‐galactosylceramide (α‐GalCer) induced potent rejection of established tumours.14, 15 Human observational studies underscore these findings, since circulating and tumour‐infiltrating iNKT cell numbers correlate with patient survival in various malignancies.16, 17 Importantly, though iNKT tend to display quantitative and qualitative defects in patients with (advanced) cancer, hampering their antitumour effect, these defects were found to be reversible.1, 6, 16 Indeed, infusion of α‐GalCer‐loaded monocyte‐derived DC (moDC) with or without adoptive transfer of ex vivo expanded iNKT has resulted in objective tumour regressions in several studies.18, 19 The iNKT‐mediated antitumour immunity is mediated either directly through presentation of self‐lipids by CD1d‐expressing tumours [e.g. multiple myeloma (MM), B‐ and T‐acute lymphoblastic leukaemia and colorectal cancer]8, 10, 20 or indirectly through iNKT–DC interactions and subsequent antitumour T‐cell activation.8, 13 Remarkably, it was demonstrated that cognate help of iNKT cells to DC can, at least in part, be mimicked by direct ligation of CD1d by CD1d‐specific monoclonal antibodies (mAbs).21 Indeed, mAb‐mediated ligation of CD1d expressed by moDC induced downstream signalling, resulting in moDC maturation and IL‐12 production, an effect that could be significantly enhanced through co‐stimulation via CD40 and Toll‐like receptors,21 indicating a potential method to bypass observed iNKT deficiencies. Interestingly, mAb ligation of CD1d expressed by tumours resulted in the induction of apoptosis in several malignancies, including B‐lymphoblastic and MM cell lines as well as in MM patient samples.22
As indicated above, iNKT cells have also been shown to be able to modulate the outcome of various autoimmune diseases. Importantly, and depending on the specific autoimmune disease that is studied, the role of iNKT cells can be either beneficial or detrimental to the host.6 In line with these observations, both activation and prevention of iNKT activation have been reported to be able to positively affect disease outcome. Indeed, in a cynomolgus macaque asthma model, blocking of CD1d resulted in significantly reduced cytokine levels and lymphocyte infiltration,23 indicating its therapeutic potential.
Many of the available anti‐CD1d mAb clones have been reported as functional in the three processes mentioned above. However, their relatively large size (~ 150 000 MW) and possible immunogenicity may limit clinical implementation in its current form. Camelid‐derived single domain antibodies (also termed variable domain of heavy‐chain‐only antibodies (VHH) or Nanobodies) have multiple advantages over conventional antibodies, as VHH are small (~ 15 000 MW) allowing deep tissue penetration, very stable, can be easily produced and re‐formatted in multi‐specific or multi‐valent molecules and are of low immunogenicity.24, 25, 26 Moreover, their single domain character allows binding to cryptic and not otherwise easily accessible epitopes in addition to the diversified and specific antigen‐binding repertoire found in conventional antibodies. Here, we describe the generation and characterization of anti‐human CD1d VHH. Twenty‐one unique CD1d‐specific VHH clones were selected, of which two clones induced efficient moDC maturation and IL‐12 production, a different clone induced signs of early apoptosis in CD1d‐transfected B‐lymphoblast and MM cells, and again one other CD1d‐specific VHH was able to inhibit CD1d‐α‐GalCer mediated iNKT cell activation. Collectively, the generated CD1d‐specific VHH have great potential for further therapeutic development in a wide variety of disorders.
Materials and methods
Cell lines
The human Epstein–Barr virus‐transformed B‐lymphoblast cell line C1R, wild‐type (WT) or stably transduced with CD1a, CD1b or CD1d4 and the human chronic myelogenous leukaemia cell line K562, transduced with either CD1c or CD1d (kind gift from Dr I. van Rhijn, Utrecht University, Utrecht, the Netherlands) were grown in Iscove's modified Dulbecco's medium (catalogue no. 12‐722F; Lonza, Basel, Switzerland) supplemented with 10% (v/v) fetal calf serum (catalogue no. SV30160.03; HyClone GE Healthcare, Chalfont, St Giles, UK), 0·05 mm β‐mercaptoethanol, 100 IU/ml sodium penicillin, 100 μg/ml streptomycin sulphate and 2·0 mm l‐glutamine (catalogue no. 10378‐016; Life Technologies, Carlsbad, CA). The human cervical adenocarcinoma cell line HeLa, WT or stably transduced with CD1d (kind gift from Dr M. Kronenberg, LIAI, San Diego, CA), was cultured in Dulbecco's modified Eagle's medium (catalogue no. BE12‐709F; Lonza) supplemented with 10% (v/v) fetal calf serum, 0·05 mm β‐mercaptoethanol, 100 IU/ml sodium penicillin, 100 μg/ml streptomycin sulphate and 2·0 mm l‐glutamine. The human myeloma cell line MM.1s, WT or stably transduced with CD1d (kind gift from Dr W. Song, Dana Farber Cancer Center, Boston, MA), was cultured in RPMI‐1640 (catalogue no. BE12‐115F; Lonza) medium supplemented with 10% (v/v) fetal calf serum, 0·05 mm β‐mercaptoethanol, 100 IU/ml sodium penicillin, 100 μg/ml streptomycin sulphate and 2·0 mm l‐glutamine. Cell lines were tested mycoplasma‐negative and frequently tested for purity (transfectants) by flow cytometry.
Flow cytometry and monoclonal antibodies
The following antibodies were used in this study: fluorescein isothiocyanate (FITC) ‐conjugated CD14 (catalogue no. 345784), phycoerythrin (PE) ‐conjugated CD1a (catalogue no. 555807), PE and allophycocyanin (APC) ‐conjugated CD25 (catalogue nos 555432 and #340907), CD83 APC (catalogue no. 551073), CD86 PE (catalogue no. 555658) and 7‐aminoactinomycin D (7‐AAD; catalogue no. 559925) were purchased from BD Biosciences (Franklin Lakes, NJ). Phycoerythrin–Cyanine 7‐conjugated Vα24 (catalogue no. PN A66907) and Vβ11 PE (catalogue no. IM2290) were purchased from Beckman Coulter (Brea, CA), anti‐MYC mAb (clone 4A6, catalogue no. 05‐724MG) from Merck Millipore, Billerica, MA, CD1d (clone 51.1, catalogue no. 12‐0016‐42) PE from eBioscience (San Diego, CA), CD1d (clone 51.1, catalogue no. 350308) APC and unconjugated low endotoxin CD1d (LEAF clone 51.1, catalogue no. 350304) from Biolegend (San Diego, CA), goat‐anti‐mouse F(ab')2 APC from Santa Cruz Biotech (Dallas, TX) (catalogue no. SC‐3818), FITC‐labelled polyclonal swine‐anti‐rabbit antibody was obtained from Dako (Glostrup, Denmark) (catalogue no. F0205), anti‐FLAG mAb (clone M2, catalogue no. F1804) from Sigma (St Louis, MO) and annexin V FITC and propidium iodide (PI) from VPS Diagnostics (Hoever, the Netherlands) (catalogue no. A700). Unconjugated IgG2b isotype control mAb was obtained from a hybridoma supernatant (clone MPC II). Rabbit‐anti‐llama sera (K976) was a kind gift from QVQ. Flow cytometry staining was performed in FACS buffer (PBS supplemented with 0·1% BSA and 0·02% sodium azide) for 30 min at 4°, unless otherwise specified. Samples were analysed on FACS Fortessa (BD Biosciences).
Generation of DC and iNKT cell lines
The moDC and primary human iNKT cells were generated as described previously.27 Briefly, monocytes were isolated from peripheral blood mononuclear cells with the use of CD14 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured in complete RPMI‐1640 medium in the presence of 1000 U/ml granulocyte–macrophage colony‐stimulating factor (Sanofi Leukine, Bridgewater, NJ) and 20 ng/ml IL‐4 (catalogue no. 204‐IL/CF; R&D Systems, Minneapolis, MN) for 5–7 days and subsequently matured with 100 ng/ml lipopolysaccharide (LPS) (catalogue no. L6529; Sigma) in the presence or absence of 100 ng/ml α‐GalCer (catalogue no. KRN7000; Funakoshi, Tokyo, Japan) for 48–72 hr. The iNKT cells were purified from peripheral blood mononuclear cells of healthy volunteers using magnetic bead sorting, and stimulated weekly with mature α‐GalCer‐loaded moDC in Yssel's medium28 supplemented with 1% human AB serum, 10 U/ml IL‐7 (catalogue no. 207‐IL/CF; R&D Systems) and 10 ng/ml IL‐15 (catalogue no. 34‐8159; eBioscience). Resting (< 50% CD25+) and pure (> 90% Vα24+ Vβ11+) iNKT cells were used for experiments.
Immunization of Lama glama and construction of VHH phage libraries
A humoral immune response against CD1d was induced by immunizing two llamas (Lama glama) subcutaneously with approximately 108 C1R‐CD1d cells four times with a 2‐weekly interval. Serum was collected before, during and after immunization (days 0, 28 and 43, respectively). Serially obtained pre‐immune and immune sera were tested for the presence of anti‐C1R‐CD1d antibodies through sequential incubation of sera with C1R‐CD1d cells, rabbit‐anti‐llama sera and FITC‐labelled swine‐anti‐rabbit antibody and analysed by flow cytometry.
One week after the last immunization, a 150‐ml blood sample was collected for peripheral blood lymphocyte isolation. Phage libraries were constructed by QVQ, Utrecht, the Netherlands. For this purpose total RNA was extracted from the peripheral blood lymphocyte, transcribed into cDNA, purified and used as a template for immunoglobulin heavy‐chain‐encoding gene amplification, as described previously.29 Agarose gel electrophoresed purified genes encoding heavy‐chain‐only immunoglobulin (~ 700 bp) were digested with SfiI and BstEII (catalogue nos R0123L, R3162L; New England Biolabs, Ipswich, MA) followed by cDNA VHH gene (~ 300–400 bp) extraction through agarose gel electrophoresis. Isolated VHH genes were subsequently ligated into the phagemid vector pUR8100 (a derivate of pHen130 with addition of an HC‐V cassette, to enable SfiI‐BstEII cloning, conferring Amp‐resistance for selection, and encoding a C‐terminal Myc and His6 tag for detection (kind gift from Dr M. El Khattabi, QVQ, Utrecht, the Netherlands) and transformed into Escherichia coli TG1 for display on filamentous bacteriophage. In this way two immune phage libraries were generated containing approximately 108 colony‐forming units (CFU) each.
Enrichment of phages that express CD1d‐specific VHH
To enrich for phages displaying CD1d‐specific VHH, multiple selection rounds were performed. Phage particles were rescued from the generated libraries as described elsewhere31 and resuspended in PBS. First, phages (approximately 1011 CFU per library) were allowed to bind to 2 × 107 Hela‐CD1d cells (2·5 × 106/ml) for 2 hr at 4°, followed by extensive washing in Hanks' balanced salt solution and PBS. Bound phages were eluted by resuspending cells in 100 mm triethylamine buffer for 15 min after which the mixture was neutralized by adding 1 m Tris–HCl (pH 7·5). Eluted phages were used to infect exponentially growing E. coli TG1, yielding approximately 106 CFU per library, which were subsequently used for phage preparation.31 Generated phages were used for either binding to captured recombinant β 2 ‐microglobulin (β 2m)‐CD1d molecules (kind gift from Prof. Dr S. Porcelli, AECOM, Bronx, NY) or for sequential incubation with C1R‐WT (negative selection) followed by selection to C1R‐CD1d cells (positive selection). For binding to captured recombinant β 2m‐CD1d, 96‐well Maxisorp plates (catalogue no. M9410‐1CS; Sigma) were coated (or not) with an anti‐FLAG antibody, washed, blocked for 30 min with 4% skimmed milk in PBS and incubated for 1 hr with recombinant FLAG‐tagged β 2m‐CD1d (5 μg/ml) diluted in 2% skimmed milk in PBS. Phages (diluted in 2% skimmed milk in PBS) were incubated with these β 2m‐CD1d coated Maxisorp plates for 2 hr at room temperature while gently shaking, after which the plates were extensively washed and bound phages were eluted with triethylamine. For sequential negative and positive selection, generated phages were incubated with 10 × 106 C1R‐WT cells (3·3 × 106/ml) for 1 hr at 4°C; unbound phages were subsequently incubated for a second time with C1R‐WT cells for 1 hr at 4°C. Residual unbound phages were then incubated with 2 × 106 C1R‐CD1d cells (0·25 × 106/ml) for 2 hr at 4°C. After extensive washing, bound phages were allowed to compete for CD1d‐binding by adding 1·9 mg/ml anti‐CD1d 51.1 mAb for 2 hr at 4°C. Remaining bound phages were eluted using triethylamine buffer. All phage fractions (approximately 106–107 CFU per library) that were now putatively enriched for CD1d‐specific phages by the above‐mentioned procedures were used for bacterial infection and plated accordingly to allow for individual colony selection.
Selection, re‐cloning and purification of unique CD1d specific VHH clones
Periplasmic extracts containing individual MYC‐HIS6‐tagged VHH were prepared as described previously32 and assessed for CD1d specificity using flow cytometry. For this purpose, carboxyfluorescein succinimidyl ester (CFSE) (catalogue no. 21888; Sigma) labelled C1R‐WT cells were mixed in a 1 : 1 ratio with C1R‐CD1d cells and incubated for 30 min with 25 μl periplasmic extracts followed by extensive washing and sequential incubation with an anti‐MYC mAb and APC‐labelled goat anti‐mouse F(ab')2 fragment and analysed by flow cytometry. Clones specifically binding to C1R‐CD1d were selected and separately screened for cross‐reactivity towards CD1a, CD1b and CD1c. To this end, CFSE labeled C1R‐CD1a, C1R‐CD1b or K562‐CD1c cells were mixed with C1R‐CD1d or K562‐CD1d cells and stained and analysed as described above. Restriction endonuclease (‘Fingerprint’) analysis with the restriction enzyme HinfI (catalogue no. R0155L; New England Biolabs) followed by conventional DNA electrophoresis was used to identify structurally different VHH genes in the selected panel of CD1d‐specific VHH. Based on these fingerprint analyses, multiple representative CD1d‐specific VHH genes were selected for sequence analysis (Baseclear BV, Leiden, the Netherlands) to identify unique CD1d‐specific VHH clones. Selected unique VHH‐encoding gene segments were re‐cloned, as SfiI‐BstEII fragments, into the expression vector pMEK219 (a derivate of pUR8100 with the deletion of gene III, a kind gift from Dr M. El Khattabi). VHH protein was purified from periplasmic extracts by means of immobilized metal ion affinity chromatography (IMAC) on Talon resin (Clontech, Mountain View, CA, catalogue no. 635503) followed by elution with 150 mm imidazole and dialysed against PBS.33 VHH protein integrity and purity were confirmed by Coomassie blue (Bio‐Rad, Hercules, CA; catalogue no.1610786) staining in SDS–PAGE gels; a Nanodrop Spectrophotometer was used for quantification (Thermo Fisher Scientific Inc., Waltham, MA). Specific binding to CD1d and non‐binding to CD1a, b, c of purified VHH protein (5 μg/ml) was confirmed.
Functional analyses of CD1d‐specific VHH
MoDC maturation
To evaluate the capacity of the generated panel of anti‐CD1d VHH to induce maturation of moDC, immature moDC were prepared as described from individual donors and seeded at 1 × 105 cells per well in a 48‐well tissue culture plate in the presence of 5 ng/ml recombinant human IL‐4, 500 U/ml recombinant human granulocyte–macrophage colony‐stimulating factor and 1000 U/ml recombinant human IFN‐γ (catalogue no. 14‐8319; eBioscience). To induce maturation, IgG2b isotype control mAb (10 μg/ml), unconjugated anti‐CD1d mAb (10 μg/ml), negative control VHH (500 nm, anti‐γδ TCR or anti‐azo‐dye RR6) or individual CD1d‐specific VHH (500 nm) were added for 72 hr. LPS (100 ng/ml, Sigma) and a cytokine cocktail consisting of IL‐1β/TNF‐α/IL‐6/prostaglandin E2 34 were used as positive controls. Polymyxin B (200 IU/ml, catalogue no. P4932; Sigma) was added to all conditions (but for the LPS condition) to inhibit the effects of any potential endotoxin contamination.35 Maturation of moDC was assessed by analysing IL‐10 and IL‐12 production in culture supernatants (t = 24 hr) using ELISA (IL‐10 ELISA, Sanquin, Amsterdam, the Netherlands, catalogue no. M1910; IL‐12p70 ELISA as described36) and by assessing the expression of CD83 and CD86 (t = 72 hr) using flow cytometry.
Apoptosis induction
For the assessment of the capacity of CD1d‐specific VHH to induce apoptosis in CD1d‐expressing tumour cells, CD1d‐transfected C1R and MM.1s cells were seeded at 1 × 105 cells per well in a 96‐well tissue culture plate and incubated for 24 hr with IgG2b isotype control mAb (5 μg/ml), unconjugated anti‐CD1d mAb (5 μg/ml), negative control VHH (100 nm), or the individual CD1d specific VHH (100 nm). This VHH concentration was at least 10‐fold higher than the minimum concentration found to be effective in inducing annexin V binding in an initial titration assay (not shown). After 24 hr, cells were stained with the combination of Annexin V FITC and PI or 7‐AAD, according to the manufacturer's protocol, and analysed by flow cytometry. Wild‐type C1R and MM.1s cell lines were used as negative controls.
Modulation of iNKT cell activation
For an evaluation of the capacity of the panel of generated CD1d‐specific VHH to block recognition of glycolipid‐loaded CD1d by iNKT cells, 5 × 104 Hela‐CD1d cells were seeded per well in a 96‐well tissue culture plate and pulsed overnight with vehicle control (DMSO 0·01%) or 100 ng/ml α‐GalCer. Cells were then washed with PBS and incubated with IgG2b isotype control mAb, unconjugated anti‐CD1d mAb, negative control VHH, or the anti‐CD1d specific VHH for 1 hr at the indicated concentrations. Subsequently, 5 × 104 pure and resting iNKT were added to each well. After 24 hr, culture supernatants were analysed for (inhibition of) cytokine production by CBA (BD Biosciences) whereas iNKT cells were harvested and analysed for CD25 expression by flow cytometry.
Statistical analysis
Statistical analyses were performed in graphpad prism version 6 (La Jolla, CA) using paired‐one‐way or two‐way analyses of variance and paired Student's t‐tests, as appropriate. Findings were considered significant when P values were < 0·05.
Results
Immunization of Lama glama with C1R‐CD1d cells induces a humoral immune response
To generate CD1d specific VHH, two Lama glama were immunized four times with approximately 108 cells of the human B‐lymphoblast cell line C1R transfected with CD1d.4 The induction of a humoral immune response was evaluated by determining the concentration of llama antibody directed towards C1R‐CD1d cells using immune sera obtained from the animals before (day 0), during (day 28) and after (day 43) immunization. In both llamas, a humoral immune response was induced (Fig. 1a,b), with llama B having a more pronounced response.
Figure 1.

Induction of a humoral immune response after immunization of two Lama glama with CD1d‐transfected C1R cells. Reactivity of differentially diluted pre‐immune (●), as well as post‐immune day 28 (■) and day 43 (▲) sera against whole C1R‐CD1d cells as detected by flow cytometry after sequential incubation of C1R‐CD1d cells with llama sera, rabbit‐anti‐llama sera and FITC‐labelled swine anti‐rabbit antibody. Data reflect mean fluorescence intensity (MFI) as determined in two individual llamas (a and b).
Selection of CD1d‐specific VHH
From each immunized llama a VHH gene library was synthesized from peripheral blood lymphocytes as described in the Materials and methods and named after their corresponding llama; A and B. Each phage library contained approximately 108 transformants. Consecutive rounds of positive and negative selection were performed to enrich for phages specific for CD1d. From both libraries, 95 individual VHH clones were randomly selected for specificity screening. For this purpose, periplasmic extracts were added to a 1 : 1 mixture of CD1d‐transfected C1R cells and CFSE‐labelled C1R‐WT cells. CFSE labelling of C1R‐WT allowed for easy discrimination of CD1d positive and negative populations and the simultaneous identification of CD1d‐specific VHH by flow cytometry (Fig. 2b). This resulted in 14 and 92 CD1d‐binding clones from respectively library A and B. Since CD1 isoforms display many structural similarities, including the expression of β 2m,37 CD1d‐specific VHH clones were then screened for cross‐reactivity to either CD1a, CD1b or CD1c. To do so, C1R‐CD1d or K562‐CD1d cells were mixed 1 : 1 with either CFSE‐labelled C1R‐CD1a, C1R‐CD1b or K562‐CD1c cells, and labelled with the putative CD1d‐specific VHH and analysed by flow cytometry. Overall, 14 (16%) and 92 (97%) of the screened clones from respectively library A and B were found to bind to CD1d. Fourteen (from library A, i.e. 100%) and 80 (from library B, i.e. 87%) of these clones were specific for CD1d and did not display reactivity towards either C1R‐WT cells or CD1a, CD1b, or CD1c.
Figure 2.

Specificity of selected variable domain of heavy‐chain‐only antibodies (VHH) for CD1d and not CD1a, CD1b or CD1c. (a) Binding of the panel of purified anti‐CD1d VHH clones, a non‐specific negative control VHH, a positive control CD1d‐specific monoclonal antibody (mAb), and a negative control IgG2b mAb against CD1a‐ (white bars), CD1b‐ (light grey bars), CD1c‐ (dark grey bars), and CD1d‐ (black bars) transfected cells. (b) Representative histograms showing binding of a negative control VHH (black) and anti‐CD1d VHH21 (light grey) to wild‐type C1R (C1R‐WT, left histogram) and CD1d‐transfected C1R cells (C1R‐CD1d, right histogram). MFI was detected by flow cytometry after sequential incubation of CD1a‐, CD1b‐, CD1c‐, and CD1d‐transfected cells with purified VHH, anti‐MYC mAb and allophycocyanin‐labelled goat‐anti‐mouse Fab2 fragment. Data represent mean + SD of three individual experiments.
To identify unique VHH, individual CD1d‐specific VHH were grouped based on the restriction profile obtained by DNA fingerprinting using the restriction enzyme HinfI. Representative clones from all groups were subsequently subjected to DNA sequencing resulting in the identification of 22 unique CD1d‐specific VHH clones. These 22 VHH clones were then recloned in the expression vector pMEK219 to enhance bacterial protein production and purified by means of IMAC to allow for VHH quantification. The specificity of these purified VHH to CD1d was confirmed by reassessment of binding to membrane‐bound CD1a, CD1b, CD1c and CD1d as described previously. As can be seen in Fig. 2(a), 21 out of 22 VHH clones were specific for CD1d, only one VHH clone (anti‐CD1d VHH 15) showed some cross‐reactivity to CD1c and was excluded from further experiments.
Based on the observed differences using DNA fingerprint analyses, we hypothesized that the individual clones would probably bind with different affinity to CD1d and/or bind to different epitopes on the CD1d molecule. These unique differences between the CD1d‐specific VHH could translate into distinct functional features of the individual VHH upon binding to CD1d. Therefore, the selected panel of CD1d‐specific VHH was assessed for their effects on different CD1d expressing cell types.
Capacity of CD1d‐specific VHH to induce moDC maturation and cytokine production
It was previously shown that ligation of CD1d using CD1d‐specific mAbs can trigger (mo)DC maturation and IL‐12 production and that this could be further enhanced in the presence of IFN‐γ, Toll‐like receptor‐triggering (e.g. by LPS) or CD40 ligation.21 To determine whether the generated CD1d‐specific VHH could exert a similar effect, we cultured immature moDC in the presence or absence of LPS (positive control), anti‐CD1d VHH, control VHH or anti‐CD1d mAb (not shown). To rule out effects of any endotoxin contamination, polymyxin B35 was added to the cultures which, as shown in Fig. 3(a,b), completely neutralized even high doses of LPS (P < 0·05). The capacity of the CD1d‐specific VHH to induce moDC maturation was determined by their IL‐12p70 production (as determined by ELISA in culture supernatants harvested after 24 hr) and by analysing expression of the maturation markers CD83 and CD86 after 72 hr by flow cytometry. Two of the CD1d‐specific VHH (anti‐CD1d VHH 2 and anti‐CD1d VHH 5) were found to induce moDC IL‐12p70 production (Fig. 3a, P < 0·01 and P < 0·001, respectively) and IL‐10 production (NS and P < 0·01, respectively, data not shown). Moreover, this was accompanied by an up‐regulation of the moDC maturation marker CD83 (Fig. 3b, P < 0·01 and NS, respectively, and Fig. 3(c) for a representative example). Though the moDC maturation marker CD86 displayed a similar pattern as observed with CD83, this increase did not reach statistical significance (not shown). Collectively these data demonstrate the ability of two of the selected CD1d‐specific VHH to induce moDC maturation and cytokine production.
Figure 3.

Induction of monocyte‐derived dendritic cell (moDC) maturation and cytokine production by CD1d‐specific variable domain of heavy‐chain‐only antibodies (VHH). Immature moDC were cultured with interferon‐γ (IFN‐γ) and either medium, lipopolysaccharide (LPS) or CD1d‐specific VHH. Polymyxin‐B was added to all conditions (except the LPS‐only condition) for 72 hr. Supernatants were harvested at 24 hr for detection of IL‐12 production (ELISA) (a). Graphical representation of CD83 expression after 72 hr (b) and representative dot‐plots showing up‐regulation of CD83 and CD86 (c). The control VHH is a representative example of the anti‐CD1d VHH panel found to have no effect upon CD1d binding. Data represent mean + SD of three individual experiments with moDC obtained from three different donors, **P < 0·01, ***P < 0·001, calculated with a one‐way analysis of variance with Dunnett's post hoc test.
Capacity to induce apoptosis in CD1d‐expressing C1R and MM1s
It was previously reported that ligation of CD1d by mAbs (e.g. clone 51.1) could trigger apoptosis of CD1d‐expressing C1R and primary MM cells.22 To address whether our panel of CD1d‐specific VHH could similarly induce early signs of apoptosis, CD1d‐transfected and untransfected C1R and MM.1s cells were cultured in the presence or absence of anti‐CD1d VHH, control VHH or anti‐CD1d mAb (clone 51.1) and 24 hr later were assessed for annexin V binding and PI or 7‐AAD staining, both markers of actual cell death.38
One CD1d‐specific VHH (anti‐CD1d VHH 17) consistently increased annexin V binding in C1R‐CD1d cells (P < 0·05), suggestive of early apoptosis (Fig. 4a). Though this effect was observed using both anti‐CD1d VHH 17 and anti‐CD1d mAb 51.1, a more pronounced shift in overall annexin V positivity was induced by anti‐CD1d VHH 17 (illustrated in Fig. 4b) suggesting a more robust effect. Of interest, and in contrast to the CD1d 51.1 mAb, which was ineffective in inducing annexin V binding in MM1s‐CD1d cells, the anti‐CD1d VHH 17 could also induce an increase in annexin V binding in MM.1s‐CD1d cells (P < 0·01), perhaps reflecting a more consistent apoptotic signal triggered by anti‐CD1d VHH 17 compared with CD1d 51.1 mAb (Fig. 4a). Importantly, no increase in annexin V binding was observed in both untransfected parental cell lines (Fig. 4a), confirming the CD1d dependency of the observed effect.
Figure 4.

Induction of annexin V binding by anti‐CD1d variable domain of heavy‐chain‐only antibodies (VHH). Either CD1d‐transfected C1R, wild‐type C1R, CD1d‐transfected MM.1s and wild‐type MM.1s were cultured for 24 hr in the presence of either medium alone, a control anti‐CD1d VHH, anti‐CD1d mAb (clone 51.1) or anti‐CD1d VHH 17. The total percentage of cells positive for annexin V was then determined by flow cytometry (a). Representative dot‐plots indicating potent induction of annexin V binding (b). The control VHH is a representative example of an anti‐CD1d VHH clone found to have no effect upon CD1d binding. Data represent mean + SD of three individual experiments, *P < 0·05, **P < 0·01, calculated with a one‐way analysis of variance with Dunnett's post hoc test.
Capacity of CD1d‐specific VHH to block glycolipid induced iNKT cell activation
Multiple anti‐CD1d mAbs have been shown to be able to block the interaction between CD1d and the iNKT TCR,23 we therefore evaluated the selected CD1d‐specific VHH for their ability to do so as well. For this purpose HeLa‐CD1d cells, loaded with α‐GalCer or vehicle control, were incubated with IgG2b mAb, anti‐CD1d mAb, control VHH or anti‐CD1d VHH for 1 hr after which iNKT cells were added in a 1 : 1 ratio for an additional 24 hr co‐culture. The capacity of the CD1d‐specific VHH to block iNKT cell activation was then determined by analysing expression of the activation marker CD25 by flow cytometry and by determining iNKT cell cytokine production (IFN‐γ and TNF‐α) by CBA. One anti‐CD1d VHH was found to induce a consistent and potent neutralizing effect. As shown in Fig. 5(a–c), the anti‐CD1d VHH 22 was able to effectively inhibit iNKT cell activation (P < 0·001) and cytokine production (P < 0·0001). Inhibition of iNKT cell activation was dose dependent with optimal inhibition in the nanomolar range (Fig. 5c), underscoring the powerful inhibitory activity of this anti‐CD1d VHH.
Figure 5.
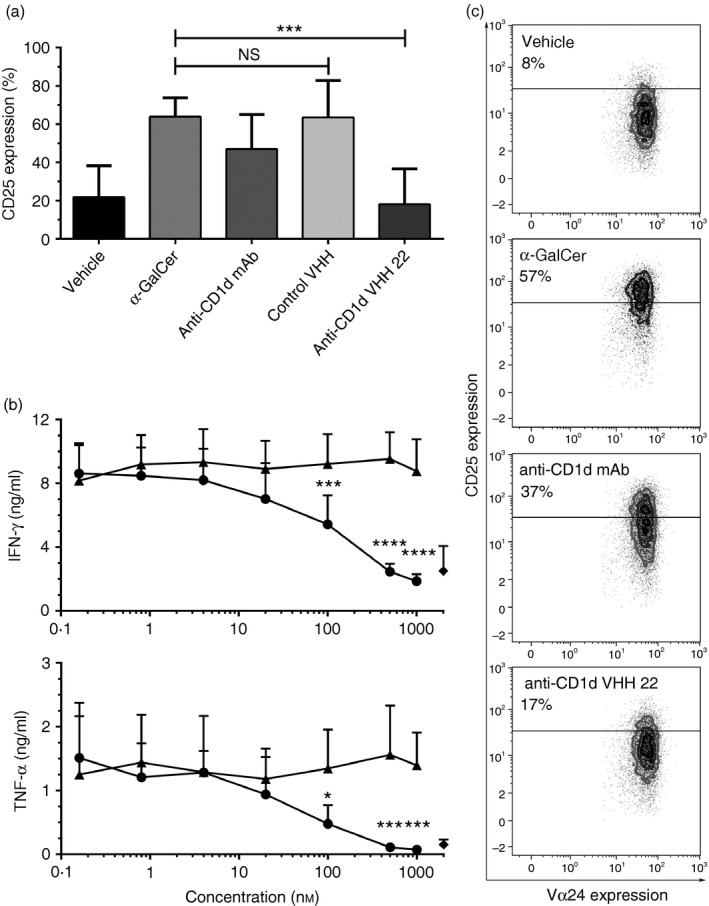
Dose‐dependent inhibition of CD1d‐α‐GalCer mediated invariant natural killer T (iNKT) cell activation. iNKT CD25 expression, interferon‐γ (IFN‐γ) and tumour necrosis factor‐α (TNF‐α) production were determined after a 24‐hr co‐culture of iNKT cells with CD1d‐transfected HeLa cells pulsed with vehicle control (vehicle) or α‐GalCer (all other conditions) and either medium alone (vehicle and α‐GalCer conditions), anti‐CD1d mAb 51.1 at 10 μg/ml (67 nm), a control anti‐CD1d variable domain of heavy‐chain‐only antibodies (VHH) (500 nm) or anti‐CD1d VHH 22 (500 nm). Bars demonstrating expression of the activation marker CD25 by iNKT cells in the various conditions (a). Effect of different concentrations of anti‐CD1d VHH 22 (●) and a control non‐inhibitory but CD1d‐specific VHH (▲) on iNKT cell IFN‐γ and TNF‐α production. ♦ symbols indicate the vehicle‐loaded negative control condition (b). Representative dot‐plots illustrating marked inhibition of iNKT cell CD25 up‐regulation by anti‐CD1d VHH 22 (c). The control VHH is a representative example of a non‐inhibitory CD1d‐specific VHH. Data represent mean + SD of three individual experiments with iNKT obtained from three different donors, **P < 0·05, **P < 0·01, ****P < 0·0001, calculated with a one‐way analysis of variance with Dunnett's post hoc test (a) or a two‐way analysis of variance with Sidak's post hoc test (b).
Discussion
Here, we describe the successful generation and isolation of a panel of CD1d‐specific VHH through the immunization of Lama glama with CD1d expressing C1R cells, phage library generation and selection of CD1d‐specific and unique VHH clones. Within the set of 21 CD1d‐specific VHH, we found substantial structural and functional variability (see Supplementary material, Table S1). Unique CD1d‐specific VHH with the capacity to either stimulate moDC maturation and cytokine production, or to induce signs of early apoptosis in CD1d‐expressing B lymphoblasts and MM cell lines, or to block the interaction between CD1d and iNKT cells were identified and characterized. Although mAbs with these features have been described,21, 22, 23 this is the first report describing the generation of CD1d‐specific VHH that are functional for one of the three mentioned processes.
Multiple ex vivo and in vivo studies indicate a role for the blocking of CD1d in controlling auto‐immune and inflammatory disorders, including systemic lupus erythematosus, asthma, sickle cell disease, psoriasis and atherosclerosis.39, 40, 41, 42 Indeed, blocking CD1d by a mAb in systemic lupus erythematosus patient‐derived peripheral blood mononuclear cells resulted in inhibition of total IgG and anti‐dsDNA IgG secretion in vitro.41 In mouse, allergic asthma models showed decreased airway hyper‐reactivity and cytokine production in both CD1d‐deficient and iNKT‐deficient mice.42 Similarly, blockade of CD1d in a cynomolgus macaque airway hyper‐reactivity model reduced cytokine production and bronchial infiltration of lymphocytes and macrophages.23 As it was shown that airborne lipid antigens were able to induce profound inflammation via activation of iNKT cells in the lung, environmental lipids presented via CD1d have been implicated in the triggering and/or support of disease progression.23, 43 Therefore, local blockade of CD1d might be effective and sufficient for the prevention of local pulmonary inflammation. Here, we identified an anti‐CD1d VHH that very effectively inhibited recognition of the CD1d‐α‐GalCer complex by iNKT cells, thereby preventing subsequent iNKT cell activation and cytokine production. As VHH are very small and stable it will be of particular interest to evaluate their therapeutic effect after local aerosol delivery.
As mentioned, interactions between iNKT TCR and CD1d expressing DC induce bidirectional activation, which is driven by CD40‐CD40 ligand interactions, iNKT cell derived IFN‐γ, and IL‐12 produced by DC.1 The mAbs against at least three CD1d epitopes were shown to trigger rapid phosphorylation of IκB, a critical step in nuclear factor‐κB pathway activation, resulting in both IL‐12 production and enhanced differentiation of DC, thereby mimicking iNKT help.21 Here, we successfully identified two CD1d‐specific VHH with the same ability to promote IL‐12 production and moDC maturation in the absence of other co‐stimulatory signals (e.g. TLR‐ and CD40‐signalling). Since CD40 and CD1d ligation were shown to have a synergistic effect on DC activation, one could envisage that a bi‐specific VHH targeting both antigens, whether or not fused to a tumour‐associated antigen, could be a powerful tool for vaccination purposes. Especially, since the lymphoid compartment of lymph nodes forms an interconnected network that maintains strict size exclusion (< 70 000 MW) criteria to prevent pathogens from entering,44 (bi‐)specific VHH (~ 15 000–30 000 MW) could easily access this conduit system and subsequently ligate the DC lining this network. This surrogate ‘cognate’ signal could possibly mimic iNKT help and equip DC to prime both antitumor CD4+ and CD8+ T cells and induce NK cell transactivation.1
Another interesting consequence of CD1d triggering was the observation that ligation of CD1d using CD1d‐specific mAbs was able to induce apoptosis in CD1d‐expressing B lymphoblasts and MM cells.22 Although reports have suggested down‐regulation of CD1d during MM disease progression22 all‐trans retinoic acid (ATRA) has been shown to be able to up‐regulate CD1d expression.45 Moreover, preclinical studies indicate a rationale for ATRA combination therapy46 and phase I/II studies (Clin. Trial. Gov NCT01985477) are currently evaluating ATRA combination therapy for relapsed/refractory MM.47 Therefore, targeting CD1d might not only be feasible in early MM but also in patients with advanced disease stages treated with ATRA. From our panel of CD1d‐specific VHH, one (anti‐CD1d VHH 17) was found to be able to induce increased cell surface expression levels of phosphatidylserine as detected by increased binding to annexin V. Interestingly, when compared with the CD1d 51.1 mAb, anti‐CD1d VHH 17 induced comparable annexin V binding levels in C1R‐CD1d cells, but superior annexin V binding to MM1s‐CD1d, suggesting a more consistent anti‐MM effect of this CD1d‐specific VHH. Apart from being a marker of early apoptosis, the presence of PS on the outer membrane of apoptotic and stressed cells can also function as an ‘eat me’ signal for macrophages48 offering at least two pathways via which ligation of CD1d may negatively impact MM survival. It should be noted that the CD1d‐specific VHH that were most effective in inducing moDC maturation and cytokine production (anti‐CD1d VHH 2 and anti‐CD1d VHH 5) were different from the one that was most effective in triggering annexin V binding on CD1d‐expressing tumour cells (anti‐CD1d VHH 17), and also differed from the one that was most effective in blocking CD1d–iNKT cell interactions (anti‐CD1d VHH 22). This contrasts with the CD1d 51.1 mAb, which functionally affects all three of these processes.21, 22, 23 Though the reason for this difference is not known, it could be related to cell‐specific differences in downstream signalling, or perhaps to differences in binding sites between the mAbs and the VHH, or may be due to clustering of CD1d by the two binding domains of the mAb. In any case, induced effects of the anti‐CD1d VHH seem more confined to a specific function, which could be an advantage when considering therapeutic applications.
In conclusion, we successfully generated a panel of CD1d‐specific VHH. Assessment of various functions resulted in the identification of two VHH that induced DC maturation and cytokine production, one VHH that triggered signs of early apoptosis in B lymphoblasts, and one VHH that blocked the interaction between CD1d and the iNKT‐TCR. The apparently more specific and more predictable effects that can be induced by the CD1d‐specific VHH can be important when considering immunotherapeutic approaches that focus on either blocking CD1d, targeting DC for vaccination purposes, or the induction of apoptosis in CD1d‐expressing tumour cells.
Authorship contributions
Roeland Lameris: performed research, collected data, analysed and interpreted data, performed statistical analysis, and wrote the manuscript. Renée C.G. de Bruin: performed research, and wrote the manuscript. Paul M.P. van Bergen en Henegouwen and Henk M. Verheul: wrote the manuscript. Sonja Zweegman: interpreted data, and wrote the manuscript. Tanja D. de Gruijl and Hans J. van der Vliet: designed research, analysed and interpreted data, and wrote the manuscript.
Disclosures
All authors have no conflicts of interest to declare.
Supporting information
Table S1. Overview of anti‐CD1d variable domain of heavy‐chain‐only antibodies (VHH) with their binding specificity and function.
Acknowledgements
This work is supported by CCA‐VICI grant 2000483 from the VU University Medical Centre, grant VU 2010‐4728 from the Dutch Cancer Society (KWF), and grant 14‐0343 from Worldwide Cancer Research.
References
- 1. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol 2013; 13:101–17. [DOI] [PubMed] [Google Scholar]
- 2. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d‐restricted antigens by natural killer T cells. Nat Rev Immunol 2012; 12:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant Vα24‐J α Q/V β 11 T cell receptor is expressed in all individuals by clonally expanded CD4–8‐ T cells. J Exp Med 1994; 180:1171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Vα24+ CD4– CD8– T cells. J Exp Med 1997; 186:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol 2004; 4:231–7. [DOI] [PubMed] [Google Scholar]
- 6. Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d‐dependent NKT cells. J Clin Invest 2004; 114:1379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B et al Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL‐2 to activate NK cells. J Immunol 2001; 167:3114–22. [DOI] [PubMed] [Google Scholar]
- 8. McEwen‐Smith RM, Salio M, Cerundolo V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol Res 2015; 3:425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Semmling V, Lukacs‐Kornek V, Thaiss CA, Quast T, Hochheiser K, Panzer U et al Alternative cross‐priming through CCL17‐CCR4‐mediated attraction of CTLs toward NKT cell‐licensed DCs. Nat Immunol 2010; 11:313–20. [DOI] [PubMed] [Google Scholar]
- 10. Chaudhry MS, Karadimitris A. Role and regulation of CD1d in normal and pathological B cells. J Immunol 2014; 193:4761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO et al Protective mucosal immunity mediated by epithelial CD1d and IL‐10. Nature 2014; 509:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benam KH, Kok WL, McMichael AJ, Ho LP. Alternative spliced CD1d transcripts in human bronchial epithelial cells. PLoS One 2011; 6:e22726, doi:10.1371/journal.pone.0022726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Der Vliet HJ, Molling JW, Von Blomberg BME, Nishi N, Kölgen W, Van Den Eertwegh AJM et al The immunoregulatory role of CD1d‐restricted natural killer T cells in disease. Clin Immunol 2004; 112:8–23. [DOI] [PubMed] [Google Scholar]
- 14. Swann JB, Uldrich AP, Van Dommelen S, Sharkey J, Murray WK, Godfrey DI, Smyth MJ. Type I natural killer T cells suppress tumors caused by p53 loss in mice. Blood 2009; 113:6382–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bellone M, Ceccon M, Grioni M, Jachetti E, Calcinotto A, Napolitano A et al iNKT cells control mouse spontaneous carcinoma independently of tumor‐specific cytotoxic T cells. PLoS One 2010; 5:e8646, doi:10.1371/journal.pone.0008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schneiders FL, De Bruin RCG, Van Den Eertwegh AJM, Scheper RJ, Leemans CR, Brakenhoff RH et al Circulating invariant natural killer T‐cell numbers predict outcome in head and neck squamous cell carcinoma: updated analysis with 10‐year follow‐up. J Clin Oncol 2012; 30:567–70. [DOI] [PubMed] [Google Scholar]
- 17. Tachibana T, Onodera H, Tsuruyama T, Mori A, Nagayama S, Hiai H, Imamura M. Increased intratumor Vα24‐positive natural killer T cells: a prognostic factor for primary colorectal carcinomas. Clin Cancer Res 2005; 11:7322–7. [DOI] [PubMed] [Google Scholar]
- 18. Kunii N, Horiguchi S, Motohashi S, Yamamoto H, Ueno N, Yamamoto S et al Combination therapy of in vitro‐expanded natural killer T cells and α‐galactosylceramide‐pulsed antigen‐presenting cells in patients with recurrent head and neck carcinoma. Cancer Sci 2009; 100:1092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamasaki K, Horiguchi S, Kurosaki M, Kunii N, Nagato K, Hanaoka H et al Induction of NKT cell‐specific immune responses in cancer tissues after NKT cell‐targeted adoptive immunotherapy. Clin Immunol 2011; 138:255–65. [DOI] [PubMed] [Google Scholar]
- 20. Ni C, Wu P, Wu X, Zhang T, Wang Z et al Thymosin α1 enhanced cytotoxicity of iNKT cells against colon cancer via upregulating CD1d expression. Cancer Lett 2015; 356(2 Pt B):579–88. [DOI] [PubMed] [Google Scholar]
- 21. Yue SC, Shaulov A, Wang R, Balk SP, Exley MA. CD1d ligation on human monocytes directly signals rapid NF‐κB activation and production of bioactive IL‐12. Proc Natl Acad Sci USA 2005; 102:11811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spanoudakis E, Hu M, Naresh K, Terpos E, Melo V, Reid A et al Regulation of multiple myeloma survival and progression by CD1d. Blood 2009; 113:2498–507. [DOI] [PubMed] [Google Scholar]
- 23. Nambiar J, Clarke AW, Shim D, Mabon D, Tian C, Windloch K et al Potent neutralizing anti‐CD1d antibody reduces lung cytokine release in primate asthma model. MAbs 2015; 7:638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tijink BM, Laeremans T, Budde M, Stigter‐van Walsum M, Dreier T, de Haard HJ et al Improved tumor targeting of anti‐epidermal growth factor receptor Nanobodies through albumin binding: taking advantage of modular Nanobody technology. Mol Cancer Ther 2008; 7:2288–97. [DOI] [PubMed] [Google Scholar]
- 25. Lameris R, de Bruin RCG, Schneiders FL, van Bergen en Henegouwen PMP, Verheul HM, de Gruijl TD, van der Vliet HJ. Bispecific antibody platforms for cancer immunotherapy. Crit Rev Oncol Hematol 2014; 92:153–65. [DOI] [PubMed] [Google Scholar]
- 26. Roovers RC, van Dongen GAMS, van Bergen en Henegouwen PMP. Nanobodies in therapeutic applications. Curr Opin Mol Ther 2007; 9:327–35. [PubMed] [Google Scholar]
- 27. Lameris R, Schneiders FL, de Gruijl TD, van der Vliet HJ. Exploiting the CD1d‐iNKT cell axis for potentiation of DC‐based cancer vaccines. Methods Mol Biol 2014; 1139:155–65. [DOI] [PubMed] [Google Scholar]
- 28. Yssel H, De Vries JE, Koken M, Van Blitterswijk W, Spits H. Serum‐free medium for generation and propagation of functional human cytotoxic and helper T cell clones. J Immunol Methods 1984; 72:219–27. [DOI] [PubMed] [Google Scholar]
- 29. Roovers RC, Laeremans T, Huang L, De Taeye S, Verkleij AJ, Revets H et al Efficient inhibition of EGFR signalling and of tumour growth by antagonistic anti‐EGFR Nanobodies. Cancer Immunol Immunother 2007; 56:303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi‐subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res 1991; 19:4133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By‐passing immunization. Human antibodies from V‐gene libraries displayed on phage. J Mol Biol 1991; 222:581–97. [DOI] [PubMed] [Google Scholar]
- 32. Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkönig A, Ruf A et al A general protocol for the generation of Nanobodies for structural biology. Nat Protoc 2014; 9:674–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roovers RC, Henderikx P, Helfrich W, van der Linden E, Reurs A, de Bruïne AP et al High‐affinity recombinant phage antibodies to the pan‐carcinoma marker epithelial glycoprotein‐2 for tumour targeting. Br J Cancer 1998; 78:1407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jonuleit H, Kühn U, Müller G, Steinbrink K, Paragnik L, Schmitt E et al Pro‐inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum‐free conditions. Eur J Immunol 1997; 27:3135–42. [DOI] [PubMed] [Google Scholar]
- 35. Duff GW, Atkins E. The inhibitory effect of polymyxin B on endotoxin‐induced endogenous pyrogen production. J Immunol Methods 1982; 52:333–40. [DOI] [PubMed] [Google Scholar]
- 36. van der Pouw Kraan TC, Boeije LC, de Groot ER, Stapel SO, Snijders A, Kapsenberg ML et al Reduced production of IL‐12 and IL‐12‐dependent IFN‐γ release in patients with allergic asthma. J Immunol 1997; 158:5560–5. [PubMed] [Google Scholar]
- 37. Ly D, Moody DB. The CD1 size problem: lipid antigens, ligands, and scaffolds. Cell Mol Life Sci 2014; 71:3069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Genderen H, Kenis H, Lux P, Ungeth L, Maassen C, Deckers N et al In vitro measurement of cell death with the annexin A5 affinity assay. Nat Protoc 2006; 1:363–7. [DOI] [PubMed] [Google Scholar]
- 39. Wallace KL, Marshall MA, Ramos SI, Lannigan JA, Field JJ, Strieter RM, Linden J. NKT cells mediate pulmonary inflammation and dysfunction in murine sickle cell disease through production of IFN‐γ and CXCR3 chemokines. Blood 2009; 114:667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simoni Y, Diana J, Ghazarian L, Beaudoin L. Lehuen a. Therapeutic manipulation of natural killer (NK)T cells in autoimmunity: are we close to reality? Clin Exp Immunol 2013; 171:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen L, Zhang H, Caimol M, Benike CJ, Chakravarty EF, Strober S, Engleman EG. Invariant natural killer T cells in lupus patients promote IgG and IgG autoantibody production. Eur J Immunol 2015; 45:612–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T et al Essential role of NKT cells producing IL‐4 and IL‐13 in the development of allergen‐induced airway hyperreactivity. Nat Med 2003; 9:582–8. [DOI] [PubMed] [Google Scholar]
- 43. Scanlon ST, Thomas SY, Ferreira CM, Bai L, Krausz T, Savage PB, Bendelac A. Airborne lipid antigens mobilize resident intravascular NKT cells to induce allergic airway inflammation. J Exp Med 2011; 208:2113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roozendaal R, Mebius RE, Kraal G. The conduit system of the lymph node. Int Immunol 2008; 20:1483–7. [DOI] [PubMed] [Google Scholar]
- 45. Li X, Garg TK, Johnson SK, Szmania S, Stivers J, Ling et al ATRA Upregulates Cell Surface CD1D on Myeloma Cells and Sensitizes Them to iNKT Cell‐Mediated Lysises. In: ASH Annual Meeting and Exposition. Vol; 2014.
- 46. Nijhof IS, Groen RW, Lokhorst HM, van Kessel B, Bloem AC, van Velzen J et al Upregulation of CD38 expression on multiple myeloma cells by all‐trans retinoic acid improves the efficacy of daratumumab. Leukemia 2015; 29:2039–49. [DOI] [PubMed] [Google Scholar]
- 47. NCT01985477. Revlimid/All‐Trans Retinoic Acid (ATRA)/Dexamethasone in Relapsed/Refractory Multiple Myeloma.
- 48. Segawa K, Suzuki J, Nagata S. Constitutive exposure of phosphatidylserine on viable cells. Proc Natl Acad Sci USA 2011; 108:19246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Overview of anti‐CD1d variable domain of heavy‐chain‐only antibodies (VHH) with their binding specificity and function.


