Abstract
Background
Recurrent legionella outbreaks at one and the same location are common. We have identified a single Legionella pneumophila genotype associated with recurrent Legionella outbreaks over 6 years.
Methods
Field emergency surveys following Legionella outbreaks were performed on a vessel in 2008, 2009 and 2013. Water samples from both the distribution and technical parts of the potable water system were analyzed with respect to L. pneumophila [Real-Time PCR, cultivation, serotyping and genotyping (PFGE)] and free-living amoebae, (FLA).
Results
Legionella pneumophila serogroup 1 was present in the ship's potable water system during every outbreak. Genotyping of the 2008 survey material showed two separate PFGE genotypes while those in 2009 and 2013 demonstrated the presence of only one of the two genotypes. FLA with intracellular L. pneumophila of the same genotype were also detected. Analyses of the freshwater system on a ship following three separate Legionella outbreaks, for L. pneumophila and FLAs, revealed a single L. pneumophila genotype and FLA (Hartmanella).
Conclusions
It is reasonable to assume that the L. pneumophila genotype detected in the freshwater system was the causal agent in the outbreaks onboard. Persistence of an apparently low-pathogenic L. pneumophila genotype and FLA in a potable water system represent a potential risk for recurrent outbreaks.
Keywords: Legionella pneumophila, Genotypes, Ships, infection outbreaks, Prevention and control, Pathogenicity, Virulence, Free-living amoeba
The legionella bacterium is present in freshwater sources all over the world (1). Even if the bacterium is usually considered to be of low pathogenicity, it is well known as a potential risk factor for serious infections (2, 3). Legionella is highly fastidious as regards the substrate it requires for growth (demands cysteine and Fe3) and can therefore be expected to survive and multiply in other organisms such as freshwater free-living amoebae (FLA) (4, 5).
FLA constitute reservoirs for many bacteria including not only well-known pathogens but also emerging pathogens responsible for respiratory diseases and contribute to the protection, survival, and dissemination of these bacteria in water systems, despite the application of disinfection or thermal treatments.
Legionella may cause serious respiratory diseases in humans through inhalation of aerosolized water containing the bacterium, for example, during showering with contaminated water (6, 7). Infections caused by high-virulence/pathogenic legionella bacteria are spontaneous and unpredictable.
Outbreaks of legionellosis on board vessels have been reported (8, 9). Studies of legionella contamination on cruise ships and ferries show that various legionella species occur in such water systems (10).
Following the Norwegian legionella regulations (11), in 2010, the Royal Norwegian Navy launched a project with the aim of preventing legionella infections on board naval vessels, and the results of this study were recently published (12). The main conclusions were that: 1) Legionella pneumophila was a frequently occurring contaminant in the potable water systems of Norwegian naval vessels (approximately 50%); 2) FLA were present in all freshwater systems with L. pneumophila, and the bacterium could also be isolated from the FLA; 3) identical L. pneumophila genotypes were present in the potable water systems of several vessels; 4) L. pneumophila genotypes found in these vessels’ water systems were also demonstrated in their water-filling stations.
No legionella infections or outbreaks of Legionnaires’ disease were seen on board the naval vessels during the 2 years that the project lasted.
In this communication, we present data from a vessel experiencing three separate legionellosis outbreaks between 2008 and 2013 and demonstrate that the freshwater system was contaminated with one and the same L. pneumophila genotype all through the observation period despite repeated efforts to eliminate the bacterium.
The findings emphasize the potential of one genotype to change qualitative attributes, for example, pathogenicity/virulence, throughout persistence in a defined environment.
Materials and methods
This study deals with one particular Offshore Supply Vessel, built for offshore construction and subsea operations all over the world. Its potable water system was in accordance with the Norwegian regulations in effect by the time for delivery and had been updated and improved before 2008 with respect to barriers (filters and UV). With a capacity of over 1000 m3, it provides an adequate supply for a crew of about 70 in some 40 cabins. The technical part of the system includes four freshwater tanks, a pressure tank, and parallel UV systems as barriers to prevent contamination entering the distribution system. The latter is the component of relevance for legionella exposure and comprises around 125 water supply point, 50 of which aerosolized (showers and high-pressure washers). During the period 2008–2013, the vessel experienced three separate outbreaks of legionellosis onboard.
Actions related to outbreaks
In our legionella survey strategies, ordinary legionella surveys are performed as screenings, that is, 10% of all outlets selected for analysis (12). When outbreaks occur, all aerosolized spots are sampled and analysed and such surveys are called ‘emergency surveys’.
Emergency surveys were carried out during the vessel's operations in Nigeria in April 2008, in UK/Norway in September 2009, and in The Netherlands in August 2013. The 2008 survey took place after a third-part employee, who joined the vessel on March 21, 2008, in Nigeria, went down with a high fever on April 8 and was transferred to a hospital in his own country on April 11. On April 18, legionellosis was confirmed. The emergency survey onboard, performed on April 24, was aimed at identifying a possible source on board and to clarify whether there was a risk of exposure to infection for the rest of the crew.
The immediate action following an outbreak is to prevent exposure to aerosolized water. During the 2008 outbreak, this was done by removing showerheads and showering using only hoses. Further efforts were in accordance with procedures described in the legionella guidance issued by the Norwegian Institute of Public Health (11), that is, concentrating on water-heating systems and chlorine.
The 2009 emergency survey, initiated on September 22 that year, was due to a hospital-confirmed case of legionellosis in a crewmember 1 week after he had left the vessel. During dry-docking in February 2009, the potable water system had been treated with hydrochloric acid. By the time for the 2009 outbreak, the recently developed disposable showerhead filters (Kleenpak™ Pall) (0.2 µm) were introduced to prevent exposure to potentially pathogenic microbes. Further efforts were in accordance to NIPH guidance (11).
Between August 8 and 19, 2013, seven cases of Legionella/pneumonia infections were confirmed among the crew; one of these, who had been on board since July 24, was confirmed with legionellosis on August 8, and six with Pontiac fever. In addition to heat treatment of the water system, a second treatment by hydrochloric acid was carried out on August 12 and 13, 2013. In view of the earlier outbreaks in 2008 and 2009, the possible persistence of the particular L. pneumophila genotype/FLA population was considered, and a third survey was carried out on the particular vessel. After the third survey, a new sanitation strategy based on chlorine dioxide was initiated, and this is still in effect.
Water sampling
Water samples were collected in 1-L sterile flasks already dosed with sodium thiosulfate, from both the technical component of the water supply system (water tanks) and the distribution network (showers).
Microbiological analysis
Detection and cultivation of Legionella pneumophila
All the samples from the water supply systems were analysed using real-time polymerase chain reaction (PCR), as described previously (13). Positive samples were cultured on legionella-selective agar (buffered charcoal yeast extract) plates to demonstrate live L. pneumophila, which is a prerequisite for infection. Legionella pneumophila growths were serotyped using MONOFLUO anti-Legionella Staining Reagent (Bio-Rad, Munich, Germany). All isolates of L. pneumophila serogroup 1 were genotyped using pulsed-field gel electrophoresis (14) in order to track potential collective routes of spread and contamination.
Free-living amoebae
The amoebic analyses were performed using cultivation and microscopic inspection, in accordance with the methods described by Winiecka-Krusnell and Linder (15).
Results
Emergency survey 2008
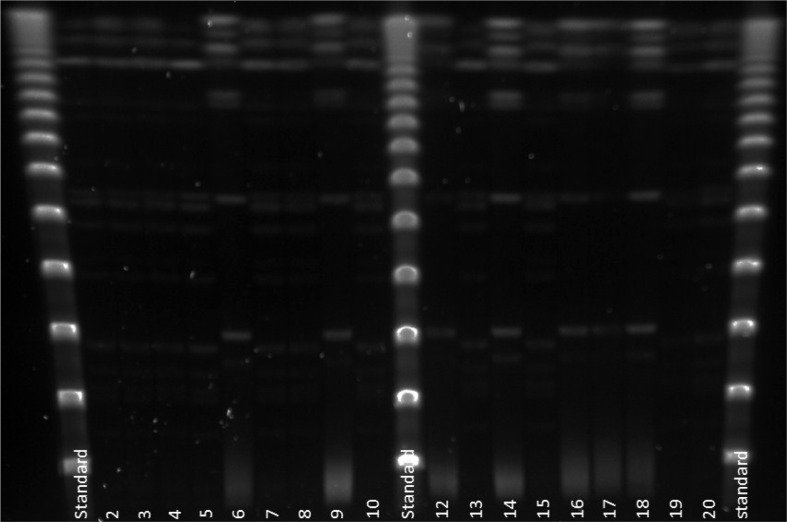
Legionella pneumophila PCR detected genetic material in 40 of the 45 water samples (89%), of which 13 showed live L. pneumophila serogroup 1. Two separate L. pneumophila genotypes (Lp-gta and Lp-gtb) were identified from the material (Fig. 1). The genotype Lp-gta was also found intracellularly in the amoeba population. A follow-up survey in mid-June, after a couple of months of treatment, showed L. pneumophila PCR positives in 12 of the 45 (27%), of which seven were live L. pneumophila serogroup 1 of the same genotype L. pneumophila (Lp-gta). Figure 1 shows the results from genotyping of the first 18 L. pneumophila isolates analysed from the 2008 survey. Two separate genotypes of L. pneumophila (Lp-gta and Lp-gtb) were identified in the potable water distribution system.
Fig. 1.
The results from genotyping of the first 18 Legionella pneumophila isolates analysed from the 2008 survey. Two separate genotypes of L. pneumophila (Lp-gta and Lp-gtb) identified in the potable water distribution system. Lane 1, 11 and 21 are Lambda Stds. Lanes 2–5, 7, 8, 10, 13, 15, 19–20 are Lp-gta. Lanes 6, 9, 12, 14, 16–18 are Lp-gtb.
Emergency survey 2009
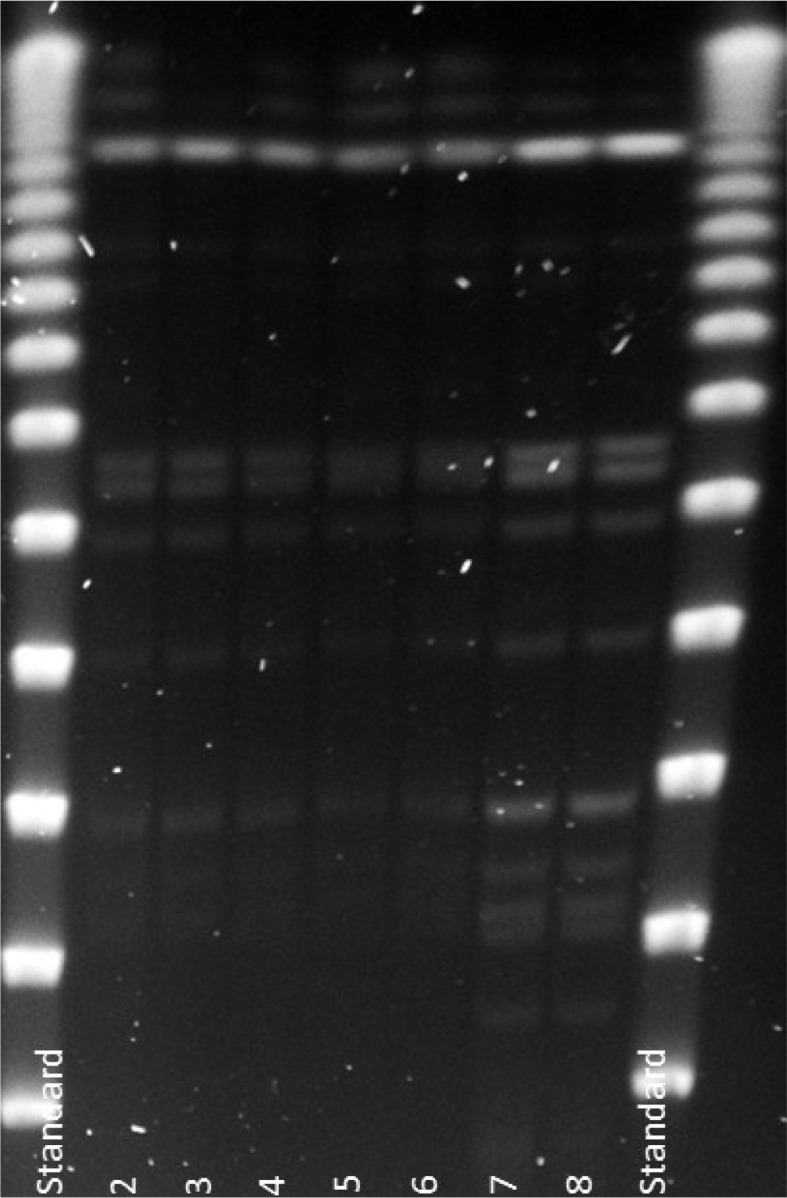
Legionella pneumophila PCR detected genetic material from five of 45 samples (11%), of which three out of five showed live L. pneumophila serotype 1. Only one of the two previously identified genotypes was present (Lp-gta) (Fig. 2). FLAs were isolated from 32 of the 45 samples (71%), and in one of these, live intracellular L. pneumophila (iLp) of the same genotype was identified. Figure 2 shows the genotype patterns of L. pneumophila population in water by the time for the second emergency survey.
Fig. 2.
The results from genotyping of the five Legionella pneumophila isolates analysed from the 2009 survey. Only one genotype of L. pneumophila (Lp-gta) was seen in the potable water distribution system by the time for outbreak. Lanes 1 and 9 are Lambda Stds. Lanes 2–6 are isolates from the 2009 survey and show Lp-gta. Lanes 7 and 8 are Lp-gta isolates from the 2008 survey.
No full follow-up study was carried out following this outbreak as the cleaning and disinfection regimens had been continuously in place since the 2008 outbreak.
Survey 2013
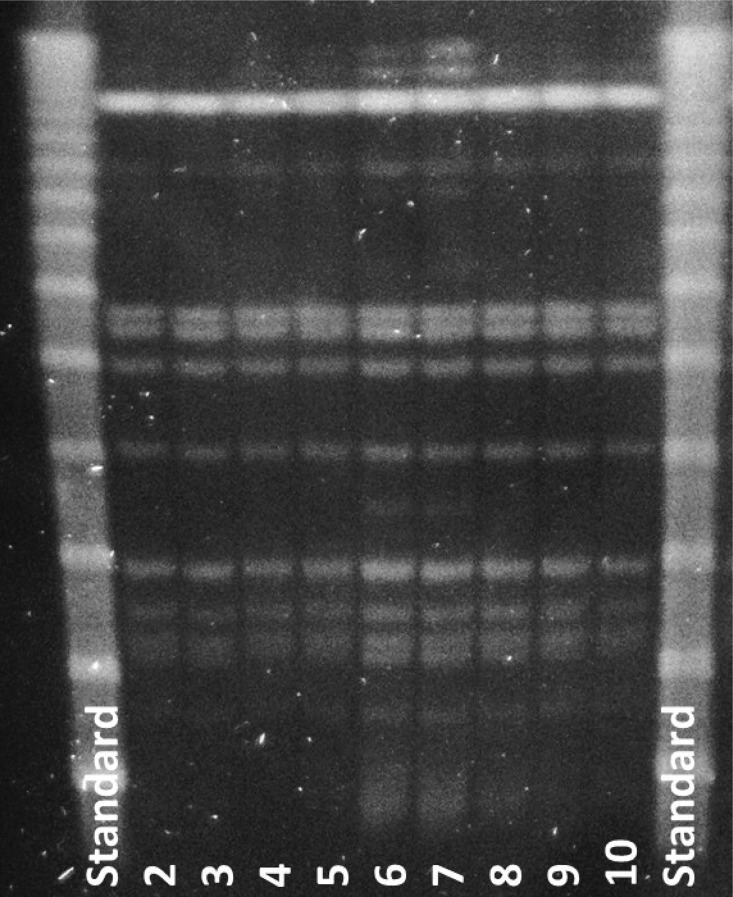
Legionella pneumophila PCR detected genetic material in 34 of 45 samples (76%), 21 of which showed live L. pneumophila, serogroup 1. FLAs were present in 19 of the 45 samples, and intracellular L. pneumophila in 16 of the 19 FLA-positive samples (84%). Among these iLp, three samples showed live L. pneumophila, serogroup 1. All L. pneumophila isolates in the 2013 survey, that is, from both water and FLAs, showed the identical genotype, Lp-gta (Fig. 3). Figure 3 shows the genotype patterns of L. pneumophila population in the potable water distribution system by the time for the third survey.
Fig. 3.
The results from genotyping of random Legionella pneumophila isolates analysed from the 2013 survey. Only one genotype of L. pneumophila (Lp-gta) was seen in the potable water distribution system by the time for the survey. Lanes 1 and 11 are Lambda Stds. Lanes 2–10 are random isolates from the 2013 survey, all isolates Lp-gta.
All legionellosis cases were caused by L. pneumophila, serogroup 1. We have not so far succeeded in tracing the sources of the particular infection isolates, mainly because individuals with legionellosis were all contract personnel working for other companies and/or from other countries.
Discussion
We present data from a vessel that experienced three separate legionella outbreaks in the course of 6 years, in spite of extensive efforts to eliminate legionella from the potable water system. Legionella infections usually present as two distinct clinical entities: Legionnaires’ disease, a severe form of pneumonia with an approximately 30% mortality rate, and Pontiac fever, an influenza-like illness of short duration (4). L. pneumophila is the dominant species in infections caused by the Legionellaceae family, being identified in 95% of outbreaks and sporadic cases of legionellosis. Of these, 85% of the L. pneumophila isolates belonged to serogroup 1 (16, 17). In all the three outbreaks reported here, legionellosis with L. pneumophila, serogroup 1 was the cause of infection.
Even if we have not had the opportunity to genotype the infectious isolates, it seems reasonable to suggest that the genotype established in the potable water system is the causal agent as the persons infected had been on board for weeks before the outbreaks. Based on this assumption, our observations indicate that one genotype of L. pneumophila may change with respect to pathogenicity over time even when growing in a defined microbial environment.
Analyses of the living L. pneumophila isolated from various aerosolized supply points in the freshwater system on every outbreak demonstrated the presence of one and the same genotype. This was also the only genotype detected by the follow-up surveys. Only one genus of FLA (Hartmanella) could be identified in the distribution system. It is worth mentioning that the vessel had been in service all over the world and had thus taken on board water from many bunkering stations. From our experiences regarding bunkering plants around the world, we often see a mix of different L. pneumophila genotypes and different FLAs (e.g. Acanthamoeba, Naegleria, and Hartmannella). The efficacy of this particular vessel's upgraded and improved barrier systems was strengthened by the fact that the microflora in the distribution system remained constant even when the bunkering water was documented to be contaminated with other legionellas and FLA than Hartmanella (data not shown).
After the first two legionella outbreaks, the potable water system was analysed for microbial contamination, followed by treatment with heat, chlorine, and detergents as well as chemical sanitation, for example, hydrochloric acid. Obviously, the treatment and efforts used after the two earlier outbreaks had had no effect on the infectious population.
After the third outbreak in August 2013, a new strategy for sanitation using chlorine dioxide was initiated. Chlorine dioxide has been reported as a treatment for legionella contamination, but to the best of our knowledge, treatment aimed at elimination of FLA from a potable water system has never been reported. Laboratory studies have shown chlorine dioxide to have biocidal effect on FLAs – including both their trophozoite and cyst forms (18).
The ClO2 sanitation process on board this vessel was started in October 2013 by using a mobile ClO2 unit (Desin-Comb). Assessment of effect of the treatment was done every third month by use of full surveys of L. pneumophila /FLA. Status upon start was L. pneumophila in 66% of the water spots, FLAs in 40%, and intracellular L. pneumophila in 80% of the FLAs. All L. pneumophila isolates of one and the same genotype.
Eighteen months later, no L. pneumophila genetic material was detectable and after another 3 months, only a couple of spots still showed FLAs but none containing intracellular genetic material from L. pneumophila. Preliminary data were presented at the 3rd ESGLI Congress, London, September 16–18, 2015 (19).
Our survey strengthens the earlier report from laboratory studies showing biocidal effect on both trophozoites and cysts of the resistant FLAs. Sanitation aimed at elimination of the hosts for potential pathogens in potable water systems is from our point of view a paradigm shift in preventive medicine.
Conclusions
Between 2008 and 2013, the vessel described here experienced three separate outbreaks of legionellosis. The individuals infected were all contract personnel working for other companies and/or from other countries, and we have not succeeded in tracing the sources of the particular infection isolates, but all isolates were L. pneumophila, serogroup 1. Nonetheless, it seems a reasonable assumption that the genotype identified in the potable water system was the cause of these outbreaks. Based on this assumption, the spontaneous outbreaks indicate that one genotype of L. pneumophila may change with respect to pathogenicity/virulence in the course of time, even when it is growing in a defined microbial environment.
Based on this study, and other legionella surveys we have conducted throughout the last decade, we suggest that potable water systems with established but apparently non-infectious populations of a single L. pneumophila serotype 1 genotype together with its survival and propagation hosts represent a potential risk for outbreak of legionellosis and should consequently be a target for elimination using products aimed to eliminate the amoeba population, for example, chlorine dioxide.
Conflict of interest and funding
The authors declare no conflicts of interest.
References
- 1.Fliermans CB, Cherry WB, Orrison LH, Smith SJ, Tison DL, Pope DH. Ecological distribution of Legionella pneumophila. Appl Environ Microbiol. 1981;41:9–16. doi: 10.1128/aem.41.1.9-16.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, et al. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–97. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 3.Brenner DJ, Steigerwalt AG, McDade JE. Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med. 1979;90:656–8. doi: 10.7326/0003-4819-90-4-656. [DOI] [PubMed] [Google Scholar]
- 4.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–26. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–8. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macfarlane JT, Worboys M. Showers, sweating and suing: Legionnaires’ disease and ‘new’ infections in Britain, 1977–90. Med Hist. 2012;56:72–93. doi: 10.1017/S0025727300000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkinham JO, III, Hilborn ED, Arduino MJ, Pruden Amy, Edwards MA. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ Health Perspect. 2015;123:749–58. doi: 10.1289/ehp.1408692. doi: http://dx.doi.org/10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goutziana G, Mouchtouri VA, Karanika M, Kavagias A, Stathakis NE, Gourgoulianis K, et al. Legionella species colonization of water distribution systems, pools and air-conditioning systems in cruise ships and ferries. BMC Public Health. 2008;8:390. doi: 10.1186/1471-2458-8-390. doi: http://dx.doi.org/10.1186/1471-2458-8-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC) Cruise-ship–associated Legionnaires disease, November 2003–May 2004. MMWR Morb Mortal Wkly Rep. 2005;54:1153–5. [PubMed] [Google Scholar]
- 10.Azara A, Piana A, Sotgiu G, Dettori M, Deriu MG, Masia MD, et al. Prevalence study of Legionella spp. contamination in ferries and cruise ships. BMC Public Health. 2006;6:100. doi: 10.1186/1471-2458-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FHI Vannrapport 118. Forebygging av legionellasmitte – en veiledning. Guidance procedures from National Institute of Public Health (in Norwegian), accessed in 2008-2010 at http://www.fhi.no, corresponding to the 1st Edition (December 2007)
- 12.Ahlén C, Aas M, Nor A, Wetteland PI, Johansen H, Sørbø T, et al. Legionella pneumophila in Norwegian naval vessels. Tidsskr Nor Legeforen. 2013;133:1445–8. doi: 10.4045/tidsskr.12.1459. doi: http://dx.doi.org/10.4045/tidsskr.12.145. [DOI] [PubMed] [Google Scholar]
- 13.Stølhaug A, Bergh K. Identification and differentiation of Legionella pneumophila and Legionella spp. with real-time PCR targeting the 16S rRNA gene and species identification by mip sequencing. Appl Environ Microbiol. 2006;72:6394–8. doi: 10.1128/AEM.02839-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann ME. Pulsed-field gel electrophoresis. Methods Mol Med. 1998;15:33–50. doi: 10.1385/0-89603-498-4:33. [DOI] [PubMed] [Google Scholar]
- 15.Winiecka-Krusnell J, Linder E. Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scand J Infect Dis. 1999;31:383–5. doi: 10.1080/00365549950163833. [DOI] [PubMed] [Google Scholar]
- 16.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis. 2002;186:127–8. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 17.European Centre for Disease Prevention and Control. Legionnaires' disease in Europe, 2014. Stockholm: ECDC; 2016. http://ecdc.europa.eu/en/publications/publications/legionnares-disease-europe-2014.pdf. [Google Scholar]
- 18.Dupuy M, Berne F, Herbelin P, Binet M, Berthelot N, Rodier MH, et al. Sensitivity of free-living amoeba trophozoites and cysts to water disinfectants. Int J Hyg Environ Health. 2013;217:335–9. doi: 10.1016/j.ijheh.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Ahlén C, Putzig Y, Krusnell J, Iversen OJ. Elimination of a six year persistent pathogenic population of Legionella pneumophila and its hosts free-living amoeba in a ship potable water system. Paper presented at: 3rd annual conference of the ESCMID Study Group for Legionella Infections (ESGLI); September 16–18, 2015; London United Kingdom. [Google Scholar]