SUMMARY
Candida albicans, the most pervasive fungal pathogen that colonizes humans, forms biofilms that are architecturally complex. They consist of a basal yeast cell polylayer and an upper region of hyphae encapsulated in extracellular matrix. However, biofilms formed in vitro vary as a result of the different conditions employed in models, the methods used to assess biofilm formation, strain differences, and, in a most dramatic fashion, the configuration of the mating type locus (MTL). Therefore, integrating data from different studies can lead to problems of interpretation if such variability is not taken into account. Here we review the conditions and factors that cause biofilm variation, with the goal of engendering awareness that more attention must be paid to the strains employed, the methods used to assess biofilm development, every aspect of the model employed, and the configuration of the MTL locus. We end by posing a set of questions that may be asked in comparing the results of different studies and developing protocols for new ones. This review should engender the notion that not all biofilms are created equal.
INTRODUCTION
Microbial biofilms represent the first and perhaps most successful attempt by organisms to employ multicellularity to succeed in an aquatic environment (1–6). This life form still remains a dominant component of the biota, in some estimates accounting for over half of the earth's present biomass (7, 8). Over 3 billion years ago, the photosynthetic cyanobacteria (blue-green algae) in the oceans converted the early toxic atmosphere to one that better supported modern terrestrial life (9–11). The cyanobacteria formed biofilms that incorporated sediment, generating stromatolites (12, 13), structures that can still be seen on the beaches in Shark Bay, Australia (14–17). These most important and ancient prokaryotes also have a lesson to teach us about the evolution and adaptability of biofilms. Today these organisms can also inhabit terrestrial niches in the form of biofilms, wreaking havoc by etching both ancient stone monuments and modern stone buildings (18–20). And bacteria, including those that colonize humans, continue to live in aquatic environments, primarily as biofilms (4, 21–24). As is the case for bacteria, the fungi, including those that are pathogenic, have also developed the capacity to form biofilms. The major yeast pathogen Candida albicans, the focus of this review, can form biofilms on a variety of host tissues, dentures, teeth, catheters, and other biomedical implants (25–32). Depending on the configuration of the mating type locus (MTL), it can also form alternative biofilms: one exhibiting traits consistent with a “pathogenic” biofilm and the other exhibiting traits consistent with a “sexual” biofilm (33, 34).
But before considering the plasticity of the biofilms formed by C. albicans, the meaning of the term “biofilm” must be considered. Can the term, as applied to prokaryotic communities that rim bodies of water, clog pipes, form on the surface of your pet's water bowl, and obstruct the respiratory tracts of cystic fibrosis patients, have the same meaning in regard to the C. albicans communities that coat dentures and form plaque on teeth, spread along the walls of intestines, and line the walls of catheters? Is the meaning of the term so general that it can apply to any population of microorganisms on a substratum, including a colony of cells on nutrient agar? Or does the term “biofilm” have specific functional and architectural connotations that apply to all species, both prokaryotic and eukaryotic? In delving into the literature on C. albicans biofilms formed in vitro and relating it to that on the more intensively studied bacterial biofilms, it became apparent that the definition of the term had indeed evolved into something more than simply a film or colony of microorganisms supported by a substratum in an aqueous environment. It connoted a community of interacting cells, anchored tightly to a substratum, made up of multiple cell phenotypes embedded in a self-generated extracellular matrix (ECM). It connoted a population in a self-established microenvironment with specialized functional and architectural characteristics and a mechanism for dispersal. And as is the case for all communities, there appeared to be the potential for a significant degree of plasticity, depending upon the genotype and different environmental pressures. It became clear that there is a lack of appreciation of the variability of C. albicans biofilms due to strain differences and the plasticity of these biofilms resulting from different conditions employed in vitro. Attempting to integrate data from different studies that employed different strains, different culture conditions, and different quantitative methods of assessment can indeed lead to problems of interpretation without considering the possibility that the biofilm preparations might differ physiologically and developmentally. There can also be a major problem in interpreting the body of knowledge that has emerged for bacterial biofilms formed in vitro (35–37). The problems for C. albicans are particularly poignant when attempts are made to generate general models of biofilm development, signal transduction pathways regulating biofilm formation, and drug resistance. The problem is further exacerbated for C. albicans by the discovery that depending upon the configuration of the mating type locus (MTL) (a/α versus a/a or α/α), cells form biofilms that have distinctly different functional characteristics (38, 39). In reviewing the literature, it became clear that researchers rarely test whether the strain they employ is representative and rarely assess the configuration of the MTL locus. For C. albicans, the latter omission has not had a profound effect on most studies, since 90% to 95% of strains are MTL heterozygous (a/α) (40–42). However, for the closely related species C. dubliniensis, which shares most of the developmental processes exhibited by C. albicans (43–46), the omission is indeed worrisome given that one-third of all natural strains are MTL homozygous (44).
The goal of this review is to consider the variability and plasticity of C. albicans biofilms formed in in vitro models, with the intent of engendering awareness that they are developmentally complex and, depending on the strain and culture conditions, not created equal. This review does not cover in detail the literature on the signal transduction pathways that regulate gene expression during biofilm formation, the genes that are differentially regulated, or the molecular mechanisms regulating matrix formation or drug resistance, unless that information pertains to the specific aim set forth. The reader is directed to a number of excellent reviews on these subjects, published in the past several years (47–55).
DEFINING A C. ALBICANS BIOFILM: LESSONS FROM BACTERIA
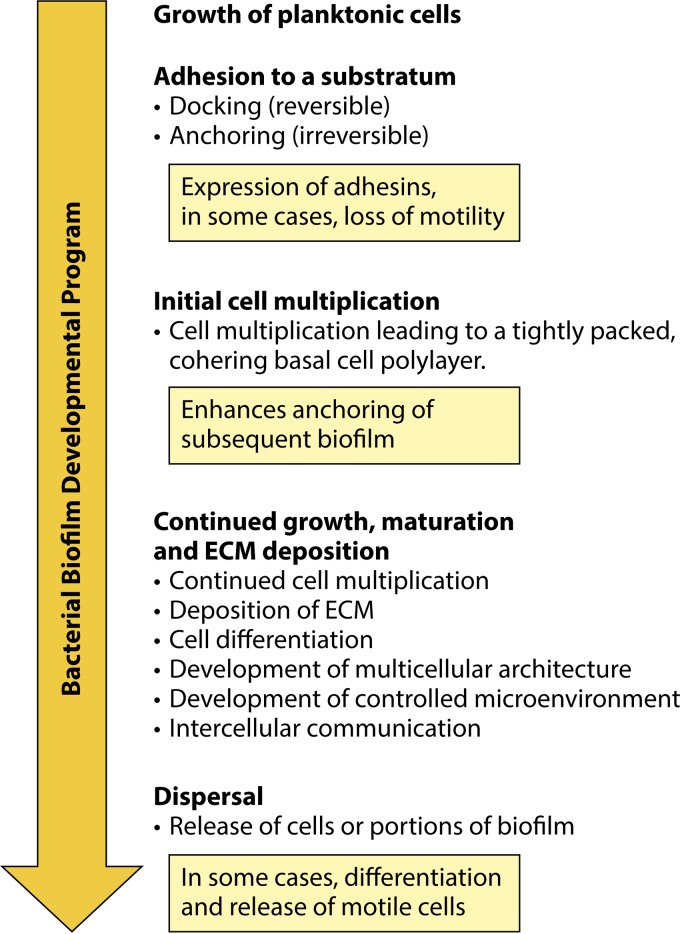
Considering what a C. albicans biofilm represents may best be accomplished by first reviewing bacterial biofilms, given that detailed studies of their formation in vitro appeared in the literature approximately 2 decades prior to studies of C. albicans biofilms. The first report of the cellular nature of a microbial biofilm was that of Antonie van Leeuwenhoek in 1684, when he described to the Royal Society of England the variety of microorganisms in dental plaques, which he examined with his newly developed microscope (56, 57). However, not until the 1940s did researchers realize that the bacterial slime covering surfaces in aquatic ecosystems represented more than disorganized detritus. Rather, they were complex and organized communities of cells (58, 59). In 1978, J. W. Costerton and his coworkers (60, 61) published the first formal description of a bacterial biofilm in researching how bacteria adhered to surfaces in aquatic systems. They defined a biofilm as a matrix-encapsulated population of microorganisms adhering to a surface in a nutrient-sufficient environment. Subsequently, they and others described the fundamental differences between bacteria in biofilms and bacteria that are free-living (planktonic) (62–64). They noted (60) that over 99% of the bacterial biomass in the natural aquatic ecosystems they first studied were in the form of biofilm communities on wet or submerged surfaces, rather than free and independent in suspension. Because the bacteria include so many species, one general biofilm description cannot encompass the variety of their forms. However, Costerton and others extracted a number of generally common steps in the formation of bacterial biofilms in vitro, as well as several shared attributes (21, 65–67), which are outlined briefly in Fig. 1. As will become evident in this review, many of the steps of bacterial biofilm formation and the attributes of these biofilms have proven relevant to the emerging descriptions of C. albicans biofilm formation in vitro. The first step in generating a bacterial biofilm in vitro is to grow cells planktonically for inoculation into the biofilm model. The concentration of cells in the inoculum can be adjusted to generate an initial monolayer on the substratum of the model employed. The second step is to add the inoculum of planktonic cells to an adherent substratum to facilitate adhesion. Strong adherence is necessary for anchoring, especially when there are fluid flow and mechanical shear forces that could release single cells not firmly attached to the substratum during initial biofilm development. It should be noted that although most in vitro models include a solid substratum for adherence, cells in natural settings may also converge at air-liquid or liquid-liquid interfaces, where differences in density, hydrophobicity, and water structure can result in a discrete physical interface that can be assessed by cell surface receptors (68, 69). Cells may also use each other as surfaces, through cohesion, to form suspended aggregates, or floccules, exhibiting properties of a biofilm on a substratum (70–73). In vitro, adherence to a substrate is basic to the establishment of a geographically confined population. Bacterial adhesion has been separated into two phases: primary, reversible adhesion, sometimes referred to as “docking,” and secondary, irreversible adhesion, sometimes referred to as anchoring or “locking” (74–77) (Fig. 1). For biofilm formation in vivo, a question arises as to whether one cell, a number of independent cells, or a fragment of a previous biofilm is responsible for dissemination, i.e., the initiation of a new biofilm in another location (60, 78, 79). In all three scenarios, however, the first step must still be adhesion to the substratum. In the process of adhesion, an adhesin or another form of a mechanoreceptor activates signal transduction pathways that effect the developmental transition from a planktonic state to a sessile, immobilized state (67, 80, 81). This transition can be associated with a change in cellular phenotype. For instance, when motile bacteria, such as Escherichia coli, adhere to a substratum (33, 82, 83), motility ceases, cell multiplication continues (84–87), and early biofilm-related genes are upregulated, including those that encode additional adhesins (88–90). Although Karatan and Watnick (91) suggested that bacteria at this point can form either an adherent monolayer biofilm or a multilayer biofilm, the former is not sufficient to encompass some of the main characteristics of a complex biofilm, most notably the formation of ECM and a three-dimensional, controlled microenvironment that facilitates biofilm growth, nutrient acquisition, control of gas and pH, and cell-cell signaling (92–97). It therefore may not be legitimate to consider an adhering cell monolayer lacking ECM a bona fide biofilm. Once cells adhere to a surface and begin to multiply, they remain aggregated both through adherence to the substratum and through coherence (cell-cell adhesion) (91, 98). Coherence may be facilitated by several mechanisms, including direct cell-cell adhesion, the formation of an encapsulating ECM that mechanically traps cells in place, or the formation of an ECM that acts as glue (99–101). The adhering cells then multiply in the z axis to form a tightly packed, cohering basal polylayer of cells that further anchors the biofilm (Fig. 1). It must be realized at this point that the final architecture of the in vitro biofilm depends on the original density of adhering cells. Low-density inocula that result in sparse, independent cells on the substratum may form patches of cells or independent cell mounds, while high-density inocula may form uniform carpets of cells that result in a uniform biofilm across the substratum. While bacteria multiply, they deposit ECM. The final thickness of the mature biofilm appears to be the result of a variety of possible pressures. These include the mass that can be anchored effectively to a substrate relative to shear force (102), the penetrability of nutrients (103), gas exchange (104), the concentrations of secreted enzymes for extracellular digestion of nutrients and for matrix modification (105), and, in vivo, the topography of the substratum to which the biofilm adheres (106–108). Intercellular signaling develops gradually and controls the size of the final biofilm (109), and it may also play a role in the development of late biofilm architecture. As the biofilm evolves, it undergoes cellular differentiation and multicellular morphogenesis. In select bacteria, there is stratification of cellular phenotypes and the development of an often complex multicellular architecture (110–113). In Pseudomonas aeruginosa, cells differentiate from a round to an elongated phenotype (113), and they exhibit changes in physiology, gene transcription, and protein synthesis (81, 114–116). One of the most dramatic phenotypic transitions is that of E. coli (80, 86) and P. aeruginosa (81, 117, 118). Both are motile when planktonic but become nonmotile during adherence, and as noted, the flagellar motor can play a direct and early role in the initial adhesion event (119–122). As P. aeruginosa biofilms grow, the cells remain nonmotile, but at maturity, mushroom-like pillars separated by water channels form within the biofilms, which contain motile cells that exit in the process of dispersal (81, 118, 123, 124). Not only the size of the biofilm but also multicellular morphogenesis is regulated by quorum sensing (125–127). Cell death, which plays a major role in vertebrate development, most notably in limb formation (128, 129), may also play a role (130–132). In the case of Bacillus subtilis, which makes wrinkled colonies on agar, areas of localized cell death underlie areas of buckling.
FIG 1.
General steps in the formation of a bacterial biofilm. The general developmental program provides us with a contextual framework for defining a C. albicans biofilm, since it includes a number of analogous steps.
During bacterial biofilm growth and maturation, cells continue to deposit an extracellular matrix, which plays roles in cell cohesion (106, 133, 134), gas exchange (104), drug resistance (21, 135–137), defense against penetration by white blood cells (138–142), and the maintenance of architecture and biofilm rigidity (106, 143, 144). The ECM is hydrated and therefore is composed primarily of water. The matrix that supports the gel is composed of structural proteins and polysaccharides and contains extracellular DNA (eDNA) (145–151). Recently, McCrate et al. (152) demonstrated that the ECM of E. coli biofilms contains cellulose and curli proteins, with the latter forming fibrous amyloid bodies. Studies combining electron microscopy, biochemistry, and solid-state nuclear magnetic resonance (NMR) spectroscopy have led to the perception that the major component of ECM is in fact protein (153–156). Studies have also demonstrated that the protein-cellulose architecture is responsible for the viscoelasticity of the ECM (154, 155). It is likely that the composition and architecture of the ECM change with developmental time. In the complex matrix of a bacterial biofilm, a microenvironment is established that has been shown to contain secreted enzymes for catalyzing the polysaccharide linkages and modifications that are basic to matrix architecture and function (106, 144, 157, 158) and secreted enzymes involved in the extracellular digestion of nutrients from the host environment or from the medium in an in vitro model (21, 159, 160). The role of eDNA is unclear, but experiments suggest that it indeed plays a role in biofilm formation in vitro (151, 161). The ECM contributes to the establishment of a microenvironment that facilitates cell-cell communication through signaling (109, 162, 163). Finally, the mature bacterial biofilm must maintain itself, rejuvenate, and release cells or biofilm fragments for dispersal (67, 106, 164). Bacterial biofilms have also been shown to facilitate conjugation, the bacterial version of mating, through the transfer of DNA between cells through direct cell-cell contact (165–167). As noted, many of the developmental steps and traits of bacterial biofilm formation in vitro have analogs in the formation of a complex C. albicans biofilm in vitro. Formation of a bacterial biofilm therefore provides a contextual framework for defining a C. albicans biofilm, as well as assessing variability and plasticity.
THE BEGINNINGS OF CANDIDA ALBICANS BIOFILM RESEARCH
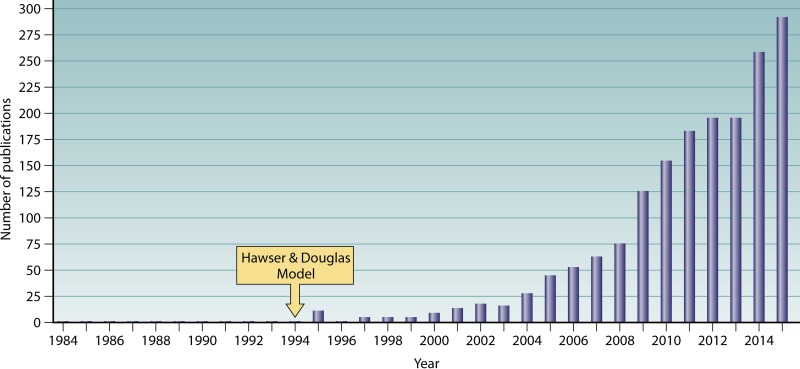
It took more than 15 years after Costerton et al.'s pioneering work on bacterial biofilms (60) for medical mycologists to recognize the possible relevance of biofilm formation in the life history of C. albicans and to develop an in vitro model (168). Even the comprehensive review and bibliography of C. albicans literature by Frank C. Odds, in his second edition of the book Candida and Candidosis: a Review and Bibliography, published in 1988, failed to have “biofilm” as a topic in the table of contents or index (169). This monumental work, which included a review of 5,796 publications, did, however, include a review of the literature on the adherence of C. albicans to host and synthetic surfaces, the first step in biofilm formation. The importance of the subject was underscored by its placement as the first comprehensively covered subject in the section “Determinants of Virulence of Candida” in chapter 26, “Pathogenesis of Candidosis,” in Frank Odds' book (169). In this review, which covered the literature up through 1986, the range of biological surfaces to which C. albicans adhered, sometimes selectively, was extensive. These surfaces included buccal epithelia, vaginal epithelia, cervical epithelia, skin cells, urinary epithelia, gastrointestinal epithelia, endothelia, and even spermatozoa (169). Synthetic surfaces included catheters, dentures, and contact lenses (169). Possibly one of the first images of a biofilm formed by a Candida species was published in 1984 by Marrie and Costerton (24). Candida parapsilosis was identified in a sample from a catheter wall. Yeast cells and extensive matrix were observed in the biofilm, but no hyphae were present, suggesting that it represented the smooth switch phenotype of this species (170). One of the first reports of the association of C. albicans with a biofilm was in a study by Ahearn and colleagues over 25 years ago (171). They assessed the efficacy of disinfectants and air drying on the clearing of biofilms from contact lens cases. They noted that C. albicans accounted for only a small fraction of the cells of the predominately bacterial biofilm. Like the report of Ahearn and coworkers (171), there were several additional reports in the early 1990s of C. albicans in mixed biofilms forming on a variety of materials, including silicone rubber prostheses (172), impression materials (173), catheters (174), and dentures (175–177). However, it was Hawser and Douglas (168) who developed the first in vitro model of C. albicans biofilm formation, in which the biofilm was formed by and composed solely of that species. The development and use of an in vitro model composed exclusively of C. albicans were the first steps in understanding how this species forms biofilms. The model was adapted from one developed in the late 1980s by Prosser et al. (178) for the formation of E. coli biofilms in vitro. However, during the 5 years following the landmark paper by Hawser and Douglas (168), the number of published studies was fewer than 10 per year, on average (Fig. 2). Medical mycologists then realized the importance of biofilm formation in C. albicans colonization, and the number of published studies increased in an exponential fashion, such that by the year 2015, the number of papers per year approached 300 (Fig. 2).
FIG 2.
The approximate number of papers published per year on Candida albicans biofilms began to increase at a nearly exponential rate after the development of the first in vitro model by Hawser and Douglas, from 1994 to 1998.
THE DOUGLAS MODEL: A GOOD STARTING POINT
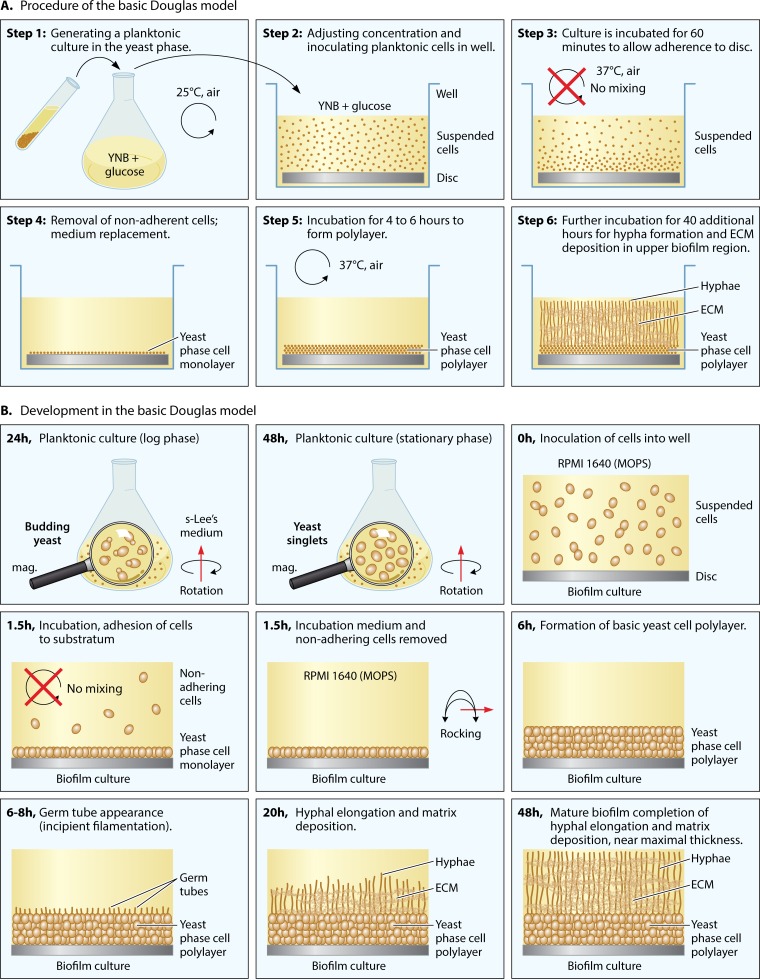
As we shall see, the original Douglas model continues to be used broadly, with lab-specific variations, because it supports the formation of a mature biofilm that is robust, thick, architecturally complex, and phenotypically and genotypically homogeneous, at least initially. In the initial protocol of Hawser and Douglas from 1994 (168), yeast-phase cells were first grown overnight in suspension (i.e., planktonically) in yeast nitrogen base (YNB) medium (Fig. 3A, step 1). The concentration of the planktonic cell culture was adjusted for optimum biofilm formation by measuring the optical density at 520 nm (OD520), and then aliquots were inoculated into YNB medium in the wells of a microtiter plate (Fig. 3A, step 2). The composition of YNB medium, which usually contains glucose as the major carbon source, is presented in Table 1. A disc cut from catheter material that had been treated with serum was inserted at the bottom of each plastic well of the multiwell plastic culture plate. The planktonic cell inoculum was incubated in the wells for 60 min at 37°C in air without mixing (i.e., static, without rotation or rocking) to facilitate adhesion to the disc (Fig. 3A, step 3). Nonadhering cells were then gently removed and fresh YNB medium added (Fig. 3A, step 4). In the original model, cultures were incubated statically for up to 72 additional hours (168). In a subsequent report (179), cultures were continuously mixed by rotation for 47 additional hours (Fig. 3A, steps 5 and 6). Aspects of this model were changed over time by other research groups. The plastic bottoms of the wells of multiwell tissue culture dishes, rather than serum-treated catheter discs, were employed as substrata for purposes of expediency, since tissue culture plastic proved to be adhesive (180–182). YNB medium was also replaced with different media to support planktonic growth and biofilm formation (38). In Table 1, the compositions of a number of media used to develop biofilms are presented, demonstrating the variety of media and the potential for biofilm variation.
FIG 3.
The Douglas model, established by Hawser and Douglas (168), has served as the basis for a number of models with variations in one or more conditions. (A) Procedure used by Hawser et al. (179). (B) Representation of the cellular and architectural development of a C. albicans biofilm, using a variation of the Douglas model in which sLee's medium was used to grow cells planktonically, RPMI 1640 (MOPS) medium was used to support biofilm development, and the cultures were rocked to mix the media (38). In panel B, cells in the yeast-phase cell monolayer and polylayer are represented by spheres, germ tubes are represented by tubes, hyphae are represented by vertical tubes, and ECM is represented by gray fibers.
TABLE 1.
Compositions of media used to grow planktonic cultures and to support biofilm formation in Candida albicans biofilm models
| Mediuma | Carbon source (per liter) | Remaining contents (per liter) |
|---|---|---|
| YNB medium | 9 g dextrose | 2 μg biotin, 400 μg calcium pantothenate, 2 μg folic acid, 2,000 μg inositol, 400 μg niacin, 200 μg p-aminobenzoic acid, 400 μg pyridoxine hydrochloride, 200 μg riboflavin, 400 μg thiamine hydrochloride, 500 μg boric acid, 40 μg copper sulfate, 40 μg potassium iodide, 200 μg ferric chloride, 400 μg manganese sulfate, 200 μg sodium molybdate, 400 μg zinc sulfate, 1 g potassium phosphate monobasic, 500 mg magnesium sulfate, 100 mg sodium chloride, 100 mg calcium chloride, 10 mg l-histidine-HCl, 20 mg methionine, 20 mg tryptophan 2 |
| YPD | 20 g dextrose | 20 g Bacto peptone, 10 g yeast extractb |
| sLee's medium | 12.5 g dextrose | 5 g NaCl, 5 g ammonium sulfate, 2.5 g potassium phosphate dibasic, 1.3 g l-leucine, 1 g l-lysine, 0.5 g l-alanine, 0.5 g l-phenylalanine, 0.5 g l-proline, 0.5 g l-threonine, 0.2 g magnesium sulfate, 0.1 g l-methionine, 70 mg l-ornithine, 70 mg l-arginine, 1 mg d-biotin, 0.1 mM zinc sulfate |
| Spider medium | 10 g mannitold | 10 g nutrient broth,c 2 g K2HPO4 |
| SD medium | 20 g dextrose | 1.45 g yeast nitrogen base without amino acids and ammonium sulfate, 5 g ammonium sulfate, 50 mg uracil, 110 mg l-tryptophan, 180 mg l-leucine, 90 mg l-histidine, 30 mg l-methionine |
| Sabouraud's medium | 20 g glucose | 10 g Bacto peptone |
| RPMI 1640 (MOPS) mediume | 2 g dextrose | 0.1 g calcium nitrate·4H2O, 48 mg magnesium sulfate, 0.4 g potassium chloride, 2 g sodium bicarbonate, 6 g sodium chloride, 0.8 g sodium phosphate dibasic, 0.2 g l-arginine, 50 mg l-asparagine, 20 mg l-aspartic acid, 65.2 mg l-cystine·2HCl, 20 mg l-glutamic acid, 10 mg glycine, 15 mg l-histidine, 20 mg hydroxy-l-proline, 50 mg l-isoleucine, 50 mg l-leucine, 40 mg l-lysine·HCl, 15 mg l-methionine, 15 mg l-phenylalanine, 20 mg l-proline, 30 mg l-serine, 20 mg l-threonine, 5 mg l-tryptophan, 28 mg l-tyrosine·2Na·2H2O, 20 mg l-valine, 0.2 mg d-biotin, 3 mg choline chloride, 1 mg folic acid, 35 mg myo-inositol, 1 mg niacinamide, 1 mg p-aminobenzoic acid, 0.25 mg d-pantothenic acid, 1 mg pyridoxine·HCl, 0.2 mg riboflavin, 1 mg thiamine·HCl, 5 mg vitamin B12, 1 mg glutathione (reduced), 5.3 mg phenol red·Na, 0.3 g l-glutamine, 35 g MOPS |
YNB, yeast nitrogen base, defined; YPD, yeast extract-peptone-dextrose, undefined; Spider medium, undefined; SD medium, synthetic defined medium, defined; Sabouraud's medium, undefined; RPMI 1640 (MOPS), defined; sLee's medium, Lee's medium (185) supplemented with arginine, biotin, and zinc according to the method of Bedell and Soll (186), defined.
Bacto peptone is produced by peptic digestion of animal tissue. Yeast extract is made by extracting the yeast cell content from the cell wall. It is undefined.
Nutrient broth is composed of the following per liter: 1 g beef extract, 2 g yeast extract, 5 g peptone, and 5 g NaCl. It is undefined.
Note that Spider medium is the only medium that contains mannitol, a sugar alcohol, rather than dextrose as a carbon source. Mannitol is produced by plants, algae, fungi, and select bacteria but is not produced in abundance by humans.
RPMI 1640 medium without MOPS is referred to as RPMI 1640, and that with MOPS is referred to as RPMI 1640 (MOPS).
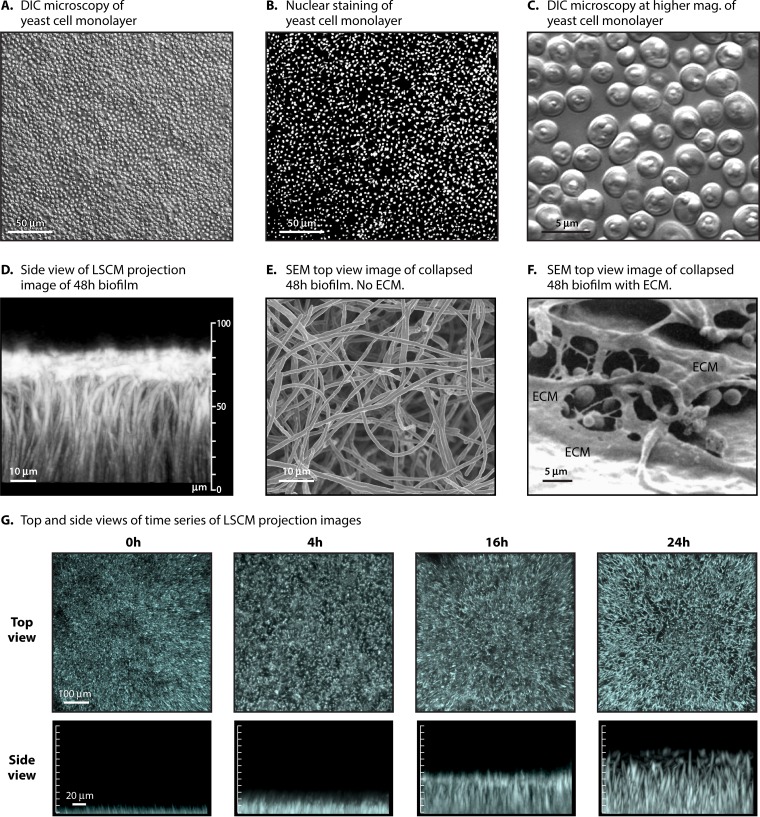
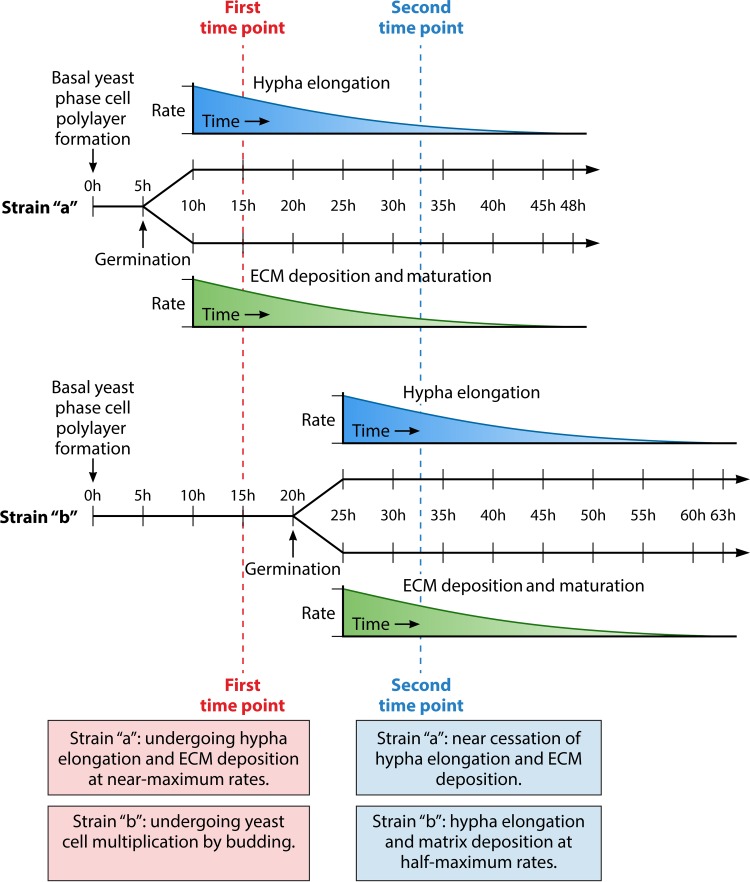
The general developmental program of biofilm formation in the Douglas model, in this case using an adaptation developed by Daniels et al. (38), is diagrammed in Fig. 3B. This variation is employed because the architecture of the biofilm formed has been assessed over time in detail (38, 45, 183, 184). In this version of the Douglas model, planktonic cells are grown to stationary phase in sLee's medium to obtain a majority of unbudded yeast-phase cells in stationary phase. sLee's medium is Lee's medium (185) supplemented with zinc and arginine, according to the method of Bedell and Soll (186) (Table 1). Since biofilm formation occurs more robustly in RPMI 1640 medium buffered with morpholinepropanesulfonic acid [RPMI 1640 (MOPS)] (Table 1), this medium was used by Daniels et al. in the actual biofilm model in the microtiter wells (38). The substratum in this model is a disc cut from silicone elastomer, a catheter material, and the incubation conditions after adhesion include air at 37°C, with gentle rocking (30° deflection, 18 cycles/min) (38, 183, 184). The cells that adhere to the catheter disc between 0 and 1.5 h of static incubation form a relatively uniform, contiguous monolayer if the cell concentration is adjusted correctly [e.g., 1 × 107 cells in 2 ml of RPMI 1640 (MOPS) medium added to a well containing a 15.6-mm-diameter silicone elastomer disc]. In Fig. 4A, a differential interference contrast (DIC) micrograph is presented of the yeast cell monolayer formed after 1.5 h (Fig. 3B). A nearly confluent, uniform sheet of adherent cells is apparent. In Fig. 4B, fixed cells are stained with the DNA dye Syto 9 (Life Technologies, Inc.) to visualize nuclei. In Fig. 4C, a higher-magnification DIC image is presented of live cells in the monolayer, which reveals the uniform unbudded yeast-phase phenotype. The cells in the monolayer then multiply in the yeast phase over the next 4 to 6 h, forming a basal yeast cell polylayer that is approximately six cells thick (∼20 μm) (Fig. 3B). Between 6 and 8 h, germ tubes (incipient hyphae) emerge at the dorsal surface of the yeast cell polylayer (Fig. 3B). During the subsequent 40 h, the germ tubes grow in the z axis, forming true hyphae (Fig. 3B). Elongation in this model is oriented vertically, slowing with time as the biofilm matures (184). In Fig. 4D, a side view is presented of a projection image of 500 scans obtained by confocal laser scanning microscopy (CLSM) of a 48-h biofilm of strain SC5314. The image was processed with Imaris 3D image processing and analysis software (Bitplane, Zurich, Switzerland) (45) to accentuate the vertical orientation of the hyphae in the mature biofilm in this model. In Fig. 4E, a scanning electron micrograph (SEM) is presented from the top of a collapsed 48-h biofilm prepared such that the extracellular matrix was removed (note the homogeneity of hyphae in the top-view image) (45). In Fig. 4F, an SEM is presented from the top of a collapsed 48-h biofilm prepared to preserve the ECM (179). Note how the hyphae are covered by the dehydrated ECM. In Fig. 4G, a time series is presented of top views and side views of CLSM projection images of biofilm formation through 24 h. Note that the hyphae appear punctate in the 16- and 24-h top-view images, since they are oriented vertically (Fig. 4G). The individual hyphae composing the upper biofilm layer each consists of a linear sequence of highly elongated cellular compartments separated by septa. Each hypha grows apically, and each of the cell compartments in the linear array preceding the dividing apical compartment contains one or more large vacuoles, a nucleus, and minimal particulate cytoplasm. These true hyphae are distinct from pseudohyphae in that they lack indentations at septal sites (187). In this model, hyphae continue to grow vertically and apically for approximately 40 h (38, 183, 184). Very little branching or lateral yeast cell formation occurs (Fig. 4D). In other media, such as Spider medium, yeast cell formation and branching occur (183), in both cases, from the distal end of internal hyphal compartments, just below the septal junction (169).
FIG 4.
Microscopic images of yeast-phase cells and nuclei in a relatively confluent monolayer, the hyphal upper layer, and both top and side images of confocal projections of biofilms forming in the Douglas model, using planktonic cells grown in sLee's medium, RPMI 1640 (MOPS) medium for biofilm formation, and a silicone elastomer disc as the substrate. (A) Differential interference contrast (DIC) microscopy image of an adhering yeast-phase cell monolayer after 90 min of incubation of planktonic cells in the model well. (B) Nuclear staining with Syto 9, imaged by fluorescence microscopy. (C) Increased magnification of the monolayer formed after 90 min, imaged by DIC microscopy. (D) Side view of a projection image of 500 CLSM scans of a 48-h C. albicans biofilm stained with calcofluor. (E) SEM image of a 48-h C. albicans biofilm prepared using methods that removed the ECM from a collapsed biofilm. (F) SEM image of a 48-h C. albicans biofilm prepared using methods that left the collapsed ECM adhering to the hyphae and substratum. (Republished from reference 179 with permission of the Society for General Microbiology; permission conveyed through Copyright Clearance Center, Inc.) (G) Top and side views of projection images, each composed of 500 CLSM scans, of developing biofilms prepared using a variation of the Douglas model, at 0, 4, 16, and 24 h.
Hawser and Douglas (168) showed in their original model that growth (assessed by dry weight), protein synthesis ([3H]leucine incorporation), and metabolic activity (measured by the reduction of tetrazolium salts) in static cultures in YNB medium at 37°C in air, is approximately 75% complete after 24 h of incubation and over 90% complete by 48 h of incubation. The kinetics of these parameters (168) correlate with the gradual decrease in the rate of biofilm growth over time in the modified model of Daniels et al. (184) (Fig. 4G). Hawser and Douglas (168) found that in YNB medium, these kinetics were similar whether 50 mM glucose or 500 mM galactose was used as the carbon source, but the final values at ≥48 h for all three parameters for biofilms formed in medium containing galactose were twice those for biofilms formed in YNB medium containing glucose. Hawser et al. (179) subsequently found that rotating (20 rpm) the biofilm culture to mix the medium increased the final dry weight. There was no effect on the reduction of tetrazolium salts, but as discussed in a later section of this review, this assay has been found to be an expedient but sometimes problematic measure of biofilm growth and development. Using SEM, Hawser et al. (179) reported that low rates of rotation during biofilm formation also increased ECM deposition. Daniels et al. (183), in their modified version of the Douglas model, showed by CLSM of calcofluor-stained preparations, that the upper portion (upper 70 to 80%) of a 48-h biofilm was composed of vertically oriented hyphae equidistant from each other, presumably separated from one another by the encapsulating ECM.
BIOFILM DISPERSAL
Because the model of biofilm development described in Fig. 3B is limited to cell phenotype and architecture, there are many aspects of biofilm formation that are not included. One such aspect is dispersal, which may have an architectural component. The first experimental demonstration of this process was by Uppuluri and López-Ribot (188, 189). Using a flow model, described in a later section of this review, they found that yeast-phase cells were released from a developing biofilm. These cells exhibited several traits that were enhanced compared to those of planktonic cells, including increased pathogenicity in a mouse model of systemic infection (188). They found that between 1 and 8 h of biofilm development, the rate of yeast-phase cells released into the medium increased by 1 order of magnitude. The rate plateaued at approximately 24 h (188). Nobile et al. (190) subsequently demonstrated, in a variation of the Douglas model using Spider medium, that the rate of accumulation of budding yeast cells in the medium decreased between 24 and 60 h. It was not clear whether the yeast-phase cells accumulating in the medium originated in the adhesive yeast cell basal layer, were released from the hyphal upper layer, or were released from yeast cell pockets within the hyphal upper region of the developing biofilm. The lowest rate of release was observed in RPMI 1640 (MOPS) medium, which is consistent with the observation that biofilms formed in this medium contain vertically oriented hyphae with few lateral yeast cells (45, 183, 184). This is therefore consistent with the suggestion that dispersed cells may be formed by hyphae in media that facilitate lateral yeast cell formation. There appears to be no time component for release of yeast-phase cells that can be correlated with an architectural transition in the developmental program of biofilm development, so dispersal was not incorporated into the temporal scheme shown in Fig. 3B.
OTHER MODELS
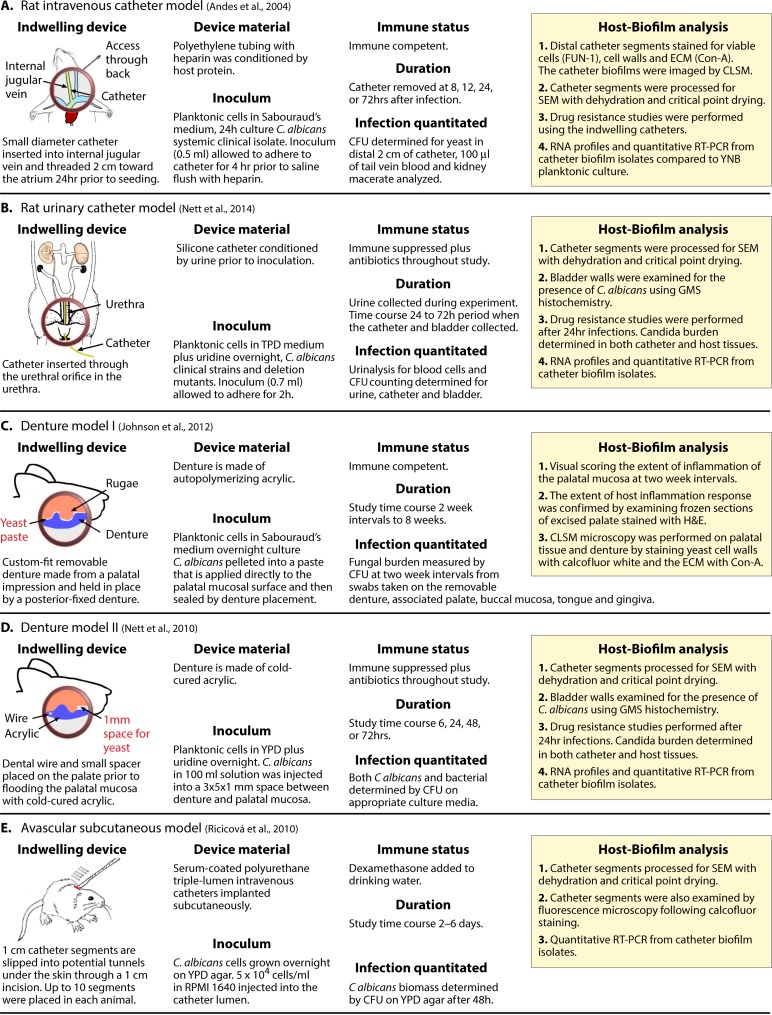
A majority of the procedures that research groups have used to generate biofilms in vitro are based on the Douglas model. However, there are several models, including those incorporating flow and those performed in an in vivo setting, that are markedly different. Andes and coworkers (192) and Ghannoum and coworkers (191) developed in vivo intravenous catheter models, and Andes and coworkers (187) developed an in vivo urinary catheter model. In the original intravenous model developed by Andes et al. (31, 192) (Fig. 5A), a polyethylene tube was inserted into the jugular vein of a rat, conditioned by back flow (without the addition of C. albicans for 24 h), and flushed. It was then inoculated with C. albicans cells obtained from colonies grown on Sabouraud's dextrose agar. Upon inoculation, the catheter was locked and incubated for up to 72 h. At the termination of the experiment, the catheter was flushed with heparinized saline to remove blood and nonadhering cells. The catheter was then assessed for biofilm formation, and the bloodstream and kidneys of the rat were assessed for fungal load. The biofilm in the catheter was visualized on the lumen surface by SEM (192), which entails fixation, critical point dehydration, and coating with palladium and gold. SEM analysis revealed that the biofilms that formed contained yeast-phase cells, pseudohyphae, and hyphae embedded in a dense ECM (192). Hyphae dominated the outer region of the biofilm, but the actual architecture was difficult to assess, in part because the hyphae and matrix were collapsed and matted, presumably as a result of SEM fixation, as was the case in SEMs of mature biofilms presented by Hawser et al. (179) for the Douglas model (Fig. 4F). Metabolically active cells in the biofilm were assessed by use of the fungal viability stain FUN-1, and the polysaccharide content was determined by staining with the lectin concanavalin A (ConA), which identifies α-d-mannosyl- and α-d-glucosyl-containing molecules. Polysaccharide localization appeared to increase in the direction of the catheter lumen. Pockets of matrix were also localized close to the catheter wall. The architectural differences between the biofilm formed in this model and those formed in the Douglas model were considered by the authors (192) but were difficult to assess, although it is safe to say that both biofilms contained the same general cellular phenotypes and ECM. The intravenous catheter model developed by Andes and coworkers (Fig. 5A) (192) has proven extremely valuable in molecular experiments and appears to be excellent for investigating a major medical problem, i.e., catheter-based C. albicans infections. The model has been used successfully to assess the role of adhesion molecules (193), drug susceptibility (194), gene expression (195, 196), and select mutants (193, 197–202). This model has been used almost exclusively by the Andes laboratory at the University of Wisconsin, in a number of cases in collaborative studies, and has not been adopted generally, probably as a result of the expertise required rather than its efficacy. This observation is in no way meant to detract from the value of this model. Schinabeck et al. (191) developed an intravenous catheter model in the jugular vein of a rabbit. The catheter was flushed with saline every day and removed at 7 days. SEMs revealed dense matrix and the contours of yeast-phase cells, but there were no signs of hyphae or pseudohyphae. This model has also not been adopted generally, for the same reasons noted for the model of Andes et al. (36), but it also appears to be an excellent one for studying catheter infections.
FIG 5.
Catheter, denture, and subcutaneous models developed for C. albicans biofilm formation. Panels cite references 192 (A), 203 (B), 211 (C), 210 (D), and 208 (E).
A urinary catheter model, also developed by Andes and coworkers (203) and diagrammed in Fig. 5B, involved inserting a silicone catheter into the urethra of an immunosuppressed rat treated with antibiotics (Fig. 5B). The catheter was conditioned by draining urine from the bladder prior to inoculation of yeast-phase cells suspended in yeast extract-peptone-dextrose (YPD) medium (Table 1). After 2 h, the inoculum with nonadhering cells was removed, the rat returned to the cage, and urine collected throughout the experimental time course. At 24 and 72 h, catheters were analyzed by SEM (187). Because of fixation, staining, and dehydration in the SEM procedure, the architecture of the biofilm formed on the catheter wall is difficult to compare with that formed in the Douglas model, but it is clear in the published images that the upper region of the biofilm was composed mainly of hyphae and matrix. Viable cells were quantitated in the collected urine and in the biofilm population adhering to the catheter. Biofilms reached confluence at 48 h, but the C. albicans burden in the urine increased through 72 h, indicating continued release and/or multiplication of released cells. The urinary catheter model warrants in vitro comparisons in which the medium in the Douglas model is replaced with synthetic urine (SU) medium (204) or fresh filtered donor urine. Using the XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxianilide] reduction assay, Uppuluri et al. (204) demonstrated that biofilms formed in SU medium were as metabolically active as those grown in RPMI 1640 medium, although in SU medium, the final concentration of cells was lower, the hyphae were shorter, the biofilm architecture was less complex, and the expression of select filamentation-associated genes (EFG1, ALS3, and HWP1) was reduced. As is the case for the intravenous catheter model, the urinary catheter model developed by Andes and coworkers (188, 203, 205, 206) is extremely effective for studying urinary catheter infections. In a subcutaneous model, van Dijck and coworkers (207, 208) seeded pieces of catheter with implanted C. albicans yeast cells subcutaneously in the back of a mouse (Fig. 5E). After 6 days, the catheters could be removed and analyzed. Cells formed a dense biofilm, composed of yeast cells, hyphae, and ECM, on the inner tube surface (208). Alternatively, biofilms could be assessed by using strains expressing a luciferase gene attached to the cell wall and noninvasive bioluminescence imaging (207). The technology for this preparation appears to be relatively simple and should be useful in in vivo studies of drug susceptibility.
Models have also been developed in which biofilms are formed on an acrylic denture placed on the palate of a rat. In 1978, Olsen and Bondevik (209) first used an inserted acrylic plate fitted to the palate of a rat to study Candida stomatitis, but in that study the biofilm that formed on the plate was not assessed. In 2010, Nett et al. (210) developed a denture model using temporary dental acrylic deposited directly onto the palate of immunosuppressed rats treated with antibiotics (Fig. 5D). The denture was secured with dental wire between molars. In this model, a spacer was placed against the hard palate prior to depositing acrylic resin. After the spacer was removed, a 1-mm-wide void between the palatal and acrylic surfaces was created that was filled with a suspension of C. albicans yeast-phase cells. After 72 h of incubation, Nett et al. (210) found that a mixed-species biofilm developed on the acrylic surface, composed of one-third C. albicans cells and two-thirds intestinal bacteria. In controls in which there was no inoculum, biofilms formed that were composed of up to one-fourth indigenous C. albicans cells and three-fourths intestinal bacteria. SEM analyses revealed budding yeast cells and hyphae but no consistent architecture. Another rat denture model (Fig. 5C) was developed by Lee and colleagues (211, 212). A denture was made from a dental impression cast for each individual rat, employing techniques similar to the prosthodontic procedures used for humans. The custom-fit, removable denture contained an embedded metal rod secured to the anterior hard palate by magnets within the fixed denture, which was in turn secured by orthodontic wires to the rear molars. Unlike the model by Nett et al. (210), the rats in this study were immunocompetent and were not treated with antibiotics. C. albicans was introduced into the tight interface between palate and denture as a paste of centrifuged C. albicans yeast-phase cells. The paste was applied directly to the mucosal surface prior to placement of the denture. In this model, biofilms formed on both the denture surface and the palate (211). Biofilms, containing yeast-phase cells, hyphae, and ECM, formed on the denture after approximately 4 weeks postinoculation and on palates after 6 weeks, even though colonization of the general oral region by C. albicans remained constant after 7 days. No mixture of C. albicans and bacteria was observed in the biofilms, possibly as a result of the tight fit of the denture to the palate. The biofilms formed on the palate and the denture, as viewed by SEM, appeared similar to those formed in vitro using the Douglas model with human saliva-conditioned acrylic (211, 213). Hence, in that study (211), the in vitro and in vivo models were architecturally similar. The reason for comparability may have been due to the flow dynamics in the area between denture and palate, which was low. Models in which C. albicans was inoculated directly into the oral cavity or under the tongue of a rodent in the absence of dentures appeared to involve primarily invasion of hyphae at the ventral surface of the tongue, and thus appear to be better as models of mucosal invasion (214–217). However, small biofilms did appear to form in the crevices between papillae on the tongue surface.
Recently, Srinivasan et al. (104, 218–220) developed a unique in vitro model, which they referred to as a “nano-biofilm,” consisting of C. albicans cells embedded in rat tail type I collagen alginate hydrogels. In the model, the nano-biofilms were printed onto microarray slides. SEM images presented by Srinivasan et al. (219, 220) revealed that the seeded yeast-phase cells within the chips formed hyphae in all directions, but an organized biofilm was not evident, presumably because of the structural impediments of the synthetic matrix. Hence, this model may more appropriately be useful for invasion rather than biofilm studies. Several additional in vitro biofilm models, distinct from the Douglas paradigm, have been developed that introduce flow across the biofilm during development. They are described in a later section of this review. Suffice it to say that the variety of models, especially those with catheters and between dentures and palate, introduce conditions quite distinct from those in the Douglas model.
METHODS FOR MEASURING BIOFILMS MAY BE A SOURCE OF VARIABILITY
Even if studies in different laboratories employ similar models, different methods of assessment have the potential to generate different results and interpretations. Therefore, before considering the variability of C. albicans biofilms resulting from the use of different strains and different model conditions in vitro, the variability inherent in the different methods of assessment must be addressed. Hawser et al. (168, 179) originally evaluated biofilm formation by measuring three parameters: dry weight, the incorporation of [3H]leucine into total acid-perceptible protein, and the reduction of tetrazolium salts to formazan. These and subsequent studies also routinely used SEM, but primarily for assessing cellular phenotype and matrix deposition, and usually for only one strain. Assessing dry weights of 48-h mature biofilms is straightforward and highly reproducible (45, 179). Dry weight, however, provides no information regarding cellular phenotype, architecture, or the contribution of the ECM. Indeed, Pujol et al. (45) showed that biofilms formed by a number of C. dubliniensis strains exhibited very different architectures but still attained similar dry weights. If used in conjunction with other assessment methods, however, dry weight remains a fundamental parameter of biofilm growth. Assessing [3H]leucine incorporation is reproducible (168) and provides a measure of the rate of protein synthesis. However, when used to assess a 48-h biofilm, it represents an assessment of the rate of translation during biofilm maintenance, not that during biofilm growth and development. It is also not an expedient method and is institutionally restricted because of the use of radioactivity. Tetrazolium salt reduction has been the most common assay, primarily for expediency, in analyses of the effects of antifungal drugs on biofilm development. This assay measures respiratory or metabolic activity. The reduction of MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] or XTT, both of which are water-soluble tetrazolium salts, by NADH-dependent oxidoreductase in the mitochondria and in specialized vesicles, produces formazan, which can be quantitated colorimetrically (221, 222). The assay was developed originally to assess the level of oxidative metabolism (223) and has proven useful for assessing cell death (181, 224, 225). However, tetrazolium salt reduction is an indirect method for measuring the extent of biofilm growth, especially for mature biofilms, for several reasons. First, stationary-phase yeast cells in the basal layer of a biofilm and the more proximal compartments of a hypha are not undergoing cell division. These compartments may therefore be far less active metabolically than the most apical compartment of hyphae. Second, such a measure does not reflect biofilm mass, volume, thickness, cellular differentiation, or architecture or, most importantly, the extent of matrix deposition. Its expediency for assessing drug susceptibility in 96-well plates has been demonstrated, and its reproducibility verified (226), but its usefulness in comparing extents of biofilm development is suspect. In a brief but revealing side-by-side comparison using the XTT reduction assay and dry weight, Kuhn et al. (227) found that while some isolates of C. albicans, from dentures, catheters, blood, bronchial tubes, urine, the vagina, and skin, formed biofilms with similarly high XTT reduction values and dry weights, others formed biofilms with relatively high dry weights but relatively low XTT reduction values, and vice versa. In an independent study, Li et al. (228) found that the variation between biofilms formed by different strains obtained from the oral cavity, the environment, and the vaginal canal was greater for crystal violet (CV) absorption than for XTT reduction. CV targets primarily mucopolysaccharides and amyloids (229), which may vary dramatically as a result of differences in matrix deposition. In an excellent analysis of the parameters that can affect XTT reduction methods for antifungal susceptibility studies, Nett et al. (230) provided a list of problems and possible reasons for those problems. They also provided methods for optimization. However, these precautions do not alter the inherent problem of the assay, namely, that it measures mitochondrion- and vesicle-based NADH-dependent oxidoreductase activity (231), which probably differs between the multiple phenotypes in a biofilm, between different phases of the budding cycle, and between the proximal and apical compartments of hyphae during elongation. Moreover, measuring XTT or MTT reduction at 48 h, after formation of the biofilm is over 90% complete and the rate of growth has decreased over 10-fold from peak values (168), does not seem to be a very direct or accurate measure of biofilm growth and development. To make things even more complicated, Liu et al. (231) reported that MTT is membrane impermeant, and they concluded that the tetrazolium salt may have to be taken up into living cells by endocytosis. The MTT and XTT reduction assays may therefore be, at least in part, a measure of uptake, not cell metabolism.
One of the most important traits for assessing biofilm development is architecture. As noted above, SEM (168, 181, 192, 232, 233) and, to a lesser extent, CLSM have been used to assess cellular phenotype, extracellular matrix, relative growth, and architecture (181, 213, 232, 234). However, both methods have limitations and artifacts and can lead to variability, depending upon how they are performed. There have been two procedures used for SEM analysis of Candida albicans. If biofilms are fixed with glutaraldehyde in cacodylate buffer, flash frozen, and then freeze-dried (179, 235), one preserves the collapsed ECM but loses resolution of the hyphal and yeast morphologies (Fig. 4F). If biofilms are fixed and dehydrated (45), hyphae and yeast cells are imaged, but the ECM is lost (Fig. 4E). As noted by Little et al. (236) for bacterial biofilms, solvent replacement of water from a biofilm removes matrix components. In addition, whether the SEM preparation is frozen or dehydrated after fixation, native architecture is lost.
CLSM (237–239) can provide information on internal architecture, cellular phenotype, and biofilm thickness. In contrast to the SEM procedure, there is no dehydration step after fixation with paraformaldehyde or other fixatives, and thus more of the native architecture is preserved (240). CLSM involves staining rather than surface shadowing, and the collection of hundreds of optical sections of a biofilm through the z axis provides information about different depths. Staining artifacts, however, can arise in CLSM. For instance, when a 48-h fixed biofilm is overlaid with a solution of calcofluor, which targets cellulose and chitin (241), staining is most intense in the upper portion of the biofilm, presumably because of disproportionate binding of the dye to hyphae at the top of the biofilm during dye penetration (45, 183, 213). This artifact is evident in side views of projection images of a stack of CLSM scans, such as those presented in Fig. 4D and G. CLSM is, however, effective at comparing the thicknesses of biofilms (183), and with advanced processing software, it can be used to image the architecture of intact hyphae in side views of projection images (45) (Fig. 4D). An additional problem arises when CLSM is used to view the ECM with dyes such as calcofluor, which binds to chitin and cellulose (242). These ECM polysaccharides are also localized in the hyphal wall. In the case of calcofluor, hyphae stain far more intensely than the diffuse ECM. One must therefore lower the laser intensity to view hyphae by CLSM, at the expense of viewing the ECM. If intensity is increased, the ECM can be visualized at the expense of resolving distinct hyphae, because hyphal walls stain so much more intensely (39, 243). Light attenuation is less problematic when multiphoton CLSM is applied (97).
Finally, transcription profiling is evolving as a major method for assessing biofilm development. Profiling can be performed by assessing genome-wide transcription by using microarrays (195, 200, 244–248), NanoString nCounter technology (249, 250), and RNA sequencing (RNA-Seq) (197, 200) or by using quantitative reverse transcription-PCR (RT-PCR) or Northern blot analysis of a smaller, select group of genes (251). Such comparisons are often made between biofilm preparations and planktonic cultures. Unfortunately, assessing the accuracy, reproducibility, and concurrence of these technologies has been hampered by the variety of genome-wide transcription platforms employed and the differing conditions used to prepare biofilms, such as different media, substrata, times of analysis, and fluid flow or mixing methods (Table 2). In two general reviews of the reliability and reproducibility of chip technology, it is noted that reproducibility can be attained for abundant transcripts but that this is not always the case for low-abundance transcripts (252, 253). Canales et al. (252) noted that the latter problem is especially true for measurements on different platforms (252). As global analyses of gene expression segue from DNA chips to RNA-Seq, new challenges arise. This is best exemplified by the elegant study of Nobile et al. (200), who used both technologies. Using a mutant library, they developed a model of a transcriptional network controlling biofilm formation, in which 1,000 target genes were regulated by six key transcription factors. In that study, they pooled data from a DNA chip analysis and an RNA-Seq analysis to determine genes differentially expressed during biofilm development, rather than selecting genes that intersected in the two studies (i.e., genes upregulated or downregulated in the same way by both methods of assessment). This was done for inclusiveness. However, the question related to our focus in this review is how comparable the results of the two methods were, given that the preparations were generated in the same study and using the same model for comparing planktonic and biofilm gene expression levels. The threshold employed for a significant difference between planktonic and biofilm expression levels for a gene was 2-fold (i.e., ≥2-fold). Of the 2,235 genes that were identified as up- or downregulated by both platforms, or by one or the other, 1,792 were actually assessed by both platforms. Of the latter genes, 405 (23%) were found to be regulated similarly by both methods (see the supplemental material in reference 200), which is a surprisingly low level of correspondence. The study by Nobile et al. (200) was followed by an additional one by the same group, using the same model and the same microarray platform but not including an RNA-Seq analysis (245). In the supplemental material of the expanded study (245), comparison of the transcription profiles of planktonic (log-phase) cells and biofilms identified 238 genes that were upregulated or downregulated ≥2-fold and were also identified as similarly regulated in the study of Nobile et al. (200). However, 845 additional genes were found to be differentially regulated ≥2-fold that were not assessed in the Nobile et al. (200) study. In the Nobile et al. (200) study, 577 genes were identified as up- or downregulated ≥2-fold that were not identified to be regulated similarly in the Fox et al. (245) study. Finally, 31 genes were oppositely regulated (upregulation versus downregulation or downregulation versus upregulation) ≥2-fold in the two studies. Some of these differences certainly arose between measurements very close to but not attaining the significance threshold. But even so, if such differences arise between sequential studies in the same laboratory, in which the same methods are used to obtain preparations and the same microarray platforms are used for assessment, one wonders what differences would be found, especially for low-abundance transcripts, between studies in different laboratories using different conditions and different platforms. Genome-wide transcription profiling does reveal differences in select abundant genes, many of them reproducible between laboratories, and when limited to such genes, it provides a powerful tool for assessing the progression and maturation of biofilm formation. It would be extremely helpful in these studies if the abundances of transcripts, especially for those not showing congruence, were noted. In addition, a threshold of ≥2-fold means that a gene must be expressed at a level only twice that of a planktonic culture, or half that of a planktonic culture, to be considered differentially regulated in a biofilm. The reliability and reproducibility of genome-wide profiling for a particular platform and the congruence between platforms need to be addressed. The use of this technology becomes even more problematic when phenotypic heterogeneity is considered. An and Parsek (254) point out, in the last line of their discussion on the use of DNA microarray technology in genome-wide assessments of transcription in bacterial biofilms, that “the real payoff might be when we are able to profile distinct spatial or functional subpopulation in the community.” This also holds true for C. albicans biofilms.
TABLE 2.
Review of the models, substrates, media, assay times, and assays employed for transcriptional profiling of Candida albicans biofilms
| Authors, yr (reference) | Model | Substrate | Mediuma | Assay time(s) (h) | Platform (source) |
|---|---|---|---|---|---|
| García-Sánchez et al., 2004 (246) | Flow | Thermanox | YNB medium | 48–72 | Microarray (Eurogentec) |
| Murillo et al., 2005 (247) | Douglas | Tissue culture plastic | Ham's F-12 medium | 0.5–6.5 | Microarray (Affymetrix) |
| Yeater et al., 2007 (248) | Douglas | Silicone elastomer, denture acrylic | YNB medium | 6, 12, 48 | Microarray (in-house) |
| Sellam et al., 2009 (292) | Flow | Silicone elastomer | YPD | 1, 3, 6 | Microarray (Biotechnology Research Institute) |
| Nett et al., 2009 (195) | Venous catheter | Catheter wall | Blood | 12, 24 | Microarray (Biotechnology Research Institute) |
| Bonhomme et al., 2011 (244) | Flow | Thermanox | SD medium | 24, 32, 40 | Microarray (Eurogentec) |
| Nobile et al., 2012 (200) | Douglas | Tissue culture plastic | Spider medium | 48 | Microarray (Agilent Technology), RNA-Seq |
| Desai et al., 2013 (197) | Douglas | Tissue culture plastic | Spider medium | 48 | RNA-Seq (Illumina GA2 solid system) |
| Fox et al., 2015 (245) | Douglas | Tissue culture plastic | Spider medium | 8, 24, 48 | Microarray (Agilent Technology) |
The compositions of the media are given in Table 1.
STRAIN VARIABILITY
The complexity of C. albicans biofilm formation in both space and time and the problems that arise in measuring its progress provide a contextual framework for assessing genetic and conditional factors responsible for biofilm variability. The first of these factors is strain variability. Do all strains form biofilms similarly? This question is fundamental to biofilm studies dealing with gene expression and mutational defects or drug resistance, since a majority of these studies employ one strain or mutants derived from a single parent strain. Single-strain studies assume that the strain employed is representative of all strains, a majority of strains, or the average strain. Hawser and Douglas (168) performed the first study of strain variability. They analyzed 15 strains, but two were mutants: one that could not form hyphae and one that could not form yeast-phase cells. Both mutant strains were derived from the same parent strain. Their results are summarized for the 13 natural strains (i.e., nonmutant strains) in their collection, along with the results of other studies of strain variability, in Table 3. The extremes of the ranges for the three traits (dry weight, [3H]leucine incorporation, and MTT reduction) analyzed by Hawser and Douglas (168) differed by 2.1-, 2.3-, and 3.2-fold, respectively, for cultures in medium containing 50 mM glucose and by 2.3-, 3.8-, and 3.2-fold, respectively, for cultures in medium containing 500 mM galactose (Table 3). The standard deviations (as percentages of the means) were 24%, 24%, and 21%, respectively, for biofilms formed in YNB medium plus glucose and 18%, 32%, and 28%, respectively, for biofilms formed in YNB medium plus galactose (Table 3). It should be noted that the strains used by Hawser and Douglas (168) and by all but one of the other studies reviewed for Table 3 were not assessed for the configuration of the mating type locus, which affects the size and functional characteristics of biofilms (39) and the susceptibility of planktonic cells to antifungals (41). As noted, however, it is reasonably safe to assume that a random collection of strains, as in the study by Hawser and Douglas (168), will usually contain no more than 10% MTL-homozygous isolates (40, 255–257). In a subsequent study of variability by Kuhn et al. (26), employing the Douglas model, biofilm formation parameters were compared among 10 strains by measuring XTT reduction and dry weights of 48-h biofilms. Isolates varied in relative XTT reduction values (A492), which were between 0.65 and 1.24, and in relative dry weights, which were between 0.55 and 1.30 mg (Table 3). For both measurements, the extremes varied approximately 2-fold. The standard deviations (as percentages of the means) were 29% and 30%, respectively (Table 3). These variability measurements were close to those obtained by Hawser and Douglas (168) (Table 3). As previously noted, Kuhn et al. (26) found that there was poor correspondence of the relative rankings in the collection for individual strains between XTT reduction and dry weight measurements.
TABLE 3.
Strain variability in studies in which collections of three or more isolates were analyzed in parallela
| Authors, yr (reference) | Mediumd (sugar) | Substrate | No. of strains | Assay | Mean ± SD | SD as % of mean | Range | Fold difference of extremes |
|---|---|---|---|---|---|---|---|---|
| Hawser and Douglas, 1994 (168) | YNB medium (glucose) | Catheter | 13 | Dry wt (mg) | 0.82 ± 0.20 | 24 | 0.52–1.10 | 2.1 |
| [3H]leucine incorporation | 4,476 ± 1,058 | 24 | 2,550–5,886 | 2.3 | ||||
| MTT A540 | 0.52 ± 0.11 | 21 | 0.38–0.66 | 3.2 | ||||
| YNB medium (galactose) | Catheter | 13 | Dry wt (mg) | 1.76 ± 0.32 | 18 | 0.51–1.99 | 2.3 | |
| [3H]leucine incorporation | 8,503 ± 2,684 | 32 | 2,672–10,225 | 3.8 | ||||
| MTT A540 | 0.98 ± 0.27 | 28 | 0.49–1.24 | 3.2 | ||||
| Kuhn et al., 2002 (26) | YNB medium | Catheter | 10 | Relative dry wt | 0.82 ± 0.23 | 29 | 0.55–1.30 | 2.4 |
| Relative XTT | 1.02 ± 0.30 | 30 | 0.65–1.24 | 1.9 | ||||
| Li et al., 2003 (228) | YNB medium | TCP | 16 (oral) | XTT A490 | 0.077 ± 0.052 | 62 | 0.053–0.329 | 62 |
| CV A570 | 0.209 ± 0.424 | 159 | 0.050–2.179 | 43.4 | ||||
| 31 (environmental) | XTT A490 | 0.079 ± 0.025 | 32 | 0.55–0.158 | 2.9 | |||
| CV A570 | 0.217 ± 0.246 | 113 | 0.087–1.032 | 11.9 | ||||
| 37 (vaginal) | XTT A490 | 0.063 ± 0.020 | 32 | 0.049–0.157 | 3.2 | |||
| CV A570 | 0.143 ± 0.081 | 57 | 0.043–0.526 | 12.2 | ||||
| Jain et al., 2007 (258) | RPMI 1640e | TCP | 63 | Relative XTT | 0.55 ± 0.05 | 9 | — | — |
| Urine | TCP | 63 | Relative XTT | 1.00 ± 0.20 | 20 | — | — | |
| Villar-Vidal et al., 2011 (259)b | RPMI 1640e | Catheter | 16 (oral) | XTT A492 | 1.01 ± 0.40 | 40 | 0.30–1.33 | 4.4 |
| Urine | 12 (blood) | XTT A492 | 0.90 ± 0.37 | 41 | 0.30–1.50 | 5.0 | ||
| Pujol et al., 2015 (45)c | RPMI 1640e | Catheter | 3 (a/α) | Thickness (mm) | 76 ± 2 | 3 | 74–78 | 1.1 |
| Catheter | 6 (a/a) | Thickness (mm) | 56 ± 3 | 5 | 51–58 | 1.2 | ||
| Catheter | 6 (α/α) | Thickness (mm) | 58 ± 2 | 3 | 56–61 | 1.1 | ||
| Catheter | 10 (a/α + α/α) | Dry wt (mg) | 0.75 ± 0.13 | 17 | 0.48–0.98 | 2.0 |
TCP, tissue culture plastic; MTT A540, MTT reduction as measured by the absorbance at 540 nm; relative dry weight, dry weight relative to the wild type or the mean; relative XTT, XTT measure relative to the wild type or the mean; CV A570, crystal violet staining as measured by the absorbance at 570 nm; XTT A490 or A492, XTT reduction as measured at 490 nm or 492 nm; —, data not provided for calculation.
The only study in which sampling occurred at 24 h, not 48 h.
The only study performed at 28°C, not 37°C, because of white cell induction at temperatures of ≥35°C.
Sugar contents for the different studies were as follows: for the study of Hawser and Douglas (168), 50 mM glucose or 500 mM galactose; for the study of Kuhn et al. (26), 50 mM glucose; for the study of Jain et al. (257), 11 mM glucose; for the study of Li et al. (227), 50 mM glucose; for the study of Villar-Vidal et al. (258), 11 mM glucose; and for the study of Pujol et al. (45), 11 mM glucose.
Buffered with MOPS.
In 2003, Li et al. (228) compared 115 isolates representing the four major clades of C. albicans. They used a variation of the Douglas model, developed by Ramage et al. (181), that employed the tissue culture plastic surface of the wells of 96-well plates, rather than a catheter disc, as the substratum. The collection was separated into isolates from the oral cavity, the vaginal canal, and the environment. Li et al. (228) measured XTT reduction activity and CV staining. Just as Kuhn et al. (26) reported, there was poor correspondence of relative rankings between the XTT and CV measurements for individual strains within a clade. Li et al. (228) found that the means, standard deviations, and ranges for XTT reduction and CV staining were statistically indistinguishable between clades. The means ± standard deviations for XTT reduction for 16 oral, 31 environmental, and 37 vaginal isolates were 0.077 ± 0.052, 0.079 ± 0.025, and 0.063 ± 0.020, respectively. The standard deviations of XTT reduction values (as percentages of the means) were 62, 32, and 32%, respectively, which are much higher than the standard deviations in both the Hawser and Douglas (168) and Kuhn et al. (26) studies (Table 3). The standard deviations (as percentages of the means) for crystal violet staining were even higher, i.e., 159%, 113%, and 57%, respectively, suggesting a problem in the reliability of the latter measurement given the parallel results for XTT reduction. Jain et al. (258) used the same variation of the Douglas model, as well as the XTT reduction assay, to compare biofilm formation levels among 63 isolates from saliva in RPMI medium or artificial urine. The standard deviations (as percentages of the means) for XTT reduction measurements were 9% for RPMI 1640 medium and 20% for urine (Table 3). The value for RPMI 1640 medium was lower than those of Hawser and Douglas (168). Jain et al. (258) did not provide individual strain data for C. albicans, so the range and extremes could not be computed. Villar-Vidal et al. (259) used the same model, but sampled the biofilms at 24 h rather than 48 h, as well as the same XTT reduction measurement, to analyze variability among 16 oral isolates and 12 blood isolates. The means ± standard deviations for XTT reduction were 1.0 ± 0.4 and 0.9 ± 0.4, respectively (Table 3). The ranges were 0.3 to 1.3 for oral isolates and 0.3 to 1.5 for blood isolates, representing 4- and 5-fold differences between extremes (Table 3). The standard deviations (as percentages of the means) were 40 and 41% for oral and blood isolates, respectively, or approximately double the values obtained in the Hawser and Douglas (168), Kuhn et al. (26), and Jain et al. (258) studies. Finally, Pujol et al. (45) performed an architectural comparison of three strains that had been typed a/α for the MTL configuration, namely, P37039, P37037, and SC5314. They found a high level of uniformity in thickness (78 ± 5 μm, 76 ± 2 μm, and 74 ± 3 μm, respectively). They also found similar basal yeast cell polylayers, hyphal upper layers, vertical orientations of hyphae, and densities of the ECM. However, the sample size was too small for valid comparisons with the previous studies reviewed here. They also presented thickness measurements for six a/a and six α/α strains, which had similarly low variability (Table 3). The differences in extremes were 1.2-fold and 1.1-fold for the a/a and α/α strains, respectively. However, the variability of dry weights for a collection of 10 a/α strains containing 5% a/a and 5% α/α cells was similar to that obtained by Hawser and Douglas for a/α strains (Table 3).
Assessing variability between strains may therefore be influenced by a scientist's technique or the assessment parameters themselves, as well as the variability due to differences between strains. The data at hand, however, suggest that variability among strains can be as great as 2-fold or more, and this must be considered in comparing data from different studies, especially those that employ only one test strain. For such studies, it would be prudent to consider whether the strain employed is representative (i.e., near the mean) of a collection of natural strains, at least for basic parameters, such as dry weight, phenotypic composition, and architecture. SC5314, the strain that is parental to a number of derivative strains used for mutant analyses, appears to be quite normal in the formation of biofilms in vitro (183, 184). However, that does not mean that all of the engineered derivatives used to select mutants form biofilms normally. Therefore, a parental strain that is a derivative of SC5314, or another natural strain, not the original natural strain, must serve as the immediate control in comparisons of biofilm formation, and it should be compared to the original natural strain, such as SC5314.
VARIABILITY DUE TO THE ORIGINAL PLANKTONIC CELL INOCULUM
In the original experiments performed by Hawser and Douglas (168), SEM images of cultures in YNB plus 500 mM galactose revealed that after a 1-h period of incubation of planktonic cells in the wells of the multiwell plate, the adhering population was far from confluent. It is not clear if this was due to too dilute an inoculum or to a loss of cells during preparation for SEM. If experiments are performed to assess adhesiveness as a function of inoculum concentration by light microscopy, conditions can be defined empirically to obtain a uniform monolayer of yeast cells after the 60- to 90-min incubation period, as in Fig. 4A and B. It is also evident in the SEMs provided by Hawser and Douglas (168) that the original planktonic cells used for inoculation were in different stages of the budding cycle. This could have been the result of heterogeneity of the yeast cell cycle in the planktonic cell population grown overnight. Growing cells planktonically in different media can lead to differences in the heterogeneity of the budding yeast cell cycle in late log phase or stationary phase. This in turn can result in phenotypic heterogeneity of the initial adhering cell population in the biofilm model. Cells can be in different phases of the yeast cell cycle (G1, S, G2, and M), heterogeneously undergoing changes in evagination, bud formation, intracellular architecture, cell separation, DNA replication, and nuclear division (260–262). Cells can also be undergoing mitochondrial duplication and migration heterogeneously (263). Heterogeneity will have an impact on the synchrony of cell division at the onset of the formation of a basal yeast cell polylayer. This in turn might affect measurements of early biofilm development. Attaining synchrony was of paramount importance more than 4 decades ago in studies related to the yeast cell cycle and cellular physiology (264–266), as well as to the bud-hypha transition in C. albicans (267).
Almost all planktonic preparations of C. albicans are grown in the yeast phase overnight, in culture tubes or flasks that are rapidly rotated. Cell density is then commonly measured by determining the optical density. Optical density is not, however, a direct measure of cell number, but rather of mass (268), and it must be normalized empirically to the cell concentration. The OD may be influenced by the heterogeneity of the budding cycle, adhesion, and flocculation and by differences in strain-specific cell size. The use of a hemocytometer for verification of concentration may be a bit more time-consuming than measuring the optical density, but it provides researchers with two benefits: an accurate count of cell number and a visual estimate of the heterogeneity of the budding yeast cell cycle, including the presence of pseudohyphae and hyphae in the preparation. For white and opaque cells in MTL-homozygous cultures, it provides a measure of phenotypic homogeneity. The concentration can then be adjusted by dilution and the adjusted planktonic suspension inoculated into test wells of the in vitro biofilm model. And if heterogeneity is a problem, another medium, strain, or preparation can be selected.
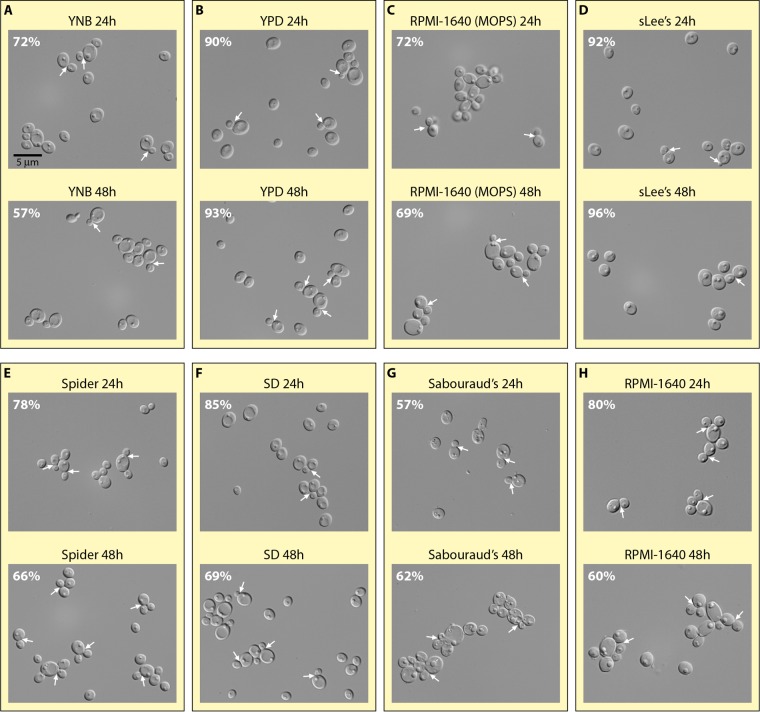
Phenotypic homogeneity of the planktonic cell preparation can be a concern, especially for measurements during the early stages of biofilm formation. Cell cycle-regulated genes have been identified in Saccharomyces cerevisiae (269), including modulation of histone messengers, first reported in 1981 (270). For C. albicans, Kusch et al. (271) used two-dimensional (2D) electrophoresis to compare proteins between log- and stationary-phase cells. Dramatic differences were observed in proteins involved in all aspects of metabolism. Unfortunately, the different media used to culture C. albicans planktonically result in quite different levels of cell cycle heterogeneity in stationary phase. In Fig. 6, representative fields of cells and the proportions of unbudded yeast-phase cells are presented for the a/α strain SC5314 cultured planktonically at 26°C for 24 and 48 h, with rotation, in a variety of media that have been used for planktonic cell growth. The compositions of these media are presented in Table 1. At 24 h, cultures in YNB, YPD, sLee's, Spider, SD, and RPMI 1640 (with or without MOPS) media consisted of over 70% unbudded yeast-phase cells (Fig. 6A to F and H). At 48 h, however, only planktonic preparations in YPD and sLee's media contained over 70% unbudded yeast-phase cells, i.e., 93% and 96%, respectively (Fig. 6B and D), suggesting that heterogeneity may increase as planktonic cultures become stationary. There are multiple reasons for why it may be advantageous to obtain a relatively uniform population of unbudded yeast cells in stationary phase for the initial planktonic inoculum of a biofilm model. First, cell cycle synchrony at the onset of a biofilm experiment allows one to interpret measurements as representative of a single cell phenotype. Second, cells in one growth phase, such as stationary phase, provide a single physiology. The differences in cell physiology between log- and stationary-phase cells have been described primarily for bacteria (272–274). In addition, the susceptibilities to antifungal agents of planktonic cells of S. cerevisiae differ between log- and stationary-phase cells (275), as do the proteomes (271). Third, the presence of a significant number of pseudohyphae or hyphae adds a second dimension to phenotypic heterogeneity. Therefore, documenting that a majority of cells in the planktonic cell inoculum have entered stationary phase, are unbudded and of uniform size, and are uniformly in the yeast phase should be considered in the development of in vitro models of biofilm formation.
FIG 6.
Comparison of budding cell phenotypes of planktonic cultures of C. albicans strain SC5314 grown in culture tubes shaken for 24 and 48 h, using eight different media to obtain planktonic yeast-phase cell cultures for in vitro C. albicans biofilm studies. Budding cells were scored as cells with buds exhibiting a diameter of less than two-thirds that of the mother cell and still attached to the mother cell. The percent unbudded cells in culture is presented in the upper left corner of each panel. Between 500 and 1,100 cells were scored for each preparation. Arrows point to examples of budding cells. Compositions of the media can be found in Table 1.
VARIABILITY DUE TO FLOW, ROTATION, OR ROCKING
In their original report, Hawser and Douglas (168) did not mix their cultures by rotation or rocking, but in a subsequent report (179), they rotated cultures at 30 rpm, which increased biofilm thickness. Mixing can affect O2 and CO2 tension in the biofilm microenvironment, as well as increase accessibility to nutrients. It can also cause shear forces that affect mechanoreceptors. Such receptors have been demonstrated to exist on the surfaces of hyphae and function in thigmotropism (276). Whether these receptors play a role in the architecture of the upper hyphal region of a biofilm is not known. However, if the amount of mixing (i.e., via rotation, shaking, rocking, or continuous flow) is too great, cell architecture, uniformity along the substratum, matrix deposition, and synchrony of the developmental program may be affected. Therefore, mixing or flow conditions must be optimized empirically for each in vitro model of biofilm formation. Because there is the reasonable perception that biofilms are challenged with constant flow in vivo, several researchers have incorporated flow into their models. For bacterial biofilms, two major flow models have been employed: a flow cell (277) and a rotating disc reactor (278, 279). The flow cell is constructed from polycarbonate or polymethyl methacrylate and contains three open tubular chambers. The chambers are covered by a glass coverslip (the substratum for the biofilm). Medium enters through an inlet port and exits through an outlet port (277). The rotating disc reactor contains a stir disc to generate flow across 18 removable polycarbonate chips with adherent cells (278). To generate flow across a C. albicans biofilm, López-Ribot and colleagues (188, 280) developed a model in which a strip of serum-coated silicone elastomer was first incubated with cells in a conical tube to promote adherence. The strip was then placed in a tube with inlet and outlet ports, and buffered RPMI 1640 medium was continuously dripped down the strip. In a second model (281), discs seeded with cells were placed in multiple wells bored into an acrylic block, through which medium was pumped. In both cases, SEM images revealed that robust biofilms formed. García-Sánchez et al. (246) developed a model in which a seeded Thermanox plastic coverslip was placed in a test tube that contained a narrower tube that released medium and air. Medium was continuously removed through an outlet port at the top. The turnover rate was 0.6 ml/min for a 40-ml chamber, or 1.5% of the volume of the medium per minute. The diagram of the model provided in the study (246) suggests that bubbles of air may have flowed across at least the upper portion of the biofilm. SEM images of wild-type biofilms formed in this flow model revealed patches on the substratum containing mixtures of tangled hyphae, pseudohyphae, and budding cells. Some of the budding cells were unusually large and possessed at least six bud scars. Results obtained with the different flow chambers may be difficult to compare given the differences in design. In developing flow chambers for bacteria, Crusz et al. (277) emphasized the importance of minimizing bubbles, since they were shown to disrupt biofilm development. More importantly, in most studies of C. albicans biofilms in which medium is either mixed or moved by flow, it is difficult to compare in vitro shear forces with those on the surfaces of host niches where C. albicans biofilms form. In addition, there have been no systematic studies of the effects of transient changes in flow on biofilm formation. Such changes do occur in several host niches. For instance, although there is normally very little flow across biofilms that form between dentures and palate, periodic movement may cause turbulence and disruption. Moreover, dentures are usually washed frequently rather than retained in place for 72 h (210) or up to 8 weeks (212). When an individual is awake, the flow rate from the salivary gland is approximately 0.7 ml/min, or 42 ml/h (282–284), but it decreases dramatically during sleep (285). Swallowing causes agitation and rapid movement of the bolus of saliva to the esophagus (286), and chewing causes extreme turbulence. In the intestine, fluid flow is approximately 10 ml/min (287), but flow rates at the topologically complex intestinal surface may be far lower, especially in the crypts of Lieberkühn and other crevices. Flow rates of pressure-injectable catheters can range from 4 to 6 ml/s, or 240 to 360 ml/min, and the pressure can be 100 to 300 lb/in2 (288). Indwelling urinary tract catheters may have relatively continuous high flow rates (289), whereas short-term indwelling intravenous catheters may have high flow rates only during therapeutic delivery or sampling (288, 290). For drainage and therapy, long-term catheters can be indwelling for more than 6 weeks. The development of in vitro flow models, especially those with the rotating or rocking conditions used in most in vitro models, may therefore not mimic any in vivo conditions. The effect of flow on bacterial biofilm formation was poignantly demonstrated by Greenberg and colleagues in a comparison of biofilms formed by wild-type Staphylococcus aureus and a mutant for the accessary gene regulator locus agr (279). Most agr isolates, which are defective in quorum sensing, were found to be defective in biofilm formation (291). The number of cells in the biofilm of the ΔagrD mutant was less than one-third that in the wild-type biofilm in the static dish model but over two times that in the wild-type biofilm in the spinning disc reactor (279). With a flow cell rather than spinning disc model, the biofilms formed by the ΔagrD mutant and the wild type were architecturally indistinguishable after 1 and 5 days, as revealed by CLSM (279). These bacterial results suggest that flow rate must be considered an important variable in biofilm formation. Hawser et al. (179) clearly showed that under their conditions, there was a significant increase in dry weight for cultures rotated at 20 rpm on an orbital rotating platform compared to undisturbed cultures. However, rotation at speeds higher than 30 rpm reduced dry weight. Based on these results, one must wonder how rotation speeds of 200 rpm, which have been employed, affect architecture. One must also consider the possibility that rotation at 200 rpm, rocking a culture 12 times/min at a 6° pitch, and constant flow across a biofilm in a flow apparatus result in different shear forces with different effects on biofilm formation. The point is that the method used may lead to major differences in biofilm formation and must therefore be taken into account in comparing data from different studies.
VARIABILITY DUE TO DIFFERENCES IN DEVELOPMENTAL TIMING
Although this is not always the case, in the majority of studies of C. albicans biofilms, the assessment of biofilm formation is performed at a single time point. This has been true for comparisons of biofilm formation between mutants and wild-type strains and for analyses of antifungal drug susceptibility. A single time point may not be sufficient. The formation of C. albicans biofilms in in vitro models must be regulated by an underlying developmental program involving cellular differentiation, the divergence of cellular phenotypes, complex cell-cell interactions, and signaling that changes with time. Time-dependent changes during biofilm formation have been demonstrated for gene expression (244–247, 292), cellular phenotype (168, 179, 183, 184), architecture (183, 184), and the deposition of ECM (293–296). Hence, by definition, there is an ordered sequence of dependent events, and in some cases parallel pathways, with a time table, in complex developmental programs (297). For C. albicans, we know that there are changes in adhesion, yeast cell multiplication, germination, hypha elongation, and ECM deposition. If there is a change resulting from a mutation in the length of time required for any of the developmental stages in the underlying program of biofilm formation, a difference in the genetic makeup between natural strains, different model conditions, or the absence or presence of an antifungal drug, a single measurement at a single time point may simply reflect a difference in when things happen, not if they happen, as outlined in the simple hypothetical example presented in Fig. 7. In this example, strain “a” forms a basal yeast cell polylayer during the first 5 h, while strain “b” does so during the first 20 h. Each then forms the upper hyphal/ECM layer over the subsequent 43 h. At 48 and 63 h, respectively, both have formed indistinguishable mature biofilms with similar pathogenic traits (Fig. 7). However, if both were sampled at 15 h, “a” would be extending hyphae and depositing ECM, while “b” would be undergoing cell multiplication in the yeast phase. And if both were sampled at 33 h, “a” would have come close to completing a mature biofilm, while “b” would be in the middle stages (Fig. 7). If “a” and “b” were compared at 4 and 15 h, respectively, or at 15 and 30 h, respectively, architecture and physiological measurements would be similar. High-frequency epigenetic switching systems that selectively affect the lengths of time between developmental stages, but not the sequence of stages that occur, have been described for the model developmental system Dictyostelium discoideum (298–300). Therefore, it seems reasonable to suggest that comparisons between different natural strains, wild-type strains and mutant derivatives, untreated and antifungal-treated cultures, and cultures developed under different model conditions should include multiple time points. Just as importantly, architectural landmarks should be assessed in parallel at multiple points to verify phenotypic comparability.
FIG 7.
Example showing why a single time point might be invalid for comparing biofilms. Strain differences, mutations, antifungal drugs, and different model conditions may affect when things happen, not if they happen. A hypothetical example is presented for a difference in timing in the first step of the biofilm development program for two strains, “a” and “b,” which results in an increase in the time it takes for the formation of the yeast-phase cell polylayer for strain “b.” Germination, hypha elongation, and ECM deposition take the normal amounts of time but are shifted in terms of the total amount of time. Data from a single time point at 15 or 33 h would reveal differences that could be interpreted as absolute, when in fact they simply reflect differences in timing.
EFFECTS OF MEDIA ON BIOFILM DEVELOPMENT
Hawser and Douglas (168) performed the first experiments on the impacts of different media on biofilm development. They first compared YNB medium containing either of two carbon sources, i.e., glucose and galactose. Although dry weight, [3H]leucine incorporation, and tetrazolium reduction increased and plateaued with similar temporal dynamics, the plateau values for galactose were approximately twice those for glucose for all parameters. The extent of the difference was also strain dependent (179). In 2001, López-Ribot and colleagues introduced RPMI 1640 medium buffered with MOPS (181). RPMI 1640 medium was developed and optimized specifically for the in vitro culture of human leukocytes (301). RPMI 1640 (MOPS) was also selected as a validated yeast susceptibility testing medium after a direct comparison of Candida growth curves obtained using six candidate media (302) and was recommended by the National Committee for Clinical Laboratory Standards (303–305). A number of media other than YNB and RPMI 1640 (MOPS) media have also been used to support C. albicans biofilm formation (Table 1). Compared to YNB medium with glucose, Lee's medium (185) supplemented with zinc and arginine (sLee's medium) (186) and YPD (yeast extract, peptone, dextrose) fail to support similarly robust 48-h biofilms (our personal observations). It may be for this reason that the two media are rarely used in models to support biofilm formation in C. albicans studies (Table 1). Spider medium, which has been used in the rat catheter model (117, 193, 202, 306–308) and the Douglas well model (309–311), was compared with RPMI 1640 (MOPS) medium for biofilm architecture (183). Spider medium supports formation of biofilms that lack a thick basal yeast cell polylayer. The biofilms are composed of a carpet of yeast cells at the substrate, from which hyphae extend in relatively random directions, with yeast cells formed throughout the upper layer. The extracellular matrix of these biofilms is less dense than that of biofilms formed in RPMI 1640 (MOPS) medium (183). Biofilms formed in RPMI 1640 (MOPS) medium differ architecturally, containing a thick basal yeast-phase polylayer, vertically oriented hyphae, and a denser matrix (183) (Fig. 3B). The lack of a thick basal yeast-phase polylayer in Spider medium may be due to the rapid germination of planktonic cells upon inoculation. Agar containing Spider medium is a known facilitator of germ tube formation and hyphal growth (312–317). Interestingly, even with the differences observed in architecture, the biofilms formed by a/α cells in Spider medium are relatively impenetrable by human white blood cells and relatively resistant to fluconazole treatment, although not to the same degrees as the more organized biofilms formed in RPMI 1640 medium (183). Spider medium becomes problematic, however, when used to support MTL-homozygous biofilms in air, as described later in this review.
VARIABILITY DUE TO THE MTL CONFIGURATION
Perhaps the most dramatic effect on the function and regulation of biofilms in C. albicans relates to the configuration of the MTL locus. MTL-heterozygous yeast-phase cells and MTL-homozygous white-phase cells, which are also in the yeast phase but differ functionally, form biofilms that are architecturally equivalent but differ in thickness and function (38, 39, 243, 318, 319). In the preceding portions of this review, we assumed that the majority of strains in nature were heterozygous (a/α) at the MTL locus, even though the configuration of the MTL locus was rarely assessed. This assumption appears reasonable, since in every major study of the MTL configuration, the proportion of MTL-homozygous strains fell between 5 and 10% of natural clinical strains (40, 42, 255, 256). In addition, the C. albicans strain which has been used as the parent strain most often in mutational and molecular studies is SC5314, which is a/α (320). However, Rustad et al. (256) found that 11 of 12 MTL-homozygous strains analyzed in a collection of 96 strains were resistant to azoles, the major family of anticandidal drugs. Over half of the collection they studied had in fact been selected for drug resistance. The disproportionate representation of resistant strains among the MTL-homozygous members of the collection was reinforced by the discovery by Odds et al. (41) that MTL-homozygous strains grown planktonically are, on average, more resistant to the major azoles than MTL-heterozygous strains. Hence, the potential exists that collections of isolates selected for a particular trait or collected from a particular host niche may be enriched for MTL-homozygous strains. Moreover, some a/α strains have a propensity to undergo homozygosis. On average, a/α strains undergo homozygosis at the MTL locus spontaneously at a low frequency in vitro. The predominant mechanism is the loss of one homolog of chromosome 5, which harbors the MTL locus, followed by duplication of the remaining homolog (321). MTL-homozygous strains that persist in nature, however, originate primarily by mitotic recombination (322). Niche conditions that may stimulate homozygosis, both in vitro and in vivo, are not known. Lockhart et al. (40) found that one of three a/α strains injected into mice in the systemic mouse model, namely, strain P75063, underwent MTL homozygosis at an extremely high rate in vivo. Wu et al. (321) showed that this strain also underwent MTL homozygosis at an extremely high rate in vitro. However, Wu et al. (322) showed that MTL-homozygous strains are less competitive than MTL-heterozygous strains in the mouse systemic infection model, and Ibrahim et al. (323) found that tetraploids resulting from a/a and α/α mating were less virulent in that model as well. Together, these observations support the assumption that most natural and laboratory strains (90 to 95%) can be assumed to be a/α, but they reveal that a significant minority of strains are MTL homozygous and that select a/α strains can spontaneously undergo MTL homozygosis at a high frequency. One environmental condition has been identified that enriches for MTL-homozygous derivatives. Indeed, Rustchenko and colleagues (324) found that on agar media in which the carbon source is sorbose, a/α derivatives that lose one copy of chromosome 5 utilize sorbose, and thus are enriched in a population growing on medium containing sorbose as the carbon source.
WHITE-OPAQUE SWITCHING, MATING, AND BIOFILM FORMATION
To understand the difference in C. albicans biofilms based on the configuration of the MTL locus, one must first understand the white-opaque transition and its role in mating (Fig. 8). When C. albicans a/α cells undergo MTL homozygosis to a/a or α/α (320), they still cannot mate (Fig. 8A). To become truly mating competent, they must then switch from a traditional budding yeast-phase cell type, referred to as the “white” phase, to an elongated, large, pimpled cell type, referred to as the “opaque” phase (Fig. 8A) (325, 326), a phenotypic transition first described in 1987 for strain WO-1, which was isolated from the bloodstream of a patient at the University of Iowa Hospitals and Clinics in Iowa City, IA (327). It was subsequently found that the great majority of MTL-homozygous strains, but not heterozygous strains, undergo this transition (40, 326). In the mating process, opaque a/a cells release a pheromone and opaque α/α cells release α pheromone. Opaque α/α and a/a cells then respond to these alternative pheromones by evaginating to form shmoos that undergo chemotropism, membrane fusion, nuclear fusion, evagination, and daughter cell formation (Fig. 8B) (38, 325, 326, 328, 329). Interestingly, white cells, which themselves cannot normally mate, play a role in mating. White cells form robust biofilms that are architecturally similar to the biofilms formed by a/α cells but differ functionally in that they support mating between minority opaque cells (24, 38, 330). Opaque cells, which themselves cannot form a biofilm, play a stimulatory role in the formation of MTL-homozygous white cell biofilms (39).
FIG 8.
To understand the profound difference in biofilms formed by MTL-heterozygous and MTL-homozygous cells, white-opaque switching, opaque cell mating, and the role of α pheromone in the formation of MTL-homozygous biofilms must be taken into account. (A) MTL homozygosis and white-opaque (Wh-Op) switching. (B) Shmoo formation induced by pheromones of opposite mating types and pheromone-directed chemotropism in the process of fusion. (C) MTL-homozygous white cell biofilms are induced by the unorthodox release of pheromone of the opposite mating type by rare opaque cells which appear through switching. In this example, white a/a cells produce minority opaque a/a cells. The latter release α pheromone in an unorthodox fashion, which induces white a/a cells to form a biofilm. Op, opaque; hy, hyphae; bp, basal cell polylayer; ECM, extracellular matrix.
ARCHITECTURE OF MTL-HOMOZYGOUS BIOFILMS AND THE ROLE OF PHEROMONE
When white MTL-homozygous cells are incubated in the model employed by Daniels et al. (38, 183, 184), a biofilm is formed that consists of a basal yeast cell polylayer, which accounts for the lower 20% of the biofilm, and an upper region of vertically oriented hyphae encapsulated in ECM, which accounts for 80% of the biofilm. However, MTL-homozygous biofilms formed by white cells are, on average, 25% thinner than MTL-heterozygous biofilms (183, 251). Most of the difference is erased if minority (10%) MTL-homozygous opaque cells of the opposite mating type are added to the white cell population (38). A mixture of 1:1 a/a and α/α opaque cells is added because opposite mating types stimulate each other to release mating factor. However, adding more than 40% opaque cells decreases biofilm thickness (38), probably because opaque cells, which do not make bona fide biofilms, physically interfere with white cell biofilm formation. Stimulation of white cell biofilm formation by minority opaque cells led to the discovery that the white-to-opaque transition regulates white cell biofilm formation (39). MTL-heterozygous biofilm formation is regulated by the RAS1/cyclic AMP (cAMP) pathway, while MTL-homozygous biofilm formation is regulated by the mitogen-activated protein (MAP) kinase pathway (39, 243, 318, 331–333). It was hypothesized that minority opaque cells added to majority white cells stimulate the thickness of MTL-homozygous biofilms by releasing pheromone, which activates the MAP kinase pathway (39). It was subsequently demonstrated that a small minority of opaque cells (i.e., <1%), which appear in a majority white cell population by spontaneous switching (327, 334, 335), release the pheromone of the opposite mating type in an unorthodox fashion (39). The secreted pheromone induces biofilm formation in a paracrine system (39). In other words, minority α/α cells in a majority a/a white cell population secrete α pheromone, which stimulates biofilm formation by a/a white cells. Deletion of the α-pheromone gene results in the formation of highly defective a/a white cell biofilms (39). The addition of 10% opaque cells (1:1 mixture of a/a and α/α cells) to an a/a or α/α white cell population stimulates biofilm thickness, presumably by providing additional pheromone of the opposite mating type (39).
PERMEABILITY, PENETRABILITY, AND DRUG SUSCEPTIBILITY OF ALTERNATIVE BIOFILMS
Although the general architecture of biofilms formed by MTL-homozygous cells is similar to that of biofilms formed by MTL-heterozygous cells, pathogenic traits and the capacity to facilitate mating differ. When a 48-h MTL-heterozygous biofilm developed in RPMI 1640 (MOPS) medium in the Douglas model was overlaid with either Sypro Ruby (1.6 kDa) or ConA (104 to 112 kDa) and incubated for an additional 30 min, there was minimal penetrance (∼15%) by either the low- or high-molecular-mass molecule (39). However, when an MTL-homozygous biofilm was treated similarly, both the low- and high-molecular-mass molecules penetrated rapidly through approximately 80% of the biofilm (39). Similarly, when 48-h biofilms were overlaid with DiI-labeled human polymorphonuclear leukocytes and incubated for 3 h, the leukocytes penetrated through only 5% of an MTL-heterozygous biofilm but through approximately 80% of an MTL-homozygous biofilm (39, 183, 264). Finally, alternative biofilms exhibit a dramatically different susceptibility to fluconazole. When a 48-h MTL-heterozygous biofilm was treated with 25 μg per ml of fluconazole for 24 additional hours, approximately 5% of cells stained with Dead Red, a dead cell nuclear stain. However, when MTL-homozygous biofilms were treated similarly, approximately 40% of cells in the biofilm stained with Dead Red (39). All of the above differences were demonstrated for three unrelated a/α strains and their a/a and α/α derivatives, the latter obtained through spontaneous homozygosis at the MTL locus (39). MTL-heterozygous and MTL-homozygous biofilms therefore represent perhaps the most extreme example of biofilms that look alike but are functionally disparate.
“SEXUAL BIOFILMS”: LESSONS FROM BACTERIA
The relationship between mating and biofilm formation is not a novel aspect of C. albicans. It has long been recognized that most bacteria live in biofilms in natural or host niches, so it was likely that gene transfer through conjugation occurred under these conditions. Hausner and Wuertz (167) demonstrated that conjugation occurs at higher frequencies in bacterial biofilms than in planktonic cultures. Conjugation in bacteria, discovered approximately 70 years ago by Lederberg and Tatum (336), involves the transfer of genetic material, usually as a plasmid or transposon, through cell-cell contact. Licht et al. (337) showed that donor cells placed on the upper surface of an E. coli biofilm transfer genetic material only to surface cells, which may be due to the restricted contact. Not only is conjugation facilitated by bacterial biofilm formation, but mating in turn can enhance biofilm formation through conjugation pili, as demonstrated for E. coli (166). Fimbriae, which are specialized pili, function in adhesion (338, 339). It therefore seemed logical to consider that if sex is facilitated by biofilms for bacteria, the same may hold true for C. albicans. Obviously, the advantage of performing mating within a biofilm relates to proximity for both bacteria and C. albicans. However, there is some question concerning the timing and location of bacterial mating in bacterial biofilms, since the encapsulation of cells in a matrix (340) may prevent contact, an issue dealt with by Molin and Tolker-Nielsen (341). Although the work on mating in bacterial biofilms began earlier than that on C. albicans biofilms, there have been no reports of the formation of functionally distinct nonsexual and sexual biofilms of bacteria, as is the case for MTL-heterozygous and MTL-homozygous C. albicans biofilms.
FACILITATION OF MATING BY SEXUAL BIOFILMS
Several observations led to the hypothesis that MTL-homozygous cells form “sexual” biofilms. First, planktonic MTL-homozygous white cells in stationary phase are induced to adhere to plastic or catheter material by pheromone (39, 319). Second, opaque cells signal white cells, through the release of pheromone, to form a white cell biofilm, employing the same pheromone receptors, trimeric G protein complex, and MAP kinase pathway as those that regulate mating between opaque cells, but with different target transcription factors (243, 319, 332). Third, and most importantly, when minority a/a and α/α opaque cells are seeded in majority a/a and/or α/α white cell biofilms, they mate 50 to 100 times more frequently than minority a/a and α/α opaque cells seeded in majority a/α biofilms (330). Furthermore, Daniels et al. (38) showed, by vitally staining a/a and α/α cells with tags of different colors, that an MTL-homozygous white cell biofilm supports chemotropism by conjugation tubes, which leads to fusion (Fig. 9).
FIG 9.
Demonstration of chemotropism in an early MTL-homozygous white cell biofilm. The seeded a/a opaque cell was stained green with fluorescein-conjugated ConA, and the seeded α/α opaque cell was stained red with rhodamine-conjugated ConA. The conjugation tubes that formed are stained blue. Note how they extended toward each other by sensing gradients of alternative mating type pheromone released by the opaque cells, in the process of chemotropism. (Reproduced from reference 38 with permission.)
The reasons for the differences in MTL-heterozygous and MTL-homozygous biofilms have been attributed to alternative functions and may be the result of differences in the ECM. The characteristics of MTL-heterozygous biofilms, including impermeability to high- and low-molecular-weight molecules, including the azole fluconazole, and impenetrability by white blood cells (39, 183, 184, 251), are compatible with a pathogenic role in the life history of this commensal but incompatible with the generation of pheromone gradients that direct chemotropism in the mating process and the directed extension of conjugation tubes during chemotropism (33, 38, 52, 82, 330). In contrast, the characteristics of MTL-homozygous biofilms, including permeability to low- and high-molecular weight molecules, including fluconazole, and penetrability by white blood cells, are incompatible with a pathogenic role but compatible with chemotropism, and hence a sexual role. It seems reasonable to suggest that in an MTL-homozygous white cell biofilm, spontaneous switching to opaque by a minority of cells results in mating, which is facilitated by the ECM of the white cell biofilm. The observation that opaque cells do not form biofilms is consistent with this scenario (38). This specialization of white cell biofilm formation may be the reason that white-opaque switching evolved in C. albicans and related species, including C. dubliniensis and Candida tropicalis (44, 342, 343).
IMPACT OF ENVIRONMENTAL CONDITIONS ON MTL-HOMOZYGOUS BIOFILMS
As is the case for MTL-heterozygous biofilms, the culture conditions used for the formation of an MTL-homozygous biofilm can affect the biofilm's architecture, phenotypic transitions, and function. Although the literature for MTL-homozygous biofilms is more limited than that for MTL-heterozygous biofilms, several recent studies focused on the effects of environmental conditions on MTL-homozygous biofilm formation, using variations of the Douglas model. In a comparison of two media used to support biofilm formation, white a/a biofilms of natural strain P37005 were found to be three times thicker when formed in RPMI 1640 (MOPS) medium than when formed in Spider medium (183). In addition, whereas MTL-homozygous white cells formed a basal yeast cell polylayer and an upper region of vertically oriented hyphae embedded in a dense ECM in RPMI 1640 medium at 37°C in air, in Spider medium they formed a loose, poorly adhesive, and poorly cohesive polylayer of yeast cells, with no significant upper hyphal region and with little ECM (183). The predominately yeast-phase cell biofilm formed in Spider medium was twice as susceptible to fluconazole as the already partially susceptible biofilm formed by MTL-homozygous cells in RPMI 1640 medium under otherwise identical conditions. MTL-homozygous biofilms formed in Spider medium were also far more penetrable by human polymorphonuclear leukocytes than the partially penetrable biofilms formed under the same conditions in RPMI 1640 medium (183). Furthermore, the biofilms formed by white MTL-homozygous cells in Spider medium did not support mating between seeded minority a/a and α/α cells, as did white MTL-homozygous biofilms formed in RPMI 1640 (MOPS) medium (183). However, white a/a cells did form biofilms in Spider medium if the air was replaced with 20% CO2. These biofilms were architecturally indistinguishable from a/α biofilms formed in Spider medium in air (183). For a/α cells in Spider medium, similar biofilms formed in either air or CO2. These results poignantly demonstrate that a single parameter, in this case the concentration of CO2, dictates whether or not MTL-homozygous white cells form bona fide biofilms in Spider medium. CO2, however, does not dictate whether a bona fide biofilm forms in RPMI 1640 medium. Daniels et al. (183) further demonstrated that a white a/a biofilm formed in Lee's medium for 24 h, a condition used by Lin et al. (344), was not comparable to a biofilm formed in RPMI 1640 medium for 48 h. The biofilm formed in Lee's medium was 1/10 as thick as the latter, formed no cohesive basal yeast cell polylayer, and had no upper layer dominated by hyphae encapsulated in ECM. Despite the dramatic decrease in thickness and lack of architecture and function, the addition of α pheromone was shown to stimulate thickness of MTL-homozygous biofilms formed in Spider medium (344). The stimulated biofilms, however, were still less than half as thick as the one formed in RPMI 1640 (MOPS) medium, and they lacked an organized architecture (183). These results demonstrate that although white-phase a/a cells are responsive to α pheromone in Lee's medium in air, they do not make bona fide biofilms and in Spider medium will not form an upper hyphal region, unless they are in an atmosphere containing high CO2. These results underscore the importance of knowing which environmental conditions are necessary for supporting formation of bona fide MTL-homozygous biofilms before experiments are performed on mutants or to determine drug susceptibility or gene expression.
STRAIN VARIABILITY OF MTL-HOMOZYGOUS BIOFILMS
Using the Douglas model with RPMI 1640 (MOPS) medium, Pujol et al. (45) analyzed six a/a and six α/α strains for biofilm architecture. They found an extraordinarily high degree of architectural uniformity (Table 3). All 12 strains formed a basal yeast cell polylayer, an upper layer of vertically oriented hyphae, and a dense extracellular matrix. More surprisingly, the means ± standard deviations for 48-h biofilm thicknesses for the six a/a and six α/α strains were 56 ± 3 μm and 58 ± 2 μm, respectively (45). The standard deviations were therefore 5 and 3%, respectively, of the means (Table 3), reflecting an extremely high level of uniformity, and therefore conservation, among strains. This level of variability was far below that observed in other studies of variability among a/α strains (Table 3). However, the mean ± standard deviation for the dry weights of biofilms formed by 10 MTL-homozygous strains was 0.75 ± 0.13 mg (Table 3) (45). The standard deviation in this case was 17% of the mean, reflecting a level of variability consistent with that observed in the majority of other studies reviewed in Table 3. This provides another example of the differences in variability that can exist between different biofilm parameters.
CONCLUDING REMARKS
This review focused on the variability of C. albicans biofilms formed in in vitro models, based on differences in strains, methods of assessment, the conditions used to develop biofilms, and the configuration of the MTL locus. It was undertaken to engender awareness that more attention must be paid to the strains employed, the methods used to assess biofilm development, and the models used to generate them in vitro. Therefore, it seems appropriate to conclude with a summary of basic questions or issues that C. albicans researchers should consider in comparing the results of different studies and developing experimental protocols for new studies. First, if one is analyzing a single strain of C. albicans in vitro, does it form a representative biofilm? One would not want to employ a strain which forms a biofilm that exhibits a basic parameter close to the lower or upper extreme for the species. Extremes can vary 2-fold or more (Table 3). Second, if one is assessing variability among a collection of natural strains, it would seem prudent to type them for the configuration of the MTL locus. It should be recognized that between 5 and 10% of clinical isolates are MTL homozygous. Moreover, some a/α strains undergo MTL homozygosis at extremely high rates, and environmental conditions, such as the use of sorbose as the carbon source, can select for MTL-homozygous derivatives. The biofilms of MTL-homozygous strains are functionally different from those of MTL-heterozygous strains. For some traits, such as drug susceptibility, a collection of strains may be distributed biphasically if the collection contains a disproportionate number of MTL-homozygous isolates. For gene regulation, the differences between MTL-homozygous and MTL-heterozygous biofilms may be profound. Third, one time point may not suffice for assessing the effects of an antifungal drug, a mutation, or an environmental condition on biofilm formation. A biofilm is developmentally programmed, and we are still ignorant of the complexity of that program. It appears that most studies do not take into account that biofilm traits, architecture, physiology, and function probably change with time during development. Therefore, multiple time points must be considered. Fourth, architectural, phenotypic, and ECM assessments should be performed in conjunction with studies on the effects of antifungal drugs, gene expression, the physiology of cells, the biochemistry of the matrix, the effects of mutation, and the effects of environmental conditions. If a mutation, an antifungal agent, or an environmental condition selectively inhibits a biofilm trait, such as hypha formation, matrix deposition, general growth, or dispersal, conclusions must be formulated with that in mind. In these cases, a measurement such as XTT reduction is not sufficient. Fifth, interpretations of data must consider the phenotypic complexity of the biofilm, and hence the possibility or probability that a measurement may represent an amalgam of phenotypes. Sixth, one must consider valid controls and comparisons in identifying biofilm-specific traits. If a measurement is made for a particular characteristic, such as gene expression on a 48-h biofilm preparation, why is a planktonic culture of yeast-phase cells used as the sole preparation for comparison? We know that hyphal growth is distinct from yeast cell multiplication in terms of cellular architecture, gene expression, molecular composition, cell cycle, and physiology. Since C. albicans biofilms are composed of hyphae as well as yeast-phase cells, why not add hyphal growth in suspension as a control? Seventh, researchers must be aware that biofilms have almost completely stopped growing at 48 h in most in vitro models. Therefore, 48-h time points provide information on biofilm development but also, later, on the final mature biofilm. Eighth, selecting a method of assessment solely for expediency may not provide a valid answer. XTT measurements are fraught with problems and have been shown, as early as 1998, not to correlate consistently with other parameters, such as dry weight. In addition, it should be realized that preparing cells for imaging, such as for SEM, can collapse a biofilm, destroy the architecture, or extract the ECM. CLSM preserves architecture but does not provide high-resolution images of cellular phenotype and can include staining artifacts. Gene expression profiles are not expedient, and their reproducibility, especially for low-abundance transcripts, has not been resolved. Measurements of dry weight provide nothing more than information on mass. Obviously, assessments of architecture, mass, cellular phenotypes, gene expression, and metabolism are synergistic in examinations of biofilm development. The more that is known about a preparation, the more impact a study will have. Ninth, perhaps the most important consideration is the model employed. Different conditions may result in different biofilms. These include the conditions used for planktonic growth, the concentration of the initial inoculum, the period of initial adhesion, the medium used to support biofilm formation, the substratum, temperature, atmosphere (air versus increasing levels of CO2), and fluid flow. Hopefully, concern for the above issues will increase the relevance of future studies and facilitate more valid comparisons of the results obtained in different in vitro studies. If nothing else, this review should engender the notion that not all biofilms are created equal.
ACKNOWLEDGMENTS
We are indebted to Claude Pujol for his suggestions and help in interpreting the data obtained in the genome-wide transcriptional profiling studies and to Sandra Beck for help in assembling the manuscript.
Work in the Soll laboratory is funded by the Developmental Studies Hybridoma Bank, a National Resource created by the NIH.
REFERENCES
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Crespi BJ. 2001. The evolution of social behavior in microorganisms. Trends Ecol Evol 16:178–183. doi: 10.1016/S0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 3.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro JA. 1988. Bacteria as multicellular organisms. Sci Am 258:82–89.2847312 [Google Scholar]
- 5.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 6.Zobell CE, Allen EC. 1935. The significance of marine bacteria in the fouling of submerged surfaces. J Bacteriol 29:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potera C. 1996. Biofilms invade microbiology. Science 273:1795–1797. doi: 10.1126/science.273.5283.1795. [DOI] [PubMed] [Google Scholar]
- 8.Reid P, Dupraz C, Visscher P, Sumner D. 2003. Microbial processes forming marine stromatolites, p 103–118. In Krumbein WE, Paterson DM, Zavarin GA (ed), Fossil and recent biofilms: a natural history of life on Earth. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 9.Berkner L, Marshall L. 1963. The history of growth of oxygen in the earth's atmosphere, p 102 In Brancazio PJ, Cameron AGW (ed), The origin and evolution of atmospheres and oceans. Proceedings of a conference, NASA, Goddard Space Flight Center, Goddard Institute for Space Studies, New York, NY, April 8, 9, 1963 Wiley, New York, NY. [Google Scholar]
- 10.Kasting JF, Siefert JL. 2002. Life and the evolution of Earth's atmosphere. Science 296:1066–1068. doi: 10.1126/science.1071184. [DOI] [PubMed] [Google Scholar]
- 11.Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC. 2013. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc Natl Acad Sci U S A 110:1791–1796. doi: 10.1073/pnas.1209927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer AG. 1965. Fossils, early life, and atmospheric history. Proc Natl Acad Sci U S A 53:1205–1215. [Google Scholar]
- 13.Krumbein WE, Paterson DM, Zavarzin GA (ed). 2003. Fossil and recent biofilms: a natural history of life on Earth. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 14.Burns BP, Goh F, Allen M, Neilan BA. 2004. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ Microbiol 6:1096–1101. doi: 10.1111/j.1462-2920.2004.00651.x. [DOI] [PubMed] [Google Scholar]
- 15.Logan BW. 1961. Cryptozoon and associate stromatolites from the Recent, Shark Bay, Western Australia. J Geol 69:517–533. doi: 10.1086/626769. [DOI] [Google Scholar]
- 16.Neilan BA, Burns BP, Relman DA, Lowe DR. 2002. Molecular identification of cyanobacteria associated with stromatolites from distinct geographical locations. Astrobiology 2:271–280. doi: 10.1089/153110702762027853. [DOI] [PubMed] [Google Scholar]
- 17.Reitner J, Quéric N-V, Arp G (ed). 2011. Advances in stromatolite geobiology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 18.Albertano P. 2012. Cyanobacterial biofilms in monuments and caves, p 317–343. In Whitton BA. (ed), Ecology of cyanobacteria II. Springer Science+Business Media, Dordrecht, The Netherlands. [Google Scholar]
- 19.Crispim C, Gaylarde C. 2005. Cyanobacteria and biodeterioration of cultural heritage: a review. Microb Ecol 49:1–9. doi: 10.1007/s00248-003-1052-5. [DOI] [PubMed] [Google Scholar]
- 20.Ljaljević-Grbić M, Vukojević J, Subakov-Simić G, Krizmanić J, Stupar M. 2010. Biofilm forming cyanobacteria, algae and fungi on two historic monuments in Belgrade, Serbia. Arch Biol Sci 62:625–631. doi: 10.2298/ABS1003625L. [DOI] [Google Scholar]
- 21.Costerton JW, Stewart PS, Greenberg E. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 22.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 24.Marrie T, Costerton J. 1984. Scanning and transmission electron microscopy of in situ bacterial colonization of intravenous and intraarterial catheters. J Clin Microbiol 19:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn D, Chandra J, Mukherjee P, Ghannoum M. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun 70:878–888. doi: 10.1128/IAI.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumamoto CA, Vinces MD. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol 59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- 28.Radford D, Challacombe S, Walter J. 1999. Denture plaque and adherence of Candida albicans to denture-base materials in vivo and in vitro. Crit Rev Oral Biol Med 10:99–116. doi: 10.1177/10454411990100010501. [DOI] [PubMed] [Google Scholar]
- 29.Ramage G, Martínez JP, López-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 30.Ramage G, Tomsett K, Wickes BL, López-Ribot JL, Redding SW. 2004. Denture stomatitis: a role for Candida biofilms. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 98:53–59. doi: 10.1016/j.tripleo.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Seneviratne C, Jin L, Samaranayake L. 2008. Biofilm lifestyle of Candida: a mini review. Oral Dis 14:582–590. doi: 10.1111/j.1601-0825.2007.01424.x. [DOI] [PubMed] [Google Scholar]
- 32.Veerachamy S, Yarlagadda T, Manivasagam G, Yarlagadda PK. 2014. Bacterial adherence and biofilm formation on medical implants: a review. Proc Inst Mech Eng H 228:1083–1099. doi: 10.1177/0954411914556137. [DOI] [PubMed] [Google Scholar]
- 33.Soll DR. 2009. Why does Candida albicans switch? FEMS Yeast Res 9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- 34.Soll DR. 2014. The evolution of alternative biofilms in an opportunistic fungal pathogen: an explanation for how new signal transduction pathways may evolve. Infect Genet Evol 22:235–243. doi: 10.1016/j.meegid.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 36.Seneviratne C, Yip J, Chang J, Zhang C, Samaranayake L. 2013. Effect of culture media and nutrients on biofilm growth kinetics of laboratory and clinical strains of Enterococcus faecalis. Arch Oral Biol 58:1327–1334. doi: 10.1016/j.archoralbio.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Webb JS, Lau M, Kjelleberg S. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J 25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR. 2011. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol 9:e1001117. doi: 10.1371/journal.pbio.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odds FC, Bougnoux M-E, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li S-Y, Tavanti A. 2007. Molecular phylogenetics of Candida albicans. Eukaryot Cell 6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J Clin Microbiol 43:5601–5613. doi: 10.1128/JCM.43.11.5601-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilfillan GD, Sullivan DJ, Haynes K, Parkinson T, Coleman DC, Gow NA. 1998. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology 144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 44.Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell 3:1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pujol C, Daniels KJ, Soll DR. 2015. Comparison of switching and biofilm formation between MTL-homozygous strains of Candida albicans and Candida dubliniensis. Eukaryot Cell 14:1186–1202. doi: 10.1128/EC.00146-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan D, Coleman D. 1998. Candida dubliniensis: characteristics and identification. J Clin Microbiol 36:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desai JV, Mitchell AP, Andes DR. 2014. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med 4:a019729. doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fox EP, Nobile CJ. 2012. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription 3:315–322. doi: 10.4161/trns.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathé L, Van Dijck P. 2013. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet 59:251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramage G, Robertson SN, Williams C. 2014. Strength in numbers: antifungal strategies against fungal biofilms. Int J Antimicrob Agents 43:114–120. doi: 10.1016/j.ijantimicag.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 52.Soll DR. 2014. The role of phenotypic switching in the basic biology and pathogenesis of Candida albicans. J Oral Microbiol 6:22993. doi: 10.3402/jom.v6.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai JV, Mitchell AP. 2015. Candida albicans biofilm development and its genetic control. Microbiol Spectr 3:MB-0005. doi: 10.1128/microbiolspec.MB-0005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce CG, Srinivasan A, Ramasubramanian AK, López-Ribot JL. 2015. From biology to drug development: new approaches to combat the threat of fungal biofilms. Microbiol Spectr 3:MB-0007. doi: 10.1128/microbiolspec.MB-0007-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandra J, Mukherjee PK. 2015. Candida biofilms: development, architecture, and resistance. Microbiol Spectr 3:MB-0020. doi: 10.1128/microbiolspec.MB-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slavkin HC. 1997. Biofilms, microbial ecology and Antoni van Leeuwenhoek. J Am Dent Assoc 128:492–495. doi: 10.14219/jada.archive.1997.0238. [DOI] [PubMed] [Google Scholar]
- 57.Van Leeuwenhoek A. 1684. Microscopical observations about animals in the scurf of the teeth. Philos Trans 1684:568–574. [Google Scholar]
- 58.Zobell CE. 1943. The effect of solid surfaces upon bacterial activity. J Bacteriol 46:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zobell CE. 1947. Marine bacteriology. Annu Rev Biochem 16:565–586. doi: 10.1146/annurev.bi.16.070147.003025. [DOI] [PubMed] [Google Scholar]
- 60.Costerton JW, Geesey G, Cheng K. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 61.Ehrlich GD, Arciola CR. 2012. From Koch's postulates to biofilm theory. The lesson of Bill Costerton. Int J Artif Organs 35:695–699. doi: 10.5301/ijao.5000169. [DOI] [PubMed] [Google Scholar]
- 62.DeLong EF, Franks DG, Alldredge AL. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 38:924–934. doi: 10.4319/lo.1993.38.5.0924. [DOI] [Google Scholar]
- 63.Fletcher M. 1984. Comparative physiology of attached and free-living bacteria, p 223–232. In Marshall KC. (ed), Microbial adhesion and aggregation. Springer, Berlin, Germany. [Google Scholar]
- 64.Middelboe M, Søndergaard M, Letarte Y, Borch N. 1995. Attached and free-living bacteria: production and polymer hydrolysis during a diatom bloom. Microb Ecol E 29:231–248. doi: 10.1007/BF00164887. [DOI] [PubMed] [Google Scholar]
- 65.Jackson CR, Churchill PF, Roden EE. 2001. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82:555–566. doi: 10.1890/0012-9658(2001)082[0555:SCIBAS]2.0.CO;2. [DOI] [Google Scholar]
- 66.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 67.Stoodley P, Sauer K, Davies D, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 68.Anderson JM, Rodriguez A, Chang DT. 2008. Foreign body reaction to biomaterials. Semin Immunol 20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Branda SS, Vik Å, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Flemming H-C, Wingender J, Griegbe T, Mayer C. 2000. Physico-chemical properties of biofilms, p 19–34. In Evans LV. (ed), Biofilms: recent advances in their study and control. Harwood Academic Publishers, Amsterdam, Netherlands. [Google Scholar]
- 71.Holmes LF. 1941. The effect of surface-tension depressants on certain serological systems. Yale J Biol Med 14:155. [PMC free article] [PubMed] [Google Scholar]
- 72.Parsek MR, Greenberg E. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Yale J Biol Med 13:27–33. [DOI] [PubMed] [Google Scholar]
- 73.Tran CS, Rangel SM, Almblad H, Kierbel A, Givskov M, Tolker-Nielsen T, Hauser AR, Engel JN. 2014. The Pseudomonas aeruginosa type III translocon is required for biofilm formation at the epithelial barrier. PLoS Pathog 10:e1004479. doi: 10.1371/journal.ppat.1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunne WM. 2002. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garrett TR, Bhakoo M, Zhang Z. 2008. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci 18:1049–1056. doi: 10.1016/j.pnsc.2008.04.001. [DOI] [Google Scholar]
- 76.Hori K. 2015. Adhesion of bacteria, p 23–34. In Kanematsu H, Barry DM (ed), Biofilm and materials science. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 77.Katsikogianni M, Missirlis Y. 2004. Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur Cell Mater 8:37–57. [DOI] [PubMed] [Google Scholar]
- 78.Characklis W. 1981. Bioengineering report. Fouling biofilm development: a process analysis. Biotechnol Bioeng 23:1923–1960. [DOI] [PubMed] [Google Scholar]
- 79.Fletcher M, Marshall K. 1982. Are solid surfaces of ecological significance to aquatic bacteria? Adv Microb Ecol 6:199–236. doi: 10.1007/978-1-4615-8318-9_6. [DOI] [Google Scholar]
- 80.Dressaire C, Moreira RN, Barahona S, Alves de Matos AP, Arraiano CM. 2015. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 6:e02352-14. doi: 10.1128/mBio.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soll DR. 2011. Evolution of a new signal transduction pathway in Candida albicans. Trends Microbiol 19:8–13. doi: 10.1016/j.tim.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 83.Mahajan A, Currie CG, Mackie S, Tree J, McAteer S, McKendrick I, McNeilly TN, Roe A, La Ragione RM, Woodward MJ. 2009. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell Microbiol 11:121–137. doi: 10.1111/j.1462-5822.2008.01244.x. [DOI] [PubMed] [Google Scholar]
- 84.Friedlander RS, Vogel N, Aizenberg J. 2015. Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31:6137–6144. doi: 10.1021/acs.langmuir.5b00815. [DOI] [PubMed] [Google Scholar]
- 85.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 86.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 87.Van Houdt R, Michiels CW. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 88.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. 2015. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol 5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chagnot C, Zorgani MA, Astruc T. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol 20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 91.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Costerton J, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. 1994. Biofilms, the customized microniche. J Bacteriol 176:2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davies D, Geesey G. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol 61:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diggle SP, Gardner A, West SA, Griffin AS. 2007. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc Lond B Biol Sci 362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Okegbe C, Price-Whelan A, Dietrich LE. 2014. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol 18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A 109:1116–1121. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vroom JM, De Grauw KJ, Gerritsen HC, Bradshaw DJ, Marsh PD, Watson GK, Birmingham JJ, Allison C. 1999. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl Environ Microbiol 65:3502–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhaskar P, Bhosle NB. 2005. Microbial extracellular polymeric substances in marine biogeochemical processes. Curr Sci 88:45–53. [Google Scholar]
- 100.Gristina AG, Costerton J. 1984. Bacterial adherence and the glycocalyx and their role in musculoskeletal infection. Orthop Clin N Am 15:517–535. [PubMed] [Google Scholar]
- 101.Karimi A, Karig D, Kumar A, Ardekani AM. 2015. Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab Chip 15:23–42. doi: 10.1039/C4LC01095G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Beer D, Stoodley P, Lewandowski Z. 1994. Liquid flow in heterogeneous biofilms. Biotechnol Bioeng 44:636–641. doi: 10.1002/bit.260440510. [DOI] [PubMed] [Google Scholar]
- 103.Bowden GH, Li YH. 1997. Nutritional influences on biofilm development. Adv Dent Res 11:81–99. doi: 10.1177/08959374970110012101. [DOI] [PubMed] [Google Scholar]
- 104.Davey ME, Caiazza NC, O'Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McNally L, Brown SP. 2015. Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos Trans R Soc Lond B Biol Sci 370:20140298. doi: 10.1098/rstb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 107.Magee B, Magee P. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 108.Renner LD, Weibel DB. 2011. Physicochemical regulation of biofilm formation. MRS Bull 36:347–355. doi: 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grandclement C, Tannieres M, Morera S, Dessaux Y, Faure DD. 2016. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev 40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 110.Lawrence J, Korber D, Hoyle B, Costerton J, Caldwell D. 1991. Optical sectioning of microbial biofilms. J Bacteriol 173:6558–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoon MY, Lee K-M, Park Y, Yoon SS. 2011. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One 6:e16105. doi: 10.1371/journal.pone.0016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abee T, Kovács AT, Kuipers OP, Van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 115.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 116.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg E. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 117.Caiazza NC, O'Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol 186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purevdorj-Gage B, Costerton WJ, Stoodley P. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151:1569–1576. doi: 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- 119.Friedlander RS, Vlamakis H, Kim P, Khan M, Kolter R, Aizenberg J. 2013. Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc Natl Acad Sci U S A 110:5624–5629. doi: 10.1073/pnas.1219662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yamamoto K, Arai H, Ishii M, Igarashi Y. 2012. Involvement of flagella-driven motility and pili in Pseudomonas aeruginosa colonization at the air-liquid interface. Microbes Environ 27:320–323. doi: 10.1264/jsme2.ME11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 124.Kim SK, Lee JH. 2016. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol 54:71–85. doi: 10.1007/s12275-016-5528-7. [DOI] [PubMed] [Google Scholar]
- 125.Kirisits MJ, Parsek MR. 2006. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell Microbiol 8:1841–1849. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- 126.von Bodman SB, Willey JM, Diggle SP. 2008. Cell-cell communication in bacteria: united we stand. J Bacteriol 190:4377–4391. doi: 10.1128/JB.00486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams P, Winzer K, Chan WC, Camara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci 362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hurle JM, Ros MA, Climent V, Garcia-Martinez V. 1996. Morphology and significance of programmed cell death in the developing limb bud of the vertebrate embryo. Microsc Res Tech 34:236–246. [DOI] [PubMed] [Google Scholar]
- 129.Zuzarte-Luís V, Hurlé JM. 2002. Programmed cell death in the developing limb. Int J Dev Biol 46:871–876. [PubMed] [Google Scholar]
- 130.Asally M, Kittisopikul M, Rué P, Du Y, Hu Z, Çağatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Süel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H, Bereswill S. 2009. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One 4:e6785. doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sadykov MR, Bayles KW. 2012. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol 15:211–215. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen X, Stewart P. 2002. Role of electrostatic interactions in cohesion of bacterial biofilms. Appl Microbiol Biotechnol 59:718–720. doi: 10.1007/s00253-002-1044-2. [DOI] [PubMed] [Google Scholar]
- 134.Cooksey K. 1992. Extracellular polymers in biofilms, p 137–147. In Melo LF, Bott TR, Fletcher M, Capdeville N (ed), Biofilms—science and technology. Springer Science+Business Media, Dordrecht, The Netherlands. [Google Scholar]
- 135.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 136.Sherrard LJ, Tunney MM, Elborn JS. 2014. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384:703–713. doi: 10.1016/S0140-6736(14)61137-5. [DOI] [PubMed] [Google Scholar]
- 137.Stewart PS. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother 40:2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cabral DA, Loh BA, Speert DP. 1987. Mucoid Pseudomonas aeruginosa resists nonopsonic phagocytosis by human neutrophils and macrophages. Pediatr Res 22:429–431. doi: 10.1203/00006450-198710000-00013. [DOI] [PubMed] [Google Scholar]
- 139.Kelly N, Kluftinger J, Pasloske B, Paranchych W, Hancock R. 1989. Pseudomonas aeruginosa pili as ligands for nonopsonic phagocytosis by fibronectin-stimulated macrophages. Infect Immun 57:3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol 175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 141.Mahenthiralingam E, Campbell ME, Speert DP. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun 62:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolcott RD, Rhoads DD, Dowd SE. 2008. Biofilms and chronic wound inflammation. J Wound Care 17:333–341. doi: 10.12968/jowc.2008.17.8.30796. [DOI] [PubMed] [Google Scholar]
- 143.Schmid T, Burkhard J, Yeo B-S, Zhang W, Zenobi R. 2008. Towards chemical analysis of nanostructures in biofilms. I. Imaging of biological nanostructures. Anal Bioanal Chem 391:1899–1905. doi: 10.1007/s00216-008-2100-2. [DOI] [PubMed] [Google Scholar]
- 144.Sutherland IW. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 145.Aguilar C, Vlamakis H, Losick R, Kolter R. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Flemming HC, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells.” J Bacteriol 189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 149.Okshevsky M, Regina VR, Meyer RL. 2015. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol 33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 150.Schlafer S, Meyer RL. 11 March 2016. Confocal microscopy imaging of the biofilm matrix. J Microbiol Methods doi: 10.1016/j.mimet.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 151.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 152.McCrate OA, Zhou X, Reichhardt C, Cegelski L. 2013. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J Mol Biol 425:4286–4294. doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Åberg V, Walker JN, Seed PC, Almqvist F. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lin Y, Sharma P, van Loosdrecht M. 2013. The chemical and mechanical differences between alginate-like exopolysaccharides isolated from aerobic flocculent sludge and aerobic granular sludge. Water Res 47:57–65. doi: 10.1016/j.watres.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 155.Wu C, Lim JY, Fuller GG, Cegelski L. 2012. Quantitative analysis of amyloid-integrated biofilms formed by uropathogenic Escherichia coli at the air-liquid interface. Biophys J 103:464–471. doi: 10.1016/j.bpj.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 157.Neu TR. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev 60:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vu B, Chen M, Crawford RJ, Ivanova EP. 2009. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Costerton JW, Cheng K, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 160.Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 161.Izano EA, Sadovskaya I, Wang H, Vinogradov E, Ragunath C, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB. 2008. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathog 44:52–60. doi: 10.1016/j.micpath.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton J, Greenberg E. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 163.Watnick P, Kolter R. 2000. Biofilm, city of microbes. J Bacteriol 182:2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Landini P, Antoniani D, Burgess JG, Nijland R. 2010. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl Microbiol Biotechnol 86:813–823. doi: 10.1007/s00253-010-2468-8. [DOI] [PubMed] [Google Scholar]
- 165.de la Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol 8:128–133. doi: 10.1016/S0966-842X(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 166.Ghigo J-M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 167.Hausner M, Wuertz S. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol 65:3710–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hawser SP, Douglas LJ. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun 62:915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Odds FC. 1988. Candida and candidosis: a review and bibliography. Bailliere Tindall, London, United Kingdom. [Google Scholar]
- 170.Laffery SF, Butler G. 2005. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 151:1073–1081. [DOI] [PubMed] [Google Scholar]
- 171.Wilson LA, Sawant AD, Ahearn DG. 1991. Comparative efficacies of soft contact lens disinfectant solutions against microbial films in lens cases. Arch Ophthalmol 109:1155–1157. doi: 10.1001/archopht.1991.01080080115043. [DOI] [PubMed] [Google Scholar]
- 172.Neu TR, Verkerke GJ, Herrmann IF, Schutte HK, Van der Mei HC, Busscher HJ. 1994. Microflora on explanted silicone rubber voice prostheses: taxonomy, hydrophobicity and electrophoretic mobility. J Appl Bacteriol 76:521–528. doi: 10.1111/j.1365-2672.1994.tb01111.x. [DOI] [PubMed] [Google Scholar]
- 173.Keyf F, Anil N, Ercan M, Etikan I, Yener O. 1995. Persistence of 99m Tc-labelled microorganisms on surfaces of impression materials. J Nihon Univ Sch Dent 37:1–7. doi: 10.2334/josnusd1959.37.1. [DOI] [PubMed] [Google Scholar]
- 174.Greenfield RA, Troutt DL, Rickard RC, Altmiller DH. 1988. Comparison of antibody, antigen, and metabolite assays in rat models of systemic and gastrointestinal candidiasis. J Clin Microbiol 26:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McCourtie J, Douglas LJ. 1981. Relationship between cell surface composition of Candida albicans and adherence to acrylic after growth on different carbon sources. Infect Immun 32:1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nikawa H, Hayashi S, Nikawa Y, Hamada T, Samaranayake L. 1993. Interactions between denture lining material, protein pellicles and Candida albicans. Arch Oral Biol 38:631–634. doi: 10.1016/0003-9969(93)90132-6. [DOI] [PubMed] [Google Scholar]
- 177.Samaranayake L, MacFarlane T. 1980. An in vitro study of the adherence of Candida albicans to acrylic surfaces. Arch Oral Biol 25:603–609. doi: 10.1016/0003-9969(80)90075-8. [DOI] [PubMed] [Google Scholar]
- 178.Prosser B, Taylor D, Dix BA, Cleeland R. 1987. Method of evaluating effects of antibiotics on bacterial biofilm. Antimicrob Agents Chemother 31:1502–1506. doi: 10.1128/AAC.31.10.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hawser SP, Baillie GS, Douglas LJ. 1998. Production of extracellular matrix by Candida albicans biofilms. J Med Microbiol 47:253–256. doi: 10.1099/00222615-47-3-253. [DOI] [PubMed] [Google Scholar]
- 180.Pierce CG, Uppuluri P, Tristan AR, Wormley FL, Mowat E, Ramage G, López-Ribot JL. 2008. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc 3:1494–1500. doi: 10.1038/nprot.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ramage G, Vande Walle K, Wickes BL, López-Ribot JL. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother 45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ramage G, VandeWalle K, López-Ribot JL, Wickes BL. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett 214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 183.Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR. 2013. Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell 12:1389–1402. doi: 10.1128/EC.00127-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Daniels KJ, Srikantha T, Pujol C, Park YN, Soll DR. 2015. Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot Cell 14:228–240. doi: 10.1128/EC.00224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee K, Buckley HR, Campbell CC. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Med Mycol 13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 186.Bedell GW, Soll DR. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun 26:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Merson-Davies L, Odds F. 1989. A morphology index for characterization of cell shape in Candida albicans. J Gen Microbiol 135:3143–3152. [DOI] [PubMed] [Google Scholar]
- 188.Uppuluri P, López-Ribot JL. 2010. An easy and economical in vitro method for the formation of Candida albicans biofilms under continuous conditions of flow. Virulence 1:483–487. doi: 10.4161/viru.1.6.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Uppuluri P, López-Ribot JL. 2016. Go forth and colonize: dispersal from clinically important microbial biofilms. PLoS Pathog 12:e1005397. doi: 10.1371/journal.ppat.1005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nobile CJ, Fox EP, Hartooni N, Mitchell KF, Hnisz D, Andes DR, Kuchler K, Johnson AD. 2014. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 5:e01201-14. doi: 10.1128/mBio.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schinabeck MK, Long LA, Hossain MA, Chandra J, Mukherjee PK, Mohamed S, Ghannoum MA. 2004. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Chemother 48:1727–1732. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun 72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nobile CJ, Nett JE, Andes DR, Mitchell AP. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nett JE, Guite KM, Ringeisen A, Holoyda KA, Andes DR. 2008. Reduced biocide susceptibility in Candida albicans biofilms. Antimicrob Agents Chemother 52:3411–3413. doi: 10.1128/AAC.01656-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nett JE, Lepak AJ, Marchillo K, Andes DR. 2009. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200:307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, López-Ribot JL, Andes D, Cowen LE. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog 7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Desai JV, Bruno VM, Ganguly S, Stamper RJ, Mitchell KF, Solis N, Hill EM, Xu W, Filler SG, Andes DR, Fanning S, Lanni F, Mitchell AP. 2013. Regulatory role of glycerol in Candida albicans biofilm formation. mBio 4:e00637-12. doi: 10.1128/mBio.00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ding C, Vidanes GM, Maguire SL, Guida A, Synnott JM, Andes DR, Butler G. 2011. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS One 6:e28151. doi: 10.1371/journal.pone.0028151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Green JV, Orsborn KI, Zhang M, Tan QK, Greis KD, Porollo A, Andes DR, Long Lu J, Hostetter MK. 2013. Heparin-binding motifs and biofilm formation by Candida albicans. J Infect Dis 208:1695–1704. doi: 10.1093/infdis/jit391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Taff HT, Marchillo K, Andes DR. 2013. Preparation of Candida albicans biofilms using an in vivo rat central venous catheter model. Biol Protoc 3:e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Uppuluri P, Nett J, Heitman J, Andes D. 2008. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother 52:1127–1132. doi: 10.1128/AAC.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nett JE, Brooks EG, Cabezas-Olcoz J, Sanchez H, Zarnowski R, Marchillo K, Andes DR. 2014. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect Immun 82:4931–4940. doi: 10.1128/IAI.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Uppuluri P, Chaturvedi AK, López-Ribot JL. 2009. Design of a simple model of Candida albicans biofilms formed under conditions of flow: development, architecture, and drug resistance. Mycopathologia 168:101–109. doi: 10.1007/s11046-009-9205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nett JE, Marchillo K, Andes DR. 2012. Modeling of fungal biofilms using a rat central vein catheter. Methods Mol Biol 845:547–556. doi: 10.1007/978-1-61779-539-8_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nett JE, Zarnowski R, Cabezas-Olcoz J, Brooks EG, Berhardt J, Marchillo K, Mosher DF, Andes DR. 2015. Host contributions to construction of three device-associated Candida biofilms. Infect Immun 83:4630–4638. doi: 10.1128/IAI.00931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kucharikova S, Van Dijck P, Lisalova M, Bujdakova H. 2010. Effect of antifungals on itraconazole resistant Candida glabrata. Cent Eur J Biol 5:318–323. [Google Scholar]
- 208.Ricicova M, Kucharikova S, Tournu H, Hendrix J, Bujdakova H, Van Eldere J, Lagrou K, Van Dijck P. 2010. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 156:909–919. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- 209.Olsen I, Bondevik O. 1978. Experimental Candida-induced denture stomatitis in the Wistar rat. Eur J Oral Sci 86:392–398. doi: 10.1111/j.1600-0722.1978.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 210.Nett JE, Marchillo K, Spiegel CA, Andes DR. 2010. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun 78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Johnson CC, Yu A, Lee H, Fidel PL, Noverr MC. 2012. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun 80:1736–1743. doi: 10.1128/IAI.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee H, Yu A, Johnson CC, Lilly EA, Noverr MC, Fidel PL Jr. 2011. Fabrication of a multi-applicable removable intraoral denture system for rodent research. J Oral Rehabil 38:686–690. doi: 10.1111/j.1365-2842.2011.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cole G, Lynn K, Seshan K. 1990. An animal model for oropharyngeal, esophageal and gastric candidosis. Mycoses 33:7–19. [DOI] [PubMed] [Google Scholar]
- 215.Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. 2001. New model of oropharyngeal candidiasis in mice. Antimicrob Agents Chemother 45:3195–3197. doi: 10.1128/AAC.45.11.3195-3197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mosci P, Pericolini E, Gabrielli E, Kenno S, Perito S, Bistoni F, d'Enfert C, Vecchiarelli A. 2013. A novel bioluminescence mouse model for monitoring oropharyngeal candidiasis in mice. Virulence 4:250–254. doi: 10.4161/viru.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Solis NV, Filler SG. 2012. Mouse model of oropharyngeal candidiasis. Nat Protoc 7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Srinivasan A, Gupta CM, Agrawal CM, Leung KP, López-Ribot JL, Ramasubramanian AK. 2014. Drug susceptibility of matrix-encapsulated Candida albicans nano-biofilms. Biotechnol Bioeng 111:418–424. doi: 10.1002/bit.25120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Srinivasan A, Leung KP, López-Ribot JL, Ramasubramanian AK. 2013. High-throughput nano-biofilm microarray for antifungal drug discovery. mBio 4:e00331-13. doi: 10.1128/mBio.00331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Srinivasan A, Uppuluri P, López-Ribot J, Ramasubramanian AK. 2011. Development of a high-throughput Candida albicans biofilm chip. PLoS One 6:e19036. doi: 10.1371/journal.pone.0019036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Berridge MV, Herst PM, Tan AS. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 222.Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, Boyd MR. 1988. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res 48:4827–4833. [PubMed] [Google Scholar]
- 223.Massa EM, Farias RN. 1983. Effect of auto-oxidized phospholipids on oxidative enzyme assays based on tetrazolium salt reduction. Biochim Biophys Acta 746:209–215. doi: 10.1016/0167-4838(83)90076-6. [DOI] [PubMed] [Google Scholar]
- 224.Roslev P, King GM. 1993. Application of a tetrazolium salt with a water-soluble formazan as an indicator of viability in respiring bacteria. Appl Environ Microbiol 59:2891–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wogulis M, Wright S, Cunningham D, Chilcote T, Powell K, Rydel RE. 2005. Nucleation-dependent polymerization is an essential component of amyloid-mediated neuronal cell death. J Neurosci 25:1071–1080. doi: 10.1523/JNEUROSCI.2381-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kuhn D, George T, Chandra J, Mukherjee P, Ghannoum M. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother 46:1773–1780. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li X, Yan Z, Xu J. 2003. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149:353–362. doi: 10.1099/mic.0.25932-0. [DOI] [PubMed] [Google Scholar]
- 229.Thompson SW, Hunt RD. 1966. Selected histochemical and histopathological methods. C C Thomas, Springfield, IL. [Google Scholar]
- 230.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Liu Y, Schubert D. 1997. Cytotoxic amyloid peptides inhibit cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction by enhancing MTT formazan exocytosis. J Neurochem 69:2285–2293. [DOI] [PubMed] [Google Scholar]
- 232.Gorman SP, Mawhinney WM, Adair CG, Issouckis M. 1993. Confocal laser scanning microscopy of peritoneal catheter surfaces. J Med Microbiol 38:411–417. doi: 10.1099/00222615-38-6-411. [DOI] [PubMed] [Google Scholar]
- 233.Nickel J, Ruseska I, Wright J, Costerton J. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27:619–624. doi: 10.1128/AAC.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Talmon Y, Steinbrecht R, Zierold K. 1987. Cryotechniques in biological electron microscopy, p 64–86. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 236.Little B, Wagner P, Ray R, Pope R, Scheetz R. 1991. Biofilms: an ESEM evaluation of artifacts introduced during SEM preparation. J Ind Microbiol 8:213–221. doi: 10.1007/BF01576058. [DOI] [Google Scholar]
- 237.Bridier A, Meylheuc T, Briandet R. 2013. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM). Micron 48:65–69. doi: 10.1016/j.micron.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 238.Khajotia SS, Smart KH, Pilula M, Thompson DM. 2013. Concurrent quantification of cellular and extracellular components of biofilms. J Vis Exp 2013:e50639. doi: 10.3791/50639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kuehn M, Mehl M, Hausner M, Bungartz HJ, Wuertz S. 2001. Time-resolved study of biofilm architecture and transport processes using experimental and simulation techniques: the role of EPS. Water Sci Technol 43:143–150. [PubMed] [Google Scholar]
- 240.Hanrahan O, Harris J, Egan C. 2011. Advanced microscopy: laser scanning confocal microscopy. Methods Mol Biol 784:169–180. doi: 10.1007/978-1-61779-289-2_12. [DOI] [PubMed] [Google Scholar]
- 241.Herth W, Schnepf E. 1980. The fluorochrome, calcofluor white, binds oriented to structural polysaccharide fibrils. Protoplasma 105:129–133. doi: 10.1007/BF01279855. [DOI] [Google Scholar]
- 242.Hamer E, Moore C, Denning D. 2006. Comparison of two fluorescent whiteners, calcofluor and blankophor, for the detection of fungal elements in clinical specimens in the diagnostic laboratory. Clin Microbiol Infect 12:181–184. doi: 10.1111/j.1469-0691.2005.01321.x. [DOI] [PubMed] [Google Scholar]
- 243.Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. 2010. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol 8:e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d'Enfert C. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol 80:995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- 245.Fox EP, Bui CK, Nett JE, Hartooni N, Mui MC, Andes DR, Nobile CJ, Johnson AD. 2015. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol 96:1226–1239. doi: 10.1111/mmi.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo J-M, d'Enfert C. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Murillo LA, Newport G, Lan CY, Habelitz S, Dungan J, Agabian NM. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell 4:1562–1573. doi: 10.1128/EC.4.9.1562-1573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yeater KM, Chandra J, Cheng G, Mukherjee PK, Zhao X, Rodriguez-Zas SL, Kwast KE, Ghannoum MA, Hoyer LL. 2007. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 153:2373–2385. doi: 10.1099/mic.0.2007/006163-0. [DOI] [PubMed] [Google Scholar]
- 249.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog 8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Xu W, Solis NV, Ehrlich RL, Woolford CA, Filler SG, Mitchell AP. 2015. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol 13:e1002076. doi: 10.1371/journal.pbio.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Srikantha T, Daniels KJ, Pujol C, Kim E, Soll DR. 2013. Identification of genes upregulated by the transcription factor Bcr1 that are involved in impermeability, impenetrability, and drug resistance of Candida albicans a/alpha biofilms. Eukaryot Cell 12:875–888. doi: 10.1128/EC.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY. 2006. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 253.Draghici S, Khatri P, Eklund AC, Szallasi Z. 2006. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet 22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.An D, Parsek MR. 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol 10:292–296. doi: 10.1016/j.mib.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 255.Legrand M, Lephart P, Forche A, Mueller FMC, Walsh T, Magee P, Magee BB. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol Microbiol 52:1451–1462. doi: 10.1111/j.1365-2958.2004.04068.x. [DOI] [PubMed] [Google Scholar]
- 256.Rustad TR, Stevens DA, Pfaller MA, White TC. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061–1072. doi: 10.1099/00221287-148-4-1061. [DOI] [PubMed] [Google Scholar]
- 257.Soll DR, Lockhart SR, Zhao R. 2003. Relationship between switching and mating in Candida albicans. Eukaryot Cell 2:390–397. doi: 10.1128/EC.2.3.390-397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Jain N, Kohli R, Cook E, Gialanella P, Chang T, Fries B. 2007. Biofilm formation by and antifungal susceptibility of Candida isolates from urine. Appl Environ Microbiol 73:1697–1703. doi: 10.1128/AEM.02439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Villar-Vidal M, Marcos-Arias C, Eraso E, Quindós G. 2011. Variation in biofilm formation among blood and oral isolates of Candida albicans and Candida dubliniensis. Enferm Infecc Microbiol Clin 29:660–665. doi: 10.1016/j.eimc.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 260.Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol 59:183–194. [DOI] [PubMed] [Google Scholar]
- 261.Hartwell LH. 1974. Saccharomyces cerevisiae cell cycle. Bacteriol Rev 38:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hartwell LH, Culotti J, Reid B. 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci U S A 66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Cottrell SF, Avers CJ. 1970. Evidence of mitochondrial synchrony in synchronous cell cultures of yeast. Biochem Biophys Res Commun 38:973–980. doi: 10.1016/0006-291X(70)90817-X. [DOI] [PubMed] [Google Scholar]
- 264.Bertoli C, Skotheim JM, de Bruin RA. 2013. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hartwell L. 1973. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp Cell Res 76:111–117. doi: 10.1016/0014-4827(73)90425-4. [DOI] [PubMed] [Google Scholar]
- 266.Manukyan A, Abraham L, Dungrawala H, Schneider BL. 2011. Synchronization of yeast. Methods Mol Biol 761:173–200. doi: 10.1007/978-1-61779-182-6_12. [DOI] [PubMed] [Google Scholar]
- 267.Soll DR. 1986. The regulation of cellular differentiation in the dimorphic yeast Candida albicans. Bioessays 5:5–11. doi: 10.1002/bies.950050103. [DOI] [PubMed] [Google Scholar]
- 268.Hulst HC, Van De Hulst H. 1957. Light scattering by small particles. Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 269.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hereford LM, Osley MA, Ludwig JR, McLaughlin CS. 1981. Cell-cycle regulation of yeast histone mRNA. Cell 24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- 271.Kusch H, Engelmann S, Bode R, Albrecht D, Morschhäuser J, Hecker M. 2008. A proteomic view of Candida albicans yeast cell metabolism in exponential and stationary growth phases. Int J Med Microbiol 298:291–318. doi: 10.1016/j.ijmm.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 272.Farrell MJ, Finkel SE. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol 185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 274.Rechsteiner M. 1988. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul 27:135–151. doi: 10.1016/0065-2571(88)90015-5. [DOI] [PubMed] [Google Scholar]
- 275.Bojsen R, Regenberg B, Folkesson A. 2014. Saccharomyces cerevisiae biofilm tolerance towards systemic antifungals depends on growth phase. BMC Microbiol 14:305. doi: 10.1186/s12866-014-0305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Watts H, Veacute A-A, Perera T, Davies J, Gow N. 1998. Thigmotropism and stretch-activated channels in the pathogenic fungus Candida albicans. Microbiology 144:689–695. doi: 10.1099/00221287-144-3-689. [DOI] [PubMed] [Google Scholar]
- 277.Crusz SA, Popat R, Rybtke MT, Cámara M, Givskov M, Tolker-Nielsen T, Diggle SP, Williams P. 2012. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling 28:835–842. doi: 10.1080/08927014.2012.716044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, López-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog 6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ramage G, Wickes BL, López-Ribot JL. 2008. A seed and feed model for the formation of Candida albicans biofilms under flow conditions using an improved modified Robbins device. Rev Iberoam Micol 25:37. doi: 10.1016/S1130-1406(08)70009-3. [DOI] [PubMed] [Google Scholar]
- 282.Belardinelli PA, Morelatto RA, Benavidez TE, Baruzzi AM, Lopez de Blanc SA. 2014. Effect of two mouthwashes on salivary pH. Acta Odontol Latinoam 27:66–71. doi: 10.1590/S1852-48342014000200004. [DOI] [PubMed] [Google Scholar]
- 283.Duckworth RM, Jones S. 2015. On the relationship between the rate of salivary flow and salivary fluoride clearance. Caries Res 49:141–146. doi: 10.1159/000365949. [DOI] [PubMed] [Google Scholar]
- 284.Rad M, Kakoie S, Brojeni FN, Pourdamghan N. 2010. Effect of long-term smoking on whole-mouth salivary flow rate and oral health. Dent Res Dent Clin Dent Prospects 4:110–114. doi: 10.5681/joddd.2010.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Dawes C. 1972. Circadian rhythms in human salivary flow rate and composition. J Physiol 220:529–545. doi: 10.1113/jphysiol.1972.sp009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Massey BT. 2001. Potential control of gastroesophageal reflux by local modulation of transient lower esophageal sphincter relaxations. Am J Med 111:186–189. doi: 10.1016/S0002-9343(01)00829-4. [DOI] [PubMed] [Google Scholar]
- 287.Brener W, Hendrix TR, McHugh PR. 1983. Regulation of the gastric emptying of glucose. Gastroenterology 85:76–82. [PubMed] [Google Scholar]
- 288.Sanelli PC, Deshmukh M, Ougorets I, Caiati R, Heier LA. 2004. Safety and feasibility of using a central venous catheter for rapid contrast injection rates. Am J Roentgenol 183:1829–1834. doi: 10.2214/ajr.183.6.01831829. [DOI] [PubMed] [Google Scholar]
- 289.Lose G, Thunedborg P, Jørgensen L, Colstrup H. 1986. A comparison of spontaneous and intubated urinary flow in female patients. Neurourol Urodyn 5:1–4. doi: 10.1002/nau.1930050102. [DOI] [Google Scholar]
- 290.Reuss M, Rudolph A. 1979. Distribution and recirculation of umbilical and systemic venous blood flow in fetal lambs during hypoxia. J Dev Physiol 2:71–84. [PubMed] [Google Scholar]
- 291.Vuong C, Saenz HL, Gotz F, Otto M. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 292.Sellam A, Al-Niemi T, McInnerney K, Brumfield S, Nantel A, Suci PA. 2009. A Candida albicans early stage biofilm detachment event in rich medium. BMC Microbiol 9:25. doi: 10.1186/1471-2180-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jin Y, Zhang T, Samaranayake YH, Fang HH, Yip HK, Samaranayake LP. 2005. The use of new probes and stains for improved assessment of cell viability and extracellular polymeric substances in Candida albicans biofilms. Mycopathology 159:353–360. doi: 10.1007/s11046-004-6987-7. [DOI] [PubMed] [Google Scholar]
- 294.Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, López-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathology 169:323–331. doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Thomas DP, Bachmann SP, López-Ribot JL. 2006. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 6:5795–5804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- 296.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, Lounes-Hadj Sahraoui A, Fontaine J, Sanchez H, Hatfield RD, Ntambi JM, Nett JE, Mitchell AP, Andes DR. 2014. Novel entries in a fungal biofilm matrix encyclopedia. mBio 5:e01333-14. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Soll DR. 1979. Timers in developing systems. Science 203:841–849. doi: 10.1126/science.419408. [DOI] [PubMed] [Google Scholar]
- 298.Kraft B, Steinbrech D, Yang M, Soll DR. 1988. High-frequency switching in Dictyostelium. Dev Biol 130:198–208. doi: 10.1016/0012-1606(88)90426-5. [DOI] [PubMed] [Google Scholar]
- 299.Soll DR. 1990. The regulation of developmental timing, p 1–82. In Roth S. (ed), Topics in developmental biology. University of Pennsylvania Press, Philadelphia, PA. [Google Scholar]
- 300.Soll DR, Kraft B. 1988. A comparison of high frequency switching in the yeast Candida albicans and the slime mold Dictyostelium discoideum. Dev Genet 9:615–628. doi: 10.1002/dvg.1020090438. [DOI] [PubMed] [Google Scholar]
- 301.Moore GE, Gerner RE, Franklin HA. 1967. Culture of normal human leukocytes. JAMA 199:519–524. [PubMed] [Google Scholar]
- 302.Radetsky M, Wheeler R, Roe M, Todd J. 1986. Microtiter broth dilution method for yeast susceptibility testing with validation by clinical outcome. J Clin Microbiol 24:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Espinel-Ingroff A, Kish C, Kerkering T, Fromtling R, Bartizal K, Galgiani J, Villareal K, Pfaller M, Gerarden T, Rinaldi M. 1992. Collaborative comparison of broth macrodilution and microdilution antifungal susceptibility tests. J Clin Microbiol 30:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.National Committee for Clinical Laboratory Standards. 1992. Performance standards for antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards, Villanova, PA. [Google Scholar]
- 305.Wanger A, Mills K, Nelson PW, Rex JH. 1995. Comparison of Etest and National Committee for Clinical Laboratory Standards broth macrodilution method for antifungal susceptibility testing: enhanced ability to detect amphotericin B-resistant Candida isolates. Antimicrob Agents Chemother 39:2520–2522. doi: 10.1128/AAC.39.11.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Nobile CJ, Andes DR, Nett JE, Smith FJ Jr, Yue F, Phan Q-T, Edwards JE Jr, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathol 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault J-S, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 310.Norice CT, Smith FJ, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot Cell 6:2046–2055. doi: 10.1128/EC.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Richard ML, Nobile CJ, Bruno VM, Mitchell AP. 2005. Candida albicans biofilm-defective mutants. Eukaryot Cell 4:1493–1502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Braun BR, Johnson AD. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Brown DH Jr, Giusani AD, Chen X, Kumamoto CA. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol 34:651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- 314.Calera JA, Zhao X-J, Calderone R. 2000. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect Immun 68:518–525. doi: 10.1128/IAI.68.2.518-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Gale CA, Bendel CM, McClellan M, Hauser M, Becker JM, Berman J, Hostetter MK. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 316.Liu H, Kohler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 317.Lo H-J, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 318.Sahni N, Yi S, Daniels KJ, Srikantha T, Pujol C, Soll DR. 2009. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog 5:e1000601. doi: 10.1371/journal.ppat.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Yi S, Sahni N, Daniels KJ, Lu KL, Huang G, Garnaas AM, Pujol C, Srikantha T, Soll DR. 2011. Utilization of the mating scaffold protein in the evolution of a new signal transduction pathway for biofilm development. mBio 2:e00237-10. doi: 10.1128/mBio.00237-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hull CM, Johnson AD. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 321.Wu W, Pujol C, Lockhart SR, Soll DR. 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 169:1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wu W, Lockhart SR, Pujol C, Srikantha T, Soll DR. 2007. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol Microbiol 64:1587–1604. doi: 10.1111/j.1365-2958.2007.05759.x. [DOI] [PubMed] [Google Scholar]
- 323.Ibrahim AS, Magee B, Sheppard D, Yang M, Kauffman S, Becker J, Edwards JE, Magee P. 2005. Effects of ploidy and mating type on virulence of Candida albicans. Infect Immun 73:7366–7374. doi: 10.1128/IAI.73.11.7366-7374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Janbon G, Sherman F, Rustchenko E. 1999. Appearance and properties of l-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Lockhart SR, Daniels KJ, Zhao R, Wessels D, Soll DR. 2003. Cell biology of mating in Candida albicans. Eukaryot Cell 2:49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 327.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol Microbiol 55:1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x. [DOI] [PubMed] [Google Scholar]
- 329.Daniels KJ, Lockhart SR, Staab JF, Sundstrom P, Soll DR. 2003. The adhesin Hwp1 and the first daughter cell localize to the a/a portion of the conjugation bridge during Candida albicans mating. Mol Biol Cell 14:4920–4930. doi: 10.1091/mbc.E03-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2013. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot Cell 12:1120–1131. doi: 10.1128/EC.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sahni N, Yi S, Pujol C, Soll DR. 2009. The white cell response to pheromone is a general characteristic of Candida albicans strains. Eukaryot Cell 8:251–256. doi: 10.1128/EC.00320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell 19:957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Yi S, Sahni N, Pujol C, Daniels KJ, Srikantha T, Ma N, Soll DR. 2009. A Candida albicans-specific region of the alpha-pheromone receptor plays a selective role in the white cell pheromone response. Mol Microbiol 71:925–947. doi: 10.1111/j.1365-2958.2008.06575.x. [DOI] [PubMed] [Google Scholar]
- 334.Anderson JM, Soll DR. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol 169:5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Rikkerink E, Magee B, Magee P. 1988. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol 170:895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lederberg J, Tatum EL. 1946. Gene recombination in Escherichia coli. Nature 158:558. [DOI] [PubMed] [Google Scholar]
- 337.Licht TR, Christensen BB, Krogfelt KA, Molin S. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145:2615–2622. doi: 10.1099/00221287-145-9-2615. [DOI] [PubMed] [Google Scholar]
- 338.Doig P, Todd T, Sastry PA, Lee K, Hodges RS, Paranchych W, Irvin R. 1988. Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect Immun 56:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rothbard JB, Fernandez R, Wang L, Teng N, Schoolnik GK. 1985. Antibodies to peptides corresponding to a conserved sequence of gonococcal pilins block bacterial adhesion. Proc Natl Acad Sci U S A 82:915–919. doi: 10.1073/pnas.82.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2013. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4:e00645-13. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 342.Porman AM, Alby K, Hirakawa MP, Bennett RJ. 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A 108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Xie Z, Thompson A, Sobue T, Kashleva H, Xu H, Vasilakos J, Dongari-Bagtzoglou A. 2012. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis 206:1936–1945. doi: 10.1093/infdis/jis607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. 2013. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog 9:e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]