SUMMARY
Heterochromatin is the transcriptionally repressed portion of eukaryotic chromatin that maintains a condensed appearance throughout the cell cycle. At sites of ribosomal DNA (rDNA) heterochromatin, epigenetic states contribute to gene silencing and genome stability, which are required for proper chromosome segregation and a normal life span. Here, we focus on recent advances in the epigenetic regulation of rDNA silencing in Saccharomyces cerevisiae and in mammals, including regulation by several histone modifications and several protein components associated with the inner nuclear membrane within the nucleolus. Finally, we discuss the perturbations of rDNA epigenetic pathways in regulating cellular aging and in causing various types of diseases.
INTRODUCTION
Heterochromatin maintains a condensed appearance throughout the cell cycle, while euchromatin undergoes condensation and decondensation as the cell cycle proceeds (1). In most organisms, including yeast, fruit flies, and mammals, heterochromatin is found at the telomeres and in regions surrounding the centromeres or ribosomal DNA (rDNA) loci. Some species-specific heterochromatic regions, such as the silent-mating-type loci in yeast or the inactive X chromosome in female mammals, also exist (1, 2). Heterochromatin has a unique ability to spread and to serve as a multipurpose platform for the recruitment of diverse regulatory proteins, thus affecting gene expression and other chromosomal processes in a region-specific, sequence-independent manner (1, 3).
Various characteristic chromatin modifications of histones in eukaryotes are known to contribute to the assembly of heterochromatin, the stability of the genome, and the restriction of heterochromatin spreading into adjacent chromatin domains (4). DNA methylation refers to a highly conserved, heritable modification that involves the methylation of cytosine into 5-methylcytosines in the CpG dinucleotides by DNA methyltransferases (DNMTs) such as DNMT1, DNMT3a, and DNMT3b (5). DNMT3a and DNMT3b establish the initial CpG methylation pattern de novo, while DNMT1 is primarily responsible for maintaining this pattern throughout each cell division (6). Although several questions remain to be answered, the epigenetic regulation of heterochromatin function, which includes regulation by several histone modifications and DNA methylation, has revived the much-debated proposition that heterochromatin silencing is important in evolution and development. Here, we focus on recent advances in the epigenetic regulation of heterochromatin silencing within the rDNA locus in the yeast Saccharomyces cerevisiae, a model organism that is widely used for studies of heterochromatin formation and function; in metazoans, especially in mammals; and briefly in Arabidopsis thaliana, a model organism for plants. This review includes recent findings on changes in histone modification, such as histone methylation, acetylation, ubiquitylation, and ADP ribosylation, as well as in DNA methylation within rDNA loci. Moreover, we present the implication of several protein components at the nuclear envelope or the noncoding RNAs in these types of epigenetic regulation of rDNA silencing. Finally, we discuss how the epigenetic pathways of rDNA are associated with cellular life span regulation and how their dysregulation leads to various types of diseases such as cancers.
PATHWAYS FOR rDNA SILENCING
In budding yeast, the rDNA region that carries rRNA is organized into a tandem array of 9.1-kb units that are repeated 100 to 200 times on chromosome XII. In particular, the rDNA region is localized to the inner nuclear membrane (INM) of the nucleolus (7, 8). In humans, each rDNA repeat is ∼43 kb and is found on the short arms of the five human acrocentric autosomal chromosomes, namely, chromosomes 13, 14, 15, 21, and 22, and in mouse, each rDNA repeat is ∼45 kb long and is located on six chromosomes, namely, chromosomes 12, 15, 16, 17, 18, and 19 (9, 10). Clusters of human rDNA repeats termed nucleolar organizer regions (NORs) are composed of 300 to 400 copies per haploid genome. In Arabidopsis, each rDNA repeat is ∼10 kb, and there are ∼570 to 750 copies in each haploid genome (11–13). Similar to human rDNA repeat clusters, the Arabidopsis rRNA gene clusters are also called NORs and are located on the short arms of chromosome 2 and chromosome 4 (NOR2 and NOR4, respectively) (11, 12).
The rDNA array is fundamentally unstable and is a target for homologous recombination. Homologous recombination within rDNA loci is one of the crucial processes that are responsible for maintaining rDNA integrity by repairing DNA double-strand breaks (DSBs); this process rescues stalled replication forks and preserves rDNA repeats (14). However, uncontrolled homologous recombination may cause the translocation of chromosomes, loss of heterozygosity, or addition/deletion of repetitive sequences (15). Indeed, the rDNA copy number remains unaltered if the equal sister chromatid exchange repairs DSBs with the nearest sister chromatid, but the rDNA array may expand or contract if unequal sister chromatid exchange (USCE) occurs during DSB repair (16, 17). Changes in rDNA repeat numbers due to aberrant recombination, such as USCE, cause genomic instability within rDNA repeats and lead to deleterious effects, such as higher sensitivity to DNA damage or impairment of the DNA repair process (16, 18, 19). Furthermore, the increased rate of this USCE at rDNA regions generates extrachromosomal rDNA circles (ERCs), which are responsible for premature cell senescence in budding yeast (20). Nevertheless, under normal conditions, rDNA repeats continue to be rather stable because rDNA recombination is negatively regulated through rDNA silencing (21).
In budding yeast, rDNA silencing occurs specifically in two regions: the nontranscribed spacer 1 (NTS1) region, which is downstream of the 5S gene and which contains the replication fork barrier (RFB), and the nontranscribed spacer 2 (NTS2) region, which is upstream of the 5S gene and which contains an autonomous replicating sequence (ARS) (22–24) (Fig. 1). In general, rDNA silencing in either the NTS1 or the NTS2 region depends on silent information regulator 2 (Sir2) (25). However, accumulating evidence further indicates that yeast rDNA is strongly associated with heterochromatin silencing, leading to rDNA silencing through dual pathways: the Sir2-dependent pathway, which involves the RENT (regulator of nucleolar silencing and telophase exit) complex, and the Sir2-independent pathway. The Sir2-independent pathway includes Tof2; two additional proteins, Csm1 and Lrs4, which are subunits of the previously identified monopolin complex that are required for coorientation during meiosis I; and the condensin complex (26, 27) (Fig. 2 and 3). In this context, it is likely that a loss of either pathway leads to reduced rDNA silencing to a similar extent, implying an overlapping mechanism within NTS regions of rDNA. However, losses of both pathways are synergistic when scoring for recombination among rDNA repeats, suggesting that the Sir2-dependent and Sir2-independent pathways work in parallel pathways for repressing recombination (26, 28).
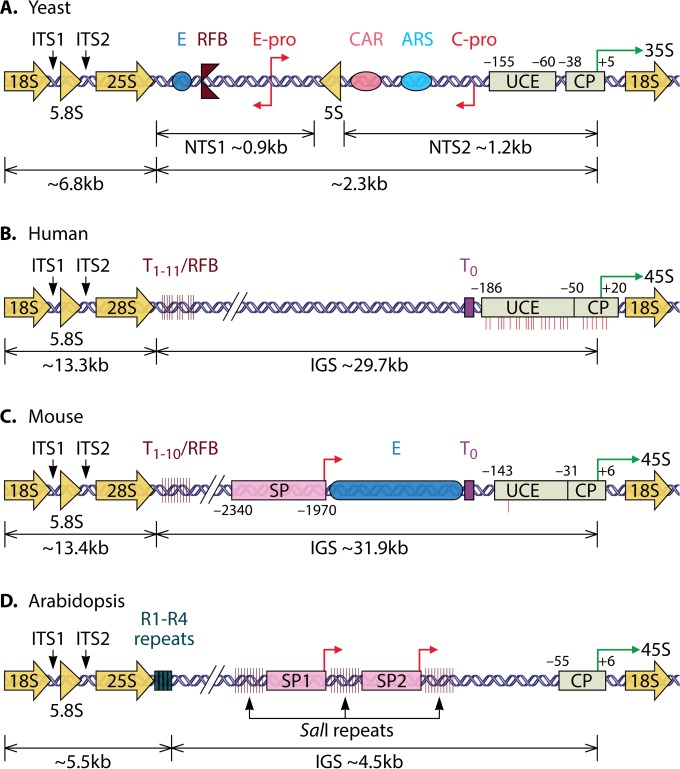
FIG 1.
rDNA structures in yeast, human, mouse, and Arabidopsis. The graphic of the yeast rRNA gene is derived from data reported under GenBank accession no. U53879, and graphics of human rRNA and mouse genes are derived from data reported under accession no. U13369 and BK000964, respectively. A graphic of the Arabidopsis rRNA gene is shown, as reported previously (11, 13, 197, 198). (A) In yeast, a single unit of rDNA (9.1 kb) consists of 5S, 5.8S, 18S, and 25S transcribed genes; internal transcribed spacers (ITS1 and ITS2); and two NTSs, NTS1 and NTS2. The repetitive sequences of rDNA undergo recombination, which consequently leads to genomic instability (199). In contrast, as the integrity and the proper function of the rDNA repeats play important roles in cell viability, rDNA recombination is generally repressed in a manner that is dependent on Sir2 (153). The yeast rDNA promoter consists of a core promoter (CP) and an upstream control element (UCE). The green arrow indicates the transcription start site (TSS) of the pre-rRNA. All nucleotide numbers are relative to the first nucleotide of the TSS (position +1). The enhancer element (E) and RFB are located near the 3′ end of the 25S rDNA in NTS1. The cohesin-associated region (CAR) and ARS are found in NTS2. RNAPII transcription of rDNA starts from two promoters, called cryptic RNAPII promoter (C-Pro) and EXP promoter (E-Pro). The intergenic cryptic transcripts are produced from C-Pro and E-Pro (200). (B and C) Human rDNA (43 kb) and mouse rDNA (45.3 kb) consist of 5.8S, 18S, and 28S transcribed genes; ITS1 and ITS2; and a long NTS called an IGS (intergenic spacer). The IGSs include multiple regulatory elements: the rDNA promoter (CP and UCE), an enhancer element (E), and upstream (T0) and downstream (T1 to T11 in human and T1 to T10 in mouse) terminators. A spacer promoter (SP) is found in the mouse IGS (201, 202). TTF-I is a transcription factor that recognizes and binds to the upstream terminator (T0) and downstream terminators to facilitate the recruitment of the NoRC or NuRD complex to the rDNA promoter (203). In mouse, DNA methylation occurs in a single CpG dinucleotide in the UCE position at position −133 relative to position +1, as shown by a red line below the UCE, while in humans, there are ∼25 CpG sites of DNA methylation in the promoter region (58, 181). (D) Arabidopsis rDNA (∼10 kb) consists of 5.8S, 18S, and 25S transcribed genes; ITS1 and ITS2; and an IGS. The Arabidopsis IGS region is composed of a gene promoter sequence (positions −55 to +6); two SPs, SP1 and SP2; and three SalI repeats. Four repeat sequences, R1 to R4, are located downstream of 25S rDNA in the 3′ external transcribed spacers (11, 13, 197, 198). In the Arabidopsis ecotype Col-0, four distinct rRNA gene variants, VAR1, VAR2, VAR3, and VAR4, are identified based on variations within R1 to R4. No UCE equivalent to yeast or mammalian UCEs has been identified in the Arabidopsis rDNA promoter.
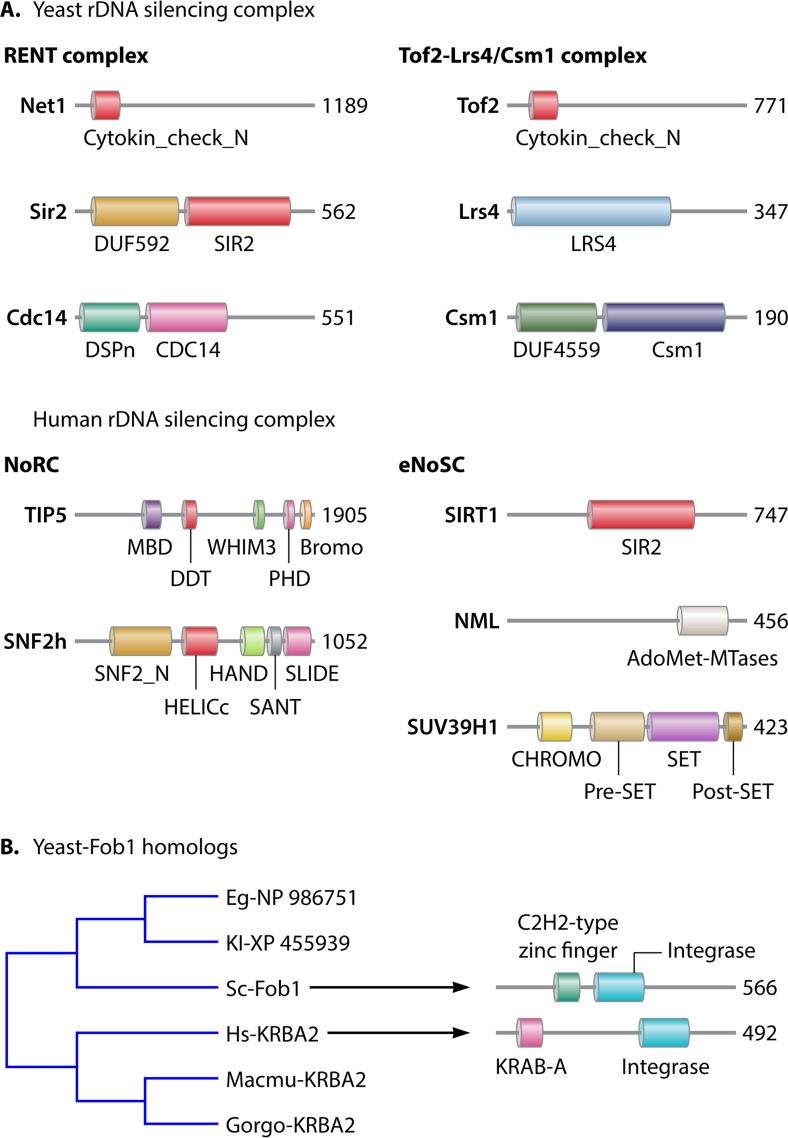
FIG 2.
rDNA-silencing components in yeast and mammals. (A) Two main rDNA-silencing complexes, RENT and Tof2-Lrs4/Csm1, are found in yeast. The RENT complex is composed of Net1, Sir2, and Cdc14 proteins. Human and mouse NoRCs are composed of TIP5 and SNF2h, while the eNoSC is composed of SIRT1, NML, and SUV39H1 (only the domain organization of human NoRC and eNoSC components is represented). Cytokin_check_N, Cdc14 phosphatase-binding protein N terminus; SIR2, silent information regulator 2; LRS4, loss of rDNA-silencing protein 4; DSPn, dual-specificity protein phosphatase, N-terminal half; CDC14, cell division control protein 14; Csm1, chromosome segregation in meiosis protein 1; MBD, methyl-CpG-binding domain; DDT, DNA-binding homeobox and different transcription factors; WHIM3, WSTF, HB1, Itc1p, and MBD9 motif 3; PHD, plant homeodomain; Bromo, bromodomain; AdoMet-MTases, S-adenosylmethionine-dependent methyltransferases; SNF2_N, SNF2 family N-terminal domain; HELICc, helicase conserved C-terminal domain; SANT, SWI3, ADA2, N-CoR, and TFIIIB; SLIDE, SANT-like ISWI domain; CHROMO, chromatin organization modifier; SET, Su(var)3-9 enhancer-of-zeste trithorax; DUF, domain of unknown function. (B) Homologs of yeast Fob1 in fungi and mammals. NCBI Blast analysis using yeast Fob1 protein sequences shows homology to human KRBA2. The sequences are derived from data reported under accession numbers XP_455939 for Kl, NP_986751 for Eg, NP_010395 for Sc, NP_998762 for Hs, XP_004058617 for Gorgo, and XP_001113012 for Macmu. Kl, Kluyveromyces lactis; Eg, Eremothecium gossypii; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Gorgo, Gorilla gorilla; Macmu, Macaca mulatta. Fob1 consists of a C2H2-type zinc finger motif and an integrase catalytic core-like structure (204). Human KRBA2 is a zinc finger protein of the C2H2 family and contains Krüppel-associated box A (KRAB-A) and integrase core domains.
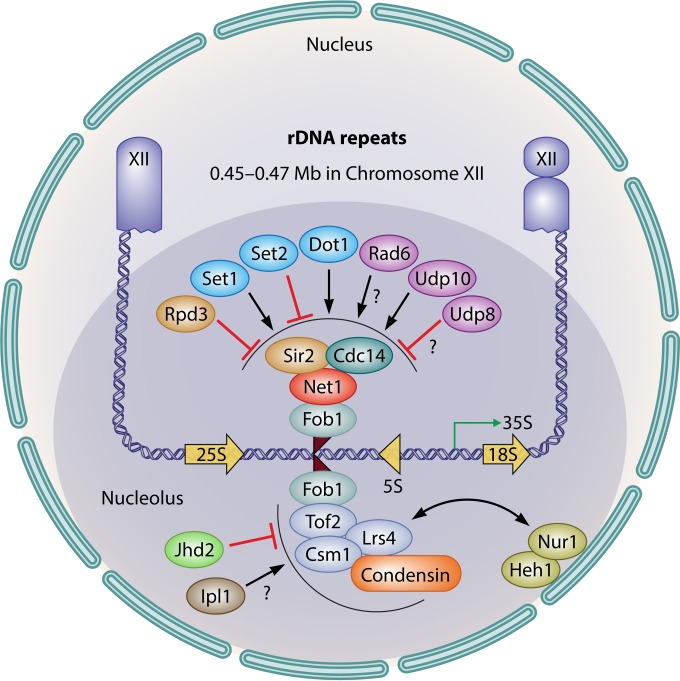
FIG 3.
Dual pathways for yeast rDNA silencing. The yeast rDNA locus is located between nucleotides 0.45 to 0.468 Mb of chromosome XII. (Top) In Sir2-dependent rDNA silencing, Fob1 binds to the RFB site of rDNA NTS1 and recruits Net1, which leads to the recruitment of Sir2 and Cdc14 to the RENT complex. The Sir2-associated histone modification that influences rDNA silencing involves various types of histone modifications during the G1 to late mitotic phases. Rpd3 is associated with the negative regulation of rDNA silencing. Set1 or Dot1 positively recruits the RENT components Net1 and Sir2, whereas Set2 has the opposite effect. Rad6 or Ubp10 positively regulates rDNA silencing by regulating Sir2 association at rDNA loci, while Ubp8 has a negative effect. (Bottom) In Sir2-independent rDNA silencing, Fob1 bound to the RFB site of rDNA mediates the hierarchical binding of Tof2, Csm1/Lrs4, and condensin to the NTS1, thereby leading to the association of rDNA with the nuclear periphery with the aid of CLIP proteins such as Heh1 and Nur1 during mitosis. In this specific stage of the cell cycle, Jhd2 preferentially regulates mitotic rDNA silencing by maintaining Csm1/Lrs4 and condensin association with rDNA regions. The aurora B kinase Ipl1 is also responsible for the hypercondensation of chromosomes during anaphase. The positive and negative regulations of the indicated histone-modifying enzymes in rDNA silencing are denoted by arrows and ⊥, respectively. Physical interaction between the indicated proteins is shown as a bidirectional arrow.
In the NTS regions of rDNA loci, Sir2 forms a RENT complex along with Net1 and Cdc14 (29). Net1 is required for rDNA silencing and Sir2 localization in the nucleolus (29, 30). In addition, Net1 functions in maintaining nucleolar integrity and in regulating telophase exit by controlling the release of the phosphatase Cdc14 (31, 32). The RENT complex is recruited to the RFB site through the physical interaction of Net1 and Sir2 with Fob1 in the NTS1 region and is most likely associated with the NTS2 region via RNA polymerase I (RNAPI) (24, 32). In cells lacking Sir2, the Fob1-dependent recombination rate is increased within rDNA repeats, leading to the accumulation of ERCs (20, 33). The association of Fob1 with the RFB inhibits replication fork progression in a single direction: following DBSs, Fob1 is required for rDNA homologous recombination with sister chromatids, which occurs at stalled forks (33–35), and FOB1 deletion reduces the rates of rDNA recombination and USCE between rDNA repeats (33). In addition, the loss of Fob1 decreases the rDNA copy number variation and consequently stabilizes the rDNA copy number repeats, suggesting that Fob1-dependent RFB activity is an important component of rDNA regulation, whose level must be tightly controlled by Sir2-dependent and -independent mechanisms (17). Moreover, interestingly, Fob1 is an indispensable protein that not only is required for the induction of rDNA recombination but also promotes rDNA silencing by recruiting the RENT complex, as described above, and the complex that is formed by Tof2, Lrs4, and Csm1 (22, 26) (Fig. 3). Tof2, Lrs4/Csm1, and the RENT complex may bind to the cohesin ring, an evolutionarily conserved multisubunit protein complex that is recruited to the chromosome and holds the sister chromatids together, thus facilitating the correction of the positions of two sister chromatids relative to each other and the inhibition of the unequal exchange between rDNA repeats (27, 28, 36). The presence of Sir2-dependent or Sir2-independent pathways for rDNA silencing appears to be attributable to the fact that cells cope efficiently with the variety of conditions that they encounter during each stage of the cell cycle. During interphase and mitosis, the RENT complex components Net1 and Sir2 colocalize to a subdomain within the nucleolus, mainly to the NTS1 and NTS2 regions of rDNA loci. However, the Sir2 protein leaves the nucleolus at the end of mitosis and scatters throughout the nucleus (29). The dispersion of Sir2 may result in an altered or unstable architecture at rDNA loci without alternative rDNA silencing pathways. Indeed, the redistribution of Sir2 from the NTS1 region to telomeres or mating-type loci in cells lacking Rif1, a telomeric DNA-binding protein, results in high rDNA instability and reduced cellular life span (37). To overcome this issue, both rDNA recombination and nucleolar silencing are regulated by the Sir2-independent pathway, which includes the Fob1-mediated hierarchical binding of Tof2, Csm1, and Lrs4 to the RFB site of the NTS1 region during mitosis.
The condensation of mitotic chromosomes requires the condensin complex, which is a multisubunit protein complex involved in regulating chromosome architecture (38). Based on the results of recent extensive studies in yeast regarding the function of mitotic condensin at the molecular level, the alteration of chromatin modifications of histones is thought to be one of the important factors that influences mitotic chromosome condensation. One important piece of evidence supporting that this epigenetic regulation of chromosome compaction comes from the study of yeast Ipl1, the aurora B kinase that phosphorylates histone H3 on the serine 10 residue (H3S10). This kinase also phosphorylates condensin during postmetaphase chromosome assembly maturation. The aurora B kinase is associated with mitosis in all eukaryotes and, in particular, drives the hypercondensation of an artificially elongated chromosome during anaphase in budding yeast, suggesting that changes in the histone modification pattern affect chromosome condensation (39).
In mammals, active or silent states of rDNA repeats in synthesizing rRNA are tightly regulated within various metabolic pathways, under different environmental conditions, or depending on each stage of the cell cycle. As in the yeast system, the maintenance of rDNA chromatin silencing in mammals is critical for rDNA genome stability: it suppresses aberrant homologous recombination within sister chromatids and maintains the rDNA in a condensed and transcriptionally refractory state. Thus, loss of silencing at rDNA loci correlates with rDNA instability, nucleolar disintegration, or even cellular senescence as well as with many types of diseases, as we discuss below (34, 40, 41). The switch from active rDNA, in which rRNA synthesis is active from this region, to its inactive state is mediated mainly by three classes of rDNA-silencing complexes (Fig. 4).
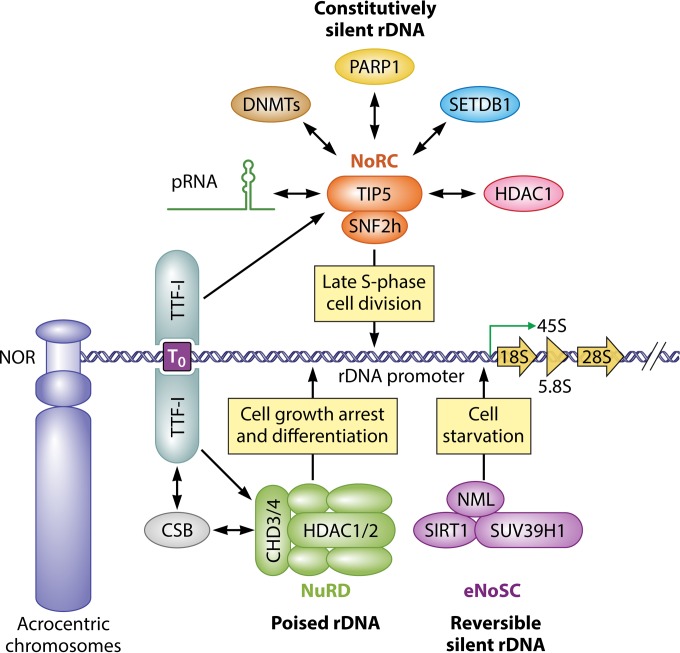
FIG 4.
Illustration of rDNA silencing pathways in mammals. Shown is a model for silent or repressed chromatin states of mammalian rDNA: first, constitutively silent rDNA repeats are organized into heterochromatin; second, reversible silent rDNA repeats are organized into an inactive structure during starvation; and third, “poised” rDNA is accessible to transcription machinery but is not actively transcribed. The transcription factor TTF-I recognizes and binds to T0, which facilitates the recruitment of the NoRC or NuRD complex to the rDNA promoter. A short noncoding pRNA (150 to 300 nt) is produced by RNAPI from the rDNA spacer promoter and is homologous to the core rDNA promoter (43). The pRNA binds to TIP5 through a hairpin structure, which facilitates and stabilizes the recruitment of the NoRC to chromatin, thus promoting rDNA silencing (205). DNMTs mediate rDNA hypermethylation with the association of the NoRC and pRNA. The eNoSC regulates rDNA silencing in response to intracellular energy levels or calorie restriction. The NuRD complex establishes the poised state for rDNA in growth-arrested and differentiated cells. The interaction of the NuRD complex with CSB and TTF-I facilitates NuRD recruitment to the rDNA promoter. Physical interaction between the indicated proteins and pRNA is shown as bidirectional arrows.
First, the NoRC (nucleolar remodeling complex) is an ATP-dependent chromatin-remodeling complex that is a member of the ISWI/SNF2h family and is composed of two subunits, the ATPase SNF2h (sucrose-nonfermenting protein 2 homolog) and the DNA-binding protein TIP5 (transcription termination factor I [TTF-I]-interacting protein 5) (42). This complex establishes rDNA silencing with the coordination of different histone-modifying enzymes, DNMTs, or promoter-associated RNA (pRNA), a small intergenic noncoding transcript required for NoRC-mediated heterochromatin formation, to block the formation of a transcription initiation complex at the rDNA promoter (43, 44). The NoRC mediates a shift of the nucleosome to 25 nucleotides (nt) downstream with respect to the transcription start site, a repressive/inactive position, to prevent transcription complex formation and to establish rDNA silencing (45). In addition, the association of the NoRC with the rDNA promoter is critical for establishing a silent rDNA chromatin structure (44), and the binding of the NoRC to the rDNA promoter via TTF-I recruits histone- or DNA-modifying proteins, such as HDAC1 (histone deacetylase 1), DNMT1, or DNMT3b (44, 46–49). In particular, the bromodomain within TIP5 plays a significant role in NoRC-mediated rDNA silencing by interacting with histone H4 acetylated at lysine 16 (H4K16ac) and consequently facilitating HDAC1-mediated deacetylation of histone H4 at the K5, K8, and K12 residues (50). Moreover, to maintain the silent state of rDNA during cell division or glucose starvation, SIRT1 (sirtuin 1) reinforces the NoRC at the rDNA chromatin by deacetylating MOF-dependent TIP5 acetylation (51). Significantly, most of the rDNA repeats in the clusters of rDNA exist in a constitutively silent and stable state that is maintained by DNA methylation and by the NoRC (5, 47). CpG DNA methylation is a highly stable modification for gene silencing and provides strong stability to the rDNA repeats, and the NoRC establishes the constitutively silent rDNA with the coordination of DNMTs and histone modification (46, 52). Moreover, rDNA methylation is associated with the inheritance of the constitutively silent and stable state of rDNA chromatin (47).
Second, the eNoSC (energy-dependent nucleolar silencing complex) is composed of SIRT1, the histone methyltransferase SUV39H1 (also known as KMT1A), and a nucleolar protein, NML (nucleomethylin). This complex mediates rDNA silencing in response to glucose starvation and protects mammalian cells from energy deprivation-dependent apoptosis (53). Elevation of the NAD+/NADP+ ratio triggers SIRT1 deacetylase activity for histone H3 at rDNA promoters, thereby facilitating energy-dependent rDNA transcriptional repression (53). Although yeast Sir2 localizes to the nucleolus and telomeres, mammalian SIRT1 is found mainly in the nucleoplasm and requires NML for its binding to rDNA after glucose deprivation (54), the binding of NML to dimethylated histone H3 at lysine 9 (H3K9me2) induces a ternary eNoSC together with SIRT1 and SUV39H1, and the loss of any of these components increases pre-RNA levels, suggesting that coordinated binding of NML, SIRT1, and SUV39H1 to the rDNA locus induces heterochromatin across the rDNA during glucose starvation (53). The eNoSC-regulated state of rDNA is unstable and reversible because eNoSC-mediated histone modification and concurrent rDNA silencing should be reversed upon the onset of suitable cellular energy levels (52). These dynamic epigenetic changes in rDNA chromatin appear to be intended to conserve energy and to increase cellular resistance in response to such limited conditions.
Third, the NuRD (nucleosome remodeling and deacetylation) complex is a large multisubunit complex with two types of catalytic subunits, including HDAC1/2 and the ATP-dependent helicase CHD3/4, and nonenzymatic proteins, including GATA zinc finger domain-containing proteins 2A and 2B (GATAD2A/2B), methyl-CpG-binding domain proteins (MBD2/3), metastasis-associated proteins (MTA1/2/3), and retinoblastoma-binding proteins (RBBP7/4) (55). The NuRD complex is activated in response to the attenuation of cell growth and establishes the poised state of rDNA, a chromatin state that is accessible for transcriptional factors but transcriptionally inactive, thereby suppressing rDNA transcription and maintaining the inactive rDNA (56). Binding of TTF-I to the T0 element and to CSB (Cockayne syndrome protein B), a DNA-dependent ATPase, facilitates the recruitment of the NuRD complex to unmethylated CpG rDNA promoters (56, 57). Interestingly, the NuRD complex also negatively regulates TIP5 expression, which leads to the suppression of CpG methylation at rDNA promoters (57). As such, the NuRD complex-mediated state of rDNA can be categorized into a poised state of rDNA chromatin or of rRNA genes, which functions in maintaining silent rDNA in growth-arrested or differentiated cells. The NuRD complex maintains this poised state of rDNA chromatin by forming a repressive nucleosome on the rDNA core promoter, which is associated with bivalent histone modifications; a heterochromatic mark, trimethylated histone H3 at lysine 27 (H3K27me3); and a euchromatic mark, trimethylated histone H3 at lysine 4 (H3K4me3), and this complex also provides an intermediate configuration of rDNA chromatin, which is not transcribed but which remains transcription permissive (56, 57).
Thus far, only a few features of regulation of rDNA silencing are common in both yeast and mammals. For example, human SIRT1, the mammalian homolog of yeast Sir2, is found in the eNoSC (Fig. 2A). In addition, although the mammalian homolog of yeast Fob1 has not yet been determined, the human KRAB-A domain-containing 2 (KRBA2) protein is likely homologous to yeast Fob1 (Fig. 2B). Furthermore, silencing proteins that mediate heterochromatin spreading across the rDNA repeats bind to NTS regions throughout eukaryotes; yeast rDNA silencing complexes primarily bind to the NTS1 or NTS2 regions; and consistently, the mammalian silencing NoRC and NuRD complexes are recruited to the rDNA promoter within the IGS region via the DNA-binding protein TTF-I that is bound to the proximal promoter element T0 (41, 47, 49, 56).
In contrast, one prominent feature of rDNA silencing in mammals that is not found in budding yeast is the existence of DNA methylation at rDNA loci. This modification is regarded as a key chromatin mark for silent and inactive chromatin (58). Several studies further suggested that DNMT-mediated hypermethylation of rDNA is one of the causes of gene silencing at rDNA loci (59, 60). Indeed, methylated DNA is found primarily in rDNA promoters and enhancers of silent rRNA genes (61). Loss of either DNMT1 or DNMT3b or treatment of mammalian cells with 5-aza-2′-deoxycytidine, a nucleoside analog mechanism-based DNMT inhibitor, reduces DNA methylation at rDNA loci with defects in both rDNA silencing and pre-rRNA synthesis (62). Also, a short noncoding pRNA has not been observed in budding yeast, and the loss of mammalian pRNA correlates with the depletion of rDNA methylation by disrupting the nucleolar localization of the NoRC and by affecting DNMT3b recruitment (63, 64).
In Arabidopsis, orthologs of yeast or mammalian rDNA-silencing complexes, such as yeast Fob1, the RENT subunit Net1/Cdc14, and Tof2-Lrs4/Csm1 proteins or the mammalian NoRC, have not been identified. In addition, although the Arabidopsis PKL (PICKLE) and PKR2 (PKL-related 2) proteins are homologs of metazoan CHD3/CHD4, their role in rDNA silencing is still not clear (65, 66). The role of Arabidopsis SRT2, a homolog of yeast Sir2, in rDNA silencing is also unknown (67). However, interestingly, in interspecific hybrids of plants, rRNA gene silencing is controlled by nucleolar dominance, a phenomenon in which rRNA genes of one species are transcriptionally dominant over the rRNA genes of other species (68–71). Notably, the regulation of nucleolar dominance in plants shares similarities with regulation by NoRC-mediated rDNA silencing in mammals, and nucleolar dominance in plants is regulated epigenetically because it requires HDA6 and DRM2, which are the homologs of mammalian Rpd3 deacetylase and DNMT3b, respectively (72–74). The role of short interfering RNA (siRNA) derived from IGS of the Arabidopsis rRNA gene is reminiscent of that of mammalian pRNA in regulating rDNA silencing (75). In addition, it was recently shown that all silenced rRNA gene subtypes map to NOR2, whereas all active rRNA gene subtypes map to NOR4 in Arabidopsis, suggesting that selective rRNA gene silencing is not regulated by gene-based mechanisms but by a subchromosomal silencing mechanism (76).
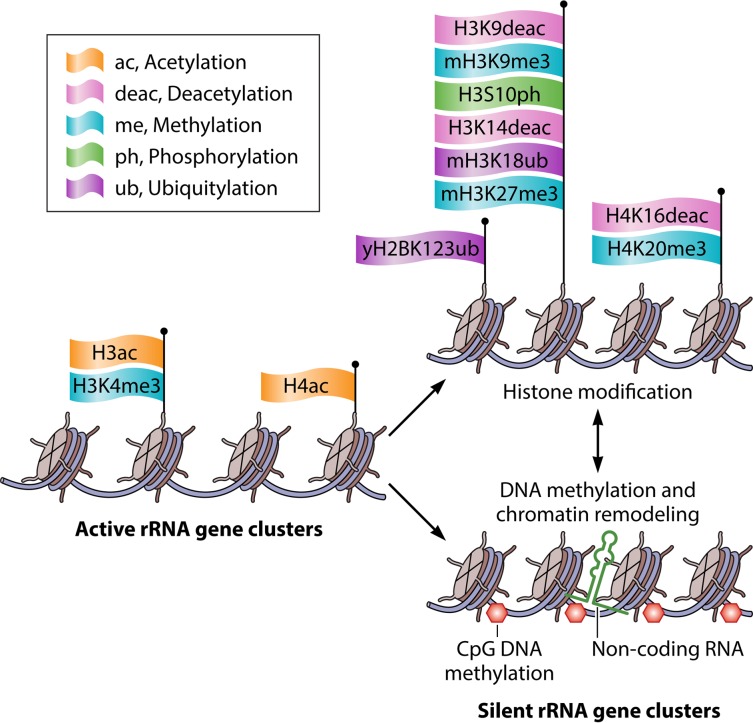
INVOLVEMENT OF HISTONE MODIFICATIONS IN rDNA SILENCING
Histone Methylation in rDNA Silencing
To date, several chromatin-modifying enzymes have been reported to affect rDNA silencing by modulating the state of DNA or histone modifications within rDNA regions in S. cerevisiae and in mammals (Fig. 5 and Tables 1 and 2). Among these modifications, histone methylation has emerged as a critical chromatin modification that regulates yeast rDNA silencing. Recently, our group has shown that histone methylation on histone H3 at lysine 4 (H3K4) and on H3 at lysine 79 (H3K79) by the histone methylases Set1 and Dot1, respectively, positively regulates the silencing within rDNA loci, whereas methylation on H3 at lysine 36 (H3K36) by Set2 methylase negatively affects rDNA silencing (26). These results indicate the bivalent epigenetic regulation of rDNA silencing by histone H3 lysine methylases. Without exception, the regulation of rDNA silencing by the three lysine methylases depends on Sir2. However, rDNA silencing is also regulated by histone demethylation, which is Sir2 independent and which is mediated by Jhd2, an evolutionarily conserved JARID1 family H3K4 demethylase that contains a Jumonji C (JmjC) domain (26). In this context, of the five JmjC-containing demethylases in yeast (Jhd1, Jhd2, Rph1, Gis1, and Ecm5), only Jhd2 and Gis1 show in vivo demethylase activity toward H3K4 and H3K36, respectively, within regions spanning rDNA loci. Surprisingly, Jhd2 demethylase affects rDNA silencing and rDNA recombination in a Sir2-independent manner by demethylating every state of methylated H3K4 within the NTS regions of rDNA. Moreover, during mitosis, Jhd2 has the ability to prevent the excessive recruitment of Tof2, Csm1/Lrs4, and condensin subunits to the RFB site within the NTS1 region, which is required for faithful mitotic chromosome segregation.
FIG 5.
Chromatin modifications during rDNA silencing. The different types of extracellular and intracellular signals modulate active rRNA gene structure and impose silent rRNA gene structure through various histone modifications, CpG dinucleotide DNA methylation, nucleosome sliding on the core promoter of the rRNA gene by the NoRC and NuRD complexes, and noncoding RNAs such as pRNA and PAPAS. m, mammals; y, yeast.
TABLE 1.
Effects of posttranslational modifications of histones on rDNA silencing and cellular aging in Saccharomyces cerevisiaec
| Histone modification | Gene | Histone target residue(s) | Effect on rDNA silencinga | Effect on life spanb |
Reference(s) | |
|---|---|---|---|---|---|---|
| RLS | CLS | |||||
| Acetylation | ESA1 | H4K5, H4K12, H4K16 | + | − | ND | 88, 89, 206 |
| NAT4 | H4 | − | ND | ND | 90 | |
| Deacetylation | SIR2 | H3K9ac, H3K14ac | + | 24, 91, 92 | ||
| H4K16ac | + | − | + | 24, 91–94, 154, 207 | ||
| RPD3 | H4K5ac, H4K12ac | − | + | ND | 93, 94, 160, 161 | |
| Methylation | SET1 | H3K4 | + | − | − | 26, 170, 171, 208 |
| SET2 | H3K36 | − | + | ND | 26, 170 | |
| DOT1 | H3K79 | + | − | − | 26, 170, 171, 209 | |
| Demethylation | JHD2 | H3K4 | − | ND | + | 26, 171 |
| GIS1 | H3K36 | NC | ND | NC | 26, 210 | |
| Phosphorylation | IPL1 | H3S10 | ND | ND | ND | 39 |
| Ubiquitylation | RAD6 | H2BK123 | + | − | ND | 25, 101, 102, 105, 173 |
| BRE1 | H2BK123 | + | − | − | 105, 172, 173, 211 | |
| Deubiquitylation | UBP8 | H2BK123 | − | + | + | 105, 173, 211–213 |
| UBP10 | H2BK123 | + | − | − | 103–105 | |
Effect of the indicated gene deletion on rDNA silencing, where “+” indicates an increase in URA3 or ADE2 expression due to the disruption of rDNA silencing and “−” indicates a decrease in URA3 or ADE2 expression due to increased silencing when a URA3 or an ADE2 reporter integrated into the NTS regions of the rDNA was used.
Effect of the indicated gene deletion on replicative life span (RLS) and chronological life span (CLS), where “+” indicates an increase in life span and “−” indicates a decrease in life span.
ND, not determined; NC, no change.
TABLE 2.
Epigenetic regulation of rDNA silencing in mammalsa
| Modification | Gene | Target residue(s) | Effect on rDNA silencing | Reference(s) |
|---|---|---|---|---|
| DNA methylation | DNMT1 | CpG | + | 60, 62 |
| DNMT3a | CpG | + | 60, 62 | |
| DNMT3b | CpG | + | 64 | |
| Acetylation | TIP60/KAT5 | H2AK5, H3K14, H4K5, H4K8, H4K12, H4K16 | ND | 96, 186, 214 |
| MOF/KAT8 | H4K16 | +/− | 51 | |
| Deacetylation | HDAC1 | H3K9ac, H4ac | + | 44, 48 |
| SIRT1 | H3K9ac, H3K14ac, H4K16ac | + | 53, 99 | |
| Methylation | EZH2/KMT6A | H3K27me3 | ND | 215 |
| PRMT5 | H3R8me2, H4R3me2 | + | 82 | |
| SETDB1/KMT1E | H3K9me1/2 | + | 77 | |
| SUV39H1/KMT1A | H3K9me1/2/3 | + | 53 | |
| SUV4-20H2/KMT5C | H4K20me3 | + | 80, 81 | |
| Demethylation | JHDM1A/KDM2A | H3K36me1/2 | + | 83 |
| JHDM1B/KDM2B | H3K4me3 | + | 84 | |
| JMJD2B/KDM4B | H3K9me3 | − | 85 | |
| PHF8/KDM7C | H3K9me2 | − | 86 | |
| PARylation | PARP1 | H3 | + | 111 |
| Ubiquitylation | UHRF1 | H3K23ub | + | 109 |
| RING1B | H2AK119ub | ND | 215 | |
| Deubiquitylation | USP21 | H2AK119ub | ND | 216 |
“+,” inducer for rDNA silencing or transcriptional repression; “−,” repressor of rDNA silencing or transcriptional repression; ND, not determined; “H2AK5,” histone H2A at lysine 5; “H2AK119ub,” ubiquitylated histone H2A at lysine 119.
rDNA silencing in mammals is closely associated with HP1 and histone methylation, such as di- or trimethylated histone H3 at lysine 9 (H3K9me2/3), H3K27me3, or trimethylated histone H4 at lysine 20 (H4K20me3) (43, 44, 46, 60). A coimmunoprecipitation experiment in human cells demonstrated that TIP5, a component of the NoRC, is associated with SETDB1, a histone methyltransferase that catalyzes mono- and dimethylation of histone H3 at lysine 9 (H3K9me1/2) (77). Consistently, TIP5 overexpression increases the presence of histone marks, such as H3K9me2/3 and H4K20me3, but decreases the dimethylation of H3K4, a histone mark for gene activation (43, 78). Moreover, SUV39H1, a component of the eNoSC, mediates H3K9me2, allowing rDNA silencing during glucose deprivation (53, 79). The level of SUV4-20H2-catalyzed H4K20me3 is increased at rDNA regions during growth arrest in NIH 3T3 cells (80). PAPAS (promoter and pre-rRNA antisense) is a long noncoding RNA that is produced in an antisense orientation by RNA polymerase II (RNAPII) and covers sequences of the pre-rRNA coding region and the rDNA promoter (81). This noncoding RNA is upregulated in growth-arrested cells and is accompanied by increased SUV4-20H2-mediated H4K20me3 and chromatin compaction (80, 81). Additionally, PRMT5, the type II arginine methyltransferase that catalyzes monomethylation or symmetric dimethylation of histones H3 and H4, plays a key role in silencing rDNA when the cells are in a stationary or nonproliferating state (82).
Moreover, mammalian histone demethylases have the ability to differentially regulate rDNA silencing: lysine-specific demethylases 2A and 2B (KDM2A and KDM2B, respectively) are the members of the JmjC family of demethylases that induce rDNA silencing, whereas other members of the JmjC family of demethylases, including JMJD2b (KDM4B) and PHF8 (KDM7C), are known to disrupt rDNA silencing. Human KDM2A (also called JHDM1A) was found to repress rDNA transcription in the nucleolus by binding to the rDNA promoter and demethylating mono- or dimethylated H3K36 during starvation (83). Human KDM2B (also called JHDM1B) also localizes to the nucleolus and facilitates rDNA silencing by demethylating H3K4me3, thereby inducing the dissociation of chromatin-bound UBF from the rDNA locus (84). In contrast, JMJD2b, which demethylates H3K9me2/3 in the pericentromeric heterochromatin, is found enriched on both rDNA promoters and the 28S rRNA coding sequence in colon and prostate cancer cells, implying that JMJD2b has a role in disrupting rDNA silencing (85). Human PHF8 also negatively regulates rDNA silencing by binding to the promoter of the rRNA gene and specifically demethylating H3K9me2 (86, 87).
Histone Acetylation in rDNA Silencing
Accumulating evidence indicates that the activities of histone acetyltransferases (HATs) and HDACs are required for regulating rDNA silencing in both yeast and mammals. Interestingly, each HAT or HDAC has a different effect on gene silencing within rDNA loci. For example, yeast Esa1, a catalytic subunit of the HAT complex NuA4, regulates the silencing of RNAPII-transcribed genes at rDNA loci (88). The loss of Esa1 results in the maximal reduction of acetylation at histone H4 lysine 5 (H4K5) as well as moderate reductions of acetylation at histone H4 lysine 12 (H4K12) and at H4K16. The increased expression of ESA1 and SIR2 reciprocally suppresses the rDNA silencing defects that are associated with esa1 and sir2 mutants, suggesting that Esa1 and Sir2 have opposite activities that contribute to achieving optimal nucleolar chromatin structure and function (89). In contrast, Nat4 negatively regulates rDNA silencing. Nat4 is a yeast histone H4 N-alpha-acetyltransferase, and a lack of its activity results in enhanced silencing of rDNA and increased deposition of asymmetric histone H4 arginine 3 (H4R3me2a) dimethylation, suggesting that the cross talk between H4 N-terminal acetylation and H4R3me2a contributes to the regulation of rDNA silencing (90).
Sir2 is undoubtedly one yeast HDAC that influences rDNA silencing. The loss of Sir2 increases acetylation on histone H3 at lysine 9 and lysine 14 residues (H3K9 and H3K14, respectively) as well as on histone H4K16 within the NTS1 region of rDNA loci, thereby disrupting rDNA silencing (24, 91, 92). However, in contrast to the positive role of Sir2 in the regulation of rDNA silencing, Rpd3 deacetylase, the catalytic subunit of a multiprotein deacetylase complex that contains Sin3 and Sap30, plays a negative role in rDNA silencing. The loss of Rpd3, Sin3, or Sap30 increases levels of acetylated H4K5 and H4K12 and enhances Sir2-dependent rDNA silencing (93–95). Therefore, Rpd3 is thought to counteract, rather than to establish or maintain, rDNA silencing (94).
Consistently, several human HATs regulate rDNA silencing in positive or negative ways. In human embryonic kidney HEK-293T cells, overexpression of TIP60 (also called KAT5), a yeast homolog of Esa1, reduces rDNA promoter activity, indicating that TIP60 downregulates rRNA gene transcription (96). Mammalian MOF (also called KAT8) is another HAT that also belongs to the MYST family and acetylates histone H4K16 preferentially at the promoter region of the rDNA loci rather than at the rRNA gene coding region (51, 97). MOF is required for the binding of TIP5 to rDNA chromatin and for the nucleolar localization of NoRC during S-phase progression; however, MOF and H4K16ac are present on both methylated and unmethylated rDNA loci, implying two roles for MOF-mediated H4K16ac in activating and silencing rRNA genes (51).
Generally, mammalian HDACs induce rDNA silencing through deacetylation of histones at the rDNA promoter. Mammalian SIN3 is a corepressor complex that belongs to a class I HDAC complex, which includes HDACs such as HDAC1/2 and other components, including SIN3A/B, SUDS3, RBAP4/7, RBBP1, SAP30, SAP130, SAP18, and SAP180, as corepressor scaffold proteins (48, 98). The SIN3 corepressor complex physically interacts with the NoRC, thereby facilitating NoRC-mediated rDNA silencing by targeting at the rDNA promoter (48). The changes in the NAD+/NADH ratio induced by the reduction of energy levels activate SIRT1 deacetylase, which mediates the deacetylation of H3K9 and H4K16 and thus leads to rDNA silencing (53). Interestingly, loss of SIRT1 by treatment with SIRT1-specific siRNA or by the addition of the SIRT1 inhibitor nicotinamide decreases SUV39H1-mediated H3K9me2 levels in human cells, and loss of DNMT1 methyltransferase decreases the recruitment of SIRT1 to rDNA regions with increased H4K16ac levels, suggesting a significant association of SIRT1 with other silencing marks, such as H3K9me2 and DNA methylation, during the establishment of mammalian rDNA silencing (53, 99).
Histone Ubiquitylation in rDNA Silencing
In budding yeast, the lysine 123 residue of histone H2B (H2BK123) is ubiquitylated by the E2 ubiquitin-conjugating enzyme Rad6 (also called Ubc2) and the E3 ligase Bre1, and this ubiquitylation is reversed by deubiquitinating enzymes such as the ubiquitin proteases Ubp8 and Ubp10 (100). Rad6 is associated with heterochromatic silencing in telomeric regions, HM loci, and rDNA regions (25, 101). In rDNA regions, the deletion of RAD6 or the expression of histone H2B with a lysine-to-arginine substitution mutation at residue 123 disrupts rDNA silencing, which elucidates the involvement of histone ubiquitylation in inducing rDNA silencing (102). Interestingly, the histone deubiquitylase Ubp10 (also called Dot4) is associated with rDNA silencing in a manner similar to that of the ubiquitylase Rad6. In cells lacking Ubp10, rDNA silencing is significantly suppressed, and a reduction of Sir2 association is consistently found with the hyperacetylation of H4K16 and the hypermethylation of H3K4 and H3K79 in NTS regions of rDNA (103–105). Therefore, the maintenance of low levels of histone ubiquitylation, as regulated by both Rad6 ubiquitylase and Ubp10 deubiquitylase, appears to be required for rDNA silencing (103, 105).
Ubp8 is another histone deubiquitylase for H2BK123 in yeast; however, in contrast to Ubp10, the loss of Ubp8 significantly increases rDNA silencing (105). The molecular mechanism by which Ubp8 regulates rDNA silencing is not known, and further studies are needed to investigate the opposite roles of the two histone deubiquitylases Ubp8 and Ubp10 in regulating rDNA silencing. In addition, the protein deubiquitylase Ubp3 does not target histone substrates but plays a role similar to that of Ubp8 in rDNA silencing, and a chromatin immunoprecipitation assay revealed that the assembly of RNAPII at rDNA loci depends on Ubp3 and that the loss of Ubp3 leads to an increase in the binding of Net1 to rDNA, which coincides with increased rDNA silencing and highly suppressed levels of USCE (106).
Few studies have addressed the role of (de)ubiquitylation in mammalian rDNA silencing. Human UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) (also called NP95) is an E3 ubiquitin ligase that mediates ubiquitylation of histone H3 at lysine 18. This E3 is required for the establishment and maintenance of DNA methylation in mammals (107, 108). A study showed that a loss of UHRF1 or mutation in the tandem tudor domain of UHRF1, a domain required for binding to H3K9me2/3, diminished DNA methylation at human IGS regions of rDNA loci (109).
Histone PARylation/Ribosylation in rDNA Silencing
Recent studies further showed the functional association between PARP1 [poly(ADP-ribose) polymerase 1], which is an ADP-ribosylating enzyme, and rDNA silencing in flies and mammals (110, 111). PARP1 is present in the nucleolus and regulates ribosomal biogenesis by maintaining the genomic integrity of rRNA repeats in Drosophila melanogaster nucleoli (110). In mammals, PARP1 binds to the rDNA promoter via TIP5, an association facilitated by noncoding pRNA, which is implicated in the maintenance of silent rDNA chromatin during cell division (111). Particularly, because silent rDNA chromatin is a specific substrate for ADP-ribosylation and because PARP1-mediated ADP-ribosylation is necessary for establishing rDNA silencing, especially in mid- to late S phase, PARP1 and ADP-ribosylation are thought to facilitate the inheritance of silent chromatin structures.
INVOLVEMENT OF INM-ASSOCIATED PROTEINS IN EPIGENETIC REGULATION OF rDNA SILENCING
Evidence from the past several years has provided a significant understanding of how the structural maintenance proteins cohesin and condensin interact with the cohibin complex and INM-associated proteins, thereby contributing to proper rDNA silencing. The nuclear envelope is composed of two membrane bilayers: the INM faces the inside of the nucleus, and the outer nuclear membrane exists in continuous contact with the endoplasmic reticulum in the cytoplasm (112). The function of the nuclear envelope is to separate the cell into two compartments, the nucleus and the cytoplasm, and to exchange RNAs or proteins through the nuclear pores that span the nuclear envelope. Large multiprotein nuclear pore complexes control the transport of macromolecules through the nucleus. However, repressive heterochromatin is surrounded by and affiliated with the INM, generally away from the nuclear pore complexes (8, 112). Particularly in budding yeast, rDNA-silent loci and telomere regions are closely associated with the INM; thus, the localization and interaction of rDNA with different types of nucleolar proteins along with the proteins at the nucleolar periphery, such as the cohibin complex, the cohesin complex, the condensin complex, and the Smc5/6 complex, provide a better understanding of epigenetic regulation of rDNA silencing (8, 113). Because the cohibin complex, which mediates the perinuclear anchoring of rDNA to INM proteins, associates with rDNA depending on changes in histone methylation, exploring additional examples of epigenetic changes that fine-tune the rDNA silencing pathways that include INM-associated proteins will be important.
The Cohibin Complex
In budding yeast, cohibin is a “V”-shaped complex that is composed of two Lrs4 and two Csm1 homodimers (27, 113). Csm1 is required for the prevention of USCE through the proper arrangement of rDNA repeats within sister chromatids, and the Lrs4 proteins physically attach the rDNA to the chromatin linkage of INM proteins (CLIP) such as Heh1 (helix extension helix 1) and Nur1 (nuclear rim 1). Both Lrs4 and Csm1 are necessary for maintaining rDNA repeat stability and for rDNA silencing (27, 113, 114). In particular, the association of rDNA with the nuclear periphery through the collaboration of RENT and the cohibin complex sequesters the recombination proteins away from rDNA repeats and prevents unequal rDNA recombination, thereby further stabilizing the repeats. This association indicates that the INM-mediated perinuclear chromosome tethering of rDNA is required to ensure rDNA repeat stability (113, 115). Moreover, the observation that the loss of either Sir2, Lrs4, or Csm1 disrupts rDNA silencing, whereas Heh1 and Nur1 are responsible for rDNA repeat stability but not essential for rDNA silencing, raises the possibility that rDNA silencing is not sufficient for proper rDNA repeat size regulation (113). In contrast to such noticeable progress in yeast systems, little is known about the roles of the mammalian cohibin complex and of CLIP proteins in regulating rDNA repeat stability. Only one report has shown that the human integral INM proteins LEM2 and MAN1, which are human homologs of the yeast CLIP protein Heh1, bind to lamins attached to the INM and tether chromatin at the nuclear periphery (116).
The Cohesin Complex
Cohesin is a ringlike four-member protein complex, which is composed of Smc1, Smc3, Scc1/Mcd1/Rad21, and Scc3/Irr1 during mitosis, whereas Scc1 is replaced by Rec8 during meiosis. This complex is required for clasping sister chromatids together during mitosis and meiosis (36, 117). Smc1 and Smc3 are members of the SMC (structural maintenance of chromosome) family, which also includes Smc2 and Smc4 as subunits of condensin and Smc5 and Smc6 as components of the Smc5/6 complex (118). The cohesin SMC proteins and non-SMC proteins are primarily conserved among vertebrates; for instance, the human cohesin complex is composed of SMC1A/B, SMC3, RAD21/SCC1, and SA1/SA2 during mitosis, whereas REC8 and STAG3 replace SCC1 and SA1/SA2 during meiosis (36). Cohesin holds the sister chromatids together, and this process is required for chromosome segregation and homologous recombination during the repair of DSBs (119). Additionally, the cohesin complex plays a role in rDNA condensation in budding yeast. At metaphase, rDNA is less condensed than most other chromosome sequences due to the persistent catenation of rDNA repeats at this locus; however, during anaphase, when the protein phosphatase Cdc14 is activated, the rDNA region condenses and is separated (120). The Sir2 protein, which prevents USCE between rDNA repeats, is necessary for cohesin binding within rDNA loci (34, 121).
There are two plausible yeast system models that may explain the recruitment of the cohesin complex to rDNA. One model suggests that Sir2 facilitates the association of cohesin with NTS regions of rDNA by silencing the conserved RNAPII promoter element E-pro near the rDNA recombination enhancer (14). Another model suggests that a direct interaction between cohesin subunits and Csm1, a cohibin subunit, accounts for the recruitment of the cohesin complex to rDNA (27). Notably, cohesin binding to the rDNA is evolutionarily conserved in eukaryotic genomes, but its role in rDNA silencing is not yet clear (122). Apart from the role of cohesin in chromosome segregation and rDNA condensation, cohesin and its associated factors have additional functions in transcription (123). Interestingly, one report suggested that the cohesin complex facilitates rRNA production and protein synthesis, which may explain a large fraction of the observed gene misregulation, such as Cornelia de Lange syndrome and Roberts syndrome, human diseases caused by mutations in cohesin (122).
Condensins
The yeast condensin complex is composed of five subunits: two core SMC subunits (Smc2 and Smc4) and three non-SMC subunits (the kleisin subunit Brn1 and the HEAT repeat subunits Ycs4 and Ycs5/Ycg1). This macromolecular complex is involved in regulating chromosome architecture and its condensation and is closely associated with silencing in heterochromatin regions, such as mating-type loci or rDNA regions (26, 38). Condensin maintains the structural and genomic stability of rDNA and aids in proper rDNA segregation during mitosis (124). In yeast, the association of condensin with rDNA is thought to reduce the topological effect that is induced by transcription, consequently facilitating the proper segregation of the rDNA (18, 125). In addition, condensin plays an important role in rDNA silencing, as it facilitates an increase in Sir2 recruitment at the rDNA loci and prevents the spreading of silenced chromatin to nonsilenced regions by forming the Sir2-enclosed rDNA chromatin loops, and the loss of condensin disrupts this rDNA chromatin loop organization, thereby leading to the spread of Sir2 to telomeres and leading to alterations in the strength of silencing at both regions (25, 126, 127). Notably, the cohibin proteins Csm1 and Lrs4 also regulate rDNA recombination by recruiting condensin proteins to the RFB site of rDNA, thus maintaining rDNA integrity (128). In this context, the RFB site in the NTS1 region of rDNA acts as a cis element for the Fob1-dependent recruitment of condensin to rDNA (128), and the H3K4 demethylase Jhd2 contributes to the regulation of condensin recruitment to rDNA by alleviating excessive condensin in the NTS1 region of rDNA during mitosis (26).
The condensation of mitotic chromosomes and the resolution of sister chromatids are two indispensable processes that are required for proper chromosome segregation during mitosis. Condensin maintains chromosome condensation during mitosis, but it also supports sister chromatid resolution during chromosome segregation in anaphase (129). Indeed, condensin and topoisomerase II are arranged properly along the rDNA and facilitate the late decatenation of rDNA sister chromatids, which regulates the late segregation of rDNA during anaphase and ensures the timely segregation of chromosomes in mitosis (130). Therefore, both cohesin and condensin are believed to be functionally linked and to serve as the major constituents of mitotic chromosomes, where cohesin directs sister chromatid cohesion from S phase until mitosis, whereas condensin is imperative for the timely compaction and resolution of chromosomal regions containing rDNA, particularly during mitosis (127, 131). To facilitate rDNA condensation, yeast condensin is assembled primarily in the nucleolus during mitosis, although it is localized across the genome during interphase (124). Of note, yeast Ipl1/aurora kinase, which phosphorylates histone H3S10, also phosphorylates the condensin subunit Ycg1, a condensin modification that is independent of cohesin and that is required for the postmetaphase chromosome assembly maturation that is important for chromosome segregation (132).
The condensin subunits are also highly conserved from yeast to humans: similarly to the yeast condensins, Xenopus laevis and human condensins are associated with the nucleolus, as their condensin subunits are localized in the granular component of the nucleolus (133). This association suggests that the conformation and function of the rDNA are closely associated with the roles of condensin in regulating rDNA condensation, recombination, silencing, or segregation in higher eukaryotes (134, 135). In humans, two analogs of condensin complexes are present, namely, condensins I and II, which are composed of two SMC proteins, SMC2 and SMC4; one kleisin protein (CAP-H for condensin I and CAP-H2 for condensin II); and two HEAT repeat proteins (CAP-D2/CAP-G for condensin I and CAP-D3/CAP-G2 for condensin II) (136). Consistent with data from yeast system studies, the findings of several reports support the implication of mammalian condensin in regulating rDNA silencing. For example, the loss of the catalytic subunit of the condensin complex SMC2 significantly increases CTCF (CCCTC-binding factor)-mediated rDNA transcription (137). Data from chromatin immunoprecipitation (ChIP) analyses suggest that SMC2 more preferentially binds to the promoter and to transcribed regions than to nontranscribed regions of the human rDNA locus, and an SMC2 knockout increases UBF recruitment and enhances H4 acetylation enrichment around the rDNA promoter (137). CTCF repression also increases SMC4 binding preferentially to the transcription initiation region and transcribed region of the rDNA locus (137). In SMC-depleted HeLa cells, sister NORs missegregate or segregate with a substantial delay, suggesting the structural instability of rDNA due to condensin depletion (138). Similar to yeast Ipl1/aurora kinase, inactivation of aurora B kinase leads to a loss of association of condensin I with chromatin in human cells (139).
The Smc5/6 Complex
The yeast Smc5/6 complex consists of the SMC group proteins Smc5 and Smc6, which are conserved throughout eukaryotes, and a number of non-SMC subunits, such as Nse1, Nse2/Mms21, Nse3, Nse4/Rad62, Nse5, and Nse6/Kre29 (15, 118). The human SMC5/6 complex includes four non-SMC subunit orthologs: NSE1, NSE2/MMS21, NSE3/MAGEG1, and NSE4A/B (118, 140). The Smc5/Smc6 complex is suggested to be essential for rDNA maintenance and segregation as well as for efficient DNA repair; however, the precise role of this complex in regulating rDNA silencing remains unclear (141). Mms21 is a small ubiquitin-related modifier (SUMO) E3 ligase that is required for sumoylation during gene silencing and regulates the binding of both the condensin and cohesin complexes to rDNA regions (142, 143). In addition, Smc5/Smc6 assists in the precise completion of DNA replication and in the proper organization of mitotic chromosomes (15, 118). Indeed, loss of human SMC5/SMC6 results in delayed DNA replication at centromeres and telomeres and in disrupted chromosome assembly and segregation (144). Therefore, based on the notion that the Smc5/6 complex is implicated in various cellular processes that are closely associated with rDNA silencing, the role of the Smc5/6 complex in regulating rDNA silencing, together with the roles of the cohesin and condensin complexes, is worth exploring.
IMPLICATION OF EPIGENETIC REGULATION OF rDNA SILENCING IN AGING AND DISEASES
Cellular Aging
Cellular aging is one of the most prominent cellular processes that are influenced by rDNA silencing. The molecular basis of cellular aging in budding yeast has been extensively studied by analyzing the replicative life span (RLS) and chronological life span (CLS). Replicative aging is measured in mitotically dividing cells and involves assessing the life span of a mother cell by observing the total number of daughter cells that are produced before death (145, 146). In contrast, chronological aging is measured in postmitotic cells and involves determining the survival time of cells in a nondividing state (147). Based on these two life span analyses, many pathways that regulate cellular life span have been described in yeast. One pathway involves the formation of ERCs, the accumulation of which shortens the life span and is thus one of the major causes of cellular aging. Another pathway involves Sir2 deacetylase, which can be activated by calorie restriction and which subsequently prolongs life span (Table 1) (148–150). In addition to the two pathways, rDNA instability or a natural polymorphism in ribosomal ARS has been shown to influence yeast life span independent of ERC accumulation or Sir2 (151, 152).
The role of Sir2 in regulating the cellular life span is observed in several examples. As mentioned above, silencing within rDNA loci plays an important role in rDNA stability and nucleolar integrity and is performed mainly by Sir2. The repression of rDNA recombination by Sir2 reduces the formation of ERCs (153, 154). The suppression of RNAPII-dependent transcription by Sir2 within rDNA loci leads to rDNA silencing, which also finally extends the replicative life span (33, 155). In addition, rapamycin treatment or nitrogen starvation inactivates TOR (target of rapamycin) complex 1 (TORC1) and increases the association of Sir2 with rDNA loci. The inhibition of TORC1 promotes the transcriptional silencing of RNAPII-transcribed genes at the rDNA locus and reduces homologous recombination between rDNA repeats, thereby extending the life span (148, 156). Several other studies have further revealed that the maintenance of a proper concentration of intracellular NAD+ through the NAD+ biosynthesis and salvage pathways is a prerequisite for the NAD+-dependent HDAC activity of Sir2 and that disruption of the NAD+ concentration shortens the replicative life span, demonstrating a significant relationship between rDNA silencing, metabolism, and cellular aging (157–159). In contrast to the role of Sir2 in rDNA silencing and cellular life span, another HDAC, Rpd3, has the opposite effect: the loss of Rpd3 not only increases rDNA silencing but also significantly extends the life span, which reflects the involvement of HDACs in regulating the cellular life span. However, the loss of Rpd3 does not overcome the shortened life span due to the loss of Sir2 and does not have an additive effect of life span extension under conditions of calorie restriction (160, 161).
Similar to yeast Sir2, metazoan Sir2 and SIRT1 regulate life span. In Caenorhabditis elegans and Drosophila, overexpression of Sir2 homologs extends the postmitotic life span (162, 163). Likewise, the expression of mouse SIRT1 decreases with age in thymus and testis tissues, leading to reduced mitotic activity and SIRT1 overexpression in brain tissue, where mitotic activity remains less changed with age, thus extending the life span and delaying the aging process (164, 165). However, one report demonstrated that SIRT1 overexpression in the entire body of mice failed to extend the life span of the mice, raising the possibility that the effect of SIRT1 on the life span in mammals is tissue specific (166). Regardless of this issue, age-dependent regulation of human SIRT1 is likely closely associated with the Sir2-dependent function in yeast, and a recent study showed that overexpression of human SIRT1 in yeast cells lacking Sir2 decreases ERC formation and histone acetylation at H4K16 within rDNA regions (167).
Beyond the role of Sir2 and SIRT1 and their concurrent histone deacetylation in regulating life span, several other epigenetic mechanisms pose a great challenge for identifying the molecular pathways that regulate the cellular life span. Indeed, the establishment of changes in histone modifications is considered an epigenetic signature of young and old cells (168, 169). As described above, our group reported that histone H3 methylation on K4/K79 and K36 acts as a bivalent marker for the regulation of Sir2-dependent rDNA recombination and rDNA silencing. This effect of histone methylation on rDNA silencing correlates well with changes in the replicative life span in budding yeast (25, 170). One study further demonstrated that the loss of H3K4 methylation leads to a decrease in the CLS and is thus responsible for enhanced cell death, whereas the suppression of Jhd2-mediated H3K4 demethylation improves cell survival, indicating that the loss of H3K4 methylation is an important cause of cell death in yeast (171). The ubiquitylation of histone H2BK123 has emerged as an important epigenetic marker, as the level of this modification is significantly elevated throughout the rDNA loci in replicatively aged yeast cells (105). Life span analyses further show that the disruption of H2B ubiquitylation by the deletion of either the H2B ubiquitylase Rad6/Bre1 or the H2B deubiquitylase Ubp10 decreases the RLS, and consistent with this finding, cells deficient in Bre1 show a decreased CLS (105, 172). Yeast Ubp10 was previously reported to be required for proper Sir2 localization at the telomeric and rDNA regions by maintaining low levels of H2BK123 ubiquitylation as well as low levels of H3K4 and H3K79 methylation within telomeric and rDNA loci (103, 104). In contrast to the role of Ubp10, cells that are deficient in the SAGA histone deubiquitylase module, including Ubp8, are exceptionally long-lived (173).
In mammals, changes in DNA methylation at rDNA loci have a potent ability to regulate cellular life span. Previously, an age-associated loss of rDNA repeats was observed in mouse brain tissues, probably due to increased homologous recombination among the tandem rDNA repeats (174). Recent studies showed that mammalian cells undergo DNA methylation drift across the genome, which involves global hypomethylation and locus-specific hypermethylation of CpG sites (169, 175). Locus-specific DNA hypermethylation is found at the rDNA locus, where methylation is frequently associated with transcription repression (175). Consistent with this finding, age-dependent DNA hypermethylation occurs specifically at rDNA regions, causing a deficiency in rRNA synthesis in somatic cells of mice (176). Another report showed that DNA hypermethylation within rDNA clusters is observed in liver tissues and spermatozoa of old rats (177). Although the Sir2- or SIRT1-engaged pathways are mainstays of the molecular basis of maintaining a normal life span in mitotically dividing or postmitotic cells so far and although most of the novel epigenetic controls are closely associated with this Sir2/SIRT1 deacetylase throughout eukaryotes, the pathways that are Sir2 or SIRT1 independent but that influence rDNA silencing also have the potential to regulate life span. For instance, the yeast Lrs4 and Cms4 proteins, which are required for rDNA silencing, regulate the cellular life span through the perinuclear anchoring of rDNA with the help of cohibin and the INM proteins Heh1 and Nur1 (114).
Disease Progression
A series of studies have interrelated epigenetic gene regulation with various diseases caused by defects in cell cycle progression, cell growth, and DNA repair and recombination. However, whether changes in epigenetic regulation within rDNA chromatin are some of the causes or consequences of aged or cancerous cells is uncertain. Generally, if rDNA transcription is abnormally repressed, cells may undergo cell cycle arrest, accompanied by senescence or apoptosis (5). In contrast, if rRNA synthesis is abnormally upregulated by a loss of silent rDNA regions primarily through DNA hypomethylation at rDNA, cells may lose rDNA stability, or rDNA transcription may be unnecessarily stimulated, thereby leading to enhanced cell proliferation or even to malignant transformation (5, 58). These possibilities strongly suggest that proper rDNA silencing acts as a barrier to suppress tumor development in mammals and that failure to impede unnecessary rDNA transcription may lead to oncogenesis (84, 178).
Recent studies suggested a significant role of rDNA hypomethylation at the onset of cancers. rDNA hypomethylation is found in a wide range of cancers, such as hepatocellular carcinoma, endometrial cancer, brain cancer, and colon cancer (178–181). Human hepatocellular carcinoma cells display DNA hypomethylation at rDNA promoters that coincides with the reactivation of silenced rDNA expression (181). In human colon carcinoma cells lacking DNMT1 or DNMT3B, the levels of the 47S primary rRNA transcript are significantly increased (181). In glioblastoma cells deficient in NPM1, a nucleolar histone chaperone, loss of DNMT3 also synergistically enhances rDNA transcription (180). Indeed, low levels of DNA methylation at rDNA genes are found in many African American women with endometrial carcinoma (178). However, notably, DNA hypermethylation at rDNA regions is also associated with certain types of cancers, such as ovarian or breast cancers (182–184). The levels of DNA methylation at 18S and 28S rRNA genes are high in patients with ovarian cancer, especially in long progression-free survival compared with short survival (183). MassARRAY EpiTYPER assays further demonstrated DNA hypermethylation at rDNA in breast cancer tissues compared to normal breast tissues (182).
Dysregulation of JmjC-containing histone demethylases is closely associated with perturbation of rDNA transcription and with various types of cancer cells (Table 3). Inhibition of the JmjC demethylase JHDM1A/KDM2A by dimethyl succinate induces pre-rRNA synthesis during starvation in breast cancer cells (83). Similarly, loss of the PHF2/KDM7B JmjC demethylase increases pre-rRNA synthesis in colon and lung cancer cells (185). Consistent with these findings, the expression of JHDM1B/KDM2B, a JmjC demethylase that is found in the nucleolus and that is required for rDNA silencing, is significantly downregulated in glioblastoma (84). In parallel, H4K20me3 at rDNA loci is downregulated in breast and colon cancer cells (80). However, in contrast to these examples, knockdown of the PHF8/JKDM7B JmjC demethylase decreases pre-rRNA synthesis and leads to reduced proliferation of osteosarcoma cells (86).
TABLE 3.
Effects of epigenetic proteins and other factors on rDNA silencing during cancer
| Factor | Gene | Disease(s) | Cell line(s) | Effect of gene loss/presence on rRNA synthesisa | Reference(s) |
|---|---|---|---|---|---|
| DNA CpG methylation | DNMT1 | Colon cancer | HCT116 | + | 181 |
| DNMT3a | Glioblastoma | U1242MG | + | 180 | |
| DNMT3b | Colon cancer | HCT116 | + | 181 | |
| Histone modification | HDAC1 | Osteosarcoma | SaOS-2 | + | 190 |
| JHDM1A/KDM2A | Breast cancer | MCF7 | + | 83 | |
| JHDM1B/KDM2B | Glioblastoma, breast cancer | T98G, MCF7, ZR-75-1, MDA-MB-231 | + | 84, 217 | |
| PHF2/KDM7B | Colon cancer, lung cancer | HCT116, A549 | + | 185 | |
| PHF8/KDM7C | Osteosarcoma | U2OS | − | 86 | |
| TIP60/KAT5 | Prostate cancer | LNCaP | +* | 186 | |
| Other | ZNF454 | Gastric cancer | MGC803 | + | 218 |
| PTEN | Prostate cancer, glioblastoma | LNCaP, U87 | + | 219 | |
| p14ARF/CDKN2A | Bronchoalveolar carcinoma | H358 | +* | 220 | |
| RUNX1/AML1 | Acute myeloid leukemia | Kasumi-1 | + | 221 | |
| RUNX2 | Osteosarcoma | SaOS-2 | + | 190 |
“+,” increase in rRNA synthesis; “−,” decrease in rRNA synthesis. “*” indicates the presence or overexpression of the gene.
The implication of changes in histone acetylation with cancers comes from studies of TIP60/KAT5 and SIRT1. In prostate cancer cells, the TIP60 acetyltransferase is recruited to the rDNA promoter upon androgen stimulation, thereby inducing rRNA transcription through acetylating both histone H4 and UBF at the rDNA promoter (186). In contrast, many studies have indicated that the expression level of SIRT1 deacetylase influences heterochromatin formation and cellular transformation (53, 187–189). SIRT1-null mouse embryos die mainly as a result of an increase in H3K9 acetylation, which is accompanied by reduced H3K9me3-associated heterochromatin formation and by increased genome instability due to an impaired DNA damage response (187). Moreover, SIRT1 levels are reduced in breast cancer and in hepatocellular carcinoma (187). One report demonstrated that HDAC1-mediated deacetylation of H3K9 and histone H4 at the rDNA promoter suppresses rRNA transcription by RUNX2, a factor that controls bone lineage commitment and cell proliferation, in human cancer osteosarcoma cells (190).
Perturbed epigenetic regulation of rRNA synthesis is also observed in other examples of diseases, such as neurodegenerative, psychiatric, hematopoiesis, autoimmune, and developmental disorders. One of the chronic neurodegenerative diseases, Alzheimer's disease (AD), occurs with the loss of neurons and synapses in the cerebral cortical tissue. Of note, the reduction of ribosomal function is one of the reasons for the development of AD pathogenesis (191). In accordance, DNA hypermethylation on the rDNA promoter is associated with patients with AD presenting relatively early stages of mild cognitive impairment (192). In addition, the promoter and 5′-regulatory regions of rDNA are significantly hypermethylated in the brains of suicidal males, consistent with decreased rRNA expression in the hippocampus (193). In CD34+ cells of patients with myelodysplastic syndrome, a clonal disorder of hematopoietic stem cells, increased DNA methylation at the rDNA promoter, together with reduced rRNA synthesis, is suggested to contribute to defective hematopoiesis and bone marrow failure in the majority of these patients (194). However, in white blood cells of patients with systemic lupus erythematosus, an autoimmune inflammatory disease, DNA methylation of rDNA is decreased (195). Similarly, in patients with X-linked alpha thalassemia/mental retardation syndrome (ATRX), hypomethylation of rDNA is associated with ATRX gene mutation (196).
CONCLUDING REMARKS
Recent advances in understanding the regulation of rDNA heterochromatin silencing reveal that specific chromatin modifications are involved in gene silencing at rDNA loci. In budding yeast, the two main epigenetic pathways by which transcription at the rDNA region is silenced suggest that a distinct set of histone modifications is required for rDNA silencing depending on cell cycle progression, by which genome stability and faithful chromosome segregation are maintained for a normal life span. In addition, rDNA is known to be strongly associated with heterochromatin in the nucleolus; however, paradoxically, these regions still allow active transcription, which affects rDNA silencing and recombination between rDNA repeats. The cell cycle-specific regulation of histone modifications on rDNA silencing may provide, in part, a plausible explanation as to how this paradoxical rDNA silencing is regulated. In mammals, the epigenetic regulation of rDNA silencing is mediated by three classes of complexes, the eNoSC, NoRC, and NuRD complex. Two complexes, the eNoSC and NuRD complex, have intrinsic activities of histone deacetylation or methylation. The NoRC also has chromatin-remodeling activity that facilitates the coordination of different histone-modifying enzymes or DNA methyltransferase to suppress rDNA transcription. In addition, there is complicated interdependency of chromatin modification by the rDNA-silencing complexes and other histone- or DNA-modifying proteins to repress rDNA transcription. For instance, coordinated binding of eNoSC components to the rDNA locus requires histone H3K9me2 during glucose starvation. TIP5, a component of the NoRC, interacts with histone H4K16ac to repress rDNA transcription. The NuRD is recruited to the unmethylated CpG rDNA promoter via TTF-I. Moreover, various types of histone modifications are closely associated with rDNA silencing, and perturbations of rDNA epigenetic pathways lead to various types of cancers or other broad ranges of diseases, such as neurodegenerative, psychiatric, hematopoiesis, and autoimmune diseases, as well as developmental disorders. Therefore, combinatorial optimization of chromatin modifications on rDNA loci is likely performed by rDNA-silencing complexes or other chromatin modifier proteins to regulate rDNA silencing depending on each stage of the cell cycle or energy-limited conditions. In addition, although fine-tuned characterization of the epigenetic pathways at rDNA regions is still required, it is encouraging that the connection between the epigenetic regulation of rDNA silencing and its effect on the regulation of cellular life span would provide promising fundamental knowledge to improve our understanding of apoptosis, cancer biology, or other age-associated diseases.
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea grant funded by the South Korean government (MSIP) (no. 20110030049) and by the research fund of Hanyang University (grant HY-2014-P).
We declare no competing financial interests.
Biographies

Rakesh Srivastava has been a postdoc in the laboratory of Seong Hoon Ahn at Hanyang University, South Korea, since 2014. He obtained his M.S. in biotechnology in 2003. He then joined a private firm and helped to develop an asthma and cancer pathogenesis database. In 2005, he became a research assistant at the National Botanical Research Institute, Lucknow, India, and was involved in the study of a stringently regulated gene expression system. He began his Ph.D. coursework at the Banaras Hindu University, Varanasi, India, in 2008. During his Ph.D. work, he studied the different core promoter elements and transcription complexes and their roles in transcriptional regulation and nucleosome remodeling of plant genes. He currently focuses on the role of histone modification during transcription initiation and gene silencing.

Seong Hoon Ahn is a Professor of Molecular Biology at Hanyang University. He studied pharmacy and obtained his Ph.D. from Sungkyunkwan University (2001), and his dissertation was on the antiproliferative activity of the histone deacetylase inhibitor apicidin. As a postdoc in the laboratory of Stephen Buratowski at Harvard Medical School (USA), he studied the role of phosphorylation of serine 2 within the RNA polymerase II C-terminal domain (CTD) in transcription, its 3′-end processing, and the link between CTD modification and histone modification. Since 2005, he has worked as a professor at Hanyang University, South Korea. The major focus of his current research is to understand how the synchronized code by the CTD and histone modifications regulates several steps in transcription and affects cellular life span.
REFERENCES
- 1.Grewal SI, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 2.Cheng TH, Gartenberg MR. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev 14:452–463. [PMC free article] [PubMed] [Google Scholar]
- 3.Talbert PB, Henikoff S. 2006. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Lawry ST, Cohen AL, Jia S. 2014. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci 71:4841–4852. doi: 10.1007/s00018-014-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grummt I, Langst G. 2013. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.O'Sullivan JM, Sontam DM, Grierson R, Jones B. 2009. Repeated elements coordinate the spatial organization of the yeast genome. Yeast 26:125–138. doi: 10.1002/yea.1657. [DOI] [PubMed] [Google Scholar]
- 8.Mekhail K, Moazed D. 2010. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson AS, Warburton D, Atwood KC. 1973. Ribosomal DNA connectives between human acrocentric chromosomes. Nature 245:95–97. doi: 10.1038/245095b0. [DOI] [PubMed] [Google Scholar]
- 10.Dev VG, Tantravahi R, Miller DA, Miller OJ. 1977. Nucleolus organizers in Mus musculus subspecies and in the RAG mouse cell line. Genetics 86:389–398. [PMC free article] [PubMed] [Google Scholar]
- 11.Copenhaver GP, Pikaard CS. 1996. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J 9:273–282. doi: 10.1046/j.1365-313X.1996.09020273.x. [DOI] [PubMed] [Google Scholar]
- 12.Saez-Vasquez J, Gadal O. 2010. Genome organization and function: a view from yeast and Arabidopsis. Mol Plant 3:678–690. doi: 10.1093/mp/ssq034. [DOI] [PubMed] [Google Scholar]
- 13.Dvorackova M, Fojtova M, Fajkus J. 2015. Chromatin dynamics of plant telomeres and ribosomal genes. Plant J 83:18–37. doi: 10.1111/tpj.12822. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T, Ganley AR. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 15.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T. 2014. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc Jpn Acad Ser B Phys Biol Sci 90:119–129. doi: 10.2183/pjab.90.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12:3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang BD, Butylin P, Strunnikov A. 2006. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ide S, Miyazaki T, Maki H, Kobayashi T. 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 20.Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033–1042. doi: 10.1016/S0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 21.Ha CW, Kim K, Chang YJ, Kim B, Huh WK. 2014. The beta-1,3-glucanosyltransferase Gas1 regulates Sir2-mediated rDNA stability in Saccharomyces cerevisiae. Nucleic Acids Res 42:8486–8499. doi: 10.1093/nar/gku570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johzuka K, Horiuchi T. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 23.Benguria A, Hernandez P, Krimer DB, Schvartzman JB. 2003. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res 31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Moazed D. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 26.Ryu HY, Ahn S. 2014. Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic rDNA condensation. BMC Biol 12:75. doi: 10.1186/s12915-014-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Moazed D. 2006. Sister chromatid cohesion in silent chromatin: each sister to her own ring. Genes Dev 20:132–137. doi: 10.1101/gad.1398106. [DOI] [PubMed] [Google Scholar]
- 29.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245–256. doi: 10.1016/S0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 30.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233–244. doi: 10.1016/S0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 31.Visintin R, Hwang ES, Amon A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 32.Shou W, Sakamoto KM, Keener J, Morimoto KW, Traverso EE, Azzam R, Hoppe GJ, Feldman R, DeModena J, Moazed D. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol Cell 8:45–55. doi: 10.1016/S1097-2765(01)00291-X. [DOI] [PubMed] [Google Scholar]
- 33.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3:447–455. doi: 10.1016/S1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117:441–453. doi: 10.1016/S0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 35.Burkhalter MD, Sogo JM. 2004. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Mehta GD, Rizvi SM, Ghosh SK. 2012. Cohesin: a guardian of genome integrity. Biochim Biophys Acta 1823:1324–1342. doi: 10.1016/j.bbamcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Salvi JS, Chan JN, Pettigrew C, Liu TT, Wu JD, Mekhail K. 2013. Enforcement of a lifespan-sustaining distribution of Sir2 between telomeres, mating-type loci, and rDNA repeats by Rif1. Aging Cell 12:67–75. doi: 10.1111/acel.12020. [DOI] [PubMed] [Google Scholar]
- 38.Hirano T. 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neurohr G, Naegeli A, Titos I, Theler D, Greber B, Diez J, Gabaldon T, Mendoza M, Barral Y. 2011. A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science 332:465–468. doi: 10.1126/science.1201578. [DOI] [PubMed] [Google Scholar]
- 40.Peng JC, Karpen GH. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santoro R. 2005. The silence of the ribosomal RNA genes. Cell Mol Life Sci 62:2067–2079. doi: 10.1007/s00018-005-5110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 44.Santoro R, Li J, Grummt I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Langst G, Grummt I. 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J 25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoro R, Grummt I. 2005. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Santoro R, Koberna K, Grummt I. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Santoro R, Grummt I. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strohner R, Nemeth A, Nightingale KP, Grummt I, Becker PB, Langst G. 2004. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol 24:1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Grummt I. 2005. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol 15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. 2009. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol 11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 52.Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 53.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Yang L, Song T, Chen L, Kabra N, Zheng H, Koomen J, Seto E, Chen J. 2013. Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol Cell Biol 33:3835–3848. doi: 10.1128/MCB.00476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen HF, Wade PA, Kutateladze TG. 2013. The NuRD architecture. Cell Mol Life Sci 70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. 2012. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A 109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling T, Xie W, Luo M, Shen M, Zhu Q, Zong L, Zhou T, Gu J, Lu Z, Zhang F, Tao W. 2013. CHD4/NuRD maintains demethylation state of rDNA promoters through inhibiting the expression of the rDNA methyltransferase recruiter TIP5. Biochem Biophys Res Commun 437:101–107. doi: 10.1016/j.bbrc.2013.06.045. [DOI] [PubMed] [Google Scholar]
- 58.McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 59.Bird A, Taggart M, Macleod D. 1981. Loss of rDNA methylation accompanies the onset of ribosomal gene activity in early development of X. laevis. Cell 26:381–390. doi: 10.1016/0092-8674(81)90207-5. [DOI] [PubMed] [Google Scholar]
- 60.Santoro R, Grummt I. 2001. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8:719–725. doi: 10.1016/S1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 61.Stancheva I, Lucchini R, Koller T, Sogo JM. 1997. Chromatin structure and methylation of rat rRNA genes studied by formaldehyde fixation and psoralen cross-linking. Nucleic Acids Res 25:1727–1735. doi: 10.1093/nar/25.9.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T. 2009. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol Cell 35:414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Mayer C, Neubert M, Grummt I. 2008. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitz KM, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, Shibata D, Kakimoto T. 2011. The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol 52:618–628. doi: 10.1093/pcp/pcr022. [DOI] [PubMed] [Google Scholar]
- 66.Ahringer J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16:351–356. doi: 10.1016/S0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 67.Wang C, Gao F, Wu J, Dai J, Wei C, Li Y. 2010. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51:1291–1299. doi: 10.1093/pcp/pcq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pikaard CS. 2014. Nucleolar dominance. eLS doi: 10.1002/9780470015902.a0005976.pub2. [DOI] [Google Scholar]
- 69.Reeder RH. 1985. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol 101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McStay B. 2006. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev 20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 71.Tucker S, Vitins A, Pikaard CS. 2010. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol 22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan SWL, Henderson IR, Jacobsen SE. 2005. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 74.Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. 2010. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, Pikaard CS. 2008. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32:673–684. doi: 10.1016/j.molcel.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandrasekhara C, Mohannath G, Blevins T, Pontvianne F, Pikaard CS. 2016. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev 30:177–190. doi: 10.1101/gad.273755.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. 2007. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell 27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 78.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. 2010. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J 29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 80.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. 2014. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 81.Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I. 2010. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harbor Symp Quant Biol 75:357–364. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 82.Majumder S, Alinari L, Roy S, Miller T, Datta J, Sif S, Baiocchi R, Jacob ST. 2010. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J Cell Biochem 109:553–563. doi: 10.1002/jcb.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y, Zhang Y, Tsuneoka M. 2010. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J 29:1510–1522. doi: 10.1038/emboj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. 2007. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 85.Bartova E, Stixova L, Galiova G, Harnicarova Horakova A, Legartova S, Kozubek S. 2011. Mutant genetic background affects the functional rearrangement and kinetic properties of JMJD2b histone demethylase. J Mol Biol 405:679–695. doi: 10.1016/j.jmb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 86.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. 2010. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol 17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 87.Zhu Z, Wang Y, Li X, Wang Y, Xu L, Wang X, Sun T, Dong X, Chen L, Mao H, Yu Y, Li J, Chen PA, Chen CD. 2010. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res 20:794–801. doi: 10.1038/cr.2010.75. [DOI] [PubMed] [Google Scholar]
- 88.Chang CS, Pillus L. 2009. Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics 183:149–160. doi: 10.1534/genetics.109.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke AS, Samal E, Pillus L. 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell 17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A, Kirmizis A. 2013. N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet 9:e1003805. doi: 10.1371/journal.pgen.1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buck SW, Sandmeier JJ, Smith JS. 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111:1003–1014. doi: 10.1016/S0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- 92.Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith JS, Caputo E, Boeke JD. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol 19:3184–3197. doi: 10.1128/MCB.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun ZW, Hampsey M. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152:921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci U S A 93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koiwai K, Noma S, Takahashi Y, Hayano T, Maezawa S, Kouda K, Matsumoto T, Suzuki M, Furuichi M, Koiwai O. 2011. TdIF2 is a nucleolar protein that promotes rRNA gene promoter activity. Genes Cells 16:748–764. doi: 10.1111/j.1365-2443.2011.01524.x. [DOI] [PubMed] [Google Scholar]
- 97.Sapountzi V, Cote J. 2011. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci 68:1147–1156. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McDonel P, Costello I, Hendrich B. 2009. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, Moreno V, Juarranz A, Stockert JC, Langst G, Grummt I, Bickmore W, Esteller M. 2007. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res 35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weake VM, Workman JL. 2008. Histone ubiquitination: triggering gene activity. Mol Cell 29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 101.Huang H, Kahana A, Gottschling DE, Prakash L, Liebman SW. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol Cell Biol 17:6693–6699. doi: 10.1128/MCB.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun ZW, Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 103.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell 17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 104.Calzari L, Orlandi I, Alberghina L, Vai M. 2006. The histone deubiquitinating enzyme Ubp10 is involved in rDNA locus control in Saccharomyces cerevisiae by affecting Sir2p association. Genetics 174:2249–2254. doi: 10.1534/genetics.106.063099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rhie BH, Song YH, Ryu HY, Ahn SH. 2013. Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochem Biophys Res Commun 439:570–575. doi: 10.1016/j.bbrc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 106.Oling D, Masoom R, Kvint K. 2014. Loss of Ubp3 increases silencing, decreases unequal recombination in rDNA, and shortens the replicative life span in Saccharomyces cerevisiae. Mol Biol Cell 25:1916–1924. doi: 10.1091/mbc.E13-10-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 108.Qin W, Wolf P, Liu N, Link S, Smets M, Mastra FL, Forne I, Pichler G, Horl D, Fellinger K, Spada F, Bonapace IM, Imhof A, Harz H, Leonhardt H. 2015. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res 25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boamah EK, Kotova E, Garabedian M, Jarnik M, Tulin AV. 2012. Poly(ADP-ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet 8:e1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. 2012. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell 45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 112.Akhtar A, Gasser SM. 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 113.Mekhail K, Seebacher J, Gygi SP, Moazed D. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan JN, Poon BP, Salvi J, Olsen JB, Emili A, Mekhail K. 2011. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell 20:867–879. doi: 10.1016/j.devcel.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 115.Moazed D. 2001. Common themes in mechanisms of gene silencing. Mol Cell 8:489–498. doi: 10.1016/S1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 116.King MC, Lusk CP, Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- 117.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature 454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 118.Jeppsson K, Kanno T, Shirahige K, Sjogren C. 2014. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol 15:601–614. doi: 10.1038/nrm3857. [DOI] [PubMed] [Google Scholar]
- 119.Sjogren C, Nasmyth K. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol 11:991–995. doi: 10.1016/S0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 120.Sullivan M, Higuchi T, Katis VL, Uhlmann F. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117:471–482. doi: 10.1016/S0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 121.Wu CS, Chen YF, Gartenberg MR. 2011. Targeted sister chromatid cohesion by Sir2. PLoS Genet 7:e1002000. doi: 10.1371/journal.pgen.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, Li H, Peak A, Ramachandran S, Seidel C, Gerton JL. 2012. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet 8:e1002749. doi: 10.1371/journal.pgen.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rudra S, Skibbens RV. 2013. Cohesin codes—interpreting chromatin architecture and the many facets of cohesin function. J Cell Sci 126:31–41. doi: 10.1242/jcs.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Freeman L, Aragon-Alcaide L, Strunnikov A. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhalla N, Biggins S, Murray AW. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell 13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J 16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Machin F, Paschos K, Jarmuz A, Torres-Rosell J, Pade C, Aragon L. 2004. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol 14:125–130. doi: 10.1016/S0960-9822(04)00002-8. [DOI] [PubMed] [Google Scholar]
- 128.Johzuka K, Horiuchi T. 2009. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell 34:26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 129.Coelho PA, Queiroz-Machado J, Sunkel CE. 2003. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J Cell Sci 116:4763–4776. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- 130.D'Ambrosio C, Kelly G, Shirahige K, Uhlmann F. 2008. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol 18:1084–1089. doi: 10.1016/j.cub.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 131.Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet 11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lavoie BD, Hogan E, Koshland D. 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev 18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF. 2013. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cabello OA, Eliseeva E, He WG, Youssoufian H, Plon SE, Brinkley BR, Belmont JW. 2001. Cell cycle-dependent expression and nucleolar localization of hCAP-H. Mol Biol Cell 12:3527–3537. doi: 10.1091/mbc.12.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uzbekov R, Timirbulatova E, Watrin E, Cubizolles F, Ogereau D, Gulak P, Legagneux V, Polyakov VJ, Le Guellec K, Kireev I. 2003. Nucleolar association of pEg7 and XCAP-E, two members of Xenopus laevis condensin complex in interphase cells. J Cell Sci 116:1667–1678. doi: 10.1242/jcs.00311. [DOI] [PubMed] [Google Scholar]
- 136.Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109–121. doi: 10.1016/S0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 137.Huang K, Jia J, Wu C, Yao M, Li M, Jin J, Jiang C, Cai Y, Pei D, Pan G, Yao H. 2013. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem 288:26067–26077. doi: 10.1074/jbc.M113.486175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Samoshkin A, Dulev S, Loukinov D, Rosenfeld JA, Strunnikov AV. 2012. Condensin dysfunction in human cells induces nonrandom chromosomal breaks in anaphase, with distinct patterns for both unique and repeated genomic regions. Chromosoma 121:191–199. doi: 10.1007/s00412-011-0353-6. [DOI] [PubMed] [Google Scholar]
- 139.Lipp JJ, Hirota T, Poser I, Peters JM. 2007. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J Cell Sci 120:1245–1255. doi: 10.1242/jcs.03425. [DOI] [PubMed] [Google Scholar]
- 140.Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. 2008. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol Cell Biol 28:1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, Dalgaard JZ, Aragon L. 2005. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol 7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- 142.Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, Strunnikov A. 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4:e1000215. doi: 10.1371/journal.pgen.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu N, Kong X, Ji Z, Zeng W, Potts PR, Yokomori K, Yu H. 2012. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev 26:1473–1485. doi: 10.1101/gad.193615.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gallego-Paez LM, Tanaka H, Bando M, Takahashi M, Nozaki N, Nakato R, Shirahige K, Hirota T. 2014. Smc5/6-mediated regulation of replication progression contributes to chromosome assembly during mitosis in human cells. Mol Biol Cell 25:302–317. doi: 10.1091/mbc.E13-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaeberlein M. 2010. Lessons on longevity from budding yeast. Nature 464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Steinkraus KA, Kaeberlein M, Kennedy BK. 2008. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol 24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kennedy BK, Smith ED, Kaeberlein M. 2005. The enigmatic role of Sir2 in aging. Cell 123:548–550. doi: 10.1016/j.cell.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 148.Kaeberlein M, Powers RW III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 149.Guarente L, Picard F. 2005. Calorie restriction—the SIR2 connection. Cell 120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 150.Sinclair DA. 2005. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev 126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 151.Ganley ARD, Ide S, Saka K, Kobayashi T. 2009. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell 35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 152.Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A. 2013. A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet 9:e1003329. doi: 10.1371/journal.pgen.1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gottlieb S, Esposito RE. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 154.Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 156.Ha CW, Huh WK. 2011. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res 39:1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McClure JM, Wierman MB, Maqani N, Smith JS. 2012. Isonicotinamide enhances Sir2 protein-mediated silencing and longevity in yeast by raising intracellular NAD+ concentration. J Biol Chem 287:20957–20966. doi: 10.1074/jbc.M112.367524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 159.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem 277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 160.Kim S, Benguria A, Lai CY, Jazwinski SM. 1999. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell 10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. 2002. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol 37:1023–1030. doi: 10.1016/S0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- 162.Whitaker R, Faulkner S, Miyokawa R, Burhenn L, Henriksen M, Wood JG, Helfand SL. 2013. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging (Albany NY) 5:682–691. doi: 10.18632/aging.100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tissenbaum HA, Guarente L. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 164.Sasaki T, Maier B, Bartke A, Scrable H. 2006. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell 5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 165.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. 2013. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab 18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. 2010. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gaglio D, D'Alfonso A, Camilloni G. 2013. Functional complementation of sir2Delta yeast mutation by the human orthologous gene SIRT1. PLoS One 8:e83114. doi: 10.1371/journal.pone.0083114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gonzalo S. 2010. Epigenetic alterations in aging. J Appl Physiol 109:586–597. doi: 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Benayoun BA, Pollina EA, Brunet A. 2015. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol 16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ryu HY, Rhie BH, Ahn SH. 6 March 2014. Loss of the Set2 histone methyltransferase increases cellular lifespan in yeast cells. Biochem Biophys Res Commun doi: 10.1016/j.bbrc.2014.02.061. [DOI] [PubMed] [Google Scholar]
- 171.Walter D, Matter A, Fahrenkrog B. 2014. Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. PLoS Genet 10:e1004095. doi: 10.1371/journal.pgen.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Walter D, Matter A, Fahrenkrog B. 2010. Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J Cell Sci 123:1931–1939. doi: 10.1242/jcs.065938. [DOI] [PubMed] [Google Scholar]
- 173.McCormick MA, Mason AG, Guyenet SJ, Dang W, Garza RM, Ting MK, Moller RM, Berger SL, Kaeberlein M, Pillus L, La Spada AR, Kennedy BK. 2014. The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep 8:477–486. doi: 10.1016/j.celrep.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Johnson R, Strehler BL. 1972. Loss of genes coding for ribosomal RNA in ageing brain cells. Nature 240:412–414. doi: 10.1038/240412a0. [DOI] [PubMed] [Google Scholar]
- 175.Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Burkle A, Caiafa P. 20 February 2015. Reconfiguration of DNA methylation in aging. Mech Ageing Dev doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 176.Swisshelm K, Disteche CM, Thorvaldsen J, Nelson A, Salk D. 1990. Age-related increase in methylation of ribosomal genes and inactivation of chromosome-specific rRNA gene clusters in mouse. Mutat Res 237:131–146. doi: 10.1016/0921-8734(90)90019-N. [DOI] [PubMed] [Google Scholar]
- 177.Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. 2003. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci U S A 100:1775–1780. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Powell MA, Mutch DG, Rader JS, Herzog TJ, Huang TH, Goodfellow PJ. 2002. Ribosomal DNA methylation in patients with endometrial carcinoma: an independent prognostic marker. Cancer 94:2941–2952. doi: 10.1002/cncr.10559. [DOI] [PubMed] [Google Scholar]
- 179.Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. 2004. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279:6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Holmberg Olausson K, Nister M, Lindstrom MS. 2014. Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription. J Biol Chem 289:34601–34619. doi: 10.1074/jbc.M114.569244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Majumder S, Ghoshal K, Datta J, Smith DS, Bai S, Jacob ST. 2006. Role of DNA methyltransferases in regulation of human ribosomal RNA gene transcription. J Biol Chem 281:22062–22072. doi: 10.1074/jbc.M601155200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Bacalini MG, Pacilli A, Giuliani C, Penzo M, Trere D, Pirazzini C, Salvioli S, Franceschi C, Montanaro L, Garagnani P. 2014. The nucleolar size is associated to the methylation status of ribosomal DNA in breast carcinomas. BMC Cancer 14:361. doi: 10.1186/1471-2407-14-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chan MW, Wei SH, Wen P, Wang Z, Matei DE, Liu JC, Liyanarachchi S, Brown R, Nephew KP, Yan PS, Huang TH. 2005. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin Cancer Res 11:7376–7383. doi: 10.1158/1078-0432.CCR-05-1100. [DOI] [PubMed] [Google Scholar]
- 184.Hsu MM, Huang PY, Lee YC, Fang YC, Chan MW, Lee CI. 2014. FT-IR microspectrometry reveals the variation of membrane polarizability due to epigenomic effect on epithelial ovarian cancer. Int J Mol Sci 15:17963–17973. doi: 10.3390/ijms151017963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shi G, Wu M, Fang L, Yu F, Cheng S, Li J, Du JX, Wong J. 2014. PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1. J Biol Chem 289:29691–29700. doi: 10.1074/jbc.M114.571653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Halkidou K, Logan IR, Cook S, Neal DE, Robson CN. 2004. Putative involvement of the histone acetyltransferase Tip60 in ribosomal gene transcription. Nucleic Acids Res 32:1654–1665. doi: 10.1093/nar/gkh296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. 2008. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. 2008. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yuan J, Minter-Dykhouse K, Lou Z. 2009. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol 185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ali SA, Dobson JR, Lian JB, Stein JL, van Wijnen AJ, Zaidi SK, Stein GS. 2012. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J Cell Sci 125:2732–2739. doi: 10.1242/jcs.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ding Q, Markesbery WR, Cecarini V, Keller JN. 2006. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem Res 31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 192.Pietrzak M, Rempala G, Nelson PT, Zheng JJ, Hetman M. 2011. Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS One 6:e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. 2008. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One 3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Raval A, Sridhar KJ, Patel S, Turnbull BB, Greenberg PL, Mitchell BS. 2012. Reduced rRNA expression and increased rDNA promoter methylation in CD34+ cells of patients with myelodysplastic syndromes. Blood 120:4812–4818. doi: 10.1182/blood-2012-04-423111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. 2010. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 197.Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, Tourmente S, Pikaard CS, Saez-Vasquez J. 2010. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6:e1001225. doi: 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Doelling JH, Pikaard CS. 1995. The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J 8:683–692. doi: 10.1046/j.1365-313X.1995.08050683.x. [DOI] [PubMed] [Google Scholar]
- 199.Reid RJ, Rothstein R. 2004. Stay close to your sister. Mol Cell 14:418–420. doi: 10.1016/S1097-2765(04)00266-7. [DOI] [PubMed] [Google Scholar]
- 200.Cesarini E, Mariotti FR, Cioci F, Camilloni G. 2010. RNA polymerase I transcription silences noncoding RNAs at the ribosomal DNA locus in Saccharomyces cerevisiae. Eukaryot Cell 9:325–335. doi: 10.1128/EC.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gonzalez IL, Sylvester JE. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320–328. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- 202.Grozdanov P, Georgiev O, Karagyozov L. 2003. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 82:637–643. doi: 10.1016/S0888-7543(03)00199-X. [DOI] [PubMed] [Google Scholar]
- 203.Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Ohrlein A. 1986. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 45:837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- 204.Kobayashi T. 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Santoro R, Schmitz KM, Sandoval J, Grummt I. 2010. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11:52–58. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lu JY, Lin YY, Sheu JC, Wu JT, Lee FJ, Chen Y, Lin MI, Chiang FT, Tai TY, Berger SL, Zhao Y, Tsai KS, Zhu H, Chuang LM, Boeke JD. 2011. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146:969–979. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. 2005. Sir2 blocks extreme life-span extension. Cell 123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 208.Fingerman IM, Wu CL, Wilson BD, Briggs SD. 2005. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem 280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Song YH, Ahn SH. 2010. A Bre1-associated protein, large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J Biol Chem 285:2361–2367. doi: 10.1074/jbc.M109.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hsu HE, Liu TN, Yeh CS, Chang TH, Lo YC, Kao CF. 2015. Feedback control of Snf1 protein and its phosphorylation is necessary for adaptation to environmental stress. J Biol Chem 290:16786–16796. doi: 10.1074/jbc.M115.639443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Picazo C, Orozco H, Matallana E, Aranda A. 2015. Interplay among Gcn5, Sch9 and mitochondria during chronological aging of wine yeast is dependent on growth conditions. PLoS One 10:e0117267. doi: 10.1371/journal.pone.0117267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kimura A, Horikoshi M. 1998. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells 3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 215.Zentner GE, Balow SA, Scacheri PC. 2014. Genomic characterization of the mouse ribosomal DNA locus. G3 (Bethesda) 4:243–254. doi: 10.1534/g3.113.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Khan A, Giri S, Wang Y, Chakraborty A, Ghosh AK, Anantharaman A, Aggarwal V, Sathyan KM, Ha T, Prasanth KV, Prasanth SG. 2015. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc Natl Acad Sci U S A 112:8338–8343. doi: 10.1073/pnas.1424705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Penzo M, Casoli L, Pollutri D, Sicuro L, Ceccarelli C, Santini D, Taffurelli M, Govoni M, Brina D, Trere D, Montanaro L. 2015. JHDM1B expression regulates ribosome biogenesis and cancer cell growth in a p53 dependent manner. Int J Cancer 136:E272–E281. doi: 10.1002/ijc.29240. [DOI] [PubMed] [Google Scholar]
- 218.Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang H, Kang W, Li X, Tao Q, Sung JJ, Yu J. 2013. Zinc-finger protein 545 is a novel tumour suppressor that acts by inhibiting ribosomal RNA transcription in gastric cancer. Gut 62:833–841. doi: 10.1136/gutjnl-2011-301776. [DOI] [PubMed] [Google Scholar]
- 219.Zhang C, Comai L, Johnson DL. 2005. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol 25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. 2006. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 25:7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- 221.Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. 2008. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J Cell Sci 121:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]