SUMMARY
Bacteria have a range of distinct immune strategies that provide protection against bacteriophage (phage) infections. While much has been learned about the mechanism of action of these defense strategies, it is less clear why such diversity in defense strategies has evolved. In this review, we discuss the short- and long-term costs and benefits of the different resistance strategies and, hence, the ecological conditions that are likely to favor the different strategies alone and in combination. Finally, we discuss some of the broader consequences, beyond resistance to phage and other genetic elements, resulting from the operation of different immune strategies.
INTRODUCTION
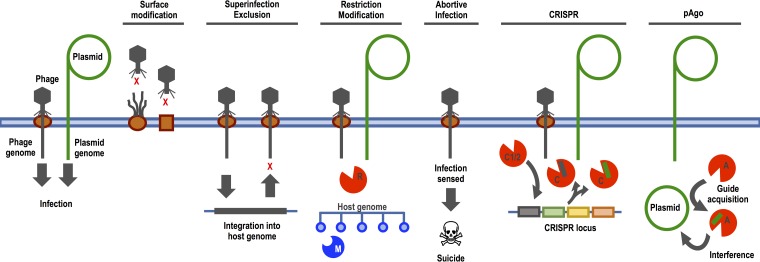
Bacteria are under constant threat by viruses (bacteriophages [phages]) and have evolved multiple immune strategies to combat phage infections (Fig. 1). Phage immune mechanisms can act at different stages of the phage life cycle, including receptor binding, genome injection into the host cell, and intracellular genome replication. First, immunity by surface modification results from masking, mutation, or loss of host receptor proteins, which serve as entry points for the phage. Second, the host can block phage DNA injection or phage replication, known as superinfection exclusion (Sie). Third, injected phage genomes can be cleaved by at least three distinct intracellular immune mechanisms: restriction-modification (RM), the CRISPR-Cas (clustered regularly interspaced short palindromic repeat–CRISPR-associated gene) system, and prokaryotic Argonaute (pAgo). Finally, bacteria can trigger a suicide reaction upon phage infection, known as abortive infection (Abi), resulting in infection failure. The molecular mechanism of these immune strategies has been the subject of several excellent reviews (see references 1–3) and are reiterated below. Below, we discuss how ecological selection pressures have driven the evolution of diverse bacterial immune mechanisms and the broader ecological and evolutionary consequences of their existence beyond resistance to viruses and other mobile genetic elements. In this review, we use the terms resistance, defense, and immunity interchangeably.
FIG 1.
Bacterial immune mechanisms. Letters indicate protein components involved in the immune mechanism (M, methylase; R, restriction enzyme; C1/2, Cas1 and Cas2; C, Cas effector-nuclease complex; A, prokaryotic Argonaute enzyme).
DIFFERENT RESISTANCE MECHANISMS OF BACTERIA
Surface Modification
The initial step of phage infection is the adsorption of a phage ligand (usually tail protein) to specific host surface receptors. To prevent phage adsorption, bacteria can either (i) lose the receptor or downregulate its expression, (ii) mutate the receptor, or (iii) block or mask the receptor.
Receptor loss is commonly seen under laboratory conditions, especially when phages use motility organelles, such as the flagellum or the pilus, to enter the host cell (4, 5) or receptors that are not essential under laboratory conditions, such as the LamB receptor, which is necessary for maltose uptake and hence is dispensable when bacteria are grown in LB medium (6). The loss of these organelles leads to complete resistance but may be associated with a large fitness cost in a natural environment (see below).
Unlike the other defense strategies described below, many examples of resistance mediated by surface modification are the result of mutation and selection rather than an inherent system that confers or predisposes bacteria toward resistance. However, there are examples where bacteria can temporarily downregulate the expression of phage receptors, which may have evolved in response to phage-imposed selection. Some bacteria alter surface receptors through “phase variation,” a process in which bacteria vary protein expression depending on environmental conditions (2, 7). By doing so, surface receptors are produced only under specific conditions or upon a specific stimulus so as to limit the risk of phage infection. In addition, some bacteria use quorum sensing to regulate phage receptor expression (8, 9). For example, Escherichia coli can reduce the expression of LamB, the receptor for phage λ, in response to quorum sensing signals (8). A recent study on the fish pathogen Vibrio anguillarum and its phage, KVP40, showed that quorum sensing is used by bacteria to alternate between two different phage protection mechanisms. At low population densities, Vibrio shows high-level expression of OmpK, the main receptor for phage KVP40, but is protected against phage infection by increased biofilm formation. As population densities increase, however, OmpK expression is downregulated through quorum sensing regulation, which renders Vibrio almost fully resistant to the phage (9).
Where receptor loss is too costly, receptor mutation, blocking, or masking may be favored by selection. Evidence for frequent receptor mutation follows from increases in the numbers of nonsynonymous mutations in phage receptors (10) and adaptive receptor mutations in response to phage in natural populations (11); a study on the human pathogen Vibrio cholerae showed that in response to phage, bacteria acquired point mutations in the outer membrane porin OmpU, the receptor for the lytic phage ICP2 (11). Many phages of Gram-negative bacteria use outer membrane lipopolysaccharides (LPSs) to enter the host cell, which are macromolecules consisting of a lipid and a polysaccharide group. Modification of LPS as a response to phage infection has been observed for a range of bacterial species, including E. coli (12) and Pseudomonas fluorescens (13).
Receptor blocking or masking occurs when bacterial molecules interfere with phage adsorption. For example, Staphylococcus aureus produces a molecule (protein A) that likely prevents phage adsorption by masking of the phage receptor (14). E. coli uses the lipoprotein TraT, encoded by the F plasmid, to modify the conformation of the OmpA protein, which is a common receptor for E. coli phages (15). The production of extracellular matrices can provide protection against phages when they form a physical barrier between the phage and the receptor. A well-known example is the production of alginate by pseudomonads. This polysaccharide causes a mucoid colony morphology that is associated with phage resistance (16).
Superinfection Exclusion
Superinfection exclusion (Sie) prevents the entry or replication of phage DNA, but as its name suggests, it is a resistance mechanism encoded by the infecting phage. Sie is commonly achieved by interfering with the injection or replication of a superinfecting phage through alterations to the cell surface or repression of replication, both of which are discussed below (reviewed in reference 1).
Relatively well-studied Sie systems include the cor gene, expressed by E. coli prophages F80 and N15 (17). This gene blocks DNA injection of related superinfecting phages by inactivating the ferrichrome uptake protein FhuA, which serves as a phage receptor (18). Another Sie system is the gp15 gene carried by the temperate E. coli phage HK97, which blocks the DNA entry of the lytic phage HK97 and the closely related phage HK95, presumably by the insertion of the Gp15 protein in the E. coli inner membrane, where it interacts with phage tail proteins (19). Further Sie systems include the ltp gene of the Streptococcus thermophilus temperate phage TP-J34. This gene encodes a lipoprotein that inhibits DNA release into the host cell during infection with a lytic phage, presumably by targeting the phage-encoded tape measure protein (TMP), which is necessary for channel formation to allow DNA passage into the cell (20, 21).
Pseudomonas aeruginosa prophage D3 mediates Sie through modification of the O-antigen of LPS on the host surface (so-called “seroconversion,” as this modification changes the host serotype). Many phages require the O-antigen to attach to the host cell and establish a successful infection, which is inhibited by this Sie mechanism (22). This is analogous to the mechanism of another phage-encoded protein (twitching-inhibitory protein [Tip]) that modifies the type IV pilus on the surface of P. aeruginosa. As many Pseudomonas phages require type IV pili for successful infection, this protein may represent a general Sie strategy (23, 24).
Although the majority of Sie systems appear prophage encoded, they are also carried by some lytic phages, such as the E. coli phage T4, which encodes one of the best-characterized Sie systems to date. This Sie system consists of the Immunity (Imm) and Spackle (Sp) proteins, which act independently and through distinct modes of action. The Imm protein blocks the translocation of phage DNA into the cytoplasm by altering the conformation of the DNA injection site on the host membrane. Sp is a membrane protein that inhibits the activity of T4 lysozyme that is contained in the phage tail and functions in creating holes in the peptidoglycan layer to facilitate phage DNA injection (reviewed in reference 25).
Apart from mechanisms that interfere with phage DNA injection, prophages can also confer repressor-mediated immunity. Repressor proteins silence phage genes in order to maintain cell viability during the lysogenic life cycle (26). These repressors bind specific DNA sequences located in intergenic regions on the phage genome, and phages vary with respect to the specificity of repressor-DNA interactions (27). A prophage can provide immunity to a phage that carries a repressor with the same specificity, as this will result in the blocking of the lytic cycle of the superinfecting phage. The phage may also capture repressor genes from an unrelated phage, presumably in order to confer immunity to superinfection (27).
Restriction-Modification
Restriction-modification (RM) systems are diverse and widespread immune mechanisms that function by cleaving nonself, unmodified DNA, while modified self DNA is left untouched. The majority of RM systems consist of two components: an enzyme that methylates DNA (methyltransferase [MT]) and an enzyme that cleaves unmethylated DNA (restriction endonuclease [RE]) by recognizing specific DNA sequences (restriction sites). Self-cleavage of the host genome is prevented by MT-catalyzed methylation of chromosomal restriction sites. Based on subunit composition and biochemical characteristics such as protein structure, restriction site recognition, cofactor requirements, and substrate specificity, RM systems are classified into four different types (types I to IV) (28). Type I systems encode a protein complex that contains restriction (HsdR), modification (HsdM), and specificity (HsdS) subunits. Unmodified target sequences trigger the DNA translocation activity of HsdR, while HsdS remains bound to the recognition sequence, leading to loop formation in the DNA. Cleavage occurs when two complexes collide, and the cleavage site can therefore be tens of thousands of base pairs away from the actual recognition site. The majority of the type II RM systems contain separate REs and MTs. Type II REs are typically homodimers or homotetramers that cleave DNA at or very close to their recognition site. These type II REs represent the restriction enzymes that became a crucial tool for DNA analysis and cloning (29) and are therefore by far the best-studied REs. The type III RM systems encode a protein complex that is usually a heterotrimer of the RE and MT (Res1Mod2) (30), and cleavage requires two recognition sites that are inversely oriented with respect to each other. Type IV systems are very different from the others, as they cleave only DNA sequences that have been modified (methylated, hydroxymethylated, or glucosyl-hydroxymethylated) and appear to have evolved independently. Based on the sequence homology, codon usage, and GC content of RM systems, it has been hypothesized that high levels of horizontal gene transfer (HGT) have contributed to the evolution and spread of RM systems (31–33). The effectiveness of RM systems in host protection against phage infection has been demonstrated in various studies reporting 10- to 108-fold protection against phage infection (reviewed in reference 34). Interestingly, the expression of many RM systems is phase variable (35–38), which has important evolutionary implications, which are discussed below.
CRISPR-Cas Systems
CRISPR-Cas systems are the adaptive immune systems of bacteria and archaea. Their molecular mechanism has been extensively reviewed elsewhere (39–43). Here we briefly explain the mechanism of these different variants of the system.
CRISPR-Cas systems consist of CRISPR-associated (cas) genes and CRISPR loci. cas genes encode the protein machinery that carries out the immune response. CRISPR loci consist of invader-derived sequences separated by direct repeats and provide a genetic memory of previous infections. CRISPR-Cas systems are extremely diverse and are currently classified into 2 distinct classes, 6 types, and 16 subtypes based on phylogeny, cas gene composition, and CRISPR sequences (44, 45). Despite differences, all systems rely on the same basic principle of spacer acquisition from foreign DNA followed by integration of these sequences into CRISPR loci on the host genome (adaptation). Next, CRISPR loci are transcribed, and the resulting RNA molecule is processed to generate mature CRISPR RNA (crRNA), which forms a complex with one or more Cas proteins (expression). Finally, crRNA-Cas complexes bind and cleave complementary nucleic acids (in some cases, this step involves additional Cas nucleases), resulting in host immunity (interference).
Class 1 CRISPR-Cas systems.
Class 1 systems encode multisubunit crRNA-Cas complexes and can be further subdivided into type I, III, and IV CRISPR-Cas systems. The latter awaits molecular characterization. Type I systems are characterized by the nuclease/helicase Cas3 and encode a Cascade (CRISPR-associated complex for antiviral defense)-like complex (46). Type III systems are typified by the presence of Cas10. In agreement with the phylogenetic relationships between type I and type III systems (44, 47, 48), their associated multisubunit crRNA-Cas complexes share key structural features (39, 49–51). Adaptation in type I systems requires only Cas1 and Cas2 (52), which form a heterotetrameric complex that binds to the leader end of the CRISPR array to catalyze the integration of spacers (53–57). During “expression,” the CRISPR is transcribed, followed by Cas6-mediated cleavage, to yield mature crRNA molecules (46, 58). Cas6 is a subunit of Cascade, which consists of multiple Cas proteins and a single crRNA molecule (59). During the “interference” stage, Cascade binds the genome of an infecting parasite and marks it for destruction by the Cas3 effector nuclease (60–65). The affinity of target DNA binding is strongly increased if the target sequence (protospacer) is flanked by a protospacer-adjacent motif (PAM) (60, 66, 67). As the PAM is absent from the CRISPR loci on the host genome, it serves to avoid autoimmunity problems (68). Some type III systems target both single-stranded RNA and transcriptionally active DNA (69–77). This RNA cleavage is important when DNA cleavage is delayed, for example, due to mismatches or because the target gene is expressed late during the phage life cycle (75). Type III-A systems also have a PAM-independent self/nonself discrimination mechanism, which relies on the inhibition of CRISPR interference when target sequences are flanked by CRISPRs (78), which avoids self-targeting of CRISPR loci on the host genome.
Class 2 CRISPR-Cas systems.
Although Class 2 systems are less common than class 1 systems (44, 79), they have received much more attention recently due to their application in genome editing. Class 2 systems are uniquely suited for this application since a single protein carries out all functions of the multisubunit crRNA-Cas complexes of class 1 systems. Class 2 systems can be subdivided into type II systems (44), which encode the Cas9 enzyme that is presently widely used for genome editing (80, 81); type V systems, which encode the Cpf1, C2c1, or C2c3 effector enzyme (44); and type VI systems, which encode the C2c2 effector enzyme (45). Type II systems require Cas1 and Cas2 for spacer acquisition as well as Cas9 for PAM specificity (82). The expression stage requires a trans-encoded crRNA (tracrRNA) molecule that pairs with pre-CRISPR RNA repeat sequences, which, in the presence of Cas9, triggers RNase III-mediated cleavage in the resulting stretch of double-stranded RNA (83). The tracrRNA remains bound to the processed crRNA and forms an essential component of the tracrRNA-crRNA-Cas9 effector complex (84). During interference, the effector complex binds the double-stranded target molecule (85), followed by PAM-dependent Cas9-mediated cleavage (85). Type V and VI systems do not encode Cas9, and their molecular details of expression and interference are distinct from those of type II systems. For example, Cpf1 and C2c1 lack a requirement for tracrRNA during the expression and interference stages (45, 86). Cpf1 also has different PAM requirements and yields different cleavage products (87).
pAgo
Argonaute (Ago) proteins are present in all domains of life and are key enzymes of the RNA interference (RNAi) pathway in eukaryotes (88). In eukaryotes, Ago plays a key role in a range of cellular functions, including gene regulation and host defense (88). Ago proteins are part of the PIWI (P-element-induced wimpy testis) protein superfamily. In addition to the PIWI domain, Ago proteins contain the N-terminal domain (N domain), the PAZ (PIWI-Argonaute-Zwille) domain, and the MID (middle) domain, and these domains are linked together by two linkers, L1 and L2. Eukaryotic Ago proteins interact with RNA molecules of well-defined lengths (ranging from 20 to 30 nucleotides [nt], varying between different paralogs), and these RNA molecules function as guides for Ago to bind (and sometimes cleave) cRNA sequences (RNA-guided RNA interference).
The discovery that Ago is also found in prokaryotic genomes (89, 90), together with the finding that prokaryotic Ago (pAgo) often colocalizes with other defense genes, led Makarova and coworkers to hypothesize that pAgo comprises a prokaryotic defense system (91). Prokaryotes have both long pAgo proteins, which carry the same domains as their eukaryotic counterparts, and short pAgo proteins, which consist of only the MID and PIWI domains (92). Of the long pAgo enzymes, only 28% are predicted to be catalytically active, and inactive enzymes often colocalize with other nucleases that may carry out target cleavage (92). Interestingly, many pAgo proteins preferentially bind DNA guides rather than RNA guides (93–96), although pAgo from Rhodobacter sphaeroides was found to associate with RNA guides (97). These guides are typically 15 to 19 nt long and have a well-conserved 5′ nucleotide (96, 97). Both the DNA-guided pAgo proteins from Thermus thermophilus and Pyrococcus furiosus and the RNA-guided pAgo protein from R. sphaeroides were reported to interfere with plasmids (96–98), and Ago deletion mutants resulted in increased plasmid gene expression (97) and plasmid transformation efficiencies (96, 98). In vitro analyses revealed that both the P. furiosus and T. thermophilus pAgo proteins carry out DNA-guided DNA cleavage (96, 98). DNA-guided T. thermophilus pAgo, but not P. furiosus pAgo (98), could also cleave cRNA in vitro (96), but the enzyme does not appear to interfere with mRNA in vivo (99). Like P. furiosus pAgo, Methanocaldococcus jannaschii pAgo is also unable to cleave cRNA (100). Hence, a picture emerges in which pAgo enzymes are generally (but not always) guided by short DNA molecules and typically interfere with cDNA by either pAgo-mediated cleavage or, possibly, the recruitment of additional nucleases if pAgo lacks catalytic residues. The enzymatic activity seems to be directed primarily against plasmids. Many key questions remain concerning the mechanism of pAgo, most prominently how the enzymes discriminate self from nonself and how guide DNA molecules are acquired from plasmid targets.
Abortive Infection
Abortive infection (Abi) systems cause programmed cell death of an infected bacterium, thereby preventing phage replication and, thus, phage spread to neighboring uninfected cells. As such, abortive infection is essentially an altruistic response to phage infection; the infected cell will die, but the rest of the bacterial population is likely to survive as phage spread is aborted (101). Several Gram-negative strains carry Abi systems, of which the rapid II (rII) exclusion (Rex) system found in phage λ-lysogenic E. coli strains is probably the most well-characterized one (1). Rex systems consist of two proteins, RexA and RexB, both of which are needed for phage protection. Upon infection, phage protein-DNA complexes are produced as replication intermediates that activate the intracellular sensor molecule RexA. Subsequently, RexA activates the membrane-anchored ion channel protein RexB. RexB activation causes a sudden drop in the cellular ATP level, thereby aborting ATP-dependent processes, including virus replication (102). Another Abi system is the late inhibitor of T4 (Lit) system, which is found in a defective prophage integrated into the genome of the E. coli K-12 strain. Upon activation by phage protein, Lit cleaves a translation elongation factor, thereby inhibiting protein synthesis (102, 103). This finally leads to abortion of virus infection and bacterial cell death. Abi systems are highly abundant in Gram-positive bacteria, in particular in the lactococci, where they are commonly encoded on plasmids (101, 104). To date, over 23 lactococcal Abi systems have been identified, all of which are thought to interfere with different steps of the phage replication cycle (1, 101). Recent studies have shown that certain toxin-antitoxin (TA) systems can also act as Abi systems upon activation by phage infection (104–106). For example, the plant-pathogenic bacterium Pectobacterium atrosepticum encodes the TA system ToxIN, which aborts phage infection. This mechanism functions through the action of the RNA antitoxin ToxI and the endoribonuclease toxin ToxN, whereby ToxI neutralizes ToxN under normal conditions (101, 107). However, upon phage infection, host gene expression is arrested, and as the antitoxin is less stable than the toxin, ToxI levels drop more rapidly than do ToxN levels. This causes ToxN activation, leading to the induction of cell death. Abi-inducing TA systems have also been identified in E. coli to prevent infection with phage T4 (108) and phage P1 (109). As TA systems are extremely widespread in bacteria, and only a few have been studied in detail, it is anticipated that many more TA systems that function through an Abi mechanism will be identified in the future.
SELECTIVE FORCES DRIVING THE EVOLUTION OF IMMUNE MECHANISMS
Why did bacteria evolve these different immune mechanisms? Despite the progress in unraveling the mechanism of bacterial defenses, we know relatively little of the selection pressures that drive their evolution. Studying the evolutionary ecology of immune mechanisms is important if we are to understand, predict, and manipulate bacterial adaptation to phage infection.
Is More Immunity Best?
Is it simply the case that having multiple defense mechanisms is best, analogous to vertebrate immune systems that are composed of both innate and adaptive mechanisms? Consistent with this idea, RM and CRISPR-Cas systems frequently cooccur (33), and their combination results in increased levels of immunity (110) and more rapid spacer acquisition (111). Moreover, immune mechanisms may even interact synergistically. For example, transcriptome analyses show that pAgo deletion impacts the expression levels of CRISPR-Cas components (99), suggesting that these systems may show genetic interactions, and synergistic interactions between Abi and CRISPR systems have been suggested based on data from in silico analyses (112).
However, if it is better to have multiple mechanisms, why is it that not all bacteria have all mechanisms? There are two possibilities: either not all mechanisms have evolved in all organisms yet or immune mechanisms are associated with a fitness cost that can select against the maintenance of the system. We can rule out the first explanation, since there are many examples of the presence and absence of mechanisms in closely related bacteria where immune mechanisms have been gained and lost or inactivated (see, for example, references 113–116). Fitness costs associated with immune mechanisms are therefore a more probable explanation as to why not all bacteria have all mechanisms.
Fitness Costs of Immunity
For obvious reasons, host immunity confers a clear selective advantage in the presence of parasites. However, investment in immunity is also typically associated with fitness costs and can therefore be selected against in the absence of parasites (117). Such costs of resistance appear to be very general among both prokaryotes and eukaryotes (reviewed in references 118 and 119). Fitness costs associated with immune systems can arise due to immunopathology (e.g., autoimmunity); due to an allocation of resources to defense, which would otherwise be used to increase reproductive success; or due to pleiotropic effects of resistance that decrease host fitness, as is often the case for surface modification (120).
Surface modification.
Surface modifications are typically associated with a constitutive fitness cost (i.e., a fixed cost that is independent of the presence or absence of phage). The cost of losing surface receptors, such as the flagellum or pilus, is likely to be lower in laboratory settings, where nutrients are readily available and broth is continuously mixed, than in natural environments. For example, pili and flagella are associated with key bacterial functions such as movement (swimming, swarming, and twitching) and biofilm formation (121), and the loss of these receptors can have a high competitive cost in spatially structured and resource-limited environments (122, 123). Moreover, modification of surface receptors such as LPS can also result in a reduction of fitness in some contexts (120).
Superinfection exclusion.
Fitness costs associated with Sie mechanisms have not been studied in great detail. One P. aeruginosa prophage was found to encode a Sie system that had a subtle effect on host motility and was cost-free during growth in soil and during infection of a Caenorhabditis elegans host (124). Prophage integration into the host genome may be associated with a constitutive fitness cost if host genes are disrupted (e.g., tRNA genes), but this cost may be compensated for by phage-borne tRNA genes and other accessory genes that confer a benefit to the host (125, 126).
Restriction-modification.
A recent study investigated fitness costs associated with RM and found that increased levels of SOS responses were elicited, suggestive of autoimmunity effects (127). Furthermore, the fitness cost of carrying RM systems was manifested under conditions of low but not high resource levels (127), which is often observed for parasite resistance (123, 128). In agreement with autoimmunity, bacterial genomes harboring RM systems generally have a decreased frequency of the cognate restriction site (129, 130). This phenomenon, named restriction site avoidance or palindrome avoidance, is even more pronounced in host than in phage genomes (129). Palindrome avoidance itself may also be associated with a fitness cost to the bacterium, as it could affect gene functioning through mutations. In addition, some (but not all) RM systems are extremely energy-consuming, with RE translocation over the DNA consuming 1 ATP per base pair (131). Whether ATP consumption provides a benefit (e.g., through more rapid detection or destruction of phage genomes) is unknown. Phase-variable expression of many RM systems may help to reduce the associated fitness costs of carrying these systems.
CRISPR-Cas.
The class 2 CRISPR-Cas system of S. thermophilus was found to be associated with both a constitutive cost of carrying the system as well as an inducible cost of using the system (132). An inducible cost of mounting a CRISPR immune response was also detected in P. aeruginosa (133). The inducible cost is consistent with the induced expression of CRISPR-Cas adaptive immune systems upon infection (134–136). A constitutive fitness cost of CRISPR-Cas systems could select for a loss of the systems in the absence of parasites (137). At present, the mechanistic basis for the observed fitness cost associated with the CRISPR-Cas system is unclear but may be related to autoimmunity (138–144) or allocation of resources to defense that would otherwise be invested in growth.
Abortive infection.
Abi was found to be associated with a constitutive cost of carrying the system (145), but the mechanistic basis of this cost is unknown. Moreover, there are inevitable individual costs associated with suicidal behavior following the induction of the Abi system.
Costs of Resistance Can Help To Explain Diversity in Immune Mechanisms
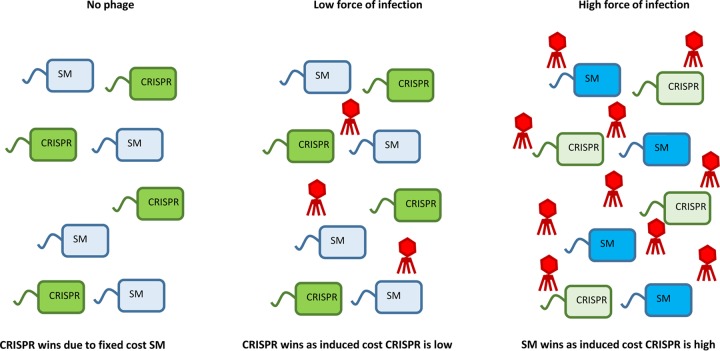
One important feature of immune mechanisms is whether they are constitutive (always active) or inducible (triggered upon infection) (Fig. 2). Inducible defenses are typically associated with an induced cost of resistance, whereas constitutive defenses are associated with a fixed cost. As a consequence, the force of infection is predicted to be a key ecological factor driving the evolution of these different immune strategies, since the overall cost of an inducible resistance strategy will depend on the frequency of infection (146). Recently, it was demonstrated that the force of infection can tip the balance from CRISPR immunity to surface modification immunity (133), and this was explained by the CRISPR-Cas system and surface modification being associated with inducible and fixed costs of resistance, respectively. Hence, depending on the risk of infection, either the CRISPR system or surface modification is favored (Fig. 3). Consistent with the idea that the CRISPR system is better when the risk of infection is low, thermophiles tend to have more and longer CRISPRs (147, 148) and have lower host and parasite population densities (149). Similar effects are expected in the context of other immune mechanisms that have inducible costs.
FIG 2.
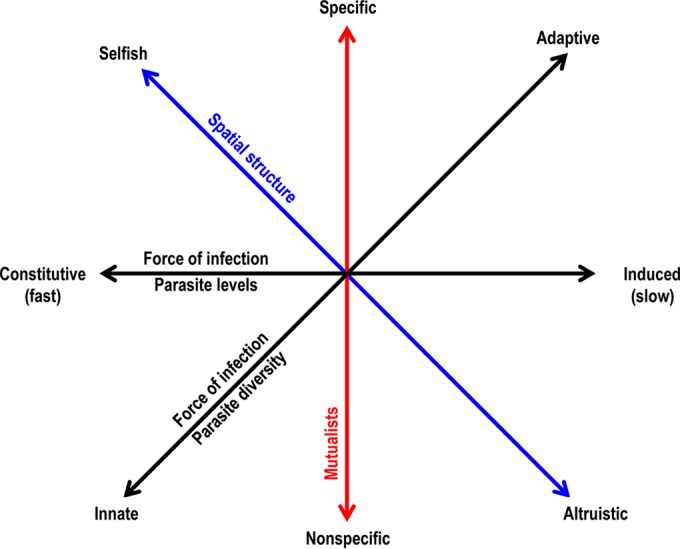
Four-dimensional space defined by the four axes that capture different features of immune mechanisms is sufficient to explain the existing diversity of immune mechanisms in nature. The ecological factors indicated drive the evolution of the feature indicated on the corresponding axis. Details are provided in the text.
FIG 3.
The force of infection is an important determinant of the relative fitness associated with CRISPRs and surface modification (SM). In the absence of phage or at a low force of infection, CRISPRs are favored over SM, since the latter is associated with a fixed cost of resistance. At high phage exposure, SM is favored over the CRISPR, because the latter is associated with an inducible cost of resistance that increases with an increasing force of infection. Empirical support for this was reported previously (133).
How Does Symbiont Diversity Contribute to Immune Diversity?
A second factor that is likely to be important in the evolution of immune strategies is the level of diversity in mobile genetic elements (here broadly referred to as symbiont diversity). Diversity of symbionts may drive diversity of host immune mechanisms in at least two ways. First, parasite diversity can drive the evolution of a division of labor, whereby different immune mechanisms coexist in the same bacterium but each one is effective against different parasites. Anecdotal evidence suggests that this may be the case. For example, surface modification is generally more effective against phages than plasmids, although mucoid phenotypes were found to confer protection against a virulent plasmid (150). The RM and CRISPR-Cas systems can target both phages and plasmids (76, 151–154), and pAgo appears to predominantly target plasmids (96), while Abi is typically induced by phages. Second, symbiont diversity is likely to also impact the evolution of stand-alone immune strategies, in particular along the innate-versus-adaptive-immunity axis and the specific-versus-nonspecific-immunity axis (Fig. 2). Common wisdom suggests that an adaptive immune system is particularly beneficial if a host is exposed to an unpredictable range of different parasites, since it allows the acquisition of immunity to all these parasites. Exposure to many parasites could also select for nonspecific immune mechanisms (Fig. 4); however, such indiscriminate, generalized immune strategies are likely to interfere with host-mutualist interactions (e.g., acquisition of plasmids that encode antibiotic resistance). Hence, the presence of mutualists may impose selection on the evolution of specific versus nonspecific defenses (Fig. 5) (see the section on mutualists, below).
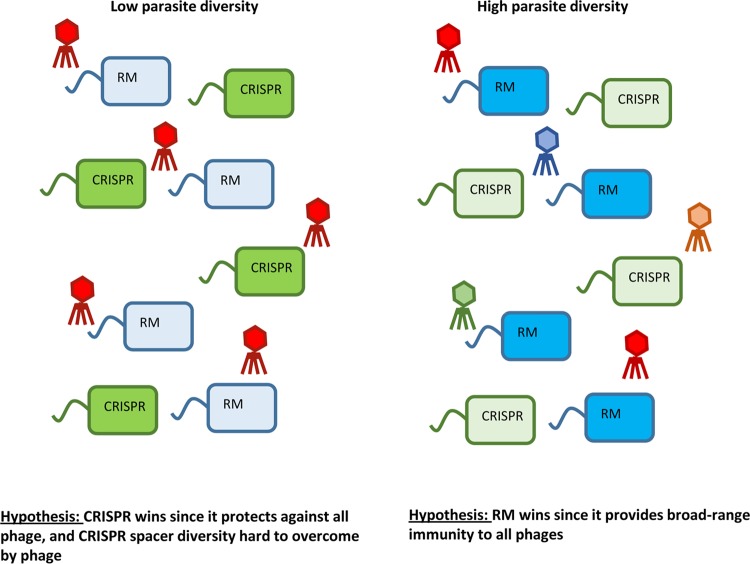
FIG 4.
Although empirical support is currently lacking, it may be the case that increasing phage diversity can select for broad-range innate immune mechanisms, such as RM, over specific adaptive immune systems, such as the CRISPR-Cas system. While the CRISPR system is extremely effective if there is low genetic variation in the phage population, theory predicts that the system becomes less effective if the host is exposed to a phage population with high levels of genetic diversity (149).
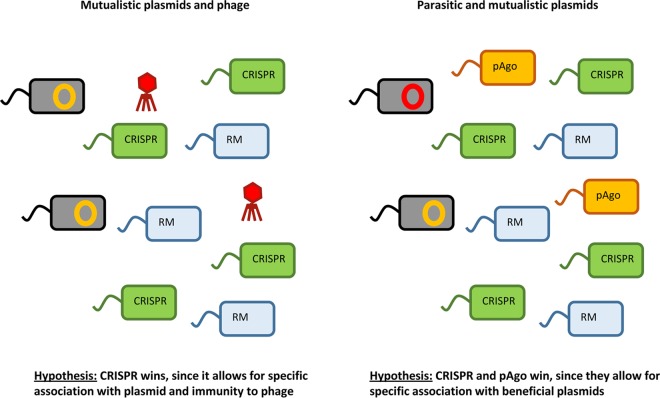
FIG 5.
Mutualists (e.g., plasmids that confer a fitness benefit to the bacterial host) can select against immune mechanisms (177). If both mutualists and parasites are present (e.g., plasmid and phage [left] or beneficial and harmful plasmids [right]), CRISPRs and pAgo may be beneficial since their specificity allows bacterial hosts to specifically acquire resistance against the parasite. Empirical data to support this idea are currently lacking.
Diverse pathogens: intraspecific diversity.
Genetic diversity in phage populations is predicted to drive the evolution of generalized defense, since phage mutants readily evolve to overcome specific defenses (137, 149). For example, it has been shown that phage can rapidly evolve to overcome CRISPR-Cas-mediated immunity by point mutation, resulting in invasion by surface mutants (155, 156). However, phage was unable to evolve infectivity in bacterial populations with high levels of CRISPR allele diversity (156). Yet, the outcome may depend on the relative levels of phage and host genetic diversities; the diversity-generating benefit of CRISPR-Cas may be more limited at increased levels of phage diversity (149), which may therefore favor more broad-range (nonspecific) defenses.
RM systems provide nonspecific immunity but are also prone to phage evolving to overcome resistance, which typically occurs through an “accidental” modification of the phage genome before cleavage by the restriction enzyme. To deal with rapid phage evolution, some RM systems also generate diversity in the specificity subunits through recombination (157, 158). As with CRISPRs, this diversity-generating property of these RM systems may limit the evolution of phage to overcome host resistance. In addition, phase-variable expression of RM systems (35–37) may also help to limit the evolution of escape phage, since infection of a bacterial clone that has the RM system switched off will result in phage progeny that are not modified and therefore subject to RM of related bacteria in the population.
Some forms of broad-range (nonspecific) defense, such as surface modification by receptor loss, seem harder to overcome by phage, but even then, phage can evolve infectivity against initially resistant hosts through the recognition of novel receptors (6). Some phages even carry mechanisms that specifically generate diversity in genes involved in host receptor recognition (159, 160). These diversity-generating retroelements (DGRs) function through a reverse transcriptase-mediated process that introduces adenine-specific substitutions in a gene that encodes a distal tail fiber protein. The DGR allows the phage to rapidly adapt to the rapidly changing host surface structures associated with phase variation (159, 160). Similar DGRs have since been discovered in a range of phages (161) and in archaeal viruses (162).
Diverse pathogens: interspecific diversity.
While intraspecific diversity may be neutralized by a sufficiently high diversity of host resistance alleles, interspecific diversity (i.e., many different viruses) is likely to select for broad-range innate immune mechanisms, such as RM, Abi, and Sie, which typically provide protection against a range of phages (124, 163) (Fig. 6). Surface modification can also provide broad-range immunity against multiple phages, but the range of immunity depends on the type of modification and the phage receptors involved; for example, a point mutation in a receptor is likely to be associated with a lower fitness cost than the complete loss of the same receptor but is also more likely to provide relatively narrow-range resistance against a single or few phage species compared to receptor loss. In agreement with this, evolution of broad-range immunity has been shown to be more costly than narrow-range immunity (164), but the mechanistic basis of immunity in these experiments was not assessed. In some cases, surface modification-based resistance against one phage can increase susceptibility to other phages, as is the case for the cyanobacterium Prochlorococcus (165).
FIG 6.
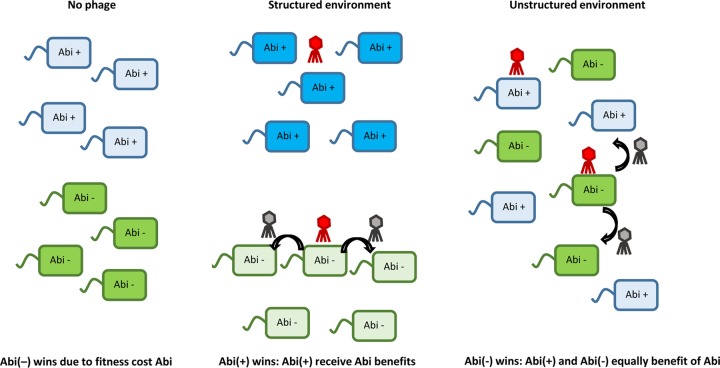
Spatial structure is an important fitness determinant for Abi, since it impacts relatedness. In the absence of phage, there will be selection against Abi because of the cost of carrying the system. In a structured environment, the benefits of Abi are directed toward related individuals that also carry the Abi gene. The phage will die out rapidly in the presence of Abi, while the phage will cause an epidemic in the absence of Abi (progeny phage is indicated in gray). In a well-mixed environment, bacteria lacking the Abi system benefit from the altruistic defense of bacteria that encode the Abi system, but they do not pay the cost. Both strains equally suffer from the epidemic that results from infection of bacteria that lack the Abi system. Empirical support for these findings was reported previously (181–183).
The benefit of the CRISPR-Cas system in the face of diverse viruses is unclear. On the one hand, an adaptive immune system allows a host to adapt to many different threats. On the other hand, the specificity of adaptive immune systems means that there is little cross-resistance. Theory and metagenomics data suggest that in the context of two different viruses, CRISPR-Cas systems may be associated with selective sweeps in host populations if a single host genotype acquires resistance against two phages that are present in the environment (166, 167). Consistent with theory (167, 168), experiments suggest that multiple phages prolong CRISPR-virus coevolution (144). Furthermore, it was found that viruses can escape CRISPRs through recombination (144, 169).
Mutualists.
Apart from inter- and intraspecific parasite diversity, the relative importance of mutualistic DNA elements is also likely to impact the relative benefits of different immune strategies. For example, accessory genes carried by plasmids can confer a fitness benefit to the host (170–173), and specific immune mechanisms, such as the CRISPR-Cas system or pAgo, which can selectively evolve immunity against parasites but not mutualists, are expected to be favored over broad-range defenses such as RM (174), which can form an important barrier for gene transfer (175) (Fig. 6). However, phase variation of RM systems may allow the uptake of beneficial DNA by bacteria in which the RM system is switched off (35). That said, there is some evidence that even the CRISPR-Cas system may restrain horizontal gene transfer. First, both in the laboratory and in nature, bacteria evolve CRISPR immunity against plasmids (76, 154), but this may be due to the fact that plasmids can act as parasites. Second, correlational studies demonstrate that the CRISPR-Cas system limits horizontal gene transfer of antibiotic resistance genes (176). It has been suggested that species can lose CRISPR-Cas systems under conditions where horizontal gene transfer is important (reviewed in reference 177). However, a recent bioinformatics analysis of >1,300 genomes failed to detect any clear effect of CRISPRs on rates of horizontal gene transfer (178).
Impact of Spatial Structure on Evolution of Immune Mechanisms
Apart from phage abundance (force of infection), phage diversity, and the relative importance of plasmids, one further ecological factor that is key for host-parasite interactions and the evolution of immune mechanisms is spatial structure (Fig. 2). Spatial structure increases the likelihood that neighboring individuals are clone mates, which in turn affects selection for different immune strategies. Most immune strategies confer an individual benefit, but Abi is a clear exception, since it protects neighboring bacteria at the expense of the individual expressing that trait. Theory suggests that altruistic behaviors (i.e., individually costly but group beneficial) such as Abi are likely to evolve when altruism preferentially benefits individuals who share the same altruism genes (179, 180). Studies using an artificially engineered suicide system in E. coli (181) and the naturally occurring E. coli Abi system Lit (145) show that spatial structuring is indeed needed for abortive infection systems to evolve, with the latter also showing that levels of mixing that are too low may prevent the evolution of abortive infection due to the absence of parasite spread (and thus epidemics) under these conditions (145). These findings were largely confirmed for the well-known E. coli Rex system (182), and this study also showed that abortive infection is likely to evolve even when genetic similarity between neighboring strains is relatively low, as long as the cost for abortive infection is also low (182). Apart from Abi, the evolution of other immune mechanisms may also be subject to spatial structure. Since spatial structure is an important factor for the evolution of lysogeny (183), it will also, indirectly, impact the evolution of superinfection exclusion, which is encoded by phages.
CONSEQUENCES OF OPERATION OF DIFFERENT MECHANISMS
While it is clear from these studies that ecology affects the evolution of immune mechanisms, it is less clear what the broader consequences of the operation of different immune mechanisms might be. As a result of the differences in the underlying genetics, different mechanisms—and their combinations—are also likely to be associated with distinct coevolutionary dynamics (184, 185). As well as affecting the extent to which bacteria are resistant to their cooccurring phage populations, different coevolutionary dynamics can have correlated effects on the evolution of important bacterial phenotypes. We first discuss what little we know about coevolution resulting from the different immune mechanisms and then discuss some of these broader consequences.
Coevolution with Different Immune Mechanisms
Surface modification.
Many laboratory studies on bacterium-phage coevolution report receptor modification-based resistance, often using B strains and T phages of the model bacterium E. coli. These studies show that coevolution in the laboratory takes place in an asymmetrical fashion; i.e., the evolutionary potentials for host and phage are different. This leads to resistance-conferring host adaptations (e.g., receptor loss or modification), which cannot be overcome by the phage on a short time scale. Loss of surface receptors is generally not associated with coevolution (186, 187), although phages could evolve to adapt to a novel receptor (1, 6). In these cases, only one or a few cycles of resistance-infectivity coevolution are typically seen, after which bacterial surface mutants emerge, which the phage is unable to infect in the short term (reviewed in reference 188). This is also seen in studies of coevolution between the cyanobacterium Plectonema boryanum and its cyanophage, LPP-1, which, after several rounds of coevolution, typically resulted in full host resistance that could not be overcome by the phage (189–191). One of the exceptions to this is the coevolution between P. fluorescens and phage ϕ2, where coevolution persists over long time spans, with bacteria modifying their receptors in response to phage infection and phage evolving to overcome bacterial resistance (192–194). By using this experimental system, it was found that bacterium-phage coevolution under high-nutrient conditions is associated with selective sweeps that lead to the fixation of bacterial resistance phenotypes (192, 195) and novel phage phenotypes that carry mutations to overcome bacterial resistance (196). This type of coevolution leads to the evolution of genotypes with increasing ranges of resistance (i.e., genotypes able to resist a wide range of phages) and infectivity (i.e., able to infect a wide range of bacterial hosts) and can be described as an arms race (197). Later studies assessing the genetics of bacterium-phage coevolution found that coevolving bacteria often have numerous mutations in the genes encoding LPS, the presumed ϕ2 receptor (13). Phages that evolved toward generalism were found to carry a number of mutations in the phage tail fiber genes (encoding proteins necessary for host receptor adsorption) (193, 194). While coevolution in nutrient-rich broth showed arms race dynamics (at least initially [196]), ecological conditions can change the dynamics. Specifically, lower nutrient levels result in fluctuations in resistance and infectivity ranges through time (198), while spatial structure results in temporal fluctuations in the frequency of different specialist resistance and infectivity genotypes (199, 200). These altered dynamics were associated with costs of increasing resistance and infectivity ranges, which started to outweigh the benefits when host-parasite encounter rates were reduced by low nutrient levels and spatial structure.
Superinfection exclusion.
Evidence that phages are able to overcome superinfection exclusion comes from a study by Bailone and Devoret, who examined the effect of the expression of the superinfection repressor protein cI from phage λ on the emergence of virulent (i.e., lytic) phages. By increasing cI expression levels in E. coli cells, phages insensitive to superinfection could be readily isolated (201). These phages had accumulated mutations in the phage λ operator (oLoR), which, when bound by cI, keeps the host cell in a lysogenic state and prevents the replication of a superinfecting phage. However, compensatory mutations in the cI gene can restore superinfection suppression (202). Based on these studies, Berngruber et al. (203) generated a theoretical model that suggests that virulence and superinfection exclusion can coevolve. Virulent phages insensitive to superinfection can infect lysogens, but their insensitivity to superinfection repression keeps them in a virulent state, which can be costly for phages. As such, selection can subsequently favor superinfection suppression through compensatory mutations in the repressor (203).
Other Sie mechanisms rely on blocking phage DNA injection (reviewed in reference 1). Mechanistic insight into how injection blocking could evolve was provided by a study by Meyer et al. (6). By using experimental evolution, this study followed the fate of phage λ that had evolved to recognize a novel receptor on the surface of its E. coli bacterial host. After shifting receptor usage (from LamB to OmpF), bacteria acquired resistance through mutations in either the manY or the manZ gene. Both of these genes encode the transmembrane channel of the ManXYZ mannose permease, which is required for λ DNA to cross the inner membrane (204–206). Thus, these mutations conferred resistance by blocking DNA transport of phage DNA, akin to some Sie mechanisms.
Restriction-modification.
In the short term (i.e., on ecological time scales), coevolution associated with RM is usually short-lived. Numerous studies have demonstrated the ability of RM systems to confer immunity against phages under laboratory conditions (reviewed in reference 34). However, even though many RM systems readily cleave unmodified phage DNA upon entry, there is a 10−2 to 10−6 probability that phage will be “accidently” modified by the host MT, which renders all host cells carrying the same RM system susceptible to that phage (207). This is likely to be a coevolutionary dead end, as a host cannot overcome this immune evasion in the short term. Hence, RM systems appear to be generally relatively unimportant for coevolution on ecological time scales compared to rapidly evolving immune responses such as surface modification or CRISPR-Cas. However, a recent theoretical study suggested that higher RM diversity may allow the stable coexistence of multiple bacterial strains due to the generation of different epigenetic variants of a phage species (due to RM-mediated modification of phage DNA) that restrict bacterial strains from dominating the population (208). Rapid evolution of the restriction site specificity of a type I RM system through DNA inversions within the HsdS-encoding gene was first described for Mycoplasma pulmonis (158), and similar mechanisms to rapidly alter type I RM specificity by DNA inversion appear to exist in Streptococcus pneumoniae (209) and Bacteroides fragilis (157, 210). This could potentially lead to RM-based bacterium-phage coevolution on ecological time scales.
Furthermore, over longer evolutionary time scales, coevolution between phages and RM systems takes place, as is evident from the extensive array of strategies that phages have evolved to avoid restriction (reviewed in references 211 and 212). Active evasion of RM systems is seen, for example, for phages that encode their own MTs (34, 213, 214). This ensures proper modification and thereby protection of the phage genome against cleavage by host REs that are compatible with that modification. Some phages have evolved the ability to modify their own DNA by adding bulky groups to it (213, 214). For example, coliphage Mu encodes a protein (Mom) that modifies adenine residues by adding an acetamide group to it, thus protecting it against cleavage by a wide range of REs (215). An intriguing example of a fierce coevolutionary arms race is seen for phage T4 and its host, E. coli K-12. Phage T4 incorporates the unusual base hydroxymethylcytosine (HMC) instead of a cytosine in its genome. As a response, hosts evolved the ability to specifically recognize and cleave this modified DNA (216), after which the phage evolved the ability to glucosylate its already modified DNA. This resulted in the evolution of RM systems that specifically recognize glucosylated DNA. In turn, phages evolved a strategy to inhibit these specialized RM systems, and recently, bacterial proteins that block these phage inhibitors were found (217). Phages can also encode proteins that mask recognition sites on the phage genome from host RE cleavage, for example, the “defense against restriction” (Dar) proteins encoded by coliphage M1 (218). Furthermore, several phages produce proteins that mimic stretches of DNA, which then bind to REs with high affinity, thereby blocking RE cleavage activity. A well-known example of such a protein is the “overcome classical restriction” (OCR) protein encoded by T7 phages (219), which binds to type I RM systems by mimicking B-form DNA to prevent cleavage activity. Similar types of mimicking proteins, named “alleviation of restriction of DNA” (Ard) proteins, are encoded by a range of plasmids, and these proteins also inhibit type I enzymes (220). Selection on phage genomes to evade restriction has led to restriction site avoidance in the phage genome (221). Furthermore, a strand bias in T7 phages ensures that all type III recognition sites are in the same orientation instead of the inverse orientation required for cleavage, which leads to immunity evasion from type III RM systems.
CRISPR-Cas.
CRISPR-Cas-mediated immunity relies on a perfect sequence match between a spacer and a virus. As a consequence, virus can overcome CRISPR-Cas-mediated resistance by a simple point mutation in the target sequence. The high specificity of the interaction between CRISPRs and escape viruses led to predictions of persistent coevolution (149, 222, 223). However, both theory and data show that CRISPR-virus coevolution can be short-lived (149, 156).
Under some conditions, phage escape mutants can be readily picked up under laboratory conditions (68, 224, 225), and metagenomics data show that escape phage can increase in abundance in vivo (226–228). These associated mutations are typically located in a confined sequence area: either in the PAM sequence or in the adjacent seed sequence (68, 225, 229). The seed sequence is a 7- to 12-nt part of the protospacer immediately adjacent to the PAM (225) and is thought to be the area where R-loop formation starts (225, 230). Single or multiple point mutations in other parts of the protospacer typically do not lead to phage escape from the immune response (225). Type III CRISPR-Cas systems appear to be generally more tolerant to mismatches, making it more difficult for phage to overcome immunity by point mutation (231, 232). A bioinformatics study showed that PAM sequences are underrepresented on phage genomes, probably as a result of CRISPR-mediated selection (233).
Under other conditions, phage cannot evolve to escape by point mutation (156). What, then, determines the ability of phage to escape CRISPR-Cas systems? Theory predicts that virus mutation rates and host spacer acquisition rates are key factors during CRISPR-virus coevolution (149). If bacterial host populations generate high levels of spacer diversity, as is the case with P. aeruginosa, the virus is driven extinct (156, 234). This is because the virus can no longer evolve infectivity against the mix of host genotypes, even though the same virus can rapidly evolve infectivity against the individual clones in monoculture (156). This shows that spacer diversity increases overall population resistance (herd immunity). The propensity to generate spacer diversity is therefore an important fitness determinant of the CRISPR-Cas system. The selective pressure imposed by diversity-generating CRISPR-Cas systems may have driven the evolution of phage-borne anti-CRISPR genes in many Pseudomonas phages (235, 236). Anti-CRISPR proteins bind CRISPR-Cas components to interfere with either target DNA recognition or DNA destruction (237). Anti-CRISPRs are encoded by an extremely diverse set of genes that are often located in a conserved locus on phage genomes (235, 236) and other mobile genetic elements (153, 236).
Interestingly, in vitro long-term bacterium-phage evolution experiments with Streptococcus thermophilus DGCC7710 and phage D2972 revealed persistent coevolution, with phage acquiring mutations in the PAM and the seed sequence and hosts acquiring novel spacers (143, 144). How are these different dynamics explained? It seems that S. thermophilus generates much less diversity (CRISPR populations are dominated by a single spacer [143, 144; our unpublished data]), which is therefore predicted to lead to ongoing coevolution (149). The lower levels of diversity may be explained the requirement for a longer PAM by S. thermophilus CRISPR-Cas systems, which reduces the putative number of protospacers by a factor of ∼10 compared to the P. aeruginosa type I-F system. An increase in the mutation fixation rate by the virus would also be expected to result in more persistent coevolution (137, 149).
Abortive infection.
Similar to restriction-modification systems, coevolution between Abi systems and phage is less important on short time scales. However, on a longer time scale, coevolution between Abi systems and phages is evident, as phages have several distinct strategies to evade Abi mechanisms (238, 239). Cell death through the action of the above-mentioned ToxIN system from P. atrosepticum can be circumvented by mutants of its phage, ΦTE, that express molecules that mimic the antitoxin (238, 239). The Rex system (described above) can be circumvented by phages that carry mutations in the motA gene (240). This gene encodes a transcription factor that directs RNA polymerase from the early to the middle promoters during the infection cycle, and mutations in this gene likely allow phages to complete their replication cycle (211). Likewise, some phages are able to escape Lit abortive infection mechanisms (see above). These phages carry mutations in the grow on Lit (Gol) gene, which normally encodes a peptide that activates the Lit system (241). For several of the numerous Abi systems present in the lactococci (Lactococcus species), phages that can overcome a particular Abi system have been identified, although the exact mechanism by which they do so is in most cases unknown (211).
Broader Consequences of Different Immune Mechanisms
The way in which bacteria evolve immunity against their viral predators can have important implications. First, resistance mechanisms can lead to different coevolutionary dynamics, which can impact microbial community composition (and therefore functioning). Phage can both increase host diversity by selecting for host defense polymorphisms (e.g., see references 242 243) and reduce diversity through population bottlenecking and selective sweeps. Generally, coevolution characterized by fluctuating selection dynamics allows the maintenance of diversity, as different genotypes coexist through negative-frequency-dependent selection (197). Arms race dynamics, on the other hand, results in low diversity levels since it is associated with selective sweeps. Furthermore, as the continuous escalation of bacterial resistance and phage virulence becomes increasingly costly, arms race dynamics and fluctuating selection dynamics may also have different impacts on microbial performance and virulence. As explained above, the fitness cost associated with resistance depends on the type of immune mechanism that evolves. For example, the acquisition of CRISPR-Cas-mediated phage resistance is cost-free in the absence of phage (133), but sm generally reduces bacterial fitness (164, 244) and may affect host colonization or immune evasion (245). Indeed, phage-mediated control of Flavobacterium psychrophilum, a fish pathogen, resulted in surface mutants with attenuated virulence (246). The human pathogen Vibrio cholerae lost the ability to spread between patients after it acquired a surface mutation that conferred resistance against phage (11), and an E. coli strain infecting calves had greatly reduced virulence following resistance evolution (247). Hence, understanding the conditions under which different types of resistance evolve may therefore be important for predicting pathogen virulence, particularly in light of the resurgent interest in the therapeutic use of phages.
Apart from effects resulting from differences in coevolutionary dynamics, resistance mechanisms can have a general impact on microbial adaptation, including the evolution of virulence, through their impact on microbial mutation rates and horizontal gene transfer. Viruses can select for bacteria with increased mutation rates (248), which increases bacterial adaptability. However, this effect may be reduced if the host carries a diversity-generating immune mechanism. In addition, immune systems can also have a long-term impact on microbial adaptation through their impact on horizontal gene transfer. Several correlational studies indicate that the CRISPR-Cas system limits gene transfer. For example, genome sizes of strains of the opportunistic pathogen P. aeruginosa, which often causes lung infections in cystic fibrosis patients, are significantly smaller if they encode CRISPR-Cas systems than if they lack CRISPR-Cas systems (153). In accordance, CRISPR-Cas systems are absent from genomes of species that rely on gene transfer, such as Streptococcus pneumoniae, which causes pneumonia and requires natural transformation for capsule switching during infection (177). Strains of Streptococcus pyogenes, which causes pharyngitis, sepsis, and necrotizing fasciitis, are more likely to carry prophage-encoded virulence factors (249, 250) if they lack CRISPR-Cas systems (177). Furthermore, the presence of CRISPR-Cas immune mechanisms is inversely correlated with antibiotic resistance in Enterococcus faecium and Enterococcus faecalis (176). Immune mechanisms can thus impact adaptation and evolution of virulence in bacteria by blocking HGT.
Alternative Function of Immune Mechanisms
Apart from their role in immunity, defense systems are also involved in other cellular processes. For example, the CRISPR-Cas system has been reported to be involved in DNA repair as well as the regulation of genes involved in virulence and group behaviors (251–253). Usually, either cas genes or CRISPR arrays are involved in these noncanonical functions, rather than the combined action of both elements (252). Alternative roles for RM systems have also been proposed (reviewed in reference 212). These roles include stabilization of genomic islands, stimulating recombination and genome rearrangements, and modulating the rate of genome evolution. In addition, it has been hypothesized that type II RM systems may behave as selfish genetic elements (254, 255), in a fashion similar to that of toxin-antitoxin addiction systems. To increase their own frequency within the population, these systems maintain themselves in the host through postsegregational killing (32), a strategy that is used by a wide variety of selfish genetic elements to render the cells dependent on (addicted to) the residing RM systems for survival. Like toxin-antitoxin systems, selfish type II systems are generally associated with mobile elements, which may allow them to easily invade new genomes (33). Theoretical modeling showed that spatial structuring is an important factor in the spread of addiction systems such as type II RM and TA systems (256). In a spatially structured environment, the frequencies of these genetic elements are likely to increase but not in an unstructured environment.
Although Sie protects hosts against phage infection, the question of whether Sie systems are selected for their role in immunity is a subject of ongoing debate. Sie systems encoded by temperate phages prevent subsequent infection by similar phages and perhaps primarily comprise a phage strategy to successfully compete with other phages, known as “phage warfare.” That said, prophage-encoded Sie systems can clearly benefit the host by either providing immunity to other (potentially virulent) phages or enhancing the competitive abilities of the lysogen over prophage-free hosts; in this context, prophages can be used as a “biological weapon.” For example, several Salmonella enterica strains carry prophages, and these populations produce low virus titers that are sufficient to eliminate competing bacterial populations devoid of such prophages (126, 257). However, this benefit disappears when the competing bacteria also become lysogenized (258). The level of lysogenization of sensitive competitors was shown to be much reduced when the lysogens carry multiple prophages, and this was associated with greatly increased host fitness when competing with phage-sensitive bacteria compared to a lysogen carrying a single prophage (259). The protective effect of lysogens may be key to explaining the observed selection for temperate phages at high bacterial densities (260).
APPLICATIONS, CONCLUSIONS, AND OUTLOOK
The ability to understand, predict, and manipulate the evolution of bacterial immune mechanisms will be extremely powerful to generate virus-resistant bacterial strains for use in industrial fermentations, agriculture, and probiotics. The type of evolved resistance could be tailored to minimize tradeoffs and unwanted coevolutionary consequences.
Apart from applications in industry, understanding bacterial resistance evolution and coevolutionary consequences is also important for phage therapy. Specifically, the ability to expose bacteria to phages in a way that results in resistance evolution associated with large tradeoffs can reduce pathogen virulence or potentially limit the acquisition of novel genes through horizontal gene transfer. Understanding the conditions under which different immune mechanisms evolve can therefore be important for manipulating the evolution of pathogen virulence.
Taken together, it is becoming increasingly clear that ecological factors are key for the evolution of distinct bacterial immune mechanisms and bacterium-phage coevolution (188). Bioinformatics analyses predict the existence of many more immune mechanisms, often clustered together in defense islands (112). One such mechanism, coined bacteriophage exclusion (BREX), is widely distributed across bacteria and appears to share some mechanistic features with RM systems (261). We are starting to understand how all these different immune mechanisms work, but the next challenge is to examine when these mechanisms are favored over one another and how they are integrated during antiviral responses.
Finally, as noted above, phages can encode a range of immune strategies themselves and are likely important vectors for moving these systems around between bacterial strains and species. In addition to Sie mechanisms, phages have been found to carry RM (262) and CRISPR-Cas (263) systems. Phage-encoded CRISPRs can play key roles in phage-phage interactions (263) and in interactions between phage and other genetic elements (264). Understanding how selection acts on diverse immune strategies will require their movement by phage to be explicitly taken into account.
ACKNOWLEDGMENTS
S.V.H. received funding from the European Union's Horizon 2020 research and innovation program under Marie Skłodowska-Curie grant agreement no. 660039. We also acknowledge the NERC, the BBSRC, the Royal Society, the Leverhulme Trust, the Wellcome Trust, and the AXA research fund for funding.
REFERENCES
- 1.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Bikard D, Marraffini LA. 2012. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol 24:15–20. doi: 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 4.Samuel ADT, Pitta TP, Ryu WS, Danese PN, Leung ECW, Berg HC. 1999. Flagellar determinants of bacterial sensitivity to chi-phage. Proc Natl Acad Sci U S A 96:9863–9866. doi: 10.1073/pnas.96.17.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Icho T, Iino T. 1978. Isolation and characterization of motile Escherichia coli mutants resistant to bacteriophage chi. J Bacteriol 134:854–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. 2012. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Woude MW. 2011. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 14:205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Hoyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362-12. doi: 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan D, Svenningsen SL, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627-15. doi: 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Roberts RJ, Kasif S. 2004. Identification of genes with fast-evolving regions in microbial genomes. Nucleic Acids Res 32:6347–6357. doi: 10.1093/nar/gkh935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, Camilli A. 2014. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife 3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qimron U, Marintcheva B, Tabor S, Richardson CC. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci U S A 103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scanlan PD, Hall AR, Blackshields G, Friman VP, Davis MR Jr, Goldberg JB, Buckling A. 2015. Coevolution with bacteriophages drives genome-wide host evolution and constrains the acquisition of abiotic-beneficial mutations. Mol Biol Evol 32:1425–1435. doi: 10.1093/molbev/msv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordstro K, Forsgren A. 1974. Effect of protein A on adsorption of bacteriophages to Staphylococcus aureus. J Virol 14:198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riede I, Eschbach ML. 1986. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett 205:241–245. doi: 10.1016/0014-5793(86)80905-X. [DOI] [PubMed] [Google Scholar]
- 16.Scanlan PD, Buckling A. 2012. Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J 6:1148–1158. doi: 10.1038/ismej.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vostrov AA, Vostrukhina OA, Svarchevsky AN, Rybchin VN. 1996. Proteins responsible for lysogenic conversion caused by coliphages N15 and phi80 are highly homologous. J Bacteriol 178:1484–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uc-Mass A, Loeza EJ, de la Garza M, Guarneros G, Hernandez-Sanchez J, Kameyama L. 2004. An orthologue of the cor gene is involved in the exclusion of temperate lambdoid phages. Evidence that Cor inactivates FhuA receptor functions. Virology 329:425–433. doi: 10.1016/j.virol.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Cumby N, Edwards AM, Davidson AR, Maxwell KL. 2012. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol 194:5012–5019. doi: 10.1128/JB.00843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X, Gohler A, Heller KJ, Neve H. 2006. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology 350:146–157. doi: 10.1016/j.virol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Bebeacua C, Lorenzo Fajardo JC, Blangy S, Spinelli S, Bollmann S, Neve H, Cambillau C, Heller KJ. 2013. X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target. Mol Microbiol 89:152–165. doi: 10.1111/mmi.12267. [DOI] [PubMed] [Google Scholar]
- 22.Newton GJ, Daniels C, Burrows LL, Kropinski AM, Clarke AJ, Lam JS. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol Microbiol 39:1237–1247. doi: 10.1111/j.1365-2958.2001.02311.x. [DOI] [PubMed] [Google Scholar]
- 23.Chung IY, Bae HW, Jang HJ, Kim BO, Cho YH. 2014. Superinfection exclusion reveals heteroimmunity between Pseudomonas aeruginosa temperate phages. J Microbiol 52:515–520. doi: 10.1007/s12275-014-4012-5. [DOI] [PubMed] [Google Scholar]
- 24.Chung IY, Jang HJ, Bae HW, Cho YH. 2014. A phage protein that inhibits the bacterial ATPase required for type IV pilus assembly. Proc Natl Acad Sci U S A 111:11503–11508. doi: 10.1073/pnas.1403537111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu MJ, Henning U. 1994. Superinfection exclusion by T-even-type coliphages. Trends Microbiol 2:137–139. doi: 10.1016/0966-842X(94)90601-7. [DOI] [PubMed] [Google Scholar]
- 26.Hershey AD, Dove W. 1983. Introduction to lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD, Anderson AG, Ang AA, Ares M Jr, Barber AJ, Barker LP, Barrett JM, Barshop WD, Bauerle CM, Bayles IM, Belfield KL, Best AA, Borjon A Jr, Bowman CA, Boyer CA, Bradley KW, Bradley VA, Broadway LN, Budwal K, Busby KN, Campbell IW, Campbell AM, Carey A, Caruso SM, Chew RD, Cockburn CL, Cohen LB, Corajod JM, Cresawn SG, Davis KR, Deng L, Denver DR, Dixon BR, Ekram S, Elgin SC, Engelsen AE, English BE, Erb ML, Estrada C, Filliger LZ, et al. 2011. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pingoud A, Wilson GG, Wende W. 2014. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res 42:7489–7527. doi: 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butterer A, Pernstich C, Smith RM, Sobott F, Szczelkun MD, Toth J. 2014. Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric methyltransferase that binds only one specific DNA. Nucleic Acids Res 42:5139–5150. doi: 10.1093/nar/gku122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeltsch A, Pingoud A. 1996. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J Mol Evol 42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res 29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira PH, Touchon M, Rocha EP. 2014. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res 42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tock MR, Dryden DTF. 2005. The biology of restriction and anti-restriction. Curr Opin Microbiol 8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Bayliss CD, Callaghan MJ, Moxon ER. 2006. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res 34:4046–4059. doi: 10.1093/nar/gkl568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaleski P, Wojciechowski M, Piekarowicz A. 2005. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology 151:3361–3369. doi: 10.1099/mic.0.28184-0. [DOI] [PubMed] [Google Scholar]
- 37.Seib KL, Peak IRA, Jennings MP. 2002. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Microbiol 32:159–165. doi: 10.1111/j.1574-695X.2002.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 38.De Bolle X, Bayliss CD, Field D, van de Ven T, Saunders NJ, Hood DW, Moxon ER. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol Microbiol 35:211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 39.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeks J, Naismith JH, White MF. 2013. CRISPR interference: a structural perspective. Biochem J 453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 42.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 43.Jiang F, Doudna JA. 2015. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koonin EV, Makarova KS. 2013. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol 10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osawa T, Inanaga H, Sato C, Numata T. 2015. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell 58:418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Jackson RN, Wiedenheft B. 2015. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol Cell 58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor DW, Zhu Y, Staals RH, Kornfeld JE, Shinkai A, van der Oost J, Nogales E, Doudna JA. 2015. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nunez JK, Lee AS, Engelman A, Doudna JA. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA. 2015. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527:535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. 18 August 2015. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y. 2015. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Charpentier E, Richter H, van der Oost J, White MF. 2015. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev 39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders AP, Dickman MJ, Doudna JA, Boekema EJ, Heck AJ, van der Oost J, Brouns SJ. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 60.Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mulepati S, Bailey S. 2013. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem 288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mulepati S, Bailey S. 2011. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J Biol Chem 286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. 2014. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A 111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163:854–865. doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJ. 2013. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet 9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B. 2015. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res 43:2216–2222. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E. 2014. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell 56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 70.Goldberg GW, Jiang W, Bikard D, Marraffini LA. 2014. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. 2015. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJ, Nogales E, Doudna JA, Shinkai A, van der Oost J. 2014. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF. 2012. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang W, Samai P, Marraffini LA. 2016. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 78.Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrangou R, van Pijkeren JP. 2016. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr Opin Biotechnol 37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, Greely HT, Jinek M, Martin GS, Penhoet E, Puck J, Sternberg SH, Weissman JS, Yamamoto KR. 2015. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 87.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ketting RF. 2011. The many faces of RNAi. Dev Cell 20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 89.Cerutti L, Mian N, Bateman A. 2000. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci 25:481–482. doi: 10.1016/S0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 90.Shabalina SA, Koonin EV. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Makarova KS, Wolf YI, van der Oost J, Koonin EV. 2009. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J. 2014. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. 2005. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang YL, Juranek S, Li HT, Sheng G, Tuschl T, Patel DJ. 2008. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, van der Oost J. 2014. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. 2013. Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol Cell 51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Swarts DC, Hegge JW, Hinojo I, Shiimori M, Ellis MA, Dumrongkulraksa J, Terns RM, Terns MP, van der Oost J. 2015. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res 43:5120–5129. doi: 10.1093/nar/gkv415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swarts DC, Koehorst JJ, Westra ER, Schaap PJ, van der Oost J. 2015. Effects of Argonaute on gene expression in Thermus thermophilus. PLoS One 10:e0124880. doi: 10.1371/journal.pone.0124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zander A, Holzmeister P, Klose D, Tinnefeld P, Grohmann D. 2014. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol 11:45–56. doi: 10.4161/rna.27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chopin MC, Chopin A, Bidnenko E. 2005. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol 8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 102.Snyder L. 1995. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol Microbiol 15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 103.Bingham R, Ekunwe SI, Falk S, Snyder L, Kleanthous C. 2000. The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J Biol Chem 275:23219–23226. doi: 10.1074/jbc.M002546200. [DOI] [PubMed] [Google Scholar]
- 104.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GPC. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GPC. 2009. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol 191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dy RL, Przybilski R, Semeijn K, Salmond GPC, Fineran PC. 2014. A widespread bacteriophage abortive infection system functions through a type IV toxin-antitoxin mechanism. Nucleic Acids Res 42:4590–4605. doi: 10.1093/nar/gkt1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, Salmond GPC. 2011. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol 18:185–190. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pecota DC, Wood TK. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol 178:2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hazan R, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics 272:227–234. [DOI] [PubMed] [Google Scholar]
- 110.Dupuis ME, Villion M, Magadan AH, Moineau S. 2013. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun 4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 111.Hynes AP, Villion M, Moineau S. 2014. Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat Commun 5:4399. doi: 10.1038/ncomms5399. [DOI] [PubMed] [Google Scholar]
- 112.Makarova KS, Wolf YI, Koonin EV. 2013. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet 8:e1002511. doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sampson TR, Weiss DS. 2013. Degeneration of a CRISPR/Cas system and its regulatory target during the evolution of a pathogen. RNA Biol 10:1618–1622. doi: 10.4161/rna.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol 193:2460–2467. doi: 10.1128/JB.01307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boots M, Haraguchi Y. 1999. The evolution of costly resistance in host-parasite systems. Am Nat 153:359–370. doi: 10.1086/303181. [DOI] [PubMed] [Google Scholar]
- 118.Graham A, Allen J, Read A. 2005. Evolutionary causes and consequences of immunopathology. Annu Rev Ecol Evol Syst 36:373–397. doi: 10.1146/annurev.ecolsys.36.102003.152622. [DOI] [Google Scholar]
- 119.Weitz J. 2015. Quantitative viral ecology. Princeton University Press, Princeton, NJ. [Google Scholar]
- 120.Buckling A, Brockhurst M. 2012. Bacteria-virus coevolution. Adv Exp Med Biol 751:347–370. doi: 10.1007/978-1-4614-3567-9_16. [DOI] [PubMed] [Google Scholar]
- 121.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 122.Hochberg ME, Baalen M. 1998. Antagonistic coevolution over productivity gradients. Am Nat 152:620–634. doi: 10.1086/286194. [DOI] [PubMed] [Google Scholar]
- 123.Lopez-Pascua L, Buckling A. 2008. Increasing productivity accelerates host-parasite coevolution. J Evol Biol 21:853–860. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- 124.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL. 3 June 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Canchaya C, Fournous G, Brussow H. 2004. The impact of prophages on bacterial chromosomes. Mol Microbiol 53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 126.Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pleska M, Qian L, Okura R, Bergmiller T, Wakamoto Y, Kussell E, Guet CC. 2016. Bacterial autoimmunity due to a restriction-modification system. Curr Biol 26:404–409. doi: 10.1016/j.cub.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 128.Boots M. 2011. The evolution of resistance to a parasite is determined by resources. Am Nat 178:214–220. doi: 10.1086/660833. [DOI] [PubMed] [Google Scholar]
- 129.Rocha EP, Danchin A, Viari A. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res 11:946–958. doi: 10.1101/gr.GR-1531RR. [DOI] [PubMed] [Google Scholar]
- 130.Rusinov I, Ershova A, Karyagina A, Spirin S, Alexeevski A. 2015. Lifespan of restriction-modification systems critically affects avoidance of their recognition sites in host genomes. BMC Genomics 16:1084. doi: 10.1186/s12864-015-2288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Seidel R, Bloom JG, Dekker C, Szczelkun MD. 2008. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J 27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vale PF, Lafforgue G, Gatchitch F, Gardan R, Moineau S, Gandon S. 2015. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proc Biol Sci 282:20151270. doi: 10.1098/rspb.2015.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A. 2015. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol 25:1043–1049. doi: 10.1016/j.cub.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 134.Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, Verberkmoes NC. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 7:e38077. doi: 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol 395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 136.Quax TE, Voet M, Sismeiro O, Dillies MA, Jagla B, Coppee JY, Sezonov G, Forterre P, van der Oost J, Lavigne R, Prangishvili D. 2013. Massive activation of archaeal defense genes during viral infection. J Virol 87:8419–8428. doi: 10.1128/JVI.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. 2012. Viral diversity threshold for adaptive immunity in prokaryotes. mBio 3:e00456–12. doi: 10.1128/mBio.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Held NL, Whitaker RJ. 2009. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol 11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 139.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 140.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet 9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 144.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. 2015. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Berngruber TW, Lion S, Gandon S. 2013. Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecol Lett 16:446–453. doi: 10.1111/ele.12064. [DOI] [PubMed] [Google Scholar]
- 146.Tollrian R, Harvell D. 1999. The ecology and evolution of inducible defenses. Princeton University Press, Princeton, NJ. [Google Scholar]
- 147.Anderson RE, Brazelton WJ, Baross JA. 2011. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol 77:120–133. doi: 10.1111/j.1574-6941.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 148.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. 2013. Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR-Cas in an explicit ecological context. J Bacteriol 195:3834–3844. doi: 10.1128/JB.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith J. 2011. Superinfection drives virulence evolution in experimental populations of bacteria and plasmids. Evolution 65:831–841. doi: 10.1111/j.1558-5646.2010.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 152.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van Belkum A, Soriaga LB, LaFave MC, Akella S, Veyrieras JB, Barbu EM, Shortridge D, Blanc B, Hannum G, Zambardi G, Miller K, Enright MC, Mugnier N, Brami D, Schicklin S, Felderman M, Schwartz AS, Richardson TH, Peterson TC, Hubby B, Cady KC. 2015. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. mBio 6:e01796-15. doi: 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Levin BR. 1988. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci 319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 156.van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, Westra ER. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532:385–388. doi: 10.1038/nature17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 158.Dybvig K, Sitaraman R, French CT. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci U S A 95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, Miller JF. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 160.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. 2004. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 161.Medhekar B, Miller JF. 2007. Diversity-generating retroelements. Curr Opin Microbiol 10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Paul BG, Bagby SC, Czornyj E, Arambula D, Handa S, Sczyrba A, Ghosh P, Miller JF, Valentine DL. 2015. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat Commun 6:6585. doi: 10.1038/ncomms7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Samson JE, Belanger M, Moineau S. 2013. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J Bacteriol 195:3947–3956. doi: 10.1128/JB.00296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koskella B, Lin DM, Buckling A, Thompson JN. 2012. The costs of evolving resistance in heterogeneous parasite environments. Proc Biol Sci 279:1896–1903. doi: 10.1098/rspb.2011.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. 2011. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474:604–608. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 166.Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10:200–207. [DOI] [PubMed] [Google Scholar]
- 167.Weinberger AD, Sun CL, Plucinski MM, Denef VJ, Thomas BC, Horvath P, Barrangou R, Gilmore MS, Getz WM, Banfield JF. 2012. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput Biol 8:e1002475. doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Haerter JO, Trusina A, Sneppen K. 2011. Targeted bacterial immunity buffers phage diversity. J Virol 85:10554–10560. doi: 10.1128/JVI.05222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 170.Lenski RE, Bouma JE. 1987. Effects of segregation and selection on instability of plasmid pACYC184 in Escherichia coli B. J Bacteriol 169:5314–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Modi RI, Wilke CM, Rosenzweig RF, Adams J. 1991. Plasmid macro-evolution: selection of deletions during adaptation in a nutrient-limited environment. Genetica 84:195–202. doi: 10.1007/BF00127247. [DOI] [PubMed] [Google Scholar]
- 172.Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165:1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol 20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 174.Gandon S, Vale PF. 2014. The evolution of resistance against good and bad infections. J Evol Biol 27:303–312. doi: 10.1111/jeb.12291. [DOI] [PubMed] [Google Scholar]
- 175.Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol 37:1052–1065. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 176.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hatoum-Aslan A, Marraffini LA. 2014. Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens. Curr Opin Microbiol 17:82–90. doi: 10.1016/j.mib.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gophna U, Kristensen DM, Wolf YI, Popa O, Drevet C, Koonin EV. 2015. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J 9:2021–2027. doi: 10.1038/ismej.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hamilton WD. 1964. The genetical evolution of social behaviour. I. J Theor Biol 7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 180.Debarre F, Lion S, Van Baalen M, Gandon S. 2012. Evolution of host life-history traits in a spatially structured host-parasite system. Am Nat 179:52–63. doi: 10.1086/663199. [DOI] [PubMed] [Google Scholar]
- 181.Fukuyo M, Sasaki A, Kobayashi I. 2012. Success of a suicidal defense strategy against infection in a structured habitat. Sci Rep 2:238. doi: 10.1038/srep00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Refardt D, Bergmiller T, Kummerli R. 2013. Altruism can evolve when relatedness is low: evidence from bacteria committing suicide upon phage infection. Proc Biol Sci 280:20123035. doi: 10.1098/rspb.2012.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Berngruber TW, Lion S, Gandon S. 2015. Spatial structure, transmission modes and the evolution of viral exploitation strategies. PLoS Pathog 11:e1004810. doi: 10.1371/journal.ppat.1004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fenton A, Antonovics J, Brockhurst MA. 2009. Inverse-gene-for-gene infection genetics and coevolutionary dynamics. Am Nat 174:E230–E242. doi: 10.1086/645087. [DOI] [PubMed] [Google Scholar]
- 185.Fenton A, Antonovics J, Brockhurst MA. 2012. Two-step infection processes can lead to coevolution between functionally independent infection and resistance pathways. Evolution 66:2030–2041. doi: 10.1111/j.1558-5646.2012.01578.x. [DOI] [PubMed] [Google Scholar]
- 186.Brockhurst MA, Buckling A, Rainey PB. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci 272:1385–1391. doi: 10.1098/rspb.2005.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lythgoe KA, Chao L. 2003. Mechanisms of coexistence of a bacteria and a bacteriophage in a spatially homogeneous environment. Ecol Lett 6:326–334. doi: 10.1046/j.1461-0248.2003.00433.x. [DOI] [Google Scholar]
- 188.Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cowlishaw J, Mrsa M. 1975. Co-evolution of a virus-alga system. Appl Microbiol 29:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cannon RE, Shane MS, Whitaker JM. 1976. Interaction of Plectonema-Boryanum (Cyanophyceae) and Lpp-cyanophages in continuous culture. J Phycol 12:418–421. doi: 10.1111/j.1529-8817.1976.tb02865.x. [DOI] [Google Scholar]
- 191.Barnet YM, Daft MJ, Stewart WDP. 1981. Cyanobacteria-cyanophage interactions in continuous culture. J Appl Bacteriol 51:541–552. doi: 10.1111/j.1365-2672.1981.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 192.Buckling A, Rainey PB. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci 269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, Quail M, Smith F, Walker D, Libberton B, Fenton A, Hall N, Brockhurst MA. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Scanlan PD, Hall AR, Lopez-Pascua LD, Buckling A. 2011. Genetic basis of infectivity evolution in a bacteriophage. Mol Ecol 20:981–989. doi: 10.1111/j.1365-294X.2010.04903.x. [DOI] [PubMed] [Google Scholar]
- 195.Hall AR, Scanlan PD, Buckling A. 2011. Bacteria-phage coevolution and the emergence of generalist pathogens. Am Nat 177:44–53. doi: 10.1086/657441. [DOI] [PubMed] [Google Scholar]
- 196.Hall AR, Scanlan PD, Morgan AD, Buckling A. 2011. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett 14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 197.Gandon S, Buckling A, Decaestecker E, Day T. 2008. Host-parasite coevolution and patterns of adaptation across time and space. J Evol Biol 21:1861–1866. doi: 10.1111/j.1420-9101.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 198.Lopez Pascua L, Hall AR, Best A, Morgan AD, Boots M, Buckling A. 2014. Higher resources decrease fluctuating selection during host-parasite coevolution. Ecol Lett 17:1380–1388. doi: 10.1111/ele.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gomez P, Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 200.Gomez P, Ashby B, Buckling A. 2015. Population mixing promotes arms race host-parasite coevolution. Proc Biol Sci 282:20142297. doi: 10.1098/rspb.2014.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bailone A, Devoret R. 1978. Isolation of ultravirulent mutants of phage lambda. Virology 84:547–550. doi: 10.1016/0042-6822(78)90273-8. [DOI] [PubMed] [Google Scholar]
- 202.Benson N, Adams C, Youderian P. 1992. Mutant lambda repressors with increased operator affinities reveal new, specific protein-DNA contacts. Genetics 130:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Berngruber TW, Weissing FJ, Gandon S. 2010. Inhibition of superinfection and the evolution of viral latency. J Virol 84:10200–10208. doi: 10.1128/JVI.00865-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Erni B, Zanolari B, Kocher HP. 1987. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J Biol Chem 262:5238–5247. [PubMed] [Google Scholar]
- 205.Saris PEJ, Palva ET. 1987. Regulation of manX (ptsL) and manY (pel) genes required for mannose transport and penetration of lambda DNA in Escherichia coli K12. FEMS Microbiol Lett 44:377–382. doi: 10.1111/j.1574-6968.1987.tb02317.x. [DOI] [Google Scholar]
- 206.Esquinas-Rychen M, Erni B. 2001. Facilitation of bacteriophage lambda DNA injection by inner membrane proteins of the bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS). J Mol Microbiol Biotechnol 3:361–370. [PubMed] [Google Scholar]
- 207.Korona R, Levin BR. 1993. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution 47:556–575. doi: 10.2307/2410071. [DOI] [PubMed] [Google Scholar]
- 208.Sneppen K, Semsey S, Seshasayee AS, Krishna S. 2015. Restriction modification systems as engines of diversity. Front Microbiol 6:528. doi: 10.3389/fmicb.2015.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 210.Patrick S, Blakely GW, Houston S, Moore J, Abratt VR, Bertalan M, Cerdeno-Tarraga AM, Quail MA, Corton N, Corton C, Bignell A, Barron A, Clark L, Bentley SD, Parkhill J. 2010. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology 156:3255–3269. doi: 10.1099/mic.0.042978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 212.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kruger DH, Bickle TA. 1983. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev 47:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu Rev Genet 25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 215.Kahmann R. 1984. The mom gene of bacteriophage Mu. Curr Top Microbiol Immunol 108:29–47. [DOI] [PubMed] [Google Scholar]
- 216.Bickle TA, Kruger DH. 1993. Biology of DNA restriction. Microbiol Rev 57:434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. 2008. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol 375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Iida S, Streiff MB, Bickle TA, Arber W. 1987. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA− phages. Virology 157:156–166. doi: 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- 219.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. 2002. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol Cell 9:187–194. doi: 10.1016/S1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 220.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu HT, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, Blakely GW, Naismith JH, Dryden DTF. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res 37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sharp PM. 1986. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol 3:75–83. [DOI] [PubMed] [Google Scholar]
- 222.Agrawal A, Lively CM. 2002. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res 4:79–90. [Google Scholar]
- 223.Vale PF, Little TJ. 2010. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc Biol Sci 277:2097–2103. doi: 10.1098/rspb.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Levin BR, Moineau S, Bushman M, Barrangou R. 2013. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet 9:e1003312. doi: 10.1371/journal.pgen.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. 2009. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One 4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Voorhies AA, Eisenlord SD, Marcus DN, Duhaime MB, Biddanda BA, Cavalcoli JD, Dick GJ. 4 March 2015. Ecological and genetic interactions between cyanobacteria and viruses in a low-oxygen mat community inferred through metagenomics and metatranscriptomics. Environ Microbiol doi: 10.1111/1462-2920.12756. [DOI] [PubMed] [Google Scholar]
- 229.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Manica A, Zebec Z, Teichmann D, Schleper C. 2011. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 233.Kupczok A, Bollback JP. 2014. Motif depletion in bacteriophages infecting hosts with CRISPR systems. BMC Genomics 15:663. doi: 10.1186/1471-2164-15-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Childs LM, England WE, Young MJ, Weitz JS, Whitaker RJ. 2014. CRISPR-induced distributed immunity in microbial populations. PLoS One 9:e101710. doi: 10.1371/journal.pone.0101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. 2014. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5:e00896-14. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. 2015. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Blower TR, Evans TJ, Przybilski R, Fineran PC, Salmond GPC. 2012. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet 8:e1003023. doi: 10.1371/journal.pgen.1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Blower TR, Short FL, Fineran PC, Salmond GP. 2012. Viral molecular mimicry circumvents abortive infection and suppresses bacterial suicide to make hosts permissive for replication. Bacteriophage 2:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shinedling S, Parma D, Gold L. 1987. Wild-type bacteriophage T4 is restricted by the lambda rex genes. J Virol 61:3790–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Champness WC, Snyder L. 1982. The gol site: a cis-acting bacteriophage T4 regulatory region that can affect expression of all the T4 late genes. J Mol Biol 155:395–407. [DOI] [PubMed] [Google Scholar]
- 242.Weitz JS, Hartman H, Levin SA. 2005. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci U S A 102:9535–9540. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Menge DN, Weitz JS. 2009. Dangerous nutrients: evolution of phytoplankton resource uptake subject to virus attack. J Theor Biol 257:104–115. doi: 10.1016/j.jtbi.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 244.Lenski RE. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. 1. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42:425–432. [DOI] [PubMed] [Google Scholar]
- 245.Hall AR, De Vos D, Friman VP, Pirnay JP, Buckling A. 2012. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol 78:5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Castillo D, Christiansen RH, Espejo R, Middelboe M. 2014. Diversity and geographical distribution of Flavobacterium psychrophilum isolates and their phages: patterns of susceptibility to phage infection and phage host range. Microb Ecol 67:748–757. doi: 10.1007/s00248-014-0375-8. [DOI] [PubMed] [Google Scholar]
- 247.Smith HW, Huggins MB, Shaw KM. 1987. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol 133:1111–1126. [DOI] [PubMed] [Google Scholar]
- 248.Pal C, Macia MD, Oliver A, Schachar I, Buckling A. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 249.Beres SB, Musser JM. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Banks DJ, Beres SB, Musser JM. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol 10:515–521. doi: 10.1016/S0966-842X(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 251.Sampson TR, Weiss DS. 2013. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog 9:e1003621. doi: 10.1371/journal.ppat.1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Westra ER, Buckling A, Fineran PC. 2014. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol 12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 253.Sampson TR, Weiss DS. 2014. CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol 4:37. doi: 10.3389/fcimb.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Naito T, Kusano K, Kobayashi I. 1995. Selfish behavior of restriction-modification systems. Science 267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 255.Nakayama Y, Kobayashi I. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci U S A 95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. 2006. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics 172:1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. 2003. Prophage contribution to bacterial population dynamics. J Bacteriol 185:6467–6471. doi: 10.1128/JB.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gama JA, Reis AM, Domingues I, Mendes-Soares H, Matos AM, Dionisio F. 2013. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One 8:e59043. doi: 10.1371/journal.pone.0059043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Burns N, James CE, Harrison E. 2015. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol Appl 8:346–351. doi: 10.1111/eva.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobian-Guemes AG, Coutinho FH, Dinsdale EA, Felts B, Furby KA, George EE, Green KT, Gregoracci GB, Haas AF, Haggerty JM, Hester ER, Hisakawa N, Kelly LW, Lim YW, Little M, Luque A, McDole-Somera T, McNair K, de Oliveira LS, Quistad SD, Robinett NL, Sala E, Salamon P, Sanchez SE, Sandin S, Silva GG, Smith J, Sullivan C, Thompson C, Vermeij MJ, Youle M, Young C, Zgliczynski B, Brainard R, Edwards RA, Nulton J, Thompson F, Rohwer F. 2016. Lytic to temperate switching of viral communities. Nature 531:466–470. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 261.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dempsey RM, Carroll D, Kong H, Higgins L, Keane CT, Coleman DC. 2005. Sau42I, a BcgI-like restriction-modification system encoded by the Staphylococcus aureus quadruple-converting phage Phi42. Microbiology 151:1301–1311. doi: 10.1099/mic.0.27646-0. [DOI] [PubMed] [Google Scholar]
- 263.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]