SUMMARY
Moonlighting proteins are multifunctional proteins that participate in unrelated biological processes and that are not the result of gene fusion. A certain number of these proteins have been characterized in yeasts, and the easy genetic manipulation of these microorganisms has been useful for a thorough analysis of some cases of moonlighting. As the awareness of the moonlighting phenomenon has increased, a growing number of these proteins are being uncovered. In this review, we present a crop of newly identified moonlighting proteins from yeasts and discuss the experimental evidence that qualifies them to be classified as such. The variety of moonlighting functions encompassed by the proteins considered extends from control of transcription to DNA repair or binding to plasminogen. We also discuss several questions pertaining to the moonlighting condition in general. The cases presented show that yeasts are important organisms to be used as tools to understand different aspects of moonlighting proteins.
INTRODUCTION
Proteins have been described as “the most versatile class of biomolecules” (1). An unexpected facet of this versatility emerged when it was shown that one protein could be used by cells to perform completely different, unrelated, functions. Piatigorsky et al. (2) demonstrated that duck δ-crystallin and argininosuccinate lyase were encoded by the same gene and termed this phenomenon gene sharing (3). Further extensive studies with crystallins from different species revealed that many of them were identical to proteins that had been identified as having a different metabolic function (4). Other cases of proteins playing dual roles continued to appear in the literature, and their number has soared in recent years (5). The term gene sharing, first proposed for these proteins (3), may present some ambiguities, as there are cases in which a gene may encode different protein forms. In yeasts, this can be due to different sites for initiation of translation (6, 7), termination (8), or splicing (9). In these cases, the corresponding protein forms also share a gene but this gene sharing is not the same as that of a single protein playing two different roles. The alternative designation “moonlighting proteins” was proposed to name those proteins, and a clear-cut definition to delimit which proteins may be considered as such was also provided (10). The limits imposed by the definition were that the diverse functions of moonlighting proteins shall not be the result of a gene fusion event, or of differential gene splicing; proteins with a single function in different subcellular localizations were excluded from the group (10). Chapple and Brun (11) have proposed to expand this definition to cover all proteins that perform dissimilar functions, therefore including proteins where separate domains are responsible for their multifunctionality. Structural proteins that serve as scaffolds and interact with different proteins are generally not considered moonlighting proteins (12), and this criterion is followed in this article. Secreted proteins are accepted in this category, as the evidence of their numerous roles in different organisms is well documented (12–15). It may be noted that the word “moonlighting” had been used already in 1978 by Freedman to refer to multifunctional proteins in an article entitled “Moonlighting Molecules” (16). Moonlighting proteins are found in diverse organisms, and current dedicated databases list several hundred of them, distributed among the different biological kingdoms (5, 17). Their noncanonical roles cover a bewildering array of functions such as control of transcription, protection of DNA, assembly of organelles, chaperoning, splicing of introns, or binding to plasminogen (12, 18–20). Such alternative functions often cause unexpected phenotypes in monogenic mutants, and it has been proposed that the complex phenotypes of some single-gene disorders could be due to unknown moonlighting functions of the main protein concerned (21).
Many moonlighting proteins are present in different yeast species, and their roles are very diverse; for example, hexokinase 2 and galactokinase act as transcriptional regulators, aconitase functions in mitochondrial DNA maintenance, and aldolase and pyruvate carboxylase participate in the assembly of some vacuolar or peroxisomal enzymes (18, 22). While most of these proteins have been described in Saccharomyces cerevisiae, this probably reflects the starring role of this yeast in research and not some basic difference between species. As awareness of the existence of moonlighting proteins has spread, more cases are being described. Since the publication of the last comprehensive review devoted to moonlighting proteins in yeasts (22), new, well-documented cases have appeared that increase the number of yeast proteins belonging to this group (Table 1).
TABLE 1.
Some moonlighting proteins in yeasts
| Protein | Canonical function | Moonlighting activity | Yeast species | Reference(s) |
|---|---|---|---|---|
| Homocitrate synthase | Acyl transferase (lysine biosynthesis) | Repair of damaged DNA | S. cerevisiae | 29, 32 |
| Superoxide dismutase | Conversion of superoxide radical into O2 and H2O2 | Regulation of genes related to oxidative stress | S. cerevisiae | 42 |
| Pyruvate decarboxylase | Pyruvate decarboxylation (glycolysis) | Regulation of PDC1/PDC5 transcription | S. cerevisiae, K. lactis | 47, 49, 50, 52 |
| Zuotin | Molecular chaperone | Activation of pleiotropic drug resistance | S. cerevisiae | 58, 60, 64 |
| Subunit Sdh3 of succinate dehydrogenase | Oxidation of succinate (citric acid cycle), electron transport | Assembly of mitochondrial translocase | S. cerevisiae | 70 |
| Enolase | Dehydration of 2-P-glycerate (glycolysis) | Binding to plasminogen | C. albicans | 79 |
| Phosphate-glycerate mutase | Isomerization of 3-P-glycerate (glycolysis) | Binding to plasminogen and to complement regulators | C. albicans | 78, 81 |
| Glycerol-3-phosphate dehydrogenase | Reduction of dihydroxyacetone phosphate to glycerol-3-phosphate (ancillary to glycolysis) | Binding to plasminogen and to complement regulators | C. albicans | 84 |
| Hgt1 | Glucose transporter (glycolysis) | Binding to complement regulators | C. albicans | 90 |
| Hal3 | Decarboxylation of phosphate-pantothenoyl–l-cysteine (CoA biosynthesis) | Inhibitor of phosphatases Ppz1 and Ppz2 | S. cerevisiae, S. pombe | 95, 99 |
Shown are the moonlighting proteins considered in the text. A table with other moonlighting proteins from yeasts is presented in reference 22.
In this article, we present several newcomers to the class of yeast moonlighting proteins, examining the experimental support to qualify them as such. The wide importance of the moonlighting phenomenon for yeast research, both basic and applied, is also discussed.
HOMOCITRATE SYNTHASE PARTICIPATES IN DNA REPAIR
The amino acid lysine plays a particular role in proteins, as its ε-amino group can be modified by different substituents, thereby altering the activity of the corresponding protein. Yeasts and other fungi synthesize lysine by the so-called α-aminoadipate pathway that starts with a condensation of α-oxoglutarate, an intermediate of the tricarboxylic acid cycle, and acetyl coenzyme A (acetyl-CoA) to produce homocitrate. This reaction is catalyzed by the enzyme homocitrate synthase. Tucci and Ceci (23) undertook the purification of the corresponding protein from S. cerevisiae and showed the existence of two enzymes by isoelectric focusing; both of them were inhibited by lysine, although not to the same extent. Ramos et al. (24) isolated a fragment of yeast DNA whose disruption greatly reduced homocitrate synthase activity; they called the corresponding gene LYS20 and demonstrated that it encoded the isoenzyme responsible for about 70% of lysine production, thus confirming the existence of another protein with homocitrate synthase activity. The corresponding gene, LYS21, was later identified by Feller et al. (25), and the protein was shown to be the main homocitrate synthase activity during growth in ethanol (26); it was also shown that Lys20 and Lys21 present different kinetic and regulatory properties (26).
Since it had been reported that not all of the steps in the biosynthesis of lysine took place in the same subcellular compartment (27), Chen et al. (28) tried to determine the localization of homocitrate synthase. Using monoclonal antibodies against nuclear proteins from S. cerevisiae, they showed that the majority of homocitrate synthase was present in the nucleus. Later on, this result was confirmed by using green fluorescent protein (GFP) fusions to Lys20 and Lys21 (25). The unexpected localization led Chen et al. (28) to raise provocative questions about the significance of the nuclear localization of the enzyme and its possible enzymatic activity in the nucleus. They speculated that in the nucleus, homocitrate synthase could have a function not directly related to lysine biogenesis, playing a role in nuclear structure and/or function. The work of Scott and Pillus (29) has shown that these questions were pertinent. In fact, Lys20 performs a function in the nucleus that is not related to lysine synthesis; it participates in the damaged DNA repair process. This was found during studies of an esa1-414 mutant affected in the activity of Esa1, an essential histone acetyltransferase in yeast (30). This mutant is impaired in the repair of DNA double-strand breaks and therefore does not grow in the presence of camptothecin, a drug that interferes with toposisomerase I (31). Looking for suppressor genes that restored growth to esa1-414 mutants on camptothecin, Scott and Pillus (29) identified LYS20; LYS21 showed a much weaker suppressor effect. The authors also showed that some lys20 mutations that abolished homocitrate synthase activity were still able to accomplish their function in DNA repair. Using homocitrate synthase versions that conserved catalytic activity but were unable either to enter the nucleus or to remain in it, the same authors (29) found that they suppressed camptothecin toxicity toward esa1-414 mutant cells less efficiently than the wild-type enzyme, thus showing that the nuclear localization of homocitrate synthase was critical for its nonmetabolic role.
Following this work, Torres-Machorro et al. (32) undertook the characterization of the mechanism of suppression of esa1 mutations by overexpression of LYS20. Testing versions of Lys20 with mutations in different amino acids and domains, they determined that the domain of Lys20 implicated in the moonlighting DNA repair function was located in the C-terminal portion of the protein, in front of a nuclear localization signal previously identified by Scott and Pillus (29); such a domain was not found in Lys21. By using chromatin immunoprecipitation preceded by a cross-linking step, it was found that Lys20 was enriched in the chromatin fraction (32). This association, however, was not detected if cross-linking was omitted, suggesting that the interaction of Lys20 with chromatin was either transient or unstable. It was also observed that Lys20 was recruited to DNA double-strand breaks and that overexpression of LYS20 caused larger amounts of Lys20 to appear at the breaks. A mutated form of Lys20 affected in the moonlighting domain was recruited at the breaks as well as a wild-type version of Lys20, but it failed to correct the defects in DNA repair that occur in esa1 mutants.
Repair of DNA double-stranded breaks is dependent on histone eviction at the damage sites, a process required for further reactions in the DNA repair pathway (33). Histone eviction, in turn, is facilitated by Esa1, which transiently acetylates histone H4 at the DNA breaks (34), and by the chromatin remodeling complex INO80 (35). Lys20 could therefore promote the recruitment at the breaks of either Esa1-414 or the INO80 complex. It has been found that Lys20 overexpression does not facilitate the binding of Esa1-414 at the damage sites but suppresses esa1 mutations by increasing the amount of the INO80 complex that accumulates at the DNA breaks (32).
SUPEROXIDE DISMUTASE Sod1 CONTROLS TRANSCRIPTION OF GENES ENCODING PROTEINS IMPLICATED IN OXIDATIVE RESPONSE AND DNA REPAIR
Organisms are exposed to injuries caused by reactive oxygen species (ROS) originated both intra- and extracellularly. ROS are potent oxidants that can damage all types of cellular components; to minimize the damage, several defense mechanisms have been selected in the course of evolution. Among the agents involved in those mechanisms are the superoxide dismutases, which catalyze the conversion of the superoxide radical O2− into O2 and H2O2. The latter molecule can, in turn, be converted into H2O and O2 by catalases. S. cerevisiae possesses two forms of superoxide dismutase, Sod1, which is cytoplasmic, and Sod2, which is mitochondrial. Sod1 is a Cu-Zn-containing enzyme, while Sod2 is a Mn-containing form. Yeast mutants affected in Sod1 present multiple phenotypes such as methionine auxotrophy, slow growth, loss of viability in stationary phase, or a decreased replicative life span (36–40). Mutants lacking Sod2 also show different phenotypes, among them decreased survival in the stationary phase (37).
While studying the regulation of yeast Sod1 under oxidative stress produced by 4-nitroquinoline-N-oxide, an oxygen radical-producing compound (41), Tsang et al. (42) did not observe changes in Sod1 activity; however, by using fluorescence microscopy, they noticed a change in the subcellular localization of Sod1. In untreated cells, Sod1 is basically cytoplasmic, while treatment with 4-nitroquinoline-N-oxide made this protein mainly nuclear. This change was also observed in response to another ROS-generating compound, H2O2, and was confirmed by subcellular fractionation of the treated cells. Experiments using different yeast mutants that spontaneously produce a high level of ROS showed that the change in localization could also take place in the absence of an external treatment, demonstrating that it also occurred when ROS were generated intracellularly.
An important consequence of oxidative stress is DNA damage that may be corrected by the DNA damage checkpoint pathway. The transmission of the DNA damage signal through a cascade of protein kinases, including Mec1 and Dun1, is affected in sod1 mutants, and this results in problems in coping with oxidative stress (36). Interestingly, the nuclear localization of Sod1 in response to ROS is blocked in the absence of Mec1 or Dun1 (42). While previous studies had shown that Sod1 and Dun1 could form a complex (43), Tsang et al. (42) found that oxidative conditions enhanced this interaction and that deletion of Dun1 blocked Sod1 in the cytoplasm. They also found that Dun1 phosphorylates Sod1 at serine residues S60 and S99 and that this phosphorylation contributed to its nuclear localization. Suspecting a regulatory role for nuclear Sod1, Tsang et al. (42) studied the effects of treatment with H2O2, the product of Sod1, in a wild-type strain and in a sod1 mutant by DNA microarray analysis. They found that induction by H2O2 of >100 genes related to different responses to oxidative stress was significantly attenuated in the sod1 mutant. Chromatin immunoprecipitation experiments showed binding of Sod1 to the promoters of two stress genes that require Sod1 for their activation but not to the promoter of ACT1, a control gene independent of Sod1. These results suggest an important role for Sod1 as a direct regulator of the transcription of genes related to oxidative stress, and it appears likely that Sod1 acts as a moonlighting protein. It should be noted, however, that it has not yet been established whether the catalytic activity of Sod1 is required for its regulatory function.
A role for Sod1 in the process of glucose repression in S. cerevisiae has also been described by Reddi and Culotta (44), who found that in sod1 mutants, glucose did not repress respiration. Sod1 acts by binding the protein kinases Yck1 and Yck2 and stabilizing them, thus allowing the repression of respiration by glucose. However, their stabilization requires the catalytic activity of Sod1, as it depends on the formation of H2O2; therefore, it is not considered a moonlighting function here (44).
PYRUVATE DECARBOXYLASE REGULATES THE TRANSCRIPTION OF ITS ENCODING GENE
In yeasts able to ferment sugars, pyruvate produced in glycolysis is located at a branching point between fermentative and oxidative metabolism. The enzymes pyruvate decarboxylase (Pdc) and pyruvate dehydrogenase, which initiate the respective metabolic branches, are finely regulated (45). Pdc, in the first step of the fermentative branch, catalyzes de decarboxylation of pyruvate to acetaldehyde. In S. cerevisiae, there are two genes, PDC1 and PDC5, that encode the bulk of the Pdc activity, with PDC1 accounting for about 90% of the protein and PDC5 accounting for about 10% (46). The expression of these genes is controlled by the carbon source available; during growth on glucose, there are high levels of Pdc1, while Pdc5 is almost undetectable (47). Although Schmitt and Zimmermann (48) isolated mutants devoid of pyruvate carboxylase activity that did not grow in glucose, Hohmann and Cederberg (47) found that a pdc1Δ mutant grew in glucose and exhibited elevated levels of Pdc. They showed that this was due to the increased expression of PDC5, which is scarcely expressed in wild-type strains, and called this type of regulation autoregulation. Eberhardt et al. (49) found that the initial mutants isolated by Schmitt and Zimmermann (48) abolished growth in glucose because they expressed a form of Pdc1 that lacked catalytic activity but was able to reduce PDC5 expression. Pdc1 also affected its own transcription, as shown by Liesen et al. (50), who observed that in a strain carrying a PDC1 deletion, the expression of the Escherichia coli lacZ gene under the control of the S. cerevisiae PDC1 promoter was increased 6-fold. They also found that in a mutant with the PDC1 gene deleted, expression of PDC1 under the control of the TPI promoter repressed the expression of PDC5. The results available indicate that autoregulation is due to an action of the Pdc1 protein not related to its catalytic activity. Deletion of PDC5 did not produce similar effects, indicating a specific role for Pdc1.
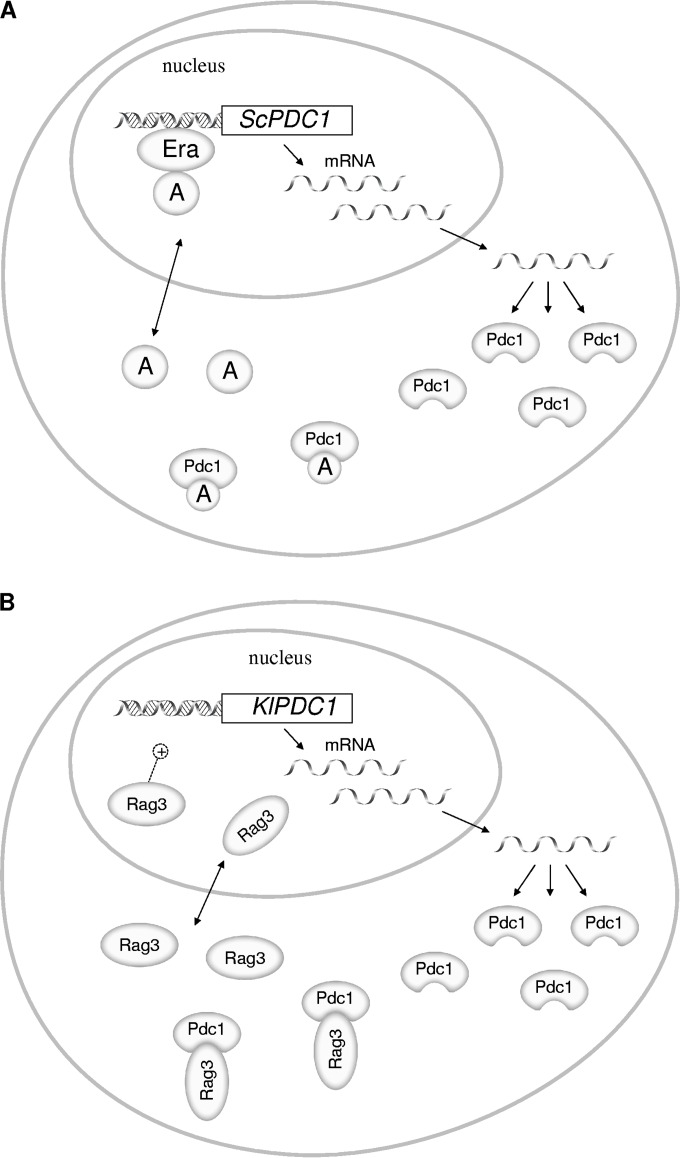
At least two possibilities may be contemplated regarding the mechanism underlying the regulatory role of Pdc1; Pdc1 could act directly as a repressor on the promoters of PDC1 and PDC5, or it may interfere with an activator of the transcription of these genes. Although Pdc1 has been found both in the cytoplasm and in the nuclei of yeast cells (51), there is as yet no evidence of its binding to DNA. Liesen et al. (50) presented a model in which a protein (Era) would interact with the ERA sites of the promoter of PDC1 and with a hypothetical activating protein whose activation capacity would depend on the concentration of Pdc1 (Fig. 1A). The activating protein could be Pdc2, a regulatory protein needed for high PDC1 expression (51). This model has received indirect support from results obtained with the respirofermentative yeast Kluyveromyces lactis (52). K. lactis has a single gene encoding Pdc, KlPDC1, that appears to be also autoregulated since a KlPDC1-lacZ fusion gene is strongly derepressed in a K. lactis mutant devoid of Pdc1 (53). ScPdc1 may replace KlPdc1, and expression of an S. cerevisiae PDC1 gene bearing a point mutation that produces an inactive Pdc1 (54) caused a decrease in KlPdc1 transcription (53), suggesting a mechanism for autoregulation in K. lactis similar to that found in S. cerevisiae. Ottaviano et al. (52), using a K. lactis strain with KlPDC1 deleted and harboring a KlPDC1-lacZ fusion gene, searched for mutants that abolished β-galactosidase production. One such mutant turned out to harbor a mutation in RAG3, a gene encoding a protein with important sequence similarity to the Pdc2 protein of S. cerevisiae (55). A two-hybrid assay showed interaction between Rag3 and KlPdc1, an interaction confirmed by coimmunoprecipitation experiments. Subcellular localization experiments showed that in glucose-grown cultures, KlPdc1 had a cytoplasmic localization and was not detected in the nucleus, while Rag3 showed a uniform distribution in the cell; in the absence of KlPdc1, Rag3 became localized predominantly in the nucleus. Pdc1 itself was not detected in the nucleus. The authors proposed a model (Fig. 1B) in which autoregulation is effected by an interaction between KlPdc1 and Rag3 that would sequester Rag3 in the cytoplasm. Abolition of this interaction will direct Rag3 to the nucleus, allowing increased transcription of KlPdc1 through direct or indirect action on the KlPDC1 promoter (52).
FIG 1.
Scheme for autoregulation of Pdc1 synthesis. (A) In S. cerevisiae, a hypothetical protein, Era, together with an activating protein A (Pdc2?), activates the transcription of PDC1. The Pdc1 protein in the cytosol would sequester the activating protein and decrease PDC1 transcription (50). (B) In K. lactis, transcription of KlPDC1 is activated by the regulatory protein Rag3, an ortholog of Pdc2. When the concentration of Pdc1 is high, most of the Rag3 protein is bound to it and trapped in the cytosol; consequently, the rate of KlPDC1 transcription is low. When Pdc1 levels decrease, Rag3 is liberated and can enter the nucleus, activating KlPDC1 transcription (52).
ZUOTIN, A COCHAPERONE THAT ACTIVATES A TRANSCRIPTION FACTOR
Zuotin was identified in nuclear extracts from S. cerevisiae by Zhang et al. (56) in a search for proteins binding to Z-DNA, the left-handed helix form of DNA. Its name is derived from the word zuo, left in Chinese (56). Shortly thereafter, while looking for tRNA-binding proteins in the nucleus of S. cerevisiae, Wilhelm et al. (57) isolated a protein that turned out to be zuotin and suggested a possible role for it in the processing or transport of tRNA to the cytoplasm. Zuotin has also been found to be associated with ribosomes by centrifugation through sucrose density gradients (58) and to coimmunoprecipitate with Pdr13 (59). Pdr13, also called Ssz1, is a chaperone that enhances the activity of the transcription factor Pdr1, which is involved in pleiotropic drug resistance (PDR) (60, 61). These observations suggest that Zuo1 may play different functional roles, and this is supported by the fact that the protein is found in different cellular compartments and that diverse zuo1 mutants do not present the same phenotype, as described below.
While Zuo1 was first identified in yeast nuclear extracts (56, 57), Yan et al. (58), using a functional Zuo1-GFP fusion, found that Zuo1 had a predominantly cytosolic localization. Later experiments carried out during a study of ribosome biogenesis demonstrated that Zuo1 had both a nuclear and a cytoplasmic localization (62). Deletion of ZUO1 produced a phenotype of slow growth at low temperature and sensitivity to salt, similar to that produced by double deletion of the chaperones Ssb1 and Ssb2. A zuo1 ssb1 ssb2 triple mutant did not present a stronger phenotype, showing that the mutations had no additive effects (58). Ssb proteins are molecular chaperones of the Hsp70 class that act in the early folding steps during protein biogenesis (63). Since the folding process is facilitated by the interaction of chaperones with DnaJ proteins and Zuo1 has the hexapeptide motif KYHPDK, which is also present in the DnaJ protein of E. coli, Yan et al. (58) proposed that Zuo1 could function together with Ssb proteins in the ribosome. They constructed a series of gene truncations and deduced that the charged region of the protein between amino acids 285 and 364 was critical for the interaction with the ribosome and that the C-terminal sequence was not necessary in the assays used.
In relation to another possible function of zuotin, the results of Eisenman and Craig (64) were important. While investigating mechanisms enhancing PDR, they found that, to activate PDR, Ssz1 needed to dissociate from the ribosomes. Following this result and considering that Ssz1 and Zuo1 form a 1:1 complex (65), the authors studied the effect of disruption of the association of Zuo1 with ribosomes on PDR. This was done with a mutant form of Zuo1 with an internal deletion of amino acids 285 to 364 that abolishes this interaction, and it was observed that resistance to antibiotics increased in this mutant. By constructing different partial deletions of ZUO1, it was established that the C-terminal domain of Zuo1 was necessary and sufficient for PDR activation. Using combinations of different mutations in SSZ1 and ZUO1, Eiseman and Craig (64) showed that PDR activation can be produced by Ssz1 or Zuo1 independently and that this activation does not require their chaperone activity.
Molecular details of the activation of Pdr1 by Zuo1 have been worked out by Ducett et al. (60), who showed that simple dissociation of Zuo1 from the ribosome was not enough to produce activation of Pdr1. They found that expression of the C-terminal fragment containing amino acids 365 to 443 allowed yeast cells to resist cycloheximide, a compound extruded from the cell by a protein whose expression is upregulated by Zuo1-activated Pdr1. Since expression of the C-terminal fragment including amino acids 348 to 443 did not allow growth in the presence of cycloheximide, an inhibitory role for a region including amino acids upstream of position 365 was suggested.
Further experiments demonstrated that the fragment including amino acids 348 to 443 formed a four-helix bundle with residues 348 to 364 located in helix 1. In vivo assays showed that resistance to cycloheximide was observed only when constructions bearing mutations that destabilize helix 1 were expressed. It appears, therefore, that Zuo1 in the correctly folded conformation associates with the ribosome and that unfolding produces release from it and the ability to activate Pdr1.
The different results show that zuotin acts as a moonlighting protein; it facilitates protein folding through its interaction with Ssb proteins on the ribosomes and participates in PDR by activating the transcription factor Pdr1. Although it has been suggested that zuotin may process tRNA or transport it to the cytoplasm (57), this idea has not been explored further.
Sdh3 PARTICIPATES IN ELECTRON TRANSFER AND IN MITOCHONDRIAL TRANSLOCASE ASSEMBLY
Succinate dehydrogenase (SDH) participates in the tricarboxylic acid cycle in the oxidation of succinate to fumarate coupled with the transfer of electrons to ubiquinone. SDH is an iron-sulfur flavoprotein located in the mitochondrial inner membrane and is composed of four subunits, Sdh1 to Sdh4. Sdh1 and Sdh2 form the active site and carry the flavin adenine dinucleotide and iron-sulfur clusters, while Sdh3 and Sdh4 are needed for ubiquinone reduction (for a review, see reference 66). Like most mitochondrial proteins, the Sdh subunits are encoded by nuclear genes and synthesized as precursors in the cytosol. Import of cytosolic protein precursors into the corresponding mitochondrial space involves different translocases; specifically, proteins directed to the mitochondrial inner membrane that do not have a cleavable signal peptide are translocated by the TIM22 complex formed by various proteins, among them Tim18 (67). The amino acid sequence of Tim18 shows 39% identity and 58% similarity to that of Sdh4 (66, 68, 69); however, Sdh4 could not substitute for Tim18 (68). Although Sdh3 has no sequence homology with Tim18, during a genetic study of the elements of the TIM22 complex, Gebert et al. (70) found that overexpression of Sdh3 could complement the defect for growth on glycerol of a temperature-sensitive TIM22 mutant at the restrictive temperature. They also showed that in an sdh3 mutant, the levels of different mitochondrial proteins were greatly reduced. In addition, by blue native electrophoresis (71), it was observed that in the mitochondria of sdh3 mutants, the TIM 22 complex was less abundant than in the wild type and showed a lower molecular mass of 250 kDa instead of the 300 kDa of the wild type (70). None of these alterations were observed in an sdh4 mutant.
Import of the Tim18 precursor into mitochondria isolated from an sdh3 mutant was not severely affected, but it could not be assembled into the TIM22 complex. This assembly was possible if the Sdh3 precursor was present in the incubation mixture, thus showing that Sdh3 is required for assembly of Tim18 into the TIM22 complex. Besides, an antibody shift analysis showed Sdh3 bound to Tim18 as an intermediate during the assembly of the TIM22 complex. Assembly of other proteins in the complex was also affected in sdh3 mutants.
Purification of the TIM22 complex by using a tag linked to Tim18 and analysis of the recovered proteins showed the presence of Sdh3 but not that of other SDH subunits (70). Purification of the SDH complex using tagged Sdh4 yielded the components of the SDH complex, with Sdh3 among them. These results indicate that Sdh3 is present in both the SDH and TIM22 complexes, and they were confirmed by further experiments. When mitochondrial proteins separated by blue native electrophoresis were exposed to antibodies to Sdh3, two complexes were identified with sizes of 200 and 300 kDa, corresponding to those of SDH and TIM22, respectively. The functional presence of Sdh3 in two complexes with very different roles, electron transfer and protein assembly, qualifies Sdh3 as a bone fide moonlighting protein.
ENZYMES RELATED TO GLYCOLYSIS THAT BIND PLASMINOGEN OR COMPLEMENT REGULATORS
During the study of different bacterial infections, several proteins well characterized biochemically as cytosolic enzymes were found extracellularly or at the bacterial surface. Although these findings were initially considered artifactual, later research has established that these proteins are part of a network of virulence factors (13, 72). As the study of infections caused by yeasts has progressed, a number of glycolytic enzymes have been found that could have important functions as virulence factors. The extracellular glycolytic enzymes described as moonlighters in yeasts usually act through their binding to plasminogen or to complement regulators.
Enzymes Binding Plasminogen
Plasminogen is a mammalian protein that may be converted to plasmin, a serine protease that participates in different events such as fibrinolysis or degradation of extracellular matrix components (73). Plasminogen is recruited by different pathogens to facilitate host invasion (74), and this appears also to be the case for the opportunistic pathogenic yeast Candida albicans. C. albicans secretes a considerable number of proteins, several of which are involved in the invasion process (14, 75). Many of those proteins are secreted not by the standard endoplasmic reticulum-Golgi pathway but by other nonconventional mechanisms (76, 77). In the cell wall of C. albicans, Crowe et al. (78) identified eight proteins that bound plasminogen with high affinity; among them were five glycolytic enzymes: fructose bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, and alcohol dehydrogenase. Other studies have found that plasminogen can also bind enolase (79) and glycerol-3-phosphate dehydrogenase from C. albicans. The interaction of some of these proteins with plasminogen has been studied in some detail, as described in the following paragraphs.
Enolase.
Enolase catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway. Jong et al. (79) found that C. albicans cells were able to bind plasminogen and plasmin and that purified enolase bound to a column or fixed to microtiter plates also interacted with those proteins. Lysine was a competitive inhibitor of this interaction, indicating that it depended on the lysine-binding domains of plasminogen (80). Besides, binding of plasminogen to enolase increased its affinity for its activator streptokinase, as occurs with its binding to other receptors. In an in vitro blood-brain barrier system, the same authors found that C. albicans cells coated with plasminogen had a 4-fold greater ability to cross that barrier and interpreted this as an indication of the importance of the interaction for infection. These results showed that enolase is a plasminogen receptor in C. albicans, although the presence of other receptors could not be excluded.
Phosphoglycerate mutase.
Phosphoglycerate mutase, Gpm1, is an enzyme that catalyzes the interconversion of 3-phosphoglycerate and 2-phosphoglycerate and participates in both glycolysis and gluconeogenesis. Using a protein microarray from the yeast S. cerevisiae to find proteins able to bind complement regulators, Poltermann et al. (81) identified Gpm1. Since C. albicans Gpm1 had an amino acid sequence about 80% similar to that of the S. cerevisiae enzyme and was among the plasminogen-binding proteins identified by Crowe et al. (78), the characteristics of this protein and its interactions were studied in detail. Using a specific antiserum against C. albicans Gpm1, Poltermann et al. (81) showed by different techniques that the protein was not only present in the cytoplasmic fraction but also on the surface of cells, appearing prominently at the tip of hyphal forms. The Gpm1-plasminogen interaction was inhibited by the lysine analog α-aminocaproic acid, thus implicating lysine-binding domains of plasminogen in this interaction (80). Plasminogen bound to Gpm1 was converted to plasmin when treated with the urokinase-type plasminogen activator, and the plasmin formed could hydrolyze a chromogenic substrate. A strain with a deletion of the GPM1 gene showed a reduction in plasminogen-binding activity (81), and strains with higher Gpm1 expression levels had greater plasminogen-binding activity (82). Gpm1 has another moonlighting role that will be considered in the next section.
Glycerol-3-phosphate dehydrogenase.
Glycerol-3-phosphate dehydrogenase catalyzes a reaction ancillary to the glycolytic pathway, the reduction of dihydroxyacetone phosphate to glycerol-3-phosphate using NADH. In S. cerevisiae, there are two isoenzymes, Gpd1 and Gpd2, that appear to work in different physiological situations (83). During a study of C. albicans proteins that might be involved in complement evasion, Luo et al. (84) identified the homologue of S. cerevisiae Gpd2 as one of them (see the next section). Using a specific antiserum, they showed that Gpd2 was found not only in the cytosol but also at the surface of C. albicans in both the yeast and hyphal morphologies. Using an enzyme-linked immunosorbent assay, these authors demonstrated that plasminogen bound to immobilized Gpd2 and that this binding was inhibited by α-aminocaproic acid, a lysine analog, implicating lysine residues of plasminogen in this interaction, as in the cases mentioned before. Plasminogen bound to Gpd2 could be activated to plasmin by the urokinase-type plasminogen activator, and the plasmin generated was active, cleaving fibrinogen or a synthetic artificial substrate. Like Gpm1, Gpd2 plays another moonlighting role that will be described in the next section.
Enzymes Binding Complement Regulators
The complement system is an important barrier against infective organisms. This system comprises about 30 proteins that participate in a cascade of proteolytic reactions that lead to the assembly of a complex of proteins that form a pore in the pathogen and cause its lysis (85). Several regulators inhibit different reactions in the cascade to avoid attack of host tissues by the proteins of the complement system (86). Among them are factor H (FH) and C4b-binding protein (C4BP), the main fluid-phase inhibitor of the classical and lectin complement pathways (86). Some invading organisms are able to recruit regulators of the complement system of the host and use them to escape from its attack (87). C. albicans cells bind different complement regulators (88, 89), and further studies have identified the sugar transporter Hgt1 and the glycolytic enzymes Gpm1 and Gpd2 as blockers of the action of the complement system through their binding to regulators.
Sugar transporter Hgt1.
To identify possible proteins of C. albicans able to bind the complement regulator FH, Lesiak-Markowicz et al. (90) screened a C. albicans expression library by incubation with serum and anti-FH antibodies. Among the positive colonies, a great number contained DNA with a sequence corresponding to the glucose transporter Hgt1 (91). Homozygous and heterozygous hgt1 mutants were constructed and studied. Using fluorescence microscopy and flow cytometry assays, the authors found that binding of FH to C. albicans was much lower in the homozygous hgt1 mutants than in the parental strain. Reintroduction of the HGT1 gene, encoding Hgt1, restored the binding to levels comparable to those in wild-type cells. In the same work, Lesiak-Markowicz et al. (90) reported that binding of C. albicans cells to another complement regulator, C4BP, was also decreased in the homozygous hgt1 mutant. To obtain evidence about the in vivo significance of those findings, deposition of terminal complement complexes in the wild type, homozygous hgt1 mutants and mutants with the reintroduced gene HGT1 was examined. After incubation of the different cells with human serum, the different populations were examined by flow cytometry. In a homozygous hgt1 mutant, increased deposition of the complement complexes was observed, a result consistent with a role for Hgt1 in blocking complement action.
It is interesting that although there are many genes in C. albicans encoding putative glucose transporters, some of them, such as Hgt2, with an amino acid sequence about 90% identical to that of Hgt1 (92), Lesiak-Markowicz et al. (90) found only HGT1 in their screening. Whether this result indicates a strong specificity for HGT1 or if it could be due to an incomplete representation of the other genes in the library used remains an open question.
Phosphoglycerate mutase.
As stated above, Poltermann et al. (81) observed that the glycolytic protein Gpm1 was also present on the C. albicans cell surface. Using an enzyme-linked immunosorbent assay, these authors showed that Gpm1 was able to bind the complement regulators FH and FHL-1 but not the regulator C4BP. Moreover, they showed that the Gpm1-bound factors retained biological activity, as shown by the appearance of degradation products of the protein C3b after adding C3b together with factor I to immobilized Gpm1 with the attached factors. A homozygous gpm1 mutant showed only a modest reduction in binding to those factors, a result interpreted as indicative of the existence of other binding proteins in the cell. By constructing several deletions in the FHL-1 and FH proteins, Poltermann et al. (81) were able to define the regions of these factors necessary to bind to Gpm1. These proteins contain short consensus repeats (SCRs) of about 60 amino acids (86); while FH uses regions in SCR6 and SCR7 and in SCR19 and SCR20 to bind Gpm1, FHL-1 utilizes only SCR6 and SCR7.
Glycerol-3-phosphate dehydrogenase.
In addition to its plasminogen-binding capacity, Gpd2 is able to bind two proteins, FH and FHL-1, that are downregulators of the complement system (84). After a C. albicans extract was passed through a column with immobilized FH and the retained proteins were eluted, a protein of 52 kDa in the eluate was identified as Gpd2. Binding of Gpd2 to FH and FHL-1 was confirmed by using procedures similar to those described above for Gpm1, and it was also demonstrated that both FH and FHL-1 bound to Gpd2 were active as complement regulators. Using deletion constructs of FH and FHL-1, the authors concluded that these proteins bind to Gpd2 mainly via SCR7.
Hal3 FROM SCHIZOSACCHAROMYCES POMBE IS A MOONLIGHTING PROTEIN AND, IN ADDITION, HAS ANOTHER FUNCTION ORIGINATED BY GENE FUSION
The proteins of S. cerevisiae that participate in the decarboxylation of phosphopantothenoyl-l-cysteine to pantotheine-4-phosphate, one of the steps in the biosynthetic pathway of CoA from pantothenate, have been identified in the first decade of the 21st century (93). A heterotrimer formed by the protein Cab3 and two monomers that can be either Hal3 or Vhs3 is required to catalyze that reaction in budding yeast. Since Hal3 and Vhs3 had been identified previously as inhibitors of S. cerevisiae serine/threonine phosphatases Ppz1 and Ppz2 (94, 95), they qualify as moonlighting proteins (96). The discovery of the function of these proteins in CoA synthesis was followed by the mapping of the functional domains of S. cerevisiae Hal3 (96). Hal3 consists of a core domain of about 250 amino acids with important sequence homology to phosphopantothenoylcysteine decarboxylases (PPCDCs) from other organisms, an N-terminal domain without marked similarity to other proteins and a C-terminal domain rich in acidic residues. While both the N- and C-terminal segments of the protein appear to be important for the inhibition of Ppz1, the core region is essential for Ppz1 binding (96). Some mutations in this region interfered with the inhibitory action on Ppz1 but did not affect the PPCDC activity (95–97), indicating that the distinct functions of the protein do not depend on separate regions of the sequence. Recent results have shown that Hal3 binds Ppz1 as a monomer with a 1:1 stoichiometry and can also readily enter into multimeric complexes, while Vhs3 tends to form stable complexes more resistant to the exchange of monomers with other multimers (98). The authors speculate that the differences in the capacity for oligomer exchange might impinge on the regulation of the moonlighting functions of these proteins.
In an interesting evolutionary study, the same group examined the properties of the corresponding protein in the fission yeast S. pombe (99). Although there has been some controversy about the precise location of S. pombe on the fungal evolutionary tree (100), it is clear that the ancestors of S. cerevisiae and S. pombe separated quite early, with estimations ranging between 420 and 330 million years ago (101). Inspection of the S. pombe genome sequence showed only a putative gene (SPAC15E1.04) whose N-terminal region showed about 40% sequence homology to ScHAL3. Surprisingly, this gene also showed a 3′ fragment with sequence homology to thymidylate synthase-encoding genes. The authors resequenced the genomic locus of S. pombe and that of the related yeast Schizosaccharomyces japonicus and confirmed the unusual structure of the gene in both cases (99). The same authors also showed that the S. pombe Hal3 protein or even its N-terminal domain could rescue an S. cerevisiae cab3 single mutant or hal3 vhs3 double mutant, indicating that this protein is sufficient to achieve PPCDC activity. The protein was also able to inhibit the in vitro activity of S. cerevisiae Ppz1 or the corresponding S. pombe homolog Pzh1. In addition, Hal3 from S. pombe or its C-terminal domain allowed the growth of a cdc21 mutant of S. cerevisiae (99) whose phenotype is due to a lack of thymidylate synthase. Thus, Hal3 from S. pombe has moonlighting activity and is also a “bifunctional” enzyme that acquired a completely unrelated activity, probably by gene fusion; according to Molero et al. (99), such fusion events are rare in S. pombe, and only 12 were found by using a dedicated algorithm.
What is the situation of PPCDCs in other yeasts? Exploration of genomic sequences within the hemiascomycetaceous yeasts suggests the existence of heterotrimeric PPCDCs with two variants. In the yeast group derived from an ancestor with a duplicated genome (102, 103), to which S. cerevisiae belongs, the enzyme would be formed by one Cab3 subunit plus two units of Hal3 and/or Vhs3. In archiascomycetaceous yeasts, to which S. pombe belongs, a homotrimeric protein formed by a Hal3-like protein is found (93). This diversity is conditioned by the structure of the active enzyme. PPCDCs are trimers in which the catalytic sites form at the interfaces between the different subunits. Specific cysteine, histidine, and asparagine residues are required for catalysis. These residues are provided by a single protein in homotrimeric PPCDCs. In S. cerevisiae, the functional histidine residue is provided by Hal3 or Vhs3 and the cysteine and asparagine residues are provided by Cab3, while in other yeasts, the residues are provided by Cab3 and Hal3- or Vhs3-like proteins. A protein with PPCDC activity similar to that of the N-terminal domain of SpHal3 could have been the primitive structure from which the others originated. This original form may have already possessed the moonlighting capacity.
FINAL REMARKS AND PERSPECTIVES
In this article, we have considered the experimental evidence that qualifies as moonlighting proteins a number of yeast proteins that were not identified as such in previous reviews. Their disparate canonical functions and the wide range of moonlighting activities are astonishing and show the power of evolution to produce new functions from extant materials. The biochemical and genetic potentialities of yeasts emphasize their importance in studying moonlighting functions, not only of yeast proteins but also of proteins from other organisms, as shown by Simonicova et al. (104), who used S. cerevisiae to study extratelomeric functions of mammalian telomerase.
Mutational Analysis and Structural Studies
Results showing the participation of a protein in two dissimilar functions might be a promising hint at a possible moonlighting protein. However, this is not always the case, as the noncanonical activity may be a secondary effect from a metabolic defect. An example of this is fumarase, a metabolic enzyme from the tricarboxylic acid cycle, whose absence may increase intracellular levels of fumarate, making yeast cells more sensitive to stresses producing double-strand breaks in DNA (105, 106). Another example is the participation of Sod1 in glucose repression of respiration in yeast, which depends on the catalytic activity of the protein (44).
An unequivocal identification of a protein as moonlighting would ideally require mutational studies. Several types of mutation could be expected in that type of protein, some affecting both the canonical and moonlighting functions, others located in the catalytic region and affecting only the canonical activity, and still others affecting only the moonlighting region and yielding a metabolically unaltered phenotype. Examples of the different types of mutations are found for yeast hexokinase (107), galactokinase (108), and some other yeast proteins (18), as well as for mammalian aldolase (109). It has been reported recently that a glyceraldehyde-3-phosphodehydrogenase may play a moonlighting role in S. cerevisiae (110). The isoenzyme Tdh3 has been shown to interact with the sirtuin Sir2 and to facilitate transcriptional silencing at the telomeres. In this work, a number of tdh3 mutants have been used and it has been observed that there is no correlation between glyceraldehyde-3-phosphodehydrogenase activity and the degree of silencing at a telomere. This is a strong indication that the moonlighting effect is not a side effect of the metabolic activity.
In some clear cases of moonlighting, such as the pyruvate carboxylase of H. polymorpha, only mutant proteins that have lost their moonlighting activity have been obtained (19). Mutational analysis of the Tps1 (trehalose-6-phosphate synthase) protein of S. cerevisiae suggests that it may be a moonlighting protein. This protein produces a regulator of glycolysis (111, 112) and appears to be implicated, independently of its metabolic activity, in resistance to different stresses (113). Reinforcing its possible moonlighting qualification is the finding that, in the rice blast fungus Magnaporthe grisea (now oryzae), the Tps1 protein itself, and not its catalytic activity, is required for efficient plant infection (114).
Mutational studies would be critical to ascertain if the moonlighting umbrella could apply to all proteins that, although usually cytosolic, are also found extracellularly and act in this form to facilitate invasion of the host by pathogenic organisms (13, 115). A detailed structural study has been made for the surface enolase of Streptococcus pyogenes and S. pneumoniae. Enolase is a glycolytic enzyme found to be able to bind to plasminogen. Derbise et al. (116) constructed different mutant forms of the enolase from S. pyogenes affected in the C-terminal region that retained normal enzymatic activity but exhibited decreased plasminogen-binding activity. Cork et al. (117) obtained a series of mutations located on the surface of the octameric protein of S. pneumoniae; some of them conserved activity but had lost the capacity to bind to plasminogen. Mutant proteins binding plasminogen but devoid of catalytic activity were not isolated.
A puzzling issue regarding proteins occasionally found extracellularly is how they reach the cell surface in the absence of a conspicuous secretion signal. Different structural characteristics appear to be involved in the process, as demonstrated by the work on the glyceraldehyde-3-phosphate dehydrogenase of S. pyogenes and on the enolase of Bacillus subtilis. Addition of a C-terminal hydrophobic tail to the glyceraldehyde-3-phosphate dehydrogenase retained it in the cytosol without affecting its enzymatic activity and the strain carrying it lost many of its adhering activities (118). Deletion of an α-helix stretch from the terminal region of the enolase resulted in a protein that was not exported; replacement of that stretch with a segment with α-helix structure from β-galactosidase restored the exportation of the protein (119).
Databases and Prediction of Moonlighting Functions
An important development since the appearance of previous reviews has been the increased use of powerful informatic tools in the moonlighting field. This could change the way in which moonlighting proteins are discovered. The development of databases specifically devoted to moonlighting proteins will increase the accessibility of data on such proteins and facilitate the tracking of new additions to this category. Two such databases have recently been made available, Moonprot (5) and MultitaskProtDB (17). When it appeared, the first one, searchable at http://www.moonlightingproteins.org, presented information on more than 200 moonlighting proteins experimentally verified to behave as such; at its publication, the second one, accessible at http://wallace.uab.es/multitask, offered different information on about 300 multitasking proteins. Critical for the usefulness of databases is the quality of annotations; care should be taken to include in them only well-authenticated cases of moonlighting, complying with the criteria defining this type of protein. Discrimination among moonlighting, pleiotropy, and secondary metabolic effects is crucial to avoid the incorporation of inadequate proteins to the databases.
Prediction of moonlighting activity is an outstanding pending task. Homology comparison is not enough, because if the moonlighting function depends on small structural changes, the global identity score may be uninformative. Attempts at and advances in moonlighting prediction have been reviewed by Khan and Kihara (120). Using Gene Ontology annotations, protein-protein interaction networks, and other omics data, Khan et al. (121) were able to predict moonlighting functions for several E. coli proteins, a prediction confirmed by results in the literature. Chapple et al. (122) used protein-protein interaction networks to predict proteins with multiple functions. They coined the term “extreme multifunctional proteins” for them and proposed that moonlighting proteins could be seen as a subset of the extreme multifunctional protein set. Using protein network information, together with current protein annotations, they identified 430 proteins in the human interactome as candidates for extreme multifunctional proteins. Among them, they looked for 39 known moonlighting proteins and found only 6. They attributed this low score to a lack of adequate information in the annotations. However, an alternative explanation could be that many moonlighting proteins do not interact with multiple proteins and therefore would not form part of the set of extreme multifunctional proteins.
Evolution of Moonlighting Proteins
The question of how moonlighting functions arose in the course of evolution is a matter of speculation, although some hints may be advanced. Because the canonical activity of many proteins is conserved between related yeast species but moonlighting activities are not, it may be assumed that they have appeared at different moments during evolution. Also, the fact that a multitude of similar enzymes perform similar reactions in disparate organisms while the moonlighting activities of the corresponding enzymes are observed in only some of them seems consistent with the previous idea.
Moonlighting proteins belong to a category different from that of pseudoenzymes, proteins whose sequences are almost identical to those of active proteins but that do not catalyze the corresponding chemical reactions; they have been shown to have regulatory functions in a number of cases (123–125). It appears likely that pseudoenzymes evolved from a protein with enzymatic activity and then lost this activity while acquiring a regulatory capacity. Nevertheless, it may be conjectured that in some cases the ancestor of a pseudoenzyme might have been a moonlighting protein that lost its enzymatic activity and conserved a regulatory function. It is important to consider that the acquisition of an additional function by a protein requires a tradeoff between improvement of the new function and conservation of the existing function. The appearance of a new function may take advantage of structural features involved in the canonical reaction. A possible example is the Pet54 protein of S. cerevisiae. The lack of Pet54 affects both the translation of the COX3 mRNA and the splicing of an intron from the COX1 pre-mRNA, each function requiring a different protein domain within Pet54 (126). Kaspar et al. (127) have shown that Pet54 is able to bind to regions in the 5′ untranslated leader of the COX3 mRNA and in the intron of COX1 pre-mRNA which show 56% sequence similarity, and that the binding involves the same surfaces of Pet54. These authors suggested that the original functions of Pet54 were to bind COX3 mRNA and to recruit Pet122 and Pet494 to form a complex to activate mRNA translation. Later on, the capacity of Pet54 to bind the intron region of COX1 pre-mRNA may have allowed the selection of variants of Pet54 able to participate in intron splicing.
In some cases, changes in the functional properties of a protein need to follow what may appear to be a tortuous trajectory, as shown by Harms and Thornton (128) in a study of the evolution of glucocorticoid receptor specificity. They discussed how, for an ancestral glucocorticoid receptor to acquire specificity for cortisol, several modifications of the protein that did not appear to modify its function were necessary before the protein could tolerate the mutations that conferred cortisol specificity. Those mutations were needed to stabilize the protein scaffold to allow the ulterior structural changes.
It should also be taken into account that the appearance of a moonlighting function may be influenced by the existence, and eventual evolution, of a partner with which to interact. An example may be the moonlighting function of pyruvate carboxylase in the assembly of methanol oxidase (129). Only pyruvate carboxylases from methylotrophic yeasts exhibit this capacity, while the enzymes from nonmethylotrophic yeasts, in spite of their great sequence similarity, lack this ability. This is probably due to the lack of selective pressure in the absence of a partner with which to interact during their evolutionary history.
Experiments to generate proteins with moonlighting properties and to infer from them the original evolutionary pathway of the naturally evolved proteins may provide insights into the regions of the protein important for the appearance of the moonlighting function (130). Nevertheless, it could be problematic to infer from results of this type of experiment the original evolutionary pathway of the naturally evolved protein. As Koschwanez et al. (131) have pointed out, results from an engineering strategy might not reflect the evolutionary pathway exactly, as other constraints may have been at work that are not mimicked in the engineered process. But even with this caveat, that type of experiment could provide hints about possible evolutionary events leading to the appearance of moonlighting functions. Jeffery (20) has pointed out that information derived from such experiments may also be useful in the design of proteins with new functions.
Some Unanswered General Questions
There are several general questions about moonlighting, not specific to yeasts, that are worth considering. One of them is how moonlighting proteins reach their partners and how they discriminate among the different tasks. Interactions of the proteins will be controlled by the intrinsic properties of the corresponding proteins and by changes in the metabolite landscape that determine their actual configuration. Modifications in folding in response to a ligand might provide opportunities to show new properties. An illustrative example of this has been provided by Ha et al. (132). They constructed a chimeric protein resulting from the fusion of a barnase and the Gcn4 DNA-binding domain in a way that prevents the fragments from being simultaneously folded and active. If a specific fragment of DNA is bound to the Gcn4 part, barnase unfolds and becomes inactive, while in the absence of DNA, the barnase domain is folded and active.
In a certain number of cases, moonlighting functions require movement of the protein to another cellular compartment. The moonlighting action of several proteins, such as Lys20, Sod1, and Zuo1, takes place in the nucleus. The increasing number of mitochondrial proteins also found in the nucleus suggests that some of them may also have moonlighting roles and participate in regulatory circuits (133). The translocation process of moonlighting proteins has not been studied in detail, except in the case of hexokinase 2, a metabolic enzyme that moonlights as a transcriptional repressor (22). Moreno and Herrero have discovered that the localization of the protein is determined by the phosphorylation state of a serine residue that, in turn, depends on the activities of the protein kinase Snf1 and the protein phosphatase Glc7 (134). It has also been reported that the phosphorylation of that serine residue is blocked in a mutant devoid of the protein kinase Tda1 (135). In addition, the traffic of hexokinase 2 between the cytoplasm and the nucleus implies the importins Kap60/Kap95 and the exportin Xpo1 (Crm1) (134).
Moonlighting action may or may not affect the canonical activity of the protein; binding of galactokinase to the inhibitory protein Gal80 blocks galactokinase activity (136), but binding of arginase to ornithine transcarbamylase does not abolish arginase activity (137).
CONCLUSIONS
Since the moonlighting protein concept was established, realization of its importance has been slowly but steadily permeating the general biochemical knowledge. As moonlighting proteins participate in diverse cellular functions and receive signals from molecules involved in different pathways, they might provide cross talk between different cellular processes, facilitating the cells' response to changes in their environment. In spite of the advances in the knowledge of moonlighting proteins, important gaps remain to be filled to understand their intimate action mechanisms and their regulation.
The implication of moonlighting proteins from pathogenic bacteria in invasive processes has added a new dimension to this class of proteins, and the finding that some pathogenic yeasts might use moonlighting proteins to help invasion extends these observations.
The different processes in which moonlighting proteins participate, not only in yeasts but also in other organisms, including higher eukaryotes, concern fields as diverse as medicine or biotechnology, making it important to widen and deepen the current knowledge about these molecules and their modes of action.
ACKNOWLEDGMENTS
We thank the reviewers for helpful suggestions and J. Ariño (Universitat Autònoma de Barcelona, Barcelona, Spain) for critical reading of the section on Hal3.
This work was supported by grant BFU2010-19628-C02-02 from the Spanish Ministry of Economy and Competitivity (MINECO).
REFERENCES
- 1.Lehninger AL. 1976. Biochemistry. Worth Publishers, Inc., New York, NY. [Google Scholar]
- 2.Piatigorsky J, O'Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF. 1988. Gene sharing by delta-crystallin and argininosuccinate lyase. Proc Natl Acad Sci U S A 85:3479–3483. doi: 10.1073/pnas.85.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piatigorsky J, Wistow GJ. 1989. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell 57:197–199. doi: 10.1016/0092-8674(89)90956-2. [DOI] [PubMed] [Google Scholar]
- 4.Piatigorsky J. 1992. Lens crystallins. Innovation associated with changes in gene regulation. J Biol Chem 267:4277–4280. [PubMed] [Google Scholar]
- 5.Mani M, Chen C, Amblee V, Liu H, Mathur T, Zwicke G, Zabad S, Patel B, Thakkar J, Jeffery CJ. 2015. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res 43:D277–D282. doi: 10.1093/nar/gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson M, Botstein D. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 7.Vivier MA, Sollitti P, Pretorius IS. 1999. Functional analysis of multiple AUG codons in the transcripts of the STA2 glucoamylase gene from Saccharomyces cerevisiae. Mol Gen Genet 261:11–20. doi: 10.1007/s004380050936. [DOI] [PubMed] [Google Scholar]
- 8.Freire-Picos MA, Lombardia-Ferreira LJ, Ramil E, Gonzalez-Dominguez M, Cerdan ME. 2001. The KlCYC1 gene, a downstream region for two differentially regulated transcripts. Yeast 18:1347–1355. doi: 10.1002/yea.787. [DOI] [PubMed] [Google Scholar]
- 9.Juneau K, Nislow C, Davis RW. 2009. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183:185–194. doi: 10.1534/genetics.109.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffery CJ. 1999. Moonlighting proteins. Trends Biochem Sci 24:8–11. doi: 10.1016/S0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 11.Chapple CE, Brun C. 2015. Redefining protein moonlighting. Oncotarget 6:16812–16813. doi: 10.18632/oncotarget.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffery CJ. 2014. An introduction to protein moonlighting. Biochem Soc Trans 42:1679–1683. doi: 10.1042/BST20140226. [DOI] [PubMed] [Google Scholar]
- 13.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorgo AG, Heilmann CJ, Brul S, de Koster CG, Klis FM. 2013. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett 338:10–17. doi: 10.1111/1574-6968.12049. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, Chen H, Zhang H, Chen W. 2014. The roles of moonlighting proteins in bacteria. Curr Issues Mol Biol 16:15–22. [PubMed] [Google Scholar]
- 16.Freedman R. 1978. Moonlighting molecules. New Sci 79:560–561. [Google Scholar]
- 17.Hernández S, Ferragut G, Amela I, Perez-Pons J, Piñol J, Mozo-Villarias A, Cedano J, Querol E. 2014. MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res 42:D517–D520. doi: 10.1093/nar/gkt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gancedo C, Flores CL. 2008. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev 72:197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huberts DH, Venselaar H, Vriend G, Veenhuis M, van der Klei IJ. 2010. The moonlighting function of pyruvate carboxylase resides in the non-catalytic end of the TIM barrel. Biochim Biophys Acta 1803:1038–1042. doi: 10.1016/j.bbamcr.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Jeffery CJ. 2015. Why study moonlighting proteins? Front Genet 6:211. doi: 10.3389/fgene.2015.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM. 2005. Single-gene disorders: what role could moonlighting enzymes play? Am J Hum Genet 76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores CL, Gancedo C. 2011. Unraveling moonlighting functions with yeasts. IUBMB Life 63:457–462. doi: 10.1002/iub.454. [DOI] [PubMed] [Google Scholar]
- 23.Tucci AF, Ceci LN. 1972. Homocitrate synthase from yeast. Arch Biochem Biophys 153:742–750. doi: 10.1016/0003-9861(72)90393-1. [DOI] [PubMed] [Google Scholar]
- 24.Ramos F, Verhasselt P, Feller A, Peeters P, Wach A, Dubois E, Volckaert G. 1996. Identification of a gene encoding a homocitrate synthase isoenzyme of Saccharomyces cerevisiae. Yeast 12:1315–1320. [DOI] [PubMed] [Google Scholar]
- 25.Feller A, Ramos F, Pierard A, Dubois E. 1999. In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur J Biochem 261:163–170. doi: 10.1046/j.1432-1327.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- 26.Quezada H, Aranda C, DeLuna A, Hernández H, Calcagno ML, Marin-Hernández A, González A. 2008. Specialization of the paralogue LYS21 determines lysine biosynthesis under respiratory metabolism in Saccharomyces cerevisiae. Microbiology 154:1656–1667. doi: 10.1099/mic.0.2008/017103-0. [DOI] [PubMed] [Google Scholar]
- 27.Jones EW, Fink GR. 1982. Regulation of amino acid and nucleotide biosynthesis in yeast, p 181–299. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 28.Chen S, Brockenbrough JS, Dove JE, Aris JP. 1997. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J Biol Chem 272:10839–10846. doi: 10.1074/jbc.272.16.10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott EM, Pillus L. 2010. Homocitrate synthase connects amino acid metabolism to chromatin functions through Esa1 and DNA damage. Genes Dev 24:1903–1913. doi: 10.1101/gad.1935910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A 95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pommier Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 32.Torres-Machorro AL, Aris JP, Pillus L. 2015. A moonlighting metabolic protein influences repair at DNA double-stranded breaks. Nucleic Acids Res 43:1646–1658. doi: 10.1093/nar/gku1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Attikum H, Fritsch O, Gasser SM. 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamburini BA, Tyler JK. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeber A, Hauer M, Gasser SM. 2013. Nucleosome remodelers in double-strand break repair. Curr Opin Genet Dev 23:174–184. doi: 10.1016/j.gde.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Carter CD, Kitchen LE, Au WC, Babic CM, Basrai MA. 2005. Loss of SOD1 and LYS7 sensitizes Saccharomyces cerevisiae to hydroxyurea and DNA damage agents and downregulates MEC1 pathway effectors. Mol Cell Biol 25:10273–10285. doi: 10.1128/MCB.25.23.10273-10285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longo VD, Liou LL, Valentine JS, Gralla EB. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys 365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- 38.Sehati S, Clement MH, Martins J, Xu L, Longo VD, Valentine JS, Gralla EB. 2011. Metabolic alterations in yeast lacking copper-zinc superoxide dismutase. Free Radic Biol Med 50:1591–1598. doi: 10.1016/j.freeradbiomed.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturtz LA, Culotta VC. 2002. Superoxide dismutase null mutants of baker's yeast, Saccharomyces cerevisiae. Methods Enzymol 349:167–172. doi: 10.1016/S0076-6879(02)49332-9. [DOI] [PubMed] [Google Scholar]
- 40.Wawryn J, Krzepilko A, Myszka A, Bilinski T. 1999. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim Pol 46:249–253. [PubMed] [Google Scholar]
- 41.Nunoshiba T, Demple B. 1993. Potent intracellular oxidative stress exerted by the carcinogen 4-nitroquinoline-N-oxide. Cancer Res 53:3250–3252. [PubMed] [Google Scholar]
- 42.Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XF. 2014. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun 5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 44.Reddi AR, Culotta VC. 2013. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 152:224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pronk JT, Yde Steensma H, Van Dijken JP. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Hohmann S. 1997. Pyruvate decarboxylases, p 187–212. In Zimmermann FK, Entian KD (ed), Yeast sugar metabolism: biochemistry, genetics, biotechnology and applications. Technomic Publishing Co., Inc., Lancaster, PA. [Google Scholar]
- 47.Hohmann S, Cederberg H. 1990. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem 188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt HD, Zimmermann FK. 1982. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol 151:1146–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eberhardt I, Cederberg H, Li H, Konig S, Jordan F, Hohmann S. 1999. Autoregulation of yeast pyruvate decarboxylase gene expression requires the enzyme but not its catalytic activity. Eur J Biochem 262:191–201. doi: 10.1046/j.1432-1327.1999.00370.x. [DOI] [PubMed] [Google Scholar]
- 50.Liesen T, Hollenberg CP, Heinisch JJ. 1996. ERA, a novel cis-acting element required for autoregulation and ethanol repression of PDC1 transcription in Saccharomyces cerevisiae. Mol Microbiol 21:621–632. doi: 10.1111/j.1365-2958.1996.tb02570.x. [DOI] [PubMed] [Google Scholar]
- 51.Mojzita D, Hohmann S. 2006. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol Genet Genomics 276:147–161. doi: 10.1007/s00438-006-0130-z. [DOI] [PubMed] [Google Scholar]
- 52.Ottaviano D, Micolonghi C, Tizzani L, Lemaire M, Wesolowski-Louvel M, De Stefano ME, Ranieri D, Bianchi MM. 2014. Autoregulation of the Kluyveromyces lactis pyruvate decarboxylase gene KlPDC1 involves the regulatory gene RAG3. Microbiology 160:1369–1378. doi: 10.1099/mic.0.078543-0. [DOI] [PubMed] [Google Scholar]
- 53.Destruelle M, Menghini R, Frontali L, Bianchi M. 1999. Regulation of the expression of the Kluyveromyces lactis PDC1 gene: carbon source responsive elements and autoregulation. Yeast 15:361–370. [DOI] [PubMed] [Google Scholar]
- 54.Schaaff I, Green JB, Gozalbo D, Hohmann S. 1989. A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet 15:75–81. doi: 10.1007/BF00435452. [DOI] [PubMed] [Google Scholar]
- 55.Prior C, Tizzani L, Fukuhara H, Wesolowski-Louvel M. 1996. RAG3 gene and transcriptional regulation of the pyruvate decarboxylase gene in Kluyveromyces lactis. Mol Microbiol 20:765–772. doi: 10.1111/j.1365-2958.1996.tb02515.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. 1992. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J 11:3787–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilhelm ML, Reinbolt J, Gangloff J, Dirheimer G, Wilhelm FX. 1994. Transfer RNA binding protein in the nucleus of Saccharomyces cerevisiae. FEBS Lett 349:260–264. doi: 10.1016/0014-5793(94)00683-0. [DOI] [PubMed] [Google Scholar]
- 58.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michimoto T, Aoki T, Toh-e A, Kikuchi Y. 2000. Yeast Pdr13p and Zuo1p molecular chaperones are new functional Hsp70 and Hsp40 partners. Gene 257:131–137. doi: 10.1016/S0378-1119(00)00381-4. [DOI] [PubMed] [Google Scholar]
- 60.Ducett JK, Peterson FC, Hoover LA, Prunuske AJ, Volkman BF, Craig EA. 2013. Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. J Mol Biol 425:19–31. doi: 10.1016/j.jmb.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hallstrom TC, Katzmann DJ, Torres RJ, Sharp WJ, Moye-Rowley WS. 1998. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol Cell Biol 18:1147–1155. doi: 10.1128/MCB.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albanèse V, Reissmann S, Frydman J. 2010. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol 189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisenman HC, Craig EA. 2004. Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol Microbiol 53:335–344. doi: 10.1111/j.1365-2958.2004.04134.x. [DOI] [PubMed] [Google Scholar]
- 65.Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci U S A 98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lemire BD, Oyedotun KS. 2002. The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553:102–116. doi: 10.1016/S0005-2728(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 67.Kutik S, Guiard B, Meyer HE, Wiedemann N, Pfanner N. 2007. Cooperation of translocase complexes in mitochondrial protein import. J Cell Biol 179:585–591. doi: 10.1083/jcb.200708199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kerscher O, Sepuri NB, Jensen RE. 2000. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol Biol Cell 11:103–116. doi: 10.1091/mbc.11.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koehler CM, Murphy MP, Bally NA, Leuenberger D, Oppliger W, Dolfini L, Junne T, Schatz G, Or E. 2000. Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol Cell Biol 20:1187–1193. doi: 10.1128/MCB.20.4.1187-1193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gebert N, Gebert M, Oeljeklaus S, von der Malsburg K, Stroud DA, Kulawiak B, Wirth C, Zahedi RP, Dolezal P, Wiese S, Simon O, Schulze-Specking A, Truscott KN, Sickmann A, Rehling P, Guiard B, Hunte C, Warscheid B, van der Laan M, Pfanner N, Wiedemann N. 2011. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell 44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 71.Schägger H, von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199:223–231. doi: 10.1016/0003-2697(91)90094-A. [DOI] [PubMed] [Google Scholar]
- 72.Henderson B. 2014. An overview of protein moonlighting in bacterial infection. Biochem Soc Trans 42:1720–1727. doi: 10.1042/BST20140236. [DOI] [PubMed] [Google Scholar]
- 73.Castellino FJ, Ploplis VA. 2005. Structure and function of the plasminogen/plasmin system. Thromb Haemost 93:647–654. [DOI] [PubMed] [Google Scholar]
- 74.Sanderson-Smith ML, De Oliveira DM, Ranson M, McArthur JD. 2012. Bacterial plasminogen receptors: mediators of a multifaceted relationship. J Biomed Biotechnol 2012:272148. doi: 10.1155/2012/272148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gil-Bona A, Llama-Palacios A, Parra CM, Vivanco F, Nombela C, Monteoliva L, Gil C. 2015. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J Proteome Res 14:142–153. doi: 10.1021/pr5007944. [DOI] [PubMed] [Google Scholar]
- 77.Nombela C, Gil C, Chaffin WL. 2006. Non-conventional protein secretion in yeast. Trends Microbiol 14:15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 78.Crowe JD, Sievwright IK, Auld GC, Moore NR, Gow NA, Booth NA. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol Microbiol 47:1637–1651. doi: 10.1046/j.1365-2958.2003.03390.x. [DOI] [PubMed] [Google Scholar]
- 79.Jong AY, Chen SH, Stins MF, Kim KS, Tuan TL, Huang SH. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J Med Microbiol 52:615–622. doi: 10.1099/jmm.0.05060-0. [DOI] [PubMed] [Google Scholar]
- 80.Plow EF, Doeuvre L, Das R. 2012. So many plasminogen receptors: why? J Biomed Biotechnol 2012:141806. doi: 10.1155/2012/141806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poltermann S, Kunert A, von der Heide M, Eck R, Hartmann A, Zipfel PF. 2007. Gpm1p is a factor H-, FHL-1-, and plasminogen-binding surface protein of Candida albicans. J Biol Chem 282:37537–37544. doi: 10.1074/jbc.M707280200. [DOI] [PubMed] [Google Scholar]
- 82.Luo S, Hipler UC, Munzberg C, Skerka C, Zipfel PF. 2015. Sequence variations and protein expression levels of the two immune evasion proteins Gpm1 and Pra1 influence virulence of clinical Candida albicans isolates. PLoS One 10:e0113192. doi: 10.1371/journal.pone.0113192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee YJ, Jeschke GR, Roelants FM, Thorner J, Turk BE. 2012. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol Cell Biol 32:4705–4717. doi: 10.1128/MCB.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo S, Hoffmann R, Skerka C, Zipfel PF. 2013. Glycerol-3-phosphate dehydrogenase 2 is a novel factor H-, factor H-like protein 1-, and plasminogen-binding surface protein of Candida albicans. J Infect Dis 207:594–603. doi: 10.1093/infdis/jis718. [DOI] [PubMed] [Google Scholar]
- 85.Sarma JV, Ward PA. 2011. The complement system. Cell Tissue Res 343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lindahl G, Sjobring U, Johnsson E. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr Opin Immunol 12:44–51. doi: 10.1016/S0952-7915(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 87.Kraiczy P, Wurzner R. 2006. Complement escape of human pathogenic bacteria by acquisition of complement regulators. Mol Immunol 43:31–44. doi: 10.1016/j.molimm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 88.Meri T, Blom AM, Hartmann A, Lenk D, Meri S, Zipfel PF. 2004. The hyphal and yeast forms of Candida albicans bind the complement regulator C4b-binding protein. Infect Immun 72:6633–6641. doi: 10.1128/IAI.72.11.6633-6641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meri T, Hartmann A, Lenk D, Eck R, Wurzner R, Hellwage J, Meri S, Zipfel PF. 2002. The yeast Candida albicans binds complement regulators factor H and FHL-1. Infect Immun 70:5185–5192. doi: 10.1128/IAI.70.9.5185-5192.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lesiak-Markowicz I, Vogl G, Schwarzmuller T, Speth C, Lass-Florl C, Dierich MP, Kuchler K, Wurzner R. 2011. Candida albicans Hgt1p, a multifunctional evasion molecule: complement inhibitor, CR3 analogue, and human immunodeficiency virus-binding molecule. J Infect Dis 204:802–809. doi: 10.1093/infdis/jir455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Varma A, Singh BB, Karnani N, Lichtenberg-Frate H, Hofer M, Magee BB, Prasad R. 2000. Molecular cloning and functional characterisation of a glucose transporter, CaHGT1, of Candida albicans. FEMS Microbiol Lett 182:15–21. doi: 10.1111/j.1574-6968.2000.tb08866.x. [DOI] [PubMed] [Google Scholar]
- 92.Fan J, Chaturvedi V, Shen SH. 2002. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J Mol Evol 55:336–346. doi: 10.1007/s00239-002-2330-4. [DOI] [PubMed] [Google Scholar]
- 93.Ruiz A, González A, Muñoz I, Serrano R, Abrie JA, Strauss E, Ariño J. 2009. Moonlighting proteins Hal3 and Vhs3 form a heteromeric PPCDC with Ykl088w in yeast CoA biosynthesis. Nat Chem Biol 5:920–928. doi: 10.1038/nchembio.243. [DOI] [PubMed] [Google Scholar]
- 94.de Nadal E, Clotet J, Posas F, Serrano R, Gómez N, Ariño J. 1998. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc Natl Acad Sci U S A 95:7357–7362. doi: 10.1073/pnas.95.13.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruiz A, Muñoz I, Serrano R, González A, Simon E, Ariño J. 2004. Functional characterization of the Saccharomyces cerevisiae VHS3 gene: a regulatory subunit of the Ppz1 protein phosphatase with novel, phosphatase-unrelated functions. J Biol Chem 279:34421–34430. doi: 10.1074/jbc.M400572200. [DOI] [PubMed] [Google Scholar]
- 96.Abrie JA, González A, Strauss E, Ariño J. 2012. Functional mapping of the disparate activities of the yeast moonlighting protein Hal3. Biochem J 442:357–368. doi: 10.1042/BJ20111466. [DOI] [PubMed] [Google Scholar]
- 97.Muñoz I, Ruiz A, Marquina M, Barcelo A, Albert A, Ariño J. 2004. Functional characterization of the yeast Ppz1 phosphatase inhibitory subunit Hal3: a mutagenesis study. J Biol Chem 279:42619–42627. doi: 10.1074/jbc.M405656200. [DOI] [PubMed] [Google Scholar]
- 98.Abrie JA, Molero C, Ariño J, Strauss E. 2015. Complex stability and dynamic subunit interchange modulates the disparate activities of the yeast moonlighting proteins Hal3 and Vhs3. Sci Rep 5:15774. doi: 10.1038/srep15774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molero C, Petrenyi K, González A, Carmona M, Gelis S, Abrie JA, Strauss E, Ramos J, Dombradi V, Hidalgo E, Ariño J. 2013. The Schizosaccharomyces pombe fusion gene hal3 encodes three distinct activities. Mol Microbiol 90:367–382. doi: 10.1111/mmi.12370. [DOI] [PubMed] [Google Scholar]
- 100.Kuramae EE, Robert V, Snel B, Boekhout T. 2006. Conflicting phylogenetic position of Schizosaccharomyces pombe. Genomics 88:387–393. doi: 10.1016/j.ygeno.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 101.Sipiczki M. 2000. Where does fission yeast sit on the tree of life? Genome Biol 1:REVIEWS1011. doi: 10.1186/gb-2000-1181-1182-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kellis M, Birren BW, Lander ES. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 103.Marcet-Houben M, Gabaldón T. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker's yeast lineage. PLoS Biol 13:e1002220. doi: 10.1371/journal.pbio.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simonicova L, Dudekova H, Ferenc J, Prochazkova K, Nebohacova M, Dusinsky R, Nosek J, Tomaska L. 2015. Saccharomyces cerevisiae as a model for the study of extranuclear functions of mammalian telomerase. Curr Genet 61:517–527. doi: 10.1007/s00294-014-0472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yogev O, Yogev O, Singer E, Shaulian E, Goldberg M, Fox TD, Pines O. 2010. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol 8:e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang Y, Qian X, Shen J, Wang Y, Li X, Liu R, Xia Y, Chen Q, Peng G, Lin SY, Lu Z. 2015. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat Cell Biol 17:1158–1168. doi: 10.1038/ncb3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peláez R, Herrero P, Moreno F. 2010. Functional domains of yeast hexokinase 2. Biochem J 432:181–190. doi: 10.1042/BJ20100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zenke FT, Engles R, Vollenbroich V, Meyer J, Hollenberg CP, Breunig KD. 1996. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272:1662–1665. doi: 10.1126/science.272.5268.1662. [DOI] [PubMed] [Google Scholar]
- 109.Ritterson Lew C, Tolan DR. 2013. Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics. J Cell Biochem 114:1928–1939. doi: 10.1002/jcb.24538. [DOI] [PubMed] [Google Scholar]
- 110.Ringel AE, Ryznar R, Picariello H, Huang KL, Lazarus AG, Holmes SG. 2013. Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet 9:e1003871. doi: 10.1371/journal.pgen.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blázquez MA, Lagunas R, Gancedo C, Gancedo JM. 1993. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett 329:51–54. doi: 10.1016/0014-5793(93)80191-V. [DOI] [PubMed] [Google Scholar]
- 112.Gancedo C, Flores CL. 2004. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res 4:351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- 113.Petitjean M, Teste MA, François JM, Parrou JL. 2015. Yeast tolerance to various stresses relies on the trehalose-6P synthase (Tps1) protein, not on trehalose. J Biol Chem 290:16177–16190. doi: 10.1074/jbc.M115.653899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Wilson RA, Jenkinson JM, Gibson RP, Littlechild JA, Wang ZY, Talbot NJ. 2007. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J 26:3673–3685. doi: 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kainulainen V, Korhonen TK. 2014. Dancing to another tune: adhesive moonlighting proteins in bacteria. Biology (Basel) 3:178–204. doi: 10.3390/biology3010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Derbise A, Song YP, Parikh S, Fischetti VA, Pancholi V. 2004. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect Immun 72:94–105. doi: 10.1128/IAI.72.1.94-105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cork AJ, Jergic S, Hammerschmidt S, Kobe B, Pancholi V, Benesch JL, Robinson CV, Dixon NE, Aquilina JA, Walker MJ. 2009. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J Biol Chem 284:17129–17137. doi: 10.1074/jbc.M109.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boël G, Jin H, Pancholi V. 2005. Inhibition of cell surface export of group A streptococcal anchorless surface dehydrogenase affects bacterial adherence and antiphagocytic properties. Infect Immun 73:6237–6248. doi: 10.1128/IAI.73.10.6237-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang CK, Ewis HE, Zhang X, Lu CD, Hu HJ, Pan Y, Abdelal AT, Tai PC. 2011. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J Bacteriol 193:5607–5615. doi: 10.1128/JB.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khan IK, Kihara D. 2014. Computational characterization of moonlighting proteins. Biochem Soc Trans 42:1780–1785. doi: 10.1042/BST20140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Khan I, Chen Y, Dong T, Hong X, Takeuchi R, Mori H, Kihara D. 2014. Genome-scale identification and characterization of moonlighting proteins. Biol Direct 9:30. doi: 10.1186/s13062-014-0030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chapple CE, Robisson B, Spinelli L, Guien C, Becker E, Brun C. 2015. Extreme multifunctional proteins identified from a human protein interaction network. Nat Commun 6:7412. doi: 10.1038/ncomms8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pils B, Schultz J. 2004. Inactive enzyme-homologues find new function in regulatory processes. J Mol Biol 340:399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 124.Adrain C, Freeman M. 2012. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol 13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- 125.Reiterer V, Eyers PA, Farhan H. 2014. Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol 24:489–505. doi: 10.1016/j.tcb.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 126.Valencik ML, McEwen JE. 1991. Genetic evidence that different functional domains of the PET54 gene product facilitate expression of the mitochondrial genes COX1 and COX3 in Saccharomyces cerevisiae. Mol Cell Biol 11:2399–2405. doi: 10.1128/MCB.11.5.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaspar BJ, Bifano AL, Caprara MG. 2008. A shared RNA-binding site in the Pet54 protein is required for translational activation and group I intron splicing in yeast mitochondria. Nucleic Acids Res 36:2958–2968. doi: 10.1093/nar/gkn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Harms MJ, Thornton JW. 2014. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature 512:203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ozimek P, van Dijk R, Latchev K, Gancedo C, Wang DY, van der Klei IJ, Veenhuis M. 2003. Pyruvate carboxylase is an essential protein in the assembly of yeast peroxisomal oligomeric alcohol oxidase. Mol Biol Cell 14:786–797. doi: 10.1091/mbc.E02-07-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gancedo C, Flores CL, Gancedo JM. 2014. Evolution of moonlighting proteins: insight from yeasts. Biochem Soc Trans 42:1715–1719. doi: 10.1042/BST20140199. [DOI] [PubMed] [Google Scholar]
- 131.Koschwanez JH, Foster KR, Murray AW. 2013. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife 2:e00367. doi: 10.7554/eLife.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ha JH, Butler JS, Mitrea DM, Loh SN. 2006. Modular enzyme design: regulation by mutually exclusive protein folding. J Mol Biol 357:1058–1062. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monaghan RM, Whitmarsh AJ. 2015. Mitochondrial proteins moonlighting in the nucleus. Trends Biochem Sci 40:728–735. doi: 10.1016/j.tibs.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 134.Fernández-García P, Peláez R, Herrero P, Moreno F. 2012. Phosphorylation of yeast hexokinase 2 regulates its nucleocytoplasmic shuttling. J Biol Chem 287:42151–42164. doi: 10.1074/jbc.M112.401679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kaps S, Kettner K, Migotti R, Kanashova T, Krause U, Rodel G, Dittmar G, Kriegel TM. 2015. Protein kinase Ymr291w/Tda1 is essential for glucose signaling in Saccharomyces cerevisiae on the level of hexokinase isoenzyme ScHxk2 phosphorylation. J Biol Chem 290:6243–6255. doi: 10.1074/jbc.M114.595074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Anders A, Lilie H, Franke K, Kapp L, Stelling J, Gilles ED, Breunig KD. 2006. The galactose switch in Kluyveromyces lactis depends on nuclear competition between Gal4 and Gal1 for Gal80 binding. J Biol Chem 281:29337–29348. doi: 10.1074/jbc.M604271200. [DOI] [PubMed] [Google Scholar]
- 137.Messenguy F, Wiame J. 1969. The control of ornithinetranscarbamylase activity by arginase in Saccharomyces cerevisiae. FEBS Lett 3:47–49. doi: 10.1016/0014-5793(69)80093-1. [DOI] [PubMed] [Google Scholar]