Abstract
The Asian tiger mosquito Aedes albopictus is currently one of the most threatening invasive species in the world. Native to Southeast Asia, the species has spread throughout the world in the past 30 years and is now present in every continent but Antarctica. Because it was the main vector of recent Dengue and Chikungunya outbreaks, and because of its competency for numerous other viruses and pathogens such as the Zika virus, A. albopictus stands out as a model species for invasive diseases vector studies. A synthesis of the current knowledge about the genetic diversity of A. albopictus is needed, knowing the interplays between the vector, the pathogens, the environment and their epidemiological consequences. Such resources are also valuable for assessing the role of genetic diversity in the invasive success. We review here the large but sometimes dispersed literature about the population genetics of A. albopictus. We first debate about the experimental design of these studies and present an up-to-date assessment of the available molecular markers. We then summarize the main genetic characteristics of natural populations and synthesize the available data regarding the worldwide structuring of the vector. Finally, we pinpoint the gaps that remain to be addressed and suggest possible research directions.
Introduction
Biological invasions, in spite of their critical impacts on native biodiversity and human societies, represent a special opportunity for population geneticists to observe in naturae phenomena that have otherwise remained mostly theoretical (Bock et al., 2015). For example, the study of a recently installed alien species represents an occasion to investigate the link between genetic diversity, whether neutral or not, and invasion success (Dlugosch and Parker, 2008; Handley et al., 2011). When such a species is also a threat to human health, as disease vectors are, the collection of empirical knowledge about population dynamics and gene flow can be used to anticipate risks (for example, modeling spread and epidemiological implications) and develop control strategies.
Indeed, the existence of multiple locally adapted vector populations could enhance the spread of parasites or arboviruses through space and time (McCoy, 2008). This has been well documented, for example, for malaria transmission in Africa: the vector system consists of several anopheline species and populations within species (for example, chromosomal forms in Anopheles gambiae sensu stricto), each of which adapted to specific bioclimatic conditions, that coexist in certain places but can also replace each other geographically or seasonally (Fontenille and Simard, 2004). The genetic variation among vector populations or individuals also diversifies their interactions with viruses and parasites, their environments and can thus alter their transmission dynamics (Barrett et al., 2008; Harris et al., 2010; Zouache et al., 2014). The intensity of gene flow among host populations is also an important point to take into account because it could influence the diffusion of key alleles such as those involved in insecticide resistance (Caprio and Tabashnik, 1992; Lenormand et al., 1999).
The Asian tiger mosquito Aedes (Stegomyia) albopictus (Skuse, 1894) has been described as one of the 100 worst invasive species in the world (Global Invasive Species Database, http://www.issg.org/database/). Originating from South and East Asia, this species has spread throughout the world mostly since the second half of the twentieth century, and it is now found on every continent except Antarctica (Kraemer et al., 2015). The new areas colonized by A. albopictus include such disparate environments as tropical South America, Africa and the mostly temperate areas of Northern America and Europe.
Although A. albopictus has long been considered a vector of secondary importance, its involvement in recent Dengue and Chikungunya outbreaks and its competence for numerous other arboviruses and nematode parasites including the threatening emerging Zika virus (Paupy et al., 2009; Grard et al., 2014) emphasizes the need to more extensively study the biology of this species (Bonizzoni et al., 2013). A. albopictus had an important role in the re-emergence of Dengue and Chikungunya in some of the recently invaded regions (Teixeira et al., 2009; Paupy et al., 2010), and it has also been implicated in new outbreaks such as the 2007 Chikungunya episode in Italy (Rezza et al., 2007). Thus, it represents a relevant case study in which invasion genetics could bring substantial improvement in vector survey and management activities.
In addition, knowledge about the genetic diversity and the genetic structure of the Asian tiger mosquito could help researchers evaluate the risk of disease and insecticide resistance spread, as well as identify the origins and frequencies of introductions. Such knowledge is also required to search for loci that could be involved in environmental adaptation.
The genetic structure of a substantial number of native and invasive populations of this species has been investigated throughout the world, multiplying the number of population genetics reports that are available. They are, however, highly disparate and range from local reports to worldwide genotype comparisons.
Regarding the increasing interest in such data about A. albopictus, we present here a comprehensive synthesis of what is known and the questions that remain to be investigated. In the following review, we first propose some guidelines to characterize the key features of the genetic structure of natural populations and exhaustively describe the genetic markers previously developed for A. albopictus. We then examine different attempts to resolve the geographical origin of invasive populations and finally suggest directions for future studies of A. albopictus.
A guideline for population genetics in mosquitoes
The sampling issue
First, it is important to notice that the boundaries used to describe ‘populations' in genetic studies are generally fuzzy, which is particularly true with A. albopictus. Most often, the term ‘population' refers to one specific sampling site (such as a point in a city or a GPS coordinate). However, it is not always clear whether this ‘sampling site' encompasses one or multiple discrete sampling points, such as one or several traps or artificial containers, and if so the average distances between them and the number of specimens sampled in each (for example, Urbanelli et al., 2000; Zhong et al., 2013 and Manni et al., 2015). In the following discussion, we will use ‘sampling site' to refer to the location in which a population is sampled (for example, neighborhood, cemetery and park) and ‘sampling point' to mean the actual unit of sampling within the sampling site (for example, a trap or an artificial container). In addition, it is crucial to clearly establish whether the collected samples are eggs, larvae or flying adults. This is important because in A. albopictus, larvae or eggs collected from the same breeding site are likely to belong to the same progeny. This potential drawback could be easily avoided by collecting only a few (ideally one) individuals per ovitrap and setting multiple traps throughout the sampling site, such as in the study by Delatte et al. (2013). Then, even if adults have been shown to have low dispersal capabilities (200–500 m on average (Liew and Curtis, 2004; Lacroix et al., 2009; Marini et al., 2010)), we could confidently expect that flying adults collected at a sample point are more likely to represent the sampling site diversity than eggs or larvae from the same breeding site. If flying adults are collected, it will also be informative to report whether one or more traps were used (or if aspirator/human sampling was performed at a precise point or in multiple places in the sampling site). Indeed, a comprehensive description of the sampling strategy is needed for the sake of data interpretation, especially for calculating the inbreeding coefficient (FIS) and for partitioning the genetic variance (analysis of molecular variance).
The genetic markers issue
Useful genetic markers are expected to have several key features: selective neutrality, ease of scoring in all specimens of the species and sufficient variability to allow for measures of genetic differentiation, and genetic clustering of individuals. They also should be robust to re-genotyping and/or allow comparison with genotypes from new samples (see for instance Lowe et al. (2004) for comprehensive guidelines about genetic markers in the field of molecular ecology). Depending on the purpose, the selected markers should be suitable for phylogeography analyses (such as mitochondrial DNA (mtDNA)), allow interpretation about the breeding structure (such as codominant allozymes or microsatellites) or be sufficiently numerous in the genome to tease apart the effects of demographic history from those of natural selection (Lowe et al., 2004; Schlötterer, 2004; Selkoe and Toonen, 2006; Avise, 2009; Garvin et al., 2010; Stapley et al., 2010).
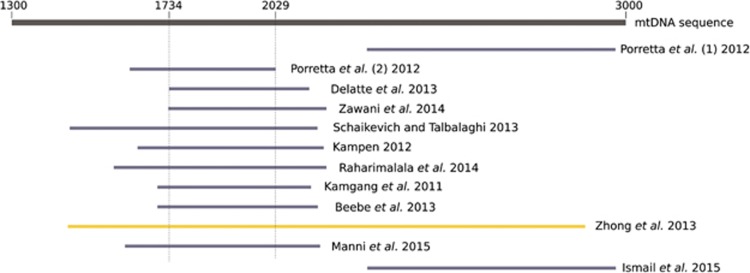
A critical comparison of the genetic markers available in A. albopictus is reported in Table 1. In addition, a discussion about the yet-to-be-applied dense markers can be found in Box 1. The first population genetics studies in this species were conducted in the late 1980s, using polymorphic enzymes. These markers have remarkable resolution and allowed to investigate genetic relationship between individuals and populations around the world using principal component analyses; they also allowed analyses of genetic variance between and within populations (Black et al., 1988a, 1988b; Kambhampati et al., 1991; Urbanelli et al., 2000; Chareonviriyaphap et al., 2004). Unfortunately, allozymes were soon withdrawn at the benefit of mtDNA. Besides rare exceptions (Beebe et al., 2013), these last showed a low level of genetic variation among samples (few haplotypes separated by only one or two mutation steps). To allow a good comparison of the studies based on mtDNA, the sequences available from data banks should be of similar size and from the same region of the gene(s). That is not the case in A. albopictus, in which a survey of the available sequences in GenBank for COI shows that only a small region of 295 bp is shared (Figure 1). Recently, extensive sequencing at the 5' and 3' ends of the COI gene displayed more polymorphism than previously sequenced parts (Table 1). However, in species with such complicated demographic histories, the use of mtDNA markers may not be the most relevant choice. Indeed, variation at mtDNA only reflect the demography in terms of the maternal line; it could be also considered as an unique locus, so the accuracy of the information given is thus more sensitive to stochastic events such as population bottlenecks. In addition, it could be affected by either direct selection on mitochondrial genes or by linkage disequilibrium with any other selected cytoplasm component such as symbiotic or parasitic bacterias (Hurst and Jiggins, 2005). In A. albopictus, the low diversity found at certain mtDNA markers, which first had been attributed to small effective population sizes of the founder populations, was later suggested to be a consequence of such a cytoplasmic sweep induced by Wolbachia infection (Armbruster et al., 2003). Phylogeographic analyses in A. albopictus may thus benefit from combining these mtDNA markers to a new one recently developed from the ribosomal DNA internal transcribed spacer 2 (ITS2, Table 1), a nuclear locus that should not be affected by this kind of issue.
Table 1. Comparison of the molecular markers available in A. albopictus.
| Type of marker | Markers/primers name | Locus in genome | Recommended usage | Comment/advice | References |
|---|---|---|---|---|---|
| Allozymes | G-3-pdh,Ldh,Hbdh,Mdh-1,Mdh-2,Mdhp-1,Mdhp-2,Idh-1,Idh-2,6-Pgdh,Sod-1,Aat-1,Hk-1,Adk-1,Acph,Aco-1,Aco-2,Gpi, Pgm, Pks-1, Pks-2, Pgi-1,Mez-1, Got-1, Got-2, Fum-1, Est-1, Aks-2, Aox-1 and Agk-2 | 30 | Population structure | Investigation of breeding structure | Black et al., 1988a; Black et al., 1988b; Kambhampati et al., 1991; Urbanelli et al., 2000; Chareonviriyaphap et al., 2004 |
| Discriminate populations at the worldwide scale | |||||
| RAPD | G06, G08, G09, G10, OPA11, OPAB01, OPB04, OPC15, OPM11, OPS01 and OPS10 | 188 | Strongly discouraged | Strongly discouraged because of the reproducibility issues with RAPD (Jones et al., 1997; Perez et al., 1998; Isabel et al., 1999) | Mutebi et al., 1997; Ayres and Romão, 2002; Gupta and Preet, 2014 |
| mtDNA | COI, ND5 and Cytb | 3 | Phylogeography | Most surveys discovered few haplotypes separated by short evolutionary distances | Birungi and Munstermann, 2002; Mousson et al., 2005; Usmani-Brown, 2009; Žitko et al., 2011; Kamgang et al., 2011; Delatte et al., 2011; Porretta et al., 2012; Bebee et al, 2013; Shaikevich and Talbalaghi, 2013; Zhong et al., 2013; Zawani et al., 2014 |
| COI (longer) | 1 | Phylogeography/population structure | Highly polymorphic with hundreds of haplotypes identified both in native and colonized areas. We would recommend for the use of COI, using the PCR primers from Zhong et al. (2013), which encompasses a large (1433 bp) sequence that contains the portions used in most of the published studies | Porreta et al., 2012; Zhong et al., 2013; Ismail et al., 2015 | |
| rDNA | ITS2 | — | Phylogeography/population structure | High polymorphism and codominant data | Higa et al., 2010; Shaikevich and Talbalaghi, 2013; Manni et al., 2015 |
| ITS2 is multi copied, thus genotyping needs several cloning and sequencing runs, and some intra-individual variation should be expected and taken into account | |||||
| EPIC | RpS8, RpS9, RPS12b, Rps16, RpS20b, RpS27A, RpL30a and RpL30b | 8 | Phylogeography/population structure | Not used yet | White et al., 2015 |
| Microsatellites | AealbA9, AealbB51, AealbB52, AealbB6, AealbD2 and AealbF3 AEDC, 34–72, alb212 and alb222 Alb-di-4, Alb-di-6, Alb-tri-3, Alb-tri-6, Alb-tri-18, Alb-tri-20, Alb-tri-21, Alb-tri-25, Alb-tri-33, Alb-tri-41, Alb-tri44, Alb-tri-45 and Alb-tri-46 | Allow intra-population discrimination of genetic membership. Genetic clustering, genetic variance partition among and within sampling sites, and landscape genetics | Porretta et al., 2006 Huber et al., 2001; Delatte et al., 2013 Beebe et al., 2013 | ||
| Aealbmic1, Aealbmic2, Aealbmic3, Aealbmic4, Aealbmic5, Aealbmic6, Aealbmic7, Aealbmic8, Aealbmic9, Aealbmic10, Aealbmic11, Aealbmic12, Aealbmic13, Aealbmic14, Aealbmic15, Aealbmic16, Aealbmic17, Aealbmic18, Aealbmic19, Aealbmic20, Aealbmic21, Aealbmic22 and Aealbmic23 | 54 | Population structure/landscape genetics/kinship | With French and Vietnamese populations, we do not recommend AealbB6, AealbD2, AealbF3, and alb212 (stuttering-like profile) and 34–72 (null allele fixation) (Minard, personal communication) | Manni et al., 2015 | |
| Ap1, Ap2, Ap3, Ap5 and AC2 | Medley et al., 2015 |
Toward dense markers in Aedes albopictus?
Genome-wide analyses such as genomic scan for selection will require the development of dense markers in A. albopictus. Restriction site-associated DNA sequencing (RADseq) is currently one of the most popular methods used to study large numbers of single-nucleotide polymorphisms in the genomes of non-model species. No study using this method has been published in A. albopictus yet, but some of us have tested its feasibility in a pilot experiment (Goubert et al., unpublished data). Briefly, we used the method described by Henri et al. (2015) to genotype eight individuals from two distinct populations from La Réunion Island, France. We used the SbfI enzyme, which has an 8 bp recognition site and is thus usually considered to be a ‘low' frequency cutter. Nevertheless, we observed a very large number of sites, producing a total amount of 149 795 RAD loci (using the Stacks pipeline (Catchen et al., 2011)). This number is almost 10 times higher than expected by in silico prediction using the genome sequence of the related species Aedes aegypti. This high number of loci implies that RADseq, at least in this simple version, cannot be performed for many individuals in most labs due to high cost. Interestingly, a RADseq experiment conducted in A. aegypti (Brown et al., 2014) also using SbfI discovered a very similar number of RAD loci (184 178) using 128 individuals from around the world. In the end, only 1504 single-nucleotide polymorphisms were effectively used in this study; this number is enough for most genetic structure analysis purposes but remains limited in the context of genomic scans, especially considering the investment needed to produce these genotypes. Investigation on other restriction enzymes or the development of double-digest RAD-seq (Peterson et al., 2012) could help us to further decrease the complexity of A. albopictus genome. The latter approach, recently developed in A. aegypti (Rašić et al., 2014), proved efficient and increased the number of single-nucleotide polymorphisms that could be genotyped up to 9369 single-nucleotide polymorphisms per individual on average (N=76; Rašić et al., 2015).
In addition, the development of such methods in A. albopictus should thus focus on the sequencing coverage issue and should be aware of several biases that have been identified in RADseq (allele drop out, uneven sequencing depending on restriction fragment length) that could lead to false estimates of critical population parameters, even with double-digest RAD-seq, such as the effective population size (Davey et al., 2012; Gautier et al., 2012; Arnold et al., 2013) and that could be thus problematic for any demographic inference made in this species.
As an alternative to RADseq, monitoring the insertion polymorphisms created by transposable elements (TEs) is a way to obtain a very large number of markers in the genome of A. albopictus, as it has been demonstrated that some TE families have several thousand well-conserved copies in this species (Goubert et al., 2015). The transposon display technique involves an enzymatic digestion of the total DNA followed by amplification of insertions of a whole TE family using a single pair of primers. TE insertion polymorphism has successfully been used in several studies involving mosquitoes (Boulesteix et al., 2007; Bonin et al., 2007; Esnault et al., 2008). Recently, transposon display using five TE families led us to amplify more than 120 000 insertion loci in 140 A. albopictus females from Vietnamese and European populations (Goubert et al., unpublished data).
Figure 1.
Comparison of the different mtDNA COI regions amplified for phylogeographic studies of Aedes albopictus (blue lines). The gray line represents the linear sequence of the COI gene (numbers are the number of base pairs from the origin (NCBI sequence NC_006817.1)). The yellow line is the PCR product amplified by Zhong et al. (2013) showing primers recommended for use.
Finally, and although they are considered as marker of choice for population genetics analysis, the development of microsatellites in the Asian tiger mosquito has been winding. Indeed, the reproducibility and efficiency of certain microsatellites could be questioned, given the fact that most authors developed their own markers for their studies. However, recent publications have provided new sets of such loci in which both their number and resolution allowed various fine-scale population genetics analysis (Table 1).
Population genetics of natural populations
With the ‘population' issue in mind, one of the most common features of genetic studies conducted in A. albopictus is that the largest part of the observed genetic variation is found at the smallest hierarchical level considered, which has been often referred as ‘within population' (that is, within the sampling site; Black et al., 1988a, 1988b; Kambhampati et al., 1991; Zhong et al., 2013; Gupta and Preet, 2014). The amount of variance attributed to this level, also called ‘between individuals in population', is remarkably high compared with the upper levels (among groups or among populations in groups) and represents more than half of the total genetic variation, regardless of the marker used (allozymes, mtDNA, microsatellites, RAPD or ITS2) and regardless of the population being invasive or native. Recently, new studies have revealed that what has also been called a ‘high local differentiation' could be caused by a lack of variation at the intra-individual level (FIS, Delatte et al., 2013; Manni et al., 2015). In the analysis of molecular variance context (Excoffier, 2004), this level would represent the covariance of alleles of a given locus within individuals within populations. For example, Manni et al. (2015) using ITS2 polymorphism showed that if the lower hierarchical level was ‘within population', the genetic variance attributed to that level was 84.78% when they added the ‘within individuals' level to the analysis of molecular variance, the first level fell to 17.51%, and individual level represented 74.36% of the genetic (co)-variance. Equivalent results were found by Delatte et al. (2013) in La Réunion Island, in which 80.766% of the variation was estimated to be at the individual level using microsatellites. It is important to highlight that these studies encompass populations from the native (Thailand) or old (La Réunion) and recent (Italy) invaded areas and thus suggest that A. albopictus populations could share this pattern globally.
The first studies that investigated the genetic structure of natural populations concluded that such a pattern of variation within the sampling sites was likely due to high genetic drift accompanying the establishment of the local population (that is, individuals found in a given sampling site), which implies that such ‘populations' were founded by small numbers of adult individuals and had low dispersion rates that restricted gene flow (Black et al., 1988a, 1988b; Kambhampati et al., 1991). This explanation makes sense in light of recent findings because a high level of genetic covariance within individuals would mean that they could have a high level of inbreeding, which is suggested by their breeding structure.
It is interesting to confront this hypothesis with the results from codominant data, such as allozyme and microsatellite data. For allozymes, most studies reported significant deviation from Hardy−Weinberg equilibrium for several loci and populations toward a heterozygotes deficit (Black et al., 1988a; Kambhampati et al., 1991; Urbanelli et al., 2000; Paupy et al., 2001; Vazeille et al., 2001; de Oliveira, 2003). Using microsatellites, the FIS has been shown to be positive, ranging from 0.1 to 0.2, supporting a high rate of inbreeding (Delatte et al., 2013; Manni et al., 2015; Minard et al., 2015). The homozygote excess found with microsatellites has sometimes been attributed to the presence of null alleles (Kamgang et al., 2011 (no FIS data), Delatte et al., 2013), but this should be interpreted with caution because estimation of null allele frequencies is more difficult in populations that are not actually at Hardy−Weinberg equilibrium and if no independent prior of the actual inbreeding level is given (Van Oosterhout et al., 2006; Chybicki and Burczyk, 2009). However, Manni et al. (2015) obtained similar results with new markers with an apparently low level of null alleles. Even if we cannot exclude the possibility that significant FIS values could reflect consequences of a Whalhund effect due to the unknown population structure, the repeated presence of such cues (either high intra-individual genetic covariance or significant FIS) among the studies considered here highly support the inbreeding hypothesis.
Thus, a credible portrait of a ‘typical' population of A. albopictus would be a network of interconnected breeding sites that each have a high level of inbreeding. Kinship analysis among individuals in sampling points and sampling sites would be very useful to test this hypothesis. This would allow to understand this original genetic structure that is also found in Aedes aegypti, the yellow fever mosquito that is also a mostly human-dispersed and invasive mosquito (Gonçalves da Silva et al., 2012; Damal et al., 2013). However, in A. aegypti, probably because its spread from Africa to America and then Asia is older than the worldwide spread of A. albopictus, there is genetic evidence for a progressive invasion along with a reduction of genetic diversity from the first to the last colonized continents (Powell and Tabachnick, 2013). This has, to the best of our knowledge, not been shown in A. albopictus. In addition, the results of two studies (Paupy et al., 2001; Vazeille et al., 2001) suggest that the urbanization gradient could positively influence the local genetic structuring (for instance at the scale of a city), but this assumption has not been formally tested yet.
Filling the gap between genetic structure and knowledge from behavioral ecology would also be extremely valuable, especially regarding the mating behavior of A. albopictus. Such studies would help to define the boundaries of what should be considered a ‘population' and could suggest a uniform sampling strategy for use for future studies. Meanwhile, as remarked earlier, it is important to carefully report the sampling strategy, especially the distance between breeding sites if sites are used as the sampling unit.
Worldwide genetic structure of native and invasive populations
It is often assumed that invasive populations are subject to founder events that leads to a reduced genetic diversity in the invaded range (Handley et al., 2011). However, the influence of such events on genetic diversity is modulated and sometimes counterbalanced by several factors such as the amount of invaders and the frequency of introduction (that is, the propagule pressure) (Handley et al., 2011; Bock et al., 2015). In A. albopictus, few studies have specifically focused on these aspects, and there is no evidence that invasions have been followed by a reduction of genetic variability. On the contrary, there are several indications of repeated and possibly massive introductions (detailed below). In addition, the colonization pattern of A. albopictus is characterized by an absence of a natural progressive wave front (that could be accompanied by allele ‘surfing' at the edge of the invasive range), but is well explained by human-mediated dissemination via the transportation infrastructure (Medlock et al., 2012; Roche et al., 2015).
Genetic diversity in the native area
To determine the origins of invasive populations, it is important to look at both genetic diversity and structure in the native area. This ranges from Southern (including India) and tropical Eastern Asia (including Indonesia) to East China and Japan (Hawley, 1988; Bonizzoni et al., 2013). One weakness of several studies looking at the origins of invasive populations is the use of a restricted sample from the native range (Birungi and Munstermann, 2002; Mousson et al., 2005; Usmani-Brown, 2009) or even the use of laboratory samples that have low genetic variation (Birungi and Munstermann, 2002). Second, most of these studies used markers with low variation such as certain mtDNA sequences (Birungi and Munstermann, 2002; Mousson et al., 2005; Usmani-Brown, 2009; Haddad et al., 2012; Shaikevich and Talbalaghi, 2013; Zawani et al., 2014). Recently, and since the development of new COI primers, two studies offered a more comprehensive view of the genetic diversity in native areas. Porretta et al. (2012) and Zhong et al. (2013) showed that the expected native area, including Japan, shows high genetic diversity (62 haplotypes found in 174 individuals, and 66/346, respectively) with little or no genetic structure. Combining phylogeography and species distribution modeling (using climate data), Porretta et al. (2012) argue that the current distribution of A. albopictus in Southeast continental Asia, Japan and the Indochinese peninsula is the result of recolonization from a single but large and ecologically diverse area, the Sundaland, which existed during the last glacial maximum (LGM, ~21 000 years BP) as an area connecting Sumatra, Java and Borneo to the Indochinese peninsula. The authors suggest that a progressive range expansion of genetically different but connected populations could be at the origin of the current genetic structure in the continental native area.
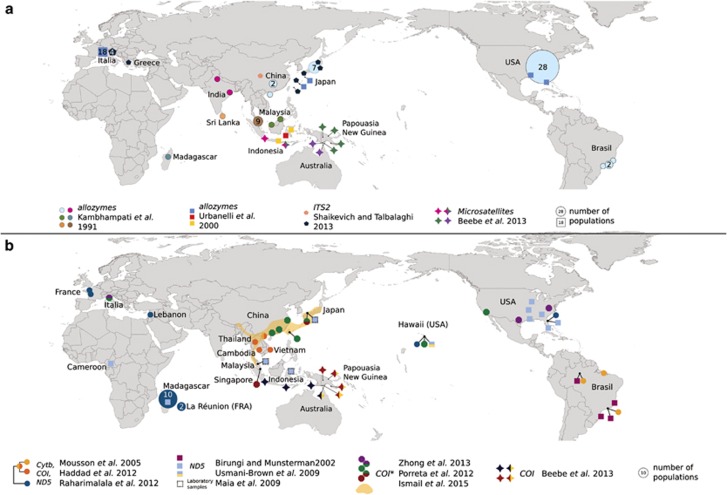
Even if no clear genetic structure has been reported, the above-mentioned studies did not include populations from the southern range of A. albopictus (such as Sumatra, Java or Borneo), with the exception of Zhong et al. (2013), who included a single sampling site from Singapore and a recent study of Ismail et al. (2015) who studied two populations sampled near Kuala Lumpur in Malaysia (Figure 2a). Thus, it would be interesting to investigate the genetic diversity and structure of these populations and compare them with continental ones. Furthermore, it is worth mentioning that allozyme studies have shown evidence for differentiation between the southern insular populations and northern ones, as well as between western (India, Sri Lanka) and eastern populations of the native area (Kambhampati et al., 1991; Urbanelli et al., 2000; Figure 2).
Figure 2.
World maps representing homogeneous genetic groups of Aedes albopictus identified using (a) nuclear markers or (b) mtDNA markers. The results of comparable studies are represented with the same shape. Colors indicate genetic groups, and the types of markers are italicized. Mixed colors for mtDNA markers indicate either intermediate haplotypes (Mousson et al., 2005) or mixed sampling sites. In (b), the orange-colored area represents the sampling of Porreta et al. (2012), who found no genetic structure. Asterisks refer to the new highly polymorphic COI sequences.
Southwest islands of the Indian Ocean (~1500 years BP to −)
SWIO (Southwest islands of the Indian Ocean) includes Madagascar, La Réunion and other smaller islands such as Mayotte, Mauritius, Rodrigue, Glorieuse Comoros and Seychelles. On the basis of anthropological research, Delatte et al. (2011) hypothesized that the first colonization of SWIO by A. albopictus could have begun 1500–2000 years ago; later SWIO were frequented by spice traders in the seventeenth to eighteenth centuries, probably favoring the spread of A. albopictus in this area. Kambampathi et al. (1991) showed that 24 individuals from a single sampling site collected in 1988 in Madagascar and genotyped at enzymatic loci formed a distinct cluster from samples of the native area including Sri Lanka, China, Japan, India, Borneo and Malaysia (Figure 2a). Using a COI polymorphism, Delatte et al. (2011) identified two distinct genetic groups in this area. The first includes numerous widespread haplotypes from samplings performed in 2006–2007, whereas the second contains much older samples (1956 and 1992) from Madagascar and La Réunion suggesting distinct waves of invasion. Attempt to identify the origin of populations sampled from 2000 and earlier in SWIO barely failed with mtDNA markers, as the lack of variation gave little statistical support about the structure reported in Figure 2b (Mousson et al., 2005; Usmani-Brown, 2009; Raharimalala et al., 2012).
Hawaii (~nineteenth century to −)
The Hawaiian islands are a location of interest in the biogeography of A. albopictus because they represent a possible source of introduction for trans-Pacific territories, especially the USA. Zhong et al. (2013) showed that the genetic diversity found in Hawaii is very similar to that of samples from continental Southeast Asia, but is well differentiated from that found in Singapore. Usmani-Brown (2009) found the highest haplotype diversity in their worldwide survey in a ND5 mtDNA fragment in Hawaii, and they hypothesized according to the low variability of this marker elsewhere that such a pattern should be the result of a longer presence time of older invasive populations in Hawaii compared with the other introduced populations.
The Americas (1985 to −)
Colonization of the Americas, and especially the USA, by A. albopictus in the 1980s was the starting point for studies of the population genetics of this species. The first invasive populations settled in Texas, and the species was quickly found in many important cities of the Southwest and Midwest United States (Sprenger and Wuithiranyagool, 1986; Black et al., 1988a); At the same time, A. albopictus was also found in Brazil (Forattini, 1986). An early enzymatic survey related the Brazilian samples to Japanese and Chinese ones (Kambhampati et al., 1991). However, using mtDNA polymorphism, Birungi and Munstermann (2002) showed that one mutation step discriminates US and Brazilian populations at the ND5 marker, and the marker later showed no variation between A. albopictus populations anywhere in the world except Brazil (Maia et al., 2009; Usmani-Brown, 2009). Mousson et al. (2005) also found evidence that Brazilian populations could belong to a separate genetic group because they formed a slightly different phylogenetic group with populations of the Indochinese peninsula at mtDNA markers (Cytb−ND5−COI; Figure 2b). However, these results are tenuous, and further investigations about the origin of Brazilian populations are needed.
In the USA, surveys suggested that a large number of individuals were repeatedly introduced due to intense maritime exchanges, which allowed A. albopictus to settle (Reiter and Sprenger, 1987; Moore, 1999; Zhong et al., 2013). For example, early after the first reports of invasion, Kambhampati et al. (1990) performed a 5-year survey of allele frequencies in several localities in the country, and they found no reduction of heterozygosity with occasional increases in certain cities and a rapid increase in effective population sizes. Most of the surveys acknowledged that US populations were mostly related to the northernmost Asian populations including Japan (Kambhampati et al., 1991; Urbanelli et al., 2000; Birungi and Munstermann, 2002; Mousson et al., 2005; Morales Vargas et al., 2013). Interestingly, some individuals sampled in Los Angeles in 2001 were related to Singapore haplotypes and were not found in 2011, suggesting that only genotypes from temperate climates could have settled successfully in the USA (Zhong et al., 2013).
Europe (1979 to −)
The first invasive population was reported in 1979 in Albania (Adhami and Murati, 1987), but the spread of the Asian tiger mosquito in this continent seems to have dramatically increased since the 1990s. Italy is probably the country of most concern because of the high density of mosquito populations, which contributed to the 2007 Chikungunya outbreak. Surveys using various types of genetic markers link the Italian samples to Japanese and the US ones (Urbanelli et al., 2000 (allozymes), Zhong et al., 2013 (COI), Shaikevich and Talbalaghi, 2013 (ITS2)), but this picture could be biased: in two of these studies, the group from which Italian populations were discriminated are either represented by one population (Zhong et al., 2013) or one genotype (Shaikevich and Talbalaghi, 2013). A recent survey by Manni et al. (2015) using microsatellites and ITS2 polymorphism compared populations from Italy, La Réunion Island and Thailand; it revealed that in spite of a high polymorphism found with those markers, no continental structuring of the populations could be found. Recent work comparing French and Vietnamese populations with microsatellites showed that a significant but small difference exists between the continents (Minard et al., 2015). Finally, the most informative study could be the older one, in which Urbanelli et al. (2000) using allozymes, found evidence for structure between populations from Italy, US and Japan vs populations from several Indonesian islands (Figure 2a). This underscores the need to include both North and Southeast Asian populations. Due to climatic similarities, it seems important to also compare European populations with those in northern and continental locations of the native range, such as China, that have been far less studied but could be a credible source of European invaders.
Africa
In central Africa, A. albopictus has been reported in Nigeria (first record in 1991), Cameroon (2000), Equatorial Guinea (2001), Gabon (2007) and Central Africa Republic (2009) (Savage et al., 1992; Fontenille and Toto, 2001; Toto et al., 2003; Coffinet et al., 2007; Diallo et al., 2010). Using COI and microsatellites, Kamgang et al. (2011) identified two distinct genetic clusters present in the same locations (microsatellites) that led the authors suspect the occurrence of multiple invasion events. In addition, COI analyses revealed a stronger identity of the Cameroonian haplotypes with tropical populations (India, Thailand, Vietnam and Brazil) than with temperate (France, Greece and USA) or Hawaii and SWIO sources. This contrasts with the findings of Usmani-Brown (2009), who found at the ND5 loci that the individuals sampled in Cameroon were mostly related to several US, Hawaii, Rome (Italy) and SWIO populations. Furthermore, a recent study in the Central African Republic highlighted the relatedness of samples with both the tropical and temperate haplotypes cited earlier (Kamgang et al., 2013). These results suggest multiple sources of introductions for Western African populations.
Other locations
On the basis of same combination of mtDNA markers used by Mousson et al. (2005), the geographical origin of individuals found in Lebanon (first record in 2003) was attributed to temperate areas (Haddad et al., 2012; Figure 2b). However, the low variation of the mtDNA markers used suggests that these results should be interpreted with caution. The geographic origin of Australian invaders found in the Torres Strait Islands region since 2004 (North Australia) was investigated using microsatellites and mtDNA, and it was found that this region was not settled from nearby Papua New Guinea (as previously thought) but rather by individuals related to Timor-Leste and Jakarta, highlighting the role of humans (probably illegal fishing) in the spread of A. albopictus (Beebe et al., 2013).
Conclusions and suggested research directions
This compilation of 30 years of research illuminates the key features of the population genetics of A. albopictus. The most striking results are the apparent lack of genetic structure according to geography and the high variability found within sampling sites, independently of whether the populations are native or invasive. The lack of isolation by distance seems to be due to a combination of low natural dispersion capabilities and a high level of human-mediated spread as recently demonstrated by Medley et al. (2015) using a landscape genomics framework. The ‘populations' of A. albopictus exhibit apparent high levels of inbreeding, which could lead to high contrasts between genotypes from different sampling points in the same site. To validate this hypothesis, more care should be taken to report the sampling strategies, and kinship analysis should be undertaken to study the genetic relationships within and between the sampling points of a sampling site.
Analysis of the available data stresses that future population genetics studies in A. albopictus should include a more exhaustive worldwide sampling from temperate (including China), sub-tropical and tropical native areas in order to infer the origin of invasive populations. It is important, in particular, to know whether restricted gene flow really exists between tropical non-diapausing and temperate-diapausing populations in the native area, and if phenotypically similar populations share a genetic kinship. Particularly, the photoperiodical diapause, which has a demonstrated genetic basis (Urbanski et al., 2012; Poelchau et al., 2013), seems to be an important component of climatic adaptation, favoring the invasive success of A. albopictus. Thus, the remaining questions in this field include how much of the genome has a low level of differentiation and is it possible to find highly differentiated regions that could be involved in this adaptation? Are there other traits involved in the local adaptation for which we can identify a genetic basis? To what extent could phenotypic plasticity be responsible for both ecological polymorphism and genetic homogeneity? In addition, another unknown is whether urbanization could also contribute to the local genetic structuring of this species, as it has been suggested in a few studies (Paupy et al., 2001; Vazeille et al., 2001).
The current availability of more informative markers (new microsatellites, ITS2) and large collections of samples including different time periods would, however, make possible the investigation and modeling of invasion routes (Cristescu, 2015), and the comprehensive study of the genetic structuring of the vector. The recent publication of genome assemblies and their forthcoming annotation for two strains of A. albopictus (Chen et al., 2015; Dritsou et al., 2015) will provide valuable information about the physical location and environment of existant markers, and will make possible the design of new markers or to investigate population structure, or detect the footprint of natural selection in candidate regions.
Another field of investigations is the evolutionary impact of the endosymbiotic bacteria Wolbachia—whose two strains are co-infecting almost all populations of A. albopictus studied (Werren et al., 1995; Kittayapong et al., 2002; De Albuquerque et al., 2011; Zouache et al., 2011; Bourtzis et al., 2014; Minard et al., 2015)—over the mtDNA polymorphism. Indeed, the hypothesis of a mitochondrial hitchhiking—and thus a reduced mtDNA diversity—due to Wolbachia could be questioned, since the discovery of a new highly polymorphic region of the COI gene in recent studies (Porreta et al., 2012; Zhong et al., 2013).
Knowledge about the genetic structure will also be valuable for the study of vector/pathogen interactions. Indeed, the genetic variability of vectors would affect the outcomes of close relationships with viruses or pathogens. In A. aegypti, the host genotype modulates the transcriptional response during infection with a strain of Dengue virus (Behura et al., 2014), and the way the mosquitoes can acquire the virus when it is present at low levels during feeding (Pongsiri et al., 2014). For A. albopictus, de Oliveira (2003) and Fernández et al. (2004) showed that the location of the strain (from USA, Caiman Island or Brazil) does not affect the competence for Dengue or Yellow Fever Virus. Artificially induced inbreeding depression did not affect also the infection rate by Plasmodium gallinaceum avian pathogen (O'Donnell and Armbruster, 2010). However, Zouache et al. (2014) demonstrated a three-level genotype X genotype X environment interaction between A. albopictus strain, CHKV strain and temperature on CHKV transmission potential, suggesting the need for research in this field. We suggest that future studies should involve multiple strains per geographic location because most of the genetic variability in A. albopictus is observed at a local scale.
The case of A. albopictus represents a concrete example of a fast and successful invasion sustained by high propagule pressure and high genetic diversity. Its expansion into the invaded areas is strongly driven by human activities that are thus actively involved in the shape of the current genetic structure. Because of its epidemiological importance, and also because of its status as an invasive species, the Asian tiger mosquito should be considered a model species for which an increase of knowledge would benefit a large community of researchers. However, standardization of sampling and genotyping methods is also recommended in order to avoid the dispersion of data that would become irrelevant for global inference.
Acknowledgments
We are grateful to Marie Fablet, who provided insightful comments, as well as three anonymous referees who contributed to improve the manuscript. We also thank Céline Toty (MIVEGEC Montpellier) for providing the A. albopictus individuals from La Réunion Island used in the pilot RAD-seq experiment and Hélène Henri for the libraries construction. CG received a grant from the French Ministry of Superior Education.
The authors declare no conflict of interest.
References
- Adhami J, Murati N. (1987). The presence of the mosquito Aedes albopictus in Albania. Rev Mjekësore 1: 13–16. [Google Scholar]
- Armbruster P, Damsky WE, Giordano R, Birungi J, Munstermann LE, Conn JE. (2003). Infection of new- and old-world Aedes albopictus (Diptera: Culicidae) by the intracellular parasite Wolbachia: implications for host mitochondrial DNA evolution. J Med Entomol 40: 356–360. [DOI] [PubMed] [Google Scholar]
- Arnold B, Corbett-Detig RB, Hartl D, Bomblies K. (2013). RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol 22: 3179–1390. [DOI] [PubMed] [Google Scholar]
- Avise JC. (2009). Phylogeography: retrospect and prospect. J Biogeogr 36: 3–15. [Google Scholar]
- Ayres CFJ, Romao TPA, Melo-Santos MAV, Furtado AF. (2002). Genetic diversity in Brazilian populations of Aedes albopictus. Mem Inst Oswaldo Cruz 97: 871–875. [DOI] [PubMed] [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ, Linde CC. (2008). Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends Ecol Evol 23: 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe NW, Ambrose L, Hill La, Davis JB, Hapgood G, Cooper RD et al. (2013). Tracing the tiger: population genetics provides valuable insights into the Aedes (Stegomyia) albopictus invasion of the Australasian region. PLoS Negl Trop Dis 7: e2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Gomez-Machorro C, deBruyn B, Lovin DD, Harker BW, Romero-Severson J et al. (2014). Influence of mosquito genotype on transcriptional response to dengue virus infection. Funct Integr Genomics 14: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birungi J, Munstermann LE. (2002). Genetic Structure of Aedes albopictus (Diptera: Culicidae) populations based on mitochondrial ND5 sequences: evidence for an independent invasion into Brazil and United States. Ann Entomol Soc Am 95: 125–132. [Google Scholar]
- Black WC, Ferrari JA, Sprengert D. (1988. a). Breeding structure of a colonising species: Aedes albopictus (Skuse) in the United States. Heredity (Edinb) 60: 173–181. [DOI] [PubMed] [Google Scholar]
- Black WC, Hawley WA, Rai KS, Craig GB. (1988. b). Breeding structure of a colonizing species: Aedes albopictus (Skuse) in peninsular Malaysia and Borneo. Heredity (Edinb) 61: 439–446. [DOI] [PubMed] [Google Scholar]
- Bock DG, Caseys C, Cousens RD, Hahn MA, Heredia SM, Hübner S et al. (2015). What we still don't know about invasion genetics. Mol Ecol 24: 2277–2297. [DOI] [PubMed] [Google Scholar]
- Bonin A, Ehrich D, Manel S. (2007). Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16: 3737–3758. [DOI] [PubMed] [Google Scholar]
- Bonizzoni M, Gasperi G, Chen X, James AA. (2013). The invasive mosquito species Aedes albopictus: current knowledge and future perspectives. Trends Parasitol 29: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulesteix M, Simard F, Antonio-Nkondjio C, Awono-Ambene HP, Fontenille D, Biémont C. (2007). Insertion polymorphism of transposable elements and population structure of Anopheles gambiae M and S molecular forms in Cameroon. Mol Ecol 16: 441–452. [DOI] [PubMed] [Google Scholar]
- Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA et al. (2014). Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop 132(Suppl): S150–S163. [DOI] [PubMed] [Google Scholar]
- Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A et al. (2014). Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution 68: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprio MA, Tabashnik BE. (1992). Gene flow accelerates local adaptation among finite populations: simulating the evolution of insecticide resistance. J Econ Entomol 85: 611–620. [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. (2011). Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 1: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareonviriyaphap T, Akratanakul P, Huntamai S, Nettanomsak S, Prabaripai A. (2004). Allozyme patterns of Aedes albopictus, a vector of dengue in Thailand. J Med Entomol 41: 657–663. [DOI] [PubMed] [Google Scholar]
- Chen X-G, Jiang X, Gu J, Xu M, Wu Y, Deng Y et al. (2015). Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci USA 112: E5907–E5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. (2009). Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100: 106–113. [DOI] [PubMed] [Google Scholar]
- Coffinet T, Mourou JR, Pradines B, Toto JC, Jarjaval F, Amalvict R et al. (2007). First record of Aedes albopictus in Gabon. J Am Mosq Control Assoc 23: 471–472. [DOI] [PubMed] [Google Scholar]
- Cristescu ME. (2015). Genetic reconstructions of invasion history. Mol Ecol 24: 2212–2225. [DOI] [PubMed] [Google Scholar]
- Damal K, Murrell EG, Juliano SA, Conn JE, Loew SS. (2013). Phylogeography of Aedes aegypti (yellow fever mosquito) in South Florida: mtDNA evidence for human-aided dispersal. Am J Trop Med Hyg 89: 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Cezard T, Fuentes-Utrilla P, Eland C, Gharbi K, Blaxter ML. (2012). Special features of RAD Sequencing data: implications for genotyping. Mol Ecol 22: 3151–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Albuquerque AL, Magalhães T, Ayres CFJ. (2011). High prevalence and lack of diversity of Wolbachia pipientis in Aedes albopictus populations from Northeast Brazil. Mem Inst Oswaldo Cruz 106: 773–776. [DOI] [PubMed] [Google Scholar]
- de Oliveira RL. (2003). Large Genetic differentiation and low variation in vector competence for dengue and yellow fever viruses of Aedes albopictus from Brazil, the United States, and the Cayman Islands. Am J Trop Med Hyg 69: 105–114. [PubMed] [Google Scholar]
- Delatte H, Bagny L, Brengue C, Bouetard A, Paupy C, Fontenille D. (2011). The invaders: phylogeography of dengue and chikungunya viruses Aedes vectors, on the South West islands of the Indian Ocean. Infect Genet Evol 11: 1769–1781. [DOI] [PubMed] [Google Scholar]
- Delatte H, Toty C, Boyer S, Bouetard A, Bastien F, Fontenille D. (2013). Evidence of habitat structuring Aedes albopictus populations in Réunion Island. PLoS Negl Trop Dis 7: e2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M, Laganier R, Nangouma A. (2010). First record of Ae. albopictus (Skuse 1894), in Central African Republic. Trop Med Int Health 15: 1185–1189. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. (2008). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17: 431–449. [DOI] [PubMed] [Google Scholar]
- Dritsou V, Topalis P, Windbichler N, Simoni A, Hall A, Lawson D et al. (2015). A draft genome sequence of an invasive mosquito: an Italian Aedes albopictus. Pathog Glob Health 109: 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Boulesteix M, Duchemin JB, Koffi AA, Chandre F. (2008). High genetic differentiation between the M and S molecular forms of Anopheles gambiae in Africa. PLoS ONE 3: e1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. (2004) Analysis of population subdivision. In: Handbook of Statistical Genetics, 2nd edn. John Wiley & Sons, Ltd: Chichester, UK. [Google Scholar]
- Fernández Z, Moncayo A, Forattini OP, Weaver SC. (2004). Susceptibility of urban and rural populations of Aedes albopictus from São Paulo State, Brazil, to infection by Dengue-1 and -2 viruses. J Med Entomol 41: 961–964. [DOI] [PubMed] [Google Scholar]
- Fontenille D, Simard F. (2004). Unravelling complexities in human malaria transmission dynamics in Africa through a comprehensive knowledge of vector populations. Comp Immunol Microbiol Infect Dis 27: 357–375. [DOI] [PubMed] [Google Scholar]
- Fontenille D, Toto JC. (2001). Aedes (Stegomyia) albopictus (Skuse), a potential new Dengue vector in southern Cameroon. Emerg Infect Dis 7: 1066–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forattini OP. (1986). Identificação de Aedes (Stegomyia) albopictus (Skuse) no Brasil. Rev Saude Publica 20: 244–245. [DOI] [PubMed] [Google Scholar]
- Garvin MR, Saitoh K, Gharrett AJ. (2010). Application of single nucleotide polymorphisms to non-model species: a technical review. Mol Ecol Resour 10: 915–934. [DOI] [PubMed] [Google Scholar]
- Gautier M, Gharbi K, Cezard T, Foucaud J, Kerdelhué C, Pudlo P et al. (2012). The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Mol Ecol 22: 3165–3178. [DOI] [PubMed] [Google Scholar]
- Gonçalves da Silva A, Cunha ICL, Santos WS, Luz SLB, Ribolla PEM, Abad-Franch F. (2012). Gene flow networks among American Aedes aegypti populations. Evol Appl 5: 664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubert C, Modolo L, Vieira C, Valiente-Moro C, Mavingui P, Boulesteix M. (2015). De novo assembly and annotation of the Asian Tiger mosquito (Aedes albopictus repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti. Genome Biol Evol 7: 1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G, Caron M, Mombo IM, Nkoghe D, Mboui Ondo S, Jiolle D et al. (2014). Zika virus in Gabon (Central Africa)—2007: a new threat from Aedes albopictus? PLoS Negl Trop Dis 8: e2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Preet S. (2014). Genetic differentiation of invasive Aedes albopictus by RAPD-PCR: implications for effective vector control. Parasitol Res 113: 2137–2142. [DOI] [PubMed] [Google Scholar]
- Haddad N, Mousson L, Vazeille M, Chamat S, Tayeh J, Osta MA et al. (2012). Aedes albopictus in Lebanon, a potential risk of arboviruses outbreak. BMC Infect Dis 12: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley L-J, Estoup A, Evans DM, Thomas CE, Lombaert E, Facon B et al. (2011). Ecological genetics of invasive alien species. BioControl 56: 409–428. [Google Scholar]
- Harris C, Lambrechts L, Rousset F, Abate L, Nsango SE, Fontenille D et al. (2010). Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog 6: e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley WA. (1988). The biology of Aedes albopictus. J Am Mosq Control Assoc Suppl 1: 1–39. [PubMed] [Google Scholar]
- Henri H, Cariou M, Terraz G, Martinez S, El Filali A, Veyssiere M et al. (2015). Optimization of multiplexed RADseq libraries using low-cost adaptors. Genetica 143: 139–143. [DOI] [PubMed] [Google Scholar]
- Higa Y, Toma T, Tsuda Y, Miyagi I. (2010). A multiplex PCR-based molecular identification of five morphologically related, medically important subgenus Stegomyia mosquitoes from the genus Aedes (Diptera: Culicidae) found in the Ryukyu Archipelago, Japan. Jpn J Infect Dis 63: 312–316. [PubMed] [Google Scholar]
- Huber K, Mousson L, Rodhain F, Failloux A-B. (2001). Isolation and variability of polymorphic microsatellite loci in Aedes aegypti, the vector of dengue viruses. Mol Ecol Notes 1: 219–222. [Google Scholar]
- Hurst GDD, Jiggins FM. (2005). Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci 272: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Thériault P, Bousquet J. (1999). Direct evidence for biased gene diversity estimates from dominant random amplified polymorphic DNA (RAPD) fingerprints. Mol Ecol 8: 477–483. [Google Scholar]
- Ismail N-A, Dom NC, Ismail R, Ahmad AH, Zaki A, Camalxaman SN. (2015). Mitochondrial cytochrome oxidase I gene sequence analysis of Aedes Albopictus in Malaysia. J Am Mosq Control Assoc 31: 305–312. [DOI] [PubMed] [Google Scholar]
- Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, van de Wiel C et al. (1997). Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3: 381–390. [Google Scholar]
- Kambhampati S, Black WC, Rai KS. (1991). Geographic origin of the US and Brazilian Aedes albopictus inferred from allozyme analysis. Heredity 67: 85–93. [DOI] [PubMed] [Google Scholar]
- Kambhampati S, Black WC, Rai KS, Sprenger D. (1990). Temporal variation in genetic structure of a colonising species: Aedes albopictus in the United States. Heredity 64: 281–287. [DOI] [PubMed] [Google Scholar]
- Kamgang B, Brengues C, Fontenille D, Njiokou F, Simard F, Paupy C. (2011). Genetic structure of the tiger mosquito, Aedes albopictus, in Cameroon (Central Africa). PLoS One 6: e20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamgang B, Ngoagouni C, Manirakiza A, Nakouné E, Paupy C, Kazanji M. (2013). Temporal Patterns of Abundance of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and Mitochondrial DNA Analysis of Ae. albopictus in the Central African Republic. PLoS Negl Trop Dis 7: : e2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittayapong P, Baimai V, O'Neill SL. (2002). Field prevalence of Wolbachia in the mosquito vector Aedes albopictus. Am J Trop Med Hyg 66: 108–111. [DOI] [PubMed] [Google Scholar]
- Kraemer MUG, Sinka ME, Duda KA, Mylne A, Shearer FM, Barker CM et al. (2015). The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4: e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix R, Delatte H, Hue T, Reiter P. (2009). Dispersal and survival of male and female Aedes albopictus (Diptera: Culicidae) on Réunion Island. J Med Entomol 46: 1117–1124. [DOI] [PubMed] [Google Scholar]
- Lenormand T, Bourguet D, Guillemaud T, Raymond M. (1999). Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature 400: 861–864. [DOI] [PubMed] [Google Scholar]
- Liew C, Curtis CF. (2004). Horizontal and vertical dispersal of dengue vector mosquitoes, Aedes aegypti and Aedes albopictus, in Singapore. Med Vet Entomol 18: 351–360. [DOI] [PubMed] [Google Scholar]
- Lowe A, Harris S, Ashton P. (2004) Ecological Genetics: Design, Analysis, and Application, 1st edn. Blackwell Publishing: Oxford, UK. [Google Scholar]
- Maia RT, Scarpassa VM, Maciel-Litaiff LH, Tadei WP. (2009). Reduced levels of genetic variation in Aedes albopictus (Diptera: Culicidae) from Manaus, Amazonas State, Brazil, based on analysis of the mitochondrial DNA ND5 gene. Genet Mol Res 8: 998–1007. [DOI] [PubMed] [Google Scholar]
- Manni M, Gomulski LM, Aketarawong N, Tait G, Scolari F, Somboon P et al. (2015). Molecular markers for analyses of intraspecific genetic diversity in the Asian Tiger mosquito, Aedes albopictus. Parasit Vectors 8: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Caputo B, Pombi M, Tarsitani G, della Torre A. (2010). Study of Aedes albopictus dispersal in Rome, Italy, using sticky traps in mark-release-recapture experiments. Med Vet Entomol 24: 361–368. [DOI] [PubMed] [Google Scholar]
- McCoy KDD. (2008). The population genetic structure of vectors and our understanding of disease epidemiology. Parasite 15: 444–448. [DOI] [PubMed] [Google Scholar]
- Medley KA, Jenkins DG, Hoffman EA. (2015). Human-aided and natural dispersal drive gene flow across the range of an invasive mosquito. Mol Ecol 24: 284–295. [DOI] [PubMed] [Google Scholar]
- Medlock JM, Hansford KM, Schaffner F, Versteirt V, Hendrickx G, Zeller H et al. (2012). A review of the invasive mosquitoes in Europe: ecology, public health risks, and control options. Vector Borne Zoonotic Dis 12: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard G, Tran F-H, Tran-van V, Goubert C, Bellet C, Lambert G et al. (2015). French invasive Asian tiger mosquito populations harbor reduced bacterial microbiota and genetic diversity compared to Vietnamese autochthonous relatives. Front Microbiol 6: 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CG. (1999). Aedes albopictus in the United-States: current status and prospects for further spread. J Am Mosq Control Assoc 15: 221–227. [PubMed] [Google Scholar]
- Morales Vargas RE, Phumala-Morales N, Tsunoda T, Apiwathnasorn C, Dujardin JP. (2013). The phenetic structure of Aedes albopictus. Infect Genet Evol 13: 242–251. [DOI] [PubMed] [Google Scholar]
- Mousson L, Dauga C, Garrigues T, Schaffner F, Vazeille M, Failloux A-B. (2005). Phylogeography of Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae) based on mitochondrial DNA variations. Genet Res 86: 1–11. [DOI] [PubMed] [Google Scholar]
- Mutebi J-P, Black WC, Bosio CF, Sweeney WP, Craig GB. (1997). Linkage map for the Asian Tiger Mosquito Aedes (Stegomyia) albopictus based on SSCP analysis of RAPD markers. J Hered 88: 489–494. [DOI] [PubMed] [Google Scholar]
- O'Donnell D, Armbruster P. (2010). Inbreeding depression affects life-history traits but not infection by Plasmodium gallinaceum in the Asian tiger mosquito Aedes albopictus. Infect Genet Evol 10: 669–677. [DOI] [PubMed] [Google Scholar]
- Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. (2009). Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect 11: 1177–1185. [DOI] [PubMed] [Google Scholar]
- Paupy C, Girod R, Salvan M, Rodhain F, Failloux AB. (2001). Population structure of Aedes albopictus from La Réunion Island (Indian Ocean) with respect to susceptibility to a dengue virus. Heredity (Edinb) 87: 273–283. [DOI] [PubMed] [Google Scholar]
- Paupy C, Ollomo B, Kamgang B, Moutailler S, Rousset D, Demanou M et al. (2010). Comparative role of Aedes albopictus and Aedes aegypti in the emergence of Dengue and Chikungunya in Central Africa. Vector Borne Zoonotic Dis 10: 259–266. [DOI] [PubMed] [Google Scholar]
- Perez T, Albornoz J, Dominguez A. (1998). An evaluation of RAPD fragment reproducibility and nature. Mol Ecol 7: 1347–1357. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. (2012). Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds Ja, Elsik CG, Denlinger DL, Armbruster PA. (2013). Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. Proc Biol Sci 280: 20130143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongsiri A, Ponlawat A, Thaisomboonsuk B, Jarman RG, Scott TW, Lambrechts L. (2014). Differential susceptibility of two field Aedes aegypti populations to a low infectious dose of dengue virus. PLoS One 9: e92971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porretta D, Gargani M, Bellini R, Calvitti M, Urbanelli S. (2006). Isolation of microsatellite markers in the tiger mosquito Aedes albopictus (Skuse). Mol Ecol Notes 6: 880–881. [Google Scholar]
- Porretta D, Mastrantonio V, Bellini R, Somboon P, Urbanelli S. (2012). Glacial history of a modern invader: phylogeography and species distribution modelling of the Asian tiger mosquito Aedes albopictus. PLoS One 7: e44515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Tabachnick WJ. (2013). History of domestication and spread of Aedes aegypti—a review. Mem Inst Oswaldo Cruz 108(Suppl): 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raharimalala FN, Ravaomanarivo LH, Ravelonandro P, Rafarasoa LS, Zouache K, Tran-Van V et al. (2012). Biogeography of the two major arbovirus mosquito vectors, Aedes aegypti and Aedes albopictus (Diptera, Culicidae), in Madagascar. Parasit Vectors 5: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rašić G, Filipović I, Weeks AR, Hoffmann AA. (2014). Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genomics 15: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rašić G, Schama R, Powell R, Maciel-de Freitas R, Endersby-Harshman NM, Filipović I et al. (2015). Contrasting genetic structure between mitochondrial and nuclear markers in the dengue fever mosquito from Rio de Janeiro: implications for vector control. Evol Appl 8: 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter P, Sprenger D. (1987). The used tire trade: a mechanism for the worldwide dispersal of container breeding mosquitoes. J Am Mosq Control Assoc 3: 494–501. [PubMed] [Google Scholar]
- Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M et al. (2007). Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370: 1840–1846. [DOI] [PubMed] [Google Scholar]
- Roche B, Léger L, L'Ambert G, Lacour G, Foussadier R, Besnard G et al. (2015). The spread of Aedes albopictus in Metropolitan France: contribution of environmental drivers and human activities and predictions for a near future. PLoS One 10: e0125600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage HM, Ezike VI, Nwankwo AC, Spiegel R, Miller BR. (1992). First record of breeding populations of Aedes albopictus in continental Africa: implications for arboviral transmission. J Am Mosq Control Assoc 8: 101–103. [PubMed] [Google Scholar]
- Schlötterer C. (2004). Opinion: the evolution of molecular markers—just a matter of fashion? Nat Rev Genet 5: 63–69. [DOI] [PubMed] [Google Scholar]
- Selkoe KA, Toonen RJ. (2006). Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9: 615–629. [DOI] [PubMed] [Google Scholar]
- Shaikevich E, Talbalaghi A. (2013). Molecular characterization of the Asian Tiger Mosquito Aedes albopictus (Skuse) (Diptera: Culicidae) in Northern Italy. ISRN Entomol 2013: 1–6. [Google Scholar]
- Sprenger D, Wuithiranyagool T. (1986). The discovery and distribution of Aedes albopictus in Harris County, Texas. J Am Mosq Control Assoc 2: 217–219. [PubMed] [Google Scholar]
- Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R et al. (2010). Adaptation genomics: the next generation. Trends Ecol Evol 25: 705–712. [DOI] [PubMed] [Google Scholar]
- Teixeira MG, Costa M, da CN, Barreto F, Barreto ML. (2009). Dengue: twenty-five years since reemergence in Brazil. Cad Saude Publica 25: S7–S18. [DOI] [PubMed] [Google Scholar]
- Toto J-C, Abaga S, Carnevale P, Simard F. (2003). First report of the oriental mosquito Aedes albopictus on the West African island of Bioko, Equatorial Guinea. Med Vet Entomol 17: 343–346. [DOI] [PubMed] [Google Scholar]
- Urbanelli S, Bellini R, Carrieri M, Sallicandro P, Celli G. (2000). Population structure of Aedes albopictus (Skuse): the mosquito which is colonizing Mediterranean countries. Heredity (Edinb) 84: 331–337. [DOI] [PubMed] [Google Scholar]
- Urbanski J, Mogi M, O'Donnell D, DeCotiis M, Toma T, Armbruster P. (2012). Rapid adaptive evolution of photoperiodic response during invasion and range expansion across a climatic gradient. Am Nat 179: 490–500. [DOI] [PubMed] [Google Scholar]
- Usmani-Brown S. (2009). Population genetics of Aedes albopictus (Diptera: Culicidae) invading populations, using mitochondrial nicotinamide adenine dinucleotide dehydrogenase subunit 5 sequences. Ann Entomol Soc Am 102: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Weetman D, Hutchinson WF. (2006). Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6: 255–256. [Google Scholar]
- Vazeille M, Mousson L, Rakatoarivony I, Villeret R, Rodhain F, Duchemin JB et al. (2001). Population genetic structure and competence as a vector for dengue type 2 virus of Aedes aegypti and Aedes albopictus from Madagascar. Am J Trop Med Hyg 65: 491–497. [DOI] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. (1995). Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci 261: 55–63. [DOI] [PubMed] [Google Scholar]
- White VL, Endersby NM, Chan J, Hoffmann AA, Weeks AR. (2015). Developing Exon-Primed Intron-Crossing (EPIC) markers for population genetic studies in three Aedes disease vectors. Insect Sci 22: 409–423. [DOI] [PubMed] [Google Scholar]
- Zawani MKN, Abu HA, Sazaly AB, Zary SY, Darlina MN. (2014). Population genetic structure of Aedes albopictus in Penang, Malaysia. Genet Mol Res 13: 8184–8196. [DOI] [PubMed] [Google Scholar]
- Zhong D, Lo E, Hu R, Metzger ME, Cummings R, Bonizzoni M et al. (2013). Genetic analysis of invasive Aedes albopictus populations in Los Angeles County, California and its potential public health impact. PLoS One 8: e68586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouache K, Fontaine A, Vega-Rua A, Mousson L, Thiberge J-M, Lourenco-De-Oliveira R et al. (2014). Three-way interactions between mosquito population, viral strain and temperature underlying chikungunya virus transmission potential. Proc Biol Sci 281: 20141078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LHR, Ravelonandro P et al. (2011). Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol 75: 377–389. [DOI] [PubMed] [Google Scholar]
- Žitko T, Kovaćić A, Desdevises Y, Puizina J. (2011). Genetic variation in East-Adriatic populations of the Asian tiger mosquito, Aedes albopictus (Diptera: Culicidae), inferred from NADH5 and COI sequence variability. Eur J Entomol 108: 501–508. [Google Scholar]