ABSTRACT
A challenging property of gammaherpesviruses is their ability to establish lifelong persistence. The establishment of latency in B cells is thought to involve active virus engagement of host signaling pathways. Pathogenic effects of these viruses during latency or following reactivation can be devastating to the host. Many cancers, including those associated with members of the gammaherpesvirus family, Kaposi’s sarcoma-associated herpesvirus and Epstein-Barr virus, express elevated levels of active host signal transducer and activator of transcription-3 (STAT3). STAT3 is activated by tyrosine phosphorylation in response to many cytokines and can orchestrate effector responses that include proliferation, inflammation, metastasis, and developmental programming. However, the contribution of STAT3 to gammaherpesvirus pathogenesis remains to be completely understood. This is the first study to have identified STAT3 as a critical host determinant of the ability of gammaherpesvirus to establish long-term latency in an animal model of disease. Following an acute infection, murine gammaherpesvirus 68 (MHV68) established latency in resident B cells, but establishment of latency was dramatically reduced in animals with a B cell-specific STAT3 deletion. The lack of STAT3 in B cells did not impair germinal center responses for immunoglobulin (Ig) class switching in the spleen and did not reduce either total or virus-specific IgG titers. Although ablation of STAT3 in B cells did not have a global effect on these assays of B cell function, it had long-term consequences for the viral load of the host, since virus latency was reduced at 6 to 8 weeks postinfection. Our findings establish host STAT3 as a mediator of gammaherpesvirus persistence.
IMPORTANCE
The insidious ability of gammaherpesviruses to establish latent infections can have detrimental consequences for the host. Identification of host factors that promote viral latency is essential for understanding latency mechanisms and for therapeutic interventions. We provide the first evidence that STAT3 expression is needed for murine gammaherpesvirus 68 to establish latency in primary B cells during an active immune response to infection. STAT3 deletion in B cells does not impair adaptive immune control of the virus, but loss of STAT3 in B cells has a long-lasting impact on viral persistence. These results indicate a potential therapeutic benefit of STAT3 inhibitors for combating gammaherpesvirus latency and, thereby, associated pathologies.
INTRODUCTION
Pathogens that cause chronic disease such as herpesviruses are a challenge to treat and eradicate because they use latency as a strategy of persistence in the host. Most gammaherpesviruses target B lymphocytes as a latency reservoir, ultimately establishing an immunologically silent form of persistence with minimal viral gene expression (1, 2). Viral gene expression during latency can promote lymphoproliferative disease, and lytic reactivation from latent reservoirs can also lead to severe pathologies. It is imperative to identify not only viral determinants but also host determinants that support gammaherpesvirus latency in order to develop novel interventions. Infections by the murine gammaherpesvirus 68 (MHV68) pathogen recapitulate many aspects of human gammaherpesvirus infection, including B cell tropism, long-term establishment of latency in class-switched B cells of the host, and a propensity for lymphomagenesis following impairment of adaptive immune control (2, 3). This model pathogen system affords an analysis of the molecular determinants of latency during the course of a natural host infection.
Signal transducer and activator of transcription 3 (STAT3) is classically activated by tyrosine phosphorylation in response to Janus kinases associated with cytokine receptors (4–6). It is a major downstream target of the interleukin-6 (IL-6) and IL-10 families of cytokines, interferons, growth factors, and oncogenic tyrosine kinases, and it functions as a transcription factor that binds consensus sequences in the regulatory regions of nuclear genes. Constitutive STAT3 activation is associated with oncogenesis (7–10). STAT3 signaling is also stimulated by human gammaherpesvirus gene products such as Kaposi’s sarcoma-associated herpesvirus (KSHV) viral IL-6 (vIL-6) (11–14), kaposin B (15), and viral-G-protein-coupled receptor (v-GPCR) (16, 17) and Epstein-Barr virus (EBV) LMP-1 (18, 19) and EBNA2 (20); and STAT3 levels influence lytic activation of these viruses in cell culture (21–23). Characterized effector responses of STAT3 include survival and proliferation via upregulation of bcl-2 and c-myc, respectively, and promotion of epithelial-mesenchymal transition via Twist (7, 24, 25).
The effect of STAT3 is difficult to predict solely on the basis of putative DNA binding sites. STAT3 binds nonconsensus DNA elements and complexes with transcription factors, including other STATs and NF-κB subunits, and STAT3 functions in phosphorylated and unphosphorylated forms to direct changes in gene expression and chromatin remodeling in a cell-specific manner (4, 26). Nuclear-cytoplasmic shuttling of STAT3 is independent of its phosphorylation status (27); however, STAT3-protein interactions and gene regulation are influenced by posttranslational modifications, in particular, tyrosine and serine phosphorylation (4, 28, 29). STAT3 has also been shown to have nongenomic effects (30, 31).
In the natural course of infection, primary B cells that are infected with a gammaherpesvirus are influenced by the cytokine and cellular interactions in the microenvironment of lymphoid tissues (32–34). Thus, to understand the biological role of STAT3 in shaping gammaherpesvirus latency, an examination of pathogenesis in the whole animal is required. We used MHV68 to examine the impact of STAT3 loss on chronic gammaherpesvirus infection in a natural rodent host. Since the STAT3 murine knockout is embryonically lethal, we used tissue-specific deletion of STAT3 with the Cre-lox recombination system (35). Mice with a B cell-specific deletion of STAT3 (CD19-Cre) were infected with MHV68. We found that MHV68 requires STAT3 to establish latency irrespective of the route of infection and that this intrinsic requirement for STAT3 is not linked to dysfunction in T cell-dependent B cell processes. The latency defect in STAT3-null B cells was sustained for up to 8 weeks after infection. Thus, we conclude that STAT3 is a critical host determinant at both early and late stages of a chronic gammaherpesvirus infection.
RESULTS
Deletion of stat3 from B cells impairs establishment of gammaherpesvirus latency.
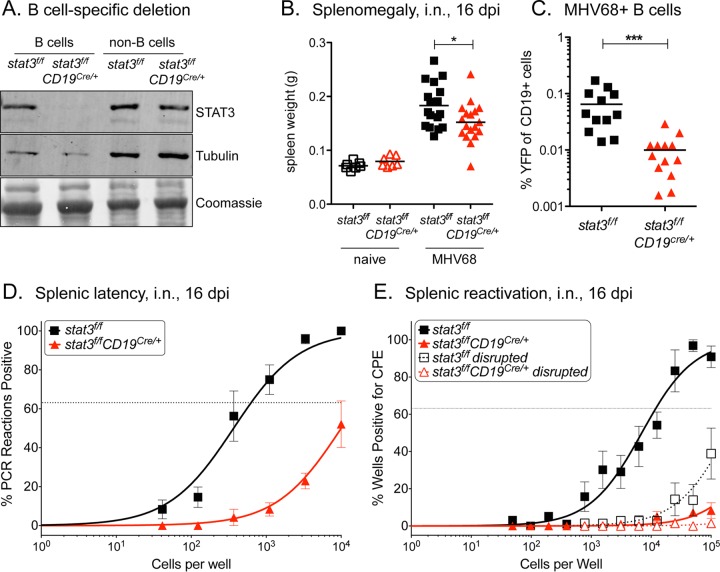
We addressed the impact of STAT3 on the ability of MHV68 to establish B cell latency by infecting mice with a tissue-specific deletion of STAT3 in B cells. Mice with a floxed STAT3 gene (stat3f/f) were crossed with mice expressing the Cre recombinase in B cells (CD19Cre/+) to drive the specific deletion of stat3 in CD19+ B cells (36). Gene knockout efficiency was demonstrated by the absence of detectable levels of STAT3 expression in B cells isolated from splenocytes of stat3f/f;CD19Cre/+ mice (Fig. 1A).
FIG 1 .
STAT3 is critical for the establishment of gammaherpesvirus latency in B cells. (A) Immunoblot of STAT3 from CD19+ B cell splenocytes of naive stat3f/f and stat3f/fCD19Cre/+ mice sorted by flow cytometry to 96% and 86% purity, respectively. (B to E) stat3f/f and stat3f/fCD19Cre/+ mice were infected with 1,000 PFU MHV68-YFP by intranasal (i.n.) inoculation and evaluated at 16 dpi. (B) Weights of spleens from uninfected and infected mice. Three independent experiments were performed with 3 to 7 mice per group. *, P < 0.05. (C) Evaluation of latency in B cells by flow cytometric evaluation of infected YFP+ CD19+ B cells. Two independent experiments were performed with 5 to 7 mice per group. ***, P ≤ 0.001. (D) Frequency of intact splenocytes harboring latent genomes. (E) Frequency of intact splenocytes that reactivated virus following explantation on fibroblasts. Dashed lines indicate disrupted splenocytes prior to quantification of preformed infectious virus. For panels B and C, each symbol represents an individual mouse. For the limiting dilution analyses whose results are shown in panels D and E, curve fit lines were determined by nonlinear regression analysis. Using Poisson distribution analysis, the intersection of the nonlinear regression curves with the dashed line at 63.2% was used to determine the frequency of cells that were positive for either the viral genome or the reactivating virus. Data are representative of the results of three independent experiments performed with 3 to 7 mice per group.
To determine the role of STAT3 in viral latency, we infected stat3f/f;CD19Cre/+ mice and their littermate stat3f/f controls with a recombinant MHV68 strain that encodes a yellow fluorescent protein (YFP) reporter gene (37). Intranasal infection with MHV68-YFP leads to an acute period of lytic replication in the nasopharynx and the lung, followed by rapid expansion of lymphocytes approximating infectious mononucleosis as the virus seeds lymphoid tissues. Within several weeks, replicating virus is cleared as the virus establishes a latent infection, with B cells serving as the predominant reservoir for latency. Splenic enlargement is a characteristic of MHV68 infection and reflects the simultaneous colonization of the spleen with MHV68 and the host response to this infection. At 16 days postinfection (dpi), the spleens of wild-type (WT) stat3f/f mice increased 2-fold by weight, but, in comparison, there was a reduction in splenomegaly in the infected stat3f/f;CD19Cre/+ mice (Fig. 1B).
As a measure of viral colonization, the frequency of B cells that expressed the YFP viral reporter gene in the spleen at 16 dpi was determined by flow cytometry. There was a significant reduction in the percentage of YFP-positive (YFP+) B cells in the stat3f/f;CD19Cre/+ mice compared to the control stat3f/f mice (0.065 ± 0.05% to 0.01 ± 0.008%, P = 0.007, respectively) (Fig. 1C). To determine the efficiency of viral latency establishment, the percentage of B cells that maintained the viral genome at 16 dpi was measured by limiting dilution of splenocytes and viral DNA PCR. Limiting dilution analysis of viral genome-positive cells enables the sensitive quantitation of intact cells that harbor the viral DNA. Approximately 1/623 splenocytes were positive for MHV68 in the control mice, while only 1/17,200 splenocytes were positive for MHV68 in mice lacking STAT3 in B cells (Fig. 1D and Table 1). Considering the reduction in splenomegaly with these results, the absence of STAT3 in B cells of stat3f/f;CD19Cre/+ mice reduced the total number of viral genome-positive splenocytes by 36-fold, from approximately 286,000 cells in the control mice to 7,910 cells in the stat3f/f;CD19Cre/+ mice (Table 1). This substantial reduction in latency was confirmed in an explant reactivation assay. The reactivation assay measures the frequency of intact splenocytes that can produce infectious virus when plated on monolayers of murine fibroblasts in tissue culture. Viral reactivation occurred at a rate of approximately 1/12,038 in the control stat3f/f mice, but reactivation events were barely detectable in the stat3f/f;CD19Cre/+ mice (Fig. 1E). These data indicate that STAT3 is essential to promote efficient establishment of gammaherpesvirus latency in the B cells of the infected animal.
TABLE 1 .
Frequencies of cell populations harboring viral genomes
| Mouse straina | Route of infectionb | Cell populationc | dpi | Total no. of cellsd | Frequency of viral-genome-positive cellse | Total no. of genome-positive cellsf |
|---|---|---|---|---|---|---|
| stat3f/f | i.n. | Splenocyte | 16 | 1.78 × 108 | 1/623 | 286,000 |
| i.n. | Splenocyte | 58 | 1.04 × 108 | 1/13,300 | 7,840 | |
| stat3f/fCD19Cre/+ | i.n. | Splenocyte | 16 | 1.36 × 108 | 1/17,200 | 7,910 |
| i.n. | Splenocyte | 58 | 1.74 × 108 | 1/79,400 | 2,190 | |
| stat3f/f | i.p. | Splenocyte | 16 | 2.62 × 108 | 1/117 | 2,060,000 |
| i.p. | CD19+ B cell | 42 | 3.79 × 107 | 1/3,950 | 9,590 | |
| stat3f/fCD19Cre/+ | i.p. | Splenocyte | 16 | 2.22 × 108 | 1/785 | 284,000 |
| i.p. | CD19+ B cell | 42 | 3.02 × 107 | 1/31,530 | 958 |
stat3f/f mice are the WT stat3 littermate controls corresponding to stat3f/fCD19Cre/+ mice that have a CD19+ B cell-specific deletion of stat3.
i.n., intranasal; i.p., intraperitoneal.
Data represent bulk unsorted splenocytes or the CD19+ B cell population isolated from spleens used for limiting dilution analysis.
Data represent total numbers of the indicated cells isolated from the infected animals. Data represent means of results from cells recovered from two to three independent experiments performed with three to six individual mice per experiment.
Data represent frequencies of the indicated cells that harbored the viral genome based on Poisson distribution analysis.
Data were derived from the experimental frequency data and the approximate total number of cells per population.
STAT3 is not required for gammaherpesvirus lytic replication.
To ensure that impairment of viral latency in the absence of STAT3 is not due to decreased acute viral replication, we first tested lytic replication in stat3−/− murine embryonic fibroblasts (MEFs) devoid of STAT3 (Fig. 2A). There was no apparent change in the kinetics of viral replication in a single-step growth curve in the stat3−/− MEFs compared to parental stat3f/f MEFs (Fig. 2B) (38). Furthermore, following intranasal infection of the mice, acute viral replication and virus expansion in the lungs of stat3f/f;CD19Cre/+ mice were indistinguishable from the results seen in stat3f/f mice (4 and 9 dpi) (Fig. 2C).
FIG 2 .
STAT3 is not essential for gammaherpesvirus replication. (A) Immunoblot of tyrosine-phosphorylated (p-STAT3) and total STAT3 protein in stat3f/f and stat3−/− murine embryonic fibroblasts (MEFs). Vertical lines indicate images joined together from the same blot to remove an independent clone. (B) Single-step growth curve in stat3f/f and stat3−/− MEFs at an MOI of 5.0 with WT MHV68-YFP. Symbols represent data from three independent wells ± standard deviations (SD). (C) Acute replication in the lungs of stat3f/f and stat3f/fCD19Cre/+ mice infected with 1,000 PFU MHV68-YFP by intranasal inoculation at the indicated dpi. (D) Acute replication in the spleens of stat3f/f and stat3f/fCD19Cre/+ mice infected with 1,000 PFU MHV68-YFP by intraperitoneal (i.p.) inoculation at the indicated dpi. Titers of lung and spleen homogenates were determined by plaque assay; the horizontal solid lines indicate the geometric mean titers. Each symbol represents an individual mouse. Dashed lines depict the limit of detection at 50 PFU/ml (log10 of 1.7).
With an intraperitoneal route of infection, the spleen is another organ site of MHV68 replication. Although the levels of acute virus replication were lower in this organ at 4 and 9 dpi, the profile of replication following intraperitoneal administration of 1,000 PFU in the stat3f/f;CD19Cre/+ spleens was similar to the profile seen with stat3f/f spleens (Fig. 2D). Increases of the dose to 10,000 and 100,000 PFU by the intraperitoneal route of infection led to higher levels of infectious virus in the spleen, but, again, the levels of acute replication seen in the stat3f/f;CD19Cre/+ and stat3f/f mice were comparable (data not shown). The results indicate that STAT3 is not essential for MHV68 acute replication, and, more importantly, that the absence of STAT3 in B cells does not impact lytic expansion in the animal prior to colonization of the spleen.
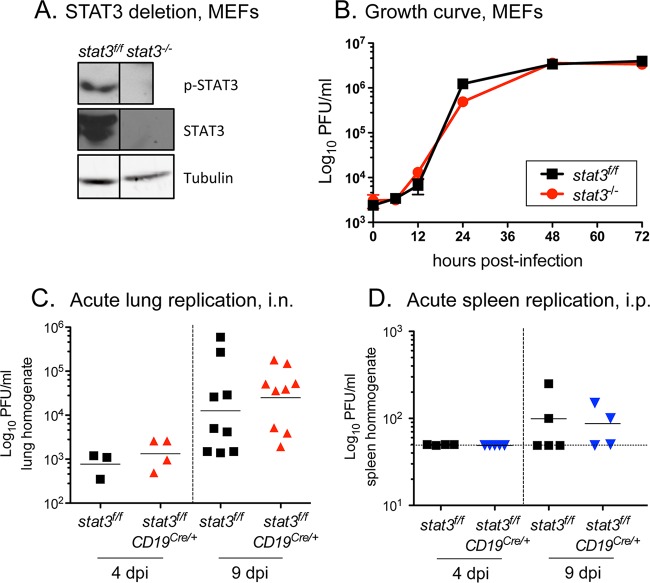
Deletion of stat3 from B cells impairs establishment of gammaherpesvirus latency independently of the infection route.
STAT3 clearly plays a critical role in the establishment of latency after intranasal infection that is not attributable to a defect in the expansion of the virus levels in the lungs (Fig. 1 and 2). Intranasal infection is commonly used as a route of infection for MHV68, since it most closely reflects a physiological relevant route of transmission. However, to understand the contribution of STAT3 to latency establishment and to eliminate the possibility that STAT3 influences B cell trafficking of the virus via the lymph nodes to the spleen, we evaluated the direct intraperitoneal route. Animals were infected with 1,000 PFU by intraperitoneal injection, and splenomegaly, viral latency establishment, and ex vivo viral reactivation were measured. The absence of STAT3 in B cells of infected mice again demonstrated a significant (P < 0.01) decrease in splenomegaly (Fig. 3A). Viral latency establishment at 16 dpi was measured by quantitation of viral genome-positive splenocytes. Limiting dilution of splenocytes and viral DNA PCR analysis showed a reduction from 1/117 cells in the control stat3f/f mice to 1/785 in the stat3f/f;CD19Cre/+ mice (Fig. 3B and Table 1). The frequency of viral reactivation from latency following explantation was also reduced by ~10-fold (P = 0.017) (Fig. 3C). This reduction in latency after intraperitoneal inoculation demonstrates that STAT3 is a critical determinant in B cells for the virus to establish latency. Therefore, the viral latency defect observed after intranasal infection (Fig. 1) is not solely a failure in dissemination and trafficking from the site of acute replication in the lung to the spleen.
FIG 3 .
STAT3 is critical for the establishment of latency in B cells by the more direct intraperitoneal route of inoculation. stat3f/f and stat3f/fCD19Cre/+ mice were infected with 1,000 PFU MHV68-YFP by intraperitoneal inoculation and evaluated 16 dpi. (A) Weights of spleens from uninfected and infected mice. Each symbol represents an individual mouse; 2 independent experiments were performed with three naive mice each, and three independent experiments were performed with 4 infected mice per group. **, P < 0.01. (B) Frequency of intact splenocytes harboring latent genomes. (C) Frequency of intact splenocytes undergoing reactivation from latency upon explantation. Dashed lines indicate splenocytes that were disrupted prior to plating to quantify preformed infectious virus. For the limiting dilution analyses whose results are shown in panels B and C, curve fit lines were determined by nonlinear regression analysis. Using Poisson distribution analysis, the intersection of the nonlinear regression curves with the dashed line at 63.2% was used to determine the frequency of cells that were positive for either the viral genome or the reactivating virus. Data represent compiled results from four independent experiments performed with 4 to 5 mice per group.
B cell STAT3 is not required for germinal center processes during gammaherpesvirus infection.
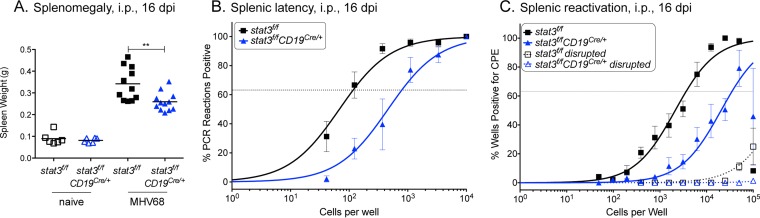
Germinal centers in the spleen are sites of B cell activation, proliferation, immunoglobulin class-switch recombination, and plasma cell and memory cell differentiation. The gammaherpesviruses take advantage of B cell differentiation processes to gain access to immunoglobulin isotype class-switched memory B cells as a long-term latency reservoir (2, 39–44). A previous study of stat3f/f;CD19Cre/+ mice found a reduction in terminal plasma cell differentiation in response to the presence of a conjugated hapten (36). However, to the best of our knowledge, there have been no reports describing the STAT3-dependent B cell response to an active, dynamic virus infection.
To address the potential contribution of STAT3 to infected B cell maturation, proliferation, and differentiation, we examined the entire B cell population in the spleens of mice at 16 dpi, representing an active phase of the host adaptive immune response, 16 days after intraperitoneal infection. The percentages of B cells (CD19+) in the spleen with infection decreased similarly in the stat3f/f and stat3f/f;CD19Cre/+ mice (Fig. 4A), consistent with the infiltration and proliferation of other cell types such as macrophages and T cells in response to gammaherpesvirus infection. Unexpectedly, the frequency of germinal center B cells (GL7+ CD95hi) in the infected stat3f/f;CD19Cre/+ mice was significantly increased in comparison to that seen with infected control stat3f/f mice (Fig. 4B). To evaluate total B cell immunoglobulin class switching, we examined expression of IgG subclasses (Fig. 4C). In germinal center B cells, IgG2b and IgG2c were the predominant immunoglobulin isotypes in B cells of both stat3f/f and stat3f/f;CD19Cre/+ mice, with a slight decrease in IgG2b levels upon the loss of STAT3 (P = 0.046) and no statistical difference in IgG2c levels. Terminal differentiation of B cells to plasma cells was found to occur at equivalent frequencies in the absence and presence of STAT3 (Fig. 4D). These analyses indicate that STAT3 is not essential in B cells for T cell-dependent germinal center formation or differentiation processes in response to intraperitoneal MHV68 infection.
FIG 4 .
Loss of STAT3 in B cells does not impair virus colonization of germinal center and post-germinal-center B cells. stat3f/f and stat3f/fCD19Cre/+ mice were infected with 1,000 PFU MHV68-YFP by intraperitoneal inoculation, and splenocytes were evaluated by flow cytometry at 16 dpi. (A) Frequency of CD19+ B cells of splenocytes gated on live lymphocytes from naive and infected mice. (B to D) Frequency of B cells that bear surface markers of germinal center B cells (GL7+ CD95hi), immunoglobulin class switching (IgG2b+ or IgG2c+ and IgD−), or plasma cells (B220hi CD138lo). Each symbol represents an individual mouse. *, P < 0.05, **, P < 0.01, ***, P ≤ 0.001. (E) Frequency of B cells infected with MHV68 determined by percentage of CD19+ B cells that express the YFP marker. (F to H) Frequency of infected YFP+ B cells that bear surface markers of germinal center B cells (GL7+ CD95hi) (F), immunoglobulin class switching (IgG2b+ or IgG2c+ and IgD−) (G), or plasma cells (B220hi CD138lo) (H). For panels A, D, E, and H, data represent compiled results from three individual experiments performed with 3 to 6 mice per group. For panels B and F, data represent compiled results from two individual experiments performed with 3 naive mice or 4 or 5 infected mice per group. For panels C and G, columns represent means ± SD of results from 4 to 6 individual mice.
In conjunction with the general profile of the B cell response to infection, we analyzed the stage-specific cell surface markers of infected B cells in the absence or presence of STAT3. As with the intranasal route of infection (Fig. 1C), flow cytometric analysis of MHV68 YFP-positive splenocytes from intraperitoneally infected animals revealed an approximately 3-fold (P < 0.001) reduction in the percentage of infected B cells that lacked STAT3 (stat3f/f;CD19Cre/+) compared to the levels seen with control stat3f/f B cells (Fig. 4E). However, over 80% of the YFP-positive cells were identified as germinal center B cells (GL7+ CD95hi), regardless of the presence or absence of STAT3 (Fig. 4F). It is known that establishment of gammaherpesvirus latency in memory immunoglobulin isotype class-switched B cells supports long-term chronic infection (43, 44). For this reason, we examined the frequency of infected YFP-positive B cells with immunoglobulin surface receptors that had switched isotypes to IgG2b or IgG2c by flow cytometry. The distributions of YFP-positive B cells in either the IgG2b+ IgD− or the IgG2c+ IgD− subsets were similar in the control stat3f/f and stat3f/f;CD19Cre/+ mice (Fig. 4G). Therefore, YFP-positive latent B cells that lack STAT3 are not impaired for isotype class switching.
Global loss of plasma cells by a B cell deletion of prdm1 (BLIMP1) reduces establishment of MHV68 latency (42). Since STAT3 has been reported to upregulate prdm1 in vitro and to facilitate antibody production by class-switched B cells in the context of T-dependent antigen in mice (36, 45, 46), we evaluated the differentiation of plasma cells lacking STAT3 in the context of an ongoing MHV68 pathogen infection. While there was a noticeable trend for a decrease in the levels of YFP-positive plasma cells (B220hi CD138lo) that lacked STAT3 (stat3f/f;CD19Cre/+), the mean values did not reach statistical significance in three independent experiments (Fig. 4H). Importantly, the lack of STAT3 did not phenocopy the absolute loss of plasma cells observed in the prdm1f/f;CD19Cre/+ mice. Taken together, STAT3 is required for MHV68 to establish latency in a significant portion of B cells, but this defect is not linked to a failure of infected B cells to participate in B cell differentiation processes initiating in the germinal center of secondary lymphoid tissues.
Deletion of B cell STAT3 does not impair plasma cell function in response to MHV68 infection.
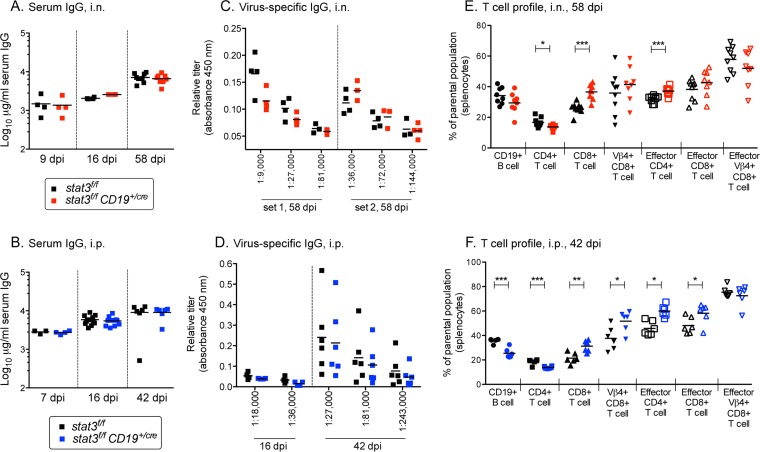
To further determine a potential role of STAT3 in plasma cell differentiation and adaptive immune control in the context of infection with gammaherpesvirus, serum immunoglobulins were measured and T cell profiles were evaluated. Total IgG levels increased with the duration of infection similarly for stat3f/f and stat3f/f;CD19Cre/+ mice, regardless of whether the route of infection was intranasal or intraperitoneal (Fig. 5A and B). Most significantly, comparable levels of virus-specific IgG were generated in the stat3f/f and stat3f/f;CD19Cre/+ mice at early and late times postinfection (Fig. 5C and D). The results indicate that STAT3 is not required for terminal plasma cell differentiation or immunoglobulin secretion in response to MHV68 infection.
FIG 5 .
STAT3 is dispensable for adaptive immune responses to gammaherpesvirus infection. stat3f/f and stat3f/fCD19Cre/+ mice were infected with 1,000 PFU MHV68-YFP by intranasal inoculation (A, C, and E) or intraperitoneal inoculation (B, D, and F) at the indicated dpi. (A and B) Total IgG production in sera of individual mice. The bars represent the geometric mean titers. (C and D) Relative titers of MHV68-specific IgG production in sera of individual mice at the indicated dilutions. (E and F) The T cell profile was evaluated by flow cytometric analysis for the frequency of splenocytes gated on live lymphocytes that bear surface markers of CD4+ T cells (CD4+ CD8− CD19−), CD8+ T cells (CD8+ CD4− CD19−), Vβ4+ CD8+ T cells (Vβ4+ CD8+ CD4− CD19−) or effector CD4+ or CD8+ T cells (CD44hi CD62Llo). Each symbol represents an individual mouse. *, P < 0.05, **, P < 0.01, ***, P < 0.001. For 42 and 58 dpi, data represent compiled results from two independent experiments performed with 3 to 5 mice per group.
CD4+ T cells and CD8+ T cells are critical components of the adaptive immune response and mediate long-term control of MHV68 during chronic infection (3, 47–52). Populations of Vβ4+ CD8+ T cells are dramatically expanded in C57BL/6 mice infected with WT MHV68, serving as an indicator of viral antigen expression and of the generation of T cell effector responses (53, 54). For this reason, we examined the T cell profile of mice by flow cytometry at 8 or 6 weeks after intranasal or intraperitoneal infection, respectively (Fig. 5E and F). There was a slight increase in the frequency of CD4+ T cells and CD8+ T cells in the stat3f/f;CD19Cre/+ mice compared to control stat3f/f mice, regardless of the route of infection. However, the effector Vβ4+ CD8+ T cell populations expanded to comparable extents in the stat3f/f;CD19Cre/+ and stat3f/f mice. Altogether, the data demonstrate that the production of antibodies or effector T cells in response to a chronic gammaherpesvirus infection does not require STAT3 in the B cell compartment.
Deletion of STAT3 in B cells has a sustained negative impact on splenic latency.
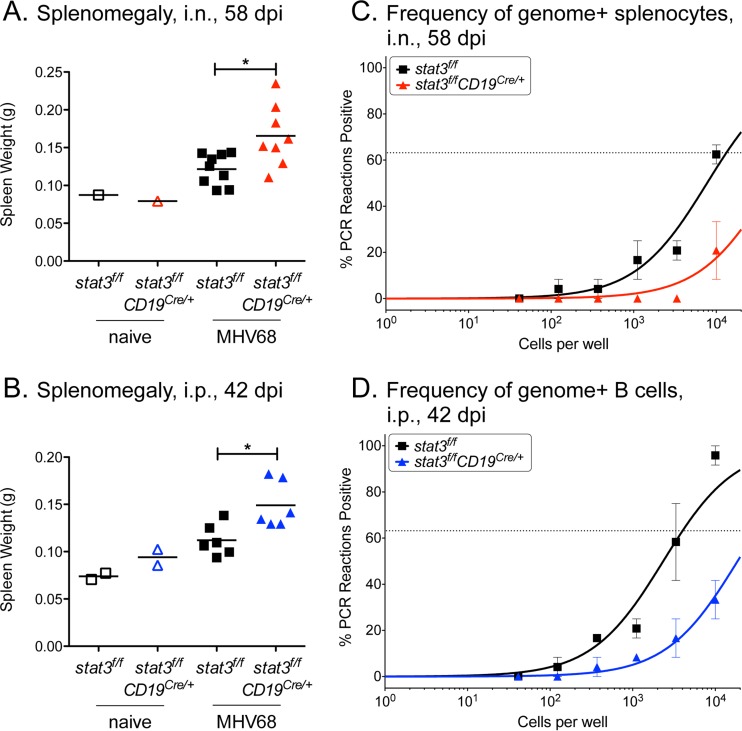
To determine if the role of STAT3 in latency establishment at 16 dpi extended into the maintenance phase of chronic infection, latency was examined 6 or 8 weeks after either intraperitoneal or intranasal infection, respectively. While splenomegaly of the infected mice was diminished at those later time points compared to 16 dpi, the infected spleens of the stat3f/f;CD19Cre/+ mice were significantly larger than those of stat3f/f mice, regardless of the infection route (Fig. 6A and B). The increased splenomegaly was not due to an expansion in the B cell compartment (Table 1) but likely involved an increase in the levels of the T cell subsets that were observed by flow cytometry (Fig. 5E and F). The deletion of STAT3 in splenocytes had a sustained negative impact on viral latency following intranasal infection at the late time point of 58 dpi (Fig. 6C). The frequency of genome-positive splenocytes was diminished by approximately 6-fold in the stat3f/f;CD19Cre/+ mice (1/13,300) compared to the stat3f/f mice (1/79,400) (Table 1). To verify that the defect in latency lay in the B cell reservoir, B cells were enriched from spleens 42 days after intraperitoneal infection. The frequency of viral latency in B cells that lacked STAT3 was ~1/31,530, an 8-fold reduction from the ~1/3,950 of B cells from the control mice (Fig. 6D and Table 1).
FIG 6 .
STAT3 loss in B cells has a sustained impact on splenic latency late during chronic infection regardless of the route of infection. stat3f/f and stat3f/fCD19Cre/+ mice were infected with 1,000 PFU MHV68-YFP by intranasal inoculation and evaluated 58 dpi (A and C) or were infected by intraperitoneal inoculation and evaluated at 42 dpi (B and D). (A and B) Weights of spleens from uninfected and infected mice. Each symbol represents an individual mouse. *, P < 0.05. (C) Frequency of intact splenocytes harboring latent genomes by limiting dilution analyses. (D) Frequency of intact CD19+ B lymphocytes harboring latent genomes determined by limiting dilution analyses. B cells from stat3f/f mice and stat3f/fCD19Cre/+ mice were enriched to 91% to 92% purity and 86% to 89% purity, respectively, by the use of magnetic beads. Curve fit lines were determined by nonlinear regression. Using Poisson distribution analysis, the intersection of the nonlinear regression curves with the dashed line at 63.2% was used to determine the frequency of cells that were positive for the viral genome. For the stat3f/fCD19Cre/+ mice, the last data point falls below 63.2%. Data represent compiled results from two independent experiments performed with 3 to 4 mice per group.
Taken together, these results indicate that MHV68 requires host STAT3 to establish B cell latency in infected animals and that this phenotype does not change with extended time. The loss of STAT3 does not appear to impair splenic B cell Ig class switching or differentiation to plasma cells that are able produce standard amounts of serum immunoglobulin (Fig. 4 and 5). We propose that STAT3 provides a critical function in supporting latency at the earliest stages of gammaherpesvirus engagement with the B cell that has a long-term impact on virus load in the host.
DISCUSSION
The interface between virus and host is influenced by specific host gene expression that can steer the course of infection from nonpathogenic to pathogenic or from lytic to latent. In a murine model of pathogenesis, we identify STAT3 as a critical host determinant of gammaherpesvirus latency in B cells, a reservoir within the microenvironment of secondary lymphoid tissues awash in cytokines known to engage JAK/STAT signaling and activate STAT3 (55). The MHV68 murine model provides a dynamic system to follow long-term latency establishment in the B cell compartment of the host, similarly to human gammaherpesviruses. To determine the contribution of STAT3 to gammaherpesvirus latency in B cells during the course of a dynamic infection, we evaluated animals with a stat3 deletion in B cells. Using this genetic system with a loss of STAT3 confined to CD19+ B cells, we found a substantial reduction in the levels of MHV68 latent DNA during chronic infection.
Plasma cell development in stat3f/fCD19Cre/+ mice appears to be intact. MHV68 is a large DNA virus that provides a myriad of antigenic epitopes, and the levels of virus-specific antibodies and the expansion of the T-cell response were not impaired in the stat3f/fCD19Cre/+ mice following infection. MHV68 reactivation from latency at the early stage of chronic infection has been shown to involve the BLIMP1 transcription factor and its ability to drive terminal plasma cell differentiation (42, 94). STAT3 contributes to BLIMP1 expression and plasma cell differentiation in coordination with IRF4 following IL-21 stimulation in murine B cells (45, 46). Strikingly, we did not observe a defect in germinal center or plasma cell formation in stat3f/fCD19Cre/+ mice. We also did not find evidence for persistent viral replication in the lungs of stat3f/fCD19Cre/+ mice at 8 weeks postinfection (data not shown), an effect previously noted in CD40−/− mice and p50−/− mice that fail to generate cellular and humoral immune responses (56, 57). Taking the data together, B cell STAT3 does not appear to be essential for terminal plasma cell IgG production or virus reactivation in the context of a complex and chronic gammaherpesvirus infection. Similarly, memory B cells from hyper-IgE patients with a spectrum of stat3 loss-of-function mutations were not impaired for plasmablast generation upon cytokine stimulation in culture (58). Thus, other known transcription factors that regulate BLIMP1 to drive B cell differentiation likely compensate for STAT3 loss (59).
Mutations in the DNA-binding or transactivation domain of STAT3 in hyper-IgE syndrome patients are known to negatively impact immune cell function and exacerbate infectious disease and EBV lymphomas (60–64). Since these STAT3 mutations are expressed in all the tissues of the patients, the global impact of the deficiency is profound and complex (65). Even with this proviso, B cells isolated from these patients are unable to support EBV-driven outgrowth (66). In tissue cell culture studies, STAT3 has been reported to contribute to the expression of EBV latency genes (18, 67–69) and to promote EBV- and KSHV-mediated cell proliferation (14, 70). STAT3 activation is associated with latent KSHV infection of endothelial cells (71) and chromatin remodeling that facilitates the latent gene expression program of herpes simplex virus 1 in neurons (72). Thus, STAT3 may serve a dual role in the promotion of a latent gene expression program in newly infected B cells while also promoting proliferation and other effector functions of the cell. These studies illustrate the complex interactions of herpesviruses with the JAK/STAT signaling pathway.
B cells are exposed to cytokines that are known to engage JAK/STAT signaling and activate STAT3 in the lymphoid tissue of MHV68-infected animals (55). Most cytokine pathway knockout studies do not use tissue-specific conditional deletions; as a result, all the cells of the animal lack the specified gene, causing systemic effects. The influence of genetic knockout of several of these cytokines or cytokine receptors has been evaluated in the context of MHV68 pathogenesis, and, notably, the phenotypes of those knockouts are distinct from those seen with the stat3f/f;CD19Cre/+ mice reported here. Mice lacking interferon (IFN) gamma or the IFN gamma receptor exhibited increased levels of viral latency and reactivation in splenocytes and even enhanced reactivation from the macrophage compartment following MHV68 infection (50, 73). IL-6, a classical cytokine activator of STAT3, was found to be dispensable for viral latency and cytotoxic T cell responses to MHV68 infection (74–76).
Two cytokines that activate STAT3 and appear to influence murine gammaherpesvirus latency are IL-10 and IL-21. IL-10 stimulates STAT3 and promotes B cell survival and yet is suppressive for inflammation and impairs T cell control of MHV68 (77–80). IL-10 knockout mice have reduced viral reactivation from splenocytes but an increase in splenic weight and leukocytosis at 15 dpi (81), unlike the infected stat3f/f;CD19Cre/+ mice (Fig. 1). Murine gammaherpesvirus colonization of germinal center B cells is a process driven in large part by IL-21 production by T follicular helper cells (33). IL-21 receptor knockout animals have a defect in latency establishment following intranasal MHV68 infection, but this is linked to a loss of germinal center formation, IgG class switching, and plasma cell differentiation (33). Mixed bone marrow chimera experiments demonstrated that the failure to support latency and participate in germinal center reactions is intrinsic to B cells that cannot sense IL-21 (33). Our infections of the stat3f/fCD19Cre/+ mice by the intraperitoneal route did not reveal a generalized defect in germinal center participation or IgG class switching for B cells lacking STAT3. Viral latency was clearly impaired in the STAT3-null B cells, but, among the cells that were infected, there was no blockade in germinal-center-dependent B cell differentiation processes (Fig. 4F to H). The effects of a single cytokine or a cytokine receptor knockout in the microenvironment of the spleen may be compensated for by other cytokines to some degree. Future pathogenesis studies of receptor knockouts crossed to the stat3f/f;CD19Cre/+ mice may determine if STAT3 is the key downstream host factor required for B cell-intrinsic responses to specific cytokines to support gammaherpesvirus latency.
We observed a relative decrease in the percentage of splenic B cells that corresponded to an increase in levels of CD4+ T cells and CD8+ T cells at 6 to 8 weeks after infection of the B cell-specific stat3-deleted mice, regardless of the route of infection (Fig. 5). The reduction in the viral load at those late times may be partially attributable to an increase in effector T cell function. T cell responses would be expected to increase in the context of antigen exposure. However, the deletion of stat3 reduced viral latency and there was no increase in reactivation from the spleen or peritoneal cavity and no evidence of recrudescence in the lungs (data not shown). Perhaps STAT3 functions to repress sporadic reactivation events during chronic infection that result in viral antigen presentation. Another consideration is the role of STAT3-regulated immunosuppressive cytokine IL-10 in suppressing T cell responses and driving regulatory T cell differentiation (82–85). IL-10 dampens T cell responses to MHV68 infection (80, 86). Since IL-10 production is driven by the viral M2 gene product in MHV68-infected B cells, a reduction in latency at 16 dpi might reduce IL-10 levels (86, 87). In addition, disregulation of the IFN-alpha/pSTAT3/IL-10 axis in systemic lupus erythematosus patients has revealed that the levels of type 1 IFN produced by plasmacytoid dendritic cells dictate the formation of regulatory B cells in a STAT3-dependent manner (88). Future studies are needed to determine whether STAT3 deletion in B cells influences IL-10 production and T cell responses during gammaherpesvirus infection.
Our study was the first to demonstrate the impact of a targeted deletion of the STAT3 transcription factor specifically in B cells, the primary latency reservoir for murine gammaherpesvirus. Given the B cell-intrinsic phenotype in the mouse model, we are now poised to dissect the specific contributions of STAT3 that enable a gammaherpesvirus to establish a latent gene expression program in vivo. STAT3 may directly regulate MHV68 gene expression or indirectly promote a cellular environment conducive to B cell latency. MHV68 targets transitional B cells and marginal-zone B cells (89–91), and STAT3 might play a critical role in the infection of these subsets prior to germinal center entry. The defects apparent in the early period of latency establishment were not ameliorated with time, indicating that STAT3 is a critical determinant of both the establishment and the maintenance of latency in the infected host. While many STAT3-responsive genes have been identified in transformed cells, the contribution of STAT3 to B cell biology and gammaherpesvirus latency in the host is poorly understood. Future studies to identify target host and viral genes in the infected B cells that require STAT3 to support gammaherpesvirus latency may provide understanding that will identify novel therapeutics that can be tested in the murine model.
MATERIALS AND METHODS
Mice and cells.
stat3f/f mice (38) and CD19-Cre mice [B6.129P2(C)-Cd19 tm1(cre)Cgn/J; The Jackson Laboratory, Bar Harbor, ME] were bred at the Stony Brook University Division of Laboratory Animal Research facility. All protocols were approved by the Institutional Animal Care and Use Committee of Stony Brook University. The stat3f/f and stat3f/fCD19Cre/+ mice used in all pathogenesis experiments were littermates derived from crossing stat3f/f mice with stat3f/f mice heterozygous for CD19-Cre. Immortalized stat3f/f and stat3−/− MEFs were as described previously (92).
Infections and organ harvests.
MHV68 expressing the H2B-YFP protein (MHV68-YFP) was used for the infections (37). WT mice (8 to 12 weeks old) were either infected by intranasal inoculation with 1,000 PFU MHV68-YFP in 20 µl or by intraperitoneal injection with 1,000 PFU in 0.5 ml under conditions of isoflurane anesthesia. Mice were sacrificed by the use of terminal isoflurane anesthesia. For determination of acute titers, mouse lungs or spleens were harvested and stored at −80°C prior to disruption in a Mini-BeadBeater (BioSpec, Bartlesville, OK). The titers of the homogenates were determined by plaque assay. For latency, reactivation, and flow cytometry experiments, mouse spleens were homogenized, treated to remove red blood cells, and passed through a 100-µm-pore-size nylon filter.
Immunoblot analysis.
Total protein lysate was harvested in lysis buffer (150 mM sodium chloride, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris [pH 8.0]) supplemented with a protease inhibitor cocktail (Sigma, St. Louis, MO) and phenylmethylsulfonyl fluoride (PMSF). Proteins were separated on 10% SDS-PAGE gels and transferred to a polyvinylidene fluoride membrane. Antibodies against STAT3 (K-15; Santa Cruz Biotechnology, Dallas, TX), tyrosine 705-phosphorylated STAT3 (Cell Signaling Technology, Danvers, MA), and antitubulin (clone B-5-1-2; Sigma) were detected by the use of secondary anti-mouse (Rockland, Limerick, PA) or secondary anti-rabbit (Invitrogen, Grand Island, NY) antibodies by immunoblot analysis with an Odyssey Imager (Li-COR Biosciences, Lincoln, NE).
Virus replication.
stat3f/f and stat3−/− MEFs were seeded in triplicate in 6-well plates, and cells were infected the following day at a multiplicity of infection (MOI) of 5 PFU per cell for single-step replication kinetics. The cells and conditioned medium were freeze-thawed four times, and NIH3T12 fibroblast cells were infected with serial dilutions of the samples for 1 h and then overlaid with a 1.5% methylcellulose solution in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum (FBS). One week later, the monolayers were washed with phosphate-buffered saline (PBS) and fixed in 100% methanol and plaques were visualized by staining using 0.01% crystal violet solution containing 20% methanol.
Limiting dilution analysis of latency and reactivation.
To determine the frequency of cells harboring the viral genome, single-cell suspensions were prepared for single-copy nested PCR. In brief, six 3-fold serial dilutions of intact cells were plated on NIH 3T12 cells and lysed overnight at 56°C with proteinase K. The samples were subjected to an 80-cycle nested PCR with primers specific for MHV68 ORF50 (93). A total of 12 replicates were analyzed at each serial dilution, and plasmid DNA was included at 0.1, 1.0, and 10 copies as a control. To determine the frequency of cells harboring latent virus capable of reactivation upon explantation, single-cell suspensions were plated in 12 serial 2-fold dilutions on a monolayer of MEF cells prepared from C57BL/6 mice. A total of 24 replicates were plated per serial dilution. Cytopathic effect (CPE) was scored 2 and 3 weeks after plating. To differentiate between preformed infectious virus and virus spontaneously reactivating upon cell explantation, parallel samples were mechanically disrupted using a Mini-BeadBeater prior to plating on the monolayer of MEFs to release preformed virus and the results were scored as CPE (93).
Flow cytometry.
For the analysis of B cells, 2 × 106 splenocytes were resuspended in 200 µl of fluorescence-activated cell sorter (FACS) buffer (PBS with 2% fetal bovine serum) and blocked with TruStain fcX (clone 93; BioLegend, San Diego, CA). The cells were washed and stained to identify B cell subsets with fluorophore-conjugated antibodies against CD19 (clone 6D5), B220 (clone RA3-682) CD138 (clone 281-2), CD95 (clone 15A7), and IgD (clone 11-26c.2a) or biotinylated antibodies against GL7 (clone GL-7), IgG1 (clone RMG1-1), IgG2b (clone R12-3), IgG2c (clone RMG2a-62), or IgG3 (clone R40-82) that were detected by the use of secondary streptavidin-conjugated allophycocyanin. T cell subsets were identified with antibodies against CD4 (clone GK1.5), CD8 (clone 53-6.7), Vβ4 (clone KT4), CD44 (clone IM7), and CD62L (clone MEL-14). For sorting, splenocytes were first enriched for B cells by depletion of non-B cells with magnetic microbeads (Pan B cell isolation kit; Miltenyi Biotec, Auburn, CA) and then sorted using CD19 conjugated to phycoerythrin and a FACSAria III sorter (BD Biosciences). All antibodies were purchased from BioLegend or BD Biosciences (San Jose, CA). The data were collected using a Dxp-8 FACScan flow cytometer (Cytek Development, Fremont, CA) and analyzed using FlowJoX v10.0.7 (Treestar Inc., Ashland, OR).
Enzyme-linked immunosorbent assay (ELISA).
To measure total IgG levels in the serum, plates were coated with 2 µg/ml of donkey anti-mouse IgG (Affinipure; Jackson ImmunoResearch Laboratories, West Grove, PA)–PBS, washed in PBS with 0.05% Tween 20, and blocked in 1% bovine serum albumin (BSA)–PBS prior to incubation with serial dilutions of serum or the mouse IgG standard (ChromPure; Jackson ImmunoResearch) in assay diluent (OptEIA assay diluent; BD Biosciences) overnight at 4°C. IgG was detected by the use of horseradish peroxidase-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch), R & D Systems (Minneapolis, MN) substrate, and stop solution, and absorbance at 450 nm was read on a FilterMax5 microplate reader (Molecular Devices, Sunnyvale, CA). To measure levels of IgG specific for viral antigens, plates were first coated with 0.5% paraformaldehyde-fixed viral antigen–PBS overnight at 4°C.
Statistical analyses.
Data were analyzed using GraphPad Prism software (Prism 5; GraphPad Software, Inc., La Jolla, CA). Statistical significance was determined using either analysis of variance (ANOVA) followed by a Tukey’s posttest or a nonpaired two-tailed t test. Using Poisson distribution analysis, the frequencies of latency establishment and reactivation from latency were determined by the intersection of nonlinear regression curves with the line at 63.2%. Significance was evaluated by paired one-tailed t tests of the log-transformed frequency values of samples from matched experiments.
ACKNOWLEDGMENTS
S.S.R., H.-C.F., N.C.R., and L.T.K. designed the experiments. S.S.R., H.-C.F., T.O.S., G.H.P., and L.T.K. executed the experiments. S.S.R., H.-C.F., N.C.R., and L.T.K. analyzed the data. N.C.R., V.P., and L.T.K. prepared the manuscript.
We thank Brandon Cieniewicz, Qiwen Dong, and Nana Minkah for technical support, the laboratory of Maurizio del Poeta for sharing critical equipment, Laurie Levine for animal husbandry, and Todd Rueb in the Flow Cytometry Core. We also thank members of the laboratories of Nicholas Carpino, L.T.K., and N.C.R. for helpful discussions.
Funding Statement
T.O.S. was supported by a Howard Hughes Medical Fellowship and the Chancellor’s Education Pipeline Biomedical Research Award from the Center for Science and Mathematics Education (CESAME) at Stony Brook University; V.P. was supported by the Italian Ministry of Research and University (MIUR PRIN); N.C.R. was supported by the NIH (NIH R56AI095268) and a Carol M. Baldwin Breast Cancer Research Award; L.T.K. was supported by an American Cancer Society Research Scholar grant (RSG-11-160-01-MPC) and the NIH (NIH RO1AI110079).
Footnotes
Citation Reddy SS, Foreman H-C, Sioux TO, Park GH, Poli V, Reich NC, Krug LT. 2016. Ablation of STAT3 in the B cell compartment restricts gammaherpesvirus latency in vivo. mBio 7(4):e00723-16. doi:10.1128/mBio.00723-16.
REFERENCES
- 1.Jha HC, Banerjee S, Robertson ES. 2016. The role of gammaherpesviruses in cancer pathogenesis. Pathogens 5:18. doi: 10.3390/pathogens5010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speck SH, Ganem D. 2010. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton E, Mandal P, Speck SH. 2011. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol 29:351–397. doi: 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- 4.Stark GR, Darnell JE Jr.. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy DE, Darnell JE. 2002. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 6.Schindler C, Levy DE, Decker T. 2007. JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE. 1999. Stat3 as an oncogene. Cell 98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 8.Barbieri I, Pensa S, Pannellini T, Quaglino E, Maritano D, Demaria M, Voster A, Turkson J, Cavallo F, Watson CJ, Provero P, Musiani P, Poli V. 2010. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res 70:2558–2567. doi: 10.1158/0008-5472.CAN-09-2840. [DOI] [PubMed] [Google Scholar]
- 9.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. 2005. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Pardoll D, Jove R. 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. 1997. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem 272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 12.Giffin L, Yan F, Ben Major M, Damania B. 2014. Modulation of Kaposi’s sarcoma-associated herpesvirus interleukin-6 function by hypoxia-upregulated protein 1. J Virol 88:9429–9441. doi: 10.1128/JVI.00511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu F, Nicholas J. 2006. Signal transduction by human herpesvirus 8 viral interleukin-6 (vIL-6) is modulated by the nonsignaling gp80 subunit of the IL-6 receptor complex and is distinct from signaling induced by human IL-6. J Virol 80:10874–10878. doi: 10.1128/JVI.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Xu Y, Mo D, Huang P, Sun R, Huang L, Pan S, Xu J. 2014. Kaposi’s sarcoma-associated herpesvirus (KSHV) vIL-6 promotes cell proliferation and migration by upregulating DNMT1 via STAT3 activation. PLoS One 9:e93478. doi: 10.1371/journal.pone.0093478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King CA. 2013. Kaposi’s sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J Virol 87:8779–8791. doi: 10.1128/JVI.02976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Guo S. 2010. p38 and STAT3 activation by vGPCR in KSHV-infected cells. Virus Adaptation Treat 2:103–113. doi: 10.2147/VAAT.S13434. [DOI] [Google Scholar]
- 17.Burger M, Hartmann T, Burger JA, Schraufstatter I. 2005. KSHV-GPCR and CXCR2 transforming capacity and angiogenic responses are mediated through a JAK2-STAT3-dependent pathway. Oncogene 24:2067–2075. doi: 10.1038/sj.onc.1208442. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Hutt-Fletcher L, Cao L, Hayward SD. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J Virol 77:4139–4148. doi: 10.1128/JVI.77.7.4139-4148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shair KH, Bendt KM, Edwards RH, Bedford EC, Nielsen JN, Raab-Traub N. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog 3:e166. doi: 10.1371/journal.ppat.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muromoto R, Ikeda O, Okabe K, Togi S, Kamitani S, Fujimuro M, Harada S, Oritani K, Matsuda T. 2009. Epstein-Barr virus-derived EBNA2 regulates STAT3 activation. Biochem Biophys Res Commun 378:439–443. doi: 10.1016/j.bbrc.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 21.King CA, Li X, Barbachano-Guerrero A, Bhaduri-McIntosh S. 2015. STAT3 regulates lytic activation of Kaposi’s sarcoma-associated herpesvirus. J Virol 89:11347–11355. doi: 10.1128/JVI.02008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, Tangye SG, Farrell PJ, Bhaduri-McIntosh S. 2013. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. J Virol 87:11438–11446. doi: 10.1128/JVI.01762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin Q, Xu Y, He T, Qin C, Xu J. 2012. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM. 2009. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28:2940–2947. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D. 2013. Distinct transcriptional regulatory modules underlie STAT3’s cell type-independent and cell type-specific functions. Nucleic Acids Res 41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, McBride KM, Reich NC. 2005. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci U S A 102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. 2010. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A 107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65:939–947. [PubMed] [Google Scholar]
- 30.Wegrzyn J, Potla R, Chwae Y-J, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu X-Y, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. 2009. Function of mitochondrial Stat3 in cellular respiration. Science 323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. 2009. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrest JC, Krug LT, Speck SH. 2008. Murine gammaherpesvirus 68 infection of mice: a small animal model for characterizing basic aspects of gamma-herpesvirus pathogenesis. In Damania B, Pipas J (ed), DNA tumor viruses. Springer Verlag, New York, NY. [Google Scholar]
- 33.Collins CM, Speck SH. 2015. Interleukin 21 signaling in B cells is required for efficient establishment of murine gammaherpesvirus latency. PLoS Pathog 11:e1004831. doi: 10.1371/journal.ppat.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins CM, Speck SH. 2014. Expansion of murine gammaherpesvirus latently infected B cells requires T follicular help. PLoS Pathog 10:e1004106. doi: 10.1371/journal.ppat.1004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akira S. 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19:2607–2611. doi: 10.1038/sj.onc.1203478. [DOI] [PubMed] [Google Scholar]
- 36.Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. 2006. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood 107:1085–1091. doi: 10.1182/blood-2005-07-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins CM, Boss JM, Speck SH. 2009. Identification of infected B-cell populations by using a recombinant murine gammaherpesvirus 68 expressing a fluorescent protein. J Virol 83:6484–6493. doi: 10.1128/JVI.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. 2001. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol 21:1621–1632. doi: 10.1128/MCB.21.5.1621-1632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babcock GJ, Thorley-Lawson DA. 2000. Tonsillar memory B cells, latently infected with Epstein-Barr virus, express the restricted pattern of latent genes previously found only in Epstein-Barr virus-associated tumors. Proc Natl Acad Sci U S A 97:12250–12255. doi: 10.1073/pnas.200366597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babcock GJ, Hochberg D, Thorley-Lawson AD. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497–506. doi: 10.1016/S1074-7613(00)00049-2. [DOI] [PubMed] [Google Scholar]
- 41.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404. doi: 10.1016/S1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 42.Siegel AM, Rangaswamy US, Napier RJ, Speck SH. 2010. Blimp-1-dependent plasma cell differentiation is required for efficient maintenance of murine gammaherpesvirus latency and antiviral antibody responses. J Virol 84:674–685. doi: 10.1128/JVI.01306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willer DO, Speck SH. 2003. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J Virol 77:8310–8321. doi: 10.1128/JVI.77.15.8310-8321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim IJ, Flaño E, Woodland DL, Lund FE, Randall TD, Blackman MA. 2003. Maintenance of long term gamma-herpesvirus B cell latency is dependent on CD40-mediated development of memory B cells. J Immunol 171:886–892. doi: 10.4049/jimmunol.171.2.886. [DOI] [PubMed] [Google Scholar]
- 45.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim H-P, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diehl SA, Schmidlin H, Nagasawa M, van Haren SD, Kwakkenbos MJ, Yasuda E, Beaumont T, Scheeren FA, Spits H. 2008. STAT3-mediated up-regulation of BLIMP1 is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol 180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obar JJ, Crist SG, Gondek DC, Usherwood EJ. 2004. Different functional capacities of latent and lytic antigen-specific CD8 T cells in murine gammaherpesvirus infection. J Immunol 172:1213–1219. doi: 10.4049/jimmunol.172.2.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usherwood EJ, Roy DJ, Ward K, Surman SL, Dutia BM, Blackman MA, Stewart JP, Woodland DL. 2000. Control of gammaherpesvirus latency by latent antigen-specific CD8(+) T cells. J Exp Med 192:943–952. doi: 10.1084/jem.192.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Z, Blackman MA, Kaye KM, Usherwood EJ. 2015. Functional heterogeneity in the CD4+ T cell response to murine gamma-herpesvirus 68. J Immunol 194:2746–2756. doi: 10.4049/jimmunol.1401928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tibbetts SA, van Dyk LF, Speck SH, Virgin H IV. 2002. Immune control of the number and reactivation phenotype of cells latently infected with a gammaherpesvirus. J Virol 76:7125–7132. doi: 10.1128/JVI.76.14.7125-7132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doherty PC, Christensen JP, Belz GT, Stevenson PG, Sangster MY. 2001. Dissecting the host response to a gamma-herpesvirus. Philos Trans R Soc Lond B Biol Sci 356:581–593. doi: 10.1098/rstb.2000.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardin RD, Brooks JW, Sarawar SR, Doherty PC. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med 184:863–871. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tripp RA, Hamilton-Easton AM, Cardin RD, Nguyen P, Behm FG, Woodland DL, Doherty PC, Blackman MA. 1997. Pathogenesis of an infectious mononucleosis-like disease induced by a murine gamma-herpesvirus: role for a viral superantigen? J Exp Med 185:1641–1650. doi: 10.1084/jem.185.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans AG, Moser JM, Krug LT, Pozharskaya V, Mora AL, Speck SH. 2008. A gammaherpesvirus-secreted activator of Vbeta4+ CD8+ T cells regulates chronic infection and immunopathology. J Exp Med 205:669–684. doi: 10.1084/jem.20071135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarawar SR, Cardin RD, Brooks JW, Mehrpooya M, Tripp RA, Doherty PC. 1996. Cytokine production in the immune response to murine gammaherpesvirus 68. J Virol 70:3264–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willer DO, Speck SH. 2005. Establishment and maintenance of long-term murine gammaherpesvirus 68 latency in B cells in the absence of CD40. J Virol 79:2891–2899. doi: 10.1128/JVI.79.5.2891-2899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krug LT, Collins CM, Gargano LM, Speck SH. 2009. NF-kappaB p50 plays distinct roles in the establishment and control of murine gammaherpesvirus 68 latency. J Virol 83:4732–4748. doi: 10.1128/JVI.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deenick EK, Avery DT, Chan A, Berglund LJ, Ives ML, Moens L, Stoddard JL, Bustamante J, Boisson-Dupuis S, Tsumura M, Kobayashi M, Arkwright PD, Averbuch D, Engelhard D, Roesler J, Peake J, Wong M, Adelstein S, Choo S, Smart JM, French MA, Fulcher DA, Cook MC, Picard C, Durandy A, Klein C, Holland SM, Uzel G, Casanova JL, Ma CS, Tangye SG. 2013. Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J Exp Med 210:2739–2753. doi: 10.1084/jem.20130323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martins G, Calame K. 2008. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 60.Casanova J-L, Holland SM, Notarangelo LD. 2012. Inborn errors of human JAKs and STATs. Immunity 36:515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L-Y, Tian W, Shu L, Jiang L-P, Zhan Y-Z, Liu W, Zhao X-D, Cui Y-X, Tang X-M, Wang M, Wu D-Q, Yang X-Q. 2013. Clinical features, STAT3 gene mutations and Th17 cell analysis in nine children with hyper-IgE syndrome in mainland China. Scand J Immunol 78:258–265. doi: 10.1111/sji.12063. [DOI] [PubMed] [Google Scholar]
- 62.Ives ML, Ma CS, Palendira U, Chan A, Bustamante J, Boisson-Dupuis S, Arkwright PD, Engelhard D, Averbuch D, Magdorf K, Roesler J, Peake J, Wong M, Adelstein S, Choo S, Smart JM, French MA, Fulcher DA, Cook MC, Picard C, Durandy A, Tsumura M, Kobayashi M, Uzel G, Casanova J-L, Tangye SG, Deenick EK. 2013. Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8(+) T-cell memory formation and function. J Allergy Clin Immunol 132:400–411.e9. doi: 10.1016/j.jaci.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. 2011. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumánovics A, Perkins SL, Gilbert H, Cessna MH, Augustine NH, Hill HR. 2010. Diffuse large B cell lymphoma in hyper-IgE syndrome due to STAT3 mutation. J Clin Immunol 30:886–893. doi: 10.1007/s10875-010-9452-z. [DOI] [PubMed] [Google Scholar]
- 65.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schäffer AA, Puck JM, Grimbacher B. 2007. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 66.Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. 2014. B lymphocytes from patients with a hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. J Virol 88:516–524. doi: 10.1128/JVI.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H, Lee JM, Wang Y, Huang DP, Ambinder RF, Hayward SD. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc Natl Acad Sci U S A 96:9339–9344. doi: 10.1073/pnas.96.16.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen H, Lee JM, Zong Y, Borowitz M, Ng MH, Ambinder RF, Hayward SD. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J Virol 75:2929–2937. doi: 10.1128/JVI.75.6.2929-2937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kis LL, Salamon D, Persson EK, Nagy N, Scheeren FA, Spits H, Klein G, Klein E. 2010. IL-21 imposes a type II EBV gene expression on type III and type I B cells by the repression of C- and activation of LMP-1-promoter. Proc Natl Acad Sci U S A 107:872–877. doi: 10.1073/pnas.0912920107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koganti S, Hui-Yuen J, McAllister S, Gardner B, Grasser F, Palendira U, Tangye SG, Freeman AF, Bhaduri-McIntosh S. 2014. STAT3 interrupts ATR-Chk1 signaling to allow oncovirus-mediated cell proliferation. Proc Natl Acad Sci U S A 111:4946–4951. doi: 10.1073/pnas.1400683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Punjabi AS, Carroll PA, Chen L, Lagunoff M. 2007. Persistent activation of STAT3 by latent Kaposi’s sarcoma-associated herpesvirus infection of endothelial cells. J Virol 81:2449–2458. doi: 10.1128/JVI.01769-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dutia BM, Clarke CJ, Allen DJ, Nash AA. 1997. Pathological changes in the spleens of gamma interferon receptor-deficient mice infected with murine gammaherpesvirus: a role for CD8 T cells. J Virol 71:4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wegenka UM, Buschmann J, Lütticken C, Heinrich PC, Horn F. 1993. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol Cell Biol 13:276–288. doi: 10.1128/MCB.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akira S, Taga T, Kishimoto T. 1993. Interleukin-6 in biology and medicine. Adv Immunol 54:1–78. doi: 10.1016/S0065-2776(08)60532-5. [DOI] [PubMed] [Google Scholar]
- 76.Sarawar SR, Brooks JW, Cardin RD, Mehrpooya M, Doherty PC. 1998. Pathogenesis of murine gammaherpesvirus-68 infection in interleukin-6-deficient mice. Virology 249:359–366. doi: 10.1006/viro.1998.9309. [DOI] [PubMed] [Google Scholar]
- 77.Riley JK, Takeda K, Akira S, Schreiber RD. 1999. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem 274:16513–16521. doi: 10.1074/jbc.274.23.16513. [DOI] [PubMed] [Google Scholar]
- 78.Rousset F, Garcia E, Defrance T, Péronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J. 1992. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci U S A 89:1890–1893. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Z, Zhang W, Usherwood EJ. 2013. Regulatory CD8+ T cells associated with erosion of immune surveillance in persistent virus infection suppress in vitro and have a reversible proliferative defect. J Immunol 191:312–322. doi: 10.4049/jimmunol.1201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molloy MJ, Zhang W, Usherwood EJ. 2011. Suppressive CD8+ T cells arise in the absence of CD4 help and compromise control of persistent virus. J Immunol 186:6218–6226. doi: 10.4049/jimmunol.1003812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peacock JW, Bost KL. 2001. Murine gammaherpesvirus-68-induced interleukin-10 increases viral burden, but limits virus-induced splenomegaly and leukocytosis. Immunology 104:109–117. doi: 10.1046/j.1365-2567.2001.01286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsu P, Santner-Nanan B, Hu M, Skarratt K, Lee CH, Stormon M, Wong M, Fuller SJ, Nanan R. 2015. IL-10 potentiates differentiation of human induced regulatory T cells via STAT3 and FoxO1. J Immunol 195:3665–3674. doi: 10.4049/jimmunol.1402898. [DOI] [PubMed] [Google Scholar]
- 83.Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HW. 2000. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J Immunol 165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 84.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. 2012. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 491:264–268. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maseda D, Candando KM, Smith SH, Kalampokis I, Weaver CT, Plevy SE, Poe JC, Tedder TF. 2013. Peritoneal cavity regulatory B cells (B10 cells) modulate IFN-γ+CD4+ T cell numbers during colitis development in mice. J Immunol 191:2780–2795. doi: 10.4049/jimmunol.1300649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siegel AM, Herskowitz JH, Speck SH. 2008. The MHV68 M2 protein drives IL-10 dependent B cell proliferation and differentiation. PLoS Pathog 4:e1000039. doi: 10.1371/journal.ppat.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rangaswamy US, Speck SH. 2014. Murine gammaherpesvirus M2 protein induction of IRF4 via the NFAT pathway leads to IL-10 expression in B cells. PLoS Pathog 10:e1003858. doi: 10.1371/journal.ppat.1003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Menon M, Blair PA, Isenberg DA, Mauri C. 2016. A regulatory feedback between plasmacytoid dendritic cells and regulatory B cells is aberrant in systemic lupus erythematosus. Immunity 44:683–697. doi: 10.1016/j.immuni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coleman CB, Nealy MS, Tibbetts SA. 2010. Immature and transitional B cells are latency reservoirs for a gammaherpesvirus. J Virol 84:13045–13052. doi: 10.1128/JVI.01455-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coleman CB, McGraw JE, Feldman ER, Roth AN, Keyes LR, Grau KR, Cochran SL, Waldschmidt TJ, Liang C, Forrest JC, Tibbetts SA. 2014. A gammaherpesvirus Bcl-2 ortholog blocks B cell receptor-mediated apoptosis and promotes the survival of developing B cells in vivo. PLoS Pathog 10:e1003916. doi: 10.1371/journal.ppat.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frederico B, Chao B, May JS, Belz GT, Stevenson PG. 2014. A murid gamma-herpesviruses exploits normal splenic immune communication routes for systemic spread. Cell Host Microbe 15:457–470. doi: 10.1016/j.chom.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 92.Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is’harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. 2002. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A 99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weck KE, Kim SS, Virgin HW IV, Speck SH. 1999. B cells regulate murine gammaherpesvirus 68 latency. J Virol 73:4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang X, Collins CM, Mendel JB, Iwakoshi NN, Speck SH. 2009. Gammaherpesvirus-driven plasma cell differentiation regulates virus reactivation from latently infected B lymphocytes. PLoS Pathog 5:e1000677. doi: 10.1371/journal.ppat.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]