ERRATUM
Volume 7, no 2, doi:10.1128/mBio.00166-16, 2016. Some chemical structures in the downstream AB- and CD-ring degradation pathways in Fig. 1 were incorrect. The revised Fig. 1 (below) shows the correct structures.
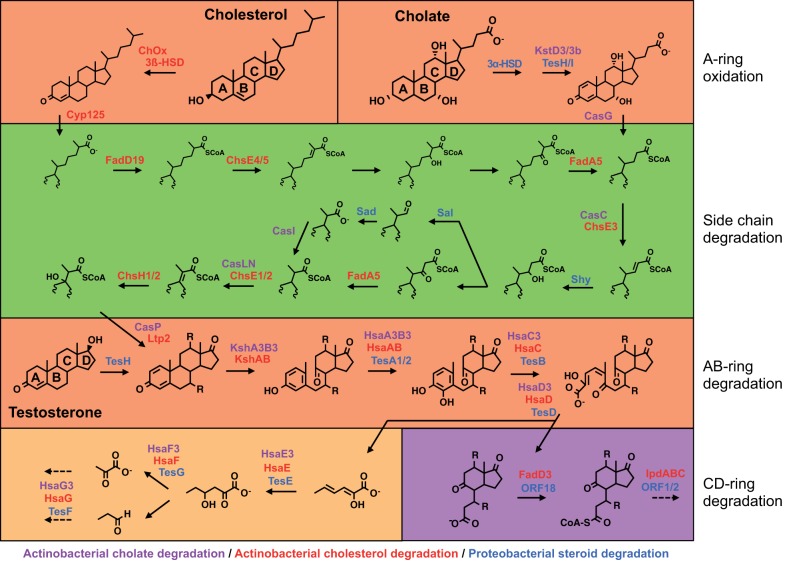
FIG 1 .
Aerobic 9,10-seco-steroid degradation pathways for cholesterol, cholate, and testosterone. The steroid ring structure is degraded by oxygen-dependent opening and subsequent hydrolytic cleavage of rings A and B. Subsequent degradation of the C and D rings occurs by a mechanism not yet reported. In Actinobacteria, side chain degradation and ring opening can occur simultaneously. Characterized or annotated enzymes involved in the degradation of cholesterol by Actinobacteria are shown in red, those involved in the degradation of cholate by Actinobacteria are shown in lilac, and those involved in the degradation of testosterone or cholate by Proteobacteria are shown in blue. Protein nomenclature is based on that for Rhodococcus jostii RHA1, Mycobacterium tuberculosis H37Rv, Comamonas testosteroni TA441, and Pseudomonas sp. strain Chol1, and not all proteins are named.