ABSTRACT
Virus genomes are prone to extensive gene loss, gain, and exchange and share no universal genes. Therefore, in a broad-scale study of virus evolution, gene and genome network analyses can complement traditional phylogenetics. We performed an exhaustive comparative analysis of the genomes of double-stranded DNA (dsDNA) viruses by using the bipartite network approach and found a robust hierarchical modularity in the dsDNA virosphere. Bipartite networks consist of two classes of nodes, with nodes in one class, in this case genomes, being connected via nodes of the second class, in this case genes. Such a network can be partitioned into modules that combine nodes from both classes. The bipartite network of dsDNA viruses includes 19 modules that form 5 major and 3 minor supermodules. Of these modules, 11 include tailed bacteriophages, reflecting the diversity of this largest group of viruses. The module analysis quantitatively validates and refines previously proposed nontrivial evolutionary relationships. An expansive supermodule combines the large and giant viruses of the putative order “Megavirales” with diverse moderate-sized viruses and related mobile elements. All viruses in this supermodule share a distinct morphogenetic tool kit with a double jelly roll major capsid protein. Herpesviruses and tailed bacteriophages comprise another supermodule, held together by a distinct set of morphogenetic proteins centered on the HK97-like major capsid protein. Together, these two supermodules cover the great majority of currently known dsDNA viruses. We formally identify a set of 14 viral hallmark genes that comprise the hubs of the network and account for most of the intermodule connections.
IMPORTANCE
Viruses and related mobile genetic elements are the dominant biological entities on earth, but their evolution is not sufficiently understood and their classification is not adequately developed. The key reason is the characteristic high rate of virus evolution that involves not only sequence change but also extensive gene loss, gain, and exchange. Therefore, in the study of virus evolution on a large scale, traditional phylogenetic approaches have limited applicability and have to be complemented by gene and genome network analyses. We applied state-of-the art methods of such analysis to reveal robust hierarchical modularity in the genomes of double-stranded DNA viruses. Some of the identified modules combine highly diverse viruses infecting bacteria, archaea, and eukaryotes, in support of previous hypotheses on direct evolutionary relationships between viruses from the three domains of cellular life. We formally identify a set of 14 viral hallmark genes that hold together the genomic network.
INTRODUCTION
A major discovery of environmental genomics and viromics over the last decade is that the most common and abundant biological entities on earth are viruses, in particular bacteriophages (1–5). In marine, soil, and animal-associated environments, virus particles consistently outnumber cells by 1 to 2 orders of magnitude. Viruses are major ecological and even geochemical agents that in large part shape such processes as energy conversion in the biosphere and sediment formation in water bodies by killing off populations of abundant, ecologically important organisms, such as cyanobacteria or eukaryotic algae (3, 5, 6). With the possible exception of some intracellular parasitic bacteria with highly degraded genomes, viruses and/or other selfish elements, such as transposons and plasmids, parasitize all cellular organisms. Complementary to their physical dominance in the biosphere, viruses collectively appear to encompass the bulk of the genetic diversity on Earth (7–9). The ubiquity of viruses in the extant biosphere and the results of theoretical modeling indicating that emergence of selfish genetic elements is intrinsic to any evolving system of replicators (10–13) jointly imply that virus-host coevolution has been the mode of the evolution of life ever since its origin (14–16).
Viruses and related mobile genetic elements (MGE) clearly have not evolved from a single common ancestor: indeed, not a single gene is conserved across the entire “greater virus world” (also known as the virosphere; here, the two terms are used interchangeably) or even in the majority of selfish elements (17, 18). However, different parts of the virosphere form dense evolutionary networks in which genomes of various selfish elements are linked through different shared genes (19–21). This type of evolutionary relationship results from extensive exchange of genes and gene modules, in some cases between widely different elements, as well as parallel capture of homologous genes from the hosts. Viruses with large genomes possess numerous genes that were acquired from the hosts at different stages of evolution; such genes are typically restricted in their spread to a narrow group of viruses. In contrast, the broader connectivity of the evolutionary network in the virus world derives from a small group of genes that have been termed virus hallmark genes, which encode key proteins involved in genome replication and virion formation and are shared by overlapping sets of diverse viruses (17–19). Virus hallmark genes have no obvious ancestors in cellular life forms, suggesting that virus-like elements evolved at a precellular stage of the evolution of life. The hallmark genes comprise only a small subset within the set of virus core genes, which for the purpose of this work will be defined as the genes that tend to have been retained in groups of related genomes along the course of evolution. The concept of core genes, which departs from the intuitive idea of genes with universal presence, accounts for the multiple evolutionary histories of distinct groups of viruses. The set of core genes also includes signature genes, i.e., genes that are highly prevalent within and specific to one particular group of viruses.
Due to the patchy gene distribution across the diversity of viruses and related MGE, standard methods of phylogenetics and phylogenomics have limited applicability in the study of the evolution of the virosphere outside relatively narrow, tight groups. Instead, methods for direct analysis of evolutionary networks are called for. These approaches benefit from the vast arsenal of mathematical concepts and tools that have been developed in different areas of network research (22–26). Recent application of network analysis methods to the comparative analysis of microbial and bacteriophage genomes has been productive, in particular, for the identification of preferred routes and patterns of horizontal gene transfer (HGT) (27–29).
The viromes and mobilomes (i.e., the supersets of viruses and other selfish elements) of the three domains of cellular life (bacteria, archaea, and eukaryotes) are fundamentally different. Although several families of double-stranded DNA (dsDNA) viruses are represented in both bacteria and archaea, no viruses are known to be shared by eukaryotes with any of the other two cellular domains, even at the family or order level (30). In bacteria and archaea, the virosphere is heavily dominated by dsDNA viruses, with relatively limited representation of single-stranded DNA (ssDNA) viruses and only a few narrow groups of RNA viruses. The eukaryotic part of the virosphere is sharply different, with a dominant presence of RNA viruses; however, dsDNA viruses of eukaryotes are also common and diverse (31). Altogether, the dsDNA viruses occupy most of the virosphere and include the largest viral genomes, thus presenting ample material for the construction of gene-sharing networks.
Bipartite networks, also known as 2-mode networks in the social sciences literature (32), are a natural way to represent, in the form of networks, complex systems that consist of two distinct classes of components (nodes). Straightforward examples from the field of biology are metabolic networks, composed of metabolites and enzymes, or pollination networks, composed of plants and their pollinators. Similarly, the network of gene sharing among viruses (due to common ancestry or HGT) calls for a bipartite network representation, where the two classes of nodes are viral genomes and homologous gene families. Historically, the lack of analytical tools to deal with bipartite networks has forced researchers to turn to monopartite projections (simplified versions of the network involving only one class of nodes), an approach that is not free from some degree of arbitrariness and can introduce biases (33). However, recent advances in network theory, in particular those related to module detection, allow for direct analysis of bipartite networks with minimal loss of information and simultaneous characterization of both classes of nodes (34).
Here we present a bipartite network analysis of all currently recognized families of dsDNA viruses and some related mobile genetic elements (MGE). Dissection of this network using formal analytical tools reveals extensive modularity of this part of the virus world, objectively identifies the set of viral hallmark genes, and quantitatively vindicates previously proposed scenarios for the origin of diverse groups of viruses.
RESULTS
Bipartite network of dsDNA viruses.
In order to develop a network representation of the relationships between all major groups of dsDNA viruses, we first had to identify the families of homologs that would become the nodes of the “gene family” class. To that end, we retrieved all predicted protein-coding sequences from more than 1,440 viral genomes and related MGE and grouped them into families by sequence similarity. Because of the large number and high diversity of the viral sequences, a multistep approach to the construction of these families was adopted. First, we developed an automated pipeline that combined sequence similarity analyses and algorithms for community detection in networks (see Materials and Methods). A comparison of the gene families obtained through this pipeline and the available clusters of orthologous genes for bacteriophages (POGs) (9, 35) and large nucleo-cytoplasmic DNA viruses of eukaryotes (NCVOGs) (36, 37) yielded a recall (average fraction of sequences in a POG or NCVOG grouped together in our analysis) of 0.92, and purity (1 minus the average fraction of false positives) of 0.89. In a subsequent step, some of the major families were manually curated to account for distant homology relations that, despite being strongly supported by previous reports, escaped automatic detection (see Materials and Methods for details).
We represented the web of relations among dsDNA viruses by means of a bipartite network in which the nodes belong to two classes: genomes and gene families. Edges connect a genome to every gene family that it contains; conversely, a gene family is linked to all genomes in which it is present. Compared to other network approaches based on genome-genome similarity metrics, the bipartite representation provides for explicit identification not only of clusters of genomes sharing gene sets but also of the genes that glue these genomes together.
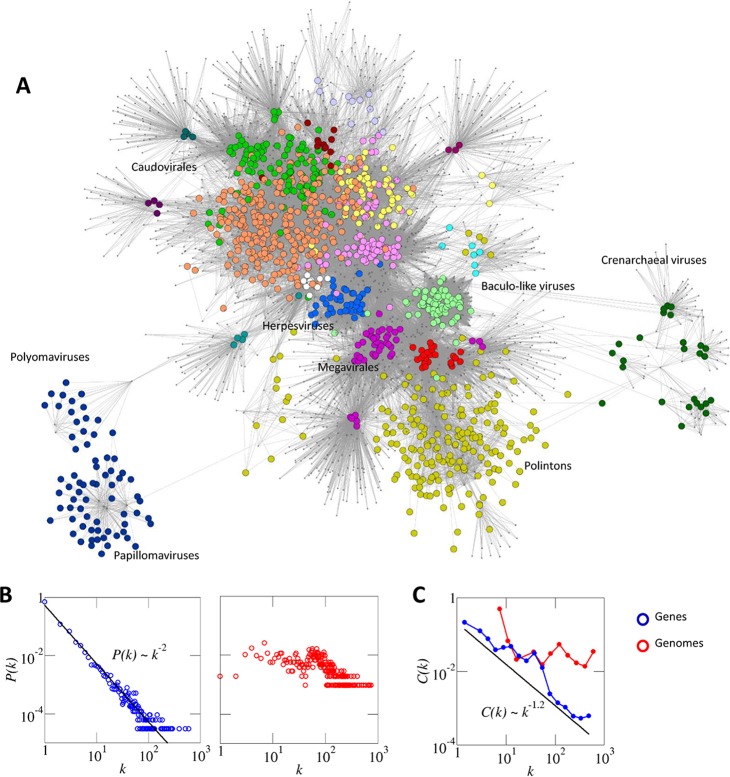
Due to the high diversity of viral genomes, the full version of the network contains a large fraction of ORFans (genes with no detectable homologs) and rare genes with a patchy distribution, which reduces the signal-to-noise ratio and makes a detailed analysis computationally unfeasible. To overcome these problems, we built a reduced version of the network that only contains core genes, making it computationally tractable and more informative in terms of evolutionary relationships among viruses. In short, core genes were identified based on their low evolutionary loss rates, according to the procedure described in the next section. Table 1 summarizes the basic statistics of the full and core gene networks. With the exception of this section, all results presented here are based on the core gene version of the bipartite network. A visual inspection of the network (Fig. 1A) indicates that (i) viruses from different taxa generally occupy different regions of the network, and (ii) despite the abundance of taxon-specific genes, there is a complex pattern of gene sharing involving, to a greater or lesser extent, all included taxa. It is also notable that Polyomaviridae and Papillomaviridae (two families of dsDNA viruses with the smallest genomes) and viruses infecting archaea (primarily hyperthermophilic Crenarchaeota) form well-defined clusters that are only weakly connected to the rest of the network.
TABLE 1 .
Basic properties of the bipartite dsDNA viral network
| Element | Value for full network | Value for core genesa |
|---|---|---|
| Genomes | 1,073 | 1,071 |
| Gene families | 33,793 | 1,576 |
| Edges | 98,343 | 30,661 |
| Edge density | 0.003 | 0.018 |
| Modules | NAb | 19 |
| Mean no. of genes per genome | 92.1 | 28.8 |
| Mean gene abundance (mean with ORFan excluded) | 2.9 (6.7) | 19.3 |
The core gene version of the full bipartite network is a bipartite subnetwork that includes core gene families and genomes with at least one core gene.
NA, not applicable.
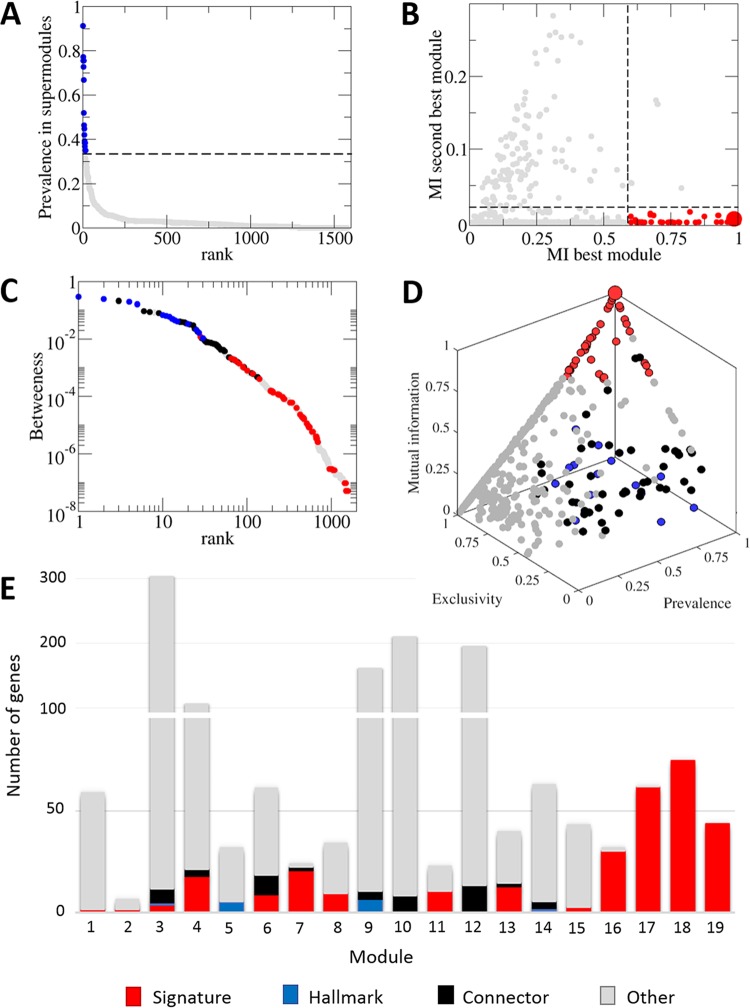
FIG 1 .
The dsDNA virus world as a bipartite network. Nodes corresponding to genomes are depicted as larger circles, and nodes corresponding to core gene families are depicted as dots. An edge is drawn whenever a genome harbors a representative of a core gene family. (A) The modular structure of the network is highlighted by coloring genome nodes according to the module to which they belong (color coding is as described for Fig. 4 to 6). The location of some major viral groups is indicated for illustrative purposes. (B) The degree distributions of genes (left) and genomes (right). In the case of genes, the best fit to a power law distribution is also shown. (C) The scaling of the clustering coefficient, C(k), with respect to the degree k (genes and genomes) suggests a hierarchical modular structure organized around high-level hallmark genes [large k and small C(k)] and low-level signature genes [small k and large C(k)].
In the context of networks, the degree of a node (k) is the number of edges connected to that node. In the bipartite network of viruses, the degree of a gene family represents the number of genomes in which the gene family is found, and it follows a power law distribution, P(k) ∼ k−γ, with the exponent γ ≈ 2 (Fig. 1B, left). Power law distributions are characterized by having a long tail, which means that the frequency of nodes with high degrees is not negligible. Thus, the power law distribution of the gene degrees implies the existence of hubs, i.e., gene families present in a large fraction of the genomes. Specifically, there are 12 gene families present in at least 20% of the analyzed genomes. In contrast, the number of gene families per genome (i.e., the genome degree) follows a roughly uniform distribution with a bulk that encompasses viruses and MGE encoding from 3 to 100 genes and a tail of highly connected nodes that corresponds to giant viruses (Fig. 1B, right). The flat shape of the degree distribution for genomes indicates that the genome network, in sharp contrast to the gene network, contains few if any prominent hubs: even the largest giant viruses only harbor a small fraction (5% or less) of all gene families.
Another useful topological measure is the clustering coefficient (C), which quantifies the extent to which the neighbors of a given node are also connected with each other. For gene families, the bipartite version of the clustering coefficient (38) decays with k as C(k) ∼ k−α, where α = 1.2 (Fig. 1C, blue). Such a scaling of the clustering coefficient is often considered an indicator of hierarchical modular organization, in which low-level modules of tightly connected nodes join through higher-level nodes to produce larger, less-cohesive supermodules, in an iterative manner (39). In the case of the virus network, low-level modules are cemented by signature genes, whereas hallmark genes, at the top of the hierarchy, bring modules together. This hierarchical structure is not observed for genomes (Fig. 1C, red). Accordingly, the bipartite network appears to be held together primarily by hallmark genes rather than chimeric genomes (see also below).
Core genes in viral genomes.
Viral gene families show the tripartite core-shell-cloud structure that has been identified as a general feature of gene frequency distributions across the diversity of life forms (40–43). Although there are no universal viral genes, the U-shaped distribution becomes evident when “coreness” is defined as the retention probability of a gene (Fig. 2; see Materials and Methods for details). Figure 2 also shows (in blue) how widespread the genes within each bin are. From this perspective, genes on the left of the distribution are either ORFans or rare genes with patchy distribution patterns. On the right side, genes with retention propensities close to unity are typically taxon-specific genes restricted to a small number of closely related genomes. The most widespread genes, i.e., the core genes according to the intuitive definition, exhibit retention propensities around 0.7 to 0.8. That this value is smaller than unity could be caused by three factors (or any combination thereof): (i) failure to detect all homologs of an ancestral gene in a set of distantly related genomes due to high sequence divergence, (ii) loss of ancestral genes in some lineages, or (iii) occasional transfer of a lineage-specific core gene to unrelated lineages where it does not tend to be retained, thus affecting the net estimate of its retention probability (although this effect is minimized by setting a similarity threshold to calculate the loss rates; see also Materials and Methods).
FIG 2 .
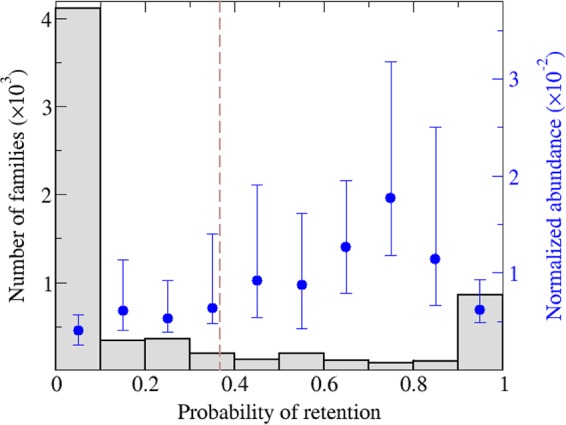
Core-shell-cloud structure of viral gene families. For each bin, the bar indicates the number of gene families with a retention probability in the range defined by the x axis. The blue dots indicate the median abundance of such families in the whole set of genomes (error bars correspond to the 25th and 75th percentiles). Family abundances were normalized so that an abundance equal to 1 means that the given family is present in each genome (the contributions of highly similar genomes were downweighted to compensate for sampling bias [see Materials and Methods]). The gene families with the highest retention probability (right-most bin) are typically restricted to a small number of genomes (median abundance, approximately 0.06). In contrast, many of the “core” genes according to the intuitive definition (i.e., those present in a large number of genomes) belong to the bin with a retention probability in the range of 0.7 to 0.8. For the purpose of this work, gene families to the right of the dashed, vertical line (i.e., those with a retention probability greater than 1/e) were considered core genes.
We defined core genes as those with a loss rate below unity [i.e., retention probability greater than exp(−1)]. Application of this criterion yielded 1,576 core gene families (see Table S1 in the supplemental material), which represented about 5% of all the families in the data set. Table 2 lists the top 25 core genes, which include some of the most conspicuous viral hallmark genes that have been previously identified qualitatively. The abundances of these genes were computed by adding the similarity-weighted contributions of genomes that contain a given gene and normalized with respect to the total number of genomes. Remarkably, this list encompasses two major classes for each of several viral hallmark proteins that are responsible for the key functions in virion morphogenesis and genome replication, in particular, icosahedral capsid proteins, DNA-packaging ATPases, DNA polymerases, and primases-helicases. The ranking of the core genes reflects the diversity of the groups of viruses in which these genes are most common. Due to the fact that tailed bacteriophages (order Caudovirales) are the most diverse (and most abundant) group of viruses on earth (2, 44, 45), the top four core genes are primarily represented in these phages. The core set combines genes that are limited in their spread but appear to be essential and are never lost in the respective groups of viruses (although in some cases could be missed due to the extreme sequence divergence), such as packaging ATPases or phage portal proteins, and genes that are represented in highly diverse groups of viruses but are lost relatively often, such as helicases (Table 2). Extending the latter trend, there are some noncore genes that are highly abundant and spread over diverse groups of viruses but failed to make it to the core set due to their high loss rates; the most widespread of these “viral mobilome” genes are listed in Table 3.
TABLE 2 .
Top 25 core genes sorted by normalized abundance
| Family no. | Annotationa | Retentionb | Abundancec | Taxon(s) with presence |
|---|---|---|---|---|
| 5 | Terminase, large subunit | 0.969 | 0.661 | Herpesvirales, Caudovirales |
| 13 | Major capsid protein, HK97-like | 0.960 | 0.532 | Herpesvirales, Caudovirales |
| 10 | XRE-family HTH domain | 0.718 | 0.327 | Caudovirales |
| 16 | Portal protein | 0.740 | 0.314 | Caudovirales |
| 8 | DEAD-like helicase | 0.522 | 0.298 | Megavirales, Caudovirales |
| 6 | DNA primase/helicase (DnaB) | 0.683 | 0.290 | Caudovirales |
| 11 | DNA polymerase B | 0.886 | 0.255 | Adenoviridae, “Megavirales,” Polintons, some virophages, Baculoviridae and related viruses, Herpesvirales, Tectiviridae, mitochondrial and cytoplasmic plasmids, Ampullaviridae, Salterprovirus, some Caudovirales |
| 24 | Integrase | 0.794 | 0.254 | Caudovirales |
| 111 | Protease (herpesvirus S21, phage U9/U35) | 0.379 | 0.254 | Herpesvirales, Caudovirales |
| 22 | Bacteriophage HK97-gp10, putative tail component | 0.904 | 0.240 | Caudovirales |
| 4 | D5-like primase-helicase | 0.594 | 0.240 | “Megavirales,” Baculoviridae, some Caudovirales |
| 27 | HNHc endonuclease | 0.600 | 0.222 | Caudovirales |
| 20 | DNA polymerase A | 0.763 | 0.205 | Caudovirales |
| 19 | Ribonucleotide reductase large subunit | 0.571 | 0.204 | “Megavirales,” Herpesvirales, some Caudovirales (mostly Myoviridae) |
| 18 | Thioredoxin | 0.371 | 0.174 | “Megavirales,” some Caudovirales |
| 2 | Ribonucleotide reductase small subunit | 0.440 | 0.169 | “Megavirales,” Herpesvirales, some Caudovirales (mostly Myoviridae) |
| 23 | Phage tail tape measure protein | 0.690 | 0.164 | Siphoviridae, some Myoviridae |
| 30 | UvrD-like helicase | 0.886 | 0.162 | “Megavirales,” Herpesvirales, some Baculoviridae, some Caudovirales |
| 35 | Portal protein | 0.794 | 0.158 | Siphoviridae |
| 12 | A32-like packaging ATPase (FtsK/HerA) | 0.868 | 0.156 | “Megavirales,” Polintons, Lavidaviridae
(virophages), Tectiviridae, Corticoviridae, Turriviridae, Sphaerolipoviridae |
| 26 | Phage mu protein F, putative minor head protein | 0.612 | 0.143 | Myoviridae, Siphoviridae |
| 68 | Double jelly roll MCP | 0.786 | 0.136 |
Adenoviridae, “Megavirales,” Polintons, Lavidaviridae (virophages), Tectiviridae, Corticoviridae, Turriviridae |
| 44 | Baseplate J family protein | 0.895 | 0.133 | Myoviridae |
| 36 | AAA family ATPase | 0.843 | 0.120 |
Bicaudaviridae, “Megavirales” (no Poxviridae), some Myoviridae/Siphoviridae |
| 47 | RuvC Holliday junction resolvase; poxvirus A22 family | 0.600 | 0.117 | “Megavirales,” some Caudovirales |
Bold text is used to denote hallmark genes.
The retention probability of a gene family is equal to exp(−r), where r is the estimated loss rate (see Materials and Methods).
The abundances were normalized to the total number of genomes, such that a family present in every genome would have an abundance equal to 1.
TABLE 3 .
Representative genes from the viral mobilome (low retention propensity, high abundance)
| Family no. | Annotation | Retentiona | Abundanceb | Taxon(s) with presence |
|---|---|---|---|---|
| 31 | HNH endonuclease | 0.002 | 0.134 |
Mimiviridae/Marseilleviridae, Phycodnaviridae, Caudovirales |
| 32 | dUTPase | 0.092 | 0.158 |
Adenoviridae, “Megavirales,” Herpesvirales, Caudovirales, Baculoviridae |
| 43 | HNH endonuclease | 0.012 | 0.103 | Mimiviridae/Marseilleviridae, Caudovirales |
| 48 | BRO protein, phage antirepressor | 0.118 | 0.121 |
Baculoviridae, Poxviridae, Ascoviridae/Iridoviridae, Caudovirales |
| 56 | DUF3310 | 0.110 | 0.124 | Caudovirales |
| 79 | DNA methylase N-4/N-6 | 0.005 | 0.109 |
Caudovirales, Phycodnaviridae, Pandoravirus, Bicaudaviridae |
| 80 | Peptidoglycan recognition protein | 0.049 | 0.105 | Caudovirales |
| 91 | ssDNA-binding protein, SSB_OBF domain, COG0629 |
0.111 | 0.122 | Caudovirales |
| 30573 | Thymidine kinase | 0.129 | 0.178 | “Megavirales,” Caudovirales |
The retention probability of a gene family is equal to exp(−r), where r is the estimated loss rate (see Materials and Methods).
The abundances were normalized to the total number of genomes, such that a family present in every genome would have abundance equal to 1.
A common debate among researchers in virus evolution is which genes represent the viral “self” and thus are most important for evolutionary reconstruction: those for structural proteins or those for components of the replication machinery (19, 46–52). The present quantification of gene “coreness” seems to resolve this dilemma by showing that members of these two functional categories are mixed in the ranked list of core genes (Table 2; see also Table S1 in the supplemental material), highlighting the equal importance of both categories of genes. Below, we return to some of the core genes when discussing connections between different groups of viruses.
Modular structure of the viral network.
To unveil the internal structure of the viral network, we applied a module identification algorithm to delineate sets of genomes and gene families that are densely connected to each other. The algorithm is aimed at finding the partition of the network that maximizes Barber’s bipartite modularity (a standard measure of the quality of the modules). Because modularity optimization is a combinatorial problem, it is generally impossible to solve it exactly for large networks. Instead, heuristic or stochastic approaches, such as simulated annealing, have to be used (see Materials and Methods for details), and different runs of the same algorithm can yield variable results due to multiple local maxima. To overcome this difficulty, we ran 100 replicates (realizations) of the algorithm which rendered 100 (not necessarily different) solutions for the module identification problem. Of these, the solution with the highest quality (evaluated as the value of Barber’s bipartite modularity) was taken as the optimal partition, and the rest of the replicates were used to assess the robustness of the modules in that optimal partition. Specifically, for each possible pair of genes or genomes assigned to the same module, we calculated the fraction of replicates in which both elements were grouped together. The average fraction over all pairs was taken as the robustness of the module. Similarly, the cross-similarity between two distinct modules is the fraction of replicates in which pairs of genomes, with one from each of the modules, appear in the same group.
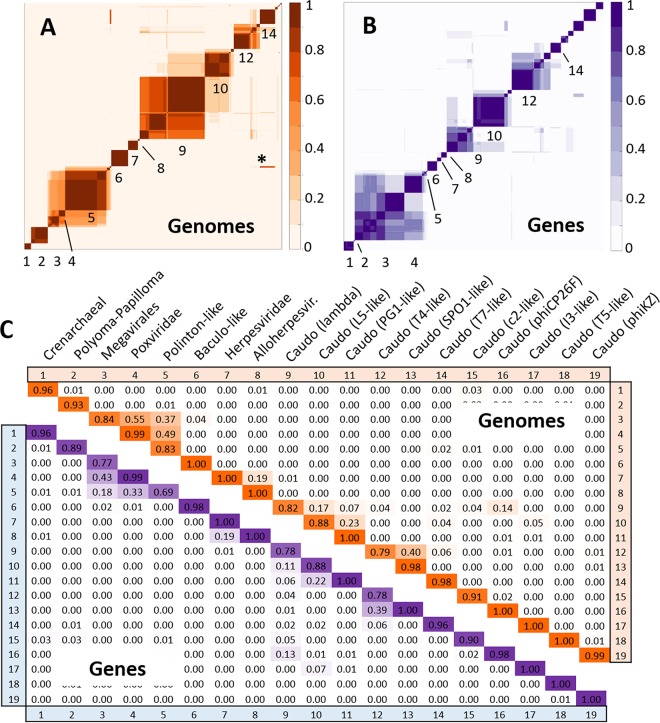
The network showed a significant modular structure, with an optimal partitioning that consisted of 19 modules (P < 0.01, compared to the Barber’s modularity of a random network with the same degree distribution) (Table 4; see also Table S3 in the supplemental material for the module composition). The modules generally were highly robust, with respect to both genes and genomes, as determined by analysis of the module composition in alternative network partitions (Table 4; Fig. 3). The number of genomes in the modules spanned 2 orders of magnitude, from 282 in the largest module (module 9) to 3 in the smallest module (module 18). Given that highly similar genomes were clustered prior to network construction, the number of genomes in a module seems to be a meaningful reflection of its heterogeneity. Each of the modules contains multiple core genes, with over 300 genes in the largest (in terms of the number of genes), module 3. However, most of these genes are limited to tight groups of viruses within a module, whereas only a few hallmark genes hold the modules together.
TABLE 4 .
Modules of the dsDNA virus network
| Module | Composition (genomes) | Representative gene product(s)a | No. of genomes | No. of genes | Robustnessc |
|
|---|---|---|---|---|---|---|
| Genomes | Genes | |||||
| 1 | Crenarcheal viruses except Turriviridae | RHH domain-containing proteins (S) | 27 | 59 | 0.98 | 0.98 |
| 2 | Papillomaviridae and Polyomaviridae | Papillomavirus L2 protein (S) | 70 | 7 | 0.93 | 0.89 |
| 3 | “Megavirales” (except Poxviridae) | D5-like primase-helicase (H) | 46 | 304 | 0.81 | 0.76 |
| 4 | Poxviridae | Virion core protein P4a, IMV membrane protein, metalloproteinase, poly(A) polymerase (S) | 26 | 107 | 0.99 | 0.99 |
| 5 | Adenoviridae, Lavidaviridae (virophages), Polintons, PLV, Tectiviridae, Corticoviridae, Sphaerolipoviridae, Turriviridae, Salterprovirus, some mitochondrial plasmidsb | pDNAP, packaging ATPase (FtsK superfamily), double and single jelly roll capsid proteins, Ulp1-like cysteine protease (H) | 183 | 29 | 0.88 | 0.77 |
| 6 | Baculoviridae, Hytrosaviridae, Nudiviridae, Nimaviridae, and some cytoplasmic plasmidsb | per os infectivity factors 0–5, capsid protein (p95/vp91) (S), S_TKc serine/threonine protein kinase (C) | 70 | 61 | 1.00 | 0.98 |
| 7 | Herpesviridae | Envelope glycoproteins H, M, B, UL73; tegument proteins UL7, UL16 (S) | 41 | 24 | 1.00 | 1.00 |
| 8 | Alloherpesviridae | Capsid triplex protein (S) | 7 | 34 | 1.00 | 1.00 |
| 9 | Multiple Caudovirales, Peduovirinae (P2-like), lambda- like, T1-like, phiC31, mu-like, P22-like, Picovirinae, Plasmaviridaee | HK97-like capsid protein, large terminase subunit, protease, tyrosine integrase (H) | 282 | 165 | 0.80 | 0.76 |
| 10 | Siphoviridae, mycobacteriophages (L5-like) | Minor tail protein (S), cutinase (lysin B) (C), PGRP (lysin A) (C) | 107 | 211 | 0.88 | 0.88 |
| 11 | Siphoviridae, mycobacteriophages (PG1-like) | Replicative helicase/primase- polymerase (S) | 12 | 23 | 1.00 | 1.00 |
| 12 | Mostly Myoviridae, Tevenvirinae (T4-like) and unclassified Myoviridae; 6 Podoviridae (N4-like), Malacoherpesviridaed | Tail completion and sheath stabilizer protein, baseplate J protein (C) | 95 | 196 | 0.79 | 0.81 |
| 13 | Myoviridae, Spounavirinae (SPO1-like) | Zn-ribbon-containing structural protein, tail tube subunit, tail assembly chaperone (S), tail sheath protein precursor (C) | 13 | 40 | 0.98 | 1.00 |
| 14 | Podoviridae, Autographivirinae (T7-like) and some other minor groups (Bpp-1-like, VP2-like, N4-like) | Head-to-tail connecting protein (S), DNA primase/helicase (DnaB) (S), DNA polymerase A (C) | 63 | 63 | 0.98 | 0.96 |
| 15 | Siphoviridae, Lactococcus phages c2-like and 936 sensu lato | Single-stranded DNA-binding protein (S) | 11 | 43 | 0.94 | 0.94 |
| 16 | Siphoviridae, Clostridium phage phiCP26F and related strains | Cytolysin, ferritin-like superfamily protein (S), phage anti- repressor (S), XRE-like regulator (C) | 4 | 32 | 1.00 | 0.98 |
| 17 | Myoviridae, I3-like mycobacteriophages | Structural proteins, lysin A (S), many uncharacterized proteins (C) | 4 | 62 | 1.00 | 1.00 |
| 18 | Siphoviridae, T5-like | Tail proteins (Pb3, Pb4, etc.), NAD- dependent DNA ligase, nicking endonuclease (S) | 3 | 74 | 1.00 | 1.00 |
| 19 | Myoviridae, phiKZ-like | RNA polymerase beta subunit, RNase H (S) | 5 | 43 | 0.97 | 1.00 |
Representative genes are presented based on their classification (in parentheses and boldface) as signature (S), hallmark (H), or connector (C) genes.
Mitochondrial plasmids from Babjeviella inositovora and Debaryomyces hansenii were assigned to module 5, and those from Kluyveromyces lactis, Lachancea kluyveri, and Millerozyma acaciae belong to module 6, although these assignations were based on a small number of widespread core genes.
The robustness is equal to the fraction of replicas in which pairs of members of a module were assigned to the same module, averaged over all possible pairs. Two measures of robustness apply to each module, depending on whether pairs of genes or pairs of genomes were considered.
Malacoherpesviridae lack the tail components listed as representative genes for module 12 (see text for further details).
The assignment of the only genome from family Plasmaviridae to module 9 is based solely on a shared integrase.
FIG 3 .
Robustness and cross-similarity of modules in the virus bipartite network. (A and B) Heat map representations of the module robustness matrices for genomes (A) and gene families (B). To generate these matrices, nodes of one class (genomes or gene families) were sorted according to the module they belong to in the optimal partition of the network. For each pair of nodes, the matrix contains the fraction of 100 replicates in which both nodes were placed in the same module. Robust modules appear as blocks in the module robustness matrix; deviations from the block structure correspond to modules that are sometimes merged or nodes without a clear module assignation. The asterisk shows the case of mitochondrial plasmids which belong to module 5 in the best partition but are often assigned to module 14. (C) Quantitative summary of the average robustness of modules at the genome and gene level (elements on the diagonal) and the cross-similarity between pairs of modules (fraction of replicates in which nodes of both modules appear together; off-diagonal elements). See Table 4 for the list of the taxa assigned to each module.
Of the 19 modules, 11 include tailed bacteriophages, emphasizing once again that the order Caudovirales is by far the largest, most diverse, extremely heterogeneous group of viruses. Most of the phage modules consist of subsets from a particular family (Siphoviridae: modules 10, 11, 15, and 16; Myoviridae: modules 13, 17, and 19; Podoviridae: module 14). However, two modules (9 and 12) include representatives of two or three families of tailed phages. It is well known that tailed phages extensively exchange genes, so the existence of robust modularity is remarkable in itself, indicating that despite this genetic fluidity, there are several partly isolated gene pools in the phage world. Unexpectedly, module 12 also includes, in addition to phages of the family Myoviridae, the only known member of the eukaryotic virus family Malacoherpesviridae.
Apart from the phages, the viruses of hyperthermophilic Crenarchaeota form a single, highly robust module 1, with the notable exception of the family Turriviridae (see discussion below). Although crenarcheal viruses contain many genes with no detectable homologs, the existence of a network of shared genes among these viruses has been noticed previously (53). The network analysis described here not only validates this conclusion but also shows that the crenarcheal virus network is a distinct module isolated from the rest of the virosphere. Other than viruses of hyperthermophilic Crenarchaeota, archaeal viruses remain poorly sampled, so that the present analysis included only the family Sphaerolipoviridae, which belongs in module 5 (see details below), and four euryarchaeal viruses, all of which share the signature genes of Caudovirales and were assigned to module 9.
The remaining 7 modules consist, with a single exception, of viruses infecting eukaryotes. As expected, papillomaviruses and polyomaviruses, the smallest known dsDNA viruses that appear to have evolved from ssDNA viruses, replicating via the rolling circle mechanism (31), form a distinct module that is only weakly connected to the rest of the network. The proposed order “Megavirales” (also known as NCLDV), which combines several families of large and giant dsDNA viruses of eukaryotes that primarily replicate in the cytoplasm of the host cell (54–56), is split between two modules that include, respectively, the poxviruses and the rest of the NCLDV. This separation is not particularly surprising because poxviruses are known as a highly derived group of the NCLDV (54). As discussed below, these two modules share several links and join at a higher level of the network hierarchy (see below).
Similarly, the order Herpesvirales is split into two modules that include, respectively, the families Herpesviridae and Alloherpesviridae, and again, are connected at a higher level. Unexpectedly, the third family of the Herpesvirales, namely, Malacoherpesviridae, has been assigned to a module containing T4-like tailed phages (see below). A separate module consists of several families of viruses that infect arthropods, including Baculoviridae, Nudiviridae, Hytrosaviridae, and Nimaviridae. An evolutionary relationship between these virus families has been suspected from previous comparative genomic studies, and it has been proposed that they could be distantly related to the NCLDV (57–59). The network analysis suggests a more complicated picture, as discussed below.
The most notable module, module 5, consists of several groups of viruses and related MGE from all three domains of life, most of which replicate their genomes via the protein-primed replication mechanism and encode the respective variety of the family B DNA polymerases (pDNAP). Previously, it was hypothesized that Polintons, the virus-like large transposons that belong to this module and have been shown to encode two capsid proteins and, accordingly, predicted to form virions (60), evolved directly from bacterial tectiviruses and then gave rise to several diverse groups of eukaryotic viruses and MGE (21, 61). Given the wide spread of the Polintons among eukaryotes and their apparent central role in the evolution of the module and beyond (see below), we denote this module Polinton-like (PL). The delineation of the PL module in the present network analysis is compatible with the previously proposed evolutionary scenario and also adds to the mix, in addition to the bacteriophage family Tectiviridae, another phage family, Corticoviridae, and archaeal viruses of the families Turriviridae and Sphaerolipoviridae. The PL module is held together by the pDNAP and a distinct suite of genes involved in virion morphogenesis that encode the major double jelly roll (DJR) capsid protein, the packaging ATPase and, in many viruses, also a minor, single jelly roll capsid protein and a maturation protease (21, 61). The pDNAP is encoded by most viruses in the PL module, with the exception of the archaeal viruses, corticovirus PM2, some virophages, and Polinton-like viruses (PLV). The cytoplasmic and mitochondrial plasmids also encode pDNAP, which is the only link between these small, capsidless genetic elements and the rest of the PL module. Similarly, Haloarchaeal viruses of the genus Salterprovirus are included in module 5 based solely on the presence of the pDNAP. The DJR major capsid protein (MCP) and the packaging ATPase are present in all viruses of the PL module except for salterproviruses. Thus, the expansive PL module is unified by both morphogenetic and replicative genes that are present in different members of the module, either separately or together. The finding that all these extremely diverse elements form a single module that is robustly supported at the genome level (although less so on the gene level), whereas at the same level of clustering both “Megavirales” and Herpesvirales are split, suggests a close relationship and evolutionary coherence among the PL elements.
Supermodular structure of the dsDNA virus network.
The existence of substantial cross-similarities between some of the modules (Fig. 3C) implies that there is a higher-order structure in the virus network, such that some of the modules can be joined to generate a hierarchy of supermodules. We implemented an iterative method to identify such supermodules by building higher-order bipartite networks in which the two classes of nodes are (primary) modules and genes that are shared by these modules.
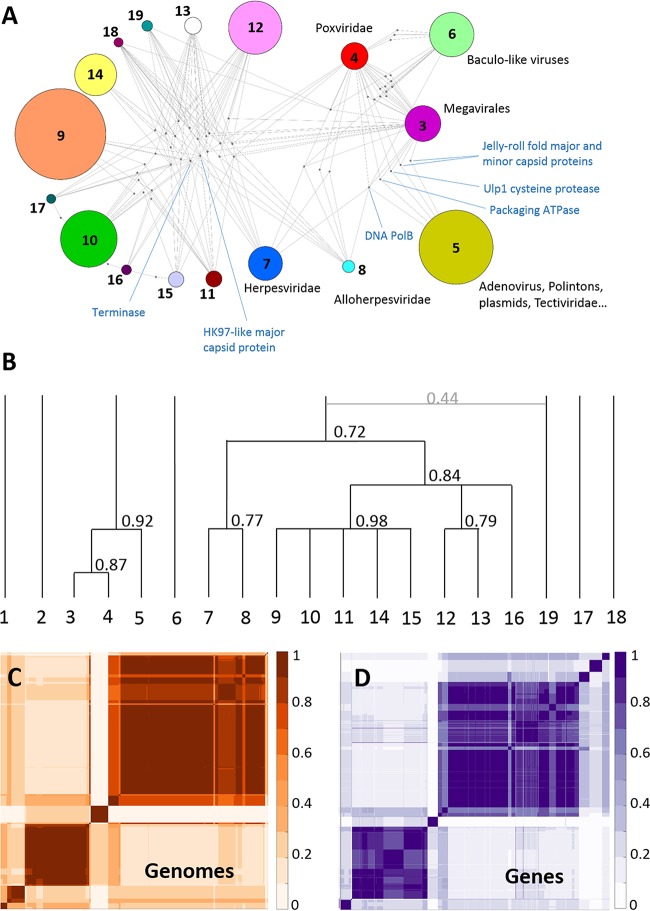
The first-order module network is depicted in Fig. 4A. Modules representing Caudovirales show a dense web of connections, with numerous shared genes, among which the HK97-like major capsid protein and the large subunit of the terminase (the genome-packaging ATPase-nuclease) are the two most prominent ones. These two genes are also shared by the Herpesvirales, which additionally connect to some of the Caudovirales modules through the S21-U9/U35 capsid maturation protease, the two subunits of the ribonucleotide reductase, and some other genes. The “Megavirales” also share genes with modules containing Caudovirales, such as the ribonucleotide reductase and some helicases. The PL module is connected to the “Megavirales” via five connector genes, namely, DNAP (at this level, all family B DNAPs, both protein primed and RNA primed, merge into a single gene cluster), A32-like packaging ATPase, Ulp1-like maturation protease, double jelly roll major capsid protein, and single jelly roll minor capsid protein. Notably, in the bipartite network, all these genes are assigned to the PL module, which appears to be compatible with the proposed central role of Polintons in the evolution of dsDNA viruses of eukaryotes (21, 31). The complete list of connector genes is provided in Table S2 in the supplemental material.
FIG 4 .
Higher-order structure of the virus network. (A) Bipartite network defined by modules (numbered as for Table 4) and connector genes. A module is linked to a connector gene if the prevalence (relative abundance) of the gene in that module is greater than exp(−1). Modules 1 (crenarcheal viruses) and 2 (polyomaviruses and papillomaviruses) that are only weakly connected to other modules are not represented. Modules are represented as colored circles, with the node size proportional to the number of genomes in the module. Connector genes are represented as dots. The position of some hallmark genes discussed in the text is shown. (B) Tree representation of the hierarchical supermodule structure of the network. At each iteration, two (super)modules were merged if their members clustered together in at least 50 of 100 replicates of the module detection algorithm. Branch lengths are proportional to the number of iterations required for two modules to merge. The number associated to each branch indicates the robustness of the respective supermodule. (C and D) Heat map representations of the supermodule robustness matrices for genomes (C) and gene families (D) after the last iteration of the higher-order supermodule search. To generate these matrices, nodes of one class (genomes or gene families) were sorted according to the supermodule they belong to in the optimal partition of the network. For each pair of nodes, the matrix contains the fraction of 100 replicates in which both nodes were placed in the same supermodule. Robust supermodules appear as blocks in the module robustness matrix.
At the highest order of the network hierarchy, the dsDNA viruses form 5 major supermodules: (i) crenarcheal viruses (except Turriviridae), (ii) Polyomaviridae-Papillomaviridae, (iii) PL elements-“Megavirales,” (iv) Baculoviridae and the related families of arthropod viruses, and (v) Caudovirales-Herpesvirales (Fig. 4B to D). There are also three minor modules encompassing I3-like, T5-like, and phiKZ-like phages that remained isolated (although the latter join the main Caudovirales supermodule in 44 of the 100 replicates). The most notable result is the unification of the PL elements and the “Megavirales” (including poxviruses) into a single module with high robustness (0.92), in agreement with the evolutionary scenario that has been proposed primarily on the basis of the shared morphogenetic genes. In parallel, Herpesvirales merge with Caudovirales in a moderately supported supermodule (robustness of 0.72).
Dissection of the PL-“Megavirales” supermodule.
We then sought to investigate in greater detail the internal structure of the large and heterogeneous supermodule that joins the “Megavirales” with the PL elements. Given the shared genes and proposed evolutionary relationships between the Baculo-like viruses and the “Megavirales,” their persistent separation in the full network analysis appeared unexpected, so we included the Baculo-like module in this additional analysis in an attempt to better characterize its relationships with the other modules. The technical advantage of analyzing (super)modules separately is that there is a resolution limit for the module detection algorithm in very large networks (34). Therefore, limiting the scope of the analysis to a subnetwork of particular interest can reveal features of its internal structure that escape the analysis of the full network.
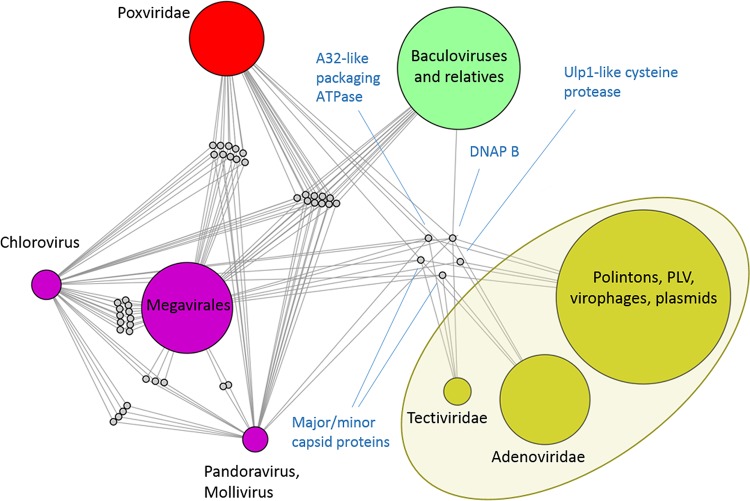
The analysis of the internal structure of the PL-“Megavirales” supermodule produced a new partitioning (Fig. 5; see also Table S3 in the supplemental material), in which the PL module remained unchanged. In contrast, the “Megavirales” module split into Poxviridae (which was already identified as a separate module in the initial analysis of the full network), Pandoravirus-Mollivirus, a group of five phycodnaviruses of the genus Chlorovirus (with Paramecium bursaria chlorella virus as the prototype), and a large module that included the remaining members of the “Megavirales.” The baculoviruses and their relatives remained a separate module as in the full network.
FIG 5 .
The internal structure of the PL-“Megavirales” supermodule. A module is linked to a connector gene if the prevalence of the gene in that module is greater than exp(−1). Modules are represented as larger circles, with sizes proportional to the number of genomes in the module; colors coding is the same as in Fig. 4. Connector genes are represented as smaller gray nodes. The PL elements, which originally formed a single module (shaded oval), were further dissected to produce the submodule structure shown. The hallmark genes are labeled.
We further analyzed the internal structure of the PL module and identified three submodules: (i) Polintons, PLV (a recently discovered group of Polinton-like viruses [62]), virophages, cytoplasmic plasmids, and one tectivirus (enterobacterial phage PRD1, which lacks several genes that are conserved in the other members of the family Tectiviridae), (ii) the rest of the Tectiviridae (with Bacillus phages AP50 and Bam35 as representative species), (iii) Adenoviridae (Fig. 5). The first submodule is the largest and most heterogeneous, as indicated by a relatively low robustness (86/100 compared to 100/100 for the rest of the modules). The submodules are connected through five hallmark genes, the same that maintain the integrity of the PL module as a whole, namely, (i) pDNAP, (ii) A32-like packaging ATPase, (iii) major capsid protein, (iv) minor capsid protein, and (v) Ulp1-like cysteine protease. The first, largest submodule is the only one that harbors all five connector genes, and moreover, they are all assigned to this submodule, again reaffirming the evolutionary centrality of Polintons and the related elements.
The case of baculoviruses and the related viruses of arthropods is of special interest. With the exception of the DNAP, these viruses do not connect directly to the PL module (although there is a small fraction of genomes in the PL group, namely, some virophages, Polintons and PLV, that possess a D5-like primase-helicase, this gene does not constitute a connector with the whole module). Instead, the Baculo-like module shows multiple connections with “Megavirales” and Poxviridae. Most of the shared genes are assigned to the Baculo-like module, apparently because they are strictly conserved in this small module but not in “Megavirales.” These shared genes include the two large subunits of the RNA polymerase, mRNA capping enzyme, transcription factor TFIIS and thiol oxidoreductase. Baculoviruses and their relatives also harbor genes assigned to other modules and shared by more diverse groups, such as PolB, ribonucleotide reductase (small and large subunits), S_TKc serine/threonine protein kinase, and D5-like primase-helicase. Despite the clear relationship with “Megavirales,” the lack of jelly roll fold capsid proteins, packaging ATPase of the FtsK/HerA family and Ulp1 cysteine protease, all of which are the connectors within the PL-“Megavirales” supermodule, explains why baculoviruses and their relatives stay as a separate module even at the highest hierarchy level.
Modularity and interconnections within the order Caudovirales.
Tailed bacteriophages constitute the most abundant and diverse group of dsDNA viruses (and all viruses). In order to obtain a more precise picture of their module structure and interconnections, we carried out a detailed analysis of the Caudovirales subnetwork (Fig. 6; see also Table S3 in the supplemental material).
FIG 6 .
Internal structure of the Caudovirales supermodule. A module is linked to a connector gene if the prevalence of the gene in that module is greater than exp(−1). Modules are represented as larger circles, with sizes proportional to the number of genomes in the module; color coding is as shown in Fig. 4. Module 15 contains Siphoviridae from the Lactococcus phage 936 sensu lato and c2-like groups. Module 16 conatins Clostridium phage phiCP26F and related strains. Connector genes are represented as smaller gray nodes. Hallmark genes are labeled.
This reanalysis rendered a finer-grained network structure in which module 9 split into four submodules: 9a, with members of the family Myoviridae, such as Mu-like and P2-like phages; 9b, with lambda-like phages and other members of the family Siphoviridae (e.g., T1-like and N15-like phages) as well as P22-like phages; 9c, with Phi31-like phages and numerous unclassified siphoviruses; 9d, containing subfamily Picovirinae and related unclassified members of the family Podoviridae. Two signature gene families are associated with submodule 9a: phage tail protein D and baseplate assembly protein V. In contrast, submodules 9b and 9c are more heterogeneous, and their integrity is maintained by a network of gene sharing which involves multiple tail components and cell lysis proteins. With the exception of the Picovirinae, submodules within module 9 are characterized by moderate to high prevalences (relative abundances) of an integrase and a protein with a helix-turn-helix DNA-binding domain of the Xre family. In agreement with the high prevalence of these genes, a survey of the lifestyle of phages in module 9 shows that it is mostly composed of temperate phages (96 of the 110 based on available data; binomial exact test, P < 10−4).
The large mycobacteriophage module 10 also splits into three submodules, each with a distinct set of signature genes (although the large majority of these genes are poorly characterized). Taking the classification in the Phamerator database (63) (http://phagesdb.org) as a reference, submodule 10a contains L5-like and related phages (Phamerator cluster A), submodule 10b corresponds to clusters E to G and I to P, and submodule 10c includes members from clusters D, H, and R. The phages in these submodules, as well as those in module 11 (PG1-like mycobacteriophages, Phamerator’s cluster B), share a set of minor tail proteins, a peptidoglycan recognition protein (lysin A), and a cutinase (lysin B). The last two genes, required to lyse the mycolic acid-rich outer membrane of the Mycobacterium host (64), also appear in the taxonomically unrelated I3-like mycobacteriophages (module 17, Phamerator cluster C). Notably, all phages from submodule 10c, as well as I3-like phages, share the gene coding for the bacterial DNA polymerase III alpha subunit. Although experimental data on the lifestyle of mycobacteriophages are scarce, the high prevalence of an integrase in submodule 10b suggests that most of the phages from this subgroup are temperate.
The genomes of I3-like phages (module 17) contain 6 to 10 genes (approximately 10% of their total gene content) assigned to submodules within the mycobateriophage module 10. Thus, although the two modules are not similar enough to merge, there seems to be a relationship between I3-like phages and other mycobacteriophages that likely reflects gene exchange, especially given that these phages share related hosts.
Of the three modules (9, 12, 14) for which there is enough information about the lifestyle of their members, modules 12 and 14 show a significant predominance of virulent phages (9 of 9 [P < 10−4] and 13 of 19 [P = 0.002], respectively). Of these two modules, the former corresponds to T4-like phages, whereas the latter includes T7-like phages and some other minor groups from the family Podoviridae. The reanalysis of the Caudovirales subnetwork resulted in refinement of the T4-like module, with approximately 30 unclassified Myoviridae moved to module 9a, and resulting in a robustness of 0.99 (compared to 0.79 in the original module reported in Table 4). Signature genes for this refined module include a set of neck, tail, and baseplate proteins, DNAP sliding clamp and clamp loader, and RNAP sigma factor. Module 14, in contrast, is defined by the central position of the head-to-tail connector protein, which is a signature of T7-like viruses and other phages of the family Podoviridae. A single-subunit RNAP is also characteristic of this module, although some phages within the module lack it. Finally, two major connector genes, the DNAP A and the DnaB primase-helicase, are also assigned to module 14.
Of the remaining modules, module 13 consists of SPO1-like phages, module 15 includes Lactococcus phages from the c-like and 936 sensu lato groups, and the small modules 16 to 19 correspond, respectively, to Clostridium phage phiCP26F (and related strains), I3-like mycobacteriophages, T5-like phages, and phiKZ-like phages. These modules, with the exception of the Lactococcus phage one, possess large sets of module-specific signature genes (Fig. 7), most of which remain uncharacterized (but see Table 4 for a list of some of these genes).
FIG 7 .
Characterization of viral hallmark genes and module-specific signature genes. (A) All core gene families sorted by their relative prevalence in the major supermodules are shown in gray. Hallmark genes are those that, besides belonging to the set of connector genes, have a relative prevalence greater than 0.35 in at least one of the two major supermodules. (B) Signature genes are those genes with mutual information greater than 0.6 to their best-matching module (x axis) and less than 0.02 to their second match (y axis). The rest of the gene families are represented in gray for comparison. (C) Betweenness-rank distribution for genes in the bipartite network. The nodes with the highest betweenness correspond to hallmark and other connector genes. Signature genes are represented in red. (D) Three-dimensional representation of core genes based on mutual information, relative prevalence, and exclusivity with respect to their assigned module (same color coding as in panel C). (E) A histogram with the number of signature, hallmark, connector (nonhallmark), and other (gray) genes per module. Reanalysis of the Caudovirales subnetwork detected 13 signature genes for module 12, which are not shown in the figure. In panels B and D, a large red point indicates the existence of 205 signature genes whose presence-absence patterns perfectly match their assigned modules.
As shown in Fig. 6, global connections across modules occur through hallmark genes, such as the HK97-like major capsid protein, portal protein, maturation protease, and DNA primase-helicase of the DnaB family. Additionally, baseplate proteins J and W connect modules within the family Myoviridae. On a local scale, there are two sets of interconnections: (i) those that bring together mycobacteriophages from submodules 10a, 10b, 10c and, to a lesser extent, I3-like phages, as discussed above; (ii) those that involve T4-like and T5-like phages. Hallmark genes excluded, there are 13 connector genes shared by T5-like and T4-like phages, of which many are related to nucleotide metabolism (multiple nucleotide and nucleoside kinases) and DNA repair (DexA exonuclease, SbcCD repair exonuclease, RNase H).
Given the multiple connections to other modules, it is clear that I3-like, T5-like, and phiKZ-like phages are bona fide members of the Caudovirales network. Therefore, it is surprising that they do not join the major bacteriophage supermodule, even at the highest level of hierarchy. The members of these modules have large genomes (phiKZ-like, 211 to 317 kb; T5-like, 111 to 122 kb; I3-like, 153 to 165 kb) which are characterized by a significant number of conserved genes without clear homologs outside the module itself (65). Our analysis showed that more than 75% of the genes that are highly retained in these genomes are module-specific signature genes. In terms of the network, the fact that these modules stay separated from the rest reflects this distinctiveness of their genome content. In contrast, network analysis places the large Vibrio phage KVP40 (245 kb) in the T4-like module, as widely accepted, and it further merges the T4-like module (also characterized by large genomes but with only 15% of signature genes) with the major Caudovirales supermodule.
Orphan modules.
A closer inspection of the modules that remain unmerged showed that crenarcheal viruses do not form strong connections with the rest of the dsDNA virosphere, although several genes are shared with individual families within other modules. The RHH DNA-binding domain that is typical of archaeal viruses also appears with low prevalence in module 5 (PL elements), because the archaeal virus families Turriviridae and Sphaerolipoviridae are assigned to that module. Archaeal viruses of the family Bicaudaviridae encode an AAA family ATPase that is common in “Megavirales” and is also present in T4-like phages. Similarly, archaeal viruses of the family Lipothrixviridae encode a DEAD-like helicase that is shared with members of the “Megavirales” as well as some tailed phages. Finally, viruses of the family Ampullaviridae encode a pDNAP.
Viruses of the families Polyomaviridae and Papillomaviridae connect with the other modules solely through the SF3 helicase domain that is present in the DNA replication proteins of these viruses (the large T-antigen of polyomaviruses and E1 protein of papillomaviruses). However, this protein as a whole (fusion of the N-terminal origin-binding domain and the C-terminal SF3 helicase domain) has no orthologs among other dsDNA viruses but rather is a derivative of Rep proteins of ssDNA viruses (31). The capsid protein of polyomaviruses and papillomaviruses is of the single jelly roll variety but shares only extremely distant similarity to the minor capsid proteins of PL elements and “Megavirales.”
Unexpected connections and hybrid viral genomes.
The network analysis revealed the hybrid nature of several groups of viruses that possess combinations of core genes from different modules as well as unexpected splits of some supposedly well-established groups. The baculoviruses and their relatives are perhaps the prime case in point, as discussed above, but there are several other notable examples of virus groups with a “split personality.”
For instance, the archaeal viruses in the family Turriviridae encode RHH domain proteins that are typical of other archaeal viruses but merge into the PL module with the eukaryotic viruses due to the shared genes for the packaging ATPase and major and minor jelly roll capsid proteins. In a similar vein, viruses of the archaeal family Ampullaviridae encode a pDNAP but are assigned to the crenarcheal module based on their sharing of a glycosyl transferase with other archaeal viruses.
Mitochondrial plasmids, which in the best solution are assigned to the PL module, often join module 14 (T7-like, Autographivirinae). These elements possess two core genes, namely, pDNAP and a single-subunit phage-type RNAP that, respectively, link them to the PL module and the T7-like module. Because of this hybrid composition, module assignment of these plasmids is uncertain.
Among the cytoplasmic plasmids, three were assigned to the Baculo-like module, whereas the rest were assigned to the PL module. Those plasmids that joined the baculoviruses and their relatives possess four core genes, namely, pDNAP, DEAD-like helicase (assigned to Caudovirales but common also among “Megavirales” and poxviruses), mRNA capping enzyme, and the largest subunit of RNAP (both assigned to baculoviruses but also nearly ubiquitous in “Megavirales” and represented in all poxviruses). This mix of widespread genes causes the split of the plasmid group between two modules, emphasizing the network character of the evolution of selfish elements.
The order Herpesvirales is split between two compact modules that duly combine within a supermodule (Fig. 4) and the phage module 12 (mostly Tevenvirinae), which includes the only available genome from the family Malacoherpesviridae (66). This herpesvirus possesses 6 core genes, namely, DNAP (module 5), large subunit of the terminase (module 9, Caudovirales), large and small subunits of the ribonucleotide reductase (assigned to module 12, Tevenvirinae, although also abundant in “Megavirales,” poxviruses, Herpesvirales, and some other phage modules), RNA ligase (assigned to Tevenvirinae but also common in “Megavirales”), and RING finger-containing ubiquitin ligase (assigned to baculoviruses, also present in poxviruses and some “Megavirales”). Although Malacoherpesvirus shares only a few core genes with Herpesvirales, its clustering with Tevenvirinae is solely based on genes that are widespread among diverse viruses. Thus, this grouping appears to be an artifact caused by the small number of core genes and the current lack of diversity among the malacoherpesviruses. It seems likely that once more genomes from this group become available, additional genes shared with Herpesvirales will be identified, likely resulting in a change of the module affinity.
Among the tailed bacteriophages, it is worth dissecting the case of the N4-like phages that are split between the T7-like and T4-like modules. Genome analysis shows that N4-like phages contain a mix of genes characteristic of each module; they share with T7-like phages the head-to-tail connecting protein, DNAP A, and a single-subunit phage-type RNAP. In contrast, UvrD-like helicase as well as rIIA and rIIB proteins are shared with T4-like phages. This chimeric gene composition of N4-like phages makes it difficult to assign them to a single module; indeed, the main difference between those assigned to the T4-like and T7-like modules is the presence or absence of thioredoxin and ribonucleotide reductase, both formally assigned to the T4-like module, although widespread among bacteriophages and eukaryotic viruses.
Members of the Picovirinae (phi29-like bacteriophages) also appear to be chimeric entities that encode a genome replication machinery shared with the viruses and related MGEs of the PL module, whereas virion structure places them among the Caudovirales. Indeed, phi29-like phages encode bona fide HK97-like major capsid proteins (67), a packaging ATPase that is a distant homolog of the large subunit of the terminase of other tailed phages as well as the portal protein (68). Thus, the phi29-like phages apparently evolved via recombination between a tailed phage and a tectivirus encoding a DJR capsid protein and pDNAP. The tailed phage contributed the genes for the major capsid protein, portal, and some additional proteins involved in virion morphogenesis and host recognition, whereas the tectivirus provided the linear genome scaffold, including the genes for pDNAP and the terminal protein.
Finally, the family Sphaerolipoviridae is peculiar in that it includes bacterial as well as euryarchaeal viruses (Alphasphaerolipovirus and Betasphaerolipovirus infect halophilic archaea, whereas Gammasphaerolipovirus infects bacteria) (69). All members of this family encode an A32-like ATPase and two major capsid proteins (typically called the small and large MCP), which correspond to two halves of the double jelly roll capsid protein characteristic of the “Megavirales”-PL supermodule. Thus, in viruses with the DJR capsid proteins, the pseudohexagonal capsomers are formed from homotrimers of one capsid protein, whereas in sphaerolipoviruses similarly shaped capsomers are heterohexamers of the small and large major capsid proteins (70, 71). Although these MCPs were treated as distinct gene families, sphaerolipoviruses joined the PL module through the packaging ATPase. This assignment appears consistent with the previous suggestion that sphaerolipoviruses diverged from the common ancestor shared with other viruses in the PL supermodule prior to the radiation of the major groups of viruses with the DJR capsid proteins (48). It is worth noting, however, that bacterial sphaerolipoviruses possess two hallmark genes from the Caudovirales supermodule, namely, the integrase and an XRE family helix-turn-helix (HTH) domain-containing protein. Thus, this subset of the sphaerolipoviruses comprises another group of viruses with hybrid genomes.
Classification of viral core genes: signature, connector, and hallmark genes, the glue of the virosphere.
The viral hallmark genes have been previously defined qualitatively as those genes that are shared by multiple, diverse groups of viruses and have no close homologs in cellular organisms (17). The present analysis allows us to quantify this concept by cataloguing and classifying the genes that connect the nodes (connector genes) in the module network. Specifically, we identified hallmark genes as the connector genes with a prevalence in any of the two major supermodules greater than 0.35 (Fig. 7A; see also Materials and Methods). This tally of the hallmark genes identified the familiar suspects, such as the double jelly roll, single jelly roll, and HK97 capsid proteins, terminases and packaging ATPases of the FtsK superfamily (A32-like), DNAP, two primase-helicase families (D5-like and phage replicative helicase DnaB), and two proteases (Ulp1-like and S21/U9/U35). The list was completed by phage portal and tail proteins, transcriptional regulators of the HTH XRE family, and phage integrases of the tyrosine recombinase superfamily. As suggested previously (17), the number of hallmark genes is small: only 14 hallmark genes identified under the above criteria account for 57% of the connections in the module network.
From the perspective of network theory, hallmark and connector genes stand out by their high betweenness (72): 44 of the 50 genes with the highest betweenness are connector or hallmark genes (Fig. 7C). In addition, although they constitute less than 1% of all core gene families, hallmark genes account for 53% of the total betweenness centrality of the network. This value increases to 89% if all connector genes are considered, although they represent less than 4% of the core gene set. Taken together, these results validate our prevalence-driven approach to identify connector and hallmark genes and highlight the role of hallmark genes in maintaining the integrity of the virosphere.
Additionally, we used mutual information to extract signature genes, i.e., genes whose presence/absence pattern is diagnostic of a module (Fig. 7B). As intuitively expected, signature genes show both high prevalence and high exclusivity among modules (Fig. 7D). As shown in Fig. 7E, the number of signature genes per module is highly variable: some modules, such as those that include poxviruses, herpesviruses, T5-like phages, and phiKZ-like phages, are well defined by large sets of signature genes; others, such as the PL and the heterogeneous lambda-like module 9, are instead characterized by a mix of shared hallmark and connector genes. In topological terms, signature genes can be viewed as module-specific hubs, as opposed to the hallmark genes which represent high-level network hubs. Accordingly, techniques for hub identification and classification would seem suitable for identification of signature genes. However, the small size of some modules strongly limits the node degree of their signature genes to the point that they are hardly detectable as hubs. It is for this reason that we chose an information theoretical approach (mutual information) instead of hub detection techniques to identify such genes.
DISCUSSION
Using bipartite network analysis, we show here that the dsDNA domain of the virosphere forms an almost fully connected network of gene sharing and that this network has a robust, hierarchical modular architecture. The dsDNA viruses and related MGE comprise the largest part of the virus world that includes the most abundant biological entities on earth, the phages of the order Caudovirales, as well as some of the most common viruses infecting eukaryotes, such as members of the putative order “Megavirales.” Furthermore, this is the part of the virosphere for which network analysis is expected to be most informative, given the large number of genes with highly variable abundances and often complex evolutionary histories.
The existence of a distinct modular architecture of the gene-genome bipartite network is far from trivial. In principle, alternative models of the virosphere can be easily envisaged, including either a set of disjointed components, or conversely, a continuous structure without robust modules. The latter possibility might be deemed especially plausible given the apparent fluidity of the virosphere, with many documented cases of gene exchange between diverse viruses as well as between viruses and hosts, and the genomic mosaicism that is particularly characteristic of bacteriophages (63, 73). The modularity of the gene-sharing networks is fairly obvious for some viruses but appears unexpectedly in other cases. For example, given the confident identification of a set of about 40 ancestral genes that, some losses notwithstanding, are represented in most members of the putative order “Megavirales” (36), it is not surprising that these viruses belong to a compact module, with the exception of the Poxviridae, a derived group that duly joins the module in the next iteration. In contrast, the robustness of the PL module, especially the inclusion of the archaeal families Turrividiae and Sphaerolipoviridae, was not at all obvious a priori, given the small number of shared genes.
The unification of the PL and “Megavirales” modules into a single supermodule, while compatible with the previously proposed evolutionary scenario (21) is even more striking considering the drastic differences between the genome sizes and gene repertoires between the viruses and MGE in the two modules. Equally nontrivial is the unification of the Herpesvirales with the Caudovirales, primarily on the strength of the shared capsid proteins, the terminase and the maturation protease.
A major advantage of the bipartite network approach is that it provides for the relationships among two categories of objects to be analyzed within the same formal framework (32, 33). This analysis showed that the coherence and robustness of the major modules and supermodules of the network hinge on the uniqueness of a small subset of the core gene set (less than 1% of the core genes and less than 20% of all connector genes), the 14 hallmark genes that are responsible for most of the intermodule connections. Previously, the hallmark genes have been identified informally by comparative analysis of viral genomes (17). Here, we defined the hallmark genes formally and quantitatively as the most prominent connectors between modules in the viral network. The fact that the small set of hallmark genes dramatically differs from the rest of the viral genes with respect to their betweenness underlies the robust modularity of the network. The hallmark genes include those coding for capsid proteins and enzymes involved in virion morphogenesis, along with genes for replication enzymes, such as DNAPs and primases-helicases. These observations seem to settle the perceived conflict between the “structural” and “replicative” perspectives on virus evolution (48, 51, 52). Analysis of the node degree distributions of the bipartite network for the gene and genome nodes clearly indicates that it is the hallmark genes, and not chimeric viral genomes, that are primarily responsible for the network cohesiveness and modularity.
We further defined the signature genes, i.e., those core genes that showed the highest prevalence within but not between modules and hold together some of the modules (the modules are also supported by distinct combinations of the hallmark and other connector genes). The hallmark and signature genes correspond, respectively, to the “date” and “party” hubs that have been identified previously in other biological contexts, such as protein-protein interaction networks, and recognized as the basis of the dynamic modularity in these networks (74–77).
The results of the network analysis revealed a surprisingly well-structured dsDNA virosphere by robustly partitioning the dsDNA viruses into two major supermodules, PL-“Megavirales” and Caudovirales-Herpesvirales, which jointly cover most of the dsDNA virosphere, and two smaller, isolated modules, the crenarcheal viruses and polyomaviruses-papillomaviruses. The small polyomaviruses and papillomaviruses, in a sense, do not rightfully belong in the dsDNA virus domain of the virosphere, given their clear evolutionary relationships with ssDNA viruses (31). The few remaining small modules can be expected to join one of the two supermodules upon expansion of the membership.
The two supermodules are defined primarily by the two distinct morphogenetic machineries, each including distinct, unrelated building blocks of icosahedral capsids, the DJR and HK97-like capsid proteins, respectively, and the accompanying genome packaging ATPases and maturation proteases, which belong to unrelated (proteases) or extremely distantly related (ATPases) protein families. Among the known viruses, the two structural modules have not been observed to recombine, i.e., a DJR capsid protein never combines with a terminase, whereas capsids made of HK97-like proteins never package DNA with the help of an FtsK superfamily (A32-like) ATPase. This strict coupling between the capsid building blocks and packaging motors is likely to have a distinct mechanistic underpinning that remains to be elucidated.
The split of the supermodules along the line separating the morphogenetic machineries of the respective viruses is compatible with the concept of the capsid structure as the “self” of a virus (46). However, the coherence of the supermodules, in particular the PL-“Megavirales” one, also depends on the replication and transcription machineries, in particular DNAP and RNAP, which bring into the module various capsidless elements, such as cytoplasmic and mitochondrial plasmids and those Polintons that lack capsid proteins (19).
The PL-“Megavirales” supermodule is by far the largest, most diverse group of eukaryotic dsDNA viruses that includes viruses and related MGE from all major eukaryotic taxa (except for land plants, some of which, however, bear imprints of past infections by members of the “Megavirales” [78]) as well as two bacterial and two archaeal virus families. The viruses and MGE in this supermodule differ by more than 3 orders of magnitude in genome size and lead vastly different lifestyles, yet they are robustly linked by a distinct set of hallmark and other connector genes as well as multiple signature genes within individual modules. The network analysis supports the central position of the Polintons and related viruses in this heterogeneous supermodule, which conceivably reflects the evolutionary potential of these elements for combining features of viruses and transposons. Notably, the PL module is the only module in the network that brings together viruses infecting hosts from all three domains of cellular life. Among the archaeal viruses, the present analysis primarily includes viruses of hyperthermophilic Crenarchaeota, whereas viruses of mesophilic archaea, in particular Lokiarchaeota, the likely ancestors of eukaryotes (79, 80), remain poorly characterized. Thus, it is currently unclear whether the Polintons, which form the link between the viruses of prokaryotes and those of eukaryotes in this supermodule, are derived from bacterial viruses (tectiviruses from the mitochondrial endosymbiont) or related archaeal viruses that remain to be identified. Given that the PL module owes its coherence to the presence of the JRC (the most common viral structural protein, if both single jelly roll and double jelly roll forms are considered), the associated small packaging ATPases, and pDNAP, a simply organized enzyme involved in the replication of relatively small DNA genomes, this viral module could be as old as cellular life itself, if not older. Indeed, from the perspective of the primordial virus world, in which capsids could have played a key role in the dissemination of the primitive, virus-like genetic elements (81, 82), the viruses of the PL module appear to be strong candidates for some of the earliest forms.
Most of the available genomes from the Herpesvirales represent the family Herpesviridae, with a rather homogeneous composition of the core genes. The rest of the herpesviruses, in the less thoroughly characterized families Alloherpesviridae and Malacoherpesviridae, show major differences in the gene repertoires and either join the Herpesvirales in a second iteration or fail to join at all, remaining in one of the phage modules. Regardless, the unification of Herpesvirales with Caudovirales is highly robust and is about as strong as that between different herpesvirus families. This result draws support from structural and biochemical studies showing that viruses from both Caudovirales and Herpesvirales execute strikingly similar programs of virion assembly and maturation and employ homologous proteins in the key steps of these processes (83–85). Notably, the Herpesvirales comprise the only offshoot of the Caudovirales (tailed viruses infecting bacteria and archaea), the most abundant group of viruses altogether, in the eukaryotic world (31). In a sense, herpesviruses that so far have been isolated only from metazoa, primarily vertebrates, can be considered “animal phages.” It is also of note that the four archaeal members of the order Caudovirales included in this analysis confidently fall into module 9 together with several groups of tailed bacteriophages (Table 4), possibly testifying to the early diversification of the Caudovirales.
Predictably, the majority of the identified modules consist of phages of the order Caudovirales that join into a supercluster together with Herpesvirales in the second iteration of network analysis. The existence of these robust modules clearly shows that despite the well-known fluidity of the phage genomes (63), the pool of phage genes is compartmentalized. The phage modules do not split along family lines, although several modules contain large subsets of phages from the same family. The modules are associated with distinctive sets of signature core genes, such as the head-to-tail connecting protein for the module that harbors most of the Podoviridae, the nicking endonuclease for T5-like viruses, and a set of minor tail proteins for the module that encompasses a large group of mycobacteriophages of the family Siphoviridae. On a larger scale, modules with taxonomic or ecological similarity are linked by connector genes, e.g., the baseplate J protein for phages of the family Myoviridae and the specialized lysins for mycobacteriophages. The partial lack of family coherence is not especially surprising given that phage families have been identified without any reference to evolutionary relationships (86).
A network approach in bacteriophage classification has been reported previously, including a comparison between phage clusters and gene modules that were defined independently (not as parts of a bipartite network of the type explored here) (87). Despite the technical differences, both approaches render similar pictures, with a core of highly interconnected temperate phages and several peripheral modules which include T4-like phages, T7-like phages, and mycobacteriophages. The two analyses also agree in some specific details, such as the subdivision of the mycobacteriophages and the isolation of phiKZ-like phages. The differences seem to stem from the increase in the amount of the genomic data during the time that elapsed since the previous analysis, which is particularly consequential for smaller groups of phages. Thus, in the study of Lima-Mendez and colleagues (87), T5-like phages, represented at the time by a single genome, appeared as a hybrid between T4-like phages and SPO1-like phages. In contrast, inclusion of two additional members in the present analysis gave rise to a distinct module which, despite the connections to T4-like phages, maintained its identity even at the highest level of hierarchy.
The only distinct module of dsDNA viruses of eukaryotes (besides the polyomavirus-papillomavirus module) that fails to join either the PL-“Megavirales” or the Herpesvirales-Caudovirales supermodule includes baculoviruses and related viruses of arthropods. This module shares connections with each of the supermodules but lacks some of the key hallmark genes, in particular either type of the capsid proteins and packaging ATPases, and apparently for this reason, it cannot be assigned to either supermodule. Nevertheless, the strongest connection of these viruses is with the “Megavirales,” suggesting that the Baculo-like module is a highly derived offshoot of the PL-“Megavirales” supermodule that, in particular, has lost the ancestral morphogenetic machinery. It should be noted that the loss of structural and morphogenetic proteins has been observed even among the “Megavirales” themselves, e.g., in Pandoraviruses (37).
The viruses of archaeal hyperthermophiles include several families, each with a number of unique features, and with the notable exception of the Turriviridae, form a distinct module in the bipartite network. In sharp contrast with the PL-“Megavirales” and Herpesvirales-Caudovirales supermodules, the archaeal module is held together not by hallmark genes coding for key proteins of viral morphogenesis and replication but rather by accessory, regulatory genes, such as those encoding RHH and HTH domain-containing proteins, which are particularly abundant in these viruses (53). These regulatory genes appear to be frequently transferred between archaeal viruses as well as between viruses and hosts, which seems to account for the coherence of this module. The extreme diversity of the structures of the crenarcheal viruses and the near lack of identifiable proteins involved in replication clearly distinguishes them from viruses of bacteria and eukaryotes. The evolutionary processes and pressures that have led to this distinct character of the crenarcheal viruses remain elusive (88). Formally, the family Turriviridae links the crenarcheal part of the virosphere with the PL module, but the implications of this weak connection are at present difficult to assess.
The results of the viral network analysis seem to reflect four interrelated but distinct processes that apparently shaped the large-scale structure of the virosphere: (i) vertical inheritance of gene ensembles that define highly cohesive groups of viruses, e.g., the “Megavirales,” (ii) sampling of hallmark and other connector genes from the partially compartmentalized but continuous in space and time pool of MGE that underlies the connectivity and modularity of the virosphere as a network, (iii) horizontal gene transfer between viruses that yields additional intermodule connections, and (iv) capture of host genes by viruses, including multiple, independent acquisitions of homologous genes.
The modules and supermodules of viruses and related MGE represent groups with coherent evolutionary histories that, given the limited applicability of traditional phylogenetic methods to the virosphere, are likely to help further evolutionary studies as well as virus taxonomy. Further developments could include extension of the bipartite network analysis to all viruses and MGE as well as their cellular hosts.
MATERIALS AND METHODS
Sequences.
Protein sequences were collected from the NCBI Genome Database for all available genomes of dsDNA viruses with an assigned family. Specifically, we collected genomes belonging to the orders Herpesvirales (families Herpesviridae, Alloherpesviridae, and Malacoherpesviridae), Caudovirales (families Siphoviridae, Podoviridae, and Myoviridae), Ligamenvirales (Lipothrixviridae and Rudiviridae), the proposed order “Megavirales” (families Ascoviridae, Asfarviridae, Iridoviridae, Marseilleviridae, Mimiviridae, Phycodnaviridae, and Poxviridae, as well as Pandoravirus and Pithovirus), the families Adenoviridae, Tectiviridae, Ampullaviridae, Bicaudaviridae, Corticoviridae, Fuselloviridae, Globuloviridae, Guttaviridae, Sphaerolipoviridae, Turriviridae, Baculoviridae, Nudiviridae, Hytrosaviridae, Nimaviridae, Papillomaviridae, Polyomaviridae, and Plasmaviridae, the genus Salterprovirus, and the available virophages (Lavidaviridae). The family Polydnaviridae was excluded given the extreme divergence of the genomes of these viruses, which consist primarily of host-derived or inactivated viral genes. This data set was complemented by several additional groups of sequences from recently reported yet unclassified viruses or MGE for which evolutionary relationships with dsDNA viruses have been demonstrated. In particular, sequences from virophages YSLV5, YSLV6, and YSLV7, Mollivirus, and Faustovirus were retrieved from the NCBI nonredundant protein database. Sequences of Polintons and Polinto-like viruses, as well as mitochondrial and cytoplasmic dsDNA plasmids, were added by using previously described custom data sets. In total, the initial data set contained 137,331 sequences from 1,442 genomes.
Automatic classification of genes into homologous families.
First, all protein sequences were clustered at 90% identity and 70% coverage by using CD-HIT (89) to generate a nonredundant data set. For each sequence in this set, a BLASTp search (90) was carried out against all other included sequences, and hit scores were used to generate a sequence similarity network. This procedure involved two steps: initially, a BLASTp search with composition-based statistics (91) and filtering of low-complexity regions was used to determine valid hits (e-val cutoff of 0.01, database size fixed to 2e7). Scores for those hits were subsequently collected from a BLASTp search with neither composition-based statistics nor a low-complexity filter. Sequences with best (reciprocal) hits to other sequences from the same genome were combined and treated as in-paralogs.
The set of BLAST hits defines a weighted sequence similarity network in which nodes are sequences and edges connect significantly similar sequences, with a weight proportional to the hit score. Preliminary groups of homologous genes were identified as communities in the sequence similarity network. In the context of network theory, a community is a set of nodes that are densely interconnected compared to the average node degree of the network. To find such communities, we employed the Infomap software (92) (100 trials, 2-level hierarchy). The communities of sequences in the sequence similarity network tend to constitute partitions of larger homologous groups, with sequences from distantly related taxa often located in separate communities. Therefore, we applied profile analysis to merge communities that consisted of homologous sequences. Sequences in each community were aligned with Muscle (93) (default parameters); the alignments were used to predict secondary structure and build profiles with the tools “addss” and “hhmake” available within the HH-suite package (94). Profile-profile comparisons were carried out using HHsearch (95). Hits were accepted or rejected based on their probability, relative coverage, and length. Specifically, hits with a probability greater than 0.90 were accepted if they covered at least 50% of the length of the profile; additionally, hits with a coverage of 20% or greater were also accepted if their probability was greater than 0.99 and their length was greater than 100 amino acids. The choice of this heuristic filtering strategy was motivated by its performance on benchmark collections of viral orthologous genes (POGs [9] and NCVOGs [37]). This pipeline rendered a total of 33,980 clusters of homologous sequences, of which 11,950 comprised multiple sequences and 22,030 were singletons (ORFans).
Manual curation.
Some homologous proteins shared among highly diverse groups of viruses (e.g., capsid proteins) have diverged to the point of becoming inaccessible to the currently available automatable sequence analysis approaches, although their homology can still be demonstrated on a case-by-case basis, through analysis of sequence motifs and structural similarity. Thus, we relied on the previous findings on these highly diverged homologous proteins to manually curate our collection of homologous sequences. The main groups that had to be manually merged included the major capsid proteins with the double jelly roll fold, their associated minor capsid proteins (pentons), Ulp1-like cysteine proteases (21, 60, 61, 96), capsid proteins with the HK97-like fold (50, 85), herpesvirus protease S21 and homologous bacteriophage prohead proteases U9 and U35 (97), and the set of proteins shared among Baculoviridae, Hytrosaviridae, and Nimaviridae (57, 98).
The concept of orthology is readily applicable to groups of viruses with similar overall genome organization, such as the NCLDV or the Polinton-like viruses and MGE. Methods for identification of orthologous genes clusters have been implemented in the construction of databases such as POGs (9, 35) and NCVOGs (36, 37), which were employed as reference sets for the present analysis. The meaning of orthology is less obvious when it comes to comparisons between widely different groups, e.g., NCLDV versus Caudovirales. Nevertheless, an effort was made to adhere to the notion of orthology as closely as possible by including in the same group only homologous genes with analogous functions, for example, capsid proteins or packaging ATPases. Accordingly, hits that were due to promiscuous conserved domains, e.g., P-loop ATPases, were considered spurious for the purpose of this analysis and discarded at this stage. In particular, manual curation involved splitting two groups that had been erroneously merged based on profile-profile comparison. The first of those false hits connected A32-like packaging ATPases to thymidine kinases (both proteins contain the P-loop NTPase domain); the second false hit involved mRNA capping enzymes from “Megavirales” and DNA ligases from Caudovirales (again, both enzymes contain homologous nucleotidyltransferase domains).
Henceforth, we use the term protein family to refer to the manually curated groups of homologous sequences.
The bipartite network of viruses.
The bipartite network of viruses was built by connecting genome nodes to protein family nodes whenever a genome contained at least one representative of a given protein family. To avoid redundancy, genomes that shared more than 90% of their protein families (including ORFans) were treated as a single pangenome. The resulting bipartite network contained a giant component, including all genomes except for two Polintons which together with 5 protein families constituted their own minor components. For subsequent analyses, only the giant component was considered.
In the context of the bipartite network, a singleton is a protein family that only appears in one genome (or the pan-genome of a set of 90% similar genomes).
Identification of core genes.
We adopted an intuitive definition of core genes as those genes that tend to be maintained in a genome for long periods of time, i.e., those with the lowest loss rates in a pure-loss evolutionary model. Let us consider a simple model in which, starting from a common ancestor, the similarity at the family content level diverges uniformly in time according to the differential equation , where Sij is the number of families shared by genomes i and j and Ni and Nj are the numbers of families in each genome. The geometric mean in this expression aims at biasing the denominator toward the smaller genome, thus avoiding artifacts caused by giant viruses (34); as it has been shown for the “Megavirales,” similar results can be obtained with min(Ni, Nj) as the normalization factor (33). This simple evolutionary model generates a natural distance between any pair of genomes: . Although a rigorous derivation of this expression from the underlying model requires that Ni and Nj remain constant, the same compositional metrics has been also applied to more general cases (e.g., analysis of evolutionary relationships within the “Megavirales”). The use of this metrics is additionally supported by the fact that the distance-based tree for the “Megavirales” that it generates shows a good correlation with the tree obtained from the concatenated sequences of the “Megavirales” universal core genes (34). For a particular protein family, the probability that it is present in any pair of genomes conditioned on its presence in the last common ancestor is , where r is the loss rate of the family relative to the average divergence rate of genomes. Similarly, the probability for the family being lost in only one genome of the pair is where Z = P10 + P11 is a normalization factor. Note that, in a pure loss model, the fact that one of the genomes contains a family implies that the last common ancestor also contained it. However, pairs of genomes that lack any representative of a family have to be discarded, because there is no guarantee that the family was present in their common ancestor. We used the expressions for P10, P11, and Dij to calculate a maximum likelihood estimate of the family-specific loss rate r. The presence of one or a few shared families in otherwise unrelated genomes due to HGT could bias loss rate estimates, thus we only considered those pairs of genomes with distances Dij < 1. This condition implies that both genomes in the pair have to share more than 35% of their gene content (relative to the size of the smaller genome), a degree of similarity that is unlikely to be due to HGT only. Only those gene families with three or more appearances were considered. Genes with relative loss rate r < 1 were assigned to the “core.” Note that different values of this threshold would lead to slightly different lists of core genes. However, because the hallmark and signature genes that underlie the multiscale modular structure of the virus network tend to be highly retained, the results are robust to the particular choice of the loss rate threshold. We additionally tested the consistency of the core gene list by defining viral groups based on the network modules (see below) and recalculating the loss rates separately for each group. The aggregated list of core genes produced in this way showed a high agreement with the original one, and in particular it contained all hallmark and signature genes. The retention probability of a gene family was calculated as exp(−r). This expression provides the probability that a family has not been lost after one time unit, with time measured in the characteristic time scale of genome composition divergence.
Gene family abundances were computed based on genome-weighted contributions as previously described (99). The purpose of weighting the contribution of a genome to the abundance of a gene is to compensate for sampling bias by assigning smaller weights to groups of closely related genomes. Distances between pairs of genomes were calculated using the same compositional metrics as described above. In the case of nonoverlapping genomes, a conservative estimate of the distance between them was calculated as , which corresponds to the time that would take for an ancestor with one more gene to lose all but that one common gene. The genome-genome distance matrix was used to build a neighbor-joining tree, and genome weights were extracted from the tree following the previously described algorithm (99). Family abundances were obtained by adding the weights of all genomes where the family is present. Therefore, an abundance equal to 1 means that the family is present in all genomes of the data set. The prevalence, or relative abundance, of a gene family in a group of genomes was calculated as the sum of weights of those genomes in the group that harbored the gene family divided by the sum of weights of all the genomes in the group.
Detection of modules in the bipartite genome: core gene network.
To detect sets of related genomes and gene families, we applied Barber’s definition of bipartite modularity (100) to the bipartite network consisting of genomes and core genes. Simulated annealing heuristics (101) was used to identify the partition of the network that optimizes the magnitude , where L is the number of links in the network, G and F represent the sets of genomes and core gene families, respectively, aij denotes the existence of a link between genome i and gene family j, is the probability that a link exists by chance (with ki and kj denoting the connectivities of nodes i and j), and δ is the Kronecker delta function, which takes the value 1 if nodes i and j belong to the same module and the value 0 otherwise. The Modular software (102) was used to carry out the modularity optimization. The significance of the resulting partition was evaluated by running 100 replicates of a null model of a randomly generated bipartite network with the same size and the same gene- and genome-degree distributions as the original network. All the results reported here correspond to partitions with a P value of <0.01. Because of the ruggedness of modularity landscapes in large, complex networks, the partition found by the module detection algorithm corresponds to only one of possibly many local maxima of modularity (34). To account for this limitation, we ran 100 replicates (realizations) of the algorithm and kept the partition with the highest modularity as the optimal partition. The robustness of the modules in the optimal partition was evaluated by comparing them with the modules obtained in the other 99 alternative partitions.
To detect supermodules, we analyzed higher-order bipartite networks consisting of modules and connector gene families. A gene family was considered a connector between two modules if its relative abundance (prevalence) in both modules was greater than exp(−1). The choice of this threshold was motivated by its correspondence with the expected abundance of a gene with a loss rate equal to 1 after a characteristic unit of time of the gene content divergence process. Prevalence thresholds from 0.35 to 0.5 were also tested, with no qualitative differences. Gene families whose abundance exceeded the threshold in a single module were also kept in the network, although they were not classified as connectors. Supermodules were identified by applying the module detection algorithm to the module-level bipartite network. This process was iterated in order to delineate a hierarchy of higher-order modules. The iterative search continued until no more mergers occurred or nonsignificant values of the modularity were obtained. As with primary modules, 100 independent replicates were carried out in order to assess the robustness of the supermodules.
The internal structures of some sets of modules (PL, “Megavirales,” and Baculovirus; PL alone; and modules that included Caudovirales) were further analyzed by extracting from the bipartite network a subnetwork consisting of the genomes assigned to each particular set of modules and the core genes connected (but not necessarily assigned) to them. Modules and connector genes in each subnetwork were identified following the same procedure as in the entire network.
Hallmark and signature genes.
The intuitive idea of “hallmark genes” was formalized as follows: (i) a hallmark gene must be a connector between first-order modules and (ii) it must be sufficiently prevalent in at least one of the two major supermodules. In order to select a suitable prevalence threshold, we searched for gaps in the distribution of relative prevalences and found a noticeable discontinuity near 0.35, which approximately agrees with the criterion used to define connector genes. Therefore, we adopted 0.35 as the minimum relative prevalence that a connector gene must attain in (at least) one of the major supermodules to be considered hallmark.
Signature genes were defined on the basis of their normalized mutual information with respect to their best and second best matching modules. For each gene and each module, their normalized mutual information (MI) was calculated as MI = , where a, b, c, and d are the added relative weights of (a) the genomes from the module that contain the gene, (b) the genomes that contain the gene but do not belong to the module, (c) the genomes from the module that lack the gene, and (d) the genomes that neither belong to the module nor contain the gene. Note that, compared to the traditional formulation of the mutual information, we did not take into account the terms associated with b and c. In doing so, we excluded the contributions from genes and modules with complementary patterns. The mutual information was subsequently normalized by the joint entropy: . For each gene, we selected the two highest values of the normalized mutual information (those that corresponded to the best and second best matching modules) and used the graphical representation in Fig. 6B to set the “signature” thresholds (>0.6 for the best match, <0.02 for the second best match).
Bacteriophage lifestyles.
Information on the temperate or virulent lifestyle of 171 tailed bacteriophages was collected from (103) (data are available at ACLAME website, http://aclame.ulb.ac.be/Classification/Phages/life_style.html). Of these phages, 57 were virulent and 114 were temperate. The number of phages with available data is much smaller than the total number of phages in our network; therefore, we discarded those modules for which there were no data for at least 10% of its members. For the remaining modules, a binomial exact test was applied to estimate the significance of the lifestyle-module association under the null hypothesis that module composition is random with respect to phage lifestyle.
SUPPLEMENTAL MATERIAL
List of core genes sorted by normalized abundance.
List of genes shared by multiple modules (connector genes).
List of genomes and module assignations.
ACKNOWLEDGMENTS
We thank David Kristensen, Sergey Mekhedov, and Natalya Yutin for expert technical advice and Koonin group members for discussions.
E.V.K. incepted the study; J.I. developed the methods and performed research; J.I., M.K., and E.V.K. analyzed the results; J.I. and E.V.K. wrote the manuscript, which was edited and approved by all authors.
Footnotes
Citation Iranzo J, Krupovic M, Koonin EV. 2016. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7(4):e00978-16. doi:10.1128/mBio.00978-16.
REFERENCES
- 1.Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat Rev Microbiol 3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 2.Rohwer F. 2003. Global phage diversity. Cell 113:141. doi: 10.1016/S0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 3.Rohwer F, Thurber RV. 2009. Viruses manipulate the marine environment. Nature 459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 5.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 7.Hendrix RW. 2003. Bacteriophage genomics. Curr Opin Microbiol 6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol 18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. 2013. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J Bacteriol 195:941–950. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szathmáry E, Demeter L. 1987. Group selection of early replicators and the origin of life. J Theor Biol 128:463–486. doi: 10.1016/S0022-5193(87)80191-1. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi N, Hogeweg P. 2007. The role of complex formation and deleterious mutations for the stability of RNA-like replicator systems. J Mol Evol 65:668–686. doi: 10.1007/s00239-007-9044-6. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi N, Hogeweg P, Koonin EV. 2011. On the origin of DNA genomes: evolution of the division of labor between template and catalyst in model replicator systems. PLoS Comput Biol 7:e1002024. doi: 10.1371/journal.pcbi.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi N, Hogeweg P. 2012. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys Life Rev 9:219–263. doi: 10.1016/j.plrev.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forterre P, Prangishvili D. 2009. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci 1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 15.Forterre P, Prangishvili D. 2013. The major role of viruses in cellular evolution: facts and hypotheses. Curr Opin Virol 3:558–565. doi: 10.1016/j.coviro.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV, Dolja VV. 2013. A virocentric perspective on the evolution of life. Curr Opin Virol 3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol Direct 1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes EC. 2011. What does virus evolution tell us about virus origins? J Virol 85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin EV, Dolja VV. 2014. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N, Raoult D, Koonin EV. 2013. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J 10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol 13:105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dagan T. 2011. Phylogenomic networks. Trends Microbiol 19:483–491. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Ponce D, Lopez P, Bapteste E, McInerney JO. 2013. Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proc Natl Acad Sci U S A 110:E1594–E1603. doi: 10.1073/pnas.1211371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, Whitfield J. 2013. Networks: expanding evolutionary thinking. Trends Genet 29:439–441. doi: 10.1016/j.tig.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Forster D, Bittner L, Karkar S, Dunthorn M, Romac S, Audic S, Lopez P, Stoeck T, Bapteste E. 2015. Testing ecological theories with sequence similarity networks: marine ciliates exhibit similar geographic dispersal patterns as multicellular organisms. BMC Biol 13:16. doi: 10.1186/s12915-015-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corel E, Lopez P, Méheust R, Bapteste E. 2016. Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol 24:224–237. doi: 10.1016/j.tim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. 2011. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res 21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popa O, Dagan T. 2011. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol 14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 29.Jachiet PA, Colson P, Lopez P, Bapteste E. 2014. Extensive gene remodeling in the viral world: new evidence for nongradual evolution in the mobilome network. Genome Biol Evol 6:2195–2205. doi: 10.1093/gbe/evu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King AMQ, Lefkowitz E, Adams MJ, Carstens B (ed). 2011. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 31.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borgatti SP, Everett MG. 1997. Network analysis of 2-mode data. Soc Networks 19:243–269. doi: 10.1016/S0378-8733(96)00301-2. [DOI] [Google Scholar]
- 33.Montañez R, Medina MA, Solé RV, Rodríguez-Caso C. 2010. When metabolism meets topology: reconciling metabolite and reaction networks. BioEssays 32:246–256. doi: 10.1002/bies.200900145. [DOI] [PubMed] [Google Scholar]
- 34.Fortunato S. 2010. Community detection in graphs. Phys Rep 486:75–174. doi: 10.1016/j.physrep.2009.11.002. [DOI] [Google Scholar]
- 35.Kristensen DM, Cai X, Mushegian A. 2011. Evolutionarily conserved orthologous families in phages are relatively rare in their prokaryotic hosts. J Bacteriol 193:1806–1814. doi: 10.1128/JB.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yutin N, Wolf YI, Koonin EV. 2014. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lind PG, González MC, Herrmann HJ. 2005. Cycles and clustering in bipartite networks. Phys Rev E Stat Nonlin Soft Matter Phys 72(5 Pt 2):056127. doi: 10.1103/PhysRevE.72.056127. [DOI] [PubMed] [Google Scholar]
- 39.Barabási AL, Oltvai ZN. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet 5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 40.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin EV. 2011. The logic of chance: the nature and origin of biological evolution. FT Press, Upper Saddle River, NJ. [Google Scholar]
- 42.Haegeman B, Weitz JS. 2012. A neutral theory of genome evolution and the frequency distribution of genes. BMC Genomics 13:196. doi: 10.1186/1471-2164-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobkovsky AE, Wolf YI, Koonin EV. 2013. Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 5:233–242. doi: 10.1093/gbe/evt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrix RW, Hatfull GF, Smith MC. 2003. Bacteriophages with tails: chasing their origins and evolution. Res Microbiol 154:253–257. doi: 10.1016/S0923-2508(03)00068-8. [DOI] [PubMed] [Google Scholar]
- 45.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev 75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bamford DH. 2003. Do viruses form lineages across different domains of life? Res Microbiol 154:231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 47.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr Opin Struct Biol 15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Krupovic M, Bamford DH. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol 6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 49.Krupovic M, Bamford DH. 2010. Order to the viral universe. J Virol 84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol 1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2009. The complexity of the virus world. Nat Rev Microbiol 7:250. doi: 10.1038/nrmicro2030-c2. [DOI] [Google Scholar]
- 52.Krupovič M, Bamford DH. 2009. Does the evolution of viral polymerases reflect the origin and evolution of viruses? Nat Rev Microbiol 7:250. doi: 10.1038/nrmicro2030-c1. [DOI] [PubMed] [Google Scholar]
- 53.Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res 117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colson P, de Lamballerie X, Fournous G, Raoult D. 2012. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology 55:321–332. doi: 10.1159/000336562. [DOI] [PubMed] [Google Scholar]
- 56.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Bininda-Emonds ORP, Jehle JA. 2012. Nudivirus genomics and phylogeny. In Rijeka GM. (ed), Molecular structure, diversity, gene expression mechanisms and host of virus interactions. InTechOpen doi: 10.5772/1346. [DOI] [Google Scholar]
- 58.Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol 101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 59.Jehle JA, Abd-Alla AM, Wang Y. 2013. Phylogeny and evolution of Hytrosaviridae. J Invertebr Pathol 112(Suppl):S62–S67. doi: 10.1016/j.jip.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 60.Krupovic M, Bamford DH, Koonin EV. 2014. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol Direct 9:6. doi: 10.1186/1745-6150-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yutin N, Shevchenko S, Kapitonov V, Krupovic M, Koonin EV. 2015. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol 13:95. doi: 10.1186/s12915-015-0207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yutin N, Kapitonov VV, Koonin EV. 2015. A new family of hybrid virophages from an animal gut metagenome. Biol Direct 10:19. doi: 10.1186/s13062-015-0054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science, Phage Hunters Integrating Research and Education, Mycobacterial Genetics Course. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Payne K, Sun Q, Sacchettini J, Hatfull GF. 2009. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol Microbiol 73:367–381. doi: 10.1111/j.1365-2958.2009.06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hendrix RW. 2009. Jumbo bacteriophages. Curr Top Microbiol Immunol 328:229–240. doi: 10.1007/978-3-540-68618-7_7. [DOI] [PubMed] [Google Scholar]
- 66.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. 2005. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell 18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 68.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pawlowski A, Rissanen I, Bamford JK, Krupovic M, Jalasvuori M. 2014. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch Virol 159:1541–1554. doi: 10.1007/s00705-013-1970-6. [DOI] [PubMed] [Google Scholar]
- 70.Rissanen I, Grimes JM, Pawlowski A, Mäntynen S, Harlos K, Bamford JK, Stuart DI. 2013. Bacteriophage P23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 21:718–726. doi: 10.1016/j.str.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gil-Carton D, Jaakkola ST, Charro D, Peralta B, Castaño-Díez D, Oksanen HM, Bamford DH, Abrescia NG. 2015. Insight into the assembly of viruses with vertical single beta-barrel Major capsid proteins. Structure 23:1866–1877. doi: 10.1016/j.str.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 72.Freeman LC. 1978. Centrality in social networks. I. Conceptual clarification. Soc Networks 1:215–239. doi: 10.1016/0378-8733(78)90021-7. [DOI] [Google Scholar]
- 73.Hatfull GF. 2010. Mycobacteriophages: genes and genomes. Annu Rev Microbiol 64:331–356. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]
- 74.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. 2002. Evolutionary rate in the protein interaction network. Science 296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 75.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. 2004. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 76.Chang X, Xu T, Li Y, Wang K. 2013. Dynamic modular architecture of protein-protein interaction networks beyond the dichotomy of “date” and “party” hubs. Sci Rep 3:1691. doi: 10.1038/srep01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiran M, Nagarajaram HA. 2013. Global versus local hubs in human protein-protein interaction network. J Proteome Res 12:5436–5446. doi: 10.1021/pr4002788. [DOI] [PubMed] [Google Scholar]
- 78.Maumus F, Epert A, Nogue F, Blanc G. 2014. Plant genomes enclose footprints of past infections by giant virus relatives. Nat Commun 5:4268. doi: 10.1038/ncomms5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koonin EV. 2015. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol 13:84. doi: 10.1186/s12915-015-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koonin EV. 2009. On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci 1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jalasvuori M, Mattila S, Hoikkala V. 2015. Chasing the origin of viruses: capsid-forming genes as a life-saving preadaptation within a community of early replicators. PLoS One 10:e0126094. doi: 10.1371/journal.pone.0126094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baker ML, Jiang W, Rixon FJ, Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol 79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rochat RH, Liu X, Murata K, Nagayama K, Rixon FJ, Chiu W. 2011. Seeing the portal in herpes simplex virus type 1-B capsids. J Virol 85:1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rixon FJ, Schmid MF. 2014. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr Opin Virol 5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 86.Krupovic M, Dutilh BE, Adriaenssens EM, et al. 2016. Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol 161:1095. doi: 10.1007/s00705-015-2728-0. [DOI] [PubMed] [Google Scholar]
- 87.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol 25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 88.Prangishvili D. 2015. Archaeal viruses: living fossils of the ancient virosphere? Ann N Y Acad Sci 1341:35–40. doi: 10.1111/nyas.12710. [DOI] [PubMed] [Google Scholar]
- 89.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. BioInformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 90.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosvall M, Bergstrom CT. 2008. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meier A, Söding J. 2015. Automatic prediction of protein 3D Structures by probabilistic Multi-template homology modeling. PLoS Comput Biol 11:e1004343. doi: 10.1371/journal.pcbi.1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 96.Maaty WS, Ortmann AC, Dlakić M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol 80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng H, Shen N, Pei J, Grishin NV. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci 13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Bininda-Emonds OR, van Oers MM, Vlak JM, Jehle JA. 2011. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes 42:444–456. doi: 10.1007/s11262-011-0589-5. [DOI] [PubMed] [Google Scholar]
- 99.Makarova KS, Wolf YI, Koonin EV. 2015. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 5:818–840. doi: 10.3390/life5010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barber MJ. 2007. Modularity and community detection in bipartite networks. Phys Rev E Stat Nonlin Soft Matter Phys 76(6 Pt 2):066102. doi: 10.1103/PhysRevE.76.066102. [DOI] [PubMed] [Google Scholar]
- 101.Guimerà R, Sales-Pardo M, Amaral LA. 2007. Module identification in bipartite and directed networks. Phys Rev E Stat Nonlin Soft Matter Phys 76(3 Pt 2):036102. doi: 10.1103/PhysRevE.76.036102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marquitti FMD, Guimarães PR, Pires MM, Bittencourt LF. 2014. Modular: software for the autonomous computation of modularity in large network sets. Ecography 37:221–224. doi: 10.1111/j.1600-0587.2013.00506.x. [DOI] [Google Scholar]
- 103.Lima-Mendez G, Toussaint A, Leplae R. 2007. Analysis of the phage sequence space: the benefit of structured information. Virology 365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of core genes sorted by normalized abundance.
List of genes shared by multiple modules (connector genes).
List of genomes and module assignations.