The controlled differentiation of embryonic stem cells into specific cell types holds promise for cell-based therapies in patients, including for the treatment of conditions affecting the heart. In recent years, the isolation of human embryonic stem cells (hESCs), the discovery of ‘induced pluripotency’ and, last but not least, the numerous studies in developing embryos, using model organisms, have brought us closer to the ultimate goal of generating cardiomyocytes in the dish.
With the view to improve existing protocols, a recent study published in Cell Stem Cell (1) has looked in detail at the genome-wide molecular responses triggered by cardiogenic signals in hESCs. In this editorial we will summarize their findings and conclusions and in addition, highlight the seminal discoveries and insights coming from developmental biology that have paved the way. We argue that studying the embryo will continue to make important contributions to stem cell science.
Clearance of roadblocks toward cardiomyocyte differentiation
Wnt/β-catenin signaling is clearly a key regulator of heart, and specifically heart muscle development [see recent reviews (2,3)]. However, experimental activation of Wnt signaling at different developmental stages causes dramatically different consequences for subsequent heart development: at an early stage, Wnt/β-catenin signaling promotes but then it restricts subsequent heart muscle (cardiomyocyte) cell differentiation. These observations in a number of model systems gave rise to the suggestion that Wnt signaling has a biphasic role in heart muscle development [see review (4)]. This biphasic role was most dramatically illustrated in human Embryonic Stem Cells, when experimental manipulation of Wnt signaling alone was essentially sufficient to drive heart muscle cell (cardiomyocyte) differentiation from pluripotent cells [as an example see ref. (5)]. In this new work, artificial activation of Wnt/β-catenin signaling (with a pharmacological agonist) followed by experimental inhibition of endogenous Wnt signaling (with a pharmacological antagonist) leads pluripotent cells to differentiate into rhythmically beating cardiomyocytes (Figure 1A). This outcome is clearly good news, but how can Wnt signaling promote and then restrict subsequent cardiomyocyte differentiation?
Figure 1.
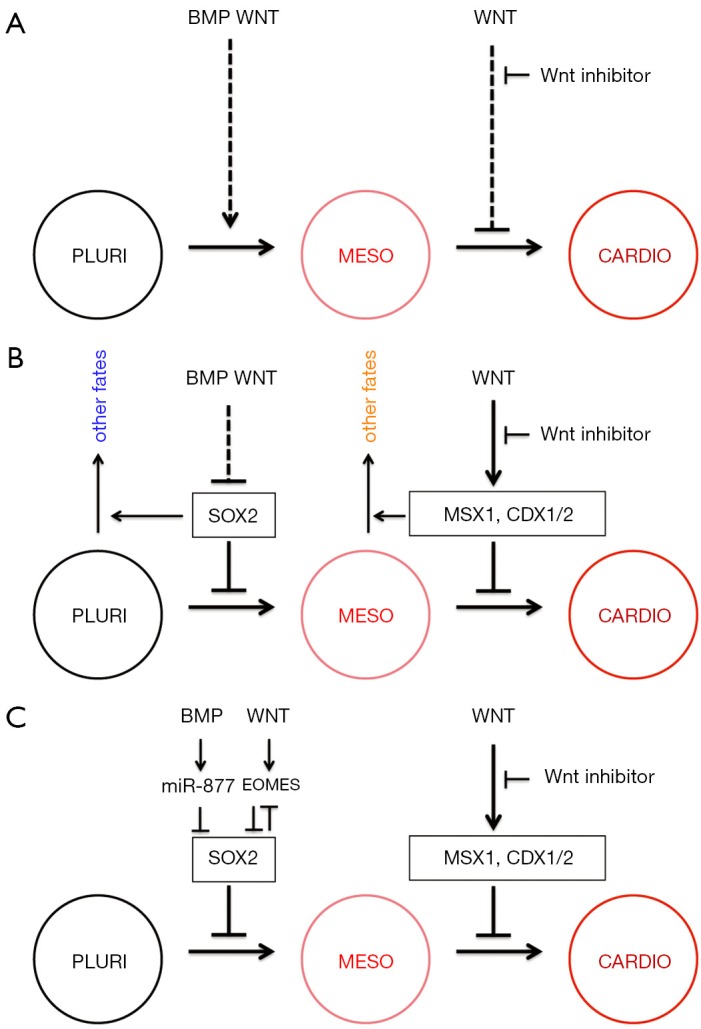
Roadblocks toward Cardiac Development. Schematic description of the cardiomyocyte differentiation protocol from pluripotent (PLURI) embryonic stem cells via mesoderm induction (MESO) to differentiated cardiomyocytes (CARDIO). (A) WNT/β-catenin signaling and BMP/SMAD1 signaling promote mesoderm induction, but WNT/β-catenin signaling subsequently restricts cardiomyocyte differentiation. Many cardiomyocyte differentiation protocols therefore combine early activation of WNT and/or BMP signaling with later inhibition of Wnt signaling; (B) SOX2 serves as a roadblock early on the path from pluripotent to mesoderm fate, then MSX1, CDX1 and CDX2 serve as roadblocks on the path from mesoderm to cardiomyocyte differentiation. Early WNT and BMP signaling remove the SOX2 roadblock, but later WNT signaling would maintain the MSX/CDX roadblock, if it were not inhibited. Rather than just roadblocks preventing cardiomyocyte development, SOX2 and MSX/CDX probably function to promote differentiation into alternative cell fates; (C) the negative regulation of SOX2 by BMP and by WNT signaling are both indirect and function in parallel. BMP regulation is faster, involving upregulation of the microRNA miR-877, which targets SOX2 transcripts directly; while WNT signaling regulates EOMES, which represses SOX2 transcription after a delay. Figure adapted from (1).
Greber and colleagues (1) solved this riddle by analyzing the transcriptional response following experimental manipulations. The expression of SOX2 is dramatically cleared following early experimental activation of Wnt/β-catenin signaling, while subsequent experimental inhibition of Wnt signaling resulted in much reduced expression particularly of CDX1, CDX2 and MSX1.
Greber and colleagues (1) developed the hypothesis that these transcription factors function as roadblocks on the path toward cardiomyocyte differentiation (Figure 1B). Indeed, experimental overexpression of SOX2 at the relevant early stages prevented progress toward subsequent cardiomyocyte differentiation, and RNAi-mediated SOX2 knockdown caused pluripotent cells to take initial steps on the path toward heart development. Correspondingly, experimental overexpression of CDX2 and MSX1 at the relevant later stage severely compromised subsequent cardiomyocyte differentiation; while CRISPR-mediated triple knockout of CDX1, CDX2 and MSX1 promoted subsequent cardiomyocyte differentiation under conditions that would otherwise not be conducive to cardiomyocyte differentiation.
From a cardiomyocyte perspective these factors can be described as roadblocks; however, from a more general perspective they represent signposts for other destinations, that is alternative cell fates (Figure 1B). Indeed, SOX2 does not only maintain pluripotency early but also regulates neural differentiation later in development [see review (6)]. Furthermore, many neural stem cell differentiation protocols use Wnt inhibitors at these early stages to steer the differentiation toward a neural fate (7). Conversely, MSX1 and CDX1 and 2 are associated with paraxial mesoderm (8-10).
Whether we see them as roadblocks toward our preferred destination or signposts for multiple alternative destinations, such factors are likely to function in concert with others. Indeed, even the triple knockout of CDX1, CDX2 and MSX1 only made subsequent cardiomyocyte differentiation a little easier, but did not completely overcome the requirement for inhibiting Wnt signaling, suggesting that there are additional Wnt-regulated genes preventing cells from differentiating into cardiomyocytes. Some of the molecular details of how the SOX2 roadblock is removed have already been worked out (Figure 1C) and see below.
Integration of BMP and Wnt signaling delineates a cardiac corridor
Several signaling pathways synergize with Wnt/β-catenin signaling to promote more efficient cardiomyocyte differentiation (11,12). Greber and colleagues found that particularly BMP signaling cooperates with Wnt/β-catenin signaling in the early stages of the differentiation protocol. Their transcriptomics analyses revealed that Wnt and BMP signaling are required for inducing an essentially identical cardiogenic gene expression signature.
Remarkably, this cardiogenic signature and subsequent beating cardiomyocytes were induced not only with medium-BMP/medium-Wnt experimental conditions, but also in low-BMP/high-Wnt or high-BMP/low-Wnt conditions. Therefore, this experiment identified what they dubbed a “cardiac corridor”, where the sum of BMP and Wnt signal is conducive to specify cardiac mesoderm (Figure 2). On either side of this corridor alternative cell fates were induced: under low-BMP/low-Wnt conditions hESCs generated mostly neural fates, whereas exposure to high-BMP/high-Wnt led to more lateral or ventral mesoderm signatures (e.g., blood, cartilage). Interestingly an endodermal signature was induced in between the cardiac corridor and the low concentrations of BMP/Wnt that induced neural fates.
Figure 2.
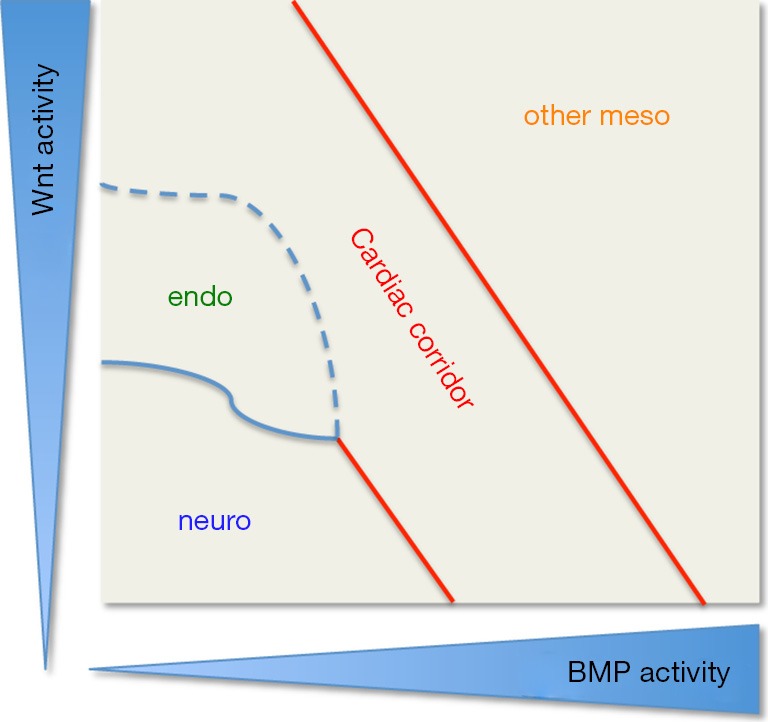
Cardiac corridor is defined by WNT and BMP input. Patterning map for hESC culture represented by a two-dimensional grid indicating high to low WNT signaling activity from top to bottom; and low to high BMP signaling activity from left to right. Cardiac induction is promoted by the combined input from WNT and BMP signaling activity during the early stages of differentiation. Cardiac development is induced by medium WNT activity combined with medium BMP activity but also with low WNT/high BMP; and high WNT/low BMP conditions. This pattern defines a diagonal corridor where cardiac induction occurs. Alternative fates are clearly delineated by high BMP and medium to high WNT activity (other mesodermal fates) and by low WNT and low BMP activity, but there is less clear delineation between cardiac and endodermal fates under low BMP and intermediate WNT activity conditions. Figure adapted from (1). hESC, human embryonic stem cell.
Wnt and BMP remove the SOX2 roadblock, but by different mechanisms
Not surprisingly the transcription factor SOX2 becomes depleted in the cardiac corridor within hours. Cooperation between Wnt and BMP signaling to downregulate SOX2 in hESCs emerges as a crucial step for mesoderm induction. Combinatorial BMP and WNT signaling is known from development of many tissues to use a variety of mechanisms to integrate combined regulation of target genes (13). Intriguingly, BMP/Smad1-mediated negative regulation of SOX2 happens much faster than repression by Wnt/β-catenin signaling, suggesting different mechanisms.
The early BMP/SMAD1 induced reduction of SOX2 turns out to be mediated post-transcriptionally by a microRNA, miR-877, which itself is upregulated after BMP4 stimulation or SMAD1 overexpression (Figure 1C). The involvement of miR-877 is entirely consistent with the proposed function of these negative regulators to provide robustness to developmental programmes and to mediate cell fate transitions (14-16), however, the details of Smad1/miR-877/Sox2 interactions remain to be resolved. One possibility suggested is that miR-877 biogenesis may be directly enhanced by SMAD1 since its precursor contains a SMAD1 binding element (17). Moreover SOX2 is a predicted target for miR-877-3p and even though the target site is poorly conserved, its potential functional importance is supported by the finding that transfection of mimics resulted in a moderate reduction of endogenous SOX2 transcripts in hESCs. Importantly, however, direct interactions between pre-miR-877 and SMAD1 on the one hand, and between miR-877-3p and SOX2 transcripts on the other will need to be confirmed.
Wnt/β-catenin signaling is normally associated with transcriptional activation of target genes [see recent review (18)]. The observed delay in downregulation of SOX2 expression following experimental activation of Wnt signaling is therefore expected and consistent with an indirect mechanism. The transcriptomics analysis identified the T-box transcription factor EOMES as being a Wnt-regulated rather than BMP-regulated gene within the cardiac corridor. CRISPR-mediated knockout of EOMES shows that EOMES is required both for WNT signaling to repress SOX2 and for subsequent cardiomyogenesis. Experimentally expressed EOMES is capable of binding to T-box binding elements in the SOX2 locus and of reducing SOX2 expression. Furthermore, it has recently been shown that the Wnt/β-catenin pathway directly regulates EOMES expression (19).
How about embryos?
Do the uncovered mechanisms in hESC all make sense in the context of whole embryos? Yes, absolutely—they do. The signaling pathways tested by Rao et al. (1) are mimicking the inductive interactions that have previously been implicated in vivo. Their work is underpinned by numerous studies in developing vertebrate embryos, particularly in experimentally accessible model systems such as avian and amphibian, but also by genetic loss-of-function approaches in mice (20-23). The characterization of a cardiac corridor for hESC differentiation suggests that there are gradients of Wnt and BMP signals that induce highly related but distinct cell fates, an idea which is indeed compatible with the expression of these signals and their inhibitors along the early embryonic axes (dorsoventral and anterior-posterior) [see examples (24-26)].
Another aspect to consider is the finding that BMP/Smad1 and Wnt/GSK3β cooperate to guide the early migration of cardiac progenitor cells as they emerge from the primitive streak (26). During embryogenesis, the processes controlling cell migration need to be coordinated with those that affect cell fate choice. This may not be necessary for hESCs differentiating into cardiomyocytes in the dish, however, the lack of morphogenesis, which deprives cells of some developmental history that can manifest as epigenetic modifications, may go some way to explaining why differentiation protocols still require optimization.
Furthermore, their findings that hESCs generated mostly neural fates under low-BMP/low-Wnt conditions, are consistent with what we know from studies in vertebrates, particularly in Xenopus laevis and chick embryos, where BMP antagonists (chordin, noggin, follistatin) and Wnt antagonists (sFRPs) are expressed in dorsal most mesoderm [recently reviewed by (27)]. More lateral or ventral mesoderm signatures (e.g., blood, cartilage) were induced in hESCs by high-BMP/high-Wnt conditions, again consistent with mesoderm patterning in early embryos.
This combinatorial patterning by WNT and BMP signaling so elegantly explored by Greber and colleagues in hESC differentiation (1) does not only find parallels in mammalian embryos, but is likely to be more deeply conserved in animal evolution (28). In addition their studies are a fine example of how to apply lessons learned in embryos to hESC differentiation (29).
Acknowledgements
Funding: A Münsterberg acknowledges research grant funding from BHF (PG/15/77/31761) and BBSRC (BB/N002326/1). Research in the SH laboratory is supported by BHF (PG/12/75/29851).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Rao J, Pfeiffer MJ, Frank S, et al. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell 2016;18:554-6. 10.1016/j.stem.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 2.Hoppler S, Mazzotta S, Kühl M. Wnt signalling in heart organogenesis. In: Hoppler S, Moon RT. editors. Wnt signalling in development and disease: molecular mechanisms and biological functions. New York: John Wiley & Sons, Inc., 2014:293-301. [Google Scholar]
- 3.Ruiz-Villalba A, Hoppler S, van den Hoff MJ. Wnt signaling in the heart fields: variations on a common theme. Dev Dyn 2016;245:294-306. 10.1002/dvdy.24372 [DOI] [PubMed] [Google Scholar]
- 4.Gessert S, Kühl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res 2010;107:186-99. 10.1161/CIRCRESAHA.110.221531 [DOI] [PubMed] [Google Scholar]
- 5.Lian X, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 2012;109:E1848-57. 10.1073/pnas.1200250109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells 2014;6:305-11. 10.4252/wjsc.v6.i3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blauwkamp TA, Nigam S, Ardehali R, et al. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun 2012;3:1070. 10.1038/ncomms2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houzelstein D, Auda-Boucher G, Chéraud Y, et al. The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development 1999;126:2689-701. [DOI] [PubMed] [Google Scholar]
- 9.Christ B, Huang R, Wilting J. The development of the avian vertebral column. Anat Embryol (Berl) 2000;202:179-94. 10.1007/s004290000114 [DOI] [PubMed] [Google Scholar]
- 10.van den Akker E, Forlani S, Chawengsaksophak K, et al. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 2002;129:2181-93. [DOI] [PubMed] [Google Scholar]
- 11.Burridge PW, Keller G, Gold JD, et al. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 2012;10:16-28. 10.1016/j.stem.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Schulte JS, Heinick A, et al. Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells 2015;33:1456-69. 10.1002/stem.1964 [DOI] [PubMed] [Google Scholar]
- 13.Itasaki N, Hoppler S. Crosstalk between Wnt and bone morphogenic protein signaling: a turbulent relationship. Dev Dyn 2010;239:16-33. [DOI] [PubMed] [Google Scholar]
- 14.Goljanek-Whysall K, Sweetman D, Abu-Elmagd M, et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci U S A 2011;108:11936-41. 10.1073/pnas.1105362108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet 2006;38 Suppl:S20-4. 10.1038/ng1803 [DOI] [PubMed] [Google Scholar]
- 16.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012;149:515-24. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BN, Hilyard AC, Nguyen PH, et al. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 2010;39:373-84. 10.1016/j.molcel.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppler S, Nakamura Y. Cell-to-cell signalling in development: Wnt signalling. eLS 2014.
- 19.Funa NS, Schachter KA, Lerdrup M, et al. β-catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 2015;16:639-52. 10.1016/j.stem.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 20.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev 1997;11:451-62. 10.1101/gad.11.4.451 [DOI] [PubMed] [Google Scholar]
- 21.Marvin MJ, Di Rocco G, Gardiner A, et al. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev 2001;15:316-27. 10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev 2001;15:304-15. 10.1101/gad.855601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lickert H, Kutsch S, Kanzler B, et al. Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev Cell 2002;3:171-81. 10.1016/S1534-5807(02)00206-X [DOI] [PubMed] [Google Scholar]
- 24.Hoppler S, Moon RT. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev 1998;71:119-29. 10.1016/S0925-4773(98)00004-5 [DOI] [PubMed] [Google Scholar]
- 25.Yue Q, Wagstaff L, Yang X, et al. Wnt3a-mediated chemorepulsion controls movement patterns of cardiac progenitors and requires RhoA function. Development 2008;135:1029-37. 10.1242/dev.015321 [DOI] [PubMed] [Google Scholar]
- 26.Song J, McColl J, Camp E, et al. Smad1 transcription factor integrates BMP2 and Wnt3a signals in migrating cardiac progenitor cells. Proc Natl Acad Sci U S A 2014;111:7337-42. 10.1073/pnas.1321764111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiecker C, Bates T, Bell E. Molecular specification of germ layers in vertebrate embryos. Cell Mol Life Sci 2016;73:923-47. 10.1007/s00018-015-2092-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niehrs C. On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 2010;137:845-57. 10.1242/dev.039651 [DOI] [PubMed] [Google Scholar]
- 29.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008;132:661-80. 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]


