Abstract
Purpose
Cancer spread to the central nervous system (CNS) often is diagnosed late and is unresponsive to therapy. Mechanisms of tumor dissemination and evolution within the CNS are largely unknown because of limited access to tumor tissue.
Materials and Methods
We sequenced 341 cancer-associated genes in cell-free DNA from cerebrospinal fluid (CSF) obtained through routine lumbar puncture in 53 patients with suspected or known CNS involvement by cancer.
Results
We detected high-confidence somatic alterations in 63% (20 of 32) of patients with CNS metastases of solid tumors, 50% (six of 12) of patients with primary brain tumors, and 0% (zero of nine) of patients without CNS involvement by cancer. Several patients with tumor progression in the CNS during therapy with inhibitors of oncogenic kinases harbored mutations in the kinase target or kinase bypass pathways. In patients with glioma, the most common malignant primary brain tumor in adults, examination of cell-free DNA uncovered patterns of tumor evolution, including temozolomide-associated mutations.
Conclusion
The study shows that CSF harbors clinically relevant genomic alterations in patients with CNS cancers and should be considered for liquid biopsies to monitor tumor evolution in the CNS.
INTRODUCTION
The treatment of human cancer has shifted toward a precision medicine paradigm that increasingly relies on the genomic annotation of each patient’s tumor tissue. This trend is supported by the clinical observation that tumor responses to signal transduction inhibitors often are greatest in tumors that harbor mutations in the targeted pathway, by the discovery of specific drug-resistance mutations in tumors that resume growth during therapy, and by the recent association between effective immunotherapy and tumor-specific missense mutations. Outgrowth of drug-resistant tumor cell clones during therapy can limit the clinical relevance of the initial tumor profile and has motivated the development of technologies that can track the evolution of the cancer genome in accessible body fluids.1
Cancers that affect the central nervous system (CNS) pose a particular challenge because of the difficulty in accessing tumor tissue and the inability to detect circulating tumor DNA in the plasma of affected patients.2 One potential source of tumor-derived DNA in patients with CNS tumors is cerebrospinal fluid (CSF), which circulates through the CNS. In patients whose primary tumor had disseminated to the CNS, several groups were able to identify selected mutations of the primary tumor in CSF by using polymerase chain reaction detection techniques.3-7 A recent study collected CSF during the resection of primary brain or spinal cord tumors and reported that 26 of 35 (74%) samples contained tumor DNA, which was defined as the presence of at least one mutation in the primary tumor.8 All patients were previously untreated, and the detection of mutations in the CSF was guided by prior profiling of the primary tumor. A separate study used targeted next-generation sequencing to reveal oncogenic mutations in tumor-derived DNA from CSF in a limited number of patients.9 Together, these studies suggest that the shedding of tumor DNA into the CSF may be a frequent occurrence in CNS cancers, but it is unclear whether comprehensive sequencing of CSF can routinely and reliably identify clinically relevant genomic alterations without prior knowledge of mutations in the primary tumor and whether this can be done successfully without a need for surgery in a large series of patients. The goal of the current study was to explore whether routine lumbar puncture and high-throughput sequencing of CSF could identify tumor-associated DNA in patients with known or suspected CNS involvement and provide clinically meaningful insights into the biology of these tumors and their treatment response.
MATERIALS AND METHODS
CSF Collection and Preparation
We collected CSF samples from 53 patients with cancer who underwent evaluation for leptomeningeal metastasis between August 2014 and February 2015. Fifty-two (98%) CSF samples were obtained by lumbar puncture and one from an Ommaya reservoir. All patients signed informed consent for use of leftover CSF for research purposes under protocols approved by the Memorial Sloan Kettering (MSK) Cancer Center Institutional Review Board. Within 2 to 3 hours of CSF collection, 5 mL of CSF was placed on ice and centrifuged at 1,000 × g at 4°C for 5 minutes. The supernatant was aseptically transferred to prelabeled cryotubes, and the cell pellet was resuspended in 1 mL of RPMI + 20% fetal bovine serum + 20% dimethyl sulfoxide. All tubes were stored at −70°C.
Extraction of Cell-Free DNA
The minimum amount of the CSF tested was 2 mL (mean, 5 mL; range, 2 to 7 mL). Stored CSF samples were thawed at room temperature and centrifuged at 10,000 × g for 30 minutes at 4°C to remove residual precipitated cellular components and various particles. The method applied for the extraction of cell-free DNA (cfDNA) was based on the manufacturer’s protocol for the QIAamp Circulating Nucleic Acid Kit (catalog #55114; QIAGEN, Valencia, CA). Briefly, 5 mL of CSF was mixed with 500 μL of protease K and 4 mL of buffer ACL. After incubation at 60°C for 30 minutes, 9 mL of buffer ACB was added and then incubated on ice for 5 minutes. The mixture was filtered through a minicolumn and rinsed by ACW1, ACW2, and ethanol. DNA was eluted in 100 μL of buffer AVE.
Targeted Capture and Sequencing
All samples were subjected to molecular analysis by using the MSK-Integrated Molecular Profiling of Actionable Cancer Targets (IMPACT) assay,10 which captures all protein-coding exons of 341 cancer-associated genes as well as 33 introns in 14 recurrently rearranged genes. The Illumina libraries were constructed with KAPA Hyper Prep Kit followed by ligation with 5 μM adaptor concentration (catalog #KK8504; Kapa Biosystems, Wilmington, MA). Libraries of this targeted capture were pooled in equimolar concentrations and sequenced on an Illumina HiSeq 2500 system (Illumina, San Diego, CA) as paired end 100–base pair reads.
Genomic Analysis
Analysis for the targeted sequencing data was performed as described previously.10 In brief, demultiplexed FASTQ files were aligned to GRCh37 reference human genome assembly by using BWA-MEM (Burrows-Wheeler Aligner software version 0.7.5a, http://arxiv.org/abs/1303.3997), and polymerase chain reaction duplicates were identified with use of the MarkDuplicates tool in Picard Tools software version 1.96 (https://github.com/broadinstitute/picard). Regions in the genome covered by more than 20× coverage were identified by using the FindCoveredIntervals from the Genome Analysis Toolkit11 and subjected to indel realignment by Assembly-Based ReAligner version 0.92 software.12 Variant calling was performed in paired tumor/normal mode by using MuTect software version 1.1.413 for single nucleotide variants and SomaticIndelDetector11 and Pindel software version 0.2.5a714 for small insertions and deletions. All variants were then annotated using ANNOVAR software version 527.15 For CSF cfDNA and cell pellets without a genetically matched normal, variants were called against a single pool of unmatched normal samples, and variants were filtered if the minor allele frequency was > 0.0004 in any subpopulation in the 1000 Genomes Project cohort16 or Exome Aggregation Consortium17 because these are more likely to be common population polymorphisms than somatic mutations. All candidate mutations and indels in the Data Supplement were called automatically by using the bioinformatics pipeline described previously and subsequently reviewed manually by using the Integrative Genomics Viewer18 to eliminate potential false-positive calls. The current framework can be found at https://github.com/rhshah/impact-pipeline.
Copy number variation was identified by analyzing sequence coverage of targeted regions in a tumor sample compared with a standard diploid normal sample after performing sample-wide LOWESS normalization for guanine-cytosine percentage across exons and normalizing for global differences in on-target sequence coverage, as previously described.10 Somatic structural aberrations were identified by using DELLY software.19 All candidate somatic structural aberrations were filtered, annotated by using in-house tools, and manually reviewed with Integrative Genomics Viewer.18 Figures were created and modified by using R for statistical computing and graphics (R Development Core Team) and Adobe Illustrator (http://www.adobe.com/products/illustrator.html) by using free templates designed by Freepik (http://freepik.com).
RESULTS
Comparison of Mutation Detection in CSF cfDNA Versus Cell-Pellet DNA
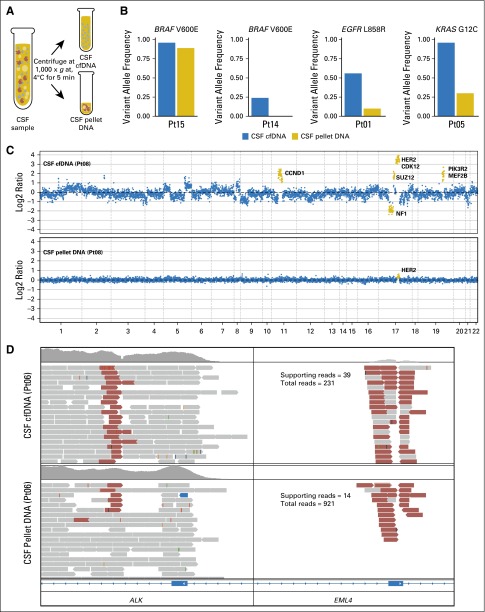
As a first step toward developing a robust mutation detection method, we examined which component of CSF was most sensitive for the detection of the most common cancer-associated genetic alterations. Because CSF in healthy individuals contains a small number of WBCs (0 to 5/μL), we were concerned that germline DNA from normal or reactive WBCs would dilute the signal from tumor-derived DNA. To address this question, we centrifuged eight freshly collected CSF samples, separated the cell pellet from the supernatant (Fig 1A), and compared DNA yields and detection of cancer-associated mutations in each aliquot. To increase the likelihood of detecting tumor-derived DNA, these pilot experiments focused on eight patients with an established diagnosis of CNS metastasis from solid tumors on the basis of typical radiographic findings (n = 8) or detection of tumor cells in the CSF (n = 7). In each patient, the primary tumor was known to harbor a clinically relevant driver mutation. There was a trend toward higher DNA yields from CSF pellets (mean, 280 ng) than from the CSF supernatants (mean, 27 ng), but this difference was not statistically significant (P = .22), and we achieved high unique sequence coverage of sequencing libraries from both pellets (mean, 746×) and supernatants (mean, 444×).
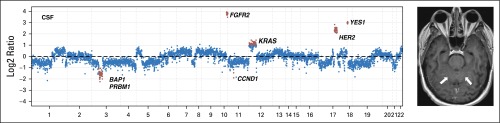
Fig 1.
Comparison of tumor-derived DNA from cerebrospinal fluid (CSF) cell pellet and supernatant. (A) Schematic of separation of CSF pellet and supernatant. Cellular DNA is isolated from the pellet, and cell-free DNA (cfDNA) is isolated from the supernatant. (B) Variant allele frequencies for known mutations in CSF cfDNA and pellet DNA. (C) Log2 ratios of normalized sequence coverage for target exons in CSF cfDNA and pellet DNA for patient 8. Greater than 10-fold amplification of HER2 was observed in CSF cfDNA, whereas HER2 amplification was barely detectable in pellet DNA. (D) Evidence of EML4-ALK gene fusion in CSF cfDNA and pellet DNA for patient 6. Read pairs supporting the fusion (red) were visualized by using the Integrative Genomics Viewer. Pt, patient ID.
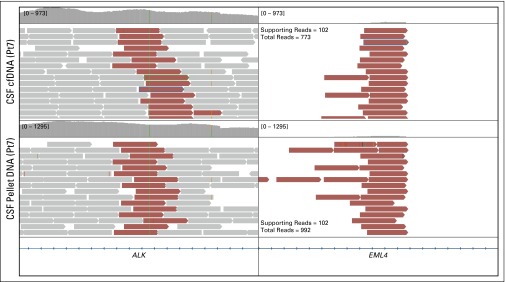
We next compared our ability to detect sequence mutations, copy number alterations, and structural rearrangements in CSF pellet DNA and CSF cfDNA with our in-house sequencing assay (MSK-IMPACT), which interrogates 341 clinically relevant cancer genes. MSK-IMPACT has been extensively validated in a cohort of > 300 distinct positive-control tumors10 and has been approved as a clinical test by the New York State Department of Health. All four patients with known single nucleotide substitutions exhibited a higher percentage of sequence reads that harbored the mutant allele in the CSF cfDNA compared with the CSF pellet DNA. These included two patients with BRAF V600E mutant melanoma, one with non–small-cell lung cancer (NSCLC) with EGFR L858R mutation, and one with NSCLC with KRAS G12C mutation (Fig 1B). In terms of gene copy number alterations, we observed an 11-fold amplification of the HER2 gene locus in CSF cfDNA from one patient with HER2-amplified breast cancer. In contrast, the copy number plot obtained from pellet DNA of the same CSF sample was markedly blunted, and the HER2 gene amplification was barely detectable (Fig 1C). In another patient with breast cancer with known loss of PTEN, we detected a homozygous deletion at the PTEN locus on chromosome 10q in the CSF cfDNA sample but not in the cell pellet DNA (Appendix Fig A1, online only). Two patients with NSCLC harbored rearrangements that involved anaplastic lymphoma kinase (ALK) in the primary tumor, which was supported by 39 and 102 DNA fragments in CSF cfDNA compared with 14 and 102 DNA fragments in the respective CSF pellet DNA (Fig 1D; Appendix Fig A2, online only). In summary, known molecular alterations from the primary tumors were readily detectable in all eight (100%) CSF cfDNA samples but only in five (63%) CSF pellet DNA samples. In every case, the evidence that supported the key alteration was greater in cfDNA than in pellet DNA (Appendix Table A1, online only), which suggests that a higher proportion of the cfDNA is tumor derived.
CSF cfDNA in Various Types of CNS Cancer
Based on results from our pilot study, we used cfDNA as the DNA source for all subsequent analyses of genomic alterations in CSF and extended the study to 45 additional CSF samples from patients with cancer referred to our neuro-oncology clinic. All patients underwent standard-of-care diagnostic testing with magnetic resonance imaging (MRI) of the brain and CSF cytology. Together with the samples from our pilot project, the current study comprised 53 CSF samples from 41 patients with solid tumors and 12 with primary brain tumors. Of the 41 with solid tumors, 32 had CNS involvement by cancer. Nine with a variety of cancer types had no evidence of CNS involvement (ie, MRI and CSF cytology negative) and had an organ-confined primary tumor (n = 6) or metastatic cancer to non-CNS sites (n = 3; Table 1).
Table 1.
Characteristics of 53 Patients With CNS Involvement and Primary Tissue Cancer for Which CSF Cell-Free DNA Was Extracted and Analyzed
| Patient No. | Primary Tumor | CNS Involvement | Original Specimen (molecular pathology) | CSF (MSK-IMPACT) |
|---|---|---|---|---|
| Solid tumors | ||||
| 1 | NSCLC | Brain metastases | EGFR L858R (bone, Sequenom) | EGFR L858R (56%) |
| 2 | NSCLC | Brain metastases | EGFR L858R (lung, Sequenom) | EGFR T790M (2.5%), EGFR L858R (76%) |
| 3 | NSCLC | Brain metastases | EGFR exon 19 del (chest wall, Sequenom) | EGFR T790M (2.8%), EGFR 745_750 del (37%) |
| 4 | NSCLC | Brain metastases | EGFR L858R (lung, Sequenom) | KRAS G12A (19%), EGFR L858R (65%) |
| 5 | NSCLC | Brain metastases | KRAS G12C (c34 G>T) (lung, Sequenom) | KRAS G12C (96%), CDKN2B del (log2, −2.9) |
| 6 | NSCLC | Brain metastases | ALK rearrangement (lung, ND) | EML4-ALK fusion (39 reads) |
| 7 | NSCLC | Leptomeningeal metastases | EML4-ALK fusion (lung, FM) | EML4-ALK fusion (102 reads) |
| 8 | Breast | Brain metastases | HER2 AMP (breast, FISH) | PIK3CA H1047R (38%), HER2 AMP (log2, 3.5) |
| 9 | Breast | Brain metastases | HER2 AMP (breast, FISH) | HER2 AMP (log2, 2.6) |
| 10 | Breast | Brain metastases | HER2 positive (breast, IHC 3+) | HER2 AMP (log2, 2.6) |
| 11 | Breast | Brain metastases | No molecular profiling performed | EGFR AMP (log2, 3.1), PIK3CA H1047R (28%) |
| 12 | Breast | Brain metastases | TP53 V272M (56%), PTEN del (log2, −2.0; lymph node, MSK-IMPACT) | TP53 V272M (81%), PTEN del (log2, −2.97) |
| 13 | Breast | Brain metastases | ER positive, PR/HER2 negative (thyroid metastases, IHC) | PIK3CA E545K (26%) |
| 14 | Melanoma | Brain metastases | BRAF V600E (skin, ND) | BRAF V600E (24%) |
| 15 | Melanoma | Leptomeningeal metastases | BRAF V600E (skin, ND) | PTEN del (log2, −XYZ), BRAF V600E (96%) |
| 16 | Melanoma | Brain metastases | BRAF V600E (skin, ND) | NRAS G12R (3%), PTEN del (log2, −3.0), BRAF V600E (47%) |
| 17 | Bladder cancer | Brain metastases | No molecular profiling performed | AKT2 AMP (log2, 3.37), TP53 R158L (43%) |
| 18 | Gastroesophageal | Leptomeningeal metastases | No molecular profiling performed (MSK-IMPACT failure) | HER2 AMP (log2, 2.4), FGFR2 (log2, 3.6) |
| 19 | Neuroendocrine | Brain metastases | No molecular profiling performed | MYCN AMP (log2, 4.1) |
| 20 | Ovarian | Brain metastases | BRCA1 insC (blood, Myriad Genetics laboratory) | BRCA1 Q1756fs (53%), CDKN2B del (log2, −2.1) |
| 21 | Ovarian | Leptomeningeal metastases | No molecular profiling performed | Negative |
| 22 | Breast | Brain metastases | No molecular profiling performed | Negative |
| 23 | Breast | Brain metastases | HER2 AMP (breast, FISH) | Negative |
| 24 | Breast | Brain metastases | ESR1 Y537S (62%), CCND1 AMP (log2, 1.5; breast, MSK-IMPACT) | Negative |
| 25 | Breast | Brain metastases | RB1 L343Sfs*3 (liver, MSK-IMPACT) | Negative |
| 26 | Breast | Brain metastases | PIK3CA R108 del (39%), CCND1 AMP (log2, 1.0; soft tissue, MSK-IMPACT) | Negative |
| 27 | NSCLC | Brain metastases | No molecular profiling performed | Negative |
| 28 | NSCLC | Brain metastases | No molecular profiling performed | Negative |
| 29 | SCLC | Brain metastases | No molecular profiling performed | Negative |
| 30 | NSCLC | None | ALK rearrangement (lung, FISH) | Negative |
| 31 | Melanoma | Brain metastases | BRAF V600E (skin, Sequenom) | Negative |
| 32 | Melanoma | Brain metastases | BRAF V600K (lymph node, Sequenom) | Negative |
| 33 | Melanoma | None | NRAS (lung, Sequenom) | Negative |
| 34 | Thyroid | Brain metastases | NRAS TP53 (thyroid, PCR) | Negative |
| 35 | Thyroid | None | No molecular profiling performed | Negative |
| 36 | Rectal | None | No molecular profiling performed | Negative |
| 37 | Prostate | None | No mutation found (prostate, MSK-IMPACT) | Negative |
| 38 | Prostate | None | NOTCH1 R1758H (13%; prostate, MSK-IMPACT) | Negative |
| 39 | Renal | None | No molecular profiling performed | Negative |
| 40 | Renal | None | No molecular profiling performed | Negative |
| 41 | Liposarcoma | None | No molecular profiling performed | Negative |
| Primary brain tumors | ||||
| 42 | Anaplastic astrocytoma | N/A | IDH1 R132H (IHC), PIK3CA H1047R | IDH1 R132H (38%), PTEN R130* (25%) |
| 43 | Glioblastoma | N/A | No molecular profiling performed | PIK3CA V344M (6%) |
| 44 | Glioblastoma | N/A | PTEN loss (IHC) | PTEN Y336_F337 delins* (14%), EGFR AMP (log2, 3.4) |
| 45 | Anaplastic oligodendroglioma | N/A | IDH1 R132H (IHC),1p/19q del (FISH) | IDH1 R132H (44%), 1p/19q del (log2, −0.8) |
| 46 | Glioblastoma | N/A | PTEN loss, CDK4 AMP, CLI1 AMP, TP53, TERT, SPTA1 (FM) | CDK4 AMP (log2, 2.4) |
| 47 | Brainstem glioma | N/A | No molecular profiling performed | PDGFRA AMP (log2, 2.0), CDKN2B del (log2, −3.0) |
| 48 | Glioblastoma | N/A | No molecular profiling performed | Negative |
| 49 | High-grade glioma | N/A | No molecular profiling performed | Negative |
| 50 | Oligodendroglioma | N/A | No molecular profiling performed | Negative |
| 51 | Anaplastic ependymoma | N/A | CDKN2A Y44* (8%; MSK-IMPACT) | Negative |
| 52 | Anaplastic oligodendroglioma | N/A | 1p/19q del (FISH) | Negative |
| 53 | Anaplastic oligodendroglioma | N/A | IDH1 R132H (IHC), 1p/19q del (FISH) | Negative |
Abbreviations: ALK, anaplastic lymphoma kinase; AMP, amplification; CNS, central nervous system; CSF, cerebrospinal fluid; del, deletion; delins, deletion/insertion; EGFR, epidermal growth factor receptor; ER, estrogen receptor; FISH, fluorescent in situ hybridization; FM, Foundation Medicine; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry; ins, insertion; MSK-IMPACT, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; N/A, not applicable; ND, not determined; NSCLC, non–small-cell lung cancer; PCR, polymerase chain reaction; PR, progesterone receptor; SCLC, small-cell lung cancer.
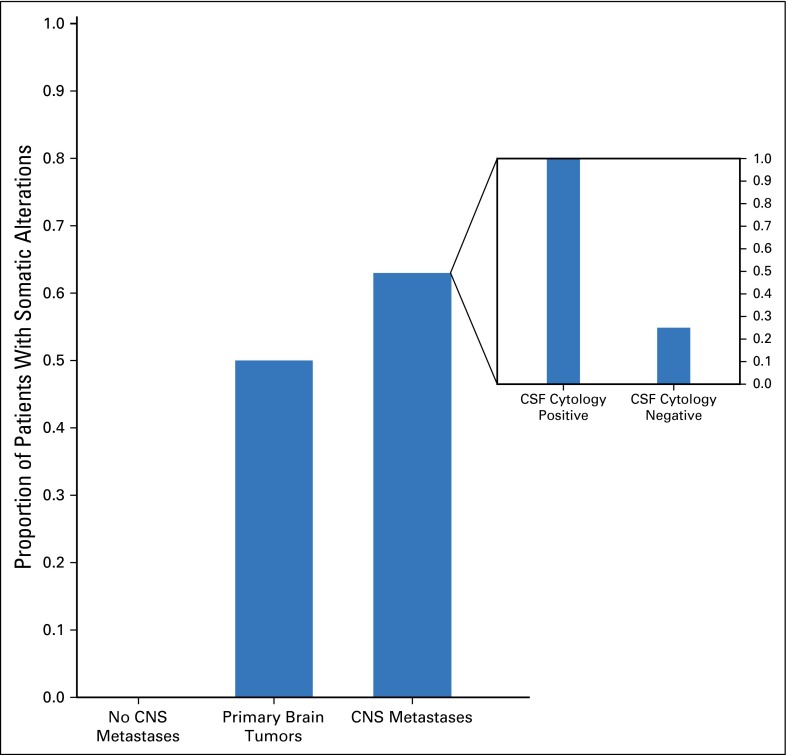
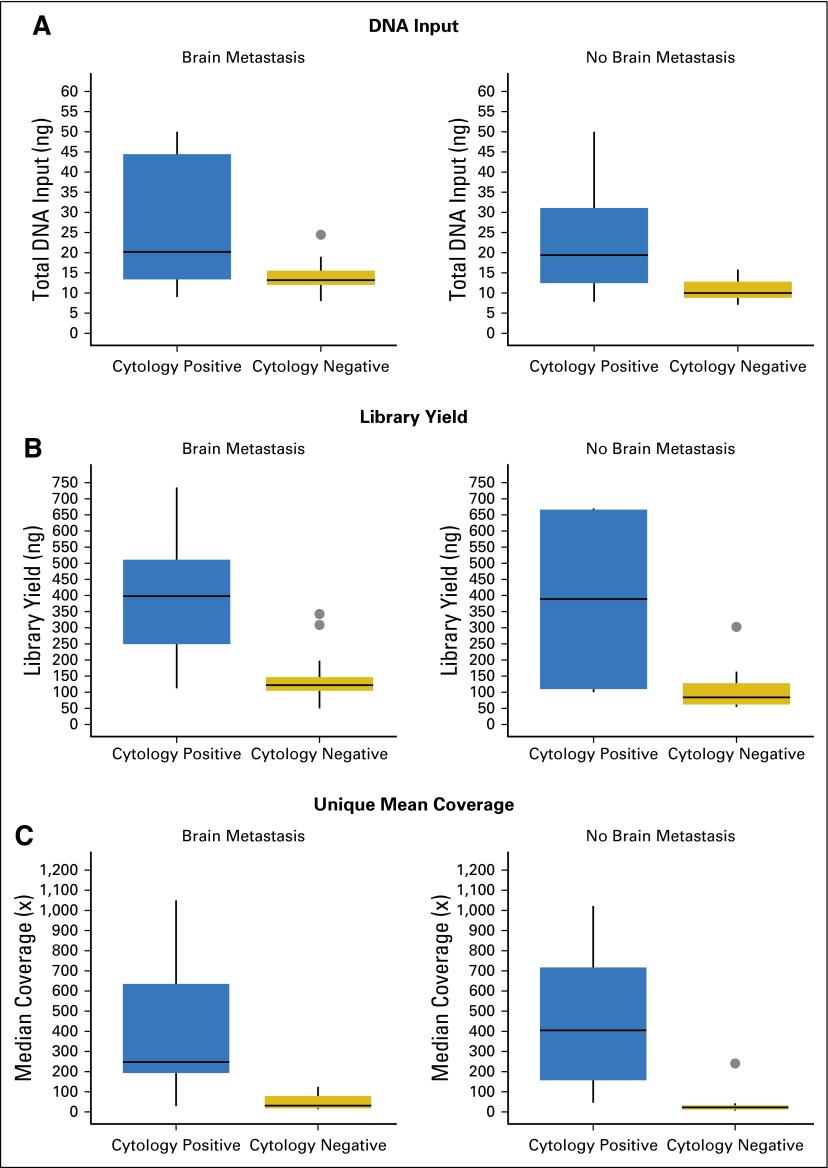
CSF from patients with brain metastases and positive CSF cytology (n = 13) showed significantly higher DNA yields, sequence library complexity, and unique sequence coverage than CSF from patients with negative CSF cytology (Appendix Fig A3, online only). Overall, we achieved ≥ 100× coverage in 12 of 16 patients with positive CSF cytology (average unique median coverage, 397×) and in four of 25 with negative CSF cytology (average unique median coverage, 175×; P < .001). The patients who did not have detectable levels of CSF cfDNA had lower overall DNA yields (13 v 90 ng) and lower unique sequence coverage (42× v 464×), which reflected the smaller quantities of tumor-derived DNA and low background levels of nontumor-derived DNA in CSF. For patients with primary brain tumors, we achieved ≥ 100× coverage in five of 12 patients (average unique median coverage, 391×). CSF from most of these patients (11 of 12) had negative cytology.
With MSK-IMPACT, we detected high-confidence somatic alterations in 63% of patients with CNS metastases and solid tumors and 50% of patients with primary brain tumors. None of the CSF samples from patients with cancer without CNS involvement (n = 9) showed tumor-derived molecular alterations. Within the group with CNS metastases, high-confidence somatic alterations were found in all 16 (100%) patients with positive CSF cytology and four of 16 (25%) with negative CSF cytology with radiographic evidence for CNS metastases (P = .1; Fig 2). No statistically significant relationship was found between the presence of tumor-associated DNA in CSF and patient age or sex, histology of the primary tumor, presence of metastases outside the CNS, prior treatment, or site of the CNS metastases (Appendix Table A2, online only).
Fig 2.
Detection of tumor-associated mutations in CSF in patients with solid tumors and primary brain tumors. Inset shows the percentage of success in finding somatic alterations in patients with central nervous system (CNS) metastasis with positive and negative cerebrospinal fluid (CSF) cytology.
Clinical Significance of Mutations in CSF cfDNA
To interpret the genetic alterations in CSF cfDNA within their clinical context, we compared them to molecular profiling results of earlier biopsy specimens taken from the same patient. For many patients, the primary tumor sample had been examined by using a range of clinically approved laboratory tests. Mutations in CSF cfDNA were called de novo on the basis of a bioinformatics pipeline without prior knowledge of the alterations seen in tissue. In all available patients in whom CSF cfDNA was detectable, the CSF cfDNA sample was concordant with the previously identified molecular alteration (Table 1; Appendix Table A3, online only).
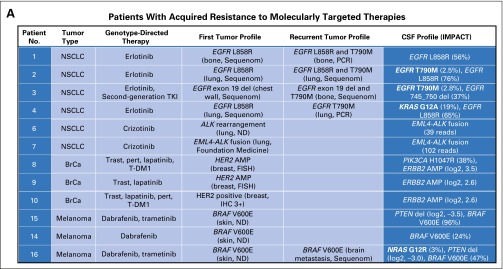
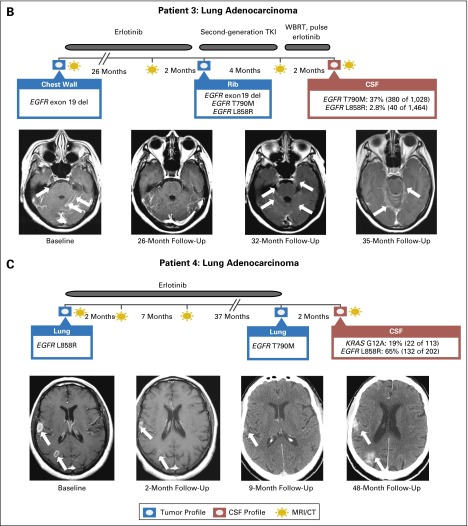
An important potential application of CSF genomic profiling is the identification of drug-resistance mechanisms in patients whose primary tumor responded to genotype-directed targeted cancer therapy but then progressed in the CNS. Of note, secondary kinase mutations can confer drug resistance at low allelic frequency and, therefore, can be difficult to detect.20 We hypothesized that the coverage of the MSK-IMPACT sequencing assay is sufficiently deep and broad to uncover a range of drug-resistance mutations in CSF cfDNA. In a group of 12 cohort patients in whom progressive CNS disease developed during treatment with inhibitors of oncogenic kinases (epidermal growth factor receptor [EGFR], ALK, human epidermal growth factor receptor 2 [HER2], or BRAF), we identified drug-resistance mutations in the CSF in four (one third). CSF cfDNA from two patients with EGFR-mutant lung cancer (patients 2 and 3 in Fig 3A) showed an EGFR T790M mutation, the most common cause of acquired resistance to first-generation EGFR tyrosine kinase inhibitors (TKIs) in NSCLC.21 Tumors obtained from both patients before EGFR TKI therapy were negative for the EGFR T790M mutation in a Sequenom-based genotyping assay, and the absence of the T790M mutation was confirmed in the pretreatment tissue by MSK-IMPACT in patient 2. In a third patient with EGFR-mutant NSCLC and acquired erlotinib resistance in the CNS, we identified an activating KRAS G12A mutation, another common cause of EGFR TKI resistance in NSCLC22,23 (Fig 3B). In a patient with BRAF V600E–mutant (and NRAS-negative) melanoma, we identified an NRAS G12R mutation in the CSF (patient 16), a genetic alteration known to promote acquired resistance to BRAF inhibition in melanoma.24-26 Both the KRAS G12A and the NRAS G12R mutations were not detected by Sequenom before therapy.
Fig 3.
Drug-resistance mutations in patients whose central nervous system (CNS) disease progresses during kinase inhibitor therapy. (A) Summary of genomic profiling results from cerebrospinal fluid (CSF) and other tumor sites in patients in whom progressive CNS disease developed during treatment with the indicated kinase inhibitors. (B) Disease timeline and brain magnet resonance images (MRIs) from a patient with EGFR-mutant NSCLC (patient 3) who presented with leptomeningeal metastasis (baseline MRI, arrows), responded to erlotinib (follow-up MRI at 26 months), was found to have a secondary EGFR mutation (T790M) in a bone metastasis, and developed progressive CNS disease (brain MRIs at 32 and 35 months) that did not respond to second-generation EGFR TKI or pulse erlotinib. CSF cell-free DNA (cfDNA) identified an EGFR T790M mutation. (C) Disease timeline and brain MRIs from a patient with EGFR-mutant NSCLC (patient 4) who presented with brain metastases (baseline MRI), responded to erlotinib (follow-up brain MRI at 2 months and brain CT scan at 9 months), and later developed progressive brain metastases. Molecular profiling of the recurrent lung tumor showed a secondary EGFR mutation (T790M), whereas CSF cfDNA identified an activating KRAS mutation (and the absence of T790M). Sequenom mass spectrometry genotyping was performed for specific mutations in eight genes: AKT1, BRAF, EGFR, ERBB2, KRAS, MEK1 (MAP2K1), NRAS, and PIK3CA. ALK, anaplastic lymphoma kinase; AMP, amplification; BrCa, breast cancer; CT, computed tomography; del, deletion; EGFR, epidermal growth factor receptor; FISH, fluorescent in situ hybridization; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry; IMPACT, Integrated Molecular Profiling of Actionable Cancer Targets; ND, not determined; NSCLC, non–small-cell lung cancer; PCR, polymerase chain reaction; pert, pertuzumab; T-DM1, trastuzumab emtansine; TKI, tyrosine kinase inhibitor; trast, trastuzumab; WBRT, whole-brain radiotherapy.
Sequencing of CSF from two additional patients identified candidate drug-resistance mutations, but the relationship to clinical drug resistance was more ambiguous. Progressive CNS disease eventually developed in one patient (patient 8) with HER2-amplified breast cancer who received multiple HER2-targeted therapies (trastuzumab, rhuMAb 2C4, and lapatinib) and who was found to harbor an activating mutation in the catalytic subunit of phosphatidylinositol 3-kinase (PI3K; PIK3CA H1047R) in CSF cfDNA, a potential cause of trastuzumab resistance in breast cancer.27,28 The allelic frequency of the PIK3CA mutation was considerably higher in the CSF sample (38%) than in the pretreatment primary tumor (4%). CSF from patient 15 with BRAF V600E–mutant melanoma showed a homozygous deletion of PTEN, but the primary tumor as not available to ascertain whether this genetic alteration occurred only in the CSF.
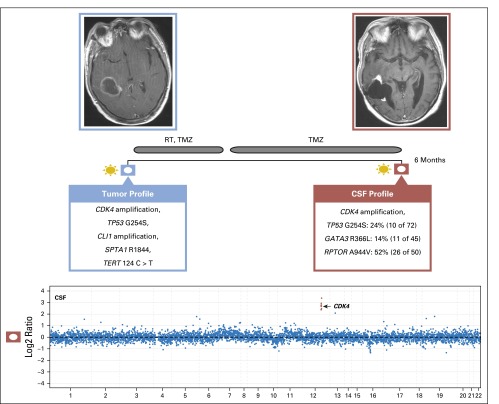
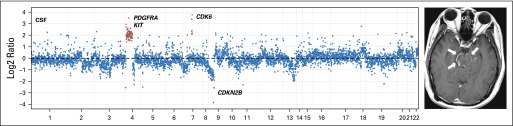
We also identified clinically relevant genomic alterations in CSF cfDNA from patients whose primary tumors could not be profiled because of limited access to tumor tissue, insufficient tumor content, or DNA quantity. For example, the CSF cfDNA from patient 18 with gastroesophageal carcinoma whose available tumor tissue was not adequate for molecular profiling (Appendix Fig A4, online only) harbored multiple somatic copy number alterations, including the loci for the receptor tyrosine kinases HER2 and fibroblast growth factor receptor 2, which are established drug targets in cancer and frequently amplified in this cancer type.29 Similarly, we identified PDGFRA amplification in a patient with a brainstem glioma30 that could not be biopsied because of the precarious tumor location (Appendix Figs A5 and A6, online only).
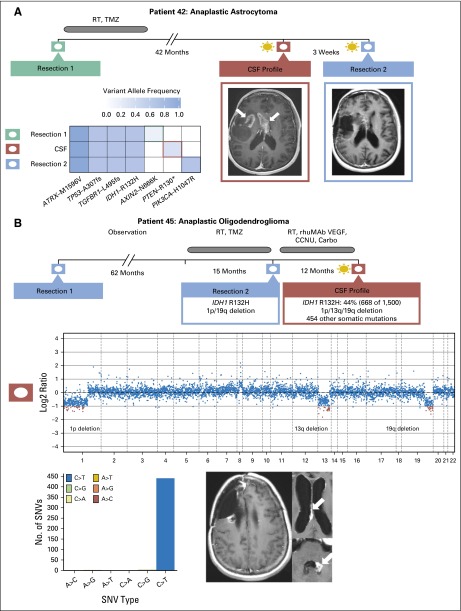
Sequencing of CSF cfDNA identified mutations in six of 12 (50%) patients with primary brain tumors despite that the CSF was negative for malignant cells in most (n = 11) of these patients. In one of these patients (Fig 4A), we were able to compare the pattern of mutations in CSF cfDNA with tumor profiling results obtained from the original tumor specimen and a second specimen obtained 3 weeks after CSF collection. Mutations in four genes (IDH1, TP53, ATRX, and TGFBR1) were detected in all three samples, but only the later samples harbored additional, but distinct mutations that activate the PI3K pathway, namely a truncating mutation in the PTEN tumor suppressor gene (CSF cfDNA) and an oncogenic hotspot mutation in the catalytic subunit of PI3K (recurrence tumor specimen). These data illustrate spatial heterogeneity within the CNS with evolutionary convergence on the PI3K signaling pathway during glioma progression. In another patient with 1p/19q codeleted oligodendroglioma31,32 (Fig 4B), CSF cDNA profiling 7.4 years after the initial diagnosis identified > 400 nonsynonymous single nucleotide variants. These mutations almost entirely represented C>T/G>A nucleotide changes, a mutation pattern that has been associated with exposure to temozolomide therapy in glioma.33
Fig 4.
Tumor evolution in patients with primary brain tumors. (A) Spatial and temporal heterogeneity among samples obtained at diagnosis, at recurrence, and from cerebrospinal fluid (CSF) in patient 42 with recurrent glioblastoma. CSF cell-free DNA harbors a PTEN R130* mutation (variant allele frequency, 0.25), whereas resection 2 harbors a PIK3CA H1047R mutation (variant allele frequency, 0.441). (B) CSF molecular profile for patient 45 with anaplastic oligodendroglioma contains the IDH1 R132H mutation and 1p/19q deletion found in tissue resection 2 as well as 454 nonsilent somatic mutations. Four hundred forty-eight SNVs represent C>T/G>A mutations that demonstrate TMZ-induced mutagenesis. Carbo, carboplatin; CCNU, lomustine; rhuMAB VEGF, bevacuzumab; RT, radiotherapy; SNV, single nucleotide variant; TMZ, temozolomide.
DISCUSSION
Cancers involving the CNS are associated with exceptional morbidity and mortality. The development of a precision medicine paradigm for these cancers is hampered by difficulty in accessing tumor tissue. The current study expands on recent technical reports9 and demonstrates the feasibility of deriving a comprehensive molecular profile from CSF collected through lumbar puncture, a procedure that often is done in the physician’s office. Key aspects of our approach included the use of CSF cfDNA, which obviated the need for additional steps to enrich for tumor cells and a clinically validated next-generation sequencing platform capable of identifying all classes of cancer-associated mutations (base substitutions, insertions, deletions, fusions, gene copy number alterations). Mutation calling and copy number analysis were performed by using an automated bioinformatics pipeline with consistent criteria and thresholds across all samples. These modifications from prior approaches help us to identify tumor-associated DNA without the need for invasive surgery, in patients with cytology-negative CSF, and without prior knowledge of molecular alterations in the primary tumor. This approach could be implemented in most health care environments that already collect and process CSF as part of routine clinical practice.
CSF represents just one of many bodily fluids from which tumor-derived cfDNA can be isolated for molecular profiling. Blood plasma has received the greatest attention in recent years for its potential to serve as a “liquid biopsy” for patients with solid tumors.34,35 Many groups have demonstrated the utility of plasma cfDNA to facilitate noninvasive mutation profiling, monitoring response to therapy, and the identification of emergent resistance mutations in patients with advanced disease.2,36-38 These studies have encompassed multiple tumor types, including breast cancer, lung cancer, and prostate cancer.37,39,40 Furthermore, plasma DNA shows promise at earlier stages of disease as a prognostic for the risk of recurrence after surgery and even as a means for early detection of cancer.41,42 Whereas early studies used targeted assays to longitudinally monitor individual prespecified mutations, such as BEAMing43 and digital droplet PCR, next-generation sequencing technologies have demonstrated the potential to reveal not only novel mutations, but also gene amplifications and fusions.44-46
One major challenge associated with plasma cfDNA sequencing is the need to detect mutations at very low allele frequencies. Tumor-derived DNA typically constitutes only a small fraction of all cfDNA in plasma due to the relatively high background of normal DNA from nonmalignant cells. Consequently, key oncogenic mutations may occur in only 1% or even 0.1% of molecules from a given genomic locus, requiring very deep sequence coverage to achieve sufficient sensitivity for detecting these mutations. This has also necessitated modifications to sequencing assays in order to maintain high specificity in the presence of sequencing errors and artifacts. Examples include Safe-SeqS47 and Duplex Sequencing,48 both of which use unique molecular identifiers to reduce errors through replicate sequence reads from the same template molecule. Importantly, we found that these approaches were not necessary for CSF cfDNA profiling. Although cfDNA yields were generally low after nucleic acid extraction from CSF, we observed that the fraction of tumor-derived cfDNA was generally high due to the relative absence of background normal DNA. As a result, we were able to readily detect somatic mutations even in cases where we only achieved modest sequence coverage (< 100×).
We found that tumor-associated DNA can be detected in CSF in a substantial number of patients with primary brain tumors, which agrees with a recent study that collected CSF intraoperatively from patients with primary brain tumors8 and suggests that collection and genomic profiling of CSF should be considered more broadly in patients with these tumors because it might provide new insights into tumor evolution and drug response. Because brain metastases and primary brain tumors are often inaccessible to surgery, we focused on an approach that did not require surgery for CSF collection. The current study also points to other scenarios where liquid biopsies and genomic profiling of CSF might guide clinical decision making, such as with leptomeningeal metastasis. This condition is notoriously difficult to diagnose with current methods and is associated with extraordinary morbidity and mortality.49,50 Similarly, CSF liquid biopsies could be informative in patients with multiple brain metastases, which are rarely biopsied but can harbor mutations not observed in the primary tumor.51
Tumor growth in the CNS despite systemic disease control remains a major clinical challenge. It is often unclear whether the CNS represents a sanctuary site, shielding malignant cells from target therapies, or whether select mutations render malignant cells more likely to successfully colonize the CNS. Our data suggest caution when using systemic genotyping results to predict the clinical response of CNS disease to targeted therapies. For extracranial sites, on-treatment biopsy specimens have shown several resistance mechanisms, including mutations that can restore signaling downstream of the target kinase, activate an alternative signaling pathway, or impair drug binding to the target kinase.52 The molecular basis of kinase inhibitor resistance in the CNS is unknown and widely attributed to inadequate drug penetration into the CNS.28-33,49-58 The current data suggest a more nuanced view of this common clinical problem because we identified a genetic explanation for drug resistance in at least four of 12 patients (33%). The current findings may explain the clinical experience that only a fraction of patients with acquired kinase inhibitor resistance in the CNS respond to an increase in the drug dose or intrathecal drug administration designed to overcome reduced penetration of drug into the CNS.59-62 They are also consistent with the recent identification of mutations in surgically resected brain tumor metastases that were not found in the primary tumors of the same patients.51
Our findings suggest that CSF sequencing could substantially increase the number of patients who are eligible for genotype-directed cancer therapy, including patients whose primary tumor could not be successfully sequenced and patients who are poor candidates for neurological surgery due to medical comorbidities or tumor location in neurologically critical areas (eg, brainstem). Our study also suggests that cfDNA analysis in the CSF could be a suitable biomarker to monitor clinical response to therapy, analogous to plasma cfDNA, an area that will warrant further prospective evaluation. Together with the cited studies, this study demonstrates that genomic analysis of CSF by using a sufficiently sensitive and comprehensive platform may be useful in facilitating the diagnosis of tumor in the CNS, monitor the evolution of the cancer genome during treatment of CNS cancers, guide the choice of second-line agents, and perhaps identify pathways that are uniquely associated with cancer spread to the CNS.
Supplementary Material
Acknowledgment
We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology at Memorial Sloan Kettering (MSK). We thank all the members of the Mellinghoff and Berger Laboratories and the MSK Department of Neurology for insightful suggestions. We also thank Mark Kris, Paul Chapman, M. Catherine Pietanza, Paul Paik, Shanu Modi, Maria Theodoulou, Michael Postow, Marjorie Zauderer, and Stuart Lichtman for helpful suggestions in the course of this work. Finally, we thank Judith Lampron for editorial assistance and Wenjing Wu for illustration assistance.
Appendix
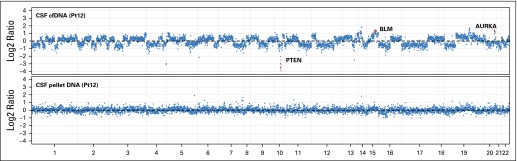
Fig A1.
Copy number plot for patient 12. cfDNA, cell-free DNA; CSF, cerebrospinal fluid; Pt, patient ID.
Fig A2.
Detection of EML4-ALK gene fusion in cerebrospinal fluid (CSF) cell-free DNA (cfDNA) and pellet DNA for patient 7. Pt, patient ID.
Fig A3.
(A) DNA input, (B) library yield, and (C) unique mean sequence coverage.
Fig A4.
Patient 18: the copy number plot (left panel) showing multiple somatic copy number alterations, including the loci for the receptor tyrosine kinases HER2 and FGFR2 and a brain MRI (right panel) with arrows pointing toward leptomeningeal metastases. CSF, cerebrospinal fluid; FGFR2, fibroblast growth factor receptor 2; HER2, human epidermal growth factor receptor 2.
Fig A5.
CSF cell-free DNA profiling of glioblastoma for patient 46. CSF, cerebrospinal fluid; RT, radiotherapy; TMZ, temozolomide.
Fig A6.
Patient 47: the copy number plot (left panel) showing amplification of the PDGFRA gene locus and a brain MRI (right panel) with arrows pointing toward the enhancing tumor in the brainstem.
Table A1.
Comparison Between cfDNA and Cell Pellet DNA From CSF to Identify Specific Genetic Alterations
| Primary Tumor | CSF cfDNA | CSF Cell Pellet | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient No. | Histology | Known Genomic Alteration | Total DNA Input (ng) | Median Sample Coverage (x) | Evidence | Detected | Total DNA Input (ng) | Median Sample Coverage (x) | Evidence | Detected |
| Pt05 | Lung adenocarcinoma | KRAS G12C | 105 | 1,050 | 96% of reads (1,111 of 1,154) | Yes | 93 | 1,252 | 30% of reads (349 of 1,165) | Yes |
| Pt01 | Lung adenocarcinoma | EGFR L858R | 13 | 247 | 56% of reads (215 of 384) | Yes | 33 | 427 | 10% of reads (45 of 439) | Yes |
| Pt07 | Lung adenocarcinoma | EML4-ALK fusion | 90 | 615 | 102 reads | Yes | 1,328 | 980 | 102 reads | Yes |
| Pt06 | Lung adenocarcinoma | ALK rearrangement | 14.5 | 177 | 39 reads | Yes | 86 | 784 | 14 reads | Yes |
| Pt08 | Breast cancer | HER2 positive | 21.5 | 430 | AMP (average log2 ratio, 3.51) | Yes | 96 | 846 | Non-AMP (average log2 ratio, 0.34) | No |
| Pt12 | Breast cancer | PTEN negative | 50 | 800 | Del (average log2 ratio, −2.97) | Yes | 5 | 78 | Non-del (average log2 ratio, −0.05) | No |
| Pt14 | Melanoma | BRAF V600E | 9 | 38 | 24% of reads (15 of 63) | Yes | 74 | 646 | 0% of reads (2 of 652) | No |
| Pt15 | Melanoma | BRAF V600E | 7.8 | 194 | 96% of reads (364 of 381) | Yes | 290 | 964 | 89% of reads (1,222 of 1,375) | Yes |
NOTE. All eight patients with cancer had clinically documented leptomeningeal metastasis from the primary tumor shown in column 2. For each patient, mutation detection focused on a specific alteration previously identified in the primary tumor tissue (column 3). Shown is the supporting evidence for the known genomic alteration in CSF. Alterations were detected in all eight CSF cfDNA samples and five CSF pellet DNA samples.
Abbreviations: ALK, anaplastic lymphoma kinase; AMP, amplification; cfDNA, cell-free DNA; del, deletion; EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor receptor 2.
Table A2.
Variables Included in Statistical Analysis
| CSF Tumor-Derived cfDNA Cohort (n = 20), No. (%) | Absent Tumor-Derived CSF cfDNA Cohort (n = 21), No. (%) | P | |
|---|---|---|---|
| Age, mean (SD) | 55.0 (11.5) | 55.4 (11.6) | .92 |
| Sex | |||
| Male | 9 (45) | 8 (38) | .65 |
| Female | 11 (55) | 13 (62) | |
| Tumor histology | |||
| Breast | 6 (30) | 5 (24) | .42 |
| Lung | 7 (35) | 4 (19) | |
| Melanoma | 3 (15) | 3 (14) | |
| Others | 4 (20) | 9 (43) | |
| Anatomic location | |||
| Brain metastasis | |||
| Supratentorial | 7 (35) | 7 (33) | .06 |
| Infratentorial | 3 (15) | 0 (0) | |
| Supra- and infratentorial | 7 (35) | 4 (19) | |
| No brain metastasis | 3 (15) | 10 (48) | |
| Distant metastasis | |||
| Yes | 19 (95) | 16 (76) | .09 |
| No | 1 (5) | 5 (24) | |
| Prior CNS surgery | |||
| Yes | 8 (40) | 3 (14) | .06 |
| No | 12 (60) | 18 (86) | |
| Chemotherapy at the time of CSF collection | |||
| Yes | 17 (85) | 14 (67) | .17 |
| No | 3 (15) | 7 (33) | |
| Radiation therapy to CNS before CSF collection | |||
| Yes | 12 (60) | 9 (43) | .27 |
| No | 8 (40) | 12 (57) |
Table A3.
Characteristics of 53 Patients, Including CSF Cytology and Primary Tissue Diagnosis, for Which CSF cfDNA Was Extracted and Analyzed
| Patient No. | Age | Sex | Tumor Type | CSF Cytology | CNS Involvement | Other Mets | Original Specimen (Molecular Path) | CSF (MSK-IMPACT) |
|---|---|---|---|---|---|---|---|---|
| Solid tumors | ||||||||
| 1 | 40 | M | Lung adenocarcinoma | Negative | BM | Yes | EGFR L858R (bone, Sequenom) | EGFR L858R (56%) |
| 2 | 52 | M | Lung adenocarcinoma | Positive | BM | No | EGFR L858R (lung, Sequenom) | EGFR T790M (2.5%), EGFR L858R (76%) |
| 3 | 51 | F | Lung adenocarcinoma | Positive | BM | Yes | EGFR exon 19 del (chest wall, Sequenom) | EGFR T790M (2.8%), EGFR 745_750 del (37%) |
| 4 | 75 | M | Lung adenocarcinoma | Negative | BM | Yes | EGFR L858R (lung, Sequenom) | KRAS G12A (19%), EGFR L858R (65%) |
| 5 | 71 | F | Lung adenocarcinoma | Positive | BM | Yes | KRAS G12C (c34 G>T) (lung, Sequenom) | KRAS G12C (96%), CDKN2B del (log2, −2.9) |
| 6 | 30 | M | Lung adenocarcinoma | Positive | BM | Yes | ALK rearrangement (lung, ND) | EML4-ALK fusion (39 reads) |
| 7 | 73 | F | Lung adenocarcinoma | Positive | LM | Yes | EML4-ALK fusion (lung, FM) | EML4-ALK fusion (102 reads) |
| 8 | 38 | F | Breast cancer | Positive | BM | Yes | HER2 AMP (breast, FISH) | PIK3CA H1047R (38%), HER2 AMP (log2, 3.5) |
| 9 | 56 | F | Breast cancer | Positive | BM | Yes | HER2 AMP (breast, FISH) | HER2 AMP (log2, 2.6) |
| 10 | 53 | F | Breast cancer | Positive | BM | Yes | HER2 positive (breast, IHC 3+) | HER2 AMP (log2, 2.6) |
| 11 | 59 | F | Breast cancer | Positive | BM | Yes | No molecular profiling performed | EGFR AMP (log2, 3.1), PIK3CA H1047R (28%) |
| 12 | 60 | F | Breast cancer | Positive | BM | Yes | TP53 V272M (56%), PTEN del (log2, −2.0; lymph node, MSK-IMPACT) | TP53 V272M (81%), PTEN del (log2, −2.97) |
| 13 | 59 | F | Breast cancer | Positive | BM | Yes | ER positive, PR/HER2 negative; thyroid metastasis, IHC) | PIK3CA E545K (26%) |
| 14 | 45 | M | Melanoma | Positive | BM | Yes | BRAF V600E (skin, ND) | BRAF V600E (24%) |
| 15 | 68 | M | Melanoma | Positive | LM | Yes | BRAF V600E (skin, ND) | PTEN del (log2, −XYZ), BRAF V600E (96%) |
| 16 | 57 | F | Melanoma | Positive | BM | Yes | BRAF V600E (skin, ND) | NRAS G12R (3%), PTEN del (log2, −3.0), BRAF V600E (47%) |
| 17 | 55 | M | Bladder cancer | Positive | BM | Yes | No molecular profiling performed | AKT2 AMP (log2, 3.37), TP53 R158L (43%) |
| 18 | 50 | M | Gastroesophageal | Positive | LM | Yes | No molecular profile (MSK-IMPACT failure) | HER2 AMP (log2, 2.4), FGFR2 (log2 3.6) |
| 19 | 54 | M | Neuroendocrine, unknown primary | Negative | BM | Yes | No molecular profiling performed | MYCN AMP (log2, 4.1) |
| 20 | 55 | F | Ovarian cancer | Negative | BM | Yes | BRCA1 insC (blood, Myriad Genetics laboratory) | BRCA1 Q1756fs (53%), CDKN2B del (log2, −2.1) |
| 21 | 70 | F | Ovarian cancer | Negative | LM | Yes | No molecular profiling performed | Negative |
| 22 | 36 | F | Breast cancer | Negative | BM | Yes | No molecular profiling performed | Negative |
| 23 | 50 | F | Breast cancer | Negative | BM | Yes | HER2 AMP (breast, FISH) | Negative |
| 24 | 49 | F | Breast cancer | Negative | BM | Yes | ESR1 Y537S (62%), CCND1 AMP (log2, 1.5; breast, MSK-IMPACT) | Negative |
| 25 | 32 | F | Breast cancer | Negative | BM | Yes | RB1 L343Sfs*3 (liver, MSK-IMPACT) | Negative |
| 26 | 40 | F | Breast cancer | Negative | BM | Yes | PIK3CA R108 del (39%), CCND1 AMP (log2, 1.0; soft tissue, MSK-IMPACT) | Negative |
| 27 | 63 | F | Lung adenocarcinoma | Negative | BM | Yes | No molecular profiling performed | Negative |
| 28 | 66 | M | Lung adenocarcinoma | Negative | BM | Yes | No molecular profiling performed | Negative |
| 29 | 66 | F | Small-cell lung cancer | Negative | BM | Yes | No molecular profiling performed | Negative |
| 30 | 58 | F | Lung adenocarcinoma | Negative | Negative | Yes | ALK rearrangement (lung, FISH) | Negative |
| 31 | 56 | M | Melanoma | Negative | BM | Yes | BRAF V600E (skin, Sequenom) | Negative |
| 32 | 54 | M | Melanoma | Negative | BM | Yes | BRAF V600K (lymph node, Sequenom) | Negative |
| 33 | 37 | F | Melanoma | Negative | Negative | Yes | NRAS (lung, Sequenom) | Negative |
| 34 | 55 | F | Thyroid cancer | Negative | BM | Yes | NRAS TP53 (thyroid, PCR) | Negative |
| 35 | 54 | F | Thyroid cancer | Negative | Negative | No | No molecular profiling performed | Negative |
| 36 | 76 | M | Rectal cancer | Negative | Negative | Yes | No molecular profiling performed | Negative |
| 37 | 58 | M | Prostate cancer | Negative | Negative | No | No mutation found (prostate, MSK-IMPACT) | Negative |
| 38 | 67 | M | Prostate cancer | Negative | Negative | Yes | NOTCH1 R1758H (13%; prostate, MSK-IMPACT) | Negative |
| 39 | 64 | M | Renal cell carcinoma | Negative | Negative | No | No molecular profiling performed | Negative |
| 40 | 58 | M | Renal cell carcinoma | Negative | Negative | No | No molecular profiling performed | Negative |
| 41 | 57 | F | Liposarcoma | Negative | Negative | No | No molecular profiling performed | Negative |
| Primary brain tumors | ||||||||
| 42 | 24 | M | Anaplastic astrocytoma | Negative | N/A | No | IDH1 R132H (IHC), PIK3CA H1047R | IDH1 R132H (38%), PTEN R130* (25%) |
| 43 | 65 | M | Glioblastoma | Negative | N/A | No | No molecular profiling performed | PIK3CA V344M (6%) |
| 44 | 63 | M | Glioblastoma | Negative | N/A | No | PTEN loss (IHC) | PTEN Y336_F337 delins* (14%), EGFR AMP (log2, 3.4) |
| 45 | 39 | F | Anaplastic oligodendroglioma | Negative | N/A | No | IDH1 R132H (IHC), 1p/19q del (FISH) | IDH1 R132H (44%), 1p/19q del (log2, −0.8) |
| 46 | 66 | M | Glioblastoma | Negative | N/A | No | PTEN loss, CDK4 AMP, CLI1 AMP, TP53, TERT, SPTA1 (FM) | CDK4 AMP (log2, 2.4) |
| 47 | 29 | M | Brainstem glioma | Positive | N/A | No | No molecular profiling performed | PDGFRA AMP (log2, 2.0), CDKN2B del (log2, −3.0) |
| 48 | 78 | M | Glioblastoma | Negative | N/A | No | No molecular profiling performed | Negative |
| 49 | 58 | F | High-grade glioma | Negative | N/A | No | No molecular profiling performed | Negative |
| 50 | 38 | M | Oligodendroglioma | Negative | N/A | No | No molecular profiling performed | Negative |
| 51 | 35 | F | Anaplastic ependymoma | Negative | N/A | No | CDKN2A Y44* (8%; MSK-IMPACT) | Negative |
| 52 | 71 | M | Anaplastic oligodendroglioma | Negative | N/A | No | 1p/19q del (FISH) | Negative |
| 53 | 65 | M | Anaplastic oligodendroglioma | Negative | N/A | No | IDH1 R132H (IHC), p19q del (FISH) | Negative |
Abbreviations: AMP, amplification; BM, brain metastasis; del, deletion; delins, deletion/insertion; EGFR, epidermal growth factor receptor; FISH, fluorescent in situ hybridization; FM, Foundation Medicine; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry; LM, leptomeningeal metastasis; Mets, metastases; MSK-IMPACT, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; N/A, not applicable; ND, not determined; PCR, polymerase chain reaction.
Footnotes
Supported by grants from the National Institutes of Health (1R01-NS080944-01, P30-CA008748), the Memorial Sloan Kettering Brain Tumor Center, the Defeat GBM Initiative of the National Brain Tumor Society, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Elena I. Pentsova, Ronak H. Shah, Ingo K. Mellinghoff, Michael F. Berger
Financial support: Lisa M. DeAngelis, Ingo K. Mellinghoff, Michael F. Berger
Administrative support: S. Duygu Selcuklu, Lisa M. DeAngelis, Ingo K. Mellinghoff, Michael F. Berger
Provision of study materials: Antonio Omuro
Collection and assembly of data: Elena I. Pentsova, Ronak H. Shah, Jiabin Tang, Adrienne Boire, Daoqi You, Samuel Briggs, Antonio Omuro, Xuling Lin, Martin Fleisher, Christian Grommes, Katherine S. Panageas, Fanli Meng, S. Duygu Selcuklu, Shahiba Ogilvie, Natalie Distefano, Larisa Shagabayeva, Marc Rosenblum, Agnes Viale, Ingo K. Mellinghoff, Michael F. Berger
Data analysis and interpretation: Elena I. Pentsova, Ronak H. Shah, Lisa M. DeAngelis, Ingo K. Mellinghoff, Michael F. Berger
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Evaluation of Cancer of the CNS Through Next-Generation Sequencing of CSF
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Elena I. Pentsova
No relationship to disclose
Ronak H. Shah
No relationship to disclose
Jiabin Tang
No relationship to disclose
Adrienne Boire
Patents, Royalties, Other Intellectual Property: Coinventor on two patents pending
Daoqi You
No relationship to disclose
Samuel Briggs
No relationship to disclose
Antonio Omuro
Consulting or Advisory Role: Stemline Therapeutics, Juno Therapeutics, Bristol-Myers Squibb, OXiGENE
Xuling Lin
No relationship to disclose
Martin Fleisher
No relationship to disclose
Christian Grommes
No relationship to disclose
Katherine S. Panageas
No relationship to disclose
Fanli Meng
No relationship to disclose
S. Duygu Selcuklu
No relationship to disclose
Shahiba Ogilvie
No relationship to disclose
Natalie Distefano
No relationship to disclose
Larisa Shagabayeva
No relationship to disclose
Marc Rosenblum
No relationship to disclose
Lisa M. DeAngelis
Consulting or Advisory Role: Juno Therapeutics
Agnes Viale
No relationship to disclose
Ingo K. Mellinghoff
No relationship to disclose
Michael F. Berger
Consulting or Advisory Role: Cancer Genetics, Sequenom
REFERENCES
- 1.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes CH, Honsinger C, Sorenson GD. PCR-detection of tumor-derived p53 DNA in cerebrospinal fluid. Am J Clin Pathol. 1995;103:404–408. doi: 10.1093/ajcp/103.4.404. [DOI] [PubMed] [Google Scholar]
- 4.Swinkels DW, de Kok JB, Hanselaar A, et al. Early detection of leptomeningeal metastasis by PCR examination of tumor-derived K-ras DNA in cerebrospinal fluid. Clin Chem. 2000;46:132–133. [PubMed] [Google Scholar]
- 5.Shingyoji M, Kageyama H, Sakaida T, et al. Detection of epithelial growth factor receptor mutations in cerebrospinal fluid from patients with lung adenocarcinoma suspected of neoplastic meningitis. J Thorac Oncol. 2011;6:1215–1220. doi: 10.1097/JTO.0b013e318219aaae. [DOI] [PubMed] [Google Scholar]
- 6.Chen WW, Balaj L, Liau LM, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan W, Gu W, Nagpal S, et al. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Springer S, Zhang M, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Mattos-Arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mose LE, Wilkerson MD, Hayes DN, et al. ABRA: Improved coding indel detection via assembly-based realignment. Bioinformatics. 2014;30:2813–2815. doi: 10.1093/bioinformatics/btu376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye K, Schulz MH, Long Q, et al. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Exome Aggregation Consortium: About ExAC. http://exac.broadinstitute.org.
- 18.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch T, Zichner T, Schlattl A, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayoola A, Barochia A, Belani K, et al. Primary and acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer: An update. Cancer Invest. 2012;30:433–446. doi: 10.3109/07357907.2012.666691. [DOI] [PubMed] [Google Scholar]
- 23.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 24.Paraiso KH, Xiang Y, Rebecca VW, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niessner H, Forschner A, Klumpp B, et al. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. Cancer Med. 2013;2:76–85. doi: 10.1002/cam4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Janiszewska M, Liu L, Almendro V, et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet. 2015;47:1212–1219. doi: 10.1038/ng.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarghooni M, Bartels U, Lee E, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28:1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 31.Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz LA, Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 36.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:827. doi: 10.1038/nm0715-827b. [DOI] [PubMed] [Google Scholar]
- 37.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 38.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 39.Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 42.Spellman PT, Gray JW. Detecting cancer by monitoring circulating tumor DNA. Nat Med. 2014;20:474–5. doi: 10.1038/nm.3564. [DOI] [PubMed] [Google Scholar]
- 43.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohan S, Heitzer E, Ulz P, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014;10:e1004271. doi: 10.1371/journal.pgen.1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azad AA, Volik SV, Wyatt AW, et al. Androgen Receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015;21:2315–2324. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- 46.Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22:915–922. doi: 10.1158/1078-0432.CCR-15-1627-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy SR, Schmitt MW, Fox EJ, et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat Protoc. 2014;9:2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke JL, Perez HR, Jacks LM, et al. Leptomeningeal metastases in the MRI era. Neurology. 2010;74:1449–1454. doi: 10.1212/WNL.0b013e3181dc1a69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: Current methods and future directions. Fluids Barriers CNS. 2011;8:14. doi: 10.1186/2045-8118-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawyers CL. Shifting paradigms: The seeds of oncogene addiction. Nat Med. 2009;15:1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 53.Heon S, Yeap BY, Britt GJ, et al. Development of central nervous system metastases in patients with advanced non-small cell lung cancer and somatic EGFR mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2010;16:5873–5882. doi: 10.1158/1078-0432.CCR-10-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toyokawa G, Seto T, Takenoyama M, et al. Insights into brain metastasis in patients with ALK+ lung cancer: Is the brain truly a sanctuary. Cancer Metastasis Rev. 2015;34:797–805. doi: 10.1007/s10555-015-9592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teplinsky E, Esteva FJ. Systemic therapy for HER2-positive central nervous system disease: Where we are and where do we go from here. Curr Oncol Rep. 2015;17:46. doi: 10.1007/s11912-015-0471-z. [DOI] [PubMed] [Google Scholar]
- 56.Jackman DM, Holmes AJ, Lindeman N, et al. Response and resistance in a non-small-cell lung cancer patient with an epidermal growth factor receptor mutation and leptomeningeal metastases treated with high-dose gefitinib. J Clin Oncol. 2006;24:4517–4520. doi: 10.1200/JCO.2006.06.6126. [DOI] [PubMed] [Google Scholar]
- 57.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 58.Dhruva N, Socinski MA. Carcinomatous meningitis in non-small-cell lung cancer: Response to high-dose erlotinib. J Clin Oncol. 2009;27:e31–e32. doi: 10.1200/JCO.2008.21.0963. [DOI] [PubMed] [Google Scholar]
- 59.Katayama T, Shimizu J, Suda K, et al. Efficacy of erlotinib for brain and leptomeningeal metastases in patients with lung adenocarcinoma who showed initial good response to gefitinib. J Thorac Oncol. 2009;4:1415–1419. doi: 10.1097/JTO.0b013e3181b62572. [DOI] [PubMed] [Google Scholar]
- 60.Kawamura T, Hata A, Takeshita J, et al. High-dose erlotinib for refractory leptomeningeal metastases after failure of standard-dose EGFR-TKIs. Cancer Chemother Pharmacol. 2015;75:1261–1266. doi: 10.1007/s00280-015-2759-y. [DOI] [PubMed] [Google Scholar]
- 61.Grommes C, Oxnard GR, Kris MG, et al. “Pulsatile” high-dose weekly erlotinib for CNS metastases from EGFR mutant non-small cell lung cancer. Neuro-oncol. 2011;13:1364–1369. doi: 10.1093/neuonc/nor121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zagouri F, Sergentanis TN, Bartsch R, et al. Intrathecal administration of trastuzumab for the treatment of meningeal carcinomatosis in HER2-positive metastatic breast cancer: A systematic review and pooled analysis. Breast Cancer Res Treat. 2013;139:13–22. doi: 10.1007/s10549-013-2525-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.