Summary
Armadillo‐related proteins regulate development throughout eukaryotic kingdoms. In the flowering plant Arabidopsis thaliana, Armadillo‐related ARABIDILLO proteins promote multicellular root branching. ARABIDILLO homologues exist throughout land plants, including early‐diverging species lacking true roots, suggesting that early‐evolving ARABIDILLOs had additional biological roles.
Here we investigated, using molecular genetics, the conservation and diversification of ARABIDILLO protein function in plants separated by c. 450 million years of evolution.
We demonstrate that ARABIDILLO homologues in the moss Physcomitrella patens regulate a previously undiscovered inhibitory effect of abscisic acid (ABA) on spore germination. Furthermore, we show that A. thaliana ARABIDILLOs function similarly during seed germination. Early‐diverging ARABIDILLO homologues from both P. patens and the lycophyte Selaginella moellendorffii can substitute for ARABIDILLO function during A. thaliana root development and seed germination.
We conclude that (1) ABA was co‐opted early in plant evolution to regulate functionally analogous processes in spore‐ and seed‐producing plants and (2) plant ARABIDILLO germination functions were co‐opted early into both gametophyte and sporophyte, with a specific rooting function evolving later in the land plant lineage.
Keywords: abscisic acid (ABA), Arabidopsis thaliana, Armadillo proteins, evolution, germination, moss, seed, spore
Introduction
Plant lifecycles undergo an alternation of generations between a haploid gametophyte and a diploid sporophyte phase (Hofmeister, 1851). In the earliest‐diverging land plants, the bryophytes, the gametophyte generation is dominant in the lifecycle, with the sporophyte being a relatively transient structure that remains attached to and largely dependent on the gametophyte (Glime, 2013). By contrast, in extant vascular plants, the sporophyte has assumed the dominant role, the most extreme example being in seed plants (spermatophytes), where the gametophyte is reduced to a few cells that are fully surrounded by sporophyte tissues (Evert & Eichhorn, 2012). Despite the different origins of the plant body in bryophytes and flowering plants, both lineages possess rooting and shooting structures, as well as dispersal structures (spores or seeds) that allow transition from one generation to the next and enable species propagation and distribution (Pires & Dolan, 2012).
The evolution of rooting systems was a key innovation enabling plants to be sessile, allowing absorption of nutrients and water, anchorage of the plant to its substrate, and responses to internal and external signals. Bryophytes possess simple hair‐like rooting structures called rhizoids (Jones & Dolan, 2012). Work in model bryophytes, the moss Physcomitrella patens (Prigge & Bezanilla, 2010) and liverwort Marchantia polymorpha (Shimamura, 2015), has demonstrated that rhizoid development has mechanistic similarity with the development of epidermal root hairs on the multicellular roots of the flowering plant Arabidopsis thaliana (Menand et al., 2007; Proust et al., 2016). This suggests that nonhomologous, but functionally similar, epidermal structures (i.e. tip‐growing cells with a rooting function) are regulated by genes that were co‐opted into both gametophyte and sporophyte (Menand et al., 2007; Proust et al., 2016). However, it is likely that a ‘rewiring’ of the root hair/rhizoid gene regulatory network occurred between the bryophyte gametophyte and the flowering plant sporophyte (Yi et al., 2010; Jang & Dolan, 2011; Jang et al., 2011; Pires et al., 2013; Tam et al., 2015).
Multicellular ‘true’ roots first evolved in vascular plants, independently in lycophytes and the fern/spermatophyte lineages (Banks, 2009; Jones & Dolan, 2012; Pires & Dolan, 2012). The most complex branched rooting systems, consisting of a primary root formed in the embryo, from which lateral roots (LRs) later arise, are present in ferns and in seed plants (Jones & Dolan, 2012; Pires & Dolan, 2012). Auxin plays a key role in promoting all stages of LR development (Peret et al., 2009; Lavenus et al., 2013), while many other plant hormones regulate various aspects of root branching (Osmont et al., 2007; Nibau et al., 2008; Fukaki & Tasaka, 2009). Moreover, ‘intrinsic’ regulators of LR development, which are not influenced by hormones, also exist (Malamy & Benfey, 1997; Nibau et al., 2008).
One example of an ‘intrinsic’ regulator of root branching in A. thaliana is the ARABIDILLO protein family, which shares structural similarity with the key animal Armadillo/β‐catenin developmental regulators (Coates, 2003). We have previously demonstrated a role for A. thaliana ARABIDILLO proteins in promoting LR formation (Coates et al., 2006). ARABIDILLO proteins are unstable, being turned over by the proteasome (Nibau et al., 2011), and their sequences are highly conserved across land plants and charophyte algae, including species that lack LRs, namely P. patens, Selaginella moellendorffii and Klebsormidium flaccidum (Nibau et al., 2011; Moody et al., 2012; Hori et al., 2014). By contrast, homologues of the ARABIDILLO‐interacting transcription factor AtMYB93, which is part of a negative feedback loop that inhibits LR development, are not found outside flowering plants (Gibbs & Coates, 2014; Gibbs et al., 2014). These data suggest that ARABIDILLO proteins had additional, early‐evolving functions in plants.
Here, we addressed this possibility by examining the function of ARABIDILLO homologues (PHYSCODILLO genes) in P. patens. Physcomitrella patens has three PHYSCODILLO genes that probably have redundant functions (Moody et al., 2012). We define novel functions for PHYSCODILLO proteins in regulating spore germination in response to abscisic acid (ABA), an ancient hormone found across eukaryotes (Hanada et al., 2011; Takezawa et al., 2011). Furthermore, we show that A. thaliana ARABIDILLO proteins perform the analogous function in seeds. Our data suggest that ARABIDILLO homologues were co‐opted into both the sporophyte and gametophyte very early in land plant evolution to regulate germination processes via a network involving ABA, and that early‐diverging ARABIDILLO homologues already had the potential to regulate multicellular root development, a later‐evolving function of this protein family requiring interaction with flowering plant‐specific proteins.
Materials and Methods
Moss growth and culture
Physcomitrella patens (Hedw.) B.S.G. ssp patens, ecotype ‘Gransden 2004’ was obtained from Dr Andrew Cuming (University of Leeds, Leeds, UK). The physcodillo2 deletion mutant has been described previously (Moody et al., 2012). Protonemata, gametophores and germinating spores were cultured as in Moody et al. (2012). The spore germination medium was additionally supplemented with 10 mM CaCl2 and 5 mM ammonium tartrate. The physcodillo2 deletion mutant was maintained on BCD medium additionally supplemented with 20 μg ml−l hygromycin B. The physcodillo1a/1b/2 triple deletion mutant was maintained on BCD medium additionally supplemented with 20 μg ml−l hygromycin B and 50 μg ml−l G418.
Arabidopsis thaliana growth and culture
For in vitro culture, Arabidopsis thaliana (L.) Heynh. seeds were sterilized in 20% Parozone bleach (Jeyes, Thetford, UK) for 15 min and then washed three times in sterile water. Seedlings were grown in long‐day conditions on 0.5× Murashige and Skoog (MS) medium, 1% agar, pH 5.7 ± 50 μg ml−l kanamycin. Mature A. thaliana plants were grown in Levington M3 compost/vermiculite (Levington Horticulture, Ipswich, UK) in the glasshouse at 22°C under long days before harvesting of mature siliques.
Physcomitrella patens transformation
Transformation was carried out as in Moody et al. (2012). Stable transformants were selected using 20 μg ml−l hygromycin B and 50 μg ml−l G418.
Preparation of DNA and RNA
Physcomitrella patens genomic DNA was prepared as in Moody et al. (2012) and used directly in PCR reactions. Transforming plasmid DNA was prepared using the Qiagen Plasmid Midi Kit according to the manufacturer's instructions. Physcomitrella patens, Selaginella moellendorffii (hieron) and A. thaliana RNA was prepared using the RNeasy plant mini‐prep kit (Qiagen). RNA was treated with TURBO™ DNase (Life Technologies, Waltham, MA, USA) and then converted to cDNA using Superscript™ II reverse transcriptase and Oligo dT (Invitrogen).
Generation and imaging of protoplast transformation constructs
pUBI::PHYSCODILLO‐GFP constructs were created by ligating the PHYSCODILLO cDNA sequences (Moody et al., 2012) into the multiple cloning site of the vector pUbi‐MCS‐GFP‐108‐II (GenBank KR297238). Generation of pHSP::PHYSCODILLO‐GFP constructs is outlined in Supporting Information Methods S1. p35S::ARABIDILLO‐GFP was used as in Coates et al. (2006). p35S::SELAGIDILLO‐GFP was derived from SELAGIDILLO cDNA (Moody et al., 2012) as in the ‘Construction of arabidillo mutant rescue lines’ subsection later. For transient expression, pUBI::PHYSCODILLO1‐GFP, pUBI::PHYSCODILLO2‐GFP, p35S::ARABIDILLO‐GFP and p35S::SELAGIDILLO‐GFP constructs were each transformed into wild‐type P. patens protoplasts multiple times. To verify nuclear localization, pHSP::PHYSCODILLO1‐GFP and pHSP::PHYSCODILLO2‐GFP were co‐transformed into P. patens protoplasts alongside pHSP::NLS‐mCherry‐10835SNPT (GenBank KP893620). Following transformations, protoplasts were incubated overnight at 22°C before capturing images using a Leica SP2 inverted confocal microscope.
Generation of PHYSCODILLO‐GFP transgenic plants
For stable expression, pHSP::GFP, pHSP::PHYSCODILLO1‐GFP and pHSP::PHYSCODILLO2‐GFP were transformed into wild‐type protoplasts and successful integration confirmed following two rounds of G418 selection. Full details of construct generation are given in Methods S1.
Protein expression analysis
For protein gel analysis, P. patens protonemal tissues expressing pHSP::GFP, pHSP::PHYSCODILLO1‐GFP and pHSP::PHYSCODILLO2‐GFP were harvested into liquid BCD protonemal medium and incubated at 22 (control), 34 or 38°C for 1 h before returning to room temperature. For MG132 experiments, tissue in liquid BCD was pretreated with 50 μM MG132 for 1 h before heat shock. Samples were collected at different times postinduction and flash‐frozen in liquid nitrogen before extraction. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and western blotting were carried out using standard procedures as in Nibau et al. (2011).
For microscopy analysis, tissues were grown on BCD medium under standard growth conditions, and protein expression was induced for 1 h at 38°C before returning to 22°C for different lengths of time. For MG132 experiments, tissue in liquid BCD was pretreated with 50 μM MG132 for 1 h before heat shock. Confocal images were captured using a Leica SP2 inverted confocal microscope. Other images were captured using a Nikon SMZ1000 dissecting microscope and nis‐elements software (Nikon, Tokyo, Japan).
Construction of physcodillo1a/1b/2 triple deletion mutants and screening procedure
The PHYSCODILLO1A/1B double deletion construct was generated by cloning 5′ and 3′ homologous flanking sequences from P. patens genomic DNA and inserting them into the pMBL10a vector either side of a G418 resistance cassette (see Fig. S3a later). The resulting construct was transformed into physcodillo2 mutant protoplasts (Moody et al., 2012). Two rounds of G418 selection were carried out to identify putative transformants. To verify the presence of a G418 resistance cassette within the PHYSCODILLO1A/1B locus and confirm the generation of physcodillo1a/1b/2 triple deletion mutants, PCR was carried out using GoTaq DNA Polymerase (Promega). 5′ integration was confirmed using the primers P1 + 3KO5′F and G418.R.319 and 3′ integration was confirmed using the primers P1 + 3KO3′R and G418.F.341. PCR products were sequenced (see Fig. S3 later) and RT‐PCR was carried out to confirm loss of PHYSCODILLO1 mRNA expression. Primer sequences are detailed in Table S1.
Confirmation of physcodillo1a/1b/2 triple deletion mutant by RT‐PCR
RNA and cDNA were prepared from 7‐d‐old protonemata (wild‐type, physcodillo2, physcodillo1a/1b/2‐8 and physcodillo1a/1b/2‐16). Gene‐specific cDNA products were amplified using GoTaq DNA Polymerase (Promega). PHYSCODILLO1 was amplified using P1‐RT.GSP.F and P1‐RT.GSP.R, PHYSCODILLO2 was amplified using P2‐RT.GSP.F and P2‐RT.GSP.R and a positive control tubulin cDNA was amplified using PptubF and PptubR.
Physcomitrella patens spore germination assays and protonemal area measurement
Sporangia were sterilized in 20% Parozone bleach for 15 min at room temperature and then washed four times in sterile water. Spores were released from sporangia by perforating them using a sterile pipette tip in a final volume of 1 ml of sterile water. Spore suspension (200 μl) was spread onto each of five Petri dishes containing cellophane‐overlaid spore regeneration medium. Spores were allowed to germinate at 22°C in long days (16 h : 8 h, light : dark). The percentage of germinating spores was calculated at regular intervals until all of the control spores had fully germinated. For each data point, > 200 spores were counted on each of three plates and the mean percentage germination ± SE of the mean was calculated. Each experiment was repeated multiple times. Plants were photographed on a Nikon SMZ1000 dissecting microscope, and the protonemal area was imaged and measured using Nikon's nis‐elements software package.
Construction of arabidillo mutant rescue lines
35S::PHYSCODILLO1‐GFP and 35S::SELAGIDILLO‐GFP fusions were made in pGreen0029 (Hellens et al., 2000) by replacing ARABIDILLO1 in the 35S::ARABIDILLO1‐GFP construct (Coates et al., 2006). Full details of construct generation are given in Methods S1. Both constructs were transformed into Agrobacterium tumefaciens GV3101 with pSoup (Hellens et al., 2000) and transformed into A. thaliana by floral dip (Clough & Bent, 1998).
Root assays
LR assays were carried out as in Coates et al. (2006). Seedling LR density was defined as the number of emerged LRs cm−1 of primary root. A minimum of 50 roots were assayed for each genotype in a single experiment and each experiment was repeated multiple times.
Arabidopsis thaliana seed germination assays
Freshly harvested seeds from wild‐type, mutant and transgenic plants were surface‐sterilized in 5% (v/v) bleach for 5 min then washed with sterile water before plating (three to four replicates; n = 50) onto water agarose (1%) supplemented with relevant concentrations of ABA (Sigma). After 4 d of chilling, seeds were incubated at 22°C under continuous light for 7 d, and germination was assessed as endosperm rupture by the radicle. Each assay was repeated multiple times.
Results
Arabidopsis thaliana ARABIDILLO proteins localize to the nucleus, where they exert their control of LR development through physically interacting with flowering plant‐specific MYB transcription factors (Coates et al., 2006; Nibau et al., 2011; Gibbs et al., 2014). However, ARABIDILLO homologues with a high degree of protein identity to one another also exist in early‐diverging land plants, which lack both LRs and relevant MYB homologues (Nibau et al., 2011; Moody et al., 2012; Gibbs et al., 2014; Fig. 1a). To analyse the behaviour of ARABIDILLO proteins in an early‐diverging land plant, we examined the localization of ARABIDILLO homologues in transiently transformed P. patens protoplasts using a series of fusion proteins with green fluorescent protein (GFP) directly fused in‐frame to the C‐terminus of ARABIDILLO homologues, driven from constitutive promoters (Fig. 1a). ARABIDILLO1‐GFP, PHYSCODILLO1A/1B‐GFP (which are identical to one another (Moody et al., 2012) and subsequently are collectively referred to as PHYSCODILLO1), PHYSCODILLO2‐GFP and SELAGIDILLO‐GFP (the S. moellendorffii ARABIDILLO homologue; Moody et al., 2012) all show considerable sequence identity across the entire length of the protein (Fig. 1a). All the proteins localize to the nucleus (Fig. 1b): this was confirmed using a red fluorescent protein marker tagged with a nuclear localization signal (NLS‐RFP; Fig. 1c), which colocalized with the PHYSCODILLO‐GFP signal.
Figure 1.
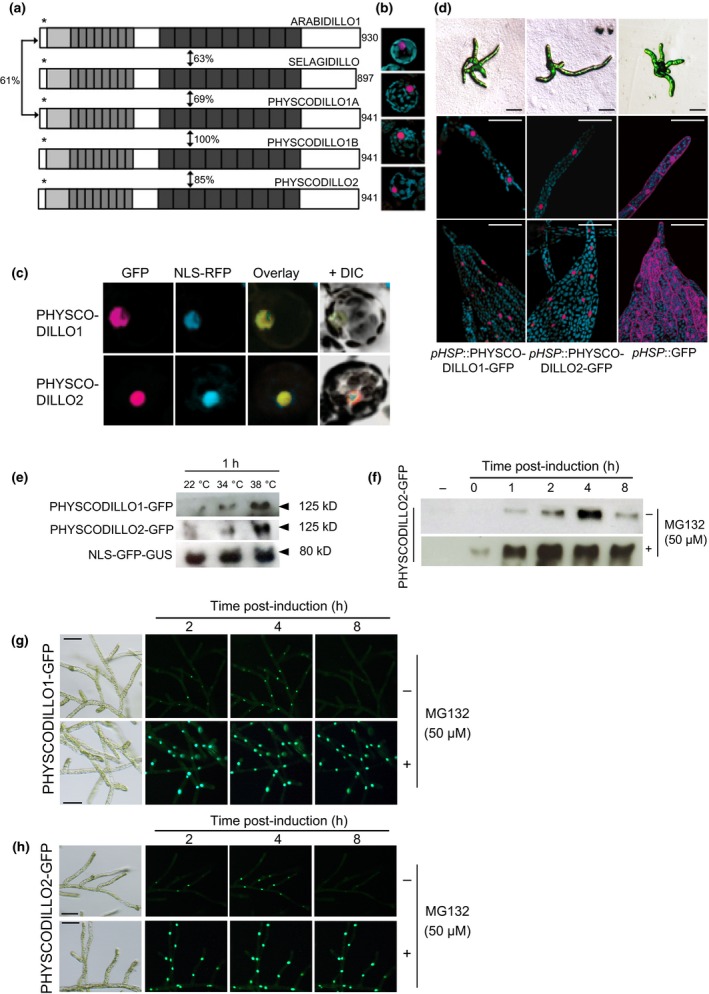
ARABIDILLO protein homologues in Physcomitrella patens and Selaginella moellendorffii. (a) ARABIDILLO homologues in Arabidopsis thaliana (ARABIDILLO1), S. moellendorffii (SELAGIDILLO) and P. patens (PHYSCODILLO1A, ‐1B and ‐2). Light grey boxes, F‐box; mid‐grey boxes, leucine‐rich repeats; dark grey boxes, Armadillo repeats. Asterisks, nuclear localization signals. Percentages indicate the degree of amino acid identity between the protein homologues, as indicated in Moody et al. (2012). Numbers at the C‐termini indicate amino acid length. (b) Constitutive expression of full‐length GFP‐tagged proteins in P. patens protoplasts. From the top, 35S::ARABIDILLO1‐GFP; 35S::SELAGIDILLO‐GFP;pUBI::PHYSCODILLO1‐GFP;pUBI::PHYSCODILLO2‐GFP. Each image is representative of 15–20 protoplasts. (c) Protoplasts co‐transformed with pUBI::PHYSCODILLO1‐GFP or pUBI::PHYSCDILLO2‐GFP and a nuclear localisation signal‐red fluorescent protein (NLS‐RFP) nuclear marker. (d) Inducible expression of full‐length green fluorescent protein (GFP)‐tagged proteins in P. patens transgenic lines. From left to right, pHSP::PHYSCODILLO1‐GFP;pHSP::PHYSCODILLO2‐GFP;pHSP::GFP. Tissues from upper panels to lower panels: protonemal filaments (low magnification); protonemal filaments (confocal sections); leafy gametophore tissue (confocal sections). Bars, 50 μm. (e) Western blot showing inducible expression of pHSP::PHYSCODILLO1‐GFP and pHSP::PHYSCODILLO2‐GFP compared with a constitutively expressed NLS‐GFP‐GUS control (β‐glucuronidase) (Bezanilla et al., 2005). (f) Western blot showing degradation of heat‐shock‐induced PHYSCODILLO2‐GFP and its stabilization in the presence of the proteasome inhibitor MG132. (g) Fluorescence micrographs showing degradation of heat‐shock‐induced PHYSCODILLO1‐GFP and its stabilization in the presence of the proteasome inhibitor MG132. Bars, 50 μm. (h) Fluorescence micrographs showing degradation of heat‐shock‐induced PHYSCODILLO2‐GFP and its stabilization in the presence of the proteasome inhibitor MG132, similarly to PHYSCODILLO1‐GFP (compare with g). Bars, 50 μm.
To further examine the spatial and temporal localization and behaviour of PHYSCODILLO proteins, we attempted to generate stable transgenic lines expressing GFP‐tagged PHYSCODILLO proteins under the control of the constitutive maize (Zea mays) Ubiquitin‐1 promoter (pUBI). However, we found that overexpression of PHYSCODILLO‐GFP in protoplasts was toxic, inhibiting the outgrowth of protonemal filaments and preventing protoplast regeneration, so no transformants could be recovered (Fig. 2). This is in accordance with the data of Nibau et al. (2011), showing the possible toxicity of overexpressed ARABIDILLO protein in A. thaliana. To circumvent this problem, we generated stable transgenic P. patens lines where inducible expression of PHYSCODILLO1 or ‐2 fused to GFP was driven from the soybean (Glycine max) heat‐shock promoter (pHSP; Saidi et al., 2005) and induced at different stages of P. patens development. In both filamentous and leafy tissue, PHYSCODILLO‐GFP is detected in the nucleus, while GFP alone localizes to both nucleus and cytosol (Fig. 1d). We determined that good induction of PHYSCODILLO‐GFP expression occurs after just 1 h at 38°C (Figs 1e, S1).
Figure 2.
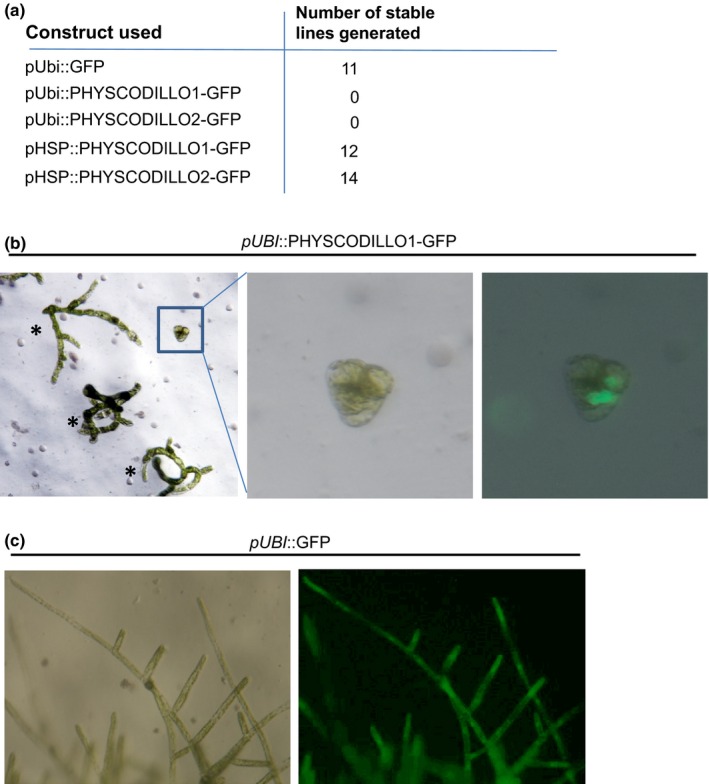
Constitutive expression of PHYSCODILLO proteins in Physcomitrella patens leads to arrested protoplast regeneration. (a) Numbers of stable transgenic lines generated as a result of transforming P. patens protoplasts with constructs encoding combinations of proteins (GFP or PHYSCODILLOs) driven from the constitutive maize UBIQUITIN promoter (pUBI) or the inducible heat‐shock promoter (pHSP). Using the constitutive promoter (pUBI) to drive PHYSCODILLO‐GFP fusion proteins results in no recovery of stable transgenic lines. pUBI driving green fluorescent protein (GFP) alone, or pHSP driving PHYSCODILLO‐GFP fusions leads to normal transgenic line generation. (b) Transient transformation of P. patens protoplasts with pUBI::PHYSCODILLO1‐GFP leads to the formation of small, arrested GFP‐expressing colonies that show no further cell growth (zoomed image) compared with untransformed protoplast regenerating colonies (asterisks). (c) A control pUBI::GFP fusion protein is not lethal, and mature regenerated transformants are obtained.
ARABIDILLOs are unstable proteins that are turned over by the proteasome (Nibau et al., 2011). Using the inducible PHYSCODILLO lines, we were able to show by confocal microscopy and western blotting that PHYSCODILLO proteins are also turned over by the proteasome (Figs 1f–h, S1), as, like ARABIDILLOs, they are stabilized by the proteasome inhibitor MG132 (Nibau et al., 2011). This fits with the previous observation that the key regions required for ARABIDILLO/PHYSCODILLO instability, namely the F‐box and leucine‐rich repeat regions, are highly conserved (Nibau et al., 2011). This demonstrates similar protein characteristics for PHYSCODILLOs and ARABIDILLOs despite c. 420 million yr of evolutionary divergence (Hedges et al., 2006).
To further investigate PHYSCODILLO behaviour and function, we investigated whether P. patens and S. moellendorffii ARABIDILLO homologues were able to substitute for A. thaliana ARABIDILLO function during LR formation. We generated stable transgenic A. thaliana lines expressing either PHYSCODILLO1 or SELAGIDILLO1 driven by the cauliflower mosaic virus (CaMV) 35S promoter, and found that both bryophyte and lycophyte ARABIDILLO homologues were able to complement the reduced LR phenotype of the arabidillo1/2 mutant (Fig. 3a,b), despite the facts that P. patens has no multicellular rooting structures, and that S. moellendorffii has tip‐bifurcating roots (Banks, 2009; Jones & Dolan, 2012). Thus, early‐diverging ARABIDILLO homologues had the capacity to affect LR development before the evolution of true, branched rooting structures occurred, suggesting that they were functionally co‐opted into pathways regulating novel structures that arose during the evolution of flowering plants.
Figure 3.
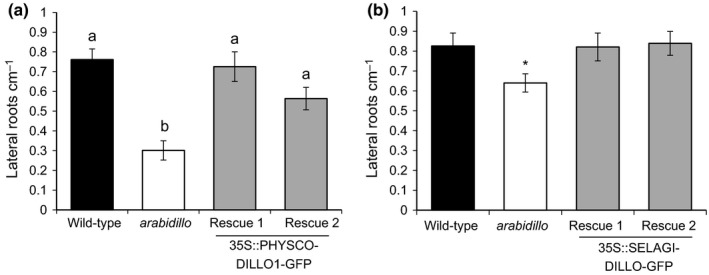
PHYSCODILLO and SELAGIDILLO can both rescue the Arabidopsis thaliana arabidillo1/2 mutant lateral root phenotype. (a) Mean lateral root density measured 8 d after germination for wild‐type (black bar), arabidillo1/2 mutant (white bar) and two independent arabidillo1/2 mutant lines constitutively expressing PHYSCODILLO1‐GFP from the 35S promoter (Rescue 1 and Rescue 2, grey bars). One‐way ANOVA shows significant differences between genotypes (P < 0.0001). A Tukey post hoc test shows that the arabidillo mutant is significantly different from wild‐type (P < 0.01) and from Rescue 1 (P < 0.01) and Rescue 2 (P < 0.05). Different lowercase letters indicate significant differences between genotypes. Error bars show ± SE of the mean. (b) Mean lateral root density 8 d after germination for wild‐type (black bar), arabidillo1/2 mutant (white bar) and two independent arabidillo1/2 mutant lines constitutively expressing SELAGIDILLO‐GFP from the 35S promoter (Rescue 1 and Rescue 2, grey bars). One‐way ANOVA indicated that differences between genotypes were not quite significant (P = 0.057), although pairwise t‐tests comparing the mutant and rescue lines with the wild‐type indicated that the wild‐type was different from the mutant (*, P < 0.05) but not different from either rescue line. Error bars show ± SE of the mean.
We previously showed that a single PHYSCODILLO2 knock‐out in moss has no obvious phenotype, suggesting that the homologues function redundantly (Moody et al., 2012), similarly to what is observed in A. thaliana (Coates et al., 2006). To investigate the function(s) of PHYSCODILLO proteins in P. patens, we therefore generated two independent triple physcodillo1a/1b/2 loss‐of‐function mutants by targeted gene replacement, deleting the entire 23‐kb PHYSCODILLO1A/1B locus in a physcodillo2 mutant background (Moody et al., 2012; Figs S2a,b, S3). The replaced locus was confirmed by sequencing across the insertion site (Fig. S3). Using RT‐PCR we confirmed the absence of PHYSCODILLO mRNAs in these knockout lines (Fig. S2c). Similarly to the physcodillo2 mutants (Moody et al., 2012), physcodillo1a/1b/2 mutant plants are overall morphologically similar to wild‐type, producing chloronema, caulonema, gametophores with rhizoids (Fig. S4a,b), antheridia, archegonia and sporophytes in a similar time frame. However, we noticed that the spores of physcodillo1a/1b/2 mutants germinated more slowly than those of wild‐type, suggesting a potential role for PHYSCODILLOs in regulating this process (Fig. 4a).
Figure 4.
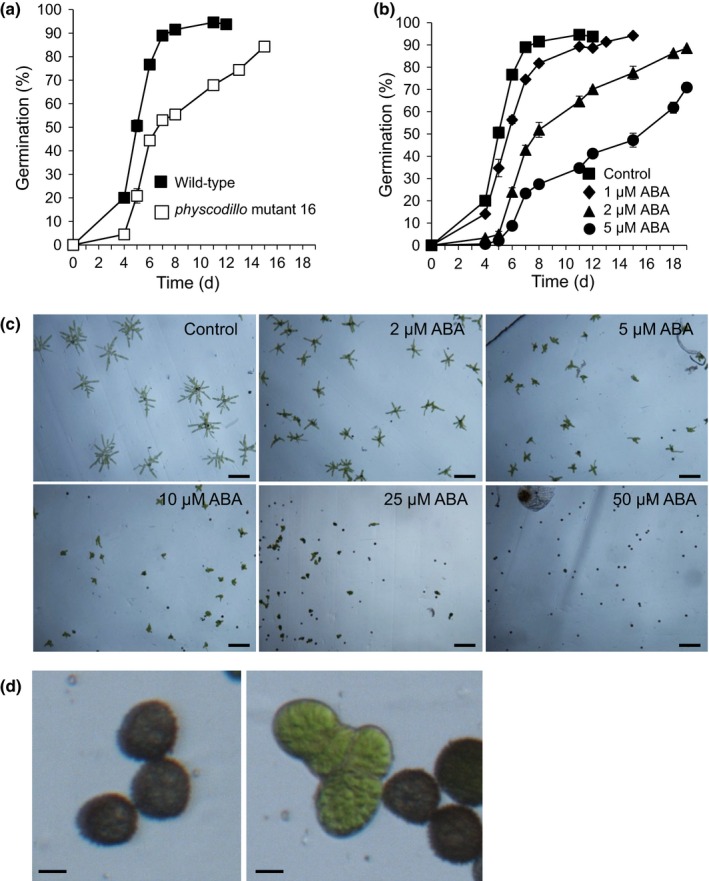
Physcomitrella patens physcodillo triple mutants show slower germination, and abscisic acid (ABA) inhibits P. patens spore germination. (a) Germination of wild‐type and physcodillo mutant spores. A Mann–Whitney test did not identify significant differences between genotypes (P = 0.08) although z‐tests suggested highly significant (P < 0.01) differences on all of days 4, 5, 6, 7, 8 and 11. (b) Inhibition of P. patens spore germination by 1–5 μM ABA. Error bars show ± SE of the mean. A Kruskal–Wallis test for differences between treatments was significant (P < 0.05) on days 4, 5, 6, 7, 8 and 12. A Dunn's test identified significant (P < 0.05) differences between control and 2 μM ABA and highly significant (P < 0.01) differences between control and 5 μM ABA on days 4, 5, 6 and 7. (c) Effect of 0–50 μM ABA on germination in a population of spores. More ungerminated spores are seen as ABA concentration increases, in addition to a reduction in plant size (cell length) occurring. Bars, 100 μm. (d) Higher magnification images of spores treated with 25 μM ABA. The left‐hand panel shows ungerminated spores, while the right‐hand panel shows a small plant on the left (with shortened cells, from a germinated spore) and ungerminated spores on the right. Bar, 10 μm.
In seed plants, ABA is a key inhibitor of germination and promotes seed dormancy (Finkelstein et al., 2008; Holdsworth et al., 2008). Some evidence also implicates ABA in the regulation of fern spore germination (Singh et al., 1990; Yao et al., 2008). However, neither the process of P. patens spore germination nor the effects of ABA on spores have previously been studied in detail (Glime, 2015). We showed a dose‐dependent inhibition of the wild‐type P. patens spore germination rate by ABA (Fig. 4b–d). This suggests that ABA has a similar negative regulatory role in both spore and seed germination, despite the different developmental origins of spores and seeds. We showed that P. patens spores showed much lower sensitivity to ABA than A. thaliana seeds, as 5 μM ABA, which would completely inhibit A. thaliana germination, significantly reduced the spore germination rate without inhibiting germination completely (Fig. 4b,c).
We also examined the response of physcodillo1a/1b/2 mutants, and found that both physcodillo1a/1b/2 mutant alleles are less sensitive to the ABA‐mediated inhibition of the spore germination rate than wild‐type, implying that PHYSCODILLOs play a role in regulating the response to ABA during this process (Fig. 5a–d). When we examined other known ABA‐mediated processes, namely response to desiccation/drought and freezing, we found that the physcodillo1a/1b/2 plants showed no obvious differences in their desiccation or freezing tolerance compared with wild‐type plants (Fig. S4c,d), indicating a specific role in germination.
Figure 5.
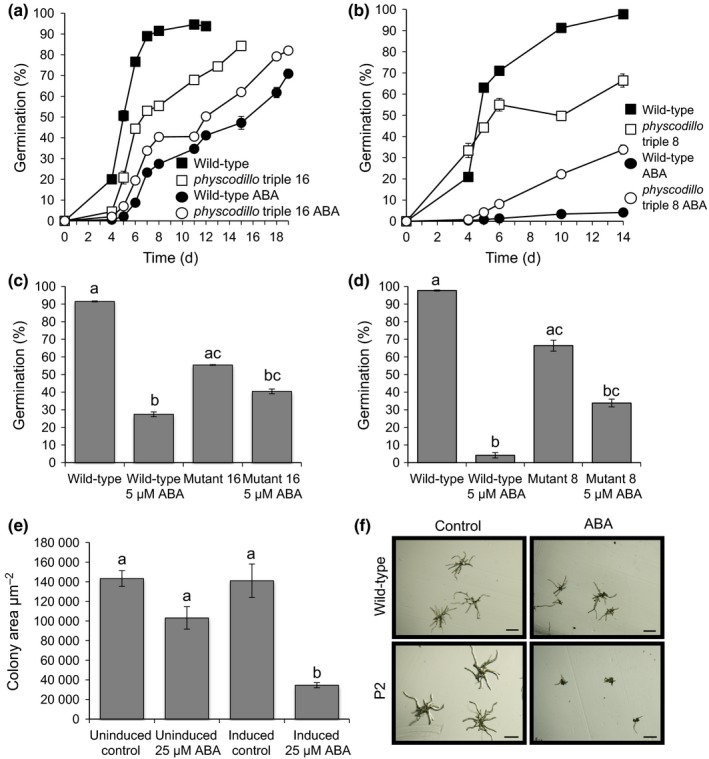
Physcomitrella patens physcodillo triple mutants show insensitivity to the inhibitory effects of abscisic acid (ABA) during germination while a P. patens PHYSCODILLO‐overexpressing line shows ABA hypersensitivity during early growth. (a) Germination of wild‐type and physcodillo triple mutant line 16 spores on medium containing 5 μM ABA or solvent‐only control. Kruskal–Wallis tests showed significant (P < 0.05) differences between genotypes/treatments on days 4, 5, 6, 7, 8 and 11. Dunn's tests showed significant (P < 0.01) differences between wild‐type spores with and without ABA on days 4, 5, 6, 7, 8 and 11, significant (P < 0.05) differences between ABA‐treated wild‐type and untreated physcodillo mutants on days 5, 6, 7, 8 and 11 and significant (P < 0.05) differences between wild‐type and ABA‐treated physcodillo mutants on days 5, 6, 7, 8 and 11. (b) Germination of wild‐type and physcodillo triple mutant line 8 mutant spores germinated on medium containing 5 μM ABA or solvent‐only control. Kruskal–Wallis tests showed significant (P < 0.05) differences between genotypes/treatments on days 4, 5, 6, 10 and 14. Dunn's tests showed significant (P < 0.01) differences between wild‐type spores with and without ABA on all days and significant (P < 0.05) differences between ABA‐treated wild‐type and untreated physcodillo mutants on all days. (c) Insensitivity of physcodillo triple mutant 16 to 5 μM ABA. Data are shown for day 8. A Kruskal–Wallis test for differences between genotypes and treatments was significant (P < 0.05). Dunn's test identified differences between groups: a and b, P < 0.001; b and c, P < 0.05; a and c, P < 0.05. Error bars show ± SE of the mean. (d) Insensitivity of physcodillo triple mutant 8 to 5 μM ABA. Data are shown for day 14. A Kruskal–Wallis test for differences between genotypes and treatments was significant (P < 0.05). Dunn's test identified differences between groups: a and b, P < 0.001; b and c, P < 0.05; a and c, P < 0.05. Error bars show ± SE of the mean. (e) Transgenic pHSP::PHYSCODILLO2‐GFP plants were either grown at 22°C on medium containing either solvent control or 25 μM ABA (left‐hand bars), or exposed to daily 1‐h heat shock and grown on medium containing either solvent control or ABA (right‐hand bars). One‐way ANOVA showed significant differences between treatments, with a Tukey's post hoc test showing that only plants with induced PHYSCODILLO2‐GFP expression grown on ABA (b) were significantly different (P < 0.05) from those in other treatments (a). Error bars show ± SE of the mean. (f) Representative morphology of wild‐type vs heat‐shocked transgenic pHSP::PHYSCODILLO2‐GFP plants germinated from spores and grown on solvent control or 25 μM ABA. Bars, 100 μm.
To extend these findings, we examined the effects of PHYSCODILLO overexpression on ABA sensitivity in P. patens. Because of the lethality of constitutive PHYSCODILLO overexpression, we used heat‐shock‐inducible lines. We showed that heat‐shock‐inducible overexpression of the PHYSCODILLO2 protein can lead to ABA hypersensitivity during early growth, when spores are germinated in the presence of ABA (Fig. 5e,f). An inducible transgenic line expressing the β‐glucuronidase (GUS) protein from the heat‐shock promoter showed no such hypersensitivity, linking the observed phenotype to PHYSCODILLO overexpression (data not shown).
We previously showed that arabidillo1/2 mutants in A. thaliana do not have altered sensitivities to ABA with regard to LR development (Nibau et al., 2011); however, we did not investigate other potential ABA functions for ARABIDILLOs in A. thaliana. To determine whether the role of PHYSCODILLOs in regulating ABA responses during germination is conserved, we examined seed germination in A. thaliana. Remarkably, we found that the A. thaliana arabidillo1/2 mutant is relatively insensitive to the ABA‐mediated inhibition of seed germination compared with the wild‐type. Moreover, ARABIDILLO1‐overexpressing and ‘rescue’ lines displayed an opposite ABA‐hypersensitive phenotype (Fig. 6a). Both mutant and overexpressing seeds germinated as wild‐type in the absence of ABA (data not shown). These data imply a conserved role for ARABIDILLO homologues in ABA‐regulated germination across land plants, as well as a unique role in LR development in flowering plants (Coates et al., 2006; Gibbs et al., 2014). To further investigate this conserved germination function, we asked whether the early‐diverging P. patens and S. moellendorffii ARABIDILLO homologues could rescue the A. thaliana arabidillo1/2 mutant germination phenotype. Reintroduction of either PHYSCODILLO or SELAGIDILLO into the arabidillo mutant led to a complementation of the ABA‐insensitive germination phenotype (Fig. 6b,c). Therefore, despite the different developmental origins of spores and seeds, P. patens and S. moellendorffii ARABIDILLO homologues can replace ARABIDILLOs during A. thaliana germination, demonstrating a novel and evolutionarily ancient function for the ARABIDILLO protein family regulating the germination of desiccation‐resistant dispersal units in response to ABA.
Figure 6.
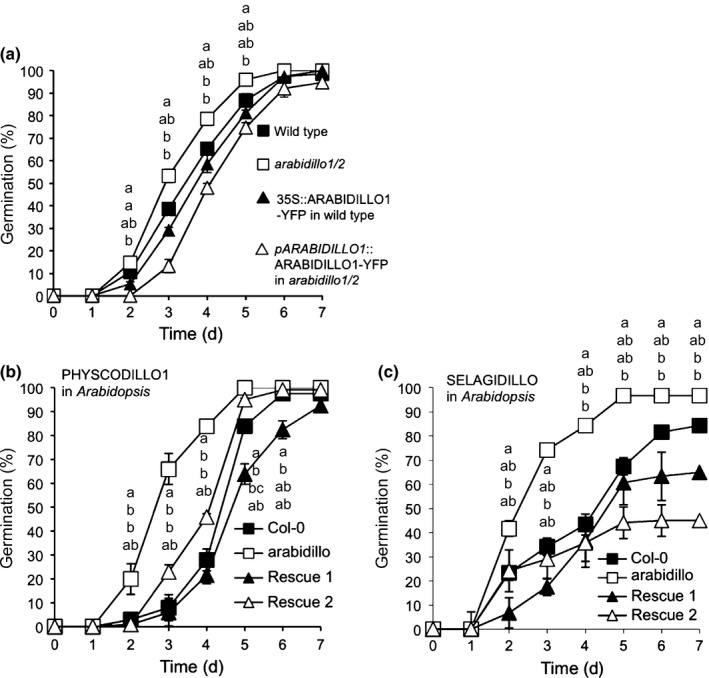
Abscisic acid (ABA) insensitivity of the Arabidopsis thaliana arabidillo mutant during seed germination and its rescue by both PHYSCODILLO and SELAGIDILLO proteins. (a) Seeds from wild‐type (Col‐0), arabidillo mutant, a 35S::ARABIDILLO‐YFP overexpressing line (Coates et al., 2006) and an arabidillo mutant stably expressing pARABIDILLO1::ARABIDILLO1‐YFP (Coates et al., 2006; Nibau et al., 2011) plated on 1 μM ABA with percentage germination assayed over 7 d. Error bars show ± SE of the mean. A Kruskal–Wallis test identified significant (P < 0.05) differences between genotypes on days 2, 3, 4 and 5. Different letters indicate significant (P < 0.05) differences between genotypes determined using Dunn's test for, top to bottom, arabidillo1/2, wild‐type, 35S::ARABIDILLO1‐YFP in wild‐type, and pARABIDILLO1::ARABIDILLO1‐YFP in arabidillo1/2. (b) Seeds from wild‐type, arabidillo mutant, and arabidillo mutant stably expressing PHYSCODILLO1 (35S::PHYSCODILLO1‐GFP; two independent lines, Rescue 1 and Rescue 2) plated on 1 μM ABA with percentage germination assayed over 7 d. A Kruskal–Wallis test identified significant differences (P < 0.05) between genotypes on days 2, 3, 4, 5 and 6. Different letters indicate significant differences between genotypes determined using Dunn's test (P < 0.05) for data points, from top to bottom, for arabidillo1/2, wild‐type, Rescue 1 and Rescue 2. (c) Seeds from wild‐type, arabidillo mutant, and arabidillo mutant stably expressing SELAGIDILLO (35S::SELAGIDILLO‐GFP: two independent lines, Rescue 1 and Rescue 2) plated on 1 μM ABA with percentage germination assayed over 7 d. A Kruskal–Wallis test identified significant differences (P < 0.05) between genotypes on days 2, 3, not 4 (P = 0.07), 5, 6 and 7. Different letters indicate significant differences between genotypes determined using Dunn's test (P < 0.05) for data points, from top to bottom, for arabidillo1/2, wild‐type, Rescue 1 and Rescue 2.
Discussion
The production of dispersal units such as spores and seeds was a critical step enabling plant survival and movement on land. Germination of such structures is tightly regulated to ensure that plants establish themselves in the right place and at the right time under favourable conditions (Holdsworth et al., 2008). In flowering plants and gymnosperms, seeds are multicellular structures that protect the diploid embryo, which gives rise to the subsequent sporophyte generation. In bryophytes, unicellular spores arise by meiosis and give rise to the haploid gametophyte, and are therefore of different developmental origin from seeds (Pires & Dolan, 2012).
Our studies reveal for the first time that the hormone ABA has evolved to regulate functionally equivalent germination processes in spore‐ and seed‐producing plants.
Furthermore, we identify conserved PHYSCODILLO/ARABIDILLO proteins as novel modulators of the germination of land plant desiccation‐resistant dispersal units in conjunction with ABA. We suggest that PHYSCODILLO/ARABIDILLO proteins represent a conserved node in a germination‐regulatory network that includes ABA, in both seeds and spores.
ABA has ancient evolutionary origins, being produced in cyanobacteria and all major eukaryote lineages: shared roles for ABA in stress tolerance have been proposed across these taxa (Takezawa et al., 2011, 2015). ABA‐mediated stomatal control has ancient origins (Chater et al., 2011; Ruszala et al., 2011; Lind et al., 2015) and ABA has also been implicated in developmental transitions in land plants, green algae and the Apicomplexan Toxoplasma (Takezawa et al., 2011).
Although ABA is widespread throughout taxa, land plants probably acquired unique ABA signalling mechanisms early during their history (Hauser et al., 2011), which have subsequently been conserved in terms of changes in gene expression (Knight et al., 1995; Kamisugi & Cuming, 2005; Cuming et al., 2007; Wang et al., 2010). ABA responses are evolutionarily conserved and have been co‐opted into both gametophyte and sporophyte tissues: ABA signalling regulates stress tolerance in the vegetative tissues of bryophytes (Cuming et al., 2007; Komatsu et al., 2009, 2013; Khandelwal et al., 2010; Richardt et al., 2010; Tougane et al., 2010) as well as stomatal opening in the sporophyte (Chater et al., 2011). ABA is well known for mediating stress responses, stomatal opening and germination in seed plants (Nambara & Marion‐Poll, 2005; Christmann et al., 2006; Wasilewska et al., 2008).
It is tempting to speculate that ARABIDILLO and PHYSCODILLO proteins have conserved protein interactors and/or transcriptional targets in spore and seed germination, especially given their highly conserved sequences, domain structure and proteasomal regulation (Nibau et al., 2011; Fig. 1). Interestingly, in charophytes, ABA can inhibit germination in light‐treated oospores (Takatori & Imahori, 1971). Whether charophyte ARABIDILLO homologues (which our searches suggest exist; Timme & Delwiche, 2010; Hori et al., 2014; Matasci et al., 2014; Wickett et al., 2014) are part of this regulation is currently unknown. We also propose that ARABIDILLOs acquired a novel ABA‐independent function in flowering plants, regulating LR development, via interaction with flowering plant‐specific MYB transcription factors (Gibbs & Coates, 2014; Gibbs et al., 2014).
Our work reveals a retention of ancient functions for plant ARABIDILLO proteins, despite them evolving to have novel additional roles in flowering plants. Furthermore, we have added a new dimension to the land plant ABA story, demonstrating the first conservation of molecular mechanisms between spore and seed germination.
Author contributions
J.C.C., L.A.M. and Y.S. conceived the experiments; L.A.M., Y.S., D.J.G., A.C., D.H., E.F.V., K.K.B., S.J.B. and J.C.C. performed the experiments; L.A.M., Y.S., D.J.G., D.H. and J.C.C. analysed data; L.A.M., Y.S., D.J.G. and J.C.C. wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 PHYSCODILLO‐GFP fusions are turned over by the proteasome.
Fig. S2 Generation of a physcodillo1A/1B/2 triple mutant by targeted gene replacement.
Fig. S3 Sequencing of the replaced physcodillo1A/1B locus in the physcodillo2 mutant background, generating triple mutants number 8 and number 16.
Fig. S4 physcodillo triple mutants show no obvious differences in rhizoid development or vegetative ABA responses.
Table S1 Primers used to generate and characterize PHYSCODILLO1A/1B gene replacement and generate arabidillo mutant rescue lines
Methods S1 Generation of PHYSCODILLO‐GFP transgenic plants, construction of arabidillo mutant rescue lines, construction of physcodillo1A/1B/2 triple deletion mutants and screening procedure.
Acknowledgements
We thank Andy Cuming and Chris Franklin for help and advice during the early stages of the project and Yoan Coudert for help with phenotypic analysis of reproductive structures. Sequencing was carried out by the University of Birmingham's Functional Genomics Facility. The research was funded by a Leverhulme Trust Research Project Grant (F/00094/BA) to J.C.C., a Royal Society‐Leverhulme Trust Senior Research Fellowship 2013‐14 awarded to J.C.C., a Birmingham Fellowship awarded to D.J.G., a BBSRC grant (BB/D007550/1) to J.C.C., a BBSRC PhD studentship awarded to L.A.M., a NERC PhD studentship awarded to E.F.V. and University of Birmingham MSci student funding provided to D.H. and K.K.B.
References
- Banks JA. 2009. Selaginella and 400 million years of separation. Annual Review of Plant Biology 60: 223–238. [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS. 2005. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol (Stuttg) 7: 251‐257. [DOI] [PubMed] [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ. 2011. Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Current Biology 21: 1025–1029. [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. 2006. Integration of abscisic acid signalling into plant responses. Plant Biology (Stuttgart, Germany) 8: 314–325. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coates JC. 2003. Armadillo repeat proteins: beyond the animal kingdom. Trends in Cell Biology 13: 463–471. [DOI] [PubMed] [Google Scholar]
- Coates JC, Laplaze L, Haseloff J. 2006. Armadillo‐related proteins promote lateral root development in Arabidopsis. Proceedings of the National Academy of Sciences, USA 103: 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS. 2007. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens . New Phytologist 176: 275–287. [DOI] [PubMed] [Google Scholar]
- Evert R, Eichhorn D. 2012. Raven biology of plants, 8th edn New York, NY, USA: WH Freeman & Co. [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59: 387–415. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. 2009. Hormone interactions during lateral root formation. Plant Molecular Biology 69: 437–449. [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Coates JC. 2014. AtMYB93 is an endodermis‐specific transcriptional regulator of lateral root development in Arabidopsis. Plant Signaling & Behavior 9: e970406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Voss U, Harding SA, Fannon J, Moody LA, Yamada E, Swarup K, Nibau C, Bassel GW, Choudhary A et al 2014. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytologist 203: 1194–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glime JM. 2013. Life cycles: surviving change In: Bryophyte ecology. Volume 1. Physiological ecology. Houghton, MI, USA: Ebook sponsored by Michigan Technological University and the International Association of Bryologists, chapter 2‐2; URL http://www.bryoecol.mtu.edu/. [Google Scholar]
- Glime JM. 2015. Ecophysiology of development: spore germination In: Bryophyte ecology. Volume 1. Physiological ecology. Houghton, MI, USA: Ebook sponsored by Michigan Technological University and the International Association of Bryologists, chapter 5‐2; URL http://www.bryoecol.mtu.edu/. [Google Scholar]
- Hanada K, Hase T, Toyoda T, Shinozaki K, Okamoto M. 2011. Origin and evolution of genes related to ABA metabolism and its signaling pathways. Journal of Plant Research 124: 455–465. [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. 2011. Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology 21: R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge‐base of divergence times among organisms. Bioinformatics 22: 2971–2972. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Molecular Biology 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hofmeister WFB. 1851. Vergleichende Untersuchungen der Keimung, Entfaltung und Fruchtbildung höherer Kryptogamen (Moose, Farrn, Equisetaceen, Rhizocarpeen and Lycopodiaceen) und der Samenbildung der Coniferen. Leipzig, Germany: Friedrich Hofmeister. Available as a free e‐book. [WWW document] URL https://play.google.com/books/reader?id=HIfC_S82j8EC&printsec=frontcover&output=reader&hl=en&pg=GBS.PP1.
- Holdsworth MJ, Bentsink L, Soppe WJ. 2008. Molecular networks regulating Arabidopsis seed maturation, after‐ripening, dormancy and germination. New Phytologist 179: 33–54. [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N et al 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G, Dolan L. 2011. Auxin promotes the transition from chloronema to caulonema in moss protonema by positively regulating PpRSL1and PpRSL2 in Physcomitrella patens . New Phytologist 192: 319–327. [DOI] [PubMed] [Google Scholar]
- Jang G, Yi K, Pires ND, Menand B, Dolan L. 2011. RSL genes are sufficient for rhizoid system development in early diverging land plants. Development 138: 2273–2281. [DOI] [PubMed] [Google Scholar]
- Jones VA, Dolan L. 2012. The evolution of root hairs and rhizoids. Annals of Botany 110: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC. 2005. The evolution of the abscisic acid‐response in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Molecular Biology 59: 723–737. [DOI] [PubMed] [Google Scholar]
- Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud PF, Pan A, Quatrano RS. 2010. Role of ABA and ABI3 in desiccation tolerance. Science 327: 546. [DOI] [PubMed] [Google Scholar]
- Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ, Coates D, Quatrano RS, Bahadur S, Stockley PG, Cuming AC et al 1995. Molecular responses to abscisic acid and stress are conserved between moss and cereals. The Plant Cell 7: 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y. 2009. Functional analyses of the ABI1‐related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens . Plant Molecular Biology 70: 327–340. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M et al 2013. Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nature Communications 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L et al 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends in Plant Science 18: 450–458. [DOI] [PubMed] [Google Scholar]
- Lind C, Dreyer I, López‐Sanjurjo EJ, von Meyer K, Ishizaki K, Kohchi T, Lang D, Zhao Y, Kreuzer I, Al Rasheid KA et al 2015. Stomatal guard cells co‐opted an ancient ABA‐dependent desiccation survival system to regulate stomatal closure. Current Biology 25: 928–935. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M et al 2014. Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Moody LA, Saidi Y, Smiles E, Bradshaw SJ, Meddings M, Winn PJ, Coates JC. 2012. ARABIDILLO gene homologues in basal land plants: species‐specific gene duplication and likely functional redundancy. Planta 236: 1927–1941. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion‐Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Bunting KA, Moody LA, Smiles EJ, Tubby JA, Bradshaw SJ, Coates JC. 2011. ARABIDILLO proteins have a novel and conserved domain structure important for the regulation of their stability. Plant Molecular Biology 75: 77–92. [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. 2008. Branching out in new directions: the control of root architecture by lateral root formation. New Phytologist 179: 595–614. [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Annual Review of Plant Biology 58: 93–113. [DOI] [PubMed] [Google Scholar]
- Peret B, Larrieu A, Bennett MJ. 2009. Lateral root emergence: a difficult birth. Journal of Experimental Botany 60: 3637–3643. [DOI] [PubMed] [Google Scholar]
- Pires ND, Dolan L. 2012. Morphological evolution in land plants: new designs with old genes. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 367: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L. 2013. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proceedings of the National Academy of Sciences, USA 110: 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Bezanilla M. 2010. Evolutionary crossroads in developmental biology: Physcomitrella patens . Development 137: 3535–3543. [DOI] [PubMed] [Google Scholar]
- Proust H, Honkanen S, Jones VA, Morieri G, Prescott H, Kelly S, Ishizaki K, Kohchi T, Dolan L. 2016. RSL Class I genes controlled the development of epidermal structures in the common ancestor of land plants. Current Biology 26: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt S, Timmerhaus G, Lang D, Qudeimat E, Corrêa LG, Reski R, Rensing SA, Frank W. 2010. Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Molecular Biology 72: 27–45. [DOI] [PubMed] [Google Scholar]
- Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, Gray JE, Hetherington AM. 2011. Land plants acquired active stomatal control early in their evolutionary history. Current Biology 21: 1030–1035. [DOI] [PubMed] [Google Scholar]
- Saidi Y, Finka A, Chakhporanian M, Zryd JP, Schaefer DG, Goloubinoff P. 2005. Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat‐shock promoter: a tool for plant research and biotechnology. Plant Molecular Biology 59: 697–711. [DOI] [PubMed] [Google Scholar]
- Shimamura M. 2015. Marchantia polymorpha; taxonomy, phylogeny, and morphology of a model system. Plant and Cell Physiology. doi: 10.1093/pcp/pcv192. [DOI] [PubMed] [Google Scholar]
- Singh PK, Bisoyi RN, Singh RP. 1990. Collection and germination of sporocarps of Azolla caroliniana . Annals of Botany 66: 51–56. [Google Scholar]
- Takatori S, Imahori K. 1971. Light reactions in the control of oospore germination of Chara delicatula 1. Phycologia 10: 221–228. [Google Scholar]
- Takezawa D, Komatsu K, Sakata Y. 2011. ABA in bryophytes: how a universal growth regulator in life became a plant hormone? Journal of Plant Research 124: 437–453. [DOI] [PubMed] [Google Scholar]
- Takezawa D, Watanabe N, Ghosh TK, Saruhashi M, Suzuki A, Ishiyama K, Somemiya S, Kobayashi M, Sakata Y. 2015. Epoxycarotenoid‐mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytologist 206: 209–219. [DOI] [PubMed] [Google Scholar]
- Tam THY, Catarino B, Dolan L. 2015. Conserved regulatory mechanism controls the development of cells with rooting functions in land plants. Proceedings of the National Academy of Sciences, USA 112: E3959–E3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF. 2010. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biology 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, Kohchi T, Takezawa D. 2010. Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1‐like type 2C protein phosphatase in the liverwort Marchantia polymorpha . Plant Physiology 152: 1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kuang T, He Y. 2010. Conservation between higher plants and the moss Physcomitrella patens in response to the phytohormone abscisic acid: a proteomics analysis. BMC Plant Biology 10: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. 2008. An update on abscisic acid signaling in plants and more. Molecular Plant 1: 198–217. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Chang C, Salmi ML, Hung YS, Loraine A, Roux SJ. 2008. Genome‐scale cluster analysis of replicated microarrays using shrinkage correlation coefficient. BMC Bioinformatics 9: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix‐loop‐helix transcription factor controls cell growth and size in root hairs. Nature Genetics 42: 264–267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 PHYSCODILLO‐GFP fusions are turned over by the proteasome.
Fig. S2 Generation of a physcodillo1A/1B/2 triple mutant by targeted gene replacement.
Fig. S3 Sequencing of the replaced physcodillo1A/1B locus in the physcodillo2 mutant background, generating triple mutants number 8 and number 16.
Fig. S4 physcodillo triple mutants show no obvious differences in rhizoid development or vegetative ABA responses.
Table S1 Primers used to generate and characterize PHYSCODILLO1A/1B gene replacement and generate arabidillo mutant rescue lines
Methods S1 Generation of PHYSCODILLO‐GFP transgenic plants, construction of arabidillo mutant rescue lines, construction of physcodillo1A/1B/2 triple deletion mutants and screening procedure.


